Abstract
Bacteroides fragilis, a human gastrointestinal commensal and an opportunistic pathogen, utilizes simple and complex sugars and polysaccharides for growth in the large intestine and at sites of infection. Because B. fragilis lacks transport-linked sugar phosphorylation systems, cytoplasmic kinase(s) was expected to be required for the phosphorylation of hexoses and hexosamines. We have now identified two hexose kinases that are important for growth of B. fragilis on glucose, mannose, and other sugars. One kinase (RokA), a member of the ROK family of proteins, was found to be the sole kinase for activation of N-acetyl-d-glucosamine (NAG). The other kinase (HexA) is responsible for the majority of the glucose kinase activity in the cell, although a hexA deletion mutant strain was not defective for growth on any substrate tested. Deletion of both the rokA and hexA kinase genes resulted in inability of the cell to use glucose, mannose, NAG, and many other sugars. We purified RokA and determined its approximate molecular mass to be 36.5 kDa. The purified RokA protein was shown to phosphorylate several substrates, including glucose, NAG, and mannose, but not N-acetylmannosamine or N-acetylneuraminic acid. Phylogenetic analysis of RokA showed that it is most similar to kinases from the Cytophaga-Flavibacterium-Bacteroides group, while HexA was most similar to other bacterial hexokinases and eukaryotic hexokinases.
The “nanaerobic” gram-negative organism Bacteroides fragilis (4) is capable of utilizing a wide variety of simple sugars and complex oligosaccharides as sources of carbon and energy for growth. This is a useful property for a bacterium that is both a gastrointestinal tract commensal and an opportunistic pathogen. In the large intestine, undigested dietary polysaccharides and host-derived glycoproteins, such as colonic mucin, can serve as growth substrates for B. fragilis (22). At sites of infection, the availability of host cell surface glycoproteins and glycolipids provides nutrients during the establishment phase and after abscess formation. As an example, the cell surface Lewis antigen contains galactose, mannose, N-acetyl-d-glucosamine (NAG), and N-acetylneuraminic acid (34, 46), all of which can serve as carbon and energy sources for B. fragilis.
Godoy et al. (17) demonstrated that the B. fragilis neuraminidase (the product of the nanH gene) catalyzes the removal of terminal sialic acids from surface polysaccharides of CHO cells in monolayer cultures and in the rat granuloma pouch model system, both of which are glucose-limited growth conditions. Accordingly, ΔnanH mutant strains are compromised for growth in these model systems. N-acetyl-d-glucosamine is an excellent carbon and energy source for B. fragilis and appears to be utilized more efficiently than glucose (8). Release of NAG from host glycoproteins and its transport into the cell can also influence anabolic cellular processes. NAG is a major component of the murein sacculus (21), and the ability to acquire or produce NAG affects cell wall production and turnover. The ability to utilize extracytoplasmic NAG allows for efficient recycling of cell wall components (30). In addition, B. fragilis strains synthesize a wide variety of extracellular capsules containing simple hexoses, amino sugars such as NAG and N-acetylmannosamine (manNAc), and more complex components, such as 2-amino-4-acetimido-2,4,6-trideoxygalactose (AAT) and 3-acetimido-3,6-dideoxyglucose (ADG) (25). It is not surprising that several capsule production operons in the two B. fragilis strains whose genomes have been sequenced, NCTC 9343 and 638R, contain genes whose products require NAG as a substrate or precursor for alteration and/or insertion into the capsule polysaccharides (10, 11).
In many bacteria, such as Escherichia coli and Bacillus subtilis, phosphoenolpyruvate (PEP)-dependent phosphotransfer systems (PTS) transport sugars into the cytoplasm of the cell with concomitant activation by addition of a phosphate group (31). As a result of the PTS system, any sugar transported across the cell membrane into the cytoplasm is able to immediately enter pathways for energy metabolism or biosynthesis. Analysis of the preliminary genome data for two B. fragilis strains (http://www.sanger.ac.uk/Projects/B_fragilis) and the published genomes of other Bacteroides and related species (29, 45) failed to identify any genes that might be part of a PTS system. Moreover, no phosphoenolpyruvate-dependent glucose and mannose uptake activities could be detected in whole cells of B. thetaiotaomicron, a close relative of B. fragilis (23). In the absence of a PTS system, B. fragilis must transport sugars into the cell and activate them via cytoplasmic kinases.
Many bacteria utilize specific hexokinases, such as glucokinases and galactokinases, capable of phosphorylating only one sugar. It has been shown that prokaryotic kinases, like those of Streptococcus mutans, B. subtilis, Zymomonas mobilis, and Rhodospirillum rubrum, are chiefly glucose kinases (13, 19, 27), as only glucose phosphorylation activity was detected. Kinases characterized in Archaea, however, have been shown to have broad substrate specificity, phosphorylating glucose, mannose, fructose, and other sugars (14, 18). Eukaryotic kinases, from yeast (13) to humans (19), have broad specificity and are capable of phosphorylating many hexoses (7), although phosphorylation of hexosamines was not tested. Broad-specificity kinases have not been characterized in many eubacteria.
Sugar kinases that are classified as members of the ROK (for repressor, open reading frame [ORF], kinase) family have been found in many bacterial species (16, 39). Members of the ROK family can be identified by two signature sequences (known as ROK boxes) in the central region of the protein. In addition, they contain an ATP binding motif (in the case of kinases) or a helix-turn-helix DNA binding motif (in the case of repressors) at the N terminus (18, 39). Previous studies focused mainly on the glucose kinase activities of these proteins, and many ROK family kinases are considered glucokinases (28, 33, 43).
In this study, we identified two kinases in B. fragilis, RokA and HexA. RokA was shown to play a key role in amino sugar utilization. HexA also has a major role in sugar metabolism in B. fragilis, as it is capable of allowing growth on glucose and mannose in the absence of RokA. Both kinases are examples of broad-specificity enzymes in bacteria. RokA, however, is an unusual eubacterial kinase based on its activity with both hexoses and hexosamines.
MATERIALS AND METHODS
Materials.
All chemicals used in this work were obtained from Sigma Chemical Co. (St. Louis, Mo.) or Fisher Scientific Co. (Agawam, Mass.) unless otherwise stated. Oligonucleotide primers were synthesized by IDT (Coralville, Iowa). The radiolabeled amino sugar [14C]NAG (specific activity, 55 mCi/mmol) was a generous gift from Eric Vimr (University of Illinois, Champain-Urbana, Ill.).
Strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are described in Table 1. For most of this work, we employed strain TM4000, a subclone of the strain 638R, which has been sequenced by the Pathogen Sequencing Group at the Sanger Centre; TM4000 is plasmid free and amenable to genetic manipulation (3, 36). B. fragilis cells were grown in an anaerobic chamber (Coy Laboratory Products) at 37°C with an atmosphere of 85% N2, 10% H2, and 5% CO2 (Airgas Northeast). Cells were grown in brain heart infusion broth (BHIS) supplemented with 0.5% yeast extract and 15 μg of hematin/ml (38). As formulated, BHIS broth always contains 0.1% glucose, therefore all strains capable of utilizing exogenous glucose are able to grow in BHIS without additional supplements. Strains with defects in the utilization of glucose as the carbon and energy source were grown on BHIS with an additional supplement of 0.5% galactose. In vitro growth experiments were performed in super anaerobic minimal medium (SAMM) (41) broth containing 0.5% of the indicated carbon source, unless otherwise stated. Thymine (200 μg/ml) was added for growth of thyA strains.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| B. fragilis | ||
| ADB77 | TM4000; ΔthyA Tpr | 3 |
| CJB100 | ADB77:ΔrokA1 | This work |
| CJB101 | ADB77:ΔrokA1ΔhexA1 | This work |
| CJB200 | ADB77:ΔhexA1 | This work |
| ATCC 25285 | Subclone of NCTC 9343 | 26 |
| TAL2480 | Cefr | 12 |
| TAL3636 | Cefr | 12 |
| ATCC 23745 | rokA+hexA+ | 26 |
| TAL21616-21633 | Clinical isolates acquired from Tufts Anaerobic Laboratory | This work |
| B. thetaiotaomicron | ||
| VPI 8582 | rokA+hexA+ | 45 |
| E. coli | ||
| DH5α | λ nonlysogen | 44 |
| HB101 | rpsL20 | 38 |
| Plasmid | ||
| RK231 | RP4 derivative, Tetr, Tra+ | 17 |
| pYT102 | p15A ori, Cmr, RP4 oriT, B. fragilis suicide vector containing B. fragilis thyA+ Tetr | 37 |
| pJST61 | E. coli-B. fragilis shuttle vector, Ampr, Clnr | 38 |
| pKDel12 | pYT102 ΔrokA1 | This work |
| pHDe12 | pYT102 ΔhexA1 | This work |
| pBFROK10 | pJST61 with 2.4-kb fragment from TM4000 containing the rokA gene | This work |
| pBFROK40 | pJST61 with 1-kb fragment from TM4000 containing the rokA gene under the λ PL promoter | This work |
| pBFHEX10 | pJST61 with 1.5-kbp fragment from TM4000 containing the hexA gene | This work |
| pBFROKHIS | pJST61 with 1.1-kb fragment from TM4000 containing the rokA gene with an oligohistidine- tagged C terminus | This work |
Ampicillin (100 μg/ml), chloramphenicol (25 μg/ml), tetracycline (2 μg/ml for B. fragilis, 10 μg/ml for E. coli), rifampin (50 μg/ml), erythromycin (8 μg/ml), gentamicin (50 μg/ml), and trimethoprim (80 μg/ml) were used as indicated.
E. coli DH5α (44) was used for cloning, and HB101/pRK231 was used for mobilization of plasmids from DH5α to B. fragilis, as previously described (38). E. coli cells were grown in Luria broth (Difco) at 37°C. Chemically competent cells were prepared and transformed as described by Chung et al. (9).
For all B. fragilis growth experiments, strains were first grown overnight in BHIS broth containing thymine, harvested by centrifugation, washed twice in modified phosphate buffered saline (MPBS) (37), and resuspended in 5 ml of MPBS. These suspensions were used to inoculate a SAMM broth culture to a starting A600 of ∼0.05. The growth of all cultures was monitored by absorbance at 600 nm.
Cloning, gene sequencing, and analysis.
Oligonucleotide primers used in this work are described in Table 2. PCR amplification was performed using Taq DNA polymerase (QIAgen, Inc., Valencia, Calif.). Oligonucleotide sequences were based on sequence data obtained from the B. fragilis NCTC 9343 preliminary genome sequence (http://www.sanger.ac.uk/Projects/B_fragilis) and from sequences of TM4000 and plasmid vectors done by our laboratory. Plasmid primers 61RAB and 1842 have been described previously (3). Plasmid primer 61FAB (Table 2) allows sequencing across the cloning site in the pJST61 vector (unpublished results). Purification of all PCR-amplified fragments and plasmids was performed using QIAprep spin columns (QIAgen). Restriction endonucleases were purchased from New England Biolabs (Beverly, Mass.), and restriction digests were performed according to the manufacturer's instructions.
TABLE 2.
List of primers
| Primer | Sequencea |
|---|---|
| KINREGF | 5′-CTCAGATCTCTTGCCGAATGGTGAGGCATCATG-3′ |
| KINREGR | 5′-CGCGGATCCCTCCAAAGGTATGAATAGTC-3′ |
| 61RAB | 5′-GGCGCGCCGTAAGGAAAGTGGCTCTCAG-3′ |
| 61FAB | 5′-GGCCGCCCTGAAGCCAAACACCAAGG-3′ |
| ROKWALK5 | 5′-CATGGAGAAGCCCTATGTGGTG-3′ |
| ROKWALKR | 5′-CTTGAGTTTTCACTGCACCGCTTG-3′ |
| Kdel1 | 5′-GCAGGATCCCGTAAAATGTCAGGAACTGTAGGCG-3′ |
| Kdel2 | 5′-GAAGCGGCCGCAGAGACTAAACGCCCTATTAAAGTG3′ |
| Kdel3 | 5′-CTGGCGGCCGCTGCTTGAATTCATGATACTC-3′ |
| Kdel4 | 5′-GACAAGCTTCGTACGGAGTTTGTTCCGCTTGATGG-3′ |
| Kdelup | 5′-GGCAATCTGGTAATCGGTTTTTGCC-3′ |
| 1842 | 5′-CAAGGCGACAAGGTGCTGATGC-3′ |
| KCHKFW | 5′-TTTGAGCTGCCGGAAATCAAC-3′ |
| KCHKRV | 5′-TTAGACGTACGGCTGATACG-3′ |
| HEX1 | 5′-CGAGGATCCGTAGGGTCCACTCTGTGC-3′ |
| HEX2 | 5′-CTTGCGGCCGCTTTGTACCTGTCCCTAC-3′ |
| HEX3 | 5′-CAAGCGGCCGCATTATCCGATATACAC |
| HEX4 | 5′-GAGAAGCTTCGGCATGCAAATCC-3′ |
| HEXCHKF | 5′-GGTTATACCCGTGAAGAGTTG-3′ |
| HEXCHKR | 5′-CGACGGGCGAGGTTTACAG-3′ |
| HEXFLA2 | 5′-GACGGCCATGCTTTGCACGGCAATAGC-3′ |
| HEXREGF | 5′-CGAAGATCTCGGTTGCTGGATAACAGGATTATG-3′ |
| HEXREGR | 5′-GATGGATCCCATCCACTCTACGCAAAATAAGTC-3′ |
| ROKCONFW | 5′-GGGTATCGGTGTTGGTGCTCCTAATGGTAACTATTATACCGG-3′ |
| ROKCONRV | 5′-GCCARACCACCGAACAGGATRATTGCTTCCGGGCTGGAGAAAGC-3′ |
| HEXCONFW | 5′-GCCTATCGGTTATTGTTTTTCTTATCCGRCCGARTCGRTACCGGG-3′ |
| HEXCONRV | 5′-GGRTAATTCATAATRGCAGTCAGTTTCCTTGCRAATTTCTCTTC-3′ |
Restriction sites are underlined.
For sequencing of the rokA gene, a 2.4-kb region of the TM4000 chromosome containing the rokA gene was amplified via PCR using primers KINREGF and KINREGR based on the rokA gene sequence of the B. fragilis NCTC 9343 genome. The PCR product was digested with BamHI and BglII and was then inserted into the unique BglII site of the E. coli-B. fragilis shuttle plasmid pJST61 to produce the plasmid pBFROK10 (Fig. 1). The rokA gene on this plasmid was sequenced using primers 61RAB, 61FAB, ROKWALK5, and ROKWALKR. For sequencing of hexA, the gene and its surrounding region were amplified from strain TM4000 by using primers HEXREGF and HEXREGR. The PCR product was purified as described above, digested with BamHI and BglII, and inserted at the BglII site of pJST61 to produce pBFHEX10. Sequencing of the hexA gene in pBFHEX10 was performed as described above, using the 61RAB and 61FAB primers. All sequencing was performed by the Tufts University Nucleic Acids and Protein Core Facility.
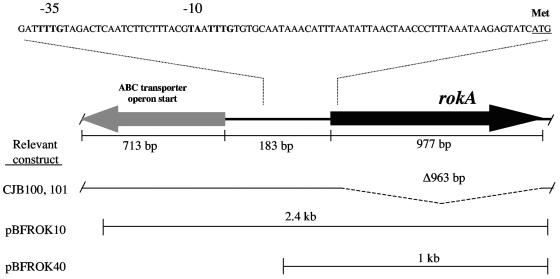
FIG. 1.
Schematic drawing of the rokA region, including the predicted promoter. The rokA gene is located upstream of and divergent from a possible ABC transporter of unknown function. A schematic of the region of the rokA deletion mutants CJB100 and CJB101 and the inserts of the rokA-containing plasmids pBFROK10 and pBFROK40 are shown. Shown above is a portion of the intergenic region containing a predicted promoter for the rokA gene, including the N-terminal methionine as confirmed by sequencing of pure RokA protein.
