Abstract
An aldehyde dehydrogenase was detected in crude cell extracts of Escherichia coli DH5α. Growth studies indicated that the aldehyde dehydrogenase activity was growth phase dependent and increased in cells grown with ethanol. The N-terminal amino acid sequence of the purified enzyme identified the latter as an aldehyde dehydrogenase encoded by aldB, which was thought to play a role in the removal of aldehydes and alcohols in cells that were under stress. The purified enzyme showed an estimated molecular mass of 220 ± 8 kDa, consisting of four identical subunits, and preferred to use NADP and acetaldehyde. MgCl2 increased the activity of the NADP-dependent enzyme with various substrates. A comparison of the effect of Mg2+ ions on the bacterial enzyme with the effect of Mg2+ ions on human liver mitochondrial aldehyde dehydrogenase revealed that the bacterial enzyme shared kinetic properties with the mammalian enzyme. An R197E mutant of the bacterial enzyme appeared to retain very little NADP-dependent activity on acetaldehyde.
Aldehyde dehydrogenases (ALDH) are members of a diverse group of related enzymes catalyzing the oxidation of aldehydes to their corresponding carboxylic acids, with some using NAD and others using NADP as the coenzyme. Their important roles in cellular metabolism have been studied extensively in organisms ranging from bacteria to humans (30). In Escherichia coli, more than 10 aldehyde dehydrogenase genes have been identified (26), and one of the genes, aldB, mapped from min 80 to 86 on the bacterial chromosome has been predicted to encode an aldehyde dehydrogenase of 512 amino acids (32). Sequence alignments of the deduced amino acid sequence showed 68% similarity and 70% identity with an aldehyde dehydrogenase from Vibrio cholerae (22) and an NAD-dependent acetaldehyde dehydrogenase from Alcaligenes eutrophus (24), respectively. Expression studies (32, 33) indicated that aldB was repressed by Fis (factor for inversion stimulation) but activated by RpoS (RNA polymerase sigma factor) and Crp (cyclic AMP receptor protein), and that the level of expression was highest in the early stationary phase. It was also shown that ethanol induced aldB expression. The latter finding, together with the high similarity to the A. eutrophus enzyme, suggests that the gene product of aldB functions as an acetaldehyde dehydrogenase and that it plays a role in ethanol metabolism through its oxidation of acetaldehyde derived from ethanol. Many free-living, gram-negative bacteria, including E. coli, express genes that are crucial to survival at the early stationary phase, and a number of these genes are RpoS dependent (12, 14, 25). The fact that aldB is positively regulated by RpoS suggests that it is an important stationary-phase gene (33). With the exception of its gene sequence, little is known about aldB at the protein level. The presumed gene product of aldB has been classified as an aldehyde dehydrogenase at the EcoCyc website (http://ecocyc.org) and is thought to catalyze some of the reactions ascribed to the gene product of aldA (13). Mapped at min 32 on the E. coli chromosome, aldA has been studied extensively at both the genetic and protein levels (2, 13). The gene product of aldA, also known as lactaldehyde dehydrogenase, is an NAD-dependent aldehyde dehydrogenase participating in various metabolic pathways (6, 16, 19). To the best of our knowledge, there is no available literature citation on the catalytic properties of the gene product of aldB. Thus, it is difficult to assess the functional relationship between aldA and aldB.
In a previous study, Heim and Strehler (11) detected a low level of NADP-dependent acetaldehyde dehydrogenase activity in a cell extract of E. coli Y1088 while studying an aldehyde dehydrogenase gene, aldH. It remains unclear whether this NADP-dependent acetaldehyde dehydrogenase is related to the gene product of aldH. We detected a similar NADP-dependent acetaldehyde dehydrogenase first in E. coli DH5α cells transformed with pGEM and later also in nontransformed cells. The NADP-dependent enzyme in the transformed cells has been isolated and characterized. Its N-terminal amino acid sequence, together with the results on growth studies, provides support for it to be the gene product of aldB. We have also prepared its R197E variant, which appears to have very little NADP-dependent activity on acetaldehyde. Here, we report our findings and discuss the relationships between the NADP-dependent enzyme and other aldehyde dehydrogenases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
DH5α(pGEM) was originated from an E. coli DH5α clone transformed with a pGEM plasmid. Transformation was conducted with E. coli DH5α competent cells (Life Technologies, Inc.). The pGEM plasmid was prepared by ligation with a pGEM vector and a control DNA insert, both of which originated from a pGEM-T Easy Vector system (Promega). A derivative of pINIII (20), pCJM was used as an expression vector.
Growth conditions.
Growth was maintained by shaking partially filled conical flasks in a 37°C incubator set at 200 rpm. Cells were grown in Luria-Bertani (LB) or M9 medium (11) containing 75 μg of ampicillin/ml.
For the effect of culture age on aldehyde dehydrogenase activity, DH5α(pGEM) grown overnight in LB was diluted into fresh medium at a ratio of 2:100 and allowed to grow over a period of 10 h. At regular intervals, growth was monitored according to the absorbance at 600 nm, and cell samples were harvested. Crude cell extracts prepared from various samples were used for enzyme assays.
