Abstract
Active detachment of cells from microbial biofilms is a critical yet poorly understood step in biofilm development. We discovered that detachment of cells from biofilms of Shewanella oneidensis MR-1 can be induced by arresting the medium flow in a hydrodynamic biofilm system. Induction of detachment was rapid, and substantial biofilm dispersal started as soon as 5 min after the stop of flow. We developed a confocal laser scanning microscopy-based assay to quantify detachment. The extent of biomass loss was found to be dependent on the time interval of flow stop and on the thickness of the biofilm. Up to 80% of the biomass of 16-h-old biofilms could be induced to detach. High-resolution microscopy studies revealed that detachment was associated with an overall loosening of the biofilm structure and a release of individual cells or small cell clusters. Swimming motility was not required for detachment. Although the loosening of cells from the biofilm structure was observed evenly throughout thin biofilms, the most pronounced detachment in thicker biofilms occurred in regions exposed to the flow of medium, suggesting a metabolic control of detachability. Deconvolution of the factors associated with the stop of medium flow revealed that a sudden decrease in oxygen tension is the predominant trigger for initiating detachment of individual cells. In contrast, carbon limitation did not trigger any substantial detachment, suggesting a physiological link between oxygen sensing or metabolism and detachment. In-frame deletions were introduced into genes encoding the known and putative global transcriptional regulators ArcA, CRP, and EtrA (FNR), which respond to changes in oxygen tension in S. oneidensis MR-1. Biofilms of null mutants in arcA and crp were severely impacted in the stop-of-flow-induced detachment response, suggesting a role for these genes in regulation of detachment. In contrast, an ΔetrA mutant displayed a variable detachment phenotype. From this genetic evidence we conclude that detachment is a biologically controlled process and that a rapid change in oxygen concentration is a critical factor in detachment and, consequently, in dispersal of S. oneidensis cells from biofilms. Similar mechanisms might also operate in other bacteria.
Most microbes in nature are believed to exist in biofilms that develop on biotic or abiotic surfaces in aqueous environments (9). The decision of an individual cell or a subpopulation of cells to transition between the surface and the planktonic compartment has critical consequences; nutrient availability, protection from predation, and differences in competitive behavior in one or the other compartment strongly determine an organism's chance for survival. While most biofilm studies so far have focused on the initial adhesion events (i.e., the transition from the planktonic to the surface compartment), the release or detachment of cells from biofilms (i.e., the transition from the surface to the planktonic compartment) is less understood.
Biofilm development is generally categorized into several phases: (i) initial attachment of bacteria to the substratum, (ii) irreversible binding and secretion of extracellular polymeric substances (EPS), (iii) biofilm maturation, and (iv) dispersal. Loss of cells from biofilms can be observed during all stages of biofilm formation. Physical forces such as abrasion, erosion, and sloughing have been recognized as significant factors causing cell loss (8, 39). However, widespread acute release of cells cannot be attributed solely to the effect of physical impact or shear stress. In particular, starvation for several hours for nutrients such as carbon and nitrogen has been shown to induce detachment in Pseudomonas spp., Escherichia coli, and Acinetobacter spp. (2, 3, 13, 19, 20, 35, 43).
Overcoming the adhesion of biofilm cells to each other and/or to the EPS is obviously critical for the release of cells, and the initial focus of detachment studies has been on factors and enzymes controlling cellular adhesion to EPS. Important EPS components include polymeric saccharides, such as alginate, colanic acid, and cellulose (11, 15, 30, 46). Extracellular DNA was also recently described to represent an important factor in biofilm formation of Pseudomonas aeruginosa (42). Additionally, surface proteins, such as Ag43 of E. coli (10), as well as cellular appendices, such as curli or pili (16, 29-31), have been reported to be involved in mediating cell-cell contact. Modification and degradation processes of EPS have been attributed almost exclusively to the activity of exopolysaccharide lyases, enabling cell dispersal (2, 7, 14, 21, 28, 44). In P. aeruginosa, rhamnolipids are thought to maintain biofilm architecture by influencing cell-cell interactions and bacterial attachment to surfaces (12).
The often long starvation periods reported in biofilm dissolution studies raise the question as to whether the transition from the surface to the planktonic compartment occurs because of some general loss of cell function under starvation conditions, such as loss of metabolic activity or energy, release of polysaccharide lyases from lysing cells, etc., or because detachment is a biologically controlled process in response to specific environmental stimuli. In support of the latter possibility, it was recently reported that carbon starvation induces rapid detachment in Pseudomonas putida biofilms, and several genes were identified to be required for the process (17). In this study, we examined the detachment of Shewanella oneidensis MR-1 cells from biofilms. S. oneidensis is an environmentally and geochemically important facultative microorganism capable of using a wide range of terminal electron acceptors under anoxic conditions, including Fe(III) and Mn(IV) minerals (26, 27). Biofilm formation in this organism was recently characterized (40). We show here that S. oneidensis MR-1 cells can be induced to rapidly disperse from biofilms in response to a sudden downshift in molecular oxygen concentration, and we provide genetic evidence that detachment in response to a specific environmental cue is a biologically controlled process.
MATERIALS AND METHODS
Growth conditions and media.