To investigate the presence of rokA and hexA in other B. fragilis strains and other Bacteroides species, we used primers ROKCONFW and ROKCONRV or HEXCONFW and HEXCONRV based upon consensus sequences in rokA and hexA, respectively. All B. fragilis strains (Table 1) were amplified by standard PCR methods using these primers. Products were analyzed by agarose gel electrophoresis. In some cases, gradient PCR analysis (from 45 to 55°C) was performed to amplify products from hexA or rokA genes in other B. fragilis strains under less stringent conditions. All PCR analysis was performed on an MJ Research PTC-200 thermal cycler.
Other nucleotide and protein sequence data described in this study were obtained from the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/). DNA and protein sequences were analyzed by using DNA STRIDER 1.2, EDITSEQ, and SEQMAN (DNAStar, Madison, Wis.), MACVECTOR 7.0 (Oxford Molecular, Madison, Wis.), and PAUP 4.0b10 (35).
Construction of in-frame deletions on the B. fragilis chromosome.
In-frame deletions in B. fragilis strain ADB77 (a derivative of TM4000; Table 1) were constructed by the two-step, double-crossover method described previously (3). The rokA gene was deleted using the plasmid pKdel12. A fragment consisting of 927 bp downstream of the chromosomal rokA open reading frame and 9 bp of the rokA C-terminal coding sequence was amplified using the oligonucleotides KDEL1 and KDEL2 as primers. A second fragment consisting of 13 bp of the N-terminal coding sequence of rokA and 968 bp upstream of the rokA open reading frame was amplified by using KDEL3 and KDEL4 as primers. The respective products were digested with BamHI and NotI as well as HindIII and NotI, and then they were ligated into BamHI/HindIII-digested pYT102 to create the ΔrokA allelic exchange plasmid pKDel12. Plasmid pKDel12 was then transferred to B. fragilis from E. coli DH5α by triparental conjugation with E. coli HB101/pRK231 as the mobilizer strain, and cointegrates were selected on BHIS medium containing rifampin, gentamicin, tetracycline, and galactose. Tetracycline-resistant colonies were screened by the colony PCR method with the oligonucleotide 61RAB (vector primer) and an upstream flanking primer of the rokA gene. Isolates that tested positive for recombination at the rokA locus were chosen for further manipulation. Integration of the deletion plasmid results in thymine prototrophy and trimethoprim sensitivity, due to the presence of an intact thyA gene on the deletion plasmid. Cointegrants were grown overnight from single colonies in BHIS plus thymine and galactose. Cultures were plated on SAMM galactose medium with thymine and trimethoprim to select for resolvants. The continuous presence of galactose in the medium during the resolution process allowed for growth of all resolvants in case a deleted rokA gene would not allow growth on glucose. Trimethoprim-resistant isolates were purified on SAMM thymine galactose plates and then screened for tetracycline sensitivity on BHIS plates containing tetracycline, thymine, and galactose. Tetracycline-sensitive isolates were screened by the colony PCR method to distinguish between wild-type resolution products and deletion resolution products.
A hexA deletion vector was constructed in the same manner as pKDel12, using HEX1 and HEX2 as well as HEX3 and HEX4 as primer pairs for the deletion fragments. The length of the PCR product of the N-terminal region of hexA plus 126 bp of upstream sequence was 747 bp. The length of the product of the downstream region, including 597 bp of downstream sequence, was 915 bp. The fragments were digested as described above and ligated into pYT102 to produce pHDel2. The plasmid pHDel2 was introduced by mating into B. fragilis, and proper cointegrants and resolvants were selected as described above.
Expression of B. fragilis RokA in E. coli.
RokA was expressed in E. coli under control of the phage λ PL promoter on plasmid pBFROK40. Strain HB101 was transformed with plasmid pBFROK40 (see Table 1 and Fig. 1) as described above. Plasmid-containing cells were grown overnight and harvested in stationary phase. Cells were washed, disrupted by sonication, and clarified by centrifugation at 2,000 × g in a microcentrifuge at 4°C, and the supernatants were transferred to fresh sterile microcentrifuge tubes and maintained on ice. Extracts were then assayed for glucose kinase activity.
Purification of RokA protein.
Because we found that RokA enzymatic activity is not affected by exposure to oxygen, all purification steps were performed under oxic conditions at 4°C. One-liter cultures of DH5α cells containing plasmid pBFROK40 (Table 1 and Fig. 1) were grown in Luria broth containing 100 μg of ampicillin/ml. Cells were harvested by centrifugation at 2,700 × g at 4°C in a Sorvall RC2-B centrifuge, washed with 10 mM potassium phosphate buffer (pH 7.6), and frozen overnight. Cells were resuspended in PED10 buffer (10 mM potassium phosphate buffer, pH 7.6; 1 mM EDTA; 1 mM dithiothreitol) and sonicated on ice for 5 min with a 30% output on a 50% duty cycle using a Branson Sonifier 250. Extracts were clarified by centrifugation for 20 min at 3,900 × g at 4°C. The supernatant was then treated with streptomycin sulfate (14 mg/ml), and the precipitate was removed by centrifugation at 100,000 × g at 4°C for 60 min. Following this treatment, the supernatant was applied at a flow rate of 4 ml/15 min to a DEAE-Sephacel column that had been equilibrated with PED10 buffer. The column was washed with PED10 and then eluted with buffer containing a gradient of 0 to 1 M NaCl. Pooled fractions containing glucose kinase activity were desalted and then applied at a flow rate of 4 ml/5 min to a hydroxyapatite column and eluted with a gradient of PED5 to PED400 (5 to 400 mM potassium phosphate, respectively). Pooled fractions with glucose kinase activity were then purified via fast-pressure liquid chromatography on a Mono-Q HR 5/5 column (Amersham Pharmacia) and eluted with a linear gradient of 0 to 1 M NaCl. Purified protein was desalted and diluted 1:10 in 10% glycerol and stored at −70°C. Desalting of protein was done by filtering through Millipore (Bedford, Mass.) Amicon Ultra centrifugal filter devices following instructions recommended by the manufacturer.
Purification of RokA6HIS from B. fragilis was performed using a Ni+-nitrilotriacetic acid (NTA) agarose purification column (QIAGEN, Inc.). Briefly, cultures of CJB101/pBFROKHIS were grown anaerobically to an A600 of approximately 0.8. Cells were harvested by centrifugation at 3,900 × g at 4°C for 10 min and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0). Cells were sonicated on ice for 1 min with a 30% output on a 10% duty cycle using a Branson Sonifier 250. Extracts were clarified by centrifugation for 20 min at 3,900 × g at 4°C. The supernatant was applied to the Ni+-NTA agarose column and eluted according to the manufacturer's instructions.
Enzyme assays.
Glucose kinase activity was measured according to methods described previously (24). Briefly, B. fragilis cells were grown in SAMM broth plus specified additions until a culture A600 of approximately 0.8. Cultures were centrifuged at room temperature in a Dupont Sorvall GLC-1 (Dupont Corp.) for 5 min at 2,800 × g, and the supernatant was removed by aspiration. The cells were then resuspended in 1 ml of chilled assay buffer (0.05 mM Tris-HCl [pH 8.0] and 13.3 mM MgCl2), transferred to 1.5-ml Eppendorf tubes, and sonicated on ice as described above. Glucose kinase activity was measured by coupling the formation of glucose-6-phosphate to the reduction of NAD+ using glucose-6-phosphate dehydrogenase (Leuconostoc mesenteroides, EC1.1.1.49; Sigma). The assay was performed using 50 μl of clarified extract or 0.75 μg of purified protein with 0.5 mM ATP, 0.25 mM NAD+, and 2 U of glucose-6-phosphate dehydrogenase brought up to a volume of 2.9 ml in assay buffer. The reaction was started by the addition of glucose. The Km and Vmax for glucose and ATP were determined by analysis of a double reciprocal (Lineweaver-Burke) plot.
