Abstract
The major regulator controlling the physiological switch between aerobic and anaerobic growth conditions in Escherichia coli is the DNA binding protein FNR. To identify genes controlled by FNR, we used Affymetrix Antisense GeneChips to compare global gene expression profiles from isogenic MG1655 wild-type and Δfnr strains grown in glucose minimal media under aerobic or anaerobic conditions. We found that 297 genes contained within 184 operons were regulated by FNR and/or by O2 levels. The expression of many genes known to be involved in anaerobic respiration and fermentation was increased under anaerobic growth conditions, while that of genes involved in aerobic respiration and the tricarboxylic acid cycle were repressed as expected. The expression of nine operons associated with acid resistance was also increased under anaerobic growth conditions, which may reflect the production of acidic fermentation products. Ninety-one genes with no presently defined function were also altered in expression, including seven of the most highly anaerobically induced genes, six of which we found to be directly regulated by FNR. Classification of the 297 genes into eight groups by k-means clustering analysis indicated that genes with common gene expression patterns also had a strong functional relationship, providing clues for studying the function of unknown genes in each group. Six of the eight groups showed regulation by FNR; while some expression groups represent genes that are simply activated or repressed by FNR, others, such as those encoding functions for chemotaxis and motility, showed a more complex pattern of regulation. A computer search for FNR DNA binding sites within predicted promoter regions identified 63 new sites for 54 genes. We suggest that E. coli MG1655 has a larger metabolic potential under anaerobic conditions than has been previously recognized.
For many bacteria, O2 plays a critical role in the regulation of numerous cellular processes. The utilization of O2 as a terminal electron acceptor during aerobic respiration results in the highest conservation of cellular energy and therefore provides a great energetic advantage to microorganisms (93). O2 in excess, however, can be detrimental to cells because of the production of reactive oxygen species (35). Many bacteria have evolved adaptive strategies to balance consumption of O2 for energy metabolism with detoxification of reactive oxygen species. In addition, it is well established that Escherichia coli and other facultative anaerobes are genetically programmed to adapt between environments of high and low O2 levels (87). In the case of E. coli, removal of O2 causes major changes in the expression of pathways that promote energy conservation (67, 93). For example, the expression of genes encoding respiratory enzymes that utilize O2 as an electron acceptor (e.g., cytochrome oxidases) is repressed, whereas the expression of genes encoding enzymes that utilize alternative electron acceptors, such as nitrate, fumarate, etc., or that promote fermentation is increased under anaerobic conditions. Nevertheless, while many cellular activities that function under anaerobic conditions have been uncovered, we still lack a comprehensive view of the anaerobic lifestyle of E. coli, even though one of its important ecological niches is the low-O2 environment of the gut.
FNR and the two-component regulatory system, ArcAB, are the two major regulatory systems that respond to decreases in O2 levels in E. coli (29). FNR has an O2-sensitive [4Fe-4S]2+ cluster that directly senses O2 and regulates site-specific DNA binding (40). In contrast, ArcAB senses signals emanating from the aerobic respiratory chain, indicating at least two divergent yet fundamental mechanisms for O2 sensing in this organism (26). Recent gene expression profiling studies have identified 55 operons of the ArcA regulon (52). In an attempt to further our understanding of the metabolic potential of E. coli under anaerobic growth conditions, we sought to identify additional genes that are controlled by the global regulator FNR through genome-wide expression studies.
Previous studies have shown that FNR controls the expression of >100 gene products in E. coli (67, 75). Known FNR-activated genes include those encoding enzymes for the anaerobic oxidation of carbon sources, such as glycerol and formate, genes encoding enzymes for the anaerobic reduction of alternate terminal electron acceptors, such as nitrate, fumarate, and dimethyl sulfoxide (DMSO), and genes encoding proteins for transport of these carbon sources or electron acceptors. FNR also represses the expression of genes that are needed for aerobic metabolism, such as the respiratory enzymes NADH dehydrogenase II and cytochrome oxidases. As a consequence, FNR-dependent gene expression allows compounds such as fumarate or nitrate to replace O2 as a terminal electron acceptor in order to sustain the energy-conserving pathways of oxidative phosphorylation under anaerobic conditions.
Genome-wide expression approaches have been invaluable in identifying genes that respond to changes in environmental conditions. In this study, we chose to analyze the gene expression profiles from strain MG1655, since this strain is considered to best represent wild-type E. coli K-12 and it is the strain from which the genome sequence was determined (7). We used Affymetrix arrays (derived from the MG1655 DNA sequence) to determine which genes are differentially expressed in response to O2 and/or FNR. Our data include many of the known FNR-regulated genes as well as many new genes not previously implicated in the anaerobic response. During preparation of this paper, Salmon et al. (73) reported results of global gene expression profiles of E. coli K-12 strain MC4100 grown under aerobic and anaerobic conditions and the effect of FNR on these global gene expression patterns. We also compare the results from these two independent studies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli K-12 strain MG1655 (seq) and an isogenic Δfnr::ΩSpr Smr strain (constructed by P1 transduction using RZ8459 [46] as the donor) were used in this study. The strains were grown under aerobic or anaerobic conditions at 37°C in MOPS (morpholinepropanesulfonic acid) minimal medium (TekNova Inc.) containing 0.1% glucose. Aerobic culture conditions were obtained by shaking 100 ml of culture in a 1-liter flask at 225 rpm in a standard laboratory shaker, while anaerobic conditions were achieved by continuously sparging in a Cytolift Glass Airlift Bioreactor (Kontes) with a 95% N2 and 5% CO2 gas mix. (No significant differences were found when comparing microarray data obtained from cells aerated in the laboratory shaker and sparged with air in the Bioreactor.)
RNA isolation and labeling.
Fifteen milliliters of mid-log-phase cultures (optical density at 600 nm of 0.2 for aerobic cultures and 0.1 for anaerobic cultures) was immediately mixed with 30 ml of Bacteria RNA protect reagent (QIAGEN). RNA was then isolated according to the Masterpure total RNA isolation kit (Epicentre). To label RNA by the method of Rosenow et al. (72), 10 μg of total RNA (measured by its absorbance at 260 nm using an ɛ of 0.025 [μg ml]−1 cm−1) was mixed with 500 ng of random hexamers and reverse transcribed with 1,200 U of Superscript II (Invitrogen) for 90 min at 42°C. Two units of RNase H and 1 μg of RNase A were added to the reaction mix and incubated at 37°C for 10 min to degrade any remaining RNA. The cDNA was purified with a Qiaquick PCR purification kit (QIAGEN) and subsequently fragmented with 0.2 U of DNase I (Epicentre) for 10 min at 37°C. The 3′ terminus of the fragmented cDNA was labeled with 2.5 μM biotin-N6-ddATP (Perkin-Elmer) at 37°C for 2 h using 20 U of terminal transferase (NEB).
DNA microarray hybridization and analysis.
Biotin-labeled fragmented cDNA (2.5 μg) was hybridized to E. coli antisense genome arrays (Affymetrix) at 45°C for 16 h as recommended in the GeneChip technical manual (Affymetrix). The probed arrays were scanned at 570 nm at a resolution of 3 μm using a confocal laser scanner (Hewlett-Packard G2500A). Gene expression levels were calculated with Microarray Suite 5.0 software (Affymetrix). The mean hybridization signal of all probe sets on each array was scaled to a common value of 1,000 (see the supplemental material). The average signal intensity value of each gene was transformed to a log2 (log base 2) value to simplify data handling. We used a log2 signal intensity value of 8 or larger rather than the Affymetrix algorithm to determine whether a gene was expressed in order to retain less highly expressed genes in the data set. Nevertheless, 90% of the genes that had an intensity value larger than 8 were also called present by the Affymetrix algorithm. The change between two experimental conditions (n-fold) was calculated by taking the ratio of the signal intensity (difference of the log2 value) between two experimental conditions. For each strain and growth condition, three independent cultures were prepared and the RNA was analyzed. The correlation coefficient between signal intensities of any replicate was ∼0.96.
ScanAce.
The algorithm ScanAce (33) was used to identify the best-matching FNR binding sites within the promoter region of genes that were differentially expressed between wild-type and fnr mutant cells under aerobic and anaerobic conditions. The 22 FNR sites (see the supplemental material) that were used to build the site matrix were chosen using published criteria that either mutagenesis of the site resulted in a loss of FNR regulation or the site was defined by footprinting. Sites were retained if they scored better than 1 standard deviation below the mean of the scores for each of the sites used to define the motif and were positioned between −1 and −400 bp upstream of each gene's translation start site. The majority of sites scoring higher than 9 had no more than 3 mismatches from the consensus sequence (TTGATNNNNATCAA).
Construction of single copy promoter-lacZ fusions.
Promoters of interest were amplified from the chromosome by using gene-specific primers flanked by XhoI or BamHI restriction sites. The positions of the fragments relative to their respective translational start sites are as follows: for ydjX, −4 to −274; for ynfE, −3 to −272; for ydhY, −57 to −331; for ydfZ, +16 to −231; for yfcZ, +16 to −312; for talA, −9 to −234; for cusC, −14 to −312; for sufD, −3 to −232; for csgD, −8 to −319; and for sufA, +5 to −229. The resulting PCR fragments were digested with XhoI and BamHI and directionally cloned into plasmid pPK7035, described below, containing XhoI and BamHI sites inserted between lacZ and the kanamycin resistance gene, so that lacZ expression is driven by the promoter fragment. This lacZ promoter construct was then recombined into the chromosomal lac operon by transformation of a second set of PCR fragments containing 36 nucleotides (nt) of lacI, the kanamycin resistance gene, a transcription terminator, the promoter fragment of interest, and ∼900 nt of lacZ in a strain overexpressing the lambda red recombinase (22); these PCR fragments were generated by a primer containing 36 nt of lacI flanked by 18 nt of the kanamycin resistance gene (5′-GGCACGACAGGTTTCCCGACGAAAGCGGGCAGTGAGCCCGGATCAATTCCCCTGCTC-3′) and a lacZ primer (5′-GACGCGATCGGCATAACC-3′).