S. oneidensis and E. coli strains used in this study (Table 1) were grown in Luria-Bertani (LB) medium at 30 and 37°C, respectively; for plates, the medium was solidified using 1.5% (wt/vol) agar. If required, the medium was supplemented with 10 μg of gentamicin/ml, 25 μg of kanamycin/ml, and/or 20 μg of tetracycline/ml. Biofilms were grown in lactate medium (LM) (40) containing 500 μM lactate without antibiotic supplementation as described previously (40).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Bacterial strain | ||
| E. coli | ||
| DH5α-λpir | φ80dlacZΔM15 Δ(lacZYA-argF)U196 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1/λpir | 23a |
| S17-1-λpir | thi pro recA hsdR [RP4-2Tc::Mu-Km::Tn7]λpir Tpr Smr | 36a |
| S. oneidensis | ||
| MR-1 | Wild type | 41a |
| AS93 | MR-1 tagged with eGFP in a Tn7 construct, Cmr Gmr | 40 |
| AS95 | Plasmid integration mutant of flhB in AS93, nonmotile, Cmr Gmr Kmr | 40 |
| AS124 | In-frame deletion of arcA in AS93, Cmr Gmr | This study |
| AS123 | In-frame deletion of crp in AS93, Cmr Gmr | This study |
| AS122 | In-frame deletion of etrA in As93, Cmr Gmr | This study |
| AS127 | AS124 harboring pME6031::arcA, Cmr Gmr Tcr | This study |
| AS125 | AS123 harboring pME6031::crp, Cmr Gmr Tcr | This study |
| AS126 | AS122 harboring pME6031::etrA, Cmr Gmr Tcr | This study |
| Plasmid | ||
| pBluescript KS+ | Apr ori ColE1 | New England Biolabs |
| pGP704-Sac28-Km | Kmr mobRP4+ori-R6K sacB, suicide plasmid for in-frame deletions | Chengyen Wu, unpublished data |
| pGP704-Sac28-Km::Farc | In-frame deletion fragment of arcA in pGP704-Sac28-Km | This study |
| pGP704-Sac28-Km::Fcrp | In-frame deletion fragment of crp in pGP704-Sac28-Km | This study |
| pGP704-Sac28-Km::Fetr | In-frame deletion fragment of etrA in pGP704-Sac28-Km | This study |
| pME6031 | repA oriVpVS1 oriVp15A oriT Tcr | 17a |
| pME6031::arcA | arcA in pME6031 | This study |
| pME6031::crp | crp in pME6031 | This study |
| pME6031::etrA | etrA in pME6031 | This study |
Strain construction in S. oneidensis: deletion and complementation of arcA, crp, and etrA.
DNA manipulations were carried out according to standard techniques (34). Enzymes were obtained from New England Biolabs (Beverly, Mass.). Kits for extracting and purifying DNA were obtained from QIAGEN (Valencia, Calif.) and used according to the manufacturer's instructions.
Mutations in the genome of S. oneidensis were introduced by in-frame deletions leaving only short N- and C-terminal sections of the target genes. For that purpose, 400- to 500-bp upstream and downstream fragments of arcA, crp, and etrA were amplified by PCR with the corresponding primer pairs (-I-fw, -II-fw, -I-rv, and -II-rv) given in Table 2. The fragments were treated with SalI (BamHI for the crp fragment) and ligated; the ligation product was used as template for a second PCR using the outer primers (-I-fw and -II-rv) to yield the truncated gene fusion product. The product was isolated from an agarose gel, digested with SacI and NcoI, and ligated into the suicide vector pGP704-Sac28-Km (C. Wu, unpublished data) treated with the same enzymes. The product was then introduced into the S. oneidensis wild type, strain AS93 (40), by mating using E. coli S17-λpir as the donor strain. Single crossover integration mutants were selected on LB plates containing gentamicin and kanamycin. Subsequently, single Kanr and Genr colonies were grown overnight in liquid LB medium without antibiotics and then plated on LB containing gentamicin and 8% (wt/vol) sucrose to select for double-crossover events. Finally, kanamycin-sensitive single colonies were subsequently checked for the targeted deletion by colony PCR using primers bracketing the location of the deletion.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′ → 3′) |
|---|---|
| arcA-I-fw | CCACGAGCTCTGAGTCATGTTGTCCATCGGTAGTC |
| arcA-I-rv | CATTGTCGACAGTTACCACATACCCTTCTGCCTCG |
| arcA-II-fw | TACTGTCGACGTGACTATCCGTCGTATCCGTAAGC |
| arcA-II-rv | TTGAGGATCCATGGTCTAAGCATTCAATGCGTGG |
| arcA-check-fw | CAACGGCGTTTGATAATGCTGCCAC |
| arcA-check-rv | GCGTTGCAGGACGAAGGCAAGTTG |
| crp-I-fw | AAACCCATGGGCCTGTATTTCACCTGGTAAC |
| crp-I-rv | GCATGGATCCAGGTGCTTTTAGCGGGATAC |
| crp-II-fw | GAAACGGATCCTCATTCAAGCACACGGTAAAAG |
| crp-II-rv | GTTCGAGCTCCAATTCGGAGACCAGCATGG |
| crp-check-fw | GCATCACCTTGCTCTGCCTGAACTTG |
| crp-check-rv | AATCGGCTTCAAGCGCTTTGTCTG |
| etrA-I-fw | ACGCGAGCTCAAGCCATCACCGCTGGATTGATATG |
| etrA-I-rv | TAATGTCGACGAGCTGATCGAGTTCATTAGCATTG |
| etrA-II-fw | CATCATCGTCGACCATCATGAACTTAATCTCTTGG |
| etrA-II-rv | CTGAGTATCCATGGTGAGTCCTAGGTGATTGTAGG |
| etrA-check-fw | CATCATGATGATCGCGACAG |
| etrA-check-rv | CACCGCTTTTAACTTGTCGTG |
| etrA-fw | GGCTGATATCGATTAACTTGAGAACCGACATGAC |
| etrA-rv | CATACTGCAGAAAAGGTGTGATTTATCTGGCGAT |
| P-ATPase-fw | TAATGTCTGTAGATTAACA |
| P-ATPase-rv | ATAAATGCGGAGAAGATGAT |