To determine the substrate specificity of RokA, kinase activity was assayed via a coupled assay with pyruvate kinase and lactate dehydrogenase (2). Assay buffer consisted of 0.1 M Tris-HCl (pH 7.0), 0.01 M MgCl2, 2 mM ATP, 0.6 mM NADH, 5 mM PEP, and excess pyruvate kinase (from rabbit muscle; EC 2.7.1.40; Sigma) and lactate dehydrogenase (from rabbit muscle; EC 1.1.1.27; Sigma). Substrate (NAG, mannose, etc.) was added from 0.67 M stock solutions. The reaction was started by adding 0.75 μg of purified RokA protein and was monitored by following the decrease in A340 as NAD+ was formed from NADH. Both kinase assays were performed at room temperature. Auxiliary enzymes in both assays were tested to ensure that they were not rate limiting. Km and Vmax were determined for all substrates as described above.
To examine the RokA gene product acting on NAG, clarified extracts of B. fragilis CJB200 were incubated with radiolabeled NAG. Reaction mixtures included 10 μl of NAG solution (5 mM stock; 14C specific activity, 0.1 μCi/μmol), 10 μl of 0.25 M Tris-HCl, 50 mM MgCl2 buffer (pH 7.5), 10 μl of 50 mM ATP, and 20 μl of cell extracts. The final reaction mixture was incubated at 37°C in an aerobic environment for up to 1 h. Assays were also performed with nonradioactive NAG at a final concentration of 5 mM under similar reaction conditions. NAG and N-acetyl-d-glucosamine 6-phosphate (NAG 6-P) were separated via descending paper chromatography on 1% sodium borate-treated paper (42) with 1-butanol:pyridine:H2O (6:4:3 ratio) as the solvent. Ten to 20 μl of reaction mixture was spotted at the origin and allowed to dry completely before chromatography. Chromatograms were run for 8 to 12 h in a sealed tank and then dried. For assays with [14C]NAG, paper chromatograms were exposed on a Kodak MD146-931 phosphor screen for 20 to 30 min and scanned on a Storm 850 PhosphorImager. Images were manipulated and spot intensities quantified using ImageQuant 1.2 imaging software (Molecular Dynamics). Phosphatase treatment of NAG 6-P was performed with calf intestinal alkaline phosphatase (New England Biolabs) for 1 h under conditions recommended by the manufacturer, and the products were analyzed on a separate chromatogram. Standards of NAG and NAG 6-P were applied to all chromatograms to gauge the separation of substrate from product. For all standards and assays using nonradioactive NAG, the paper chromatogram was dried, baked at 95°C for 5 min, and sprayed thoroughly with a fine mist of Ehrlich's reagent (acetic acid, HCl, p-aminobenzaldehyde):acetic acid (1:9) solution (32) by using an air-driven spraying apparatus (Sigma). Chromatograms were baked at 95°C for another 5 to 10 min, and spots from reaction mixtures were compared to the standards.
Analysis of RokA and HexA activity in other B. fragilis strains.
Extracts of B. fragilis TM4000, ATCC 25285 (a subclone of the strain NCTC 9343), and TAL2480 (a clinical isolate) (12) were prepared as described above. Glucose kinase activity was assayed using the coupled assay described above. Total glucose kinase activity was measured by NADH generation for the first 10 min of incubation. NAG was then added to each reaction mixture to a final concentration of 114 mM, and the rate of NADH production was redetermined. RokA activity was defined as the NAG-inhibitable fraction of the total activity.
Phylogenetic analysis.
Sequences of RokA homologs and HexA homologs were aligned by the CLUSTALW method. Phylogenetic trees were calculated with PAUP4.0b10 (34) using the bootstrap method with heuristic search. Confidence limits were estimated using 100 bootstrapping replicates. Phylogenetic trees were also calculated with MACVECTOR 7.0 using the neighbor-joining method, and confidence limits were estimated using 100 neighbor-joining replicates.
Nucleotide and protein sequence accession numbers.
The sequences of rokA and hexA have been deposited in the DDJB, EMBL, and GenBank databases under the accession numbers AY662335 and AY664812, respectively. The sequences of the rokA and hexA deletion alleles have been deposited under the accession numbers AY672841 and AY672840, respectively.
RESULTS
Identification of the B. fragilis rokA gene.
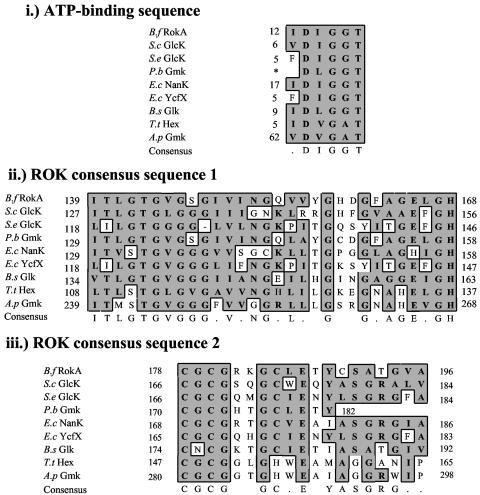
A search for potential kinase genes in the unfinished B. fragilis NCTC 9343 genome led us to a gene, rokA, whose putative protein product shares sequence similarities with several previously characterized ROK family proteins. The putative B. fragilis RokA gene product, in addition to showing extensive conservation of many residues present in the ROK consensus sequences, also contains the conserved ATP-binding domain (Fig. 2). The rokA gene is approximately 250 bp upstream of, and divergently transcribed from, an operon of unknown function that contains genes with similarities to ABC transporter genes (Fig. 1).
FIG. 2.
Comparison of the B. fragilis RokA protein with recently characterized ROK family members. Shown are the conserved ATP binding sequence (i) and the ROK domains 1 (ii) and 2 (iii). B.f, Bacteroides fragilis; S.c, Streptomyces coelicolor; S.e, Salmonella enterica; P.b, Prevotella bryantii; E.c, Escherichia coli (NanK and YcfX); B.s, Bacillus subtilis; T.t, Thermoproteus tenax; A.p, Aeropyrum pernix. An asterisk indicates the start of the known protein sequence. Identical or similar residues are boxed in gray.
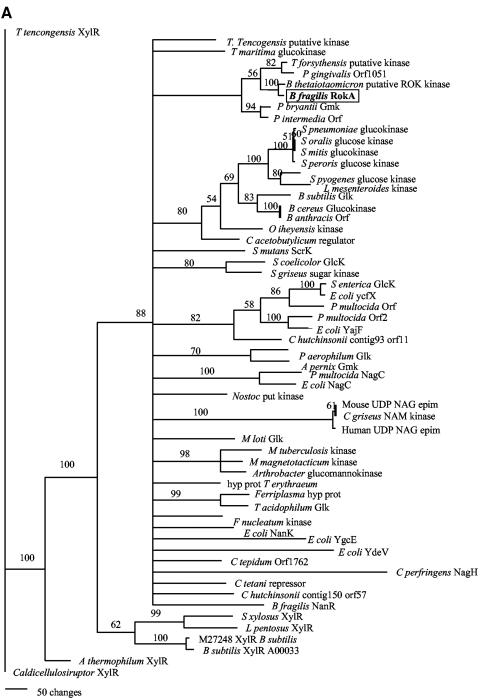
Our reference strain, B. fragilis TM4000 (a subclone of strain 638R), is not 100% identical to B. fragilis NCTC 9343 (25). Thus, the rokA gene from TM4000 was sequenced as described above and found to be 99.9% identical at the DNA level, and 100% identical at the protein level, to the rokA gene and its predicted protein product, respectively, in NCTC 9343. In addition, PCR analysis was performed, with primers ROKCONFW and ROKCONRV (Table 2), on B. fragilis strains TM4000, NCTC 9343, TAL2480, and ATCC 23745 (26) as well as on clinical isolates of B. fragilis obtained from the Tufts Anaerobic Laboratory (TAL21616 to TAL21633). Identical-sized products were obtained from PCR analysis of all the B. fragilis strains, indicating that they contain a rokA gene. A sequence comparison of B. fragilis RokA and other ROK family proteins using the BLAST algorithm at the NCBI nonredundant database showed that this protein is similar to many previously characterized kinases. RokA shares closest similarity with the characterized glucomannokinase from Prevotella bryantii strain B14, especially in the ROK consensus domain I (Fig. 2).
