Abstract
Sinorhizobium meliloti dctA encodes a transport protein needed for a successful nitrogen-fixing symbiosis between the bacteria and alfalfa. Using the toxicity of the DctA substrate fluoroorotic acid as a selective agent in an iterated selection procedure, four independent S. meliloti dctA mutants were isolated that retained some ability to transport dicarboxylates. Two mutations were located in a region called motif B located in a predicted transmembrane helix of the protein that has been shown in other members of the glutamate transporter family to be involved in cation binding. A G114D mutation was located in the third transmembrane helix, which had not previously been directly implicated in transport. Multiple sequence alignment of more than 60 members of the glutamate transporter family revealed a glycine at this position in nearly all members of the family. The fourth mutant was able to transport succinate at almost wild-type levels but was impaired in malate and fumarate transport. It contains two mutations: one in a periplasmic domain and the other predicted to be in the cytoplasm. Separation of the mutations showed that each contributed to the altered substrate preference. dctA deletion mutants that contain the mutant dctA alleles on a plasmid can proceed further in symbiotic development than null mutants of dctA, but none of the plasmids could support symbiotic nitrogen fixation, although they can transport dicarboxylates, some at relatively high levels.
In order for rhizobia to be able to establish an effective nitrogen-fixing (Fix+) symbiosis with a legume plant, the plant must provide the bacteria with energy, which is ultimately derived from plant photosynthesis. Dicarboxylic acids, such as malate, fumarate, and succinate, are thought to be a major carbon and energy source for the nitrogen-fixing bacteria (25). Importing these compounds requires the activity of the bacterial dicarboxylate transport protein, DctA. DctA belongs to the glutamate transporter family which, in addition to the prokaryotic dicarboxylate transporters, includes glutamate, aspartate, and neutral amino acid transporters found in bacteria and eukaryotes. Sinorhizobium meliloti DctA is typical of the bacterial C4-dicarboxylate transporters. Although it was originally identified by its ability to transport dicarboxylic acids, recent studies revealed that DctA exhibits a much broader substrate specificity (27). In addition to succinate, malate, fumarate, oxaloacetate, and aspartate, DctA can transport d-malate and compounds that are not dicarboxylates, such as orotate and succinamic acid. It also does not appear to transport some dicarboxylates, such as maleate, the cis-isomer of fumarate.
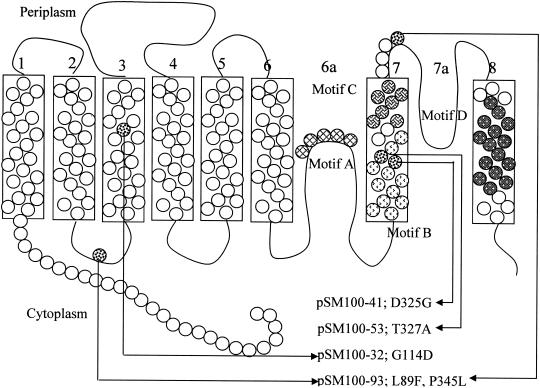
Glutamate and aspartate transporters are found in neurons and are involved in the postsynaptic reuptake of glutamate, an excitatory neurotransmitter. Defects in these transporters have been implicated in several neurological disorders and, as a result, they have attracted much attention. Several studies predicted six hydrophobic segments in the N-terminal part of the protein (12, 22), but the C-terminal part of the protein does not contain clear alternating regions of high and low hydrophobicity. In the most generally accepted model of these proteins (Fig. 1), the six N-terminal α-helices are followed by a cytoplasmic “reentrant” loop that can be accessed by extracellular labeling reagents, a seventh membrane-spanning helix, an extracellular reentrant loop, and an eighth membrane-spanning segment, leaving the C terminus on the cytoplasmic side (12). This predicted structure (12) differs somewhat from previous models for the DctA protein (8) in which the reentrant loops were each speculated to be two transmembrane helices.
FIG. 1.
Membrane topology of DctA. The positions of mutations described in the present study are indicated on the diagram of the protein. The model is modified from that of Slotboom et al. (22). Motifs A, B, C, and D are indicated as labeled in the figure by different types of shading.
There are several motifs that are conserved to some degree in all members of the glutamate transporter family. The most conserved of these is the serine-rich motif A in reentrant loop 6a, which appears to be important for the function of the transporter. In some glutamate transporters, one or more of the residues in this motif can be modified by reagents that cannot penetrate the cell. Mutagenesis studies in Bacillus stearothermophilus GltT confirmed that the serine-rich Motif A in the reentrant loop is extremely important for the transporter's function and may be part of the substrate-binding site (22). Motif B, which has been implicated in cation binding (12), is located in the inner, cytoplasm-proximal section of membrane helix 7 and is present in all functionally characterized transporters. Motif C, in the periplasmic section of transmembrane helix 7, is thought to be involved in binding a carboxylate group of the substrate (15). Motif D, in transmembrane helix 8, has similar features in most members of the family, but its exact amino acid composition correlates fairly closely with the substrate specificity of the transporters and suggests that helix 8 contains part of the substrate binding site or translocation pore (2, 14, 22).
Because S. meliloti DctA can transport a range of physiologically significant substrates, there are potentially a variety of genetic and physiological tools for carrying out mutant isolation and analysis. These could make DctA a good system for investigating structure-function questions in this transporter family. More directly, isolation and analysis of S. meliloti DctA mutants with partial ability to transport dicarboxylates might also clarify the operation of DctA in symbiosis, where it is very difficult to distinguish between various models of bacterial physiology (9). To better understand the mechanism of substrate recognition by the DctA protein and the relationship of DctA transport to bacterial function in symbiosis, we have isolated S. meliloti dctA mutants that retain some ability to transport at least one of the normal dicarboxylate substrates. The results suggest that malate and fumarate are more important in symbiotic function than succinate.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacteria and plasmids used in the present study are listed on Table 1. S. meliloti strains were grown at 30°C either on YMB medium or minimal mannitol medium containing NH4 (MM-NH4) (23), minimal medium with 0.2% of carbon sources other than mannitol (Min-NH4 plus “carbon source”), or on M9 minimal medium with modifications (27). Escherichia coli strains were grown at 37°C in Luria-Bertani or M9 minimal medium. Antibiotics for S. meliloti were added at 200 μg/ml (streptomycin) and 10 μg/ml (tetracycline) for plasmid-carried tetracycline resistance and at 1 μg/ml for chromosomal resistance. For E. coli, tetracycline was used at 25 μg/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| S. meliloti | ||
| Rm1021 | Wild type | 11 |
| RmF726 | Rm1021 ΔΩ5079-5149::Tn5-233 | 26 |
| WSUb20611-II | Rm1021 ΔdctA | This study |
| E. coli S17-1 | pro hsdR recA [RP4-2(Tc::Mu) (Km::Tn7)] | 20 |
| Plasmids | ||
| pSM100 | pCPP33 (1.5-kb BamHI dctA) | 27 |
| pK19mob-dctA-100 | pK19mob (dctA from pSM100) | This study |
| pSM100-32 | pSM100 (dctA G340T) | This study |
| pSM100-41 | pSM100 (dctA A975G) | This study |
| pSM100-53 | pSM100 (dctA C979T) | This study |
| pSM100-93 | pSM100 (dctA C265T C1034T) | This study |
| pK 19mob-dctA-93 | pK19mob (dctA from pSM100-93) | This study |
| pSM100-93-265 | pSM100 (dctA C265T) | This study |
| pSM100-93-1034 | pSM100 (dctA C1034T) | This study |
| pSYDCTII | 0.5-kb flanking dctA fragments cloned into pJPA22 in the BamHI site | This study |
| pJPA22 | pJQ200KS (BglII Tn10:tet) | 7 |
| pK19mob | pUC19 derivative lacZα, mob; Kmr | 19 |
Kmr, kanamycin resistance.
DNA manipulation and sequencing.
DNA manipulations were carried out according to standard procedures (18). Plasmid DNA was conjugated into rhizobia by biparental mating by using E. coli S17-1 as the donor strain (20). DNA sequencing was carried out by the DyeDeoxy terminator cycle protocol with synthetic primers synthesized by Invitrogen (Carlsbad, Calif.). Sequencing reactions were analyzed on an Applied Biosystems 373 DNA Sequencer at the Washington State University Laboratory of Bioanalysis and Biotechnology.
Hydroxylamine mutagenesis.
The mutagenesis was carried out as described previously (7). Briefly, 100 μl (20 μg) of plasmid DNA was added to 500 μl of 0.1 M sodium phosphate buffer (pH 6.0) containing 1 mM EDTA and 400 μl of hydroxylamine hydrochloride (1 M, adjusted to pH 6.0 with NaOH). The mixtures were incubated at 70°C, and 200-μl samples were removed at 2-h intervals starting from 2 h of incubation. The total length of the incubation was 10 h. Reactions were terminated by ethanol precipitation.
Iterated selection.
Mutagenized plasmids were electroporated into E. coli S17-1 and then mated into RmF726, selecting for the tetracycline resistance on the plasmid. The cells that carried mutagenized plasmid DNA were grown for 48 h in 50 ml of MM-NH4 that contained 1 μg of 5-fluoorotic acid (FOA)/ml. Survivors were harvested by centrifugation, resuspended in 50 ml of Min-NH4 plus 0.2% succinate plus 0.2% fumarate plus 0.2% malate and shaken for 72 h at 30°C. Pools of mutants were cycled through three FOA-dicarboxylate enrichment steps. To ensure that the mutations were on pSM100, plasmid DNA was isolated from the cultures and electroporated into E. coli S17-1, and the population of plasmids was mated into RmF726. We then screened for altered DctA-related phenotypes, such as FOA resistance.
Growth measurements.
To evaluate growth on various carbon substrates, S. meliloti strains were first grown to late log phase in MM-NH4 medium at 30°C. Cells were then diluted 50-fold into Min-NH4 medium containing 0.2% of the tested carbon source. Growth during 48 h was monitored at regular intervals by measuring the absorbance at 600 nm by using a SPECTRAmax 250 Microplate Spectrophotometer System (Molecular Devices).
Plant tests.
Alfalfa (Medicago sativa cv. Champ) was used for all nodulation studies. Seeds were surface sterilized by using concentrated sulfuric acid, washed several times in sterile water, and soaked in bleach (5% sodium hypochlorite) for 5 min. Seeds were again rinsed several times in sterile distilled water and then spread evenly on YMB agar plates and allowed to germinate at 30°C. Seedlings showing no signs of contamination were moved to sterile growth boxes consisting of two Magenta (Sigma GA-7 vessel) plant tissue boxes with the top box inverted to act as an aseptic barrier and containing a mixture of sand and LECA clay aggregate (Eco Enterprises, Shoreline, Wash.). Six seedlings were used per box, and each strain was evaluated in at least three boxes. Tested strains were grown on petri dishes with appropriate antibiotics for 48 h and then resuspended in sterile water to an optical density at 600 nm of 0.8. Each growth box was inoculated with the tested strains by applying 1 ml of cell suspension. Plants were grown in a walk-in growth room at 22°C as previously described (13). At 5 to 6 weeks after inoculation, plants were harvested and examined for root nodule formation. The shoot dry mass of the plants was measured, and the nitrogenase activity was assayed by using the acetylene reduction technique (23) on whole individual nodules.
Transport assays with whole cells of S. meliloti.
The uptake rates of radioactive compounds were measured by filtration of cells as previously described (27). Cells were grown in 2 ml of M9 mannitol medium for 48 h. Then, 1 ml of this culture was transferred into flasks containing 50 ml of M9 mannitol medium and grown for another 24 h. Cells were then washed three times in M9 buffer (M9 salts without a carbon source) and finally resuspended in 5 ml of M9 buffer. Vmax and Km values were determined by using at least two sampling times when a series of substrate concentrations was being assayed. The velocity of the substrate transport was measured at least at seven substrate concentrations. Experiments were repeated at least three times.
Microscopy.
Root nodules were excised from alfalfa roots 5 weeks after inoculation. These were fixed overnight in 2 to 2.5% glutaraldehyde-2% paraformaldehyde in 1 ml of 50 mM PIPES buffer and then washed three times (10 min/wash) in 1 ml of 50 mM PIPES buffer. Nodules were dehydrated by incubating the nodules in 1 ml of 30, 50, 60, 70, 80, and 90% ethanol for 10 min/incubation, followed by three successive incubations in 100% ethanol. Embedding was done in LR White resin (London Resin Company, Ltd.) by incubating the nodules in 1:3, 1:2, 1:1, and 3:1 ratios of LR White resin to ethanol at room temperature each time for 24 h and then in 100% LR White at 4°C for 24 h. The resin was solidified by incubating the perfused nodules overnight at 68°C. Sections (0.4 μm thick) were stained with basic fuchsin and/or iodine, examined by light microscopy, and photographed under ×200 magnification by using an Olympus BH-2 light microscope with a Nikon CoolPix 990 digital camera.
Separation of the mutations carried by pSm100-93.
BamHI dctA fragments from pSM100 and pSM100-93 were first recloned into multipurpose cloning vector pK19mob (19), resulting in plasmids pK19mob-dctA-100 and pK19mob-dctA-93, respectively. The correct orientation of the insertions was checked by digestion the plasmids with SmaI. The two mutations in pSM100-93 are separated by an NcoI site, and there is an NcoI site in the Tn5 Kmr gene. Digestion of pK19mob-dctA-100 and pK19mob-dctA-93 with NcoI resulted in two fragments for each plasmid with the sizes 1.7 and 3.45 kb. The smaller fragment from one plasmid was ligated to the larger fragment from the other, resulting in plasmids pK19mob-93-265 and pK19mob-93-1034, each containing a single mutation of the dctA-93 open reading frame (ORF). The presence of each single mutation was confirmed by DNA sequencing. Finally, the dctA-containing BamHI fragments from pK19mob-93-265 and pK19mob-93-1034 were recloned back into pCPP33 resulting the plasmids pSM100-93-265 and pSM100-93-1034.
Construction of the S. meliloti dctA deletion mutant WSUb20611-II.
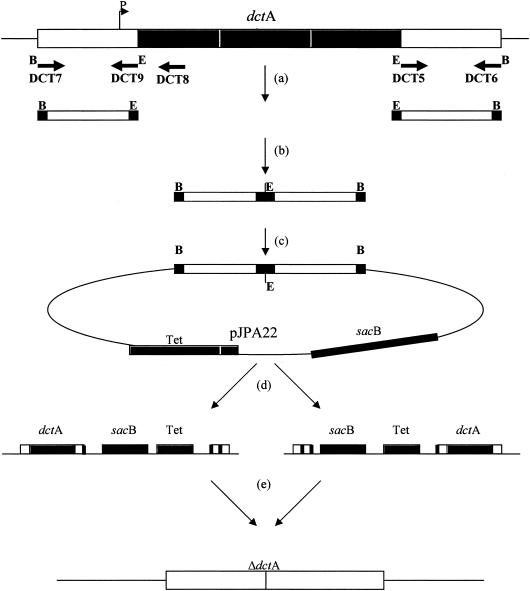
A mutant with a precisely defined deletion in S. meliloti dctA was constructed so that point mutations could not be rescued by recombination. First, 0.5-kb regions flanking dctA were amplified from chromosomal DNA by PCR with primers: DCT7 (5′-CATGGATCCCGGAAGGCATAGTCGTTGCC-3′) and DCT9 (5′-CATGAATTCTGCGTGCCAGTTTGCCGCGG-3′) for the upstream flanking region, and DCT5 (5′-CATGAATTCTGCGTGCCAGTTTGCCGCGG-3′) and DCT6 (5′-CATGGATCCGCACATTATCCACGAACGGG-3′) for the downstream flanking region (Fig. 2a). Each primer had a 20-bp overlap with the S. meliloti sequences flanking dctA and a 9-bp 5′ extension, which contained a BamHI (GGATCC) restriction site for external ends and an EcoRI (GAATTC) site for internal ends (underlined in the sequences above). After amplification, the PCR products were digested with EcoRI and ligated together (Fig. 2b). The resulting DNA fragment, which was expected to contain the DNA regions flanking dctA but with the dctA gene deleted, was then digested with BamHI and ligated to pJPA22 (6) that had been previously digested with BamHI (Fig. 2c). Plasmid pJPA22 is a tetracycline-resistant suicide plasmid carrying the sacB gene, which codes for the enzyme levansucrase and leads to sucrose sensitivity in S. meliloti. The resulting plasmid was named pSYDCTII. Plasmid pSYDCTII was conjugated into S. meliloti strain Rm1021 and, since pSYDCTII cannot replicate in S. meliloti, we expected that pSYDCTII integration into the S. meliloti Rm1021 chromosome to yield tetracycline-resistant (1 μg/ml) colonies would occur by recombination within one of the regions flanking the dctA ORF (Fig. 2d). These single recombinants were subcultured on minimal mannitol ammonia agar supplemented with 5% sucrose. Only rhizobia that lose the sacB gene in pSYDCTII by mutation or recombination can grow on this medium. If a second recombination occurs in the other flanking region, the allele with the deletion of the dctA ORF would remain and the resulting strain would be tetracycline sensitive and Dct− (Fig. 2e).
FIG. 2.
Construction of WSUb20611-II. (a) Fragments flanking dctA ORF were amplified by using PCR. (b) The end of the upstream PCR fragment was ligated to the beginning of downstream PCR fragment. (c) The resulting fragment was ligated into the BamHI site of pJPA22 to yield pSYDCTII. (d) pSYDCTII was transferred into S. meliloti Rm1021, and single recombinants were selected on MM-NH4 plates containing 1 μg of tetracycline per ml. (e) Double recombinants were selected on MM-NH4-5% sucrose plates.
Colonies with this phenotype contained DNA with the expected structure. PCR amplification of Rm1021 DNA with the DCT7 and DCT6 primers (Fig. 2a) resulted in a 2.5-kb fragment. The PCR product from the presumed deletion mutants was about 1 kb, indicating the expected deletion of a 1.5-kb fragment containing the dctA ORF. DNA from Rm1021 and newly constructed mutant strains were amplified by using DCT7 and the DCT8 primer located 200 bp downstream of the first dctA start codon. Amplification of Rm1021 DNA resulted in a 0.7-kb PCR product but, as expected, there was no PCR product from the constructed strains (data not shown). One of these deletion mutant strains, WSUb20611-II, was used in experiments as described.
RESULTS
Isolation of dctA mutants with altered ability to transport dicarboxylates. To identify dctA mutants with altered transport properties, we used FOA, a toxic analog of orotate that can be transported by both E. coli and S. meliloti DctA (1, 27). Our aim was to use the toxicity of FOA to select for mutant DctA proteins unable to recognize FOA but, by also growing potential mutants on a mixture of dicarboxylates, to find mutants that retained DctA function. To avoid possible interactions between DctA and DctB (17, 26), dctA expressed from the E. coli lacZ promoter (27) was carried on pSM100, and selection was done in S. meliloti strain Rm726, which has a deletion of the dct region, including many adjacent genes (3).
Direct selection for FOA resistance recovered resistant mutants unable to grow on any dicarboxylates tested. These presumably lack DctA function. To carry out a more sophisticated selection, we mutagenized pSM100 with hydroxylamine in vitro and used an iterated selection procedure with FOA and dicarboxylate media. Selection in the presence of 1 μg of FOA/ml, which is near the MIC for this strain (28), allowed us to enrich for mutants of DctA with impaired FOA transport. Subsequent selection with a mixture of dicarboxylates enriched for mutants that retained the ability to transport at least one substrate. Separating the enrichments into two steps was needed since dicarboxylates directly inhibit FOA transport (27).
We then used several approaches to screen for DctA mutants. In the best of these, potential mutants were screened for growth in the presence of FOA (20 to 100 μg/ml) on a mixture of dicarboxylates (Min-NH4 plus 0.1% succinate, fumarate, malate, and maleate). Colonies that grew under these conditions were then tested on MM-NH4 plus 0.1 or 1 μg of FOA/ml and on Min-NH4 medium supplemented with 0.2% of each of the dicarboxylic acid substrates (Table 2). The dicarboxylate-FOA plates represent marginal growth conditions, where it appears that growth was significantly affected by small changes in the affinity of DctA for either a dicarboxylate or FOA.
TABLE 2.
Growth properties of dctA mutants
| Strain | Colony size (score)a at 72 h on:
|
||||||
|---|---|---|---|---|---|---|---|
| MM-NH4 | Min-NH4 + succinate | Min-NH4 + fumarate | Min-NH4 + l-malate | Min-NH4 + d-malate | MM-NH4 + FOA (0.1 μg/ml) | MM-NH4 + FOA (1 μg/ml) | |
| Rm1021 (wild-type) | 4 | 4 | 4 | 4 | 4 | 0 | 0 |
| WSUb20611-II dctA deletion | 4 | 0 | 0 | 0 | 0 | 4 | 4 |
| RmF726 (∼200-kb deletion) | 4 | 0 | 0 | 0 | 0 | 4 | 4 |
| WSUb20611-II (pSM100) | 4 | 4 | 4 | 4 | 4 | 0 | 0 |
| RmF726(pSM100) | 4 | 4 | 4 | 4 | 4 | 0 | 0 |
| WSUb20611-II(pSM100-32) | 4 | 2 | 2 | 2 | 1 | 2 | 2 |
| RmF726(pSM100-32) | 4 | 2 | 2 | 2 | 1 | 2 | 2 |
| WSUb20611-II(pSM100-41) | 4 | 1 | 1 | 1 | 1 | 2 | 1 |
| RmF726(pSM100-41) | 4 | 1 | 1 | 1 | 1 | 2 | 1 |
| WSUb20611II(pSM100-53) | 4 | 2 | 2 | 2 | 1 | 2 | 2 |
| RmF726(pSM100-53) | 4 | 2 | 2 | 2 | 1 | 2 | 2 |
| WSUb20611-II(pSM100-93) | 4 | 1 | 1 | 1 | 1 | 0 | 0 |
| WSUb20611-II(pSM100-93-265) | 4 | 3 | 3 | 3 | 1 | 0 | 0 |
| WSUb20611-II(pSM100-93-1034) | 4 | 2 | 2 | 2 | 1 | 0 | 0 |
| RmF726(pSM100-93) | 4 | 1 | 1 | 1 | 1 | 0 | 0 |
Colony size was scored by numbers from 4 to 0 at 72 h. Scores: 4, colony size equal to wild-type S. meliloti Rm1021 growing on the same medium or, in the case of FOA medium, Rm1021 growing on medium not containing FOA; 3, diameters of single colonies were reduced, but more than half were formed by wild-type strain; 2, diameters of single colonies were less than half those of colonies formed by wild-type strain; 1, formed hardly visible colonies; 0, no growth.
Several potential mutants were identified. Plasmids were isolated from each mutant, transferred into RmF726, and the phenotypes of the resulting strains were confirmed. The phenotypes of RmF726 carrying four of the mutated plasmids, pSM100-32, pSM100-41, pSM100-53, and pSM100-93 are presented in Table 2. Strain RmF726(pSM100) was similar to wild-type S. meliloti Rm1021; RmF726(pSM100-53) and RmF726(pSM100-32) were FOA resistant and grew slower than RmF726(pSM100) on succinate, malate, and fumarate; RmF726(pSM100-93) was FOA sensitive but grew poorly on succinate, malate, and fumarate; RmF726(pSM100-41) grew very poorly on dicarboxylates and was slightly sensitive to increased concentrations of FOA. In contrast to strains containing pSM100, none of the strains carrying mutant plasmids could grow on d-malate, a relatively low-affinity substrate for the wild-type protein (26, 27).
Location of mutations in dctA.
The DNA sequences of the dctA regions in the mutant plasmids were determined (Fig. 1). The DctA protein expressed from pSM100-32 was predicted to have a G114D mutation, altering a highly conserved glycine residue located on the third transmembrane domain of the protein (Fig. 3). Of all of the glutamate family transport proteins, only the bacterial serine transporters and eukaryotic alanine/serine/cysteine/threonine transporters lack glycine in this position, and in these families the substitution is always to alanine. The DctA proteins predicted to be made by pSM100-41 and pSM100-53 contain substitutions of a single residue in motif B in positions D325G and T327A, respectively. This region in the seventh transmembrane domain of the protein is thought to be involved in cation binding (22). The aspartate corresponding to D325 is highly conserved in the glutamate family of proteins and is missing only in the bacterial serine transporters (Fig. 3).
FIG. 3.
Multiple sequence alignments of transmembrane domains 3 and 7. A representative set of 31 members of the glutamate transporter family is shown. Boldface letters and asterisks in the diagram at the top refer to fully conserved amino acids; boldface italic letters and colons refer to strongly conserved amino acids; italic letters and periods refer to weakly conserved amino acids. Underlined letters in the first row refer to the residues corresponding to the mutations located on S. meliloti DctA. Abbreviated organism names: RHIME, Sinorhizobium meliloti; ECOLI, Escherichia coli; SALTY, Salmonella enterica serovar Typhimurium; YERPE, Yersinia pestis; PSEAE, Pseudomonas aeruginosa; AGRT5, Agrobacterium tumefaciens (strain C58/AACC 33970); XYLFA, Xylella fastidiosa; RALSO, Ralstonia solanacearum; BACSU, Bacillus subtilis; BACCA, Bacillus caldotenax; BACST, Bacillus stearothermophilus; MOUSE, Mus musculus (mouse); HUMAN, Homo sapiens; BOVIN, Bos taurus; RABIT, Oryctolagus cuniculus; CAEEL, Caenorhabditis elegans; AMBTI, Ambystoma tigrinum (tiger salamander); RAT, Rattus norvegicus (rat); ONCVO, Onchocerca volvulus.
The DctA protein predicted in pSM100-93 had a L85F/P345L double mutation. The L85F mutation was in the short cytoplasmic loop between the second and third transmembrane domains and the P345L mutation was located on the external surface of the membrane following the seventh transmembrane domain. A strong consensus of these residues within the glutamate transporter family was not observed. In addition to analyzing the double mutant in pSM100-93, the mutations were separated onto two plasmids and analyzed separately.
Influence of the DctB and DctD proteins on dicarboxylate transport.
A pSM100 derivative of RmF726 was ineffective (Fix−). Since RmF726 has a deletion of the entire dct region that includes about 200 kb around dctABD (3), there was a strong possibility that this deletion would affect the symbiotic phenotype. Existing point mutations were judged unsuitable for complementation studies since these might recombine with a mutant dctA or produce defective proteins that could interact with the mutated DctA. We therefore constructed S. meliloti strain WSUb20611-II, which contains a precise deletion of the dctA ORF. WSUb20611-II was FOA resistant and was not able grow on any of dicarboxylates (Table 2 and Fig. 4). When wild-type pSM100 and plasmids containing the mutant versions of dctA were introduced into WSUb20611-II, the free-living phenotype of the strains was similar to the phenotype of RmF726 carrying the corresponding plasmids (Table 2). The Km for succinate transport was similar when DctA was expressed from pSM100 in RmF726 and in WSUb20611-II (Table 3). However, the Vmax for succinate uptake of strain WSUb20611-II(pSM100) was about twice that for RmF726(pSM100) (Table 3). Significantly, the pSM100 derivative of WSUb20611-II was Fix+.
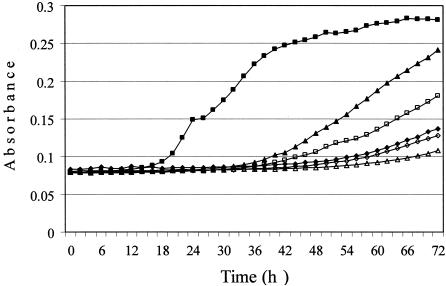
FIG. 4.
Growth of S. meliloti strains on 0.2% succinate. Cultures were grown at 30°C, and the absorbance at 600 nm was monitored by using a kinetic microplate reader. Symbols: ▵, WSUb20611-II; ▪, WSUb20611-II(pSM100); ⋄, WSUb20611-II(pSM100-93); ▪, WSUb20611-II(pSM100-93-1034); ▴, WSUb20611-II(pSM100-93-265); ♦, WSUb20611-II(pSM100-41).
TABLE 3.
Transport of dicarboxylates by S. meliloti dctA mutants
| Strain |
Vmax (nmol [min mg of protein]−1) and Km (μM) valuesa on medium plus:
|
|||||
|---|---|---|---|---|---|---|
| Succinate
|
Fumarate
|
Malate
|
||||
| Vmax | Km | Vmax | Km | Vmax | Km | |
| RmF726(pSM100) | 32 | 51 | ND | ND | ND | ND |
| WSUb20611-II(pSM100) | 61 | 66 | 45 | 79 | 36 | 62 |
| WSUb20611-II(pSM100-32) | 30 | 457 | 21 | 145 | 17 | 706 |
| WSUb20611-II(pSM100-41) | 0.86 | 24 | 0.85 | 103 | 0.56 | 65 |
| WSUb20611-II(pSM100-53) | 40 | 321 | 40 | 155 | 40 | 771 |
| WSUb20611-II(pSM100-93) | 56 | 95 | 19 | 58 | 16 | 45 |
| WSUb20611-II(pSM100-93-1034) | 82 | 112 | 35 | 70 | 31 | 72 |
| WSUb20611-II(pSM100-93-265) | 81 | 92 | 37 | 46 | 29 | 72 |
Mutants were grown on M9 mannitol medium without induction. Km values are the micromolar concentrations of the corresponding substrate. ND, not determined.
Dicarboxylate uptake by dctA mutants.
The uptake of dicarboxylates by the mutants at different substrate concentrations was measured and used to determine Vmax and Km values (Table 3). We have been unable to raise an antibody to DctA and therefore cannot quantify the amount of protein present in these strains, so decreases in Vmax could be the result of faulty protein processing decreasing the amount of DctA present in the mutants. WSUb20611-II carrying the mutant allele on pSM100-32 had a twofold-lower Vmax for succinate, fumarate, and malate uptake than strain WSUb20611-II(pSM100), which contains the wild-type allele. The DctA protein expressed from pSM100-32 had significantly lower affinities for dicarboxylates than wild-type DctA, but the extent of this decrease was greater for malate (11-fold) than for succinate (7-fold) or fumarate (2-fold) (Table 3). The mutant grew on these dicarboxylates (Table 2), which are present in the growth medium at concentrations of ca. 10 mM—many times the Km. However, WSUb20611-II(pSM100-32) was significantly more resistant to FOA; cells grew on agar medium containing 1 μg of FOA/ml, whereas 0.1 μg of FOA/ml completely blocked the growth of WSUb20611-II(pSM100).
WSUb20611-II(pSM100-53) transported succinate, fumarate, and malate with about the same Vmax as the corresponding strain containing pSM100 (Table 3). The affinity of the protein expressed from pSM100-53 for dicarboxylates was significantly lower. As for WSUb20611-II(pSM100-32), WSUb20611-II(pSM100-53) recognized fumarate relatively better than succinate or malate. Transport by DctA encoded on pSM100-53 was able to support growth on the dicarboxylates, and deletion strains carrying pSM100-53 were resistant to 1 μg of FOA/ml (Table 2).
WSUb20611-II(pSM100-41) transported succinate, malate, and fumarate only 1 to 2% as well as wild-type DctA (Table 3), which correlates with the observation that dctA deletion strains carrying pSM100-41 grew poorly on dicarboxylates (Table 2 and Fig. 4). The growth rate was scored as “1,” very close to the rate of dctA deletion strains that do not utilize dicarboxylates (Table 2 and Fig. 4). However, dctA deletion strains carrying pSM100-41 were still sensitive to FOA at the higher concentration (1 μg/ml) tested (Table 2). Thus, the DctA produced by pSM100-41 still retains partial ability to transport FOA.
Strain WSUb20611-II(pSM100-93) exhibited a high level of dicarboxylate uptake (Table 3), and the affinities of WSUb20611-II(pSM100-93) and WSUb20611-II(pSM100) for various dicarboxylates were similar. The apparent Vmax for succinate uptake by WSUb20611-II(pSM100-93) was like that of WSUb20611-II(pSM100), but the Vmax for fumarate and malate uptake by WSUb20611-II(pSM100-93) was about half that of WSUb20611-II(pSM100) (Table 3). Despite its high level of dicarboxylate uptake and its sensitivity to 0.1 μg of FOA/ml, strain WSUb20611-II(pSM100-93) did not grow well on Min-NH4 plates amended with any of dicarboxylates or in liquid media (Table 2 and Fig. 4).
One possible explanation for this phenomenon might be that the mutant protein expressed from pSM100-93 cannot maintain a concentration of dicarboxylates inside cells adequate to supply the cells with energy. However, the concentration of dicarboxylates in the medium was about 10 mM, which should be high enough to saturate tricarboxylic acid cycle enzymes such as succinate dehydrogenase (5), even if the mutant DctA was unable to couple transport to a membrane potential and concentrate the carbon source.
Another possibility we considered was that the interaction of dicarboxylates with the pSM100-93 mutant DctA causes a conformational change that leads to cell damage. In this scenario, DctA could retain the ability to transport dicarboxylates during the short time needed for a transport assay, but in the presence of the substrate, the mutant protein would harm the cells. A situation like this was described by Choi and Chiu (4), in which a L325H mutation in the EAAT1 glutamate transporter caused cell death when the mutant protein was expressed in Xenopus oocytes. We tested the ability of wild-type Rm1021 and dctA mutants RmF726 and WSUb20611-II carrying pSM100-93 to grow on MM-NH4 medium containing mannitol and dicarboxylic acids in various combinations. All strains grew well in the presence of dicarboxylates, indicating that the DctA from pSM100-93 was not toxic.
Although the transport rate of WSUb20611-II(pSM100-93) was similar to wild-type under standard conditions, preliminary experiments have shown that succinate transport by the mutant was much lower early in the growth curve. This would suggest that a difference in the stability or assembly of the mutant DctA could account for the slow growth of WSUb20611-II(pSM100) on dicarboxylates.
To better understand pSM100-93, we separated the two mutations in this plasmid as described in Materials and Methods. DctA carrying the L85F mutation was expressed from plasmid pSM100-93-265, the number “265” indicating the position of the mutation in the dctA DNA sequence. The plasmid carrying DctA with the P345L mutation located on the external surface of the membrane following the seventh transmembrane domain was similarly named pSM100-93-1034.
Both derivatives of WSUb20611-II(pSM100-93) that carried a single mutation from pSM100-93 grew better on Min-NH4 plates supplied with any of dicarboxylates than WSUb20611-II(pSM100-93) (Table 2). WSUb20611-II(pSM100-93-265) grew almost as well as WSUb20611-II(pSM100) on Min-NH4 plates supplied with dicarboxylates (Table 2) but still showed impaired growth in liquid (Fig. 4). The P345L mutation significantly decreased the ability of the strain to grow on dicarboxylates (Table 2), including slow growth in liquid (Fig. 4). Each mutation confers the same level of FOA sensitivity as the double mutation in pSM100-93. WSUb20611-II(pSM100-93-265) and WSUb20611-II(pSM100-93-1034) had levels of dicarboxylate uptake similar to WSUb20611-II(pSM100); the only difference we could detect was that both mutants had slightly lower Vmax values for fumarate and malate uptake (Table 3). These results indicated that neither of the single mutations in pSM100-93 were individually responsible for the impaired ability of WSUb20611-II(pSM100-93) to grow on dicarboxylates, but the mutation in the periplasmic loop had a stronger impact on the growth rate of plasmid-containing cells (Fig. 4).
Symbiotic phenotypes of dctA mutants.
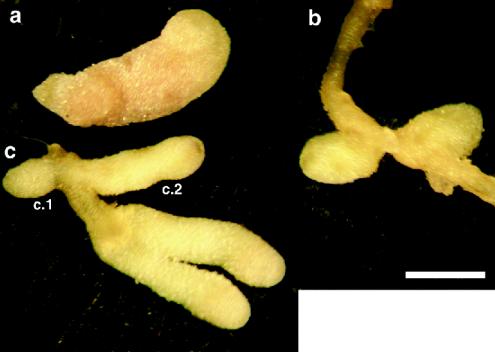
A comparison of the symbiotic phenotypes of the S. meliloti dctA mutants is shown in Table 4. The dctA deletion strain WSUb20611-II formed Fix− nodules on alfalfa (Fig. 5b) that were similar in phenotype to those induced by the previously described S. meliloti mutant RmF726, which carries a longer deletion encompassing the dct genes (26) (Table 4). Nodules formed by WSUb20611-II and RmF726 were small and white and did not have any nitrogenase (acetylene reduction) activity. Light microscopy revealed that the nodules contained a large number of empty, uninfected cells and that the infected cells contain few developed bacteroids (data not shown). After 5 weeks, the dry weight of shoots from plants inoculated with WSUb20611-II was less than 50% of shoots from plants inoculated with Rm1021 (Table 4). Plants inoculated with WSUb20611-II and RmF726 resembled uninoculated controls, which were yellow and much smaller than plants nodulated by S. meliloti Rm1021.
TABLE 4.
Symbiotic phenotype of S. meliloti dctA mutants
| Strain | Mean shoot dry wt (mg/plant) ± SD | Mean acetylene reduction (nmol [g of nodule × min]−1) ± SD |
|---|---|---|
| Rm1021 | 11.9 ± 3.8 | 57.0 ± 6.2 |
| Uninoculated | 5.6 ± 0.7 | 0 |
| RmF726 | 5.3 ± 0.6 | |
| RmF726(pSM100) | 5.4 ± 1.0 | |
| WSUb20611-II | 5.0 ± 1.0 | 0 |
| WSUb20611-II(pSM100) | 10.8 ± 1.9 | 34.8 ± 15.9 |
| WSUb20611-II(pSM100-32) | 4.6 ± 0.5 | 0 |
| WSUb20611-II(pSM100-41) | 4.9 ± 0.4 | 0 |
| WSUb20611-II(pSM100-53) | 5.0 ± 0.3 | 0 |
| WSUb20611-II(pSM100-93) | 4.8 ± 0.7 | 0 |
| WSUb20611-II(pSM100-93-265) | 6.0 ± 0.9 | 0 |
| WSUb20611-II(pSM100-93-1034) | 5.8 ± 0.3 | 0 |
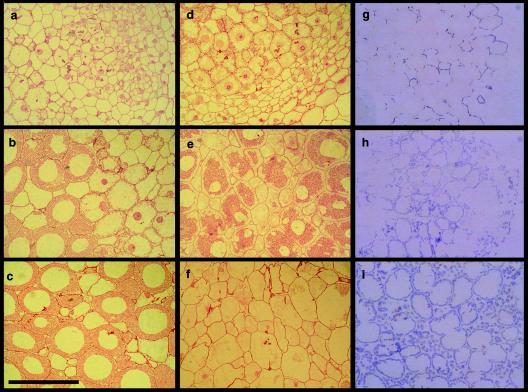
FIG. 5.
Alfalfa nodules induced by wild-type and dctA mutant strains of S. meliloti. Nodules were induced by wild-type S. meliloti Rm1021 (a), dctA deletion strain WSUb20611-II (b), and WSUb20611-II(pSM100-53) (c). c.1, Type A nodules; c.2, type B nodules. Bar, 1 mm.
The presence of the wild-type allele of dctA carried on pSM100 restored an effective symbiotic phenotype to WSUb20611-II. Plants nodulated with WSUb20611-II(pSM100) were a rich green and appeared to be healthy. The shoot dry mass of the plants inoculated by WSUb20611-II(pSM100) was almost (90%) as high as the plants inoculated with the wild-type strain, Rm1021. WSUb20611-II(pSM100) induced pink nodules (Fig. 5a) that were able to reduce acetylene, an index of nitrogenase activity, at a level slightly lower then nodules induced by Rm1021 (Table 4). Histological analysis revealed that alfalfa nodules induced by Rm1021 and WSUb20611-II(pSM100) were similar, with well-developed meristematic (I), infection (II), nitrogen-fixing (III) zones (24). Cells within the nitrogen-fixing zone were packed with bacteroids. Longitudinal cross sections from a nodule induced by WSUb20611-II(pSM100) are shown in Fig. 6a to c.
FIG. 6.
Nodules induced by wild-type and dctA mutant strains of S. meliloti. (Left column) Longitudinal cross sections from the meristematic (a), infection (b), and nitrogen-fixing (c) zones of a nodule induced by the wild-type strain of S. meliloti Rm1021; (middle column) longitudinal cross sections from the meristematic (d), pseudo-nitrogen-fixing (e), senescing (f) zones of a B-type nodule induced by WSUb20611-II(pSM100-53); (right column) starch accumulation in the nodules induced by Rm1021 (g), WSUb20611-II (h), and WSUb20611-II(pSM100-53) (i). Bar, 100 μm.
In contrast to pSM100, plasmids pSM100-32, pSM100-41, pSM100-53, pSM100-93, pSM100-93-1034, and pSM100-93-265 did not permit WSUb20611-II to establish an active nitrogen-fixing symbiosis with alfalfa. Plants inoculated with WSUb20611-II strains containing these plasmids were small and yellow. The mass of the plants inoculated with the mutants was similar to uninoculated plants or plants inoculated with WSUb20611-II, and nodules formed by these strains had no acetylene reduction activity (Table 4).
There was a symbiotic phenotype of most of these strains that differed from the phenotype of the deletion mutant. The majority of the nodules formed by strains WSUb20611-II(pSM100-32), WSUb20611-II(pSM100-53), WSUb20611-II(pSM100-93), WSUb20611-II(pSM100-93-1034), and WSUb20611-II(pSM100-93-265) and all of the nodules formed by WSUb20611-II(pSM100-41) were the “type A” ineffective nodules we observed on a plasmid-free strain (Fig. 5c, c.1). These were small and white and contained few bacteroids. However, with all mutants except WSUb20611-II(pSM100-41) we observed a second, “type B,” nodule. These nodules were elongated, bulb shaped, and slightly green, with some pink color near the tip approximately where the start of the nitrogen-fixing zone would be in a normal nodule (Fig. 5c, c.2). A longitudinal cross section from a type B nodule elicited by WSUb20611-II(pSM100-53) is shown in Fig. 6d to f. Cells contained in the pseudo-nitrogen-fixing zone of the nodule elicited by WSUb20611-II(pSM100-53) were morphologically different from those in the nitrogen-fixing zone of a wild-type nodule, as illustrated by comparison under higher magnification in Fig. 6c and e. The apparent separation of the bacteroids from the plant cell wall (Fig. 6e) was due to the large amount of starch that lines the cell walls (see below). The more mature regions of type B nodules appeared to be undergoing early senescence, and these regions were green, a phenotype that is thought to be due to the accumulation of heme degradation products.
Starch accumulation by dctA mutants.
When the normal energy flow toward nitrogen fixation is disrupted, sucrose transported to the nodules can accumulate as starch. Starch deposition in the nodules formed by the dctA mutants was visualized by staining with iodine. An iodine-stained cross section from a normal nodule induced by WSUb20611, an ineffective nodule induced by WSUb20611-II, and a type B nodule induced by WSUb20611-II(pSM100-53) are shown in Fig. 6g to i.
The nodules induced by the dctA deletion strain contained a much higher level of starch than was seen in nodules induced by Rm1021 or WSUb20611-II(pSM100) and the distribution of the starch was different. Starch in nodules formed by bacteria containing a wild-type allele of dctA was found primarily in infected cells in the interzone II/III region and was located near the cell wall. In nodules formed by the dctA deletion mutant, the area of starch deposition was more extensive, including more infected cells and considerable accumulation in the uninfected cells (Fig. 6.h). The pattern of starch accumulation found in type A nodules induced by WSUb20611-II(pSM100-32), WSUb20611-II(pSM100-53), WSUb20611-II(pSM100-41), WSUb20611-II(pSM100-93), WSUb20611-II(pSM100-93-1034), and WSUb20611-II(pSM100-93-265) was similar to that in the dctA mutant. In contrast, the type B nodules induced by these strains accumulate enormous amounts of starch in virtually every cell with almost a continuous line of amyloplasts at the periphery of infected cells and prominent granules in the uninfected cells (Fig. 6i).
DISCUSSION
Mutants of S. meliloti dctA partially altered in their ability to transport dicarboxylic acids were isolated by using iterated selection. We report on several dctA mutants with distinctive phenotypes. In most cases these mutants retained the ability to transport succinate at a level that was sufficient for growth. Two mutations, in plasmids pSM100-41 and pSM100-53, were in DctA protein motif B that is located within the predicted transmembrane helix 7. In other members of the glutamate transporter family this motif has been shown to be involved in cation binding. Mutagenesis studies of motif B in the rat glutamate transporter EAAT2 demonstrated that residues Y403 and E404 (corresponding to S. meliloti dctA positions 330 and 331, Table 5) are involved in binding potassium (30, 10). Residues 396 to 400 (323 to 327 in S. meliloti DctA) were suggested to be part of the Na+-binding site(s). Replacing each of the five residues by cysteine in EAAT2 abolished transport activity (29). The proteins expressed from plasmids pSM100-41 and pSM100-53 contain mutations in positions 325 and 327, respectively.
TABLE 5.
Sequence motifs in the glutamate transporter family
| Motif | Sequence |
|---|---|
| B (all members) | 317a PxGx (TS) xN (ML) DbGxcx (LI) (FY) 330a |
| D | |
| Bacterial C4-dicarboxylate transporters | DRFMSExRxxxNxxGN |
| Bacterial glutamate transporters | DRxxDMARTxxNxxG (NH) |
| Eukaryotic Ala/Ser/Cys/Thr transporters | DWxVDRxxTxxNVEGD |
| Eukaryotic neutral amino acid transporters | DWxVDRxxTxxNVEGD |
| Eukaryotic glutamate transporters | DWxLDRxRTxxNVxGD |
| Third transmembrane domain | |
| Bacterial C4-dicarboxylate transporters | 99a AxxY---Fxxx(ST)TxA---LxxGdL 115b |
| Bacterial glutamate transporters | TII(YW)FExIT---TI----xxxGL |
| Eukaryotic Ala/Ser/Cys/Thr transporters | AVAY---FGLTT---LSA---SALAV |
| Eukaryotic neutral amino acid transporters | ALLF---FLVTT---LLA---SALGV |
| Eukaryotic glutamate transporters | AxxY---YxxTT---xxA---xxxG (IV) |
Position on S. meliloti DctA.
Position of mutation in pSM100-41 D325G.
Position of mutation in pSM100-53 T327A.
Position of mutation in pSM100-32 G114D.
Although these mutations are physically very close to each other, the phenotypes of the mutants are different. Substitution of negatively charged aspartate with glycine at residue 325 almost completely abolished transport of dicarboxylic acids by the protein encoded by pSM100-41 (Table 3 and Fig. 4). Replacement of aspartate with cysteine, asparagine, glycine, or glutamate in the corresponding position in EAAT2 also blocks transport, and these mutant proteins were found in the cells (29, 16). D325 is present in all members of the glutamate transporter family except the bacterial serine transporters (Fig. 3).
In contrast, WSUb20611-II(pSM100-53) retained its ability to transport dicarboxylates with a maximum velocity close to that of wild-type DctA but the affinity of the protein for substrates was significantly decreased. This phenotype differs from those of T400C GLT-1 mutant, which completely lost its ability to transport aspartate (29). The T327 residue, which was replaced by A327 in DctA expressed from pSM100-53 (Table 5), is not conserved in all members of the glutamate transporter family. Bacterial dicarboxylate transporters and eukaryotic glutamate transporters have threonine, but the neutral amino acid transporters have alanine at this position, and the bacterial glutamate transporters have serine (Fig. 3).
Isolation of partially active S. meliloti DctA mutants carrying mutations in positions previously described as being important for function in eukaryotic glutamate transporters suggests that the selection used in isolating the mutants was adequate and can be used further to identify residues important in protein function. These mutants also suggest that the operation of the bacterial dicarboxylate and eukaryotic glutamate transporters are similar and that it is appropriate to use the well-analyzed members of the family to guide experimentation with S. meliloti DctA.
The G114D mutation on DctA expressed from pSM100-32 was located in the third transmembrane helix, which does not appear to have been previously implicated in transport. This substitution changed the character of dicarboxylate transport significantly. S. meliloti cells carrying the mutation had a slower growth rate on dicarboxylates but were partially sensitive to FOA. The mutant protein had half of the wild-type Vmax for dicarboxylate uptake. The affinity of the protein for substrates was significantly decreased and resembled those of DctA expressed from pSM100-53. These findings indicate that this mutation did not severely disrupt function of the protein but changed it substrate specificity.
A multiple sequence alignment of more than 70 members of the glutamate transporter family revealed that glycine was located at this position in all members of the family except the bacterial serine and eukaryotic alanine/serine/cysteine/threonine transporters, which have alanine in this position (Fig. 3). The region that forms membrane helix 3 is conserved to some degree in most members of the family, but its exact amino acid composition varies with the substrate specificity of the members of the subfamilies (Table 5). These differences in helix 3 may reflect the historical relatedness of the proteins but also suggests that transmembrane helix 3 could be involved in substrate recognition and/or substrate binding (Table 5), as suggested for motif D on the eighth transmembrane domain (21). Preliminary studies with site-specific mutations at this position suggests that various phenotypes can result from substitution of different amino acids at this position (31).
None of the mutant dctA alleles carried on plasmids could support symbiotic nitrogen fixation. However, some of the nodules formed by the dctA mutants had an unusual morphology. These type B nodules were more developed than the usual ineffective nodule, i.e., they were longer, had more extensive regions of infected cells, and were pink near the meristem, which is characteristic of nodules producing leghemoglobin. We interpret this to mean that, because these mutants were able to import dicarboxylates at a low level, they could transform to partially functional bacteroids and proceed further in the normal communication with the plant. As a result of bacterial signals, it appears that the plant provides the nodules with more photosynthate than in a typical ineffective nodule, resulting in the accumulation of considerable starch in the plant cells. Each of the mutants except WSUb20611-II(pSM100-41) formed both types of nodules.
Because their transport of succinate under standard conditions was near normal (Table 3), it was puzzling that strain WSUb20611-II(pSM100-93) and its derivatives were unable to support symbiotic nitrogen fixation. Although the DctA protein expressed from pSM100-93 transported dicarboxylates at a level close to the level of wild-type DctA, free-living bacteria containing pSM100-93 did not grow well on any of the dicarboxylates tested (Table 2 and Fig. 4). Strains carrying separated point mutations grew better but were still impaired (Fig. 4). Further research is needed to characterize biochemically how the defects in DctA proteins produced by WSUb20611-II(pSM100-93), WSUb20611-II(pSM100-93-1034), and WSUb20611-II(pSM100-93-265) lead to such a dramatic effect on the plant-associated phenotype and a more modest but significant effect on the growth rate.
In the present study we demonstrated the use of a toxic substrate to help enrich for mutants of DctA, a member of the glutamate transport family. The S. meliloti dctA system, with its relatively broad substrate specificity and the potential involvement of many of these substrates in cell growth, may provide a good system for investigating structure-function relationships in this unusual class of transporters. More detailed analysis of the transport properties of the mutants reported here may also give some insight into the requirements for successful operation of the transporter during symbiosis.
Acknowledgments
We thank the Washington State University Electron Microscopy Center for assistance with the microscopy.
This study was supported by grants 98353056553 from the U.S. Department of Agriculture-Competitive Research Grants Office and MCB-0131376 from the National Science Foundation. We also acknowledge support from the Agriculture Research Center at Washington State University.
REFERENCES
- 1.Baker, K. E., K. P. Ditullio, J. Neuhard, and R. A. Kelln. 1996. Utilization of orotate as a pyrimidine source by Salmonella typhimurium and Escherichia coli required the dicarboxylate transport protein encoded by dctA. J. Bacteriol. 178:7099-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendahan, A., A. Armon, N. Madani, M. P. Kavanaugh, and B. I. Kanner. 2000. Arginine 447 plays a pivotal role in substrate interactions in a neuronal glutamate transporter. J. Biol. Chem. 275:37436-37442. [DOI] [PubMed] [Google Scholar]
- 3.Charles, T. C., and T. M. Finan. 1991. Analysis of a 1,600-kilobase Rhizobium meliloti megaplasmid using defined deletions generated in vivo. Genetics 127:5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, I., and S. Y. Chiu. 2000. Loss of cell viability by histidine substitution of leucine 325 of the glutamate transporter EAAAT1. Biochem. Biophys. Res. Commun. 275:382-385. [DOI] [PubMed] [Google Scholar]
- 5.Gardiol, A., A. Arias, C. Cervenansky, and G. Martinez-Drets. 1982. Succinate dehydrogenase mutant of Rhizobium meliloti. J. Bacteriol. 15:1621-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grzemski, W., J. P. Akowski, and M. L. Kahn. Altering bacterial metabolism in a nitrogen fixing symbiosis using conditional and impaired mutations in Sinorhizobium meliloti citrate synthase. Mol. Plant Microbiol. Int., in press.
- 7.Humphreys, G. O., G. A. Willshaw, H. R. Smith, and E. S. Anderson. 1976. Mutagenesis of plasmid DNA with hydroxylamine: isolation of mutants of multi-copy plasmids. Mol. Gen. Genet. 145:101-108. [DOI] [PubMed] [Google Scholar]
- 8.Jording, D., and A. Puhler. 1993. The membrane topology of the Rhizobium meliloti C4-dicarboxylate permease (DctA) as derived from protein fusions with Escherichia coli K12 alkaline phosphatase (PhoA) and β-galactosidase (LacZ). Mol. Gen. Genet. 241:106-114. [DOI] [PubMed] [Google Scholar]
- 9.Kahn, M. L., T. R. McDermott, and M. K. Udvardi. 1998. Carbon and nitrogen metabolism in rhizobia, p. 461-485. In H. P. Spaink, A. Kondorosi, and P. J. J. Hooykaas (ed.), The Rhizobiaceae. Kluwer Academic Publisher, Dordrecht, The Netherlands.
- 10.Kavanaugh, M. P., A. Bendahan, N. Zerangue, Y. Zhang, and B. I. Kanner. 1997. Mutation of an amino acid residue influencing potassium coupling in the glutamate transporter GLT-1 induces obligate exchange. J. Biol. Chem. 272:1703-1708. [DOI] [PubMed] [Google Scholar]
- 11.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lolkema, J. S., and D. J. Slotboom. 1998. Estimation of structural similarity of membrane proteins by hydropathy profile alignment. Mol. Membr. Biol. 15:33-42. [DOI] [PubMed] [Google Scholar]
- 13.McDermott, T. R., and M. L. Kahn. 1992. Cloning and mutagenesis of the Rhizobium meliloti isocitrate dehydrogenase gene. J. Bacteriol. 174:4790-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitrovie, A. D., S. G. Amara, G. A. Johnston, and R. J. Vandenberg. 1998. Identification of functional domains of the human glutamate transporters EAAT1 and EAAT2. J. Biol. Chem. 273:14698-14706. [DOI] [PubMed] [Google Scholar]
- 15.Pines, G., N. C. Danbolt, M. Bjoras, Y. Zhang, A. Bendahan, L. Eide, H. Koepsell, J. Storm-Mathisen, E. Seeberg, and B. I. Kanner. 1992. Cloning and expression of a rat brain l-glutamate transporter. Nature 360:464-467. [DOI] [PubMed] [Google Scholar]
- 16.Pines, G., Y. Zhang, and B. I. Kanner. 1995. Glutamate 404 is involved in the substrate discrimination of GLT-1, a (Na+ + K+)-coupled glutamate transporter form rat brain. J. Biol. Chem. 270:17093-17097. [DOI] [PubMed] [Google Scholar]
- 17.Reid, C. J., and P. S. Poole. 1998. Roles of DctA and DctB in signal detection by the dicarboxylic acid transport system of Rhizobium leguminosarum. J. Bacteriol. 180:2660-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 19.Schafer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 20.Simon, R., U. Priefer, and A. Pühler. 1983. Vector plasmids for in-vivo and in-vitro manipulations of gram-negative bacteria, p. 98-106. In A. Pühler (ed.), Molecular genetics of the bacterial-plant interaction: the Rhizobium meliloti-Medicago sativa system. Springer-Verlag, Berlin, Germany.
- 21.Slotboom, D. G., J. S. Lolkema, and W. N. Konings. 1996. Membrane topology of the C-terminal half of the neuronal, glial, and bacterial glutamate transporter family. J. Biol. Chem. 271:31317-31321. [DOI] [PubMed] [Google Scholar]
- 22.Slotboom, D. G., I. Sobczak, W. N. Konings, and J. S. Lolkema. 1999. A conserved serine-rich stretch in the glutamate transporter family forms a substrate-sensitive loop. Proc. Natl. Acad. Sci. USA 96:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somerville, J. E., and M. L. Kahn. 1983. Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J. Bacteriol. 156:168-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasse, J., F. DeBilly, S. Camut, and G. Truchet. 1990. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172:4295-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson, R. J., Y.-K. Chan, R. Wheatcroft, A.-F. Yang, and S. Han. 1988. Rhizobium meliloti genes required for C4-dicarboxylate transport and symbiotic nitrogen fixation are located on a megaplasmid. J. Bacteriol. 170:927-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarosh, O. K., T. C. Charles, and T. M. Finan. 1989. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol. Microbiol. 3:813-823. [DOI] [PubMed] [Google Scholar]
- 27.Yurgel, S., M. W. Mortimer, K. N. Rogers., and M. L. Kahn. 2000. New substrates for the dicarboxylate transport system of Sinorhizobium meliloti. J. Bacteriol. 182:4216-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yurgel, S. N., and M. L. Kahn. 2000. Unpublished observations.
- 29.Zarbiv, R., M. Grunewald, M. P. Kavanaugh, and B. I. Kanner. 1998. Cysteine scanning of the surroundings of an alkali-ion binding site of the glutamate transporter GLT-1 reveals a conformationally sensitive residue. J. Biol. Chem. 273:14231-14237. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, T., A. Bendahan, R. Zarbiv, M. P. Kavanaugh, and B. I. Kanner. 1998. Molecular determinant of ion selectivity of a (Na+ + K+)-coupled rat brain glutamate transporter. Proc. Natl. Acad. Sci. USA 95:751-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziegler, M. A. 2004. Personal communication.