Abstract
The expression of extracellular virulence factors in various species of the Bacillus cereus group is controlled by the plcR and papR genes, which encode a transcriptional regulator and a cell-cell signaling peptide, respectively. A processed form of PapR, presumably a pentapeptide, specifically interacts with PlcR to facilitate its binding to its DNA targets. This activating mechanism is strain specific, with this specificity being determined by the first residue of the pentapeptide. We carried out in vivo complementation assays and compared the PlcR-PapR sequences of 29 strains from the B. cereus group. Our findings suggested that the fifth amino acid of the pentapeptide is also involved in the specificity of activation. We identified four classes of PlcR-PapR pairs, defining four distinct pherotypes in the B. cereus group. We used these findings to look at the evolution of the PlcR-PapR quorum-sensing system with regard to the phylogeny of the species forming the B. cereus group.
The Bacillus cereus group includes six highly related species of gram-positive, spore-forming, AT-rich bacteria: Bacillus anthracis, Bacillus cereus (sensu stricto), Bacillus mycoides, Bacillus pseudomycoides, Bacillus thuringiensis, and Bacillus weihenstephanensis (15, 18, 21). These species were originally identified on the basis of phenotypic traits, such as the production of a capsule and toxins (B. anthracis), the rhyzoid morphology of the colonies (B. mycoides and B. pseudomycoides), the production of parasporal inclusions (B. thuringiensis), and psychrotolerance (B. weihenstephanensis). The B. cereus species (sensu stricto) comprises all bacteria belonging to the B. cereus group that do not belong in any of the other species due to the absence of any distinctive traits. This phenotypic species definition may appear highly subjective and poorly rigorous. The limitations of this definition are highlighted by the example of B. thuringiensis, in which the genes encoding parasporal inclusions are harbored on conjugative plasmids (24). This means that any B. thuringiensis strains lacking such plasmids will be identified as B. cereus. Conversely, a B. cereus strain that receives this plasmid by conjugation becomes a B. thuringiensis strain.
The objective definition of the species forming the B. cereus group requires the analysis of a large set of genes and the characterization of specific chromosomal markers. Several molecular and biochemical methods have been used to examine the diversity of the strains composing the B. cereus group. DNA-DNA hybridization methods do not generally make it possible to distinguish precisely between the six species belonging to the B. cereus group (18). Analysis of amplified fragment length polymorphisms (AFLP) shows that B. anthracis is a monomorphic species (12) but that B. thuringiensis and B. cereus exhibit a high degree of variability (7, 10). Multilocus enzyme electrophoresis (MEE), sequence analysis of housekeeping genes, and multilocus sequence typing (MLST) suggest that these three species are highly related and genetically indistinguishable (5, 6). This result is strengthened by the comparison of the nucleotide sequences of B. anthracis strain Ames, B. cereus ATCC 10987, and B. cereus strain ATCC 14579, showing that about 75% of the genes are common to these three strains and that B. cereus ATCC 10987 is closer to B. anthracis than to B. cereus strain ATCC 14579 (9, 22, 23). However, genetic analysis of sympatric populations has shown that the B. thuringiensis and B. cereus populations are clustered in distinct groups, suggesting limited genetic exchanges between these two species (28).
The expression of various chromosomal genes encoding extracellular factors (i.e., phospholipases C, proteases, cell wall proteins, enterotoxins and hemolysins) is activated by a pleiotropic regulator, PlcR, that is specific to the B. cereus group (1, 16, 19). The activity of PlcR depends on the presence of PapR, a small signaling peptide that acts as a quorum-sensing effector (25). PapR is exported by the bacterial cell, processed, presumably as a pentapeptide, and then reimported into the cell, where it interacts with PlcR to facilitate its binding to its DNA targets. This activating mechanism is strain specific, and this specificity is determined by the first residue of the pentapeptide (25). Alignment of PapR sequences from various strains and PlcR activation assays led to the identification of three specificity groups: those with a PapR pentapeptide with a leucine, a valine, or a methionine as first residue. Here, we describe a fourth class of pentapeptides within the B. cereus group, those with a leucine at the first residue and a histidine at the fifth position instead of an aromatic residue (a phenylalanine or a tyrosine) as in the other groups. In addition, we show that there is a perfect correlation between the specificities of the pentapeptides and the peptide sequences of the corresponding PlcR regulators. These results suggest that the B. cereus group can be divided into four classes based on the sequence and the specificity of the PlcR-PapR pair. Thus, these two genes might be used, in association with other genetic markers, to provide new insight into the different species forming the B. cereus group.