A rokA deletion mutant exhibits growth defects on several substrates.
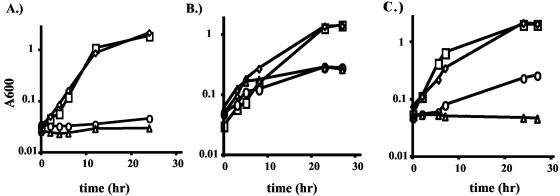
To determine if the rokA gene has a role in sugar utilization in B. fragilis, this gene was deleted as described above. The resulting rokA deletion strain, CJB100, had a growth defect with glucose as the main carbon source (doubling time of 8 h, as opposed to 2.5 h for the wild type; Fig. 3C). Because the kinase found in Prevotella bryantii has been shown to phosphorylate glucose and mannose (16), we also tested the response of the B. fragilis ΔrokA mutant to the presence of mannose as the main carbon and energy source. Indeed, growth of CJB100 was also impaired in mannose minimal medium (doubling time of 8.5 h, compared to a WT doubling time of 2.5 h; data not shown). Although the rokA mutant is impaired for growth in glucose and mannose, there must be another kinase present to activate these two sugars for growth, albeit at a reduced rate. The slow growth rate suggests that the remaining kinase is unable to fully compensate for the lack of functional RokA. Strikingly, the ΔrokA mutant was unable to grow in minimal medium with NAG as the carbon and energy source (Fig. 3A). Strain CJB100 was able to grow on lactose but was unable to grow on maltose. Also, maltotriose and maltotetraose could not be used as carbon and energy sources (data not shown).
FIG. 3.
Growth of ADB77 (wild type; squares), CJB100 (ΔrokA; circles), CJB200 (ΔhexA; diamonds) and CJB101 (ΔrokAΔhexA; triangles) in SAMM broth supplemented with 100 μg of thymine/ml and 0.5% NAG (A), 0.5% NANA and 0.05% galactose (B), and 0.5% glucose (C). Graphs in this figure are representative of results from three separate growth experiments.
In species capable of catabolizing N-acetylneuraminic acid (NANA), NAG and NAG 6-P are possible downstream products of NANA catabolism (1). Strain CJB100 exhibited a severe growth deficit with NANA as the main carbon and energy source (Fig. 3B), implying that RokA is involved in phosphorylating NANA or a breakdown product of NANA utilization. These results lead to the conclusion that RokA is the principal kinase in the catabolism of NAG and NANA.
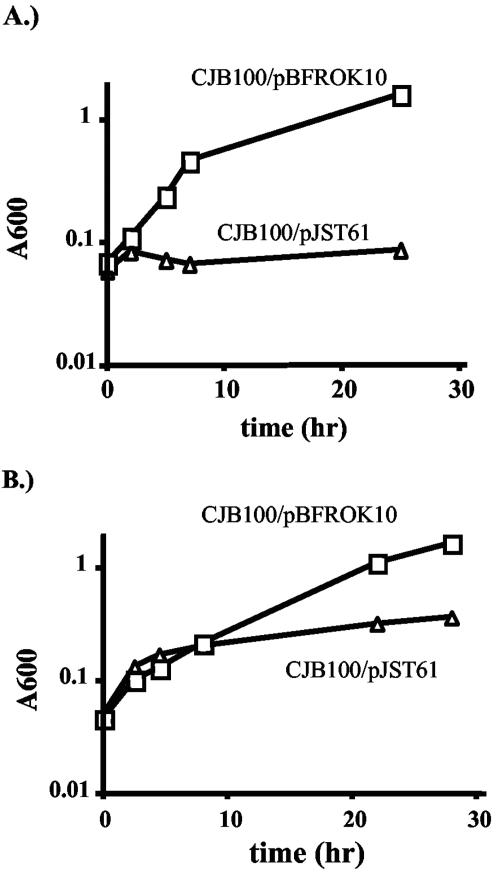
The defects of CJB100 could be complemented in trans by a plasmid, pBFROK10, containing the intact rokA gene. Growth of CJB100/pBFROK10 on NAG and NANA was compared to that of a strain containing vector alone. B. fragilis CJB100/pBFROK10 was able to grow with a doubling time similar to that of the wild type in minimal medium plus NAG or NANA, whereas CJB100 containing vector alone still exhibited growth defects on these substrates (Fig. 4). At the end of each growth experiment, the cultures were tested to determine if the plasmid-borne rokA gene had integrated into the chromosome; no such recombination was detected.
FIG. 4.
Growth of the ΔrokA strain CJB100 containing pBFROK10 (boxes) or vector alone (triangles) in minimal medium with (A) NAG or (B) NANA as the main carbon and energy sources. Graphs in this figure are representative of results from three separate growth experiments.
Characterization and deletion of the B. fragilis hexA gene.
Our search of the B. fragilis NCTC 9343 genome yielded a gene, designated hexA, whose putative protein product shares sequence similarities with the poorly characterized mammalian general hexokinase C (7). The hexA gene from TM4000 was sequenced as described above and found to be 99.9% identical at the DNA level, and 100% identical at the protein level, to the hexA gene and its predicted protein product, respectively, in NCTC 9343. PCR analysis was performed, with primers HEXCONFW and HEXCONRV (Table 2), on several B. fragilis strains (Table 1). Identical-sized products were obtained from most strains by PCR, indicating that they contain a hexA gene. For some strains, such as TAL2480 and TAL3636, it was necessary to perform a gradient PCR with temperatures ranging from 45°C (less stringent) to 55°C (more stringent). PCR amplification in these strains gave a band of expected size under the less stringent temperatures (from 45 to 50.9°C), indicating that these strains contain a hexA gene which is similar but not identical in DNA sequence to that of B. fragilis TM4000. The product of the hexA gene was investigated as a potential candidate for a glucose-phosphorylating enzyme. To establish a role for HexA, an in-frame deletion of the hexA gene was created. The resulting hexA deletion strain, CJB200, exhibited no growth defects in minimal medium with any sugar tested (Fig. 3).
This result could be explained if either the RokA kinase or the HexA kinase were sufficient to allow growth on glucose and mannose. To test this hypothesis, a mutant strain lacking both rokA and hexA was created (CJB101) and tested for growth in minimal medium in the presence of various substrates. Indeed, CJB101 was unable to grow using NAG and NANA as carbon and energy sources (Fig. 3A and B). In addition, the strain with deletions in both kinases was unable to grow with glucose (Fig. 3C) or mannose as carbon and energy sources. Thus, under our growth conditions, RokA and HexA are the only two glucose and mannose kinases in the cell. Furthermore, these results suggest that RokA is the only NAG kinase.
RokA expression in E. coli.
For purification of RokA, it was beneficial to overexpress the gene in E. coli. Therefore, we compared the glucose kinase activity of HB101/pBFROK40 (Table 1) and plasmid-free HB101. Glucose kinase activity of the cell extracts was measured as described above, and a specific activity of 0.93 ± 0.02 μmol/min/mg was obtained for HB101/pBFROK40, compared to 0.02 ± 0.01 μmol/min/mg for plasmid-free HB101.
Purification of RokA.
RokA was purified aerobically in three steps from HB101/pBFROK40 extract prepared as described above. RokA glucose kinase activity was monitored before and after each step (Table 3). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) indicated that the RokA protein sample was ∼99% pure, and the protein had an apparent molecular mass of 36.5 kDa. The N-terminal sequence of RokA purified from E. coli was determined to be MNSSMEKPYVV by automated Edman degradation at the Tufts University Nucleic Acids and Protein Core Facility. We also determined the N-terminal sequence of oligohistidine-tagged RokA expressed in B. fragilis CJB101 containing pBFROKHIS after purification on a Ni+-NTA agarose column. The N terminus of RokA expressed in B. fragilis was the same as the sequence of the protein purified from E. coli, thus confirming that the same translation initiation signal, as indicated in Fig. 2, was used in both species.
