Abstract
Zinc is an essential trace metal ion for growth, but an excess of Zn is toxic and microorganisms express diverse resistance mechanisms. To understand global bacterial responses to excess Zn, we conducted transcriptome profiling experiments comparing Escherichia coli MG1655 grown under control conditions and cells grown with a toxic, sublethal ZnSO4 concentration (0.2 mM). Cultures were grown in a defined medium lacking inorganic phosphate, permitting maximum Zn bioavailability, and in glycerol-limited chemostats at a constant growth rate and pH. Sixty-four genes were significantly up-regulated by Zn stress, including genes known to be involved in Zn tolerance, particularly zntA, zraP, and hydG. Microarray transcriptome profiling was confirmed by real-time PCR determinations of cusF (involved in Ag and Cu efflux), ais (an Al-inducible gene), asr (encoding an acid shock-inducible periplasmic protein), cpxP (a periplasmic chaperone gene), and basR. Five up-regulated genes, basR and basS [encoding a sensor-regulator implicated in Salmonella in Fe(III) sensing and antibiotic resistance], fliM (flagellar synthesis), and ycdM and yibD (both with unknown functions), are important for growth resistance to zinc, since mutants with mutations in these genes exhibited zinc sensitivity in liquid media and on metal gradient plates. Fifty-eight genes were significantly down-regulated by Zn stress; notably, several of these genes were involved in protection against acid stress. Since the mdt operon (encoding a multidrug resistance pump) was also up-regulated, these findings have important implications for understanding not only Zn homeostasis but also how bacterial antibiotic resistance is modulated by metal ions.
Zinc plays a vital role in biology, and more than 300 Zn-containing proteins have been identified. Zn is an essential trace metal ion for growth of all organisms (22). The chemical properties of Zn allow stable association with proteins and make Zn highly adaptable to meeting the needs of proteins and enzymes that carry out diverse biological functions (4). Zn acts as a strong Lewis acid; in these reactions, either substrates are coordinated, polarized, and activated or a Zn-bound water molecule is ionized to Zn(OH) and attacks the substrate. Zn also has a structural role and is coordinated with four cysteine residues (42). Some of the most important such roles are in Zn finger motifs found in many transcription factors, which facilitate specific interactions with target DNA sequences and ultimately gene regulation (4). Also, Zn is important in situations where it cross-links proteins, taking the place of disulfide bridges to form knots within structures like RNA polymerase (12). Thus, Zn is not simply a cofactor for enzyme catalysis but also is the structural factor for folding domains involved in protein-protein and protein-DNA interactions (42).
Although Zn is classified as essential, excess Zn is toxic. The diverse toxic effects have been attributed to the interactions with sulfydryl groups in a wide range of proteins, especially those proteins involved with respiration in electron transport systems (22). In Escherichia coli, membrane uptake systems, including the primary importer ZnuABC, a high-affinity periplasmic binding protein-dependent transport system for Zn (32), and ZupT (zinc uptake transporter) (16), a member of the ZIP (ZRT, IRT-like protein) family, provide adequate levels of Zn, even in the face of variable extracellular concentrations. Intracellular pools must be regulated, in part by sequestration. However, information on the E. coli proteins ZraP (zinc resistance-associated protein) (31) and YdaE (5) that may process or chaperone Zn for intracellular functions is rudimentary. In other bacteria, metallothioneins play a role in detoxification of Zn (34). The levels of Zn are also maintained by the export of excess ions from the cell by both ZntA, a P-type Zn-exporting ATPase (3), and members of the CDF (cation diffusion facilitator) family of proteins, including ZitB (formerly YbgR), a metal ion transporter (15).
Genome-wide transcriptional analysis with microarray technology is a powerful tool for determining how gene expression within the cell is affected by the external growth conditions. To date, genome-wide microarray studies on the effect of zinc on E. coli have been carried out only with batch cultures (8) by using zinc-adapted cells. The problem with these cultures is that the growth conditions cannot be strictly monitored and maintained at constant levels as the pH and the dissolved oxygen and nutrient concentrations change during growth. The regulation of gene expression is therefore affected not only by the difference in growth conditions between a control and an experimental culture but also by culture variables, including, perhaps most importantly, the specific growth rate. This variation in gene regulation can obscure the interpretation of gene expression in response to a supposedly single difference in culture growth conditions.
In this study, E. coli cultures were grown in chemostats at a fixed specific growth rate, temperature, and pH so that the effects on the transcriptional profile in the presence and absence of additional ZnSO4 in the medium could be compared by microarray analysis. We found significant changes in gene transcription in response to elevated levels of Zn. The transcriptional response to zinc stress involves not only established zinc transport systems but also membrane transporters and sensors previously not linked to zinc metabolism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. Cells were grown at 37°C in a defined medium containing 10 mM glycerol as the sole carbon source. The glycerol was added to a solution containing 40 mM MES (morpholineethanesulfonic acid), 18.69 mM NH4Cl, 13.41 mM KCl, 4.99 mM K2SO4, 68 μM CaCl2, and trace elements (18.5 μM FeCl3 · 6H2O, 600 nM ZnO, 400 nM CuCl2 · 2H2O, 344 nM CoNO3 · 6H2O, 16 μM H3BO3 [all final concentrations]) in distilled water. After the pH was adjusted to 7.4 and autoclaving, 1 mM MgCl2 and 7.6 mM (final concentration) β-glycerophosphate (as a phosphate source) were added to the medium before use (3).
TABLE 1.
E. coli K-12 strains
| Straina | Description |
|---|---|
| MG1655 | Wild type |
| FB21676 | MG1655 basS::Tn5 |
| FB21678 | MG1655 basR::Tn5 |
| FB20544 | MG1655 fliM::Tn5 |
| FB21603 | MG1655 metA::Tn5 |
| FB21630 | MG1655 qor::Tn5 |
| FB22122 | MG1655 ycdM::Tn5 |
| FB21346 | MG1655 yibD::Tn5 |
All strains were obtained from F. R. Blattner, E. coli Genome Project, University of Wisconsin-Madison.
E. coli strain MG1655 was grown in custom-built chemostats constructed entirely of nonmetal parts and based on Sigma Proculture Dynalift spinner flasks. The flasks were modified by addition of another side arm that acted as an overflow to maintain the culture volume (125 ml). Holes were drilled in the plastic screw lid of the flask and filled with Subaseals (Fisher). In one of these seals, a needle attached to a Hepa-Vent filter (Whatman) was pushed through the seal, and this needle acted as an air vent to allow waste gas to be expelled. The other seal was used as an inoculation port for addition of inoculum with a needle and syringe. Cultures were fed with the defined minimal medium in which glycerol was the growth-limiting nutrient. This was pumped (Minipuls 2; Gilson) from a reservoir via a flow-back trap into the flask side arm through a silicone bung. The growth conditions in the chemostats were kept constant; the temperature was maintained at 37°C by using a silicone tube water jacket attached to a circulating water bath. The cultures were aerated by using a vortex (33) aided by the three baffles of the spinner flasks. To this end, the flasks were placed on KMO 2 Basic IKA-Werke stirrers (arbitrary setting, 600; Scientific Laboratory Supplies), which gave maximum stirring without splashing. Twice daily, a 2-ml sample was taken and used to test the pH (which stayed constant at pH 6.9), glycerol limitation, and contamination on nutrient agar plates. To check for glycerol limitation, culture supernatant was subjected to an enzymatic glycerol assay described by Garland and Randle (14). The working volume was 125 ml, and the dilution rate (which at steady state equaled the specific growth rate) was 0.1 h−1.