To complement the mutants, the corresponding genes were amplified from wild-type chromosomal DNA using the outer primer pairs (-I-fw and -II-rv) for arcA and crp and the primer pair etrA-fw and etrA-rv for etrA. For arcA and crp, the fragments were treated with NcoI and SacI and ligated into pME6031 treated with the same enzymes. To achieve constant expression of etrA, apparently part of a larger operon with an unknown promoter region, the gene was cloned behind the promoter of the genes encoding the ATPase subunits. The putative ATPase promoter region was amplified with the primer pair P-ATPase-fw and P-ATPase-rv and ligated into the EcoRV restriction site of pBluescript KS+ (New England Biolabs), and the gene fragment of etrA was inserted by using the EcoRV and PstI sites of the vector. The merged fragments were finally released by EcoRI and KpnI and ligated into pME6031 treated with the same enzymes. The resulting plasmids were introduced into the corresponding S. oneidensis mutant by electroporation (24) and selected on LB medium containing gentamicin and tetracycline.
Biofilm cultivation and induced detachment.
S. oneidensis biofilms were grown as described previously (40). All biofilm characterizations were conducted in triplicate in at least two independent experiments.
For flow-stop experiments, the flow was arrested by clamping the inflow tubing immediately upstream of the flow chamber, and the pump tubing was released from the peristaltic pump to avoid a buildup of pressure. To resume the flow, the clamp was taken off before the tubing was reinserted into the peristaltic pump. In all experiments, biofilms in control channels not subjected to a flow stop were treated the same as the test biofilms to ensure that differences in biomass before and after the experiment were not due to bleaching or changes of signal gain. Confocal images were taken immediately prior to clamping the inflow tubing and 15 min after resuming the flow, independent of the flow stop duration.
For the nutrient downshift experiment, the flow was arrested as described above. Prior to connecting the inflow tubing to the new medium, the medium present in the inflow reservoir and the bubble trap was replaced in order to avoid a nutrient gradient formation in the bubble trap reservoir. This replacement process took less than 1 min. In control channels, the flow was arrested for the same period to exclude detachment due to the stop of flow rather than the new medium conditions.
The oxygen downshift experiment required a modification in the setup and made use of the rapid diffusion of molecular oxygen through the silicone tubing (S. Kirkelund-Hansen and S. Molin, unpublished data). The inflow tubing upstream of the flow chamber was extended by 2 m and inserted through a rubber septum into a vacuum flask. The silicone connection tubing between flask and flow chamber was replaced by glass tubing, leaving only short joints that allowed handling of the flow chamber setup on the microscope stage. A drop in oxygen concentration by diffusion through the silicone tubing walls was then induced by applying vacuum to the flask using a Gast G608X vacuum pump (Benton Harbor, Mich.). No flow stop was applied during this experiment.
Image acquisition.
Microscopic visualization of biofilms was carried out at the Stanford Biofilm Research Center using an upright LSM510 confocal laser scanning microscope (CLSM; Carl Zeiss, Jena, Germany). The following objectives were used: 10×/0.3 Plan-Neofluar, 20×/0.5 W Achroplan, and 40×/1.2 W C-Apochromat. For displaying biofilm images, CLSM images were processed with the IMARIS software package (Bitplane AG, Zürich, Switzerland) and Adobe Photoshop. Biofilm parameters, such as biomass, and average biofilm thickness were quantified with the program COMSTAT (18).
RESULTS
Stop of flow induces detachment of cells from S. oneidensis MR-1 biofilms.
During studies on S. oneidensis MR-1 biofilms grown hydrodynamically in a flow chamber system, we noticed that arresting the medium flow triggered substantial dispersal of the biofilm. To understand this cell detachment, we developed a quantitative biofilm detachment assay using CLSM in conjunction with image quantification by COMSTAT (18) (see Materials and Methods). Biofilms of constitutively gfp-expressing S. oneidensis MR-1 wild-type strain AS93 were grown aerobically for 12 to 14 h on the surface of glass coverslips in flow chambers irrigated with LM containing 0.5 mM lactate as electron donor as described previously (40). Randomly chosen locations in the biofilm from the first third of the channel were imaged by CLSM. The flow of medium was then arrested, typically for 15 min, and subsequently resumed for 15 min before a second series of CLSM images were taken at exactly the same positions (Fig. 1). The corresponding before-stop-of-flow and after-stop-of-flow image stacks were quantified for total biomass using the computer program COMSTAT (18) (Fig. 2). The biomass detached was calculated as the difference between the biomass before stop of flow minus the biomass after stop of flow. Prior control experiments confirmed that bleaching of the green fluorescent protein (GFP) fluorescence was not significant within the parameters used for this assay (data not shown).