For the effect of ethanol on aldehyde dehydrogenase activity, DH5α(pGEM), grown overnight in LB, was diluted into M9 medium (1:100) and supplemented with 0.1% yeast extract (Difco) and various amounts of ethanol. After 12 h of growth, cells were harvested to prepare crude cell extracts for enzyme assays.
For enzyme purification, batch cultures of DH5α(pGEM) were grown in 4-liter flasks containing 1 liter of LB. When the optical density at 600 nm reached 2 to 2.5, the cultures were harvested and used for purifying the aldehyde dehydrogenase encoded by aldB, henceforth referred to as AldB.
Preparation of crude cell extracts.
Cells were harvested by centrifugation at 3,000 × g for 10 min at 4°C. The cell pellets were washed once in 0.9% sodium chloride solution and resuspended in 10 mM sodium phosphate or Tris-HCl buffer, pH 7.5, containing 1 mM EDTA and 1 mM dithiothreitol. The cell suspensions were kept at −70°C until they were ready to use. After thawing, the cell suspensions were disrupted by a French Press cell (three cycles at 15,000 lb/in2) prechilled on ice. The insoluble fractions were removed by centrifugation at 30,000 × g for 30 min at 4°C. The remaining soluble fractions were filtered through a 0.8/0.2-μm-thickness Acrodisc Supor membrane (Gelman) and used as the crude cell extracts.
Purification of AldB.
All steps were conducted at 4°C. The crude cell extract, prepared from 40 g of wet cell paste in Tris-HCl buffer, was loaded onto a DEAE-Sephacel column (2 by 22 cm) preequilibrated with the same buffer. After washing with 50 ml of Tris-HCl buffer, the column was eluted with a 500-ml linear gradient of 0.1 to 0.5 M NaCl in the same buffer. Fractions containing a majority of the activity were pooled and dialyzed for several hours against 10 mM sodium phosphate, pH 7.5, containing 1 mM EDTA and 5 mM NaCl. The dialysate was clarified by centrifugation at 30,000 × g for 15 min and loaded onto an affinity column (0.9 by 12.5 cm) of p-hydroxylacetophenone (HAP) (10) preequilibrated with sodium phosphate buffer. The AldB-containing fractions were not retained on the column and were collected for ammonium sulfate fractionation. The proteins precipitated at 20 to 60% saturation were resuspended in Tris-HCl buffer and centrifuged as before. The resulting supernatant was loaded onto a Q-Sepharose column (0.8 by 10 cm) preequilibrated with Tris-HCl buffer. The Q-Sepharose column, connected to a fast protein liquid chromatographic system (Amersham Pharmacia Biotech), was eluted with a 400-ml linear gradient of 0 to 20% NaCl, generated by mixing buffer A (Tris-HCl buffer) with buffer B (1 M NaCl in Tris-HCl buffer). Active fractions were pooled, concentrated, and exchanged into 10 mM sodium phosphate, pH 7.5, and 100 mM NaCl using an Amicon cell concentrator fitted with a YM100 membrane. The concentrated sample was loaded onto a 5′AMP-Sepharose column (0.5 by 5 cm) equilibrated with 10 mM sodium phosphate, pH 7.5, containing 100 mM NaCl and 0.5 mM EDTA. The bound enzyme was eluted with a 200-ml linear gradient of 10 to 30 mM sodium phosphate, pH 7.0, containing 100 mM NaCl. Fractions containing homogeneous AldB, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), were pooled for subsequent studies.
Enzyme assays.
AldB activity was measured at 25°C by determining spectrophotometrically or fluorometrically the reduction of NADP or NAD (9, 28). Measurement of ALDH activity was performed in a 1-ml mixture containing 50 mM sodium phosphate (pH 7.5), 50 to 100 μl of crude cell extract or 10 to 15 μg of purified enzyme, 500 μM NADP or NAD, and various aldehyde concentrations (0.05 to 1 mM). Malate dehydrogenase activity was measured in the same crude cell extracts used for studying the effect of culture age on AldB activity. Measurement of malate dehydrogenase activity with 1 mM oxaloacetic acid and 0.1 mM NADH was performed by a method of Park et al. (21).
Electrophoresis.
SDS-PAGE was performed on 10% (wt/vol) slab gels with a discontinuous buffer system (15) in a Bio-Rad Mini-PROTEAN II electrophoresis cell (Bio-Rad). Protein bands were stained with Coomassie brilliant blue (3) and recorded by a photo imaging system (Chemilmager 5500; Alpha Innotech Corporation).
Molecular mass determination.