Interspecies activation of the PlcR regulon within the B. cereus group.
We studied the PlcR-PapR specificities of strains belonging to the five major species of the B. cereus group. In addition to the 18 strains already assayed (25), we determined the ability of 11 new strains to activate the expression of the PlcR regulon in a reporter strain: the B. cereus type strain ATCC 14579, B. cereus strain ATCC 10987, B. cereus strain 569 (laboratory stock), two B. cereus environmental strains (Bc90 and Bcc; Institute National de la Recherche Agronomique [INRA] collection), two B. thuringiensis strains from the Institut Pasteur collection (B. thuringiensis serovar canadensis serotype 5 and B. thuringiensis serovar roskoldiensis serotype 45), three environmental B. thuringiensis strains from INRA collection (Bt12, Bt44, and Bt51), and the B. weihenstephanensis type strain. PlcR is highly representative of the B. cereus group, and the plcR gene was found in all the strains we have tested. However, to avoid bias due to the lack of PlcR production, we selected only strains that displayed hemolytic activity and/or lecithinase activity, both of which depend on PlcR-regulated proteins. Indeed, it was shown that about 1% of the strains of the B. cereus group produced an inactive PlcR protein (26). Nevertheless, an exception was made for B. anthracis, which does not produce a functional PlcR protein (1, 17).
We performed extracellular complementation assays on Luria-Bertani (LB) agar plates supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 100 μg · ml−1) using the B. thuringiensis 407 Cry− (ΔpapR plcA′-lacZ) mutant as the reporter strain, as previously described (25). This strain carries a chromosomal transcriptional fusion between the plcA promoter and the lacZ gene. As plcA belongs to the PlcR regulon, lacZ expression directly reflects PlcR activity in the cell. Thus, a strain producing a diffusible PapR peptide specifically able to activate the PlcR regulator of B. thuringiensis 407 Cry− (ΔpapR plcA′-lacZ) is able to restore β-galactosidase production, and the complemented colonies are blue on plates supplemented with X-Gal. Figure 1 shows the results obtained after streaking a sample (see the legend) of the 29 strains near the reporter strain. Only B. thuringiensis 407 Cry− and B. thuringiensis serotype 14 were able to restore expression of the plcA′-lacZ transcriptional fusion in the papR mutant strain. Thus, these are the only strains of the nine strains on this plate that produce the specific peptide needed for PlcR activation in the reporter strain. The results obtained with the 29 Bacillus strains are presented in Fig. 2. Twelve of the 29 strains activated the expression of the PlcR regulon in the papR mutant. These results show that the ability of a strain to restore expression of the PlcR regulon in the papR mutant strain does not depend on the species to which the strain belongs.
FIG. 1.
Intercellular activation of the PlcR regulon in bacteria of the B. cereus group. The indicated numbers refer to the following strains: B. thuringiensis serovar thuringiensis strain 407 Cry− (serotype 1) (1), B. thuringiensis serovar kurstaki strain HD1 (serotype 3) (2), B. thuringiensis serovar entomocidus type strain (serotype 6) (3), B. thuringiensis serovar aizawai type strain (serotype 7) (4), B. thuringiensis serovar thompsoni type strain (serotype 12) (5), B. thuringiensis serovar israelensis type strain (serotype 14) (6), B. thuringiensis serovar roskoldiensis (serotype 45) (7), B. cereus strain 569 (8), B. thuringiensis Bt51 (9), and B. thuringiensis strain 407 Cry− (plcA′-lacZ) ΔpapR (10), used as reporter strain for the detection of PlcR activity (blue phenotype). The bacterial cells were grown on LB plates containing X-Gal for 24 h at 37°C.
FIG. 2.