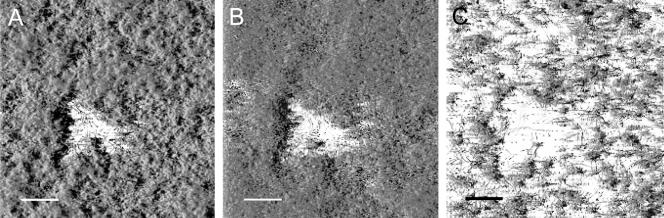
FIG. 1.
Detachment induced by stop of flow. The experiment was conducted as a standard stop-of-flow assay as described in Materials and Methods. Images are shadow projections of 12-h-old CLSM files taken of AS93 biofilms before (A), 10 min after (B), and 30 min after (C) the initial stop of medium flow. The scale bar in each image represents 40 μm.
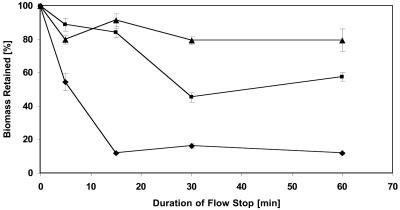
FIG. 2.
Effect of duration of stop of flow on extent of detachment of biofilms of different thicknesses. The graph represents quantified CLSM images generated by COMSTAT. Detachment assays were conducted as described in Materials and Methods, but the duration of stop of flow was varied as indicated by the x axis. ▴, 48-h-old biofilm; ▪, 18-h-old biofilm; and ⧫, 12-h-old biofilm. Each data point is the mean from six independent images taken from two channels. Error bars represent one standard deviation.
Loss of biomass was observed and was found to comprise up to 80% of the total biomass for young (e.g., 12-h-old) biofilms (Fig. 2). Closer examination of the CLSM images as well as time-lapse movies of detachment in 12-h-old biofilms revealed some characteristic features of the detachment process: rather than large segments of biomass separating from the biofilm, stop of flow resulted in a more or less uniform loosening of the entire biofilm structure and an even release of individual cells or small aggregates (Fig. 3A to C). This made the biofilm appear as if it was thinning out.
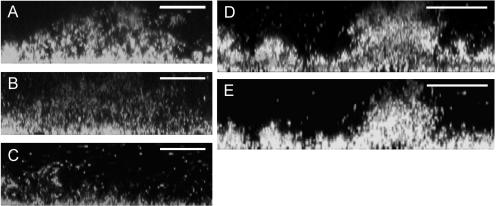
FIG. 3.
Induction of detachment in 12- and 48-h-old biofilms. Images display cross sections of stop-of-flow-induced detachment in 12-h-old (A to C) and 48-h-old (D and E) biofilms. Images A, B, and C were obtained immediately before, during (15 min), and after the stop of flow, respectively; images D and E were obtained before and after the stop of flow. Each scale bar represents 25 μm in A to C and 50 μm in D and E.
To determine whether biofilm thickness and the duration of the flow stop interval were important parameters for inducing detachment, biofilms were grown for 12, 18, and 48 h, and the detachment assay was applied with flow stop intervals lasting 5, 15, 30, or 60 min (Fig. 2). The studies revealed an inverse correlation between the percentage of biomass detached and the thickness of the biofilm (Fig. 2): the thicker the biofilm, the smaller the overall degree of cell release. It was further determined that not only the degree of detachment but also the onset of detachment varied with biofilm thickness (Fig. 2). In 12-h-old biofilms, up to 80% of the cell mass was detached after 15 min, and extension of the time interval beyond 15 to 30 min resulted in only minor additional biomass loss. In contrast, in 18-h-old biofilms only about 50% detached under the same conditions (Fig. 2 and 3D and E). The nondetached biomass was found to be viable, and the biofilms regrew into the S. oneidensis typical architecture (40) when medium flow was provided again (data not shown).
We hypothesized that this observed differential degree of detachment, or detachability, in the upper layers of older biofilms could lead to exposure of the nondetached, previously deeper cell layers to higher concentrations of nutrients and oxygen. Consequently, a subsequent increase in metabolic activity might render those cells detachable again. Therefore, we subjected a 20-μm-thick biofilm that did not completely detach after a stop-of-flow treatment to a second cycle of flow stop of 15 min. Between the two stop-of-flow treatments, the biofilm was exposed to regular medium flow for 45 min. After the second stop of flow, substantial detachment of former nondetached cells was observed (data not shown). This result implies that detachability can be induced. Nondetached cells are not locked irreversibly in a detachment-incompetent state but are capable of reversing to a detachment-competent state, probably by increasing their metabolic activity.