Recombinants were selected for Knr and screened for the Lac+ phenotype. To verify that the promoter-lacZ fusions recombined at the lac operon, this region of chromosomal DNA was amplified by PCR with gene-specific primers: lacI (5′-GCAATCAGCTGTTGCCCGTCTCACTGG-3′) was used pairwise with either kan (5′-GCTGCCGCAAGCACTCAGG-3′) or lacZ (5′-CCGCACGATAGAGATTCGG-3′). After confirming the constructs by PCR and DNA sequencing, P1 transduction was used to move the Knr promoter-lacZ fusion to RZ4500 and its FNR− derivative RZ8459 (46). The rpoS-lacZ fusion was constructed by Sledjeski et al. (80).
pPK7035 was constructed by ligation of the kanamycin resistance gene, including the sequence corresponding to 600 bp upstream of the translational start site and 200 bp downstream of the translational stop site, from pHP45Ω (25), and 900 bp of the lacZ sequence from pRS1553, including 100 bp upstream of the start codon (78), into the BamHI-NdeI site of pBR322 (9). To minimize read-through transcription from the Knr gene, the orientation was constructed to be opposite to that of lacZ, and the strong transcriptional terminator adjacent to the upstream end of the Knr gene from pHP45Ω was retained in the construct. When a promoterless lacZ construct was recombined into the lac operon, only 5 U of β-galactosidase (β-Gal) was obtained from strains grown under aerobic conditions, while 7 U was obtained from strains grown under anaerobic conditions.
Assay of β-Gal.
Assays of β-Gal from whole cells were carried out as described previously (46). Strains were grown anaerobically in M9 minimal liquid medium (60) containing 0.2% (wt/vol) glucose, 10 μM ferric ammonium citrate, and 2 μM ammonium molybdate to an A600 of 0.2 to 0.4.
In vitro transcription assays.
For in vitro transcription analysis, the chromosomal PCR fragments used to make the promoter-lacZ fusions were ligated into a pUC19-spf derivative (24) in which the SalI site was replaced with an XhoI site by site-directed mutagenesis. In vitro transcription assays were performed using 2 nM template DNA, 1 μM FNR-D154A (46), and 50 nM RNAP holoenzyme as previously described (46). Transcripts labeled with [32P]UTP were quantified with a Molecular Dynamics phosphorimager and ImageQuant version 1.2 software.
RESULTS AND DISCUSSION
Strategy for identifying genes regulated by O2 and/or FNR.
To identify which genes of E. coli K-12 are differentially expressed under anaerobic conditions and to determine the role of FNR in this process, we compared changes in transcript profiles of an FNR− mutant with its wild-type parent, MG1655, grown under both aerobic and anaerobic conditions. By using comparisons of pairwise combinations of the four different experimental conditions (aerobic and anaerobic growth of both FNR+ and FNR− strains), we identified 297 genes that showed a greater-than-threefold change in gene expression with a P value of <0.05 (Student's t test) and had at least an average log2 signal intensity greater than 8 when at least one experimental variable was changed. We also calculated the q value for each gene in the pairwise comparison (see the supplemental material). The P value is a measure of the false positive rate, and the q value is a measure of the false discovery rate (86). These data also showed that a slightly greater number of MG1655 genes were expressed under anaerobic growth conditions (2,257) than under aerobic conditions (2,157). Many of the 297 genes are located in operons with other genes that fell below the threshold established in this analysis. If all genes in an operon are taken into account, then 184 operons comprised of 465 total genes are regulated by FNR and/or O2.
The 297 genes fall into 23 functional groups based on the E. coli gene annotation by Serres et al. (77) (Table 1), indicating that a diverse set of functions is regulated by O2. Nevertheless, a large number (23.6%) of these genes function in energy and central intermediary metabolism, consistent with the metabolic changes previously observed between aerobically and anaerobically grown cells (Fig. 1). In particular, the expression of genes known to be involved in anaerobic respiration and fermentation was increased under anaerobic growth conditions, while those involved in aerobic respiration and the tricarboxylic acid (TCA) cycle were repressed under anaerobic conditions. Surprisingly, the expression of many of the genes encoding functions involved in chemotaxis, motility, and surface structures was increased under anaerobic conditions, a finding we believe has not been previously reported. Of particular significance was the observation that 30.6% of the genes identified have no presently defined function, and seven genes (ynfEFG, ydfZ, ydhV, yhiE, and yliH) of this gene class were the most highly induced under anaerobic conditions. Taken together, these data suggest that we have a limited view of the functions regulated in response to changes in environmental O2 in E. coli.
TABLE 1.
Functional gene groups
| Functional groupa | No. of genes | wt N2:wt O2b
|
Δfnr N2:Δfnr O2b
|
Δfnr N2:wt N2b
|
|||
|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | ||
| Energy metabolism, carbon | 61c | 29 | 21 | 18 | 12 | 13 | 15 |
| Central intermediary metabolism | 12 | 3 | 5 | 3 | 1 | 7 | 2 |
| Fatty acid and phosphatidic acid biosynthesis | 2 | 1 | 1 | 1 | |||
| Global regulatory functions | 3 | 1 | 1 | 1 | |||
| Macromolecule synthesis; modification: polysaccharides (cytoplasmic) | 1 | 1 | |||||
| Ribosomal proteins: synthesis, modification | 1 | 1 | |||||
| Purine ribonucleotide biosynthesis | 1 | 1 | 1 | ||||
| Protein, peptide secretion | 2 | 2 | |||||
| Cell division | 1 | 1 | |||||
| Amino acid biosynthesis | 3 | 1 | 1 | 1 | |||
| Biosynthesis of cofactors; carriers: thiamine | 1 | 1 | |||||
| 2′-Deoxyribonucleotide metabolism | 3 | 1 | 2 | ||||
| Degradation of proteins, peptides, glyco | 1 | 1 | 1 | ||||
| Degradation of small molecules, carbon compounds | 4 | 2 | 1 | 1 | |||
| Detoxification | 3 | 2 | 1 | 1 | |||
| Drug and/or analog sensitivity | 2 | 2 | 2 | ||||
| Outer membrane constituents | 3 | 1 | 2 | 1 | 1 | ||
| Osmotic adaptation | 4 | 1 | 1 | 4 | |||
| Chemotaxis and mobility | 10 | 4 | 10 | ||||
| Surface structures | 37 | 5 | 16 | 2 | 32 | ||
| Transport of small molecules | 22 | 7 | 9 | 2 | 7 | 4 | 6 |
| Unknown | 119 | 32 | 27 | 39 | 12 | 47 | 24 |
| Total | 297 | 78 | 69 | 75 | 55 | 90 | 92 |
Functional group assignments are those described by Serres et al. (77). wt, wild type.
The change (n-fold) between two experimental conditions was calculated by taking the ratio of the signal intensity (difference of the log2 value) between two experimental conditions. Genes that showed at least a threefold increase in mRNA abundance relative to the control and had an average log2 signal intensity greater than 8 were considered upregulated. Conversely, genes whose transcript abundance decreased more than threefold and had an average log2 signal intensity greater than 8 in the control were considered downregulated.
The same genes could appear in more than one comparison using this classification system.
FIG. 1.
Changes in expression of energy-generating pathways of E. coli under anaerobic conditions. Changes in expression of fermentation pathways, TCA cycle, and electron transport systems from the global gene expression data are represented by the different color intensities of lines and protein names, where dark blue are the most anaerobically repressed functions and dark red are the most anaerobically activated functions. The three major branch points that determine the fluxes of carbon and electrons through the various pathways are labeled in bold and larger font. This figure is adapted from Spiro and Guest (82) and Alexeeva et al. (1). The direction of the triangles indicates whether the gene product is positively (Δ) or negatively (▿) regulated by FNR.
Clustering of regulated genes into eight groups by gene expression patterns.
k-means clustering (30) provided a tool to classify the 297 genes by gene expression patterns into eight major groups (Fig. 2). k-means clustering was applied with k running from 1 to 16, and the total within-cluster sum of Euclidean distance squares for each clustering was computed. A kink in the sum of squares curve located at a k of 8 indicates that 8 is the optimal number of clusters (32). The expression patterns of groups I, II, III, and V were similar to those that have previously been observed with FNR-regulated genes, supporting the notion that other genes in these classes may be directly regulated by FNR. Groups VII and VIII, in contrast, appeared to be regulated in a manner that was independent of FNR. In addition, the expression profile of some groups (IV, VI) did not fit into any characterized models of FNR regulation, and thus, further analysis is required to determine the mechanism of regulation of these groups of genes. Nevertheless, genes that clustered by common gene expression patterns also showed a strong functional relationship, which may provide clues to the function of unknown genes in each group.