TABLE 3.
Purification of RokA protein
| Purification step | Total protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Fold purifi- cation |
|---|---|---|---|---|---|
| Streptomycin sulfate- treated cell extract | 588 (441)a | 410.3 (307.7)b | 0.93 | 100 | 1 |
| DEAE | 28.7 (9.7)a | 334.9 (113.2)b | 34.5 | 38 | 37.2 |
| Hydroxyapatite | 3.7 (1.3)a | 62.4 (21.4)b | 48 | 19 | 51.6 |
| Mono-Q | 0.29 | 20 | 68.6 | 93 | 73.9 |
Numbers in parentheses indicate the amount of protein used for the next purification step.
Numbers in parentheses indicate the total activity used for the next purification step.
RokA-dependent phosphorylation of glucose, NAG, and other substrates.
Glucose kinase activity can be assayed by coupling the kinase reaction with the glucose-6-phosphate dehydrogenase reaction (see Materials and Methods) and measuring the production of NADH from NAD+ by the increase in A340. Using this method, glucose kinase activity of CJB100 was compared to that of wild-type B. fragilis. The rokA mutant exhibited only 36% as much glucose kinase activity in vitro as did ADB77 (Table 4). This result is consistent with the idea that RokA plays a role in phosphorylation of glucose. Surprisingly, the ΔhexA strain CJB200 exhibited a glucose kinase activity that was 14% of the level seen in the wild-type strain (Table 4), even though CJB200 showed no growth defect with glucose as a carbon and energy source. The ΔrokAΔhexA mutant had no detectable glucose kinase activity in the coupled assay, indicating that there are only two kinases in B. fragilis with glucose phosphorylation abilities.
TABLE 4.
Glucose kinase activities of ADB77, CJB100, CJB200, and CJB101 B. fragilis cell extractsa
| Strain | Activity (μmol of NAD+ reduced/mg/min) | % Activityb |
|---|---|---|
| ADB77 (Wild type) | 0.095 ± 0.001 | 100 |
| CJB100 (ΔrokA) | 0.035 ± 0.001 | 36 |
| CJB200 (ΔhexA) | 0.013 ± 0.001 | 14 |
| CJB101 (ΔrokAΔhexA) | NDc | 0 |
Extracts of each strain were assayed at least five times.
Percent activity compared to ADB77 activity.
ND, not detected.
Highly purified RokA protein was used to determine substrate specificity and kinetic parameters. Glucose, NAG, and mannose were preferred substrates, with Km values of 0.58, 0.85, and 1.75 mM, respectively (Table 5). The Km and Vmax for ATP in a glucose kinase reaction were determined to be 0.23 mM and 69 μmol/min/mg, respectively. The glucose analog 2-deoxyglucose could be phosphorylated with a Km of 1.82 mM but with a Vmax of 2.9 μmol/min/mg, much lower than that of glucose. No phosphorylation of N-acetylmannosamine (manNAc) or N-acetylneuraminic acid (NANA) was detected. RokA-dependent phosphorylation of galactose was also not detected (Table 5). Phosphorylation of maltose was also undetectable. This result is surprising, because the ΔrokA mutant strain CJB100 lost the ability to utilize maltose. Furthermore, manNAc, NANA, galactose, and maltose were incapable of inhibiting the glucose kinase activity of either purified RokA or any of the cell extracts tested above. This indicates that none of these sugars competes for the active site of either RokA or HexA, reinforcing the conclusion that they are not substrates of either kinase.
TABLE 5.
Km and Vmax values of highly purified RokA kinase activity with various substrates
| Substrate | Km (mM) | Vmax (μmol/min/mg) |
|---|---|---|
| Glucose | 0.58 | 69 |
| NAG | 0.85 | 37 |
| Mannose | 1.75 | 50 |
| 2-Deoxyglucose | 1.82 | 1.4 |
| Fructose | 44.5 | 2.9 |
| ATP | 0.23a | 69a |
| manNAcc | NDb | ND |
| NANAc | ND | ND |
| maltosec | ND | ND |
| galactosec | ND | ND |
Values of Km and Vmax for ATP were measured with 6.7 mM glucose.
ND, not detected.
Concentration of sugars tested to measure the kinase activities was >50 mM.
A direct assay of NAG kinase activity in extracts of B. fragilis.
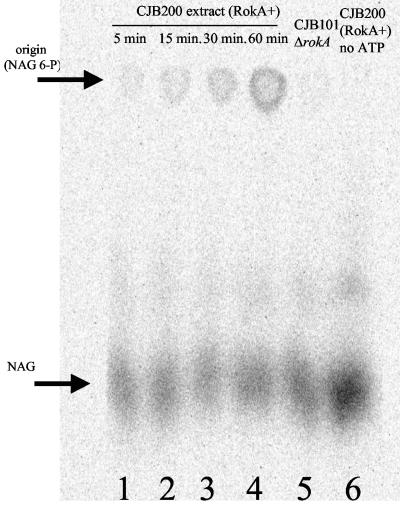
To compare the levels of NAG kinase activity in wild-type versus rokA mutant strains, we incubated cell extracts with [14C]NAG and ATP as described above and analyzed the results via descending paper chromatography. As shown in Fig. 5, a spot at the origin increased in intensity upon incubation of a reaction mixture containing RokA and ATP, indicating the appearance of phosphorylated product. Extracts of CJB101 (ΔrokA ΔhexA) were unable to phosphorylate NAG to any appreciable extent. Also, a control reaction mixture containing labeled NAG and CJB200 (ΔhexA) cell extract but lacking ATP showed no phosphorylation of the substrate (Fig. 5). Standards of NAG and NAG 6-P were included on the chromatogram and were developed by spraying with Ehrlich's reagent (Materials and Methods) to provide a reference for locating the labeled NAG and NAG 6-P. To prove that the spots at the origin represented NAG 6-P, the spots were eluted from the paper with H2O and treated with calf intestine alkaline phosphatase. The dephosphorylated product was analyzed via descending paper chromatography and shown to travel the same distance as authentic NAG (data not shown). The above results suggest that extracts containing RokA were able to phosphorylate NAG in a time-dependent, ATP-dependent fashion.
FIG. 5.
RokA converts NAG to NAG 6-P. (A) Extracts of B. fragilis containing RokA phosphorylate NAG in a time-dependent, ATP-dependent manner (lanes 1 to 4). Extracts were incubated with ATP and [C14]NAG, as described in Materials and Methods, for the time indicated on each lane. Lanes 5 and 6 were incubated for 60 min at 37°C. Lane 5 is labeled NAG incubated with buffer, ATP, and an extract of the double kinase mutant CJB101. Lane 6 is labeled substrate incubated with CJB200 (ΔhexA) extract and no ATP. Arrows represent standards of NAG and NAG 6-P run on the same chromatogram and developed as described in Materials and Methods.
RokA activity is present in other B. fragilis strains.
To determine the presence of RokA in other strains of B. fragilis, extracts of strains ATCC 25285 (a subclone of NCTC 9343), TAL2480 (a cefoxitin-resistant clinical isolate obtained from the Tufts Anaerobic Laboratory [12]), and TM4000 were assayed for glucose kinase activity as described in Materials and Methods. All strains tested had a similar proportion of NAG-inhibitable activity (data not shown). These results, coupled with the presence of rokA consensus regions, suggests that the other B. fragilis strains contain a functional rokA gene.
Phylogenetic analysis.
We performed phylogenetic analyses on both kinases by using two different methods described above. Figure 6Ashows that the B. fragilis RokA protein groups most closely with the glucomannokinase of P. bryantii as well as with ORFs from Bacteroides thetaiotaomicron, Porphyromonas gingivalis, and Tannerella forsythus. Other members of the ROK family shown in Fig. 6A include the characterized glucokinases from many bacterial species, Archaea, and also some kinase/epimerase gene products from higher eukaryotes. This ROK family phylogenetic analysis suggests that ROK proteins are ubiquitously distributed among the three domains of life. For the ROK family kinases, phylogenetic analyses performed by both methods yielded similar results.