Three methods were used to determine the molecular mass of the purified AldB. First, SDS-PAGE was conducted as described above. The marker proteins (Low Range Control, catalog no. 161-0305; Bio-Rad Laboratories) included phosphorylase B (103 kDa), bovine serum albumin (77 kDa), ovalbumin (50 kDa), carbonic anhydrase (34.3 kDa), and soybean trypsin inhibitor (28.8 kDa). Second, gel filtration chromatography was conducted on a Bio-Rad Bio-Sil SEC250-5 column (300 by 7.8 mm) by using an SP 8800/8810 LC pump (Spectra-Physics). The column was washed with 100 mM sodium phosphate (pH 7.0) and 150 mM NaCl and 20-μl samples containing ∼5 μg of the purified protein were chromatographed at a flow rate of 1 ml/min. The effluent was monitored by following the absorbance at 280 nm. The column was calibrated by marker proteins, including ferritin (443 kDa), β-amylase (200 kDa), yeast alcohol dehydrogenase (150 kDa), human class 3 aldehyde dehydrogenase (92.5 kDa), bovine serum albumin (66.2 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). Third, mass spectrometry was conducted with a Voyager-DE PRO matrix-assisted laser desorption ionization time of flight instrument (PerSpective Biosystem Inc.). Pinapinic acid was used as a matrix, and measurement was conducted in a linear mode combined with delayed extension. The scale of the molecular mass was calibrated with bovine serum albumin.
N-terminal sequence and amino acid composition analysis.
For amino acid sequencing, a purified sample of AldB was blotted onto a polyvinylidene difluoride membrane (Bio-Rad) after SDS-PAGE (29). Analysis was carried out by an automated protein sequenator (Applied Biosystems Model 477A) in accordance with the manufacturer's instructions. The BLAST program (1), provided by the NCBI website (http://www.ncbi.nlm.nih.gov), was used to search for proteins with sequence similarity. Amino acid composition analysis was performed after hydrolysis of proteins with distilled 6 M HCl in evacuated sealed tubes at 150°C for 24, 48, and 96 h. The residues were determined using a Durrum D-500 amino acid analyzer according to the manufacturer's instructions.
Cloning of AldB, oligonucleotide-directed mutagenesis, and gene expression.
AldB was amplified by PCR using genomic DNA from E. coli ATCC 700926D (American Type Culture Collection). Primers corresponding to the N terminus and C terminus of AldB were designed according to the published gene sequence (4). The forward (F) and reverse (R) primers, 5′GAATTCCATATGACCAATAATCCCCCTTCAGCACAGATTAAGC-3′ and 5′-CGGATCCTCAGAACAGCCCCAACGGTTTATCCGAGTAGC-3′, contained an NdeI and a BamHI site, respectively. PCR was conducted at an annealing temperature of 57°C. A 15-kbp PCR fragment containing AldB was cloned into a pGEM-T easy vector (Promega). AldB in the recombinant plasmid, pGEMAldB, was amplified by PCR using the F and R primers, digested with NdeI and BamHI, and then cloned into pCJM. To construct the gene for the mutated protein AldB R187E, two other primers, 5-GTGGTGCTGAAACCTGCAGAGCTTACTCCGCTTTCT GTAC-3′ and 5′-GTACAGAAAGCGGAGTAAGCTCTGCAGGTTTCAGCACCAC-3′, were used. These primers were used for PCR with pGEMAldB based on the instructions of a QuikChange XL site-directed mutagenesis kit (Stratagene). The altered AldB was then amplified using the F and R primers, digested with NdeI and BamHI, and cloned into pCJM. Proteins were expressed in E. coli DH5α cells by growing the transformed cells at 37°C and inducing them with isopropyl-β-d-thiogalactopyranoside overnight at 16°C when the optical density at 600 nm of the culture reached 0.4 to 0.5.
RESULTS
Crude cell extracts corresponding to different culture ages were measured for aldehyde dehydrogenase activities on acetaldehyde and propionaldehyde with NADP. Malate dehydrogenase activity was measured for the purpose of comparison. The results (Fig. 1) indicate that malate dehydrogenase activity increased initially to a maximum at 4 to 6 h after subculturing and then began to decline. In contrast, aldehyde dehydrogenase activities on acetaldehyde or propionaldehyde initially declined to a minimum at 2 h after subculturing and then increased to a maximum 8 to 9 h after subculturing, during the stationary phase. Among the different cell cultures, the level of enzyme activity with acetaldehyde was consistently higher than that with propionaldehyde. The change in aldehyde dehydrogenase activity on either substrate followed a pattern similar to that of aldB expression during the growth course from early log phase to stationary phase (32), suggesting that the gene product of aldB is responsible for the enzyme activity. Our second finding, that addition of 0.5 to 2% ethanol increased aldehyde dehydrogenase activity in the crude extracts by twofold, agrees with this suggestion since ethanol has been reported to induce aldB expression (32). In the latter study, addition of 2.1% ethanol in the growth medium resulted in about a twofold increase in AldB-LacZ protein fusion activity. It should be noted that an ethanol concentration above 0.25% appeared to slow bacterial growth initially since cultures exposed to ethanol were found to take longer than the control culture to reach the stationary phase (data not shown). In order to ensure that the cell cultures at different ethanol concentrations reached the stationary phase before cell harvesting, a predetermined growth period of 12 h was applied to all cultures. Heim and Strehler (11) have conducted a similar study, growing E. coli Y1088 cells on 0.3% ethanol for 18 h, and found an increase of NADP-dependent acetaldehyde dehydrogenase activity in the crude cell extract. These authors also reported NAD-dependent acetaldehyde dehydrogenase activity in the extract. However, this NAD-dependent activity was either too low for detection or absent in our extracts.
FIG. 1.