Growth in liquid batch cultures was measured in sidearm flasks by using a Klett colorimeter and a red filter (number 66; Manostat Corporation). The colorimeter was zeroed by using minimal medium.
ZnSO4 chemostat experiments and RNA isolation.
Cells were grown in two parallel chemostats under batch conditions overnight. ZnSO4 (final concentration, 0.2 mM) was then added to one medium reservoir before continuous culturing began. Cells in both chemostats were grown under the conditions described above to the steady state, as verified by collecting at least 5 culture volumes and measuring the optical density at 600 nm with a Jenway 6100 spectrophotometer (path length, 1 cm) before cells were collected by centrifugation. Samples were taken for subsequent microarray analysis by harvesting 10 ml of culture directly into RNA Protect (QIAGEN) to stabilize the RNA. Total RNA was purified by using a QIAGEN RNeasy Mini kit as recommended by the supplier. The RNA concentration and purity were determined by using a Beckman DU 650 spectrophotometer (37).
Preparation of labeled cDNA and hybridization.
Equal quantities of RNA from control and ZnSO4-supplemented cultures were labeled by using nucleotide analogues of dCTP containing either the Cy3 or Cy5 fluorescent dye. For each microarray slide, one sample was labeled with Cy3-dCTP, while another sample was labeled with Cy5-dCTP. Dye swap experiments were performed for each pair of samples to compensate for the different efficiencies of incorporation of the labeled nucleotides (41). The slides used were E. coli K-12 Pan arrays purchased from MWG Biotech. These slides contained 4,288 gene-specific oligonucleotide probes representing the complete E. coli K-12 genome.
cDNA synthesis (with MWG Biotech's protocol) was carried out by using 12 μg of starting material, which was primed with 9 μg of pd(N)6 random hexamers (Amersham Biosciences). Reaction mixtures (20 μl) containing 0.5 mM dATP, 0.5 mM dTTP, 0.5 mM dGTP, 0.2 mM dCTP, and 1 mM Cy3- or 1 mM Cy5-dCTP were incubated for 2 h at 42°C with 200 U of Superscript II RNase H reverse transcriptase (Invitrogen). Following synthesis, cDNA was purified by using a PCR purification kit (QIAGEN) to remove the unincorporated deoxynucleoside triphosphates, fluorescent dye, and primers. Equal volumes of cDNA were combined and evaporated for approximately 45 min with an SPD121P Speed Vac (Thermo Savant). For hybridization to the microarray slides, cDNA was resuspended in an appropriate volume of salt-based hybridization buffer (provided by MWG Biotech). Prior to addition to the slides, cDNA samples were heated to 95°C for 3 min. The slides were placed in MWG Biotech hybridization chambers and incubated for 16 to 24 h in a water bath at 42°C.
Washing and scanning of slides.
Following incubation, the slides were washed with decreasing salt concentrations, as recommended by the supplier, ranging from 2× SSC-1% sodium dodecyl sulfate to 0.5× SSC at 37°C with gentle agitation (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The slides were dried by centrifugation at 1,200 rpm (254 × g) for 5 min and subsequently scanned with an Affymetrix 428 scanner.
Data analysis.
The average signal intensity and local background correction were obtained by using commercially available software from Biodiscovery, Inc. (Imagene, version 4.0, and GeneSight, version 3.5). The mean values from each channel were log2 transformed and normalized by using the subtract-by-mean method to remove intensity-dependent effects in the log2 (ratio) values. The Cy3/Cy5 fluorescence ratios were calculated from the normalized values. Biological experiments (i.e., chemostat growth with and without 0.2 mM ZnSO4) were carried out three times, and a dye swap analysis was performed for each experiment, which provided three technical repeats, one for each of the three biological repeats. Data from the independent experiments were combined. Genes that were differentially regulated ≥twofold and displayed a P value of ≤0.05 (as determined by a t test) were defined as being statistically significantly differentially transcribed. This experiment (raw data only) has been loaded into ArrayExpress with accession number E-MEXP-222 (http://www.ebi.ac.uk).
Real-time PCR.
RNA was extracted as described above. cDNA synthesis was carried out by using 4 μg of starting material primed with 9 μg of pd(N)6 random hexamers (Amersham Biosciences). Reaction mixtures (20 μl) containing 0.5 mM dATP, 0.5 mM dTTP, 0.5 mM dGTP, and 0.5 mM dCTP were incubated for 2 h at 42°C with 200 U of Superscript II RNase H reverse transcriptase (Invitrogen). Following synthesis, cDNA was purified by using a PCR purification kit (QIAGEN) to remove unincorporated deoxynucleoside triphosphates and primers.
Gene-specific primers were designed to amplify 50- to 150-nucleotide fragments of target genes by using the Primer 3 software (36). Each reaction was carried out in a 25-μl (total volume) mixture in a 96-well optical reaction plate (Applied Biosystems). Each well contained 50% SYBR green PCR master mixture (AB Applied Biosystems), 12.5 pmol of each of the two primers, and 5 μl of cDNA sample. PCR amplification was carried out by using an ABI 7700 thermocycler (PE Applied Biosystems) and the following thermal cycling conditions: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The data were analyzed by using the SEQUENCE DETECTOR SYSTEM software (PE Applied Biosystems) and were further processed with Microsoft EXCEL. A standard curve was established by using genomic DNA for each gene studied to confirm that the primers amplified at the same rate and to validate the experiment. The relative levels of expression of genes of interest in zinc-grown E. coli compared with untreated controls were calculated by using the protocol for the standard curve method in User Bulletin #2 (ABI Prism 7700 sequence detection system) supplied by Applied Biosystems. No-template reactions were included as negative controls.
RESULTS
Batch cultivation in a medium lacking inorganic phosphate.
Cells were grown in glycerol-glycerophosphate medium (GGM) from which all inorganic phosphates were omitted, since these phosphates form insoluble precipitates with divalent cations and reduce their bioavailability (3, 21). The concentrations of ZnSO4 that inhibited the growth of wild-type strain MG1655 in GGM were determined (Fig. 1). Below a ZnSO4 concentration of 0.15 mM the specific growth rate was not affected and was 0.35 h−1, and the stationary phase was reached at 90 Klett units. Growth was reduced with 0.15 mM ZnSO4 as the stationary phase was reached at 60 Klett units, but the specific growth rate again was not affected. Growth in 0.25 mM ZnSO4 was further reduced as the stationary phase was reached at 22 Klett units and the growth rate was 0.25 h−1. Growth was completely inhibited at 0.35 mM ZnSO4. A preliminary experiment carried out to identify a suitable medium for growth demonstrated that this inhibitory concentration is much lower than the MICs of Zn(II) in complex media, like Luria-Bertani (LB) medium. Brocklehurst and Morby (8) showed that in LB medium the MIC of ZnSO4 for E. coli strain TG1 was 2.2 mM, which was considerably higher than the MIC in our GGM.
FIG. 1.
Growth of E. coli MG1655 in minimal medium in the absence of additional ZnSO4 (○) and in the presence of 0.05 mM ZnSO4 (▴), 0.15 mM ZnSO4 (□), 0.25 mM ZnSO4 (▪), and 0.35 mM ZnSO4 (•). The values are means for triplicate cultures; the error bars indicate standard deviations.
Transcriptome changes induced by ZnSO4.
The genome-wide transcriptional patterns of strain MG1655 chemostat cultures grown with and without additional 0.2 mM ZnSO4 were analyzed by using microarray technology. In total, of the 4,288 genes arrayed, 64 displayed statistically significant >twofold increases in mRNA levels (significance levels were identified by using the t test, where the cutoff P value was 0.05). Genes involved in cell membrane structure and membrane transport and binding were highly represented in the up-regulated genes. However, the largest class of genes that were up-regulated was the genes with unknown functions. Fifty-eight genes showed significant decreases in mRNA levels of more than twofold, and there were more unclassified genes in this group than in the up-regulated group. Transport and binding and cell membrane structure genes were equally represented, as were many genes whose products are located in the inner membrane.