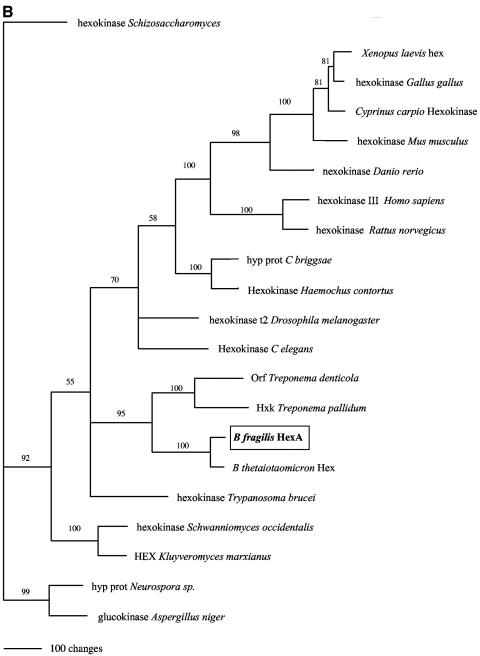
FIG. 6.
(A) A phylogram of ROK family members, including B. fragilis RokA (boxed). (B) A phylogram of HexA and its closest relatives. The trees were calculated by using the parsimony method and were visualized with PAUP 4.0b10 (34). epim, epimerase; hyp prot, hypothetical protein.
Sequence comparison analysis of the other kinase described in this work, HexA (Fig. 6B), has shown that it bears little similarity to bacterial kinases. Other than a hexA homolog in the genome of Bacteroides thetaiotaomicron (also a member of the Cytophaga-Flavibacterium-Bacteroides group) (45), as well as hexokinase homologs in Treponema denticola and Treponema pallidum, the closest homologs are mammalian hexokinase C proteins. Mammalian hexokinase C has not been well characterized, but it is known that such kinases are not found in every tissue type (7). The significance of the relationship between B. fragilis HexA and mammalian hexokinase C is unclear at this time.
DISCUSSION
The RokA kinase from B. fragilis is the first eubacterial ROK family kinase that shows high and similar affinities for both glucose and NAG. The NagK protein from E. coli phosphorylates NAG and glucose, but the Km for glucose is at least two orders of magnitude higher than that for NAG (2, 40). Other broad-specificity kinases are not able to phosphorylate NAG efficiently (14, 18, 19). Our findings imply a central role of the RokA protein in metabolism of hexoses and aminosugars in B. fragilis. As the only NAG kinase in the cell, it may act as an activator for the synthesis of capsule components, the recycling of cell wall material, and for activation of sugars in the cytoplasm for growth. It is likely that RokA plays a role for growth of B. fragilis in vivo, because B. fragilis utilizes sugar (including NANA, NAG, and mannose) released from polysaccharides on the surfaces of epithelial cells as growth substrates (17). The rokA mutant is also incapable of utilizing maltose and higher order oligomers of glucose. This is an interesting observation, because the mutant is still capable of growth on glucose, albeit at a reduced rate.
Because the B. fragilis RokA protein is able to phosphorylate hexoses, such as glucose and mannose, and amino sugars, such as NAG, with similar efficiency and Vmax, the active site must have the ability to accommodate glucose or NAG. The glucose kinase of B. subtilis has been shown to contain a consensus glucose binding sequence located in the C-terminal half of the protein, described as CXCGXXGCXE (27). B. fragilis RokA contains a similar glucose binding sequence (CGCGRKGCLE) located in ROK consensus sequence 2 (Fig. 2). Little information is available as to what distinguishes a glucose kinase from a NAG kinase. For murine NAG kinase, Berger et al. (6) described the importance of two cysteines, one for ATP binding and one for NAG binding. RokA contains five cysteines (C53, C179, C181, C186, and C191), but it is unclear whether any of these cysteines are functionally analogous to cysteines found in the murine NAG kinase. Furthermore, it is not known if the putative active-site cysteines play the same roles as those in the murine NAG kinase. A mutational dissection of rokA may separate the specificity determinants for glucose and NAG, allowing us to identify amino acid residues for amino sugar binding.
Figure 2 indicates that the region upstream of the rokA open reading frame contains a possible B. fragilis promoter, but this promoter sequence deviates from the published consensus sequence (5) at the −10 region. Where the canonical −10 region is described as TAnnTTTG, the rokA promoter is TAATTTG (Fig. 1). While this sequence may indeed be the rokA promoter, it is possible that trans-acting factors are required to facilitate transcription initiation at this nonstandard promoter.
Table 4 shows that extracts of the hexA deletion mutant, CJB200, are capable of phosphorylating glucose at a level only 14% of that of the wild-type extract and that the extract of the rokA deletion mutant, CJB100, phosphorylates glucose at 36% of wild-type levels. It is possible that expression from the rokA and hexA genes is regulated by an as-yet unidentified factor in the wild-type B. fragilis cell. Alternatively, the two kinases may play a role in regulating each other's expression and/or activity.
The characterization of RokA also helps to define the pathway for sialic acid utilization in B. fragilis. Because strains CJB100 and CJB101 are unable to utilize NANA as the main carbon and energy source, we can postulate that RokA phosphorylates NANA or a downstream product of the breakdown of NANA, most notably manNAc or NAG. Although we have shown that pure RokA is capable of phosphorylating NAG, it shows no activity with manNAc or NANA. This implies that after B. fragilis NANA aldolase cleaves NANA into manNAc and pyruvate, another reaction must convert manNAc into a compound that is a RokA substrate. Recent results from our laboratory (R. E. Caughlan, M. H. Malamy, et al., unpublished data) confirmed this prediction.
Phylogenetic analysis shows that the B. fragilis RokA protein groups closely with the orthologs from the dental pathogens Porphyromonas gingivalis and Tannerella forsythus (Fig. 6A). P.gingivalis is unable to utilize external sugars as significant sources of carbon and energy (20); thus, the ROK kinase in P. gingivalis may only be used for phosphorylating the NAG liberated in the cell wall recycling process. Many of the characterized ROK kinases have been shown to be specific glucose or glucose/mannose kinases (15, 16, 28, 33), but none have been cited as NAG kinases. It is interesting that some mammalian epimerase/kinases are members of the ROK family while similar proteins have not been found in groups such as yeast, nematodes, or insects. It is unclear from the phylogenetic analysis in Fig. 6A when the first ancestral ROK protein appeared, as the evolutionary distance between the modern ROK family members cannot be resolved in most cases. However, because ROK proteins are ubiquitously distributed among the three domains of life, it is probable that a ROK family protein was present in the last universal common ancestor.
Sequence analysis of HexA (Fig. 6B) reveals that its closest homologs are broadly distributed in eukaryotes. It is interesting that very few Eubacteria and no Archaea possess a HexA homolog, and the bacterial species that do possess a homolog are mammalian commensals (B. fragilis and B. thetaiotaomicron) or pathogens (T. denticola and T. pallidum). Phylogenetic analyses of HexA and its orthologs (Fig. 6B) does not give a clear picture as to the location of the clade of bacterial orthologs in relation to the other clades. Sequence analysis suggests that these hexokinases were present very early in the eukaryotic lineage. It is possible, given the HexA phylogenetic analyses, that a hexokinase homolog was present in the last universal common ancestor and was lost in most Bacteria and all Archaea. This suggests gene loss in a significant number of known bacterial species, insects, and many lower eukaryotes. However, we cannot rule out the possibility of horizontal gene transfer from bacteria to eukaryotes or vice versa.
Acknowledgments
This work was supported by Public Health Grant AI 19497 from The National Institute of Allergy and Infectious Disease of The National Institutes of Health.