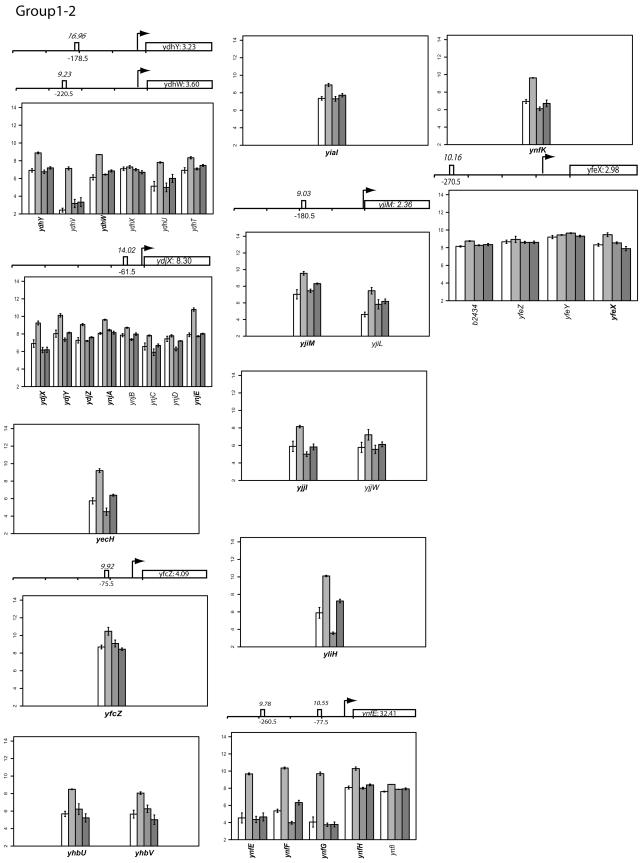
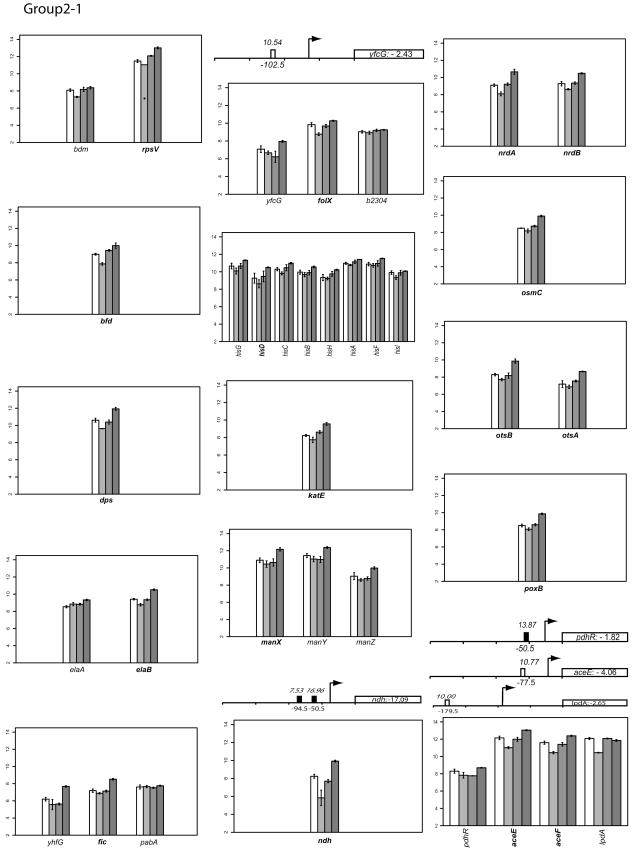
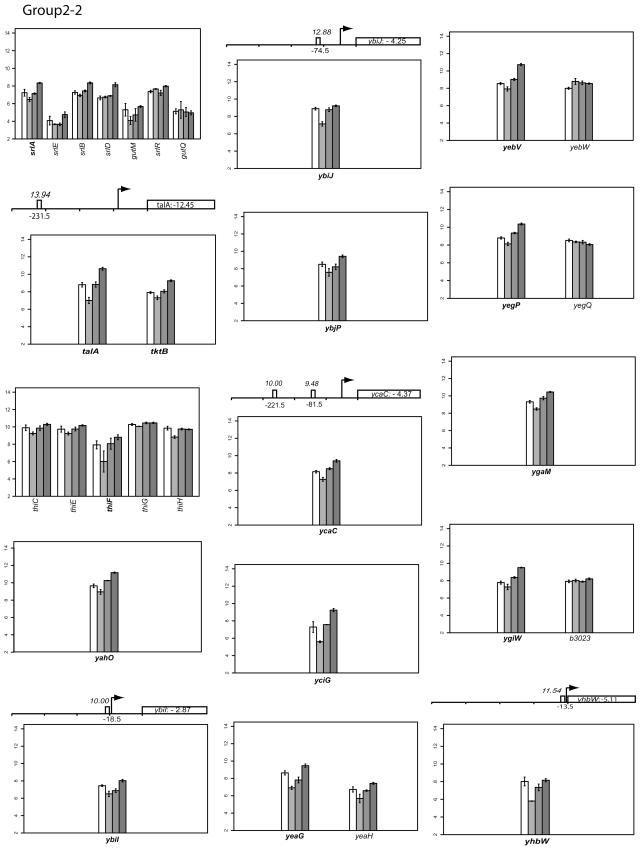
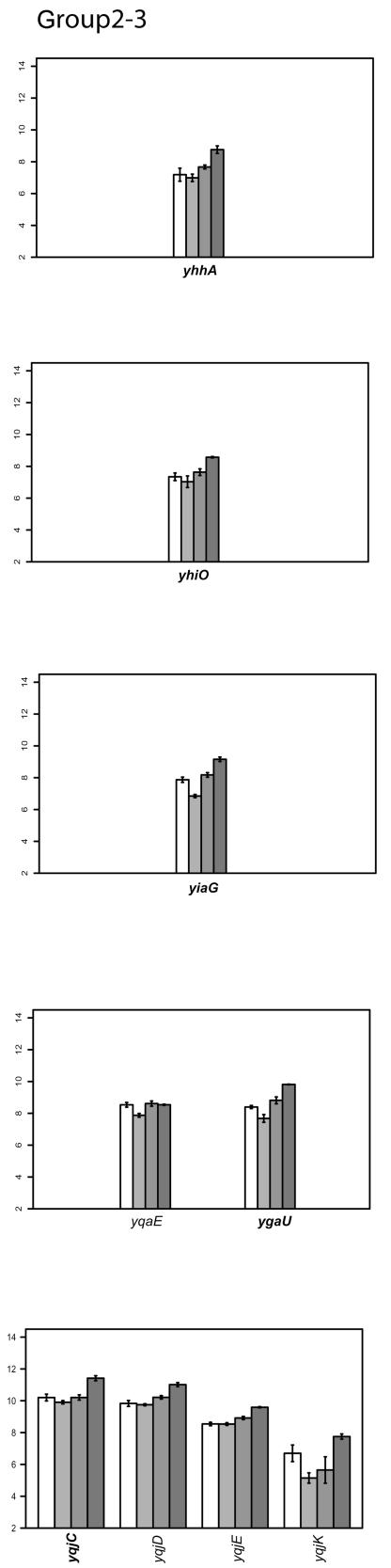
FIG.2.
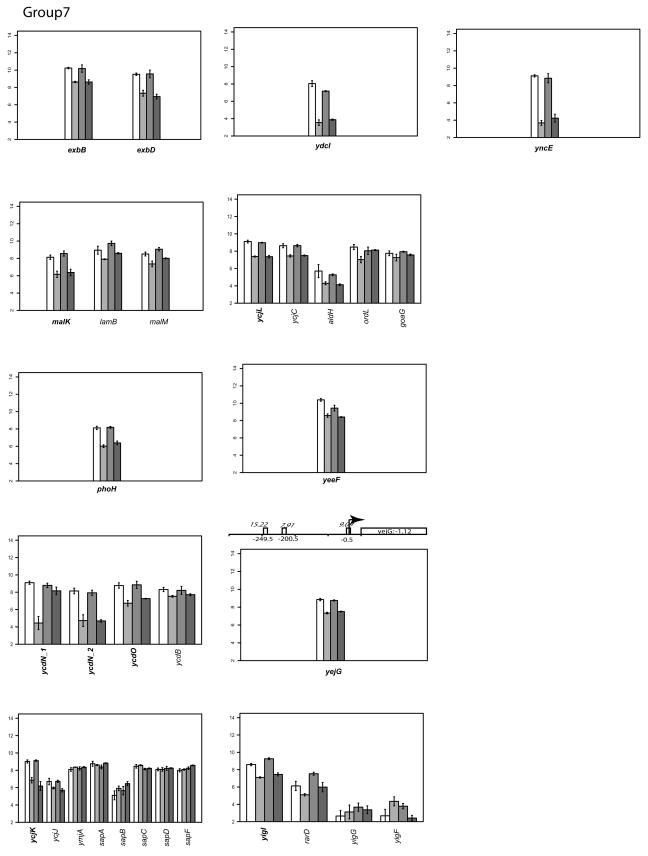
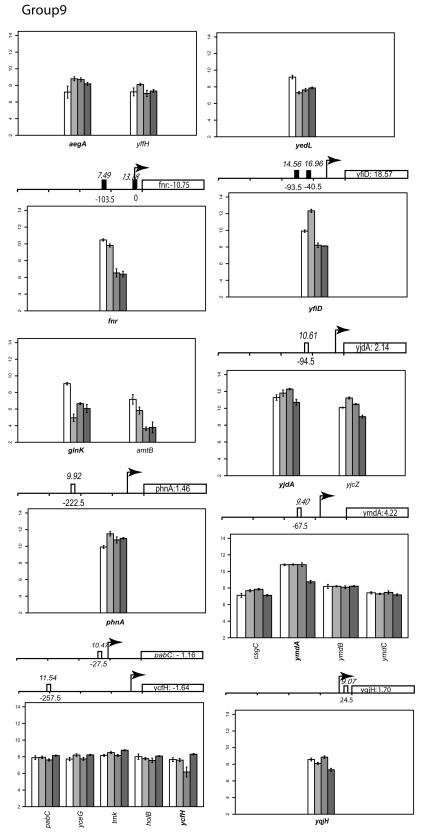
Classifying gene expression patterns by k-means clustering analysis. The log2 transcription levels are plotted as histograms for each gene (the wild type [left two bars] and the FNR− strain [right two bars] under aerobic and anaerobic conditions, respectively). Any member of a known or predicted polycistronic operon that meets our criteria for differential expression is indicated in bold. The whole operon is shown for comparison. FNR binding sites in the upstream region of a gene are represented by a bar (solid, documented; open, predicted). The italicized number above the box is the ScanAce quality score for each FNR site; the number below is the distance from the FNR site to the transcriptional start site. Black arrows indicate the position and the orientation of transcriptional start sites of each operon either documented or predicted by RegulonDB or by Bockhorst et al. (8). Tick bars are spaced at 100-bp intervals. Error bars represent standard deviations of results from three replicates.
The log2 intensity of the transcript signal determined for the FNR+ and FNR− strains grown under either aerobic or anaerobic conditions is presented for each group in Fig. 2. Where there are known or predicted polycistronic operons, the other operon members are included for comparison. The transcription start sites are those either predicted by RegulonDB (http://www.cifn.unam.mx/Computational_Genomics/regulondb/), Bockhorst et al. (8), or the Ecocyc database (http://ecocyc.org) or taken from the literature.
Group I: genes whose expression is increased under anaerobic conditions and whose activation is FNR dependent.
Transcript levels of the 53 genes in Group I showed a pattern characteristic of genes that are activated by FNR (Fig. 2). In this case, transcript levels were higher under anaerobic conditions than under aerobic growth conditions in the wild-type strain, and this increase in transcript levels was dependent on FNR. This group also contains the most-studied promoters of the FNR regulon. Several of these promoters have previously been shown to be directly regulated by FNR, and therefore, there is a high probability that other genes in this group have a similar mode of regulation.
The functions encoded by this group of genes are primarily associated with the unique aspects of anaerobic metabolism of E. coli, such as anaerobic respiration using alternative reductases for nitrite (nirBD and nrfABC) (62), DMSO (dmsABC) (4), and fumarate (frdABCD) (13, 37), enzymes associated with anaerobic carbon utilization (glpABC and pepT) (17, 88), proteins that facilitate transport of these compounds (dcuB and dppB) (2, 79), maturation and cofactor synthesis of hydrogenase (hypABC and nikABCDE) (55, 61), and enzymes that functionally replace an “aerobic” counterpart (nrdD [89], which encodes the O2-sensitive anaerobic ribonucleoside triphosphate reductase). For many of the above genes, the promoter that is increased under anaerobic growth conditions is known, and in most cases, the FNR binding sites have been defined by mutagenesis (23, 36, 59, 60, 92) and/or by footprinting (28, 56). The typical location of the FNR binding site is at a position centered ca. −41.5 bp upstream of the transcription start site (49).