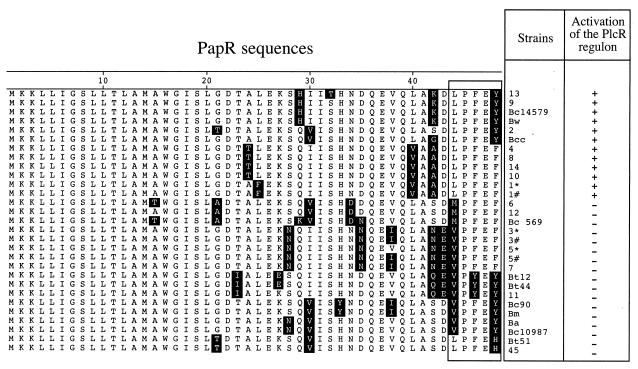
Comparison of the PapR peptide sequences of various B. weihenstephanensis, B. thuringiensis, B. mycoides, B. cereus, and B. anthracis strains. Numbers 2, 4, 6, 7, 8, 9, 10, 11, 12, 13, 14, and 45 refer to the serotypes of the B. thuringiensis strains as defined by Lecadet and colleagues (14). The type strains of these serotypes were used in this study. 1* and 1#, B. thuringiensis serotype 1 strain 407 Cry− and strain Berliner 1715, respectively; 3* and 3#, B. thuringiensis serotype 3 strains kurstaki HD1 and kurstaki KT0, respectively; 5* and 5#, B. thuringiensis serotype 5 serovars canadensis and galleriae, respectively; Bc14579, B. cereus ATCC 14579; Bc10987, B. cereus ATCC 10987; Bw, B. weihenstephanensis (type strain); Bcc and Bc90, B. cereus environmental strains; Bc 569, B. cereus strain 569; Bt12, Bt44, and Bt51, B. thuringiensis environmental strains; Bm, B. mycoides; Ba, B. anthracis strain Ames. The sequences of PlcR-PapR from B. anthracis and from B. cereus strains ATCC 14579, ATCC 10987, and 569 were determined from published data (9, 20, 22, 23). PlcR-PapR sequences from B. thuringiensis serotypes 1, 2, 3 (kurstaki KT0), 4, 5 (serovar galleriae), 6, 7, 8, 9, 10, 11, 12, 13, 14, and 45 have been described previously, as well as PapR from B. mycoides (25, 26). The other PlcR-PapR sequences were determined in this study, as indicated in the text. The symbol “+” indicates that the strain was able to activate the PlcR regulon in the reporter strain B. thuringiensis 407 Cry− (plcA′-lacZ) ΔpapR, as described in the legend to Fig. 1. The symbol “-” indicates that the strain was unable to activate the PlcR regulon.
Identification of four PapR pentapeptide classes on the basis of their peptide sequence.
We analyzed the PapR sequences of the additional 11 strains. The origins of these new PapR sequences are given in the legend of Fig. 2. The DNA regions including the unknown papR genes were amplified and sequenced using specific primers as previously described (25, 26). The 11 deduced PapR peptide sequences were aligned with the 18 sequences already available (Fig. 2). Amino acid variations were found throughout the sequence. However, we previously showed that the minimal fragment required for an active PapR is a pentapeptide corresponding to the last 5 carboxy-terminal amino acids, with the first residue determining the specificity. In the 29 PapR sequences analyzed in this study, the pentapeptides were distributed in the three previously described classes (with a leucine, methionine, or valine at position 1 of the pentapeptide) plus a fourth class. The pentapeptides within this new group possessed a leucine in the first position and a histidine in position 5, instead of a tyrosine or a phenylalanine as found in the three previously described classes. The strains carrying papR genes encoding pentapeptides with a leucine as first residue and a tyrosine or a phenylalanine at position 5 of the pentapeptide were able to activate PlcR in the 407 Cry− (ΔpapR plcA′-lacZ) mutant strain (Fig. 2). In contrast, the PlcR of this strain was not activated by strains carrying papR genes with the following pentapeptides: MPFEF, VP(F/Y)E(F/Y), and LPFEH.