We thank J. Mecsas, J. T. Park, D. RayChaudhuri, T. Uehara, and A. Wright for critical reading of the manuscript. We thank J. T. Park and T. Uehara for technical assistance in protein purification and for helpful discussion regarding NAG utilization and cell wall recycling. We thank E. Vimr for supplying labeled NAG and separation protocols, as well as for many helpful discussions. We thank A. Baughn for guiding us through the phylogenetic analyses.
REFERENCES
- 1.Angata, T., and A. Varki. 2002. Chemical diversity of sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439-469. [DOI] [PubMed] [Google Scholar]
- 2.Asensio, C., and M. Ruiz-Amil. 1966. N-acetyl-D-glucosamine kinase, II. Escherichia coli. Methods Enzymol. 9:421-425. [Google Scholar]
- 3.Baughn, A. D., and M. H. Malamy. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. USA 99:4662-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baughn, A. D., and M. H. Malamy. 2004. Nanaerobes: “strict” anaerobes that grow and benefit from the presence of nanomolar O2. Nature 427:441-444. [DOI] [PubMed] [Google Scholar]
- 5.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 6.Berger, M., H. Chen, W. Reutter, and S. Hinderlich. 2002. Structure and function of N-acetylglucosamine kinase-identification of two active site cystienes. European J. Biochem. 269:4212-4218. [DOI] [PubMed] [Google Scholar]
- 7.Cardenas, M. L., A. Cornish-Bowden, and T. Ureta. 1998. Evolution and regulatory role of the hexokinases. Biochim. Biophys. Acta 1401:242-264. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H.-C., C.-C. Chang, W.-J. Mau, and L.-S. Yen. 2002. Evaluation of N-acetylchitooligosaccharides as the main carbon source for the growth of intestinal bacteria. FEMS Microbiol. Lett. 209:53-56. [DOI] [PubMed] [Google Scholar]
- 9.Chung, C. T., S. L. Niemela, and R. H. Miller. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. USA 86:2172-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comstock, L. E., M. J. Coyne, A. O. Tzianabos, and D. L. Kasper. 1999. Interstrain variation of the polysaccharide B biosynthesis locus of Bacteroides fragilis: characterization of the region from strain 638R. J. Bacteriol. 181:6192-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comstock, L. E., M. J. Coyne, A. O. Tzianabos, A. Pantosti, A. B. Onderdonk, and D. L. Kasper. 1999. Analysis of a capsular polysaccharide biosynthesis locus of Bacteroides fragilis. Infect. Immun. 67:3525-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuchural, G. J., M. H. Malamy, and F. P. Tally. 1986. Beta-lactamase-mediated imipenem resistance in Bacteroides fragilis. Antimicrob. Agents Chemother. 30:645-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delvalle, J. A., and C. Asensio. 1978. Distribution of adenosine 5′-triphosphate (ATP)-dependent hexose kinases in microorganisms. BioSystems 10:265-282. [DOI] [PubMed] [Google Scholar]
- 14.Dorr, C., M. Zaparty, B. Tjaden, H. Brinkmann, and B. Seibers. 2003. The hexokinase of the hyperthermophile Thermoproteus tenax. J. Biol. Chem. 278:18744-18753. [DOI] [PubMed] [Google Scholar]
- 15.Fields, M. W., and J. B. Russell. 2001. The glucomannokinase of Prevotella bryantii B14 and its potential role in regulating beta-glucanase expression. Microbiology 147:1035-1043. [DOI] [PubMed] [Google Scholar]
- 16.Fields, M. W., and J. B. Russell. 2002. The glucomannokinase of the gram-negative ruminal bacterium Prevotella bryantii B14 and its sequence conservation with regulatory glucokinases of gram-positive bacteria. Anaerobe 8:69-74. [Google Scholar]
- 17.Godoy, V. G., M. M. Dallas, T. A. Russo, and M. H. Malamy. 1993. A role for Bacteroides fragilis neuraminidase in bacterial growth in two model systems. Infect. Immun. 61:4415-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen, T., B. Reichstein, R. Schmid, and P. Schonheit. 2002. The first archaeal ATP-dependent glucokinase, from the hyperthermophilic crenarchaeon Aeropyrum pernix, represents a monomeric, extremely thermophilic rok glucokinase with broad hexose specificity. J. Bacteriol. 184:5955-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, T., and P. Schonheit. 2003. ATP-dependent glucokinase from the hyperthermophilic bacterium Thermotoga maritima represents an extremely thermophilic ROK glucokinase with high substrate specificity. FEMS Microbiol. Lett. 226:405-411. [DOI] [PubMed] [Google Scholar]
- 20.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed., vol. 1. Williams and Wilkins, Baltimore, Md.
- 21.Holtje, J.-V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus in Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 23.Hylemon, P. B., J. L. Young, R. F. Roadcap, and J. P. V. Phibbs. 1977. Uptake and incorporation of glucose and mannose by whole cells of Bacteroides thetaiotaomicron. Appl. Environ. Microbiol. 34:488-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi, M. D., and V. Jagannathan. 1966. Hexokinase I. Brain. Methods Enzymol. 9:371-376. [Google Scholar]
- 25.Kalka-Moll, W. M., Y. Wang, L. E. Comstock, S. E. Gonzalez, A. O. Tzianabos, and D. L. Kasper. 2001. Immunochemical and biological characterization of three capsular polysaccharides from a single Bacteroides fragilis strain. Infect. Immun. 69:2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macy, J., I. Probst, and G. Gottschalk. 1975. Evidence for cytochrome involvement in fumarate reduction and adenosine 5′-triphosphate synthesis by Bacteroides fragilis grown in the presence of hemin. J. Bacteriol. 123:436-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mesak, L. R., F. M. Mesak, and M. K. Dahl. 2004. Bacillus subtilis GlcK activity requires cystienes within a motif that discriminates microbial glucokinases into two lineages. BMC Microbiology. 4:6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer, D., C. Schneider-Fresenius, R. Horlacher, R. Peist, and W. Boos. 1997. Molecular characterization of glucokinase from Escherichia coli K12. J. Bacteriol. 179:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reissig, J. L., J. L. Strominger, and L. F. LeLoir. 1955. A modified method for the estimation of N-acetylamino sugars. J. Biol. Chem. 217:959-966. [PubMed] [Google Scholar]
- 33.Skarlatos, P., and M. K. Dahl. 1998. The glucose kinase of Bacillus subtilis. J. Bacteriol. 180:3222-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanley, P. 1984. Glycosylation mutants of animal cells. Annu. Rev. Genet. 18:525-552. [DOI] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 2002. Phylogenetic analysis using parsimony and other methods. Sinauer, Sunderland, Md.
- 36.Tally, F. P., D. R. Snydman, M. J. Shimell, and M. H. Malamy. 1982. Characterization of pBFTM10, a clindamycin-erythromycin resistance transfer factor from Bacteroides fragilis. J. Bacteriol. 151:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, Y. P., and M. H. Malamy. 2000. Isolation of Bacteroides fragilis mutants with in vivo growth defects by using Tn4400′, a modified Tn4400 transposition system, and a new screening method. Infect. Immun. 68:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson, J. S., and M. H. Malamy. 1990. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus beta-lactamase II. J. Bacteriol. 172:2584-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Titgemeyer, F., J. Reizer, A. Reizer, and H. Saier. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349-2354. [DOI] [PubMed] [Google Scholar]
- 40.Uehara, T., and J. T. Park. 2004. The N-acetyl-d-glucosamine kinase of Escherichia coli and its role in murein recycling. J. Bacteriol. 186:7273-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varel, V. H., and M. P. Bryant. 1974. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl. Microbiol. 18:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vimr, E. R., and F. A. Troy. 1985. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 164:845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, E., S. Marcandier, O. Egeter, J. Deutscher, F. Gotz, and R. Bruckner. 1995. Glucose kinase-dependent catabolite repression in Staphylococcus xylosus. J. Bacteriol. 177:6144-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Widener, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 46.Yarema, K. J., and C. R. Bertozzi. 2001. Characterizing glycosylation pathways. Genome Biol. 2:0004.1-0004.10. [DOI] [PMC free article] [PubMed] [Google Scholar]