Bright-field microscopic studies of the detachment process suggested that cell release was not linked to cellular motility, since the majority of the cells detaching from the biofilm did not show visible swimming motility (data not shown). In order to test whether swimming motility is a crucial prerequisite for detachment, a stop-of-flow experiment was carried out using S. oneidensis AS95, a nonswimming mutant strain generated by plasmid insertion in flhB (40). Twelve-hour-old biofilms of AS95 were very similar to those of the wild type in size and shape (biomass of 12-h-old AS95 biofilm was 4.45 ± 1.85 μm3/μm2, compared to 5.16 ± 1.39 μm3/μm2 for the wild type) and were found to lose at least 40% of biomass under flow stop conditions (data not shown), indicating that swimming motility is not a critical factor for the observed detachment process.
Stop-of-flow-induced detachment is triggered by a decrease in oxygen concentration.
Arresting the flow of medium over biofilms grown in flow chambers results in several major changes in the biofilm environment. First, while the rate of nutrient transfer from the medium drops sharply to 0, the rate of nutrient consumption by the biofilm cells will continue until the electron donor or acceptor becomes limiting, which depends on the medium composition. Such nutrient downshifts have been implicated to lead to detachment in a number of organisms (2, 3, 13, 19, 20, 35, 43). Second, removal of excreted compounds, such as metabolic end products, and/or potential EPS-degrading enzymes and signaling molecules is arrested, which would lead to an accumulation of compounds that could cause the observed biofilm dispersal. Third, stopping the medium flow eliminates the shear stress acting on the biofilm. In order to identify the trigger(s) for detachment, we modified our assay to uncouple these numerous changes associated with the stop of flow.
In a first modification, biofilms were grown in LM as described previously; however, rather than stopping the flow, the composition of the medium was changed by switching to a medium without a usable electron donor. This switch required an interruption of flow of less than 1 min, which did not induce any significant detachment (Fig. 2). The substituting medium solution contained only the medium's buffer base (HEPES and NaHCO3), which was previously determined not to support cell growth. After more than 1 h of irrigation with a medium devoid of any usable carbon source, more than 90% of the biomass was still attached (Fig. 4A). This observation demonstrates that a downshift in electron donor concentration can be ruled out as the dominant signal for the stop-of-flow-induced detachment.
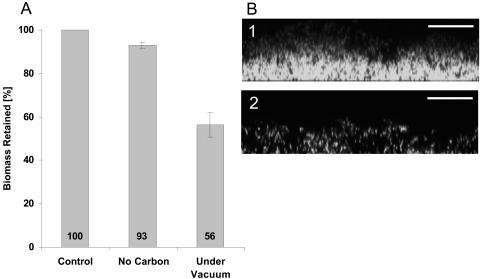
FIG. 4.
Induction of detachment by nutrient downshifts. The graph (A) represents quantified CLSM images generated by COMSTAT. Modified detachment assays were conducted as described in Materials and Methods. The nutrient downshift was accomplished either by switching to a medium containing buffer only or by removal of oxygen through applying vacuum to the medium inflow tubing. Each data point is the mean from at least three independent images. Error bars represent one standard deviation. Images B1 and B2 are biofilm cross sections 45 and 90 min after oxygen downshift, respectively. Each scale bar represents 25 μm.
We then examined the depletion of molecular oxygen as an inducer of detachment in the biofilm. As the silicone tubing used for medium intake is permeable to molecular oxygen, we made use of this diffusion property and inserted the inflow tubing upstream of the flow chamber into a flask to which vacuum could be applied. After allowing the biofilm to develop for 12 to 16 h under regular conditions, oxygen removal from the inflowing LM medium was initiated by applying vacuum to the flask. This procedure did not require any stop of flow and did not result in a significant pH change (<0.15). As shown in Fig. 4, the downshift in oxygen resulted in a loss of biomass. Significant dispersal of cells began approximately 45 min after application of the vacuum, and cell loss leveled off after approximately 75 min. After 2 h, about 50% of the biomass had detached, and the appearance of the depleted biofilm strongly resembled that observed in the stop-of-flow experiments (Fig. 3A to C and 4). Therefore, we concluded that a decrease in molecular oxygen concentration is the major trigger for the observed biomass detachment from S. oneidensis biofilms. Since this set of experiments was conducted without ever arresting the flow, it was also concluded that the accumulation of extracellular compound(s) during stop of flow is not required to induce detachment.
As a rapid decrease in oxygen concentration was identified in this respiring microorganism as the major factor inducing detachment, we wondered whether the presence of an anaerobic electron acceptor, such as fumarate, would attenuate the detachment response. Fumarate is readily used by S. oneidensis as an electron acceptor with lactate as an electron donor under anaerobic conditions (25). The detachment experiment was repeated with standard LM that was amended with 2.5 mM fumarate. The detachment response of S. oneidensis MR-1 to the flow stop under these conditions was found to be unaltered compared to standard detachment conditions. (data not shown).
ArcA, CRP, and EtrA (FNR) are potential regulators of oxygen-dependent detachment.
Since a rapid decrease in oxygen concentration was identified as the main trigger for detachment, we examined whether regulators known in other gamma-proteobacteria to mediate cellular responses to changing oxygen tension might also be involved in the detachment response. EtrA, the homolog of the E. coli fumarate-nitrate reduction regulator FNR, and CRP have been shown to be involved in regulation of the shift between aerobic and anaerobic metabolism in S. oneidensis MR-1 (32, 33). ArcA, a response regulator, is the analog of the aerobic respiration control protein of E. coli (4).