Effect of culture age on malate dehydrogenase (10−1) (▿) and aldehyde dehydrogenase (acetaldehyde, ▵; propionaldehyde, ○) activities in crude cell extracts. Samples were removed at 1- or 2-h intervals for measuring cell densities (▪) and enzyme activities. DH5α(pGEM) was grown as described in Materials and Methods. All values represent the means of three measurements, and similar results were obtained in a duplicate experiment. OD, optical density.
The scheme for purifying AldB is summarized in Table 1. It involved a number of steps, including ammonium sulfate precipitation and chromatographic separation on different gel matrices. HAP affinity chromatography, originally designed to bind NAD-dependent dehydrogenases (10), was found to be important. Omission of this step resulted in a partially purified AldB with two contaminating proteins as determined by SDS-PAGE, and numerous attempts to remove them by other methods proved unsuccessful. AldB failed to bind to the HAP affinity column, and this tends to support our latter finding, that AldB has a preference for NADP. The homogeneity of AldB was verified by both SDS-PAGE and N-terminal amino acid sequencing. The first 10 amino acid residues from the N terminus were determined to be MTNNPPSAQI. This unique sequence was identical to that inferred from the nucleotide sequence of aldB (32). The amino acid composition of AldB was also similar to that inferred from the deduced amino acid sequence of the aldB (data not shown). Throughout the purification, the AldB activities with acetaldehyde and propionaldehyde maintained a ratio of about 1.2:1.7 in favor of acetaldehyde. This near-constant relationship implies that both activities were catalyzed by the same enzyme. An apparent mass of about 58 kDa was determined for AldB by SDS-PAGE. A more accurate molecular mass of 56,352 Da was determined by mass spectrometry. This value is close to the one calculated from the deduced amino acid sequence of AldB (56,306 Da) (32). Under nondenatured conditions, the apparent size of AldB was about 220 ± 8 kDa as determined by gel filtration chromatography on a calibrated column. The elution time (8.2 min) was not affected by protein concentrations ranging from 0.25 to 2.5 μg/μl. All these data are consistent with the presence in solution of a stable tetrameric AldB with presumed identical subunits.
TABLE 1.
Purification of AldB from DH5α(pGEM-T) cells
| Purification step | Vol (ml) | Protein (mg) | Total activity (U)a | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|---|
| Cell extract (after filtration) | 60 | 11,250 | 175 | 0.015 | 100 |
| DEAE-Sephacel | 52 | 350 | 75 | 0.2 | 42 |
| HAP affinity column | 24 | 210 | 70 | 0.33 | 40 |
| 20-60% (NH4)2SO4 pellet | 30 | 89 | 49 | 0.55 | 28 |
| Q-Sepharose | 35 | 41 | 37 | 0.9 | 21 |
| 5′ AMP-Sepharose | 4 | 20 | 25 | 2.0 | 14 |
One unit is defined as the amount of enzyme required to produce 1 μmol of NADPH per minute.
The substrate specificity of AldB was investigated at an optimum pH of 7.0, using different aldehydes and coenzymes (Table 2). AldB oxidized a number of tested aldehydes with NADP. Their relative activities increase in the following order: chloroacetaldehyde > acetaldehyde > propionaldehyde > benzaldehyde > mafosfamide > 4-hydroperoxycyclophosphamide. Other tested aldehydes, including glycolaldehyde and glyceraldehyde, were oxidized not at all or insignificantly by AldB with NADP. None of the tested aldehydes were sufficiently oxidized by the enzyme in the presence of NAD. For instance, with 1 to 2 mM NAD and 250 μM acetaldehyde or propionaldehyde, the enzyme activity obtained was only about 1% of that obtained with the same NADP and substrate concentrations. When assayed with 0.25 to 0.5 mM MgCl2, NADP-dependent AldB activity on acetaldehyde, propionaldehyde, or benzaldehyde increased by about 100% (Table 2). In the case of chloroacetaldehyde, the enzyme activity remained the same regardless of the presence or absence of MgCl2. These results are very different from those reported for the substrate specificity of lactaldehyde dehydrogenase (2). To further characterize the substrate specificity of AldB, its kinetic properties with different substrates were determined by bisubstrate kinetics analysis. Several concentrations of NADP (0.05 to 0.5 mM) and aldehyde (0.1 to 1 mM) were used for analysis of the kinetic data. The kinetics constants obtained by a graphic procedure (7) are summarized in Table 3. The results indicated that chloroacetaldehyde and acetaldehyde were preferable substrates for AldB and that acetaldehyde appeared to have a lower Km, 2.5 μM. The kcat/Km values also indicated that the catalytic efficiencies for the two substrates were similar. Another bacterial enzyme reported to have a low Km for acetaldehyde is A. eutrophus NAD-dependent acetaldehyde dehydrogenase (Km = 4 μM) (24). Thus, it appears that the two enzymes share not only a high degree of sequence similarity but also a preference for acetaldehyde.
TABLE 2.