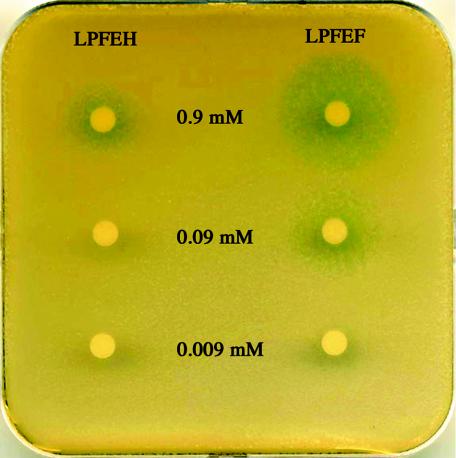
Previous in vivo and in vitro analyses showed that pentapeptides starting with a methionine or a valine are unable to activate the PlcR protein of strain 407 as effectively as those carrying a leucine in the first position (25). However, the inefficiency of pentapeptides ending in a histidine was not demonstrated. The inability of the two B. thuringiensis strains (serotype 45 and Bt51) to activate the PlcR protein of strain 407 might be due to the specificity of the pentapeptide LPFEH or to the lack of production of the PapR peptide in these two strains. Synthetic pentapeptides (LPFEF and LPFEH) were used to determine whether a histidine in the fifth position of the pentapeptide affects PlcR activation in the 407 Cry− (ΔpapR plcA′-lacZ) mutant strain (Fig. 3). A stationary-phase ΔpapR culture (optical density at 600 nm, 9) was diluted 10 times in LB medium, and 5 ml of this dilution were spread onto an LB plate containing X-Gal (100 μg · ml−1). The liquid medium remaining on the plate was removed, and the plate was dried. Ten microliters of each peptide at various concentrations was loaded onto Whatman paper disks and placed on the plate. The plate was then incubated at 37°C for 24 h. A blue coloration near the paper disk indicated the activation of the transcriptional plcA′-lacZ fusion in the reporter strain. We found that LPFEF was at least 10-fold more efficient than LPFEH at activating PlcR in the 407 strain. Indeed, no induction of the transcriptional fusion was detected when LPFEH was loaded onto the disks at a concentration of 0.009 or 0.09 mM, whereas a blue coloration (diameters of diffusion, 1 and 2.5 cm, respectively) was detected when LPFEF was loaded. Induction was detected when LPFEH was used at a concentration of 0.9 mM, but the blue coloration was less widespread than with the same concentration of LPFEF (1.7- and 3-cm diameters, respectively). This result indicates that the last residue of the pentapeptide is important for activity and might be involved in the specificity of the interaction between PapR and PlcR. The low but positive cross-reactivity of the pentapeptide LPFEH (at a concentration of 0.9 mM) with PlcR of strain 407 has been previously observed with the other heterologous pentapeptides, VPFEF and MPFEF (25). Moreover, it was shown that the heterologous pentapeptides had no antagonist effect on the expression of the PlcR regulon in strain 407. This finding is in contrast with the inhibitory interactions reported for the Staphylococcus aureus Agr system (11).
FIG. 3.
Pentapeptide LPFEH is less active than pentapeptide LPFEF towards PlcR from the reporter strain B. thuringiensis 407 (plcA′-lacZ) ΔpapR. The sequence and the concentration of each peptide loaded on the paper disks are indicated. B. thuringiensis 407 (plcA′-lacZ) ΔpapR cells were spread on LB plates containing X-Gal. The plate was then incubated at 37°C for 24 h.
The PapR classes correlate with PlcR clusters.
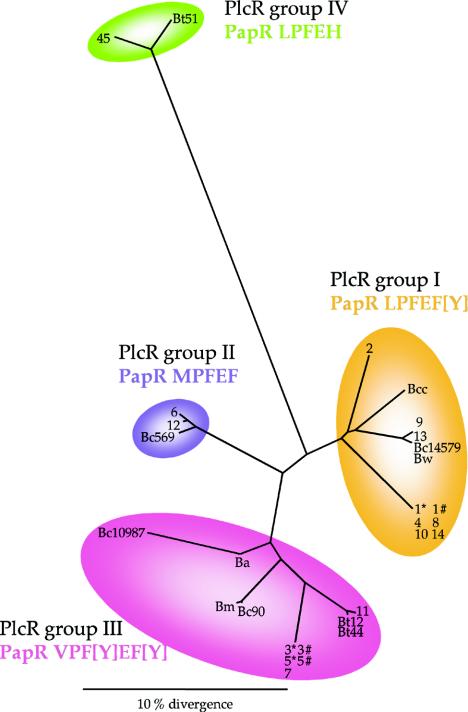
We then investigated whether the four distinct groups of PapR pentapeptides, LPFE(F/Y), LPFEH, MPFEF, and VP(F/Y)E(F/Y), were correlated with four distinct groups of PlcR proteins. We used ClustalX and Treeview to build a phylogenetic tree (Fig. 4) based on the deduced PlcR amino acid sequences of the 29 Bacillus strains (see the legend of Fig. 2 for the origins of the PlcR sequences). The sequences appeared to be clustered in four groups, with group IV being most distant from the three other PlcR groups. For example, PlcR from B. anthracis presents a 27.7% divergence from PlcR from Bt51 but just 12 and 10.7% divergences from PlcR from B. cereus ATCC 14579 and B. thuringiensis serotype 6, respectively. The PlcR sequences from the last two strains differ by 12.7%. We also studied the association between each PlcR group and the corresponding PapR pentapeptide found in the bacteria falling in this group (Fig. 4). Each PapR pentapeptide class corresponds to a PlcR group, as represented by the colored areas. PlcR groups I, II, and III are associated with PapR pentapeptides LPFE(F/Y), VP(F/Y)E(F/Y), and MPFEF, respectively. PlcR group IV is associated with the new PapR class, LPFEH. A perspective of this work would be to demonstrate experimentally that each PapR class activates the PlcR regulator belonging to the corresponding class, with the ultimate aim of identifying the PlcR regions involved in the specific interaction with PapR.