To determine whether any of the unknown genes in this group are regulated in a similar manner, we cloned a DNA fragment containing the region upstream of what was predicted to be the first gene in the operon into vectors for promoter-lacZ fusions. The plasmids containing promoter-lacZ fusions were amplified by specific primers, and those PCR fragments were recombined into the lac operon as described in Materials and Methods, such that these promoters replaced the chromosomal lac promoter located between lacI and lacZ. This method allowed for the study of the in vivo expression of these genes. Of the 22 unknown genes in this group, we chose to study the regions upstream of ynfE, ydfZ, ydhY, and ydjX because these genes showed some of the largest increases in expression when aerobically and anaerobically grown cells were compared (from 44- to 3.5-fold). In agreement with the array data, anaerobic growth of the strains containing the single copy promoter-lacZ fusions (Fig. 3A) showed that the expression of ynfE, ydfZ, ydhY, and ydjX was strongly dependent on FNR, since an 86- to 13-fold reduction in β-Gal was observed by comparing FNR+ and FNR− strains. With all four promoter constructs, we were able to restore activity to each FNR− strain with a plasmid-encoded copy of FNR (data not shown). We also chose a gene, yfcZ, on whose expression FNR showed a small effect. The in vivo analysis of a yfcZ-lacZ fusion strain revealed that the expression of yfcZ was not as strongly regulated by FNR, in agreement with the microarray data (data not shown). Of the unknown genes that are highly regulated by FNR, ynfE is part of an operon that encodes a putative DMSO reductase (54), ydhY is predicted to be an Fe-S-containing oxidoreductase, ydfZ is postulated to be involved in selenium delivery (44), and ydjX is a putative membrane protein.
FIG. 3.
In vivo expression of selected promoters. FNR+ or FNR− strains carrying promoter-lacZ fusions in single copy were grown anaerobically and assayed for β-Gal. The activities shown are the averages of results from two independent determinations. The promoters of the genes analyzed are indicated on the x axis. A promoterless lacZ fusion produced 7 U of activity.
To establish whether FNR directly regulated the transcription of these promoters, we measured FNR-dependent transcription activation in a purified system using DNA templates containing the analogous region of DNA that was used to construct the promoter-lacZ fusions. Results from the in vitro transcription assays for promoters ynfE, ydjX, ydhY, and ydfZ were in agreement with the in vivo data and showed that FNR directly activated transcription of these promoters (Fig. 4). For yfcZ, little to no difference in transcription was observed upon addition of FNR, suggesting that either other factors are required for initiation of transcription or additional DNA sequences within the yfcZ promoter are needed for transcription. In addition, the approximate location of the transcription start site of these activated promoters was estimated from the size of the transcript. Inspection of the DNA sequence in the upstream region of these promoters with the ScanAce program revealed the presence of putative FNR sites. Three promoters (PynfE, PydhY, and PydfZ) had a putative FNR site centered at ∼41.5 bp upstream of the transcription start site, a position typical of previously characterized FNR class II promoters (49). One promoter (PydjX) contained an FNR site at a class I spacing (60.5 bp upstream of the transcription start site), making this the first naturally occurring class I FNR-dependent promoter characterized by in vitro transcription assays.
FIG. 4.
In vitro transcription assays of selected promoters. Autoradiograph of RNA transcripts generated from plasmid templates carrying the ynfE, ydjX, ydhY, and ydfZ promoters. Incubations containing FNR are indicated, and the RNA-1 transcript serves as an internal control. Transcripts that appeared to be regulated directly by FNR are marked (•).
Using the ScanAce algorithm, we also found putative FNR sites in the regions upstream of other operons of unknown function (yjiM and yfeX), suggesting that these genes may also be directly regulated by FNR (Fig. 2). No putative FNR sites were found upstream of yjjI, yiaI, yecH, ynfK, yliH, and yhbUV. Further analysis is necessary to determine whether FNR directly regulates the expression of these genes.
In summary, we identified 53 genes in group I that are upregulated by FNR. Twenty-seven of these genes have functions associated with anaerobic metabolism and have previously been established to be members of the FNR regulon. However, members of the FNR regulon (e.g., nap [85], narG [51], narK [42], and fdn [50] operons) that were previously shown to be strongly nitrate dependent were below the threshold of detection ability in our analysis. These genes are also known to require the nitrate-responsive two-component regulatory system (84, 94), thus indicating the need for further studies in the presence of nitrate to identify members of this subgroup of genes regulated by FNR. In addition, we found that four operons containing genes of unknown function were directly regulated by FNR, suggesting that these genes encode new anaerobic metabolic functions.
Group II: genes whose expression appears to be repressed by FNR under anaerobic conditions.
Under anaerobic growth conditions, transcript levels of 39 genes, representing 35 operons (group II) (Fig. 2), were increased in the strain lacking FNR relative to wild-type strain MG1655, indicating that FNR repressed their transcription under anaerobic growth conditions. Repression by FNR has not been as well studied as activation and, as a consequence, not as many genes were known to be repressed by FNR. In addition, repression of genes under anaerobic conditions has largely been attributed to the ArcAB system (29).
Nevertheless, among this group of genes is ndh, which codes for the aerobic respiratory enzyme NADH dehydrogenase II (10). Repression of ndh by FNR has been well characterized and requires two FNR binding sites, located at −50.5 and −94.5 relative to the transcription start site, for maximal repression (58). Expression of the pdh operon (pdhR-aceEF-lpdA and aceFA-lpdA encode subunits of the pyruvate dehydrogenase complex) is also known to be repressed under anaerobic conditions (70). We found that the expression pattern of aceEF was indicative of the group II genes, but lpdA had an expression pattern similar to those genes found in group III. These results may be explained by the finding that the pdh operon contains two major promoters: Ppdh, which generates a pdhR-aceEF-lpdA transcript, and Plpd, which produces an independent lpdA transcript. Plpd is anaerobically repressed by ArcA, whereas the Ppdh promoter was not strongly regulated by FNR or ArcA. Lastly, the expression of katE (76) (encoding catalase), nrdAB (11) (aerobic ribonucleoside diphosphate reductase), and poxB (14) (pyruvate oxidase) is also known to be reduced under anaerobic conditions through as-yet-undetermined mechanisms.
The functions of the remaining 33 genes in this group, including 15 genes of unknown function, have not been previously examined under anaerobic conditions. They represent different functional groups, such as intermediary metabolism (talA and tktB), biosynthesis of cofactors (thiF and folX), ribosomal protein synthesis and modification (rpsV), Fe-containing proteins (bfd and dps), cell division (fic), transport of small molecules (srlA and manX), osmotic adaptation (otsAB and osmC), and amino acid biosynthesis (hisD). To determine whether FNR directly effected the expression of talA, encoding transaldolase A of the nonoxidative branch of the pentose phosphate pathway, we constructed a talA′-lacZ fusion and measured its activity in both the presence and the absence of fnr. In a strain lacking fnr, β-Gal activity was twofold greater than in the wild-type strain, suggesting that FNR represses talA expression (Fig. 3B). However, even in the absence of FNR, we were unable to detect a transcript in our in vitro transcription system using a template that contained the same upstream sequences, suggesting that expression of the talA promoter may require additional factors or that it is a weak promoter (data not shown). The ScanAce algorithm identified putative FNR binding sites with a score higher than 9 for talA as well as aceE, ybiJ, yhbW, ybiI, and ycaC; ycaC possesses two putative FNR sites, consistent with another well-characterized member of this group, ndh. Thus, these latter genes are also candidates for direct repression by FNR.
Finally, some of the genes in this group, such as rpsV, fic, otsAB, poxB, dps, and katE, also belong to the RpoS (σS) regulon (53). In order to test whether FNR specifically represses genes in the rpoS regulon or whether FNR acts on rpoS directly, we measured the β-Gal activity from fnr+ and Δfnr derivatives of strains containing an rpoS′-lacZ fusion grown under anaerobic conditions (80) (Fig. 3B). The FNR+ strain had half as much β-Gal activity as the strain lacking FNR, suggesting that FNR does repress rpoS expression under anaerobic conditions. In addition, ScanAce revealed the presence of multiple FNR sites within the rpoS promoter region. This loss of repression of rpoS expression by FNR may contribute to the increase in transcript levels of the RpoS-dependent genes in group II in the strain lacking FNR. However, we are also aware of recent studies that show a prominent role of growth rate in the regulation of RpoS (34). Therefore, it is also possible that the differential expressions of genes in the RpoS regulon observed here might be attributed to the complex regulation of RpoS rather than a direct effect of FNR.
Group III: genes whose expression is decreased under anaerobic conditions and whose repression is partially FNR dependent.
Transcript levels of 55 genes (Fig. 2) were decreased in wild-type strain MG1655 under anaerobic growth conditions compared to aerobic conditions. Furthermore, in a strain lacking FNR, transcript levels were increased, although not always to the same level found in aerobic growth conditions. These data suggest that factors, in addition to FNR, repress these genes.
The majority of the genes in this group have known “aerobic” functions, providing a rationale for their repression under anaerobic conditions. Several gene products function in facilitating energy production under aerobic conditions (TCA cycle, aerobic respiration, etc.) and include the enzymes of the TCA cycle (gltA, acnB, icdA, sucABCD, lpdA, sdhCDAB, fumA, mdh, and mqo) (29), glyoxylate bypass (aceA) (6), cytochrome oxidase (cyoABCDE) (57), and aldehyde dehydrogenase (aldA) (68). Previous studies have shown that the expression of these genes is reduced under anaerobic conditions. In addition, the expression of cyoABCDE (18), sucABCD, sdhCDAB (63), sodA (31), tpx (41), fumA (65), and icdA (15) was shown to be negatively regulated by both FNR and ArcA under anaerobic conditions. ArcA and not FNR has previously been shown to negatively regulate gltA (66), acnB (20), lpdA (21), mdh (64), mqo (95), and aldA (68). Since FNR has also been reported to enhance arcA expression under anaerobic conditions (although this result was not observed in our studies), it raises the question of whether the effects of FNR observed with this group are all direct. Nevertheless, the ScanAce algorithm predicted that mdh, mqo, gltA, aldA, and icdA have at least one FNR site. Thus, these data indicate an interesting but complex relationship between FNR and ArcA in controlling the expression of genes in group III.