Activities of AldB relative to activities of acetaldehyde in the presence and absence of 0.4 mM MgCl2a
| Substrate | Relative activity (%)
|
|
|---|---|---|
| −MgCl2 | +MgCl2 | |
| Acetaldehyde | 100 | 100 |
| Chloroacetaldehyde | 193 | 100 |
| Propionaldehyde | 65 | 57 |
| Glyceraldehyde | 1 | 0 |
| Glycolaldehyde | 0 | 0 |
| Benzaldehyde | 40 | 37 |
| Mafosfamide | 21 | 16 |
| 4-Hydroperoxycyclophosphamide | 12 | 20 |
All values represent means of results from duplicate experiments.
TABLE 3.
Kinetic properties of AldBa
| Substrate | Vmax (U/mg) | Km (μM) | Kcat/Km (mM−1 · min−1) |
|---|---|---|---|
| Acetaldehyde | 2 | 2.5 | 225 |
| Chloroacetaldehyde | 3.3 | 3.6 | 206 |
| Propionaldehyde | 1 | 5.8 | 39 |
| Benzaldehyde | 0.6 | 56.8 | 2 |
| 4-Hydroperoxycyclophosphamide | 0.2 | 900 | 0.05 |
All values represent means of results from duplicate experiments.
The effect of MgCl2 on the kinetic properties of AldB was also studied with acetaldehyde. The results (Table 4) indicated that the Km of AldB for acetaldehyde was not affected by MgCl2, since the Km for the substrate varied very little in the presence or absence of MgCl2. In contrast, the results showed a twofold increase of the dissociation constant (Kia) for NADP in the presence of MgCl2, suggesting a decreased affinity of the enzyme for the coenzyme.
TABLE 4.
Effect of MgCl2 on the properties of AldB as analyzed by two substrate kineticsa
| Kinetic property (μM) | Value in the presence of:
|
|
|---|---|---|
| 0 mM MgCl2 | 0.4 mM MgCl2 | |
| Km, NADP | 65 | 102 |
| Km, acetaldehyde | 3.5 | 2.7 |
| Kia, NADP | 35 | 80 |
| Vmax, acetaldehyde | 2 | 4.5 |
All values represent means of results from duplicate experiments. Kia, dissociation constant.
Sequence alignment comparisons (Fig. 2) show that the NAD-dependent aldehyde dehydrogenases have a glutamate at a position corresponding to R197 in AldB. To determine the importance of R197 in the coenzyme specificity of AldB, the genes coding for the wild-type enzyme and the R197E mutant were cloned and expressed. Expression of the wild-type enzyme in DH5α E. coli produced a high level of enzyme activity in the crude cell extract. When the R197E mutant was expressed under similar conditions, a low level of enzyme activity was detected in the crude extract, prepared from approximately the same wet cell mass as the wild-type enzyme. When expressed in terms of per-unit protein, the R197E extract had less than 10% of the NADP-dependent acetaldehyde dehydrogenase activity found in the wild-type enzyme's extract. There was no detectable NAD-dependent activity in either cell extract. The AldB activity from individual extracts was further purified by a DEAE-Sephacel step described for the native enzyme from DH5α(pGEM). The amount of AldB used in individual purifications was adjusted to contain the same enzyme activity. The elution profiles for both enzymes were essentially the same, each being characterized by the presence of an activity peak and an overlapping protein peak. The fractions containing the majority of the activity were then pooled from individual preparations and assayed for enzyme activity and protein concentration. Based on the amount per unit of protein, the NADP-dependent acetaldehyde dehydrogenase activity of the R197E preparation was only a few percent of that found with the wild-type AldB preparation. Again, there was no detectable NAD-dependent activity in either preparation. SDS-PAGE revealed no significant differences between the two preparations in their protein profiles when the same amount of protein from each was electrophoresed. Each had a dominant 58-kDa band with a subunit mass identical to that of the native enzyme from DH5α(pGEM). The wild-type AldB was purified to homogeneity by the method established for the native enzyme from DH5α(pGEM), and its specific activity was comparable to that of the purified preparation made from DH5α(pGEM). Due to the low level of enzyme activity recovered after the DEAE-Sephacel step, we did not further purify the mutant enzyme.
FIG. 2.
Details of the multiple sequence alignment for AldB (E. coli) with aldehyde dehydrogenase sequences of AcoD (A. eutrophus), AldA (V. cholerae), ALDH1 (human liver cytosol, NP-000680) and ALDH2 (human liver mitochondria, NP-000681) (National Center for Biotechnology Information database; www.ncbi.nlm.nih.gov). Dashes (−) indicate the amino acids lying close to the adenosine ribose. Asterisks (*), colons (:), and periods (.) indicate residues with identity, strong similarity, and weak similarity, respectively. The alignment represents the region where all the sequences exhibit identities greater than 60%.
DISCUSSION
Our results clearly indicate that AldB is different from lactaldehyde dehydrogenase in both coenzyme and substrate specificities (2). While AldB is NADP dependent, lactaldehyde dehydrogenase is NAD dependent. Lactaldehyde dehydrogenase is also characterized by high activity on either glycolaldehyde or glyceraldehyde. Under our assay conditions, AldB was unable to oxidize either substrate. Instead, it showed a high activity with acetaldehyde, which was not oxidized by the other enzyme. Another major difference between the two enzymes is their response to MgCl2. With AldB, the enzyme activities on selected substrates were increased by MgCl2. In contrast, such activating effect of MgCl2 was not reported with lactaldehyde dehydrogenase (2). All these findings suggest that the two enzymes are not functionally related. The finding that acetaldehyde is a major substrate for AldB, together with our growth studies results, is consistent with those of other workers (32), suggesting that AldB is a stationary-phase enzyme and that it can help in the removal of toxic alcohols and aldehydes in cells growing in severe conditions. E. coli and other bacteria (5) are known to carry out mixed-acid fermentation of sugars under anaerobic conditions, resulting in the formation of ethanol.