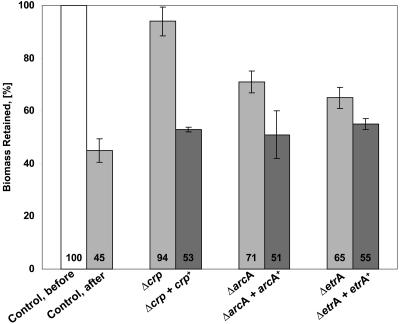
We constructed in-frame deletions of arcA (strain AS124), crp (strain AS123), and etrA (strain AS122) and tested these strains for biofilm growth and detachability. Twelve- and sixteen-hour-old biofilms of the mutants AS123 and AS122 were grown and subjected to the standard 15-minute flow stop experiment. Since the mutation ΔarcA resulted in a growth phenotype, biofilms of AS124 were allowed to grow for 24 to 28 h to reach a corresponding thickness before being subjected to the detachment assay. All three deletion mutants were found to give rise to biofilms that were defective in the detachment response (Fig. 5). The most pronounced effect was observed in the Δcrp mutant, where about 90% of the total biomass was retained after arresting the medium flow. It should be noted that the biofilms formed by AS124 and AS123 were significantly altered in appearance compared to wild-type AS93. ΔarcA biofilms grew sparsely, while Δcrp biofilms consisted of smaller, more-densely-packed cells. The average biomasses of 12-h-old biofilms of AS124 were 1.2 ± 0.7 μm3/μm2 and 1.97 ± 0.47 μm3/μm2 for AS123, compared to a wild-type biofilm biomass of 5.16 ± 1.39 μm3/μm2. Complementation of ΔarcA and Δcrp with the corresponding wild-type genes expressed from a stable plasmid restored cellular detachment to wild-type levels (Fig. 5) but did not change the altered biofilm appearance (data not shown). Deletion mutants carrying the plasmid without insert retained the mutant phenotype (data not shown). Notably, the detachment phenotype of the ΔetrA mutant was less pronounced than that of the Δcrp and ΔarcA mutants. Since biofilms of the complemented strains exhibited detachment to levels equivalent to that of wild-type biofilms, we concluded that the altered biofilm architecture in the null mutants did not cause the strongly reduced detachment response. We conclude that the regulators ArcA and CRP, and to a lesser extent EtrA, are likely candidates to be directly or indirectly involved in the regulation of flow stop-induced detachment of S. oneidensis MR-1.
FIG. 5.
Induced detachment in biofilms of strains with in-frame deletions in crp, arcA, and etrA. The graph represents quantified CLSM images generated by COMSTAT. Detachment assays were conducted as described in Materials and Methods on biofilms of strains with either null mutations (AS122, AS123, and AS124) or null mutations complemented with the corresponding wild-type allele in trans (AS126, AS125, and AS127). Values are the means from at least six independent images taken from two channels. Error bars represent one standard deviation.
DISCUSSION
Few reports so far have addressed active detachment in microbial biofilms, and in most cases the inducing signal had been attributed to changes in electron donor or carbon source, such as in an Acinetobacter sp., Aeromonas hydrophila, E. coli, P. aeruginosa, Pseudomonas fluorescens, or Pseudomonas sp. strain S9 (2, 13, 19, 20, 35, 43). In contrast, oxygen as an environmental trigger has received little attention, and a rapid response to a change in oxygen concentration was not shown to trigger detachment (2, 3), despite the fact that oxygen is a limiting factor in most environments. In this study, we demonstrated for the first time that a sudden decrease in molecular oxygen concentration is a dominant trigger that induces rapid detachment in biofilms. Moreover, depending on the biofilm external thickness, the fraction of detachable cells is variable (Fig. 2). The identification of mutants that are defective in the detachment response demonstrates that detachment is a biologically controlled process (Fig. 5).
Presumably, the oxygen depletion-induced detachment and the stop-of-flow-induced detachment we reported here are related. First, the microscopic features—the thinning out of the biofilm by individual cells separating from the matrix—are very similar in both detachments. Second, the stop of flow is likely to lead to rapid local decrease in oxygen concentration. Complete mineralization of 0.5 mM lactate in the LM is limited by oxygen, as the air-saturated medium contains a maximum oxygen concentration at room temperature of about 250 μM. Therefore, a restriction of transport of both electron donor and oxygen into a biofilm, as caused by a stop of flow, will result in a “self-induced,” rapid oxygen depletion. The different degrees of biomass detachment in thin versus thicker biofilms can be explained similarly on the same basis by oxygen metabolism. If the signal for detachment were simply the decrease of oxygen concentration below a certain threshold, then there should be a limiting thickness in S. oneidensis biofilms above which the biofilm's oxygen consumption would decrease the oxygen concentration below the detachment-specific threshold. However, the thickness of S. oneidensis biofilms has not been observed to level off at a particular biofilm thickness (40). We hypothesize that under standard biofilm growth conditions, a slow, metabolically driven decrease in local oxygen concentration leads to an adaptation of the biofilm cells to the new conditions of oxygen limitation, and only minor detachment from the biofilm occurs, while most of the cells stay within the community. Cells, especially in the lower layers of thicker (e.g., >18-h-old) biofilms, have presumably been adapting gradually to a reduced metabolic activity due to low oxygen concentration as the biofilm has increased in thickness. As demonstrated for P. putida, the more metabolically active cells are those that are in direct contact with the medium flow (38). The existence of cell layers in S. oneidensis biofilms with lower internal metabolic activity is supported by the observation that GFP fluorescence increased in lower biofilm layers after the top layers were removed by induced detachment (Fig. 3D and E). Once such internal cell layers are exposed to a higher oxygen concentration, their metabolic activity is expected to increase, and the cells would adapt to a metabolism at high oxygen concentration. Such cells would have the ability to respond to the sudden decrease in oxygen by detaching, and we observed such response upon applying another stop of flow (see above). Therefore, the absence of an apparent threshold for oxygen concentration and biofilm thickness, together with the restoration of detachment competence, suggest that an oxygen decrease that is more rapid than a metabolic adaptation can induce detachment in metabolically active S. oneidensis cells. A rapid drop in oxygen concentration rather than low oxygen concentration per se might be the direct trigger inducing detachment.