Genes up-regulated by ZnSO4.
Table 2 lists all 64 genes that were significantly up-regulated along with the functions of their products and their levels of regulation. Encouragingly, several genes that we expected to see up-regulated because they are known from other studies to be induced by zinc were found to be highly responsive. These included zntA, whose product is the primary zinc ATPase transport pump (3), zraP, which encodes a possible zinc chaperone as it is known to bind to zinc in the periplasm (25), and hydG, which is one component of a two-component response regulator for zraP (25). More interestingly, there were many genes involved in transport, sensing, or regulation that have not previously been implicated in zinc tolerance but were highly up-regulated in response to zinc in this study.
TABLE 2.
Genes whose mRNA levels displayed >twofold increases after addition of 0.2 mM ZnSO4 to minimal medium
| Genea | Gene product and/or functionb | Fold increase | P value (<0.05) |
|---|---|---|---|
| Transport and Binding | |||
| acrD | Aminoglycoside efflux pump | 6.2 | 6.9 × 10−4 |
| amtB | Ammonium/methylammonium transporter | 2.5 | 2.9 × 10−4 |
| cusC | Ag and Cu efflux, outer membrane component; confers Cu and Ag resistance | 11 | 6.8 × 10−7 |
| cusF | Ag and Cu efflux, periplasmic binding protein; Cu and Ag resistance | 48 | 5.2 × 10−9 |
| cusB | Cation efflux membrane fusion protein; Cu and/or Ag efflux | 6.8 | 4.8 × 10−5 |
| cusA | Ag and possibly backup for Cu efflux | 3.3 | 3.7 × 10−6 |
| mdtA | MdtABC-TolC efflux pump; multidrug resistance | 2.1 | 1.1 × 10−2 |
| mdtC | MdtABC-TolC efflux pump; multidrug resistance | 2.8 | 2.9 × 10−2 |
| mdtD | Member of MFS efflux transporter family | 2.2 | 1.1 × 10−2 |
| ompC | Outer membrane porin | 2.1 | 1.3 × 10−2 |
| yfiK | Cysteine and O-acetylserine exporter; integral membrane protein | 2.6 | 6.1 × 10−3 |
| zntA | Zn(II), Cd(II), and Pb(II) translocating P-type ATPase | 2.4 | 7.5 × 10−3 |
| Other genes implicated in responses to metals | |||
| ais | AI-inducible protein; cell protection | 16 | 5.7 × 10−7 |
| basR | Response regulator; resistance to Fe(III) killing | 3.4 | 3.4 × 10−6 |
| basS | Sensor kinase; Salmonella ortholog confers polymyxin B resistance | 2.4 | 3.4 × 10−4 |
| cusR | Response regulator with CusS sensor for Cu and possibly Ag ion-induced expression of cusCFBA | 2.2 | 5.8 × 10−5 |
| hydG | Two-component response regulator for zraP | 3.3 | 3.5 × 10−6 |
| mgrB | Mg(II) starvation-stimulated gene; function unknown; evgAS phoPQ regulon | 2.5 | 1.8 × 10−5 |
| spy | Induced by Zn and envelope stress; cpxR and baeSR regulon | 21 | 2.7 × 10−9 |
| zraP | Zn-binding periplasmic protein; responsive to Zn(II) and Pb(II); Zn tolerance | 47 | 3.3 × 10−9 |
| Genes implicated in response to stresses other than metals | |||
| asr | Acid shock-inducible periplasmic protein required for acid tolerance | 25 | 2.5 × 10−7 |
| glnK | Potent activator of NRII (GlnL/NtrB) phosphatase | 4.1 | 6.2 × 10−7 |
| pspA | Negative regulatory gene for phage shock protein psp operon; binds PspB and PspC | 2.1 | 2.2 × 10−3 |
| rstA | Putative response regulator | 2.1 | 2.1 × 10−3 |
| rstB | Putative sensor kinase | 2.0 | 2.3 × 10−3 |
| rsxA | Required for the reduction of SoxR; membrane protein | 2.3 | 1.3 × 10−3 |
| Motility | |||
| flgB | Motility; flagellar basal body rod subunit | 3.2 | 6.4 × 10−3 |
| fliM | Flagellar synthesis; motor switching and energizing | 4.1 | 1.6 × 10−2 |
| motB | Flagellar regulon member; flagellar rotation | 2.3 | 3.6 × 10−2 |
| Periplasmic products | |||
| cpxP | Possible chaperone and involved in feedback inhibition of Cpx signal transduction | 11 | 2.1 × 10−7 |
| degP | Membrane-associated serine endoprotease; high-temperature growth | 8.1 | 9.3 × 10−7 |
| ppiA | Rotamase; peptidylprolyl-cis-trans-isomerase A | 2.6 | 8.7 × 10−5 |
| yccA | Membrane-associated protein that binds to FtsH (HflB) and HflKC proteins | 2.7 | 2.9 × 10−4 |
| ycfJ | Putative periplasmic protein; UmoD homolog | 2.3 | 4.0 × 10−4 |
| Biosynthesis | |||
| argA | N-Acetylglutamate synthase | 3.6 | 1.8 × 10−5 |
| cysK | Cysteine synthase; O-acetylserine sulfhydrylase A | 2.2 | 9.2 × 10−3 |
| Nucleic acid metabolism | |||
| mutL | Methyl-directed mismatch repair | 3.5 | 2.8 × 10−7 |
| Intermediary metabolism | |||
| argI | Ornithine transcarbamylase | 2.9 | 1.4 × 10−3 |
| gcvT | Aminomethyl transferase, tetrahydrofolate dependent | 2.1 | 1.3 × 10−3 |
| sucA | 2-Oxoglutarate dehydrogenase; E1 component | 2.1 | 2.9 × 10−3 |
| sucD | Succinyl coenzyme A synthase alpha subunit | 2.2 | 2.4 × 10−2 |
| thrB | Homoserine kinase | 2.1 | 2.4 × 10−3 |
| Membrane structure | |||
| arnA | UDP-glucuronate dehydrogenase; basR regulon | 2.2 | 2.5 × 10−2 |
| arnB | Involved in aminoarabinose modification of lipid A | 6.2 | 3.4 × 10−4 |
| arnC | Salmonella ortholog in an operon that is regulated by PmrAB (BasRS) | 11 | 5.3 × 10−6 |
| yfbH | Salmonella ortholog in an operon that is regulated by PmrAB (BasRS) | 4.5 | 9.0 × 10−7 |
| yfbJ | Salmonella ortholog in an operon that is regulated by PmrAB (BasRS) | 2.0 | 5.3 × 10−4 |
| eptA | Lipid A phosphoethanolamine transferase; associated with polymyxin resistance | 3.5 | 6.4 × 10−6 |
| ugd | Probable UDP-glucose 6-dehydrogenase | 5.0 | 9.1 × 10−6 |
| Phage-related function | |||
| yeeV | Overexpression inhibits growth; mitigated by yeeU coexpression | 2.8 | 2.4 × 10−2 |
| Unclassified | |||
| b3913 | Function unknown | 10 | 6.8 × 10−7 |
| ybjG | Function unknown; pgpB paralog | 3.0 | 8.1 × 10−5 |
| ybjX | Function unknown | 2.6 | 2.2 × 10−4 |
| ycdL | Function unknown | 2.0 | 3.3 × 10−2 |
| ycdM | Function unknown | 3.5 | 4.6 × 10−2 |
| ycfS | Function unknown | 3.6 | 2.2 × 10−5 |
| yeaM | Function unknown | 2.2 | 5.7 × 10−3 |
| yecD | Function unknown | 2.7 | 2.6 × 10−4 |
| yebE | Function unknown | 2.5 | 5.3 × 10−4 |
| yfaZ | Function unknown | 2.9 | 4.0 × 10−2 |
| ygiV | Function unknown | 2.2 | 3.9 × 10−3 |
| yibD | Function unknown | 2.7 | 2.9 × 10−6 |
| yijP | Function unknown | 4.7 | 4.9 × 10−4 |
| yncJ | Function unknown | 2.2 | 2.4 × 10−2 |
Gene names obtained from http://bmb.med:miami.edu/EcoGene/EcoWeb/.