Other gene products in group III also function primarily under aerobic growth conditions and include betT, which is involved in the O2-dependent synthesis of the osmoprotectant glycine-betaine from choline, and gcd, which encodes the pyrroloquinoline-dependent glucose dehydrogenase. Transcription of both gcd and betT has been reported to be repressed under anaerobic conditions (45, 96). Neither gene is predicted to have an upstream FNR site, and ArcA was implicated in the negative regulation of betT.
Group III genes also include functions induced by oxidative stress conditions. Previous studies have shown that the transcription of the genes encoding Mn-dependent superoxide dismutase (sodA), thiol peroxidase (tpx), and thioredoxin 2 (trxC) is repressed under anaerobic conditions, in agreement with the results obtained here. FNR binding sites were predicted with ScanAce in the control regions of tpx and sodA. Recently, another group III operon, sufABCDSE, which encodes functions that facilitate biosynthesis of iron-sulfur clusters, has also been shown to be upregulated under oxidative stress conditions (47). In addition to our finding that transcript levels of sufA, the first gene in the operon, were decreased under anaerobic conditions, we found that sufD showed an even greater decrease in transcript levels than that of the three preceding genes in the operon. To determine whether the decreased transcript levels reflected regulation of distinct promoter regions, we constructed lacZ fusions to the region upstream of sufA and to the region upstream of sufD. In strains lacking FNR, β-Gal activity from both sufA′-lacZ and sufD′-lacZ was greater than that of the FNR+ strain (Fig. 3B). Transcripts of sufA or sufD were poorly detected in the in vitro transcription system, precluding a test of FNR in their repression. Nevertheless, the in vivo expression data support the view that FNR represses expression of this operon and also suggest that there is an internal promoter within the operon controlling at least the expression of sufD. We also tested another member of this group, iscR, which is the first gene of the isc operon encoding another pathway for iron-sulfur cluster biogenesis. However, in this case, β-Gal activity of an iscR′-lacZ fusion showed that FNR has little to no effect on the expression of the iscR promoter (data not shown). This result was also confirmed by in vitro transcription assays (data not shown).
The remaining genes in this expression group have not been previously studied under anaerobic conditions, yet all function almost exclusively in metabolism, particularly in transport. These genes are psiF (induced by phosphate starvation), glpFK (glycerol facilitator and glycerol kinase), mglAB (galactose transport), argT (lysine-, arginine-, ornithine-binding periplasmic protein), cstA (carbon starvation protein), gltI (glutamate/aspartate transport protein), kgtP (α-ketoglutarate permease), fhuF (Fe3+ transport), and ndk (nucleoside diphosphate kinase). The ScanAce algorithm predicted that kgtP has one FNR binding site, whereas glpF and fhuF each have two FNR sites, indicating that FNR could be acting directly to repress their transcription.
Group IV: genes that are upregulated by FNR under anaerobic conditions but are only somewhat O2 regulated.
Fifty-five genes showed a decrease in expression in a strain lacking FNR, although surprisingly, their expression in a wild-type strain was only slightly increased under anaerobic conditions (Fig. 2). Expression of feoB, encoding the iron(II) transporter, has previously been shown to be mediated by FNR, as an FNR− strain was unable support feoAB transcription (39). A putative FNR binding site was identified in the feoAB promoter region, suggesting that regulation by FNR is direct.
The majority (49 of 55) of the genes in this group function in flagellar biosynthesis, chemotaxis, or motility, which have not previously been found to be regulated by FNR. The master regulator known to control expression of these classes of genes is FlhD/FlhC; however, levels of flhDC transcripts remained constant under all conditions tested. In addition, no FNR sites were predicted in the region upstream of any of these genes, including FlhD/FlhC. Thus, while it seems likely that FNR does not directly effect transcription of these genes, the decrease in expression in Δfnr strains remains unexplained.
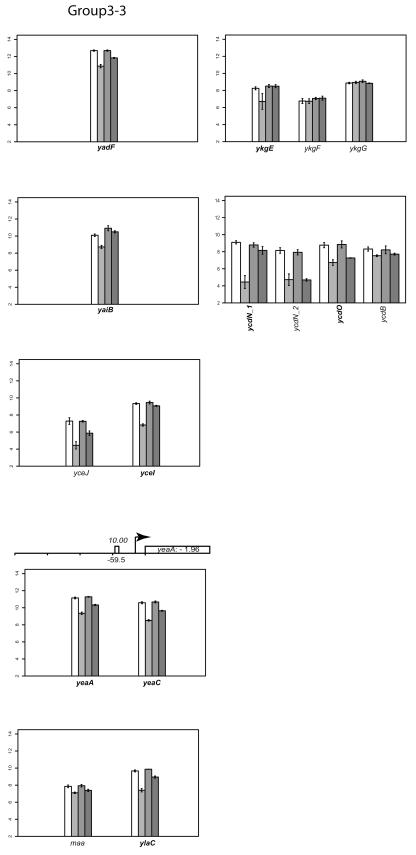
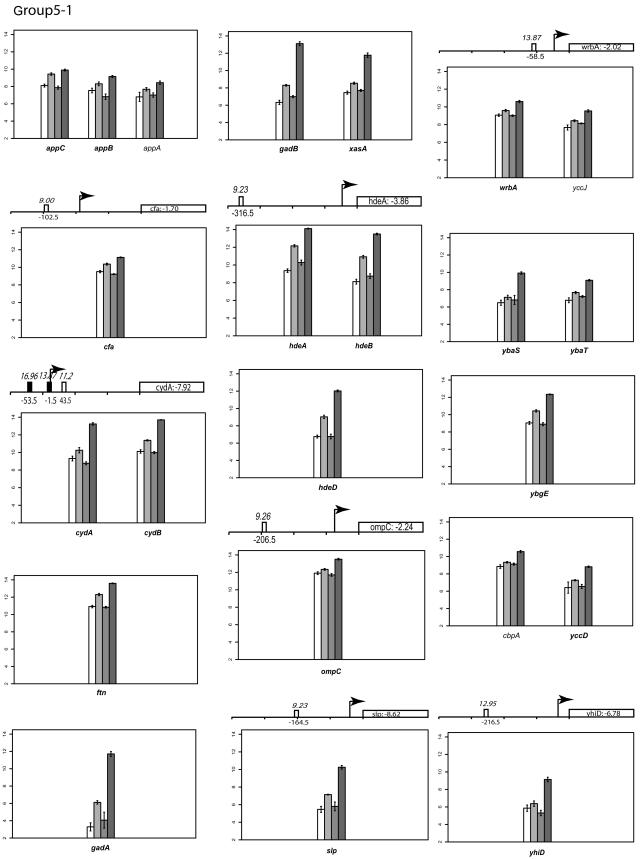
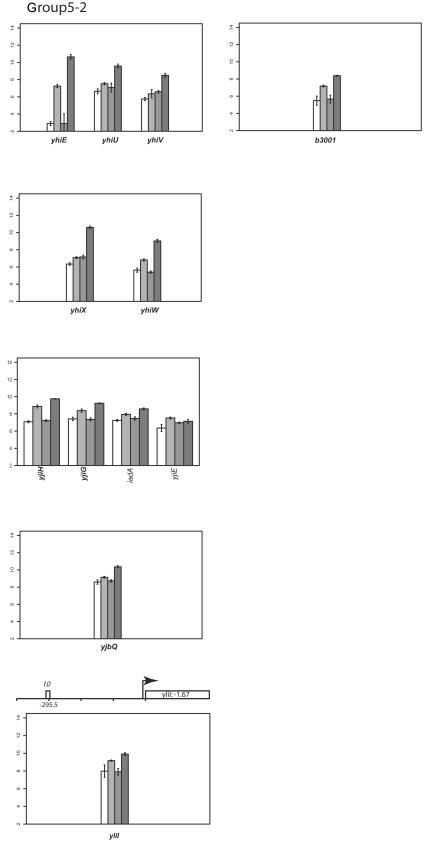
Group V: genes that are slightly upregulated under anaerobic conditions but repressed by FNR.
Transcription of genes in this group appears to be slightly upregulated under anaerobic conditions (Fig. 2). However, in anaerobically grown strains lacking FNR, expression of these genes was increased, suggesting that anaerobic expression of genes in group V is repressed by FNR and that the increase in expression under anaerobic conditions must be from another factor.
One of the best-characterized FNR-regulated genes in this group is cydAB, encoding the terminal cytochrome d oxidase that reduces O2 to H2O. Cytochrome d oxidase has a higher affinity for O2 than cytochrome o oxidase, and accordingly, it is the more highly expressed cytochrome oxidase under O2-limiting conditions. Transcription of genes encoding cytochrome d oxidase (cydAB) has previously been shown to be maximal under microaerobic conditions, whereas under anaerobic conditions, transcription is intermediate between microaerobic and fully aerobic conditions (19). This pattern of expression results from ArcA activation as O2 becomes limited, while FNR represses cydAB as O2 is depleted. Thus, the slight increase in cydAB transcripts observed under anaerobic growth conditions in the microarray data can be explained by ArcA activation, and the increase in cydAB transcripts in the absence of FNR is due to the loss of FNR repression.
Many of the genes in this group (cfa, gadAB, ompC, slp, hdeABD, wrbA, ybaST, yhiEWX, and xasA) are also regulated by a decrease in pH, called the acid stress response (91). Of these genes, only gadAB and ompC have been previously shown to be induced anaerobically (3, 6). Other genes in group V include iadA (isoaspartyl dipeptidase), accBA (acetyl-coenzyme A carboxylase subunits), ftn (iron storage), and a large number of genes of unknown function (yjiHGE, yhiD, yccJ, yccD, yliI, ybgE, yghZ, and yjbQ). ScanAce revealed putative FNR sites for cfa, hdeA, slp, ompC, wrbA, yccD, yliI, and yhiD, suggesting that FNR may act directly at these promoters. All of the known genes in this group, except slp and ftn, also belong to the RpoS (σS) regulon (53). As with group II genes, the loss of repression of rpoS expression by FNR may contribute to the increase in transcript levels of rpoS-dependent genes in group V or may be an indirect effect of regulation of RpoS. Thus, regulation of this group of genes appears to be complex.