In an earlier study of aldH in E. coli Y1088 (11), two aldehyde dehydrogenase activities with acetaldehyde, one dependent on NADP and the other on NAD, have been detected in the cell extracts. When ethanol was included in the growth medium, the level of NADP-dependent activity in the cell extract increased. In contrast, the level of NAD-dependent activity in the same cell extract appeared to decrease. This suggests that the two activities belong to different enzymes. The NADP-dependent activity is probably related to aldB in E. coli Y1088S, since the ethanol effect on this activity concurs with that on AldB activity in DH5α(pGEM).
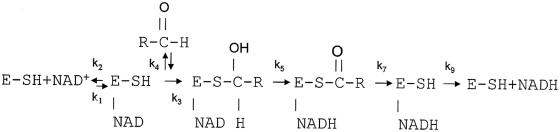
Although AldB is a prokaryotic enzyme, it has some of the properties found in human liver mitochondrial aldehyde dehydrogenase. First, like the human enzyme (17), AldB is a tetramer with a preference for acetaldehyde. Second, their coenzyme binding region and amino acid residues responsible for enzyme catalysis appear to be similar, as indicated by sequence alignments (32). Third, when assayed with MgCl2, AldB was found to increase activity on various substrates. This Mg2+ ion-activating effect occurs in the human class-2 mitochondrial aldehyde dehydrogenase (18) and is related to a rate-limiting step in a postulated enzyme mechanism (31) depicted in Fig. 3. Basically, the reaction involves binding of an aldehyde to an initial enzyme-NAD complex to form a thiohemiacetal intermediate, which is then oxidized to a thioacyl intermediate. Mg2+ ions are thought to enhance deacylation of the thioacyl intermediate (k7), the rate-limiting step prior to release of NADH. The same rate-limiting step may occur in AldB, as suggested by the Mg2+ ion-activating effect on the enzyme. Consistent with this suggestion is the finding (Table 2) that without Mg2+ ions, AldB oxidized chloroacetaldehyde faster than acetaldehyde. In fact, the ratio between AldB activities with chloroacetaldehyde and acetaldehyde was about two. Finally, it was found that AldB activity on chloroacetaldehyde was not increased by Mg2+ ions, whereas the enzyme activity on acetaldehyde was increased twofold, matching that with chloroacetaldehyde (Table 2). This finding has also been obtained with the human enzyme (A. Allali-Hassani and H. Weiner, unpublished data). One would expect that chloroacetaldehyde is a better substrate than acetaldehyde for AldB or the human enzyme with or without Mg2+ ions, due to the electron-withdrawing effect of its chlorine atom. Instead, both substrates were found to oxidize equally well in the presence of Mg2+ ions. This finding suggests that the chloroacyl intermediate might be hydrolyzed at a rate that probably could not be increased further by Mg2+ ion activation of the general base. It is worth mentioning that the Mg2+ ion-activating effect on enzyme activity has been reported for a yeast cytosolic aldehyde dehydrogenase (8). The enzyme was reported to show a dramatic 100-fold increase in its affinity for NADP as a result of the Mg2+ ion activation that appeared to be accompanied by a conformational change in the enzyme from an inactive form to an active form. The inactive form was found to have only about 3% of the Mg2+ ion-activated rate with acetaldehyde. With AldB, we noticed that only a small change in the enzyme affinity for NADP with Mg2+ ions and that the enzyme activity on acetaldehyde without Mg2+ ions was more than half of that with Mg2+ ions. Such findings show that AldB does not behave like the yeast enzyme.
FIG. 3.
Scheme showing the reaction pathway for the aldehyde dehydrogenase-catalyzed oxidation of an aldehyde (31).