Gene product descriptions obtained from http://bmb.med.miami.edu/EcoGene/EcoWeb/.
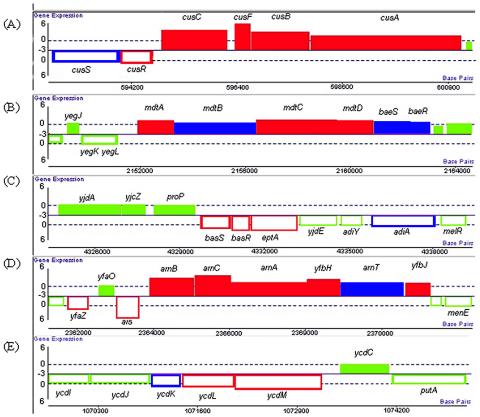
The microarray results revealed that the most up-regulated gene was cusF (48-fold increase). Previously, this gene was believed to encode a protein that has only been considered to be involved in copper and possibly silver binding in the periplasm (10). It is believed that this protein might have a function in improving the efficiency of the CusCBA system, acting as a periplasmic copper chaperone shuttling Cu(I) and Cu(II) [after reduction to Cu(I)] to the CusCBA efflux system, thereby increasing the accessibility of periplasmic copper (11). All other genes in the cus operon (encoding a metal efflux-detoxification system involved in copper and possibly silver transport) were also significantly up-regulated in this study (Fig. 2A). These genes included cusC (11-fold increase), encoding a putative outer membrane component; cusB (6.8-fold increase), encoding a member of the membrane fusion protein family; and cusA (3.3-fold increase), encoding a member of the resistance-nodulation-division (RND) protein superfamily (11). In addition to this operon, cusR, which encodes the cytoplasmic response regulator, was significantly up-regulated (2.2-fold increase), and cusS (encoding a membrane-bound histidine kinase) was found to be 1.8-fold up-regulated (data not shown in Table 2 but indicated in Fig. 2). These two genes encode the CusRS two-component signal transduction system that is required for copper-inducible expression of cusC (29).
FIG. 2.
Sections of the E. coli K-12 chromosomal map showing the locations and operon structures for up-regulated genes. Green represents the genes that are nonresponsive, purple represents the genes that are up-regulated more than 1.5-fold (excluding mdtB, which is up-regulated 1.28-fold), and red represents the genes that are up-regulated more than twofold. The shaded boxes indicate genes transcribed in one direction, and the open boxes indicate genes transcribed in the opposite direction. (A) cusRS and cusCFBA operons; (B) mdtABCD and baeRS operons; (C) basRS and eptA; (D) BasR-regulated operons, arnBCA, yfbH, arnT, yfbJ, and ais; (E) ycdM, ycdL, and ycdK. Genes shown in panels C and D are suggested to be operons as the homologs in Salmonella are known to be under the control of a single promoter.
The cus genes are not the only genes up-regulated in this work that are known to be transporters. Little is known about the mdtABC genes (formerly yegMNO), which encode an RND-type efflux system. These genes, like the cus genes, all are in the same operon, and the whole operon appeared to be up-regulated (Fig. 2B). Both mdtA (2.1-fold increase), encoding a membrane fusion protein, and mdtC (2.8-fold increase), encoding a transmembrane heteromultimer with MdtB, were significantly up-regulated, whereas mdtB was up-regulated only 1.3-fold (data not shown). This system was implicated in conferring resistance to novobiocin, deoxycholate, and bile salts following mdtABC stimulation by overproduction of the response regulator BaeR in a ΔacrAB mutant (2, 30). It is interesting that MdtAC alone confers bile salt resistance but not novobiocin resistance, suggesting that MdtB extends the substrate specificity of the transporter (30). mdtD was also up-regulated; although this gene is a member of the same operon, it is not involved in novobiocin and deoxycholate resistance, but it does encode a transporter of the major facilitator type.
Other up-regulated transporters include acrD (6.2-fold increase), encoding an aminoglycoside efflux pump. It has been suggested that AcrD can become associated with a member of the membrane fusion protein family and an outer membrane channel, encoded by genes elsewhere on the chromosome, to function as part of a multisubunit transporter complex (35). Also, amtB was induced (2.5-fold increase); this gene encodes an ammonium-methylammonium transporter and is linked with another up-regulated gene, glnK (4.1-fold increase), encoding a small cytoplasmic signal transduction protein (39).
Three genes encoding proteins involved with flagella were significantly up-regulated. These genes were fliM, which is involved in flagellar synthesis, motor switching, and energizing (4.1-fold increase), flgB, which encodes the basal body rod subunit (3.2-fold increase), and motB, whose product is involved in flagellar rotation (2.3-fold increase). These genes were not known to be regulated by zinc, but fliC has previously been shown to be up-regulated by Al (17).
The requirement in zinc tolerance for BasR-BasS, a two-component sensor regulation system, is unclear. Both genes were significantly up-regulated in this study (Fig. 2C), but to date little is known about this system in E. coli. The equivalent system in Salmonella, PmrA-PmrB, has been better studied and is required for resistance to polymyxin B and other microbial compounds. This two-component system controls transcription of several loci (Fig. 3), including pmrF (pbgPE operon) and pmrG, whose products modify lipopolysaccharide (LPS) and therefore are essential for polymyxin B resistance. Transcription of pmrCAB is also controlled, and the system is autoregulated (24). The up-regulation of basR (3.4-fold increase) and basS (2.4-fold increase) was confirmed by the up-regulation of numerous genes, all of which are E. coli equivalents of the Salmonella pmrF, pmrE, and pmrCAB genes. Unlike the Salmonella pmrF operon, which contains seven genes, the gene set in E. coli consists of six genes, all of which were shown to be up-regulated in this study (Fig. 2D) and some of which are known to be involved in lipopolysaccharide modification in E. coli. The first gene, arnB (6.2-fold increase), encodes an aminotransferase that is involved in aminoarabinose modification of lipid A (6). arnA (2.2-fold increase) encodes a UDP-glucuronate dehydrogenase that converts UDP-glucuronic acid to UDP-Ara4O (uridine 5′-β-L-threo-pentapyranosyl-4"-ulose diphosphate) (7), and arnT (1.9-fold up-regulated) (data not shown) encodes a 4-amino-4-deoxy-l-arabinose:lipid A transferase that utilizes the novel glycolipid donor undecaprenyl phosphate-l-4-amino-4-deoxy-l-arabinose (40). The other three genes in the predicted operon, arnC (11-fold increase), yfbH (4.5-fold increase), and yfbJ (2.0-fold increase), were all up-regulated, but at present their functions are unknown. Like Salmonella pmrC, the homologue in E. coli, eptA was also up-regulated (3.5-fold increase) and encodes a lipid A phosphoethanolamine transferase. The homolog of pmrE in E. coli, ugd, was up-regulated (5.0-fold increase) and is believed to encode a UDP-glucose 6-dehydrogenase.
FIG. 3.
(A) Model for the activation of the PmrA-PmrB two-component system based on the PhoP-PhoQ two-component system in Salmonella. Transcription of the PmrA-activated genes can be induced either by growth in the presence of low magnesium concentrations via the PhoP-PhoQ two-component system that induces transcription of PmrD to activate PmrA or by growth in the presence of high Fe(II) concentrations (or slight acid conditions) independent of PhoP-PhoQ. (Reprinted from reference 24 with permission of the publisher.) (B) Suggested model for activation of the BasR-BasS two-component system in E. coli by a high concentration of extracellular Zn(II). During growth in the presence of high concentrations of Zn(II), BasS senses the cation and activates BasR to induce transcription of a number of genes, some of which are known to be involved in the modification of lipopolysaccharides and, hence, resistance to polymyxin B.
Transcription of Salmonella PmrA-activated genes is thought to be promoted by one of two stimuli: (i) growth in the presence of low concentrations of extracellular magnesium in a process that requires PhoP-PhoQ and the small protein PmrD, which controls the activity of the PmrA-PmrB system at a posttranscriptional level, and (ii) growth with high iron concentrations or mild acid pH in a process that does not require PhoP-PhoQ (24). Note that none of the pmrA, pmrB, or pmrD homologues were up-regulated in this study. However, our results with E. coli clearly revealed that at constant magnesium concentrations and a constant pH (pH 7.0; therefore, we presume similar iron bioavailability), zinc alone in chemostat cultures was sufficient to increase basR and basS transcription.
Interestingly, up-regulation in most of the operons or groups of adjacent genes identified resulted in similar patterns of expression. For the majority of genes in operons, all genes showed up-regulation, although the induction ratios decreased for the downstream genes (Fig. 2).
In addition to the various genes mentioned above, there are many “y” genes of unknown function that either are unclassified or have unknown functions.
Genes down-regulated by ZnSO4.
Table 3 lists all 58 significantly down-regulated genes along with the functions of their products and their levels of regulation. The percentage of genes whose functions are unclassified or unknown that were down-regulated was larger than the percentage of genes whose functions are unclassified or unknown that were up-regulated, and the most down-regulated gene was yrbL (7.7-fold decrease). Although cells in the chemostat were constantly growing at a steady state and growth was limited by a single nutrient (glycerol in this case), some down-regulated genes are known to be induced under stationary-phase conditions; these genes include csiE (2.7-fold decrease), osmE (2.9-fold decrease), osmC (2.6-fold decrease; involved in the detoxification of organic hydroperoxides [1]), uspB (2.1-fold decrease; required for stationary-phase resistance to ethanol [9]), and gadE (formerly yhiE) (2.7-fold decrease; encodes a protein that activates glutamate decarboxylase-dependent acid resistance [26]).
TABLE 3.
Genes whose mRNA levels displayed >twofold decreases after addition of 0.2 mM ZnSO4 to minimal medium
| Genea | Gene product and/or functionb | Fold decrease | P value (<0.05) |
|---|---|---|---|
| Transport and binding | |||
| actP | Acetate permease | 3.1 | 4.4 × 10−3 |
| csgF | Possible assembly or transport protein for curli | 6.2 | 2.5 × 10−3 |
| csgG | Possible assembly or transport protein for curli; novel lipoprotein | 2.7 | 4.3 × 10−4 |
| gadC | Putative glutamate-γ-aminobutyrate antiporter. | 5.9 | 1.7 × 10−5 |
| lsrB | Part of a system for autoinducer 2 uptake in Salmonella | 2.5 | 4.4 × 10−3 |
| hdeD | Putative membrane transporter, H-NS repressed | 2.8 | 2.4 × 10−4 |
| lsrD | Part of a system for autoinducer 2 uptake in Salmonella | 2.1 | 1.0 × 10−2 |
| lsrG | Function unknown; part of an operon for autoinducer 2 uptake in Salmonella | 2.8 | 1.1 × 10−3 |
| ompW | Outer membrane protein; colicin S4 receptor | 2.4 | 2.9 × 10−2 |
| ugpA | sn-Glycerol-3-phosphate transport system; integral membrane protein | 2.2 | 3.5 × 10−2 |
| ugpE | sn-Glycerol-3-P transport system | 2.4 | 5.0 × 10−3 |
| uidB | Glucuronide permease | 3.7 | 8.1 × 10−3 |
| ydcU | Function unknown; putative ABC transporter permease protein | 2.2 | 8.0 × 10−4 |
| ygjU | Na+/serine (threonine) symporter | 2.3 | 6.0 × 10−4 |
| Other gene implicated in responses to metals | |||
| yrbL | Mg(II) starvation-stimulated gene; function unknown; phoPQ regulon | 7.7 | 3.0 × 10−7 |
| Genes implicated in response to stresses other than metals | |||
| csgD | Transcriptional regulator for curli operon csgBA | 2.3 | 2.0 × 10−3 |
| csiE | Stationary-phase inducible protein; sigma S-dependent promoter | 2.7 | 5.0 × 10−4 |
| cstA | Starvation-induced protein involved in peptide utilization during carbon starvation | 2.3 | 8.3 × 10−4 |
| dps | Stress response DNA-binding protein | 2.1 | 1.3 × 10−3 |
| gadE | Required for stationary-phase induction; pH 5.5 growth medium induced; overexpression confers acid resistance | 2.7 | 1.7 × 10−4 |
| hdeB | Acid resistance; periplasmic protein; repressed by H-NS; rpoS regulon | 5.8 | 1.7 × 10−4 |
| osmE | Regulated by growth phase as well as osmotic pressure | 2.9 | 3.5 × 10−4 |
| osmC | Osmotically inducible, stress-inducible membrane protein C; involved in defense against oxidative compounds | 2.6 | 7.2 × 10−7 |
| psiF | pho regulon member, requiring PhoRB system | 2.4 | 2.9 × 10−3 |
| rssB | Two-component response regulator, affecting RpoS regulon | 2.2 | 3.6 × 10−2 |
| sodC | Superoxide dismutase; Cu, Zn, periplasmic | 2.6 | 2.3 × 10−5 |
| uspB | Ethanol resistance in stationary phase | 2.1 | 5.5 × 10−3 |
| Periplasmic protein | |||
| hdeA | Periplasmic chaperone of acid-denatured proteins | 5.4 | 3.6 × 10−5 |
| Extracellular protein | |||
| hlyE | Latent hemolysin | 2.9 | 2.0 × 10−2 |
| Intermediary metabolism | |||
| astA | Putative arginine succinyltransferase | 3.9 | 1.1 × 10−4 |
| gadB | Glutamate decarboxylase B, vitamin B6 dependent | 5.3 | 3.7 × 10−6 |
| otsB | Trehalose phosphate phosphatase; cold and heat induced | 2.3 | 5.6 × 10−3 |
| talA | Transaldolase A; creBC regulon | 2.1 | 2.1 × 10−2 |
| Membrane structure | |||
| frdD | Fumarate reductase membrane anchor polypeptide | 2.0 | 3.5 × 10−3 |
| ychH | Function unknown; putative membrane protein transcribed divergently from pth | 2.6 | 1.0 × 10−2 |
| ynfA | Inner membrane protein; function unknown | 2.5 | 8.9 × 10−4 |
| Phage-related function | |||
| sgcC | Putative phosphotransferase enzyme IIC | 5.3 | 6.4 × 10−4 |
| sgcX | Putative gene in sgc cluster; function unknown | 3.5 | 1.3 × 10−4 |
| stfR | Phage lambda stf gene homolog in prophage Rac | 2.0 | 4.9 × 10−2 |
| Unclassified | |||
| elaB | Function unknown | 2.4 | 1.2 × 10−4 |
| yagU | Function unknown | 5.1 | 4.5 × 10−5 |
| yahO | Function unknown | 3.1 | 5.3 × 10−3 |
| ybaY | Function unknown | 2.2 | 1.1 × 10−3 |
| ycfH | Putative DNAse; tatD paralog; no Tat phenotype | 2.2 | 5.1 × 10−3 |
| ycgK | Function unknown | 5.2 | 5.0 × 10−3 |
| ydeI | Function unknown | 2.2 | 1.0 × 10−2 |
| ydhZ | Function unknown | 2.7 | 4.8 × 102− |
| yeaQ | Function unknown | 2.1 | 2.6 × 10−4 |
| yebV | Function unknown | 2.4 | 1.2 × 10−3 |
| yhcO | Function unknown | 2.5 | 3.5 × 10−2 |
| yhfG | Function unknown | 2.4 | 1.9 × 10−4 |
| yjbJ | Function unknown | 2.0 | 1.3 × 10−2 |
| yjfO | Function unknown | 2.4 | 3.3 × 10−2 |
| yoaE | Function unknown | 2.3 | 9.8 × 10−4 |
| yodD | Function unknown | 2.1 | 4.9 × 10−3 |
| ytfR | Function unknown | 2.4 | 4.2 × 10−4 |
| ytfT | Function unknown | 3.0 | 3.7 × 10−2 |
| ytfR | Function unknown | 2.4 | 4.2 × 10−4 |
| ytfT | Function unknown | 3.0 | 3.7 × 10−2 |
Gene names obtained from http://bmb.med.miami.edu/EcoGene/EcoWeb/.
Gene product descriptions obtained from http://bmb.med.miami.edu/EcoGene/EcoWeb/.
A number of genes whose expression is regulated by the two-component system EvgAS were shown to be down-regulated. These genes all encode proteins that are known to provide protection against acid stress in E. coli. gadB (5.3-fold decrease) encodes an isozyme of glutamate decarboxylase, which catalyzes the conversion of glutamate to γ-aminobutyrate, and gadC (5.9-fold decrease) is predicted to encode a putative glutamate-γ-aminobutyrate antiporter (28). It has been proposed that these two proteins function together to help maintain the intracellular pH when cells are exposed to extremely acidic conditions (38). The expression of gadBC, however, requires gadE, which is also down-regulated (26). hdeA (5.4-fold decrease) encodes a periplasmic chaperone of acid-denatured proteins, and hdeB encodes a structural homologue of HdeA, which may form heterodimers with HdeA in order to prevent aggregation of the periplasmic proteins that become denatured in acidic conditions (13). hdeD (2.8-fold decrease) was also repressed; this gene encodes a putative membrane transporter that is suggested to be involved in acid stress resistance (27).
Other transport systems shown to be down-regulated include csgF (6.2-fold decrease), csgG (2.7-fold decrease), ompW (2.4-fold decrease) encoding an outer membrane protein, and lsrB (2.5-fold decrease), which is part of a system for autoinducer 2 uptake in Salmonella. Also, ugpA (2.2-fold decrease) and ugpE (2.4-fold decrease) were down-regulated; these genes encode hydrophobic integral inner membrane proteins that form a substrate-translocating complex for the uptake of sn-glycerol-3-phosphate (20).
Validation of the microarray gene profiling by quantitative real-time PCR (RT-PCR).
The fact that our analysis identified a number of genes (e.g., zntA, zraP, hydG, and spy) previously known to be induced (directly or indirectly) by Zn(II) in E. coli validates the ability of the microarray experiments to identify candidate genes important in resistance to toxic concentrations of extracellular Zn(II).
To verify further the results obtained by the microarray analysis, several genes that exhibited induction with 0.2 mM ZnSO4 were examined by quantitative real-time PCR to independently determine relative mRNA levels. The levels of up-regulation determined by RT-PCR (mean ± standard deviation) were as follows; cusF, 89 ± 0.7; basR, 4.6 ± 0.2; ais, 19 ± 0.02; asr, 38 ± 0.03; and cpxP, 14 ± 0.3. These values correspond to increases in the microarray analysis of 48-, 3-, 16-, 25-, and 11-fold, respectively. The mRNA levels from asnS (internal control) were unchanged as determined by both RT-PCR and the array analysis. The values obtained by RT-PCR strongly support the fidelity of the cDNA microarray analysis, taking into account any variation expected due to the differences between the two methodologies.
Hypersensitivity to ZnSO4 of strains deficient in genes up-regulated in the microarray experiments.
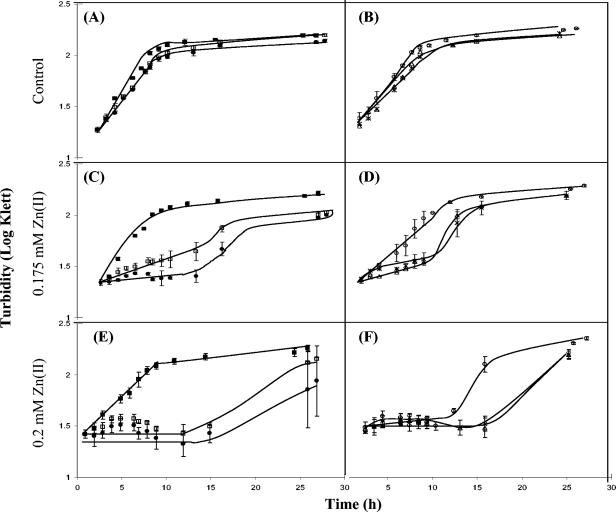
In this study we identified many genes that were up-regulated by zinc, and some of these genes (notably zntA) are known to be important for conferring resistance to high zinc concentrations. To assess the importance of other up-regulated genes, a selection of knockout mutants with mutations in genes shown to be up-regulated in the microarray work were obtained. These mutants included strains lacking basR, basS, fliM, ycdM, and yibD. Triplicate liquid culture growth curves (Fig. 4) revealed that all five mutants were hypersensitive to ZnSO4 when their growth was compared to wild-type growth. In GGM containing no additional ZnSO4 (Fig. 4A and B) there was no difference in the growth between any of the mutant strains and the wild type. When 0.15 mM (final concentration) ZnSO4 was added (data not shown), slight hypersensitivity was revealed by a small reduction in growth for the basR and basS mutants compared to the other strains. Addition of 0.175 mM (final concentration) ZnSO4 (Fig. 4C and D) to the medium resulted in a large reduction in growth (particularly for the basS mutant strain) and a much longer lag phase for the basR and basS mutants. Mutants with mutations in ycdM and yibD also showed hypersensitivity to ZnSO4, and there was an extended lag phase of growth similar to that observed for the basR and basS mutants. Finally, when ZnSO4 was added to a final concentration of 0.2 mM (Fig. 4E and F), all mutants showed hypersensitivity. All mutants except the fliM mutant exhibited extended lag phases during growth and reductions in the amount of growth. The fliM mutant had an extended lag phase that was shorter than those of the other mutants but showed no reduction in the amount of growth. To eliminate the possibility that a Tn5 insertion anywhere in the chromosome does not result in Zn hypersensitivity, growth curves for metA::Tn5 and qor::Tn5 mutant strains in liquid culture were determined. Both strains showed no heightened sensitivity (compared to wild-type strain MG1655) to a range of ZnSO4 concentrations up to 0.2 mM.
FIG. 4.
Growth of MG1655 (▪), FB21678 (basR mutant) (□), FB21676 (basS mutant) (•), FB20544 (fliM mutant) (○), FB22122 (ycdM mutant) (▴), and FB21346 (yibD mutant) (×) in the absence of additional ZnSO4 in the minimal medium (A and B) and in the presence of 0.175 mM ZnSO4 (C and D) and 0.2 mM ZnSO4 (E and F). The values are means for triplicate cultures; the error bars indicate standard deviations.
To support these findings in liquid cultures, zinc gradient plates were used to confirm the comparisons of hypersensitivity to ZnSO4 (Fig. 5). Overnight liquid cultures were streaked across a ZnSO4 gradient (maximum concentration, 0.5 mM) on minimal medium agar plates, which were then incubated for 2 days at 37°C. Compared to wild-type strain MG1655 growth, the basS mutant strain was the most hypersensitive to ZnSO4, followed closely by the basR mutant. The other three mutant strains were hypersensitive in the following order: yibD mutant, ycdM mutant, and fliM mutant.
FIG. 5.
Growth of MG1655, FB21678 (basR mutant), FB21676 (basS mutant), FB20544 (fliM mutant), FB22122 (ycdM mutant), and FB21346 (yibD mutant) across a concentration gradient of Zn(II). Freshly grown biomass was streaked across minimal medium plates containing concentration gradients of ZnSO4 (maximum concentration, 0.50 mM). The plates were incubated at 37°C for 2 days.
DISCUSSION
The present work revealed that in response to zinc stress (0.2 mM ZnSO4), 64 genes were up-regulated more than twofold and 54 genes were down-regulated more than twofold in chemostat-grown E. coli. Many of the genes that were up-regulated were involved in membrane structure, transport, sensing, and regulation, and most of them have not previously been implicated in zinc tolerance.
Previous work on metal ion tolerance in E. coli by Brocklehurst and Morby (8) involved microarray analysis of preadapted wild-type cells grown (under batch conditions) to the stationary phase. The work was carried out in LB medium in which the exact amounts of bioavailable metal ions were unknown. The use of a minimal medium, particularly a medium lacking inorganic phosphate that might form insoluble products with metal ions, allows maximum availability of added metal ions (3, 21). We did not observe any of the genes that were shown to be zinc responsive in the study of Brocklehurst and Morby (8), nor did the experiments of these authors reveal any genes that are known through other studies to be responsive to zinc. However, Brocklehurst and Morby (8) did find insA and insB genes that were up-regulated, which was observed with many different stresses, not just with stress due to excess zinc. In this work, the dilution rate, nutrient limitation, and pH were all controlled and/or monitored and were shown to be equal for both control and zinc-treated cultures. In agreement with Hayes et al. (19), we consider the use of chemostat cultures to be highly desirable in microarray analysis for identifying a gene expression change as a result of a single change in the growth conditions.
The microarray analysis demonstrated that the mdtABC operon was up-regulated in response to stress caused by excess zinc. This RND-type efflux system has been implicated in conferring resistance to certain antibiotics, including novobiocin and the bile salt component deoxycholate (2). Our finding that mdt is up-regulated in response to zinc therefore has potentially important implications for the influence of metal stresses on bacterial resistance to antibiotics. The presumed transport function of Mdt might indicate that it transports zinc directly, resulting in net export of the metal. An alternative possibility (of many) is that the transported solute moderates zinc toxicity, perhaps by binding the metal. The regulation of mdt is thought to be achieved by the two-component BaeRS system, encoded by genes immediately downstream of mdtABC. The signals sensed by BaeRS are not known. However, baeRS and ultimately mdt are clearly up-regulated by zinc, but it is possible that mdt transcription is not dependent solely on BaeRS.
Zinc supplementation in chemostat cultures was sufficient to increase expression of basR and basS (pmrA and pmrB, respectively, in Salmonella). We propose that high extracellular concentrations of zinc ions may be sensed by BasS (Fig. 3B) in a way that is similar to way that high iron concentrations are sensed by PmrB in Salmonella (Fig. 3A). This sensing of high zinc concentrations by BasS could in turn (as it does in Salmonella) cause phosphorylation of BasR and ultimately up-regulation of numerous genes (Fig. 3B). The up-regulation of these genes is an important finding, since this two-component system regulates, for example, resistance to polymyxin B and cationic peptides in Pseudomonas aeruginosa and Salmonella and also controls virulence in Erwinia carotovora (23). Interestingly, the importance of zinc in extracellular polysaccharide synthesis has recently been independently described by Hagiwara et al. (18). The Rcs signaling system in E. coli that is involved in capsular polysaccharide synthesis has recently been shown to be zinc responsive. Propagation of the zinc-responsive Rcs system was also largely dependent on PhoPQ, while in Salmonella the same two-component regulator system is implicated in the PmrAB mechanism. Thus, there is a recent confluence of data that strongly point to links among the sensing of and tolerance to divalent metal cations, including Zn and Mg(II), extracellular polysaccharide synthesis, the sensitivity of LPS to metal ions, and ultimately the sensitivity to antibiotics. Teleologically, a reason for basRS up-regulation by zinc ions might be related to the requirement for this system in modifying LPS, which is stabilized by divalent cations. Indeed, for yfb and arn (pbg in Salmonella), whose products are required for the modification of lipid A, there was a dramatic increase in expression, which was induced by zinc in these microarray experiments. Millimolar concentrations of magnesium stabilize the outer membrane (as judged by decreased susceptibility of periplasmic proteins to loss), while very high concentrations of calcium (20 mM) and magnesium (100 mM) increase the outer membrane permeability (and are commonly used, of course, in preparing cells that are competent for transformation by exogenous DNA). We speculate that the role of increased basRS transcription is to increase synthesis or effect modification of the LPS either to stabilize it in the face of high zinc concentrations or to act as a biosorbent of free metal ions.
The results of the hypersensitivity experiments with E. coli strains lacking some of the up-regulated genes identified, including basR, basS, yibD, and ycdM, support the hypothesis that cells grown with excess zinc need these up-regulated genes to respond to the stress. These mutant strains all showed hypersensitivity to zinc compared to the wild-type strain, with extended lag phases and lower growth yields, and it can therefore be suggested that these genes are important for zinc homeostasis within the cell. On the basis of present knowledge, it is not clear why ycdM and yibD (or fliM [Fig. 4]) appear to have roles in Zn tolerance. However, microarray analyses provide investigators with clues to the roles of genes with unknown functions or new roles for previously studied genes.
In summary, our results endorse the power of the microarray approach for identifying new genes required for zinc tolerance. Many genes have been identified as genes that are up-regulated in response to zinc, some of which were expected; however, the vast majority of the genes are not currently known to be involved in zinc homeostasis in E. coli. These findings may have important implications for understanding not only zinc homeostasis within the cell but also how bacterial resistance to antibiotics is modulated by metal ions.
Acknowledgments
This work was supported by BBSRC grant P10354 and by a University of Sheffield Krebs Studentship.
We thank Julie Scholes (Department of Plant and Animal Sciences, University of Sheffield) for her help and for use of resources for the RT-PCR experiments. We also thank F. Blattner and the E. coli Genome Project, University of Wisconsin-Madison, for kindly providing the MG1655 mutant strains and Tony Gordon and Pam Trickett for assistance with microarray profiling. We are grateful to Martin Hughes (King's College, London, United Kingdom) for his critical reading of the manuscript.
REFERENCES
- 1.Atichartpongkul, S., S. Loptasert, P. Vattanaviboon, W. Whangsuk, J. D. Helmann, and S. Mongkolsuk. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775-1782. [DOI] [PubMed] [Google Scholar]
- 2.Baranova, N., and H. Nikaido. 2002. The BaeSR two-component regulatory system activates transcription of the yegMNOB (mdtABCD) transporter gene cluster in Escherichia coli and increases its resistance to novobiocin and deoxycholate. J. Bacteriol. 184:4168-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard, S. J., R. Hashim, J. Membrillo-Hernández, M. N.Hughes, and R. K. Poole. 1997. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol. Microbiol. 25:883-891. [DOI] [PubMed] [Google Scholar]
- 4.Berg, J. M., and Y. Shi. 1996. The galvanization of biology: a growing appreciation for the roles of zinc. Science 271:1081-1085. [DOI] [PubMed] [Google Scholar]
- 5.Blindauer, C. A., M. D. Harrison, A. K. Robinson, J. A. Parkinson, P. W. Bowness, P. J. Sadler, and N. J. Robinson. 2002. Multiple bacteria encode metallothioneins and SmtA-like zinc fingers. Mol. Microbiol. 45:1421-1432. [DOI] [PubMed] [Google Scholar]
- 6.Breazeale, S. D., A. A. Ribeiro, and C. R. H. Raetz. 2003. Origin of lipid A species modified with 4-Amino-4-deoxy-l-arabinose in polymyxin-resistant mutants of Escherichia coli. J. Biol. Chem. 278:24731-24739. [DOI] [PubMed] [Google Scholar]
- 7.Breazeale, S. D., A. A. Ribeiro, and C. R. H. Raetz. 2002. Oxidative decarboxylation of UDP-glucuronic acid in extracts of polymyxin-resistant Escherichia coli. J. Biol. Chem. 277:2886-2896. [DOI] [PubMed] [Google Scholar]
- 8.Brocklehurst, K. R., and A. P. Morby. 2000. Metal-ion tolerance in Escherichia coli: analysis of transcriptional profiles by gene-array technology. Microbiology 146:2277-2282. [DOI] [PubMed] [Google Scholar]
- 9.Farewell, A., K. Kvint, and T. Nyström. 1998. uspB, a new σs-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J. Bacteriol. 180:6140-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke, S., G. Grass, and D. H. Nies. 2001. The product of the ybdE gene of the Escherichia coli chromosome is involved in the detoxification of silver ions. Microbiology 147:965-972. [DOI] [PubMed] [Google Scholar]
- 11.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraústo da Silva, J. J. R., and R. J. P. Williams. 2001. Zinc: Lewis acid catalysis and regulation, p. 321. In The biological chemistry of the elements. The inorganic chemistry of life, 2nd ed. Oxford University Press. Oxford, United Kingdom.
- 13.Gajiwala, K. S., and S. K. Burley. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 14.Garland, P. B., and P. J. Randle. 1962. A rapid enzymatic assay for glycerol. Nature 196:987-988. [DOI] [PubMed] [Google Scholar]
- 15.Grass, G., B. Fan, B. P. Rosen, S. Franke, D. H. Nies, and C. Rensing. 2001. ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J. Bacteriol. 183:4664-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grass, G., M. D. Wong, B. P. Rosen, R. L. Smith, and C. Rensing. 2002. ZupT is a Zn(II) uptake system in Escherichia coli. J. Bacteriol. 184:864-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzzo, A., C. Diorio, and M. DuBow. 1991. Transcription of the Escherichia coli fliC gene is regulated by metal ions. App. Environ. Microbiol. 57:2255-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagiwara, D., M. Sugiura, T. Oshima, H. Mori, H. Aiba, T. Yamashino, and T. Mizuno. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayes, A., N., Zhang, J. W., P. R. Butler, N. C. Hauser, J. D. Hoheisel, F. L. Lim, A. D. Sharrocks, and S. G. Oliver. 2002. Hybridization array technology coupled with chemostat culture: tools to interrogate gene expression in Saccharomyces cerevisiae. Methods 26:281-290. [DOI] [PubMed] [Google Scholar]
- 20.Hekstra, D., and J. Tommassen. 1993. Functional extrachangeability of the ABC proteins of the periplasmic binding protein-dependent transport systems Ugp and Mal of Escherichia coli. J. Bacteriol. 175:6546-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes, M. N., and R. K. Poole. 1991. Metal speciation and microbial growth—the hard (and soft) facts. J. Gen. Microbiol. 137:725-734. [Google Scholar]
- 22.Hughes, M. N., and R. K. Poole. 1989. Metals and micro-organisms. Chapman and Hall, London, United Kingdom.
- 23.Hyytiäinen, H., S. Sjöblom, T. Palomäki, A. Tuikkala, and E. T. Palva. 2003. The PmrA-PmrB two-component system responding to acidic pH and iron controls virulence in the plant pathogen Erwinia carotovora ssp. carotovora. Mol. Microbiol. 50:795-807. [DOI] [PubMed] [Google Scholar]
- 24.Kox, L. F. F., M. M. S. M. Wösten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leonhartsberger, S., A. Huber, F. Lottspeich, and A. Böck. 2001. The hydH/G genes from Escherichia coli code for a zinc and lead responsive two-component regulatory system. J. Mol. Biol. 301:93-105. [DOI] [PubMed] [Google Scholar]
- 26.Ma, Z., S. Gong, H. Richard, D. L. Tucker, T. Conway, and J. W. Foster. 2003. GadE (YhiE) activates a glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol. Microbiol. 49:1309-1320. [DOI] [PubMed] [Google Scholar]
- 27.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:669-712. [DOI] [PubMed] [Google Scholar]
- 28.Masuda, N., and G. M. Church. 2002. Escherichia coli gene expression responsive to levels of the response regulator EvgA. J. Bacteriol. 184:6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munson, G. P., D. L. Lam, F. W. Outten, and T. V. O'Halloran. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J. Bacteriol. 182:5864-5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagakubo, S., K. Nishino, T. Hirata, and A. Yamaguchi. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noll, M., K. Petrukhin, and S. Lutsenko. 1998. Identification of a novel transcription regulator from Proteus mirabilis, PMTR, revealed a possible role of YjaI protein in balancing zinc in Escherichia coli. J. Biol. Chem. 273:21393-21401. [DOI] [PubMed] [Google Scholar]
- 32.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 33.Pirt, S. J. 1975. Aeration and agitation methods, p. 95-106. In Principles of microbe and cell cultivation. Blackwell Scientific Publications, Oxford, United Kingdom.
- 34.Robinson, N. J., S. K. Whitehall, and J. S. Cavet. 2001. Microbial metallothioneins. Adv. Microb. Physiol. 44:183-213. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg, E. Y., D. Ma, and H. Nikaido. 2000. AcrD of Escherichia coli is an aminoglycoside pump. J. Bacteriol. 182:1754-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Small, P. L. C., and S. R. Waterman. 1998. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 6:214-216. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, G., G. Coutts, and M. Merrick. 2000. The glnKamtB operon. Trends Genet. 16:11-14. [DOI] [PubMed] [Google Scholar]
- 40.Trent, M. S., A. A. Ribeiro, S. Lin, R. J. Cotter, and C. R. H. Raetz. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A. J. Biol. Chem. 276:43122-43131. [DOI] [PubMed] [Google Scholar]
- 41.Tseng, G. C., M. Oh, L. Rohlin, J. C. Liao, and W. H. Wong. 2001. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res. 29:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams, R. J. P. 1984. Zinc: what is its role in biology? Endeavour New Ser. 8:65-70. [DOI] [PubMed] [Google Scholar]