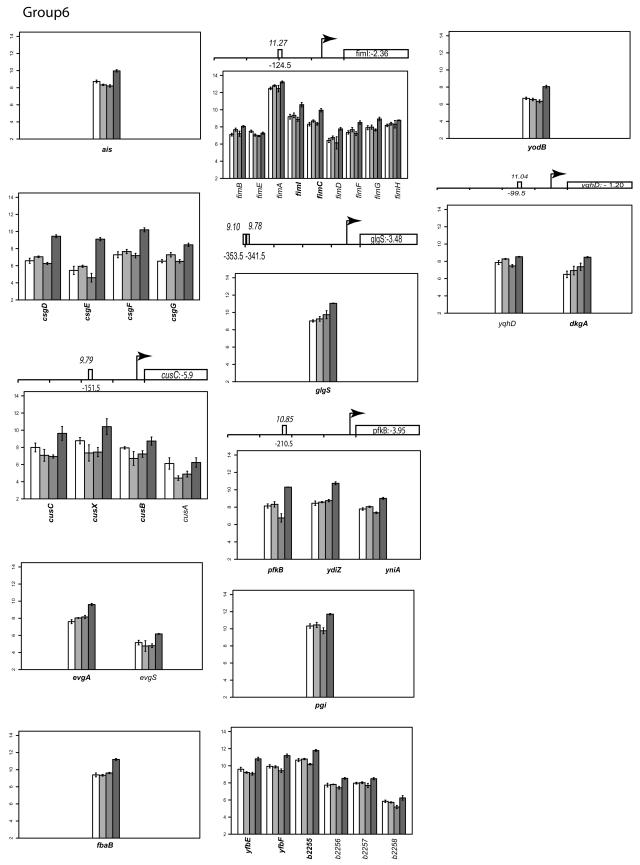
Group VI: genes that are repressed by FNR but that do not show any O2-dependent changes in gene expression.
Twenty-two genes clustered into group VI have similar levels of transcripts in both anaerobically and aerobically grown wild-type cells (Fig. 2). However, expression of these genes was upregulated under anaerobic growth conditions in the absence of FNR. None of these genes have previously been studied under anaerobic conditions. The 18 known genes belong to different functional groups, such as carbon metabolism (pgi, pfkB, glgS, and fbaB), surface structures (csgDEFG and fimICDFGH), and resistance to copper and silver (cusCFBA). Analysis with ScanAce revealed that five genes (glgS, cusC, pfkB, ydhD, and fimI) contain putative FNR sites. To determine whether the effects of FNR on cusC and csgD were direct, we constructed lacZ fusions to the region upstream of these genes. FNR had very little effect on the expression of either cusC′-lacZ or csgD′-lacZ (data not shown), suggesting that the effects of FNR on cusC and csgD expression may not be direct or, alternatively, that an unrecognized regulatory element was not present in the DNA region selected to construct the promoter-lacZ fusions.
Group VII: genes that are repressed under anaerobic conditions independently of FNR.
Expression of the 12 genes in group VII was decreased under anaerobic conditions in both wild-type and FNR mutant strains, indicating that the regulation of these genes is independent of FNR (Fig. 2). The genes of known function are malK (maltose transport), phoH (phosphate starvation-inducible protein), and exbBD (transport of iron compounds). The rest of the genes in this group are of unknown function and include yigL, ydcI, ycjK, yejG, ycdN_2, yeeF, ychL, and yncE. ScanAce analysis revealed that only yejG contains a putative FNR binding site, supporting the hypothesis that genes in group VII are regulated in an FNR-independent manner.
Group VIII: genes whose expression is increased under anaerobic conditions and whose activation is FNR independent.
Transcript levels of the 23 genes in group VIII were increased anaerobically (Fig. 2). This increase in anaerobic expression does not appear to be FNR dependent, as shown by the fact that strains lacking FNR showed no differences from the FNR+ strain in transcript levels. Several of the genes within this group have previously been shown to be upregulated anaerobically.
Among this group are genes with key functions in fermentative pathways. pflB (pyruvate formate-lyase) catalyses the nonoxidative cleavage of pyruvate to formate (74). Located in the same operon as pflB is focA, the formate transporter (90). Previous studies have shown that the focApfl operon is upregulated anaerobically and that transcription is directed by multiple promoters that are controlled by ArcA and FNR (38). Thus, while our analysis did not indicate a role for FNR in focApflB expression, potential changes in transcript levels in the absence of FNR may have been masked by the presence of multiple promoters.
Other fermentative functions that showed an FNR-independent increase in transcript levels were adhE, encoding an alcohol dehydrogenase (16, 48), ackA (43), encoding acetate kinase, and hycG (5), encoding a subunit of formate hydrogenlyase or hydrogenase-3. Previous studies did not indicate a role for FNR in the anaerobic expression of these genes (5, 48), in agreement with the findings here.
Other members of this group include the genes coding for the Ni-Fe hydrogenases 1 and 2, hyaABCDFE and hybOABCDEFG, respectively. Both of these operons have previously been reported to be induced under anaerobic conditions (71). However, the factors controlling the anaerobic transcription of hya and hyb have not been completely established, since neither FNR nor ArcA could completely account for the anaerobic induction of these operons. In addition, anaerobic induction of hya expression was partially dependent on AppY, while hybO expression was partially dependent on NarP even in the absence of nitrate. Thus, the regulation of these two promoters is complex, and differences in strains and/or growth conditions may explain why we did not observe FNR-dependent regulation in our experiments.
The other seven members of this group include genes involved in zinc resistance (zraP) and genes of unknown function (ydiH, yfhM, ybcW, ycbJ, ygjU, and yfjO). Analysis of the genes in group VIII with ScanAce revealed that, despite the FNR-independent induction of genes in this group, seven genes contain putative FNR sites: adhE, hycA, focA, ybcW, ycbJ, ackA, and yfjN. Thus, it remains an open question whether FNR contributes to expression of these genes, particularly if they have complex promoters with multiple transcription initiation sites, as observed with focApflB.
Genes that do not fit any expression group.
There are 10 genes whose expression pattern did not cluster with any of the other groups (Fig. 2). In the case of fnr, the reason can be explained by the deletion of the gene in the mutant strain. In agreement with previous observations, the expression of fnr in the wild-type strain shows a slight decrease under anaerobic conditions, as expected from a gene that is negatively autoregulated (83). Levels of transcripts of three genes, ymdA, yqjH, and yjdA, were unchanged in the wild-type strain under the experimental conditions tested but were decreased in a strain lacking FNR under anaerobic conditions.
Although previous studies demonstrated that FNR is inactivated by the presence of O2, expression of glnK and yedL decreased under aerobic growth conditions in an fnr mutant compared to the wild-type strain. In contrast, transcript levels of aegA, phnA, and ycfH were increased under aerobic conditions in an fnr mutant strain compared to the wild type, although this aerobic effect of FNR was not observed with an aegA′-lacZ fusion (12). yfiD expression was anaerobically induced in the wild-type strain, while in the absence of FNR, both the aerobic and anaerobic expression levels were decreased. Previous studies have shown that YfiD is an acid-inducible protein, and its expression is regulated by two FNR sites at −40.5 (FNR I) and −93.5 (FNR II) upstream of its transcriptional start sites (6, 27). FNR binding at FNR II impairs the ability of FNR I to activate transcription anaerobically. No data are available to explain why yfiD expression was altered by FNR under aerobic conditions in this study.
Phenotype of null mutants of genes of unknown function.
To test whether the disruption of representative genes of unknown function caused a general defect in anaerobic respiration, 39 mutants were screened for growth on glycerol nitrate-containing agar under anaerobic conditions. There was no observed difference in growth for any of these strains except for the disruption in yjjI. Under anaerobic conditions, this mutant produces small colonies on glucose and no growth on glycerol and nitrate. This defect was specific to anaerobic conditions, since growth was normal when this strain was incubated on the same medium under aerobic conditions.
Comparison of microarray data to other data in the literature.
During preparation of this paper, Salmon et al. (73) reported results of global gene expression profiles of E. coli K-12 strain MC4100 grown under aerobic and anaerobic conditions and the effect of FNR on these global gene expression patterns. A comparison of the two data sets using the statistical program reported by Salmon et al. (http://visitor.ics.uci.edu/genex/cybert/) indicated differences in the genes that were regulated by these conditions; the number of genes from MG1655 whose expression is changed between anaerobic and aerobic conditions with 95% confidence is 962, whereas the number for MC4100 is 1,445. However, only 334 genes are common to both data sets, and of those, 123 genes are regulated in the opposite direction, leaving only 211 genes that show similar patterns of regulation by FNR and/or anaerobiosis. In addition, three of the most highly expressed genes found in our study, which we also showed to be directly regulated by FNR, were not found in the data set of 1,445 genes.
While we cannot definitively say why the results differ, it is possible that strain backgrounds could account for at least some of the differences between the two data sets. We analyzed MG1655 (seq), the K-12 (F− λ− ilvG rfb-50 rph-1 eut) strain from the Yale stock center that was sequenced in the Blattner lab (7), reisolated from cultures frozen while sequencing was in progress, verified to be FNR+, and redeposited in the stock center after sequencing (www.genome.wisc.edu). We are also aware that an FNR− variant of MG1655 was unknowingly in use in the scientific community (81), which likely resulted from distribution of a mixed culture by the E. coli stock center (cgsc.biology.yale.edu), an issue that was corrected in 2003. MG1655 is considered to be a good approximation to the original E. coli K-12 isolate. In contrast, MC4100 has a different set of mutations [F− araD139(argF-lac)U169 rspL150 relA1 flbB5301 fruA25 deoC1 ptsF25], including a large deletion (97 kb) from argF to lac. Recently, the endpoints of four deletions of MC4100 were mapped by comparing the genomes of MC4100 and MG1655 (69), which demonstrated the loss of 133 open reading frames, including 10 putative regulatory proteins from MC4100. In addition, the other known mutations of MC4100, in relA (stringent response) and pts (a mediator of energy-coupled transport), might indirectly affect the response of E. coli to anaerobiosis. Although, we cannot rule out that differences in array platforms and/or growth conditions also contributed to differences between the two data sets (73), it seems that differences in the genotype between MG1655 and MC4100 is at least one possibility. Additional experiments will be needed to resolve this question.
Conclusion.
The studies described here reveal the global changes in gene expression of E. coli K-12 strain MG1655 that are dependent on environmental O2 and/or the anaerobic regulator FNR. A total of 297 genes, which belong to 184 operons, show significant changes in transcript levels by either of these variables. Taking all of the genes in the 184 operons into account, we propose that up to 465 genes may be regulated by FNR and/or anaerobic conditions in strain MG1655. Of the 184 operons, 47 or 26% (86 genes) agree well with previously published work on individual genes and operons, confirming the accuracy of the global gene expression array method.