FIG. 4.
PlcR peptide sequences can be divided into four clusters to which the four PapR classes correspond. This phylogenic tree was built on the basis of PlcR peptide sequences from strains belonging to the PlcR group, using the ClustalX and Treeview programs. The sequences were determined and the abbreviations used are as described in the legend of Fig. 2. The colors designate individual PlcR clusters with which a PapR class is associated: orange indicates PapR LPFE(F/Y), pink indicates PapR VP(F/Y)E(F/Y), purple indicates PapR MPFEF, and green indicates PapR LPFEH.
Evolution of the PlcR-PapR system in B. cereus.
Our results reveal the polymorphism of the PlcR-PapR quorum-sensing system within the B. cereus group and show a phylogenic relationship between the regulator (PlcR) and its signaling peptide (PapR). These findings strongly suggest that PlcR and PapR coevolved, as was expected from their mode of action, which appears to involve protein-protein interaction (25). Polymorphisms in the quorum-sensing systems and coevolution of pheromones and their receptor proteins have already been observed in other gram-positive bacteria. The competence quorum-sensing systems in B. subtilis and Streptococcus species and the Agr virulence system in Staphylococcus species exhibit similar characteristics (2-4, 11, 27). The polymorphism of these genetic systems and the existence of specificity groups within a species or a genus define pherotypes based on the pheromone-receptor association. Thus, the four PlcR-PapR pherotypes might enable us to classify better the various strains of the B. cereus group. Comparison of these pherotypes with the phylogenetic trees established by using MEE (5), MLST (6), and AFLP (7) analyses gives both common and divergent results. In agreement with the MEE and MLST analyses showing that B. anthracis and B. cereus ATCC 10987 are closely related, we found that these two strains belong to the same pherotype (PlcR group III). Our finding that B. thuringiensis 407, B. thuringiensis (serotype 4), and B. cereus ATCC 14579 belong to PlcR group I is also consistent with the results of MEE and MLST analyses, which showed that these three strains are highly related. Similarly, AFLP analysis showed that B. thuringiensis serotypes 3, 5, and 7 are clustered in PlcR group III and are closely related. However, several results are divergent. For example, MEE, MLST, and AFLP analyses showed that the B. thuringiensis serotype 6 strain (type strain) clustered with the B. thuringiensis serotype 3 strain (type strain). However, we found that these two strains have different pherotypes (PlcR group II and PlcR group III, respectively). MLST analysis showed that B. weihenstephanensis (type strain) was in a group different from that of B. thuringiensis strain 407 and B. cereus ATCC 14579. In contrast, the three strains have the same pherotype (PlcR group I).
A B. cereus strain (G9241) possessing the anthrax toxin genes and a functional PlcR regulon was recently isolated and characterized (8). Based on its sequence, the PlcR and PapR peptide sequences of this B. cereus strain are identical to those of B. thuringiensis serovar roskoldiensis (serotype 45). Thus, the PlcR and PapR proteins of B. cereus G9241 belong to PlcR group IV. In contrast, MLST analysis based on seven housekeeping genes indicated that this B. cereus strain is closely related to B. anthracis and B. cereus ATCC 10987 (8). As the PlcR and PapR peptides of B. anthracis and B. cereus ATCC 10987 belong to PlcR group I, this result shows the lack of congruence between the evolution of housekeeping genes and the evolution of the PlcR-PapR system. Unfortunately, no data concerning the phylogeny of B. thuringiensis strain Bt 51 or strains belonging to serovar roskoldiensis are available. It would be interesting to know whether analysis of housekeeping genes reveals that these B. thuringiensis strains are related