Knowledge of the regulation of detachment is even scarcer. Only in E. coli were detachment and central carbon flux correlated experimentally. In this microorganism, the global carbon storage regulator CsrA was demonstrated to strongly affect not only attachment but also dispersal of biofilm cells (19). In Xanthomonas campestris, the process of cell dispersal is controlled by cell-cell signaling involving a small diffusible signal factor (14). The signal was demonstrated to regulate an endo-β-mannanase that degrades cell-cell interconnection formed by polysaccharides.
In the present study, we presented strong evidence that global regulators of anaerobic metabolism in S. oneidensis control factors mediating biofilm detachment in response to sudden oxygen depletion. EtrA is the homolog of E. coli FNR, and its regulating role in response to anaerobic conditions has been well established for S. oneidensis MR-1 (6, 23, 32). CRP has also been demonstrated to play a role in the regulation of anaerobic respiration in S. oneidensis and to be required for activation of expression of terminal electron acceptor reductases, such as fumarate reductase and Fe(III) reductase (33). However, complete understanding of the CRP regulon is lacking. Notably, CRP and its homologs in other microorganisms were found to control factors that could affect biofilm formation: twitching motility, elastase production, and quorum sensing in Pseudomonas (1, 5), type IV pili in Vibrio cholerae (37), and flagella and/or motility in E. coli and Salmonella enterica serovar Typhimurium (45). The third regulator targeted in our study was the homolog of the E. coli ArcA response regulator (79% identity and 86% similarity). Expression of ArcA was found to be up-regulated in S. oneidensis MR-1 during anaerobic growth on insoluble ferric oxides as electron acceptor (41). This result indicates a similar role in redox or oxygen sensing, although a direct homolog to the corresponding sensor kinase, ArcB, is not present in the S. oneidensis genome. An ΔarcA mutation in S. oneidensis was found to result in a growth phenotype (data not shown), indicating a more general role for this regulator in this organism.
We showed here for S. oneidensis that ΔarcA and Δcrp mutations, and to a lesser extent an ΔetrA mutation, negatively affected biofilm detachment. This result might indicate that the factor(s) for sensing and/or the machinery of detachment are within the regulon of these regulators and suggests regulatory cross talk. The regulatory networks of FNR and ArcA have been shown to overlap in E. coli (22), and concerted action of ArcA and CRP appears to regulate the production of cholera toxin and the toxin-coregulated pilus in V. cholerae (36, 37).
All three regulators can act as repressors as well as activators and might control both up- and down-regulation of such factors. So far, neither potential release factors, such as EPS lyases, nor the exact composition of the extracellular matrix are known for S. oneidensis MR-1. Whether the regulatory function is directly linked to the observed dispersal remains to be elucidated. Further characterization of the corresponding regulons might help to directly identify important players in the detachment process and is the subject of current studies by our group.
Acknowledgments
We are grateful to Søren Molin, Tim Tolker-Nielsen, and the DTU biofilm group for helpful discussions and for providing excellent technical advice. We thank the group of Gary Schoolnik for kindly providing the vector pGP704-Sac28-Km.
This work was supported by grants from the National Science Foundation and a Powell Foundation faculty award to A.M.S. Part of this work was initiated while A.M.S., supported by a grant from the Otto Moensted Foundation, was on sabbatical in Søren Molin's laboratory.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, D. G., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 3.Applegate, D. H., and J. D. Bryers. 1991. Effects of carbon and oxygen limitations and calcium concentrations on biofilm removal processes. Biotechnol. Bioeng. 37:17-25. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53:495-523. [DOI] [PubMed] [Google Scholar]
- 5.Beatson, S. A., C. B. Whitchurch, J. L. Sargent, R. C. Levesque, and J. S. Mattick. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beliaev, A. S., D. K. Thompson, M. W. Fields, L. Wu, D. P. Lies, K. H. Nealson, and J. Zhou. 2002. Microarray transcription profiling of a Shewanella oneidensis etrA mutant. J. Bacteriol. 184:4612-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, A., and A. M. Chakrabarty. 1994. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 60:2355-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryers, J. D. 1988. Modeling biofilm accumulation, p. 45-88. In M. Bazin and J. I. Prosser (ed.), Biofilms II: process analysis and application. Wiley-Liss, New York, N.Y.