A striking outcome of this study is the number of genes regulated by O2 and/or FNR that were not previously implicated in the anaerobic response. For 97 of the 206 genes of known function, we are unaware of any previous reports of O2 levels affecting their regulation or function. We also found that 73 genes of unknown function are regulated by O2 and/or FNR. Interestingly, the seven most highly anaerobically induced genes are unknown genes: ynfEFG (putative DMSO reductase), ydfZ (a selenium binding protein), ydhV (hypothetical protein), yhiE (hypothetical protein), and yliH (putative receptor). These results reinforce the view that while the anaerobic lifestyle of E. coli has been well studied, we still lack a complete understanding of the cellular functions that facilitate growth under O2-limiting conditions.
While differences in the pattern of gene expression were used to cluster genes into eight major groups, the genes within each group also showed a strong functional similarity. Functions required for aerobic energy conservation, such as the TCA cycle and aerobic respiratory enzymes, belong largely to the repressed genes of group III. Most genes involved in anaerobic respiration fall into group I, which are upregulated by FNR anaerobically. The genes found in group IV are almost exclusively involved in flagellar biosynthesis, chemotaxis, and motility. The fact that genes within a group show a strong functional relationship should facilitate identifying the role of genes of unknown function within the same group. For example, the ynfEFG operon, located in group I with many well-characterized genes of anaerobic respiration, encodes the subunits of a putative DMSO reductase (54). Therefore, it is very likely that other genes of unknown function in this group are also involved in anaerobic respiration.
The data reported here also indicate that gene regulation under anaerobic conditions involves complex integration with other pathways. For example, most of the genes in group III are repressed by the O2-sensing system mediated by ArcA (49, 70), and although not directly demonstrated, the data in this paper suggest that these genes can also be repressed by FNR. In addition, we also observed overlap between the RpoS regulon and some of the FNR-repressed genes in group II and group V; approximately one-half of the total genes in the RpoS regulon (37 genes) have altered expression patterns in our experiments, although the significance of this finding remains to be established. We also observed overlap with regulons involved in other environmental sensing responses, such as acid stress response (group V) and oxidative stress (group III). A common feature of this subset of FNR-regulated genes is that they appear to be repressed by FNR. Therefore, an emerging theme from these studies is that FNR-repressed genes are subject to a number of regulatory inputs.
In conclusion, these data have enhanced our understanding of gene regulation under anaerobic conditions. Our findings suggest that as many as 465 genes of MG1655 are regulated by changes in environmental O2 and/or FNR. As shown in Fig. 1, a major consequence of these changes in gene expression is a reprogramming of energy metabolism to utilize alternative mechanisms of energy conservation in the absence of O2. Further characterization of these genes will allow us to gain even greater insight not only into the FNR regulon but also other functions required for adaptation to O2-limited environments.
Supplementary Material
Acknowledgments
We thank Robert Mau, Jr., Guy Plunkett III, and Jeremy Glasner for advice with data analysis. We also thank Karen Wassarman for the rpoS′-lacZ strain.
This work was supported by National Institutes of Health Grants GM-45844 (to P.J.K.) and GM35682 (to F.R.B.).
F.R.B. has financial interest in NimbleGen System, Inc.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alexeeva, S., B. de Kort, G. Sawers, K. J. Hellingwerf, and M. J. de Mattos. 2000. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J. Bacteriol. 182:4934-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, J. C., and S. A. Short. 1986. opp-lac operon fusions and transcriptional regulation of the Escherichia coli trp-linked oligopeptide permease. J. Bacteriol. 165:434-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhriain, N. N., C. J. Dorman, and C. F. Higgins. 1989. An overlap between osmotic and anaerobic stress responses: a potential role for DNA supercoiling in the coordinate regulation of gene expression. Mol. Microbiol. 3:933-942. [DOI] [PubMed] [Google Scholar]
- 4.Bilous, P. T., S. T. Cole, W. F. Anderson, and J. H. Weiner. 1988. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol. Microbiol. 2:785-795. [DOI] [PubMed] [Google Scholar]
- 5.Birkmann, A., F. Zinoni, G. Sawers, and A. Bock. 1987. Factors affecting transcriptional regulation of the formate-hydrogen-lyase pathway of Escherichia coli. Arch. Microbiol. 148:44-51. [DOI] [PubMed] [Google Scholar]
- 6.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181:2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Bockhorst, J., Y. Qiu, J. Glasner, M. Liu, F. Blattner, and M. Craven. 2003. Predicting bacterial transcription units using sequence and expression data. Bioinformatics 19(Suppl. 1):i34-i43. [DOI] [PubMed] [Google Scholar]
- 9.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 10.Calhoun, M. W., and R. B. Gennis. 1993. Demonstration of separate genetic loci encoding distinct membrane-bound respiratory NADH dehydrogenases in Escherichia coli. J. Bacteriol. 175:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casado, C., M. Llagostera, and J. Barbe. 1991. Expression of nrdA and nrdB genes of Escherichia coli is decreased under anaerobiosis. FEMS Microbiol. Lett. 67:153-157. [DOI] [PubMed] [Google Scholar]
- 12.Cavicchioli, R., T. Kolesnikow, R. C. Chiang, and R. P. Gunsalus. 1996. Characterization of the aegA locus of Escherichia coli: control of gene expression in response to anaerobiosis and nitrate. J. Bacteriol. 178:6968-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cecchini, G., C. R. Thompson, B. A. Ackrell, D. J. Westenberg, N. Dean, and R. P. Gunsalus. 1986. Oxidation of reduced menaquinone by the fumarate reductase complex in Escherichia coli requires the hydrophobic FrdD peptide. Proc. Natl. Acad. Sci. USA 83:8898-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, Y. Y., A. Y. Wang, and J. E. Cronan, Jr. 1994. Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS(katF) gene. Mol. Microbiol. 11:1019-1028. [DOI] [PubMed] [Google Scholar]
- 15.Chao, G., J. Shen, C. P. Tseng, S. J. Park, and R. P. Gunsalus. 1997. Aerobic regulation of isocitrate dehydrogenase gene (icd) expression in Escherichia coli by the arcA and fnr gene products. J. Bacteriol. 179:4299-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, Y. M., and E. C. Lin. 1991. Regulation of the adhE gene, which encodes ethanol dehydrogenase in Escherichia coli. J. Bacteriol. 173:8009-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole, S. T., K. Eiglmeier, S. Ahmed, N. Honore, L. Elmes, W. F. Anderson, and J. H. Weiner. 1988. Nucleotide sequence and gene-polypeptide relationships of the glpABC operon encoding the anaerobic sn-glycerol-3-phosphate dehydrogenase of Escherichia coli K-12. J. Bacteriol. 170:2448-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotter, P. A., and R. P. Gunsalus. 1992. Contribution of the fnr and arcA gene products in coordinate regulation of cytochrome o and d oxidase (cyoABCDE and cydAB) genes in Escherichia coli. FEMS Microbiol. Lett. 70:31-36. [DOI] [PubMed] [Google Scholar]
- 19.Cotter, P. A., S. B. Melville, J. A. Albrecht, and R. P. Gunsalus. 1997. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol. Microbiol. 25:605-615. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham, L., M. J. Gruer, and J. R. Guest. 1997. Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli. Microbiology 143:3795-3805. [DOI] [PubMed] [Google Scholar]
- 21.Cunningham, L., and J. R. Guest. 1998. Transcription and transcript processing in the sdhCDAB-sucABCD operon of Escherichia coli. Microbiology 144:2113-2123. [DOI] [PubMed] [Google Scholar]
- 22.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eiglmeier, K., N. Honore, S. Iuchi, E. C. Lin, and S. T. Cole. 1989. Molecular genetic analysis of FNR-dependent promoters. Mol. Microbiol. 3:869-878. [DOI] [PubMed] [Google Scholar]
- 24.Erickson, J. W., and C. A. Gross. 1989. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 25.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 26.Georgellis, D., O. Kwon, and E. C. Lin. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314-2316. [DOI] [PubMed] [Google Scholar]
- 27.Green, J., M. L. Baldwin, and J. Richardson. 1998. Downregulation of Escherichia coli yfiD expression by FNR occupying a site at −93.5 involves the AR1-containing face of FNR. Mol. Microbiol. 29:1113-1123. [DOI] [PubMed] [Google Scholar]
- 28.Green, J., A. S. Irvine, W. Meng, and J. R. Guest. 1996. FNR-DNA interactions at natural and semi-synthetic promoters. Mol. Microbiol. 19:125-137. [DOI] [PubMed] [Google Scholar]
- 29.Gunsalus, R. P., and S. J. Park. 1994. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res. Microbiol. 145:437-450. [DOI] [PubMed] [Google Scholar]
- 30.Hartigan, J. 1975. Clustering algorithms. John Wiley & Sons, New York, N.Y.
- 31.Hassan, H. M., and H. C. Sun. 1992. Regulatory roles of Fnr, Fur, and Arc in expression of manganese-containing superoxide dismutase in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:3217-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hastie, T., R. Tibshirani, and J. H. Friedman. 2001. The elements of statistical learning: data mining, inference, and prediction. Springer, New York, N.Y.