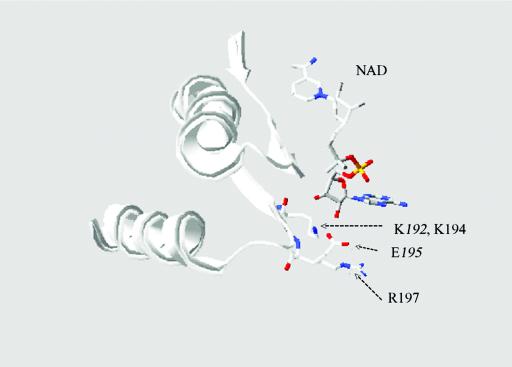
Most of the NAD-dependent aldehyde dehydrogenases studied have two conserved residues, lysine and glutamate, lying close to the adenosine ribose (23). The lysine residue, also conserved in NADP-dependent aldehyde dehydrogenases, interacts with the adenosine ribose in NAD and the 2′ phosphate in NADP. The glutamate residue helps coordinate the adenine ribose 2′ hydroxyl of NAD and is thought to be responsible for the exclusion of NADP. The alignment of the aino acid sequences of AldB and other NAD-dependent aldehyde dehydrogenases reveals that in AldB, lysine (K194) and arginine (R197) correspond to the conserved residues lysine and glutamate, respectively (Fig. 2). Using the Swiss-Model software at the ExPASy website (http://swissmodel.expasy.org/), a three-dimensional (3D) model of the region extending from H158 to D212 of AldB has been constructed using bovine mitochondrial aldehyde dehydrogenase (27) as a template. This region showed a significant 60% sequence identity when compared with the selected region from H156 to E210 of the template. A comparison of their 3D structures (Fig. 4) confirms that K194 and R197 of AldB are located at positions corresponding to those of the conserved K192 and E195 in the bovine mitochondrial aldehyde dehydrogenase, respectively. The arginine residue at position 197 was mutated to a glutamate to learn whether this residue was responsible for the enzyme being an NADP-dependent dehydrogenase. Though the mutant was not purified to homogeneity, it was found to possess approximately 10% of the catalytic activity of the wild-type enzyme when assayed with NADP and acetaldehyde. Unexpectedly, it did not exhibit any detectable NAD-dependent activity. This residue, while interacting with the coenzyme, is important for overall activity, but does not seem to be the sole determinant for coenzyme specificity. Others have found that they can change the coenzyme specificity of different aldehyde dehydrogenases by making point mutations. Replacement of T175 by a negatively charged amino acid resulted in a change of coenzyme specificity from NAD to NADP and an increase of catalytic efficiency for NAD in Vibrio harveyi fatty aldehyde dehydrogenase (34). Another example of changing NAD/NADP specificity preference as a result of replacing just one amino acid (E140) is found in class-3 rat aldehyde dehydrogenase (23). For the respective enzymes, T175 and E140 correspond to R197 of AldB based upon the sequence alignment. Thus, it was surprising to find that the coenzyme specificity did not change in the R197E mutant. A change in the enzyme's catalytic properties can be affected when changing a residue that is in contact with the coenzyme. Mutation of the conserved lysine at position 192 of the human class-2 aldehyde dehydrogenase caused the rate-limiting step of the enzyme to change from deacylation, k7, to hydride transfer, k5 (Fig. 3) (18). Thus, a small alteration in coenzyme binding may have been the cause of the R197E mutant of AldB having so little activity compared to the native enzyme.
FIG. 4.
A comparison of the region from H156 to E210 of HALD2 (human liver mitochondria, aldehyde dehydrogenase, NP-000681) with the region from H158 to D212 of the 3D-structure model derived for AldB (E. coli). The overlapping structures show that K194 and R197 from AldB are located at positions resembling those of K192 and E195 from HALD2.
Both mammals and E. coli possess many different isozymes of aldehyde dehydrogenase. Though some share little identity in sequence or properties, the human mitochondrial enzyme and AldB have much in common. Most significant is the Mg2+ ion effect. Unfortunately, structural data of the human enzyme (17, 27), with respect to the NAD binding domain, cannot be used to explain why AldB uses NADP instead of NAD.
Acknowledgments
This work was supported in part by grant AA05812 from the National Institutes of Health.
Footnotes
This is journal paper 17531 from the Purdue University Agricultural Experiment Station.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldoma, L., and J. Aguilar. 1987. Involvement of lactaldehyde dehydrogenase in several metabolic pathways of Escherichia coli K12. J. Biol. Chem. 262:13991-13996. [PubMed] [Google Scholar]
- 3.Blakesley, R. W., and J. A. Boezi. 1977. A new staining technique for protein in polyacrylamide gels using Coomassie brilliant blue G250. Anal. Biochem. 82:580-582. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Lett. 63:223-234. [DOI] [PubMed] [Google Scholar]
- 6.Cocks, G. T., J. Aguilar, and E. C. C. Lin. 1974. Evolution of the l-1,2-propanediol catabolism in Escherichia coli by recruitment of enzymes for l-fucose and l-lactate metabolism. J. Bacteriol. 118:83-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalziel, K. 1957. Initial steady state velocities in the evaluation of enzyme-coenzyme-substrate reaction mechanisms. Acta Chem. Scand. 11:1706-1723. [Google Scholar]
- 8.Dickinson, F. M. 1996. The purification and some properties of the Mg2+-activated cytosolic aldehyde dehydrogenase of Saccharomyces cerevisiae. Biochem. J. 315:393-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman, R. I., and H. Weiner. 1972. Horse liver aldehyde dehydrogenase. II. Kinetics and mechanistic implications of the dehydrogenase and esterase activity. J. Biol. Chem. 247:267-272. [PubMed] [Google Scholar]
- 10.Ghenbot, G., and H. Weiner. 1992. Purification of liver aldehyde dehydrogenase by p-hydroxyacetophenone-sepharose affinity matrix and the coelution of chloramphenicol acetyl transferase from the same matrix with recombinantly expressed aldehyde dehydrogenase. Protein Expr. Purif. 3:470-478. [DOI] [PubMed] [Google Scholar]
- 11.Heim, R., and E. E. Strehler. 1991. Cloning an Escherichia coli gene encoding a protein remarkably similar to mammalian aldehyde dehydrogenases. Gene 99:15-23. [DOI] [PubMed] [Google Scholar]
- 12.Hengge-Aronis, R. 1993. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell 72:165-168. [DOI] [PubMed] [Google Scholar]
- 13.Hidalgo, E., Y. M. Chen, E. C. C. Lin, and J. Aguilar. 1991. Molecular cloning and DNA sequencing of the Escherichia coli K-12 ald gene encoding aldehyde dehydrogenase. J. Bacteriol. 173:6118-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolter, R., D. A. Siegele, and A. Tormo. 1993. The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47:855-874. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.LeBlanc, D., and R. P. Mortlock. 1971. Metabolism of d-arabinose: a new pathway in Escherichia coli. J. Bacteriol. 106:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, N., J. Z. Zhou, T. D. Hurley, and H. Weiner. 1999. Human liver mitochondrial aldehyde dehydrogenase: three dimensional structure and the restoration of solubility and activity of chimeric forms. Protein Sci. 8:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, N., S. Sheikh, and H. Weiner. 1997. Involvement of glutamate 399 and lysine 192 in the mechanism of human liver mitochondrial aldehyde dehydrogenase. J. Biol. Chem. 272:18823-18826. [DOI] [PubMed] [Google Scholar]
- 19.Limon, A., E. Hidalgo, and J. Aguilar. 1997. The aldA gene of Escherichia coli is under the control of at least three transcriptional regulators. Microbiology 143:2085-2095. [DOI] [PubMed] [Google Scholar]
- 20.Masui, Y., T. Mizuno, and M. Inouye. 1984. Novel high-level expression cloning vehicles: 104-fold amplification of Escherichia coli minor protein. Nat. Biotechnol. 2:81-85. [Google Scholar]
- 21.Park, S. J., P. A. Cotter, and R. P. Gunsalus. 1995. Regulation of malate dehydrogenase (mdh) gene expression in Escherichia coli in response to oxygen, carbon, and heme availability. J. Bacteriol. 177:6652-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsot, C., and J. J. Mekalanos. 1991. Expression of the Vibrio cholerae gene encoding aldehyde dehydrogenase is under control of ToxR, the cholera toxin transcription activator. J. Bacteriol. 173:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perozich, J., I. Kuo, B. C. Wang, J. S. Boesch, R. Lindahl, and J. Hempel. 2000. Shifting the NAD/NADP preference in class 3 aldehyde dehydrogenase. Eur. J. Biochem. 267:6197-6203. [DOI] [PubMed] [Google Scholar]
- 24.Priefert, H., N. Kruger, D. Jendrossek, B. Schmidt, and A. Steinbuchel. 1992. Identification and molecular characterization of the gene coding for acetaldehyde dehydrogenase II (acoD) of Alcaligenes eutrophus. J. Bacteriol. 174:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schellhorn, H. E., J. P. Audia, L. I. C. Wei, and L. Chang. 1998. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J. Bacteriol. 180:6283-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sophos, N. A., and V. Vasiliou. 2003. Aldehyde dehydrogenase gene superfamily: the 2002 update. Chem. Biol. Interact. 143-144:5-22. [DOI] [PubMed] [Google Scholar]
- 27.Steinmetz, C. G., P. G. Xie, H. Weiner, and T. D. Hurley. 1997. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure 5:701-711. [DOI] [PubMed] [Google Scholar]
- 28.Svanas, G. W., and H. Weiner. 1985. Aldehyde dehydrogenase activity as the rate-limiting factors for acetaldehyde metabolism in rate liver. Arch. Biochem. Biophys. 236:36-46. [DOI] [PubMed] [Google Scholar]
- 29.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vasiliou, V., A. Pappa, and D. R. Petersen. 2000. Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem. Biol. Interact. 129:1-19. [DOI] [PubMed] [Google Scholar]
- 31.Weiner, H., J. Farres, U. J. Rout, X. P. Wang, and C. F. Zheng. 1995. Site directed mutagenesis to probe for active site components of liver mitochondrial aldehyde dehydrogenase, p. 1-7. In H. Weiner, R. S. Holmes, and B. Wermuth (ed.), Enzymology and molecular biology of carbonyl metabolism, vol. 5. Plenum Press, New York, N.Y. [DOI] [PubMed]
- 32.Xu, J., and R. C. Johnson. 1995. aldB, an RpoS-dependent gene in Escherichia coli encoding an aldehyde dehydrogenase that is repressed by Fis and activated by Crp. J. Bacteriol. 177:3166-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, J., and R. C. Johnson. 1995. Identification of genes negatively regulated by Fis: Fis and RpoS comodulate growth-phase-dependent gene expression in Escherichia coli. J. Bacteriol. 177:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, L., B. Ahvazi, R. Szittner, A. Vrielink, and E. Meighen. 1999. Change of nucleotide specificity and enhancement of catalytic efficiency in single point mutants of Vibrio harveyi aldehyde dehydrogenase. Biochemistry 38:11440-11447. [DOI] [PubMed] [Google Scholar]