- 9.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 10.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 11.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davey, M. E., N. C. Caiazza, and G. A. O'Toole. 2003. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 185:1027-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delaquis, P. J., D. E. Caldwell, J. R. Lawrence, and A. R. McCurdy. 1989. Detachment of Pseudomonas fluorescens from biofilms on glass surfaces in response to nutrient stress. Microb. Ecol. 18:199-210. [DOI] [PubMed] [Google Scholar]
- 14.Dow, J. M., L. Crossman, K. Findlay, Y. Q. He, J. X. Feng, and J. L. Tang. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. USA 100:10995-11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gacesa, P. 1998. Bacterial alginate biosynthesis—recent progress and future prospects. Microbiology 144:1133-1143. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 17.Gjermansen, M., and T. Tolker-Nielsen. 2003.Programmed dissolution of Pseudomonas putida biofioms, poster no. 211(A). ASM Conference: Biofilms 2003.
- 17a.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O’Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in Gram-negative, plant-associated bacteria. Mol. Plant Microbe Interact. 13:232-337. [DOI] [PubMed]
- 18.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, D. W., K. Suzuki, L. Oakford, J. W. Simecka, M. E. Hart, and T. Romeo. 2002. Biofilm formation and dispersal under the influence of the global regulator CsrA of Escherichia coli. J. Bacteriol. 184:290-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James, G. A., D. R. Korber, D. E. Caldwell, and J. W. Costerton. 1995. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 177:907-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan, J. B., C. Ragunath, N. Ramasubbu, and D. H. Fine. 2003. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185:4693-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, X., and P. De Wulf. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588-12597. [DOI] [PubMed] [Google Scholar]
- 23.Maier, T. M., and C. R. Myers. 2001. Isolation and characterization of a Shewanella putrefaciens MR-1 electron transport regulator etrA mutant: reassessment of the role of EtrA. J. Bacteriol. 183:4918-4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed]
- 24.Myers, C. R., and J. M. Myers. 1997. Replication of plasmids with the p15A origin in Shewanella putrefaciens MR-1. Lett. Appl. Microbiol. 24:221-225. [DOI] [PubMed] [Google Scholar]
- 25.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 26.Nealson, K. H., A. Belz, and B. McKee. 2002. Breathing metals as a way of life: geobiology in action. Antonie Leeuwenhoek 81:215-222. [DOI] [PubMed] [Google Scholar]
- 27.Nealson, K. H., and D. Saffarini. 1994. Iron and manganese in anaerobic respiration: environmental significance, physiology, and regulation. Annu. Rev. Microbiol. 48:311-343. [DOI] [PubMed] [Google Scholar]
- 28.Ott, C. M., D. F. Day, D. W. Koenig, and D. L. Pierson. 2001. The release of alginate lyase from growing Pseudomonas syringae pathovar phaseolicola. Curr. Microbiol. 42:78-81. [DOI] [PubMed] [Google Scholar]
- 29.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 30.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 31.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 32.Saffarini, D. A., and K. H. Nealson. 1993. Sequence and genetic characterization of etrA, an fnr analog that regulates anaerobic respiration in Shewanella putrefaciens MR-1. J. Bacteriol. 175:7938-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saffarini, D. A., R. Schultz, and A. Beliaev. 2003. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J. Bacteriol. 185:3668-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, K., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sawyer, L. K., and S. W. Hermanowicz. 2000. Detachment of Aeromonas hydrophila and Pseudomonas aeruginosa due to variations in nutrient supply. Water Sci. Technol. 41:139-145. [Google Scholar]
- 36.Sengupta, N., K. Paul, and R. Chowdhury. 2003. The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect. Immun. 71:5583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791.
- 37.Skorupski, K., and R. K. Taylor. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 94:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sternberg, C., B. B. Christensen, T. Johansen, A. T. Nielsen, J. B. Andersen, M. Givskov, and S. Molin. 1999. Distribution of bacterial growth activity in flow-chamber biofilms. Appl. Environ. Microbiol. 65:4108-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoodley, P., S. Wilson, L. Hall-Stoodley, J. D. Boyle, H. M. Lappin-Scott, and J. W. Costerton. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thormann, K. M., R. M. Saville, S. Shukla, D. A. Pelletier, and A. M. Spormann. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186:8096-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanrobaeys, F., B. Devreese, E. Lecocq, L. Rychlewski, L. De Smet, and J. Van Beeumen. 2003. Proteomics of the dissimilatory iron-reducing bacterium Shewanella oneidensis MR-1, using a matrix-assisted laser desorption/ionization-tandem-time of flight mass spectrometer. Proteomics 3:2249-2257. [DOI] [PubMed] [Google Scholar]
- 41a.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed]
- 42.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 43.Wrangstadh, M., U. Szewzyk, J. Ostling, and S. Kjelleberg. 1990. Starvation-specific formation of a peripheral exopolysaccharide by a marine Pseudomonas sp., strain S9. Appl. Environ. Microbiol. 56:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xun, L. Y., R. A. Mah, and D. R. Boone. 1990. Isolation and characterization of disaggregatase from Methanosarcina mazei LYC. Appl. Environ. Microbiol. 56:3693-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yokota, T., and J. S. Gots. 1970. Requirement of adenosine 3′,5′-cyclic phosphate for flagella formation in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 103:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Römling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]