- 33.Hughes, J. D., P. W. Estep, S. Tavazoie, and G. M. Church. 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296:1205-1214. [DOI] [PubMed] [Google Scholar]
- 34.Ihssen, J., and T. Egli. 2004. Specific growth rate and not cell density controls the general stress response in Escherichia coli. Microbiology 150:1637-1648. [DOI] [PubMed] [Google Scholar]
- 35.Imlay, J. A. 2002. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 46:111-153. [DOI] [PubMed] [Google Scholar]
- 36.Jayaraman, P. S., J. A. Cole, and S. J. Busby. 1989. Mutational analysis of the nucleotide sequence at the FNR-dependent nirB promoter in Escherichia coli. Nucleic Acids Res. 17:135-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, H. M., and R. P. Gunsalus. 1985. Transcription of the Escherichia coli fumarate reductase genes (frdABCD) and their coordinate regulation by oxygen, nitrate, and fumarate. J. Bacteriol. 164:1100-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaiser, M., and G. Sawers. 1997. Overlapping promoters modulate Fnr- and ArcA-dependent anaerobic transcriptional activation of the focApfl operon in Escherichia coli. Microbiology 143:775-783. [DOI] [PubMed] [Google Scholar]
- 39.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiley, P. J., and H. Beinert. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6:181-185. [DOI] [PubMed] [Google Scholar]
- 41.Kim, S. J., Y. H. Han, I. H. Kim, and H. K. Kim. 1999. Involvement of ArcA and Fnr in expression of Escherichia coli thiol peroxidase gene. IUBMB Life 48:215-218. [DOI] [PubMed] [Google Scholar]
- 42.Kolesnikow, T., I. Schroder, and R. P. Gunsalus. 1992. Regulation of narK gene expression in Escherichia coli in response to anaerobiosis, nitrate, iron, and molybdenum. J. Bacteriol. 174:7104-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwan, H. S., H. W. Chui, and K. K. Wong. 1988. ack::Mu d1-8 (Apr lac) operon fusions of Salmonella typhimurium LT2. Mol. Gen. Genet. 211:183-185. [DOI] [PubMed] [Google Scholar]
- 44.Lacourciere, G. M., R. L. Levine, and T. C. Stadtman. 2002. Direct detection of potential selenium delivery proteins by using an Escherichia coli strain unable to incorporate selenium from selenite into proteins. Proc. Natl. Acad. Sci. USA 99:9150-9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamark, T., T. P. Rokenes, J. McDougall, and A. R. Strom. 1996. The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J. Bacteriol. 178:1655-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamberg, K. E., and P. J. Kiley. 2000. FNR-dependent activation of the class II dmsA and narG promoters of Escherichia coli requires FNR-activating regions 1 and 3. Mol. Microbiol. 38:817-827. [DOI] [PubMed] [Google Scholar]
- 47.Lee, J. H., W. S. Yeo, and J. H. Roe. 2004. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 51:1745-1755. [DOI] [PubMed] [Google Scholar]
- 48.Leonardo, M. R., P. R. Cunningham, and D. P. Clark. 1993. Anaerobic regulation of the adhE gene, encoding the fermentative alcohol dehydrogenase of Escherichia coli. J. Bacteriol. 175:870-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, B., H. Wing, D. Lee, H. C. Wu, and S. Busby. 1998. Transcription activation by Escherichia coli FNR protein: similarities to, and differences from, the CRP paradigm. Nucleic Acids Res. 26:2075-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, J., and V. Stewart. 1992. Localization of upstream sequence elements required for nitrate and anaerobic induction of fdn (formate dehydrogenase-N) operon expression in Escherichia coli K-12. J. Bacteriol. 174:4935-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, S. F., and J. A. DeMoss. 1987. Promoter region of the nar operon of Escherichia coli: nucleotide sequence and transcription initiation signals. J. Bacteriol. 169:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu, X., and P. De Wulf. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588-12597. [DOI] [PubMed] [Google Scholar]
- 53.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 54.Lubitz, S. P., and J. H. Weiner. 2003. The Escherichia coli ynfEFGHI operon encodes polypeptides which are paralogues of dimethyl sulfoxide reductase (DmsABC). Arch. Biochem. Biophys. 418:205-216. [DOI] [PubMed] [Google Scholar]
- 55.Lutz, S., A. Jacobi, V. Schlensog, R. Bohm, G. Sawers, and A. Bock. 1991. Molecular characterization of an operon (hyp) necessary for the activity of the three hydrogenase isoenzymes in Escherichia coli. Mol. Microbiol. 5:123-135. [DOI] [PubMed] [Google Scholar]
- 56.Melville, S. B., and R. P. Gunsalus. 1996. Isolation of an oxygen-sensitive FNR protein of Escherichia coli: interaction at activator and repressor sites of FNR-controlled genes. Proc. Natl. Acad. Sci. USA 93:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Melville, S. B., and R. P. Gunsalus. 1990. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J. Biol. Chem. 265:18733-18736. [PubMed] [Google Scholar]
- 58.Meng, W., J. Green, and J. R. Guest. 1997. FNR-dependent repression of ndh gene expression requires two upstream FNR-binding sites. Microbiology 143:1521-1532. [DOI] [PubMed] [Google Scholar]
- 59.Messenger, S. L., and J. Green. 2003. FNR-mediated regulation of hyp expression in Escherichia coli. FEMS Microbiol. Lett. 228:81-86. [DOI] [PubMed] [Google Scholar]
- 60.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 61.Navarro, C., L. F. Wu, and M. A. Mandrand-Berthelot. 1993. The nik operon of Escherichia coli encodes a periplasmic binding-protein-dependent transport system for nickel. Mol. Microbiol. 9:1181-1191. [DOI] [PubMed] [Google Scholar]
- 62.Page, L., L. Griffiths, and J. A. Cole. 1990. Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch. Microbiol. 154:349-354. [DOI] [PubMed] [Google Scholar]
- 63.Park, S.-J., G. Chao, and R. P. Gunsalus. 1997. Aerobic regulation of the sucABCD genes of Escherichia coli, which encode α-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J. Bacteriol. 179:4138-4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Park, S.-J., P. A. Cotter, and R. P. Gunsalus. 1995. Regulation of malate dehydrogenase (mdh) gene expression in Escherichia coli in response to oxygen, carbon, and heme availability. J. Bacteriol. 177:6652-6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park, S. J., and R. P. Gunsalus. 1995. Oxygen, iron, carbon, and superoxide control of the fumarase fumA and fumC genes of Escherichia coli: role of the arcA, fnr, and soxR gene products. J. Bacteriol. 177:6255-6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park, S. J., J. McCabe, J. Turna, and R. P. Gunsalus. 1994. Regulation of the citrate synthase (gltA) gene of Escherichia coli in response to anaerobiosis and carbon supply: role of the arcA gene product. J. Bacteriol. 176:5086-5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Patschkowski, T., D. M. Bates, and P. J. Kiley. 2000. Mechanisms for sensing and responding to oxygen deprivation, p. 61-78. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 68.Pellicer, M. T., A. S. Lynch, P. De Wulf, D. Boyd, J. Aguilar, and E. C. Lin. 1999. A mutational study of the ArcA-P binding sequences in the aldA promoter of Escherichia coli. Mol. Gen. Genet. 261:170-176. [DOI] [PubMed] [Google Scholar]
- 69.Peters, J. E., T. E. Thate, and N. L. Craig. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quail, M. A., D. J. Haydon, and J. R. Guest. 1994. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 12:95-104. [DOI] [PubMed] [Google Scholar]
- 71.Richard, D. J., G. Sawers, F. Sargent, L. McWalter, and D. H. Boxer. 1999. Transcriptional regulation in response to oxygen and nitrate of the operons encoding the [NiFe] hydrogenases 1 and 2 of Escherichia coli. Microbiology 145:2903-2912. [DOI] [PubMed] [Google Scholar]
- 72.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:E112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salmon, K., S. P. Hung, K. Mekjian, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2003. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J. Biol. Chem. 278:29837-29855. [DOI] [PubMed] [Google Scholar]
- 74.Sawers, G., and A. Bock. 1988. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J. Bacteriol. 170:5330-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sawers, R. G., E. Zehelein, and A. Bock. 1988. Two-dimensional gel electrophoretic analysis of Escherichia coli proteins: influence of various anaerobic growth conditions and the fnr gene product on cellular protein composition. Arch. Microbiol. 149:240-244. [DOI] [PubMed] [Google Scholar]
- 76.Schellhorn, H. E., and H. M. Hassan. 1988. Transcriptional regulation of katE in Escherichia coli K-12. J. Bacteriol. 170:4286-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Serres, M. H., S. Gopal, L. A. Nahum, P. Liang, T. Gaasterland, and M. Riley. 2001. A functional update of the Escherichia coli K-12 genome. Genome Biol. 2:research0035.1-0035.7. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 79.Six, S., S. C. Andrews, G. Unden, and J. R. Guest. 1994. Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport system (Dct). J. Bacteriol. 176:6470-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 81.Soupene, E., W. C. van Heeswijk, J. Plumbridge, V. Stewart, D. Bertenthal, H. Lee, G. Prasad, O. Paliy, P. Charernnoppakul, and S. Kustu. 2003. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185:5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spiro, S., and J. R. Guest. 1991. Adaptive responses to oxygen limitation in Escherichia coli. Trends Biochem. Sci. 16:310-314. [DOI] [PubMed] [Google Scholar]
- 83.Spiro, S., and J. R. Guest. 1987. Regulation and over-expression of the fnr gene of Escherichia coli. J. Gen. Microbiol. 133:3279-3288. [DOI] [PubMed] [Google Scholar]
- 84.Stewart, V. 2003. Biochemical Society special lecture. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 31:1-10. [DOI] [PubMed] [Google Scholar]
- 85.Stewart, V., Y. Lu, and A. J. Darwin. 2002. Periplasmic nitrate reductase (NapABC enzyme) supports anaerobic respiration by Escherichia coli K-12. J. Bacteriol. 184:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Storz, G., and R. Hengge-Aronis. 2000. Bacterial stress responses. ASM Press, Washington, D.C.
- 88.Strauch, K. L., J. B. Lenk, B. L. Gamble, and C. G. Miller. 1985. Oxygen regulation in Salmonella typhimurium. J. Bacteriol. 161:673-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sun, X., J. Harder, M. Krook, H. Jornvall, B. M. Sjoberg, and P. Reichard. 1993. A possible glycine radical in anaerobic ribonucleotide reductase from Escherichia coli: nucleotide sequence of the cloned nrdD gene. Proc. Natl. Acad. Sci. USA 90:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suppmann, B., and G. Sawers. 1994. Isolation and characterization of hypophosphite-resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol. Microbiol. 11:965-982. [DOI] [PubMed] [Google Scholar]
- 91.Tucker, D. L., N. Tucker, and T. Conway. 2002. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 184:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tyson, K. L., J. A. Cole, and S. J. Busby. 1994. Nitrite and nitrate regulation at the promoters of two Escherichia coli operons encoding nitrite reductase: identification of common target heptamers for both NarP- and NarL-dependent regulation. Mol. Microbiol. 13:1045-1055. [DOI] [PubMed] [Google Scholar]
- 93.Unden, G., S. Achebach, G. Holighaus, H. G. Tran, B. Wackwitz, and Y. Zeuner. 2002. Control of FNR function of Escherichia coli by O2 and reducing conditions. J. Mol. Microbiol. Biotechnol. 4:263-268. [PubMed] [Google Scholar]
- 94.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 95.van der Rest, M. E., C. Frank, and D. Molenaar. 2000. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Escherichia coli. J. Bacteriol. 182:6892-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamada, M., S. Asaoka, M. H. Saier, Jr., and Y. Yamada. 1993. Characterization of the gcd gene from Escherichia coli K-12 W3110 and regulation of its expression. J. Bacteriol. 175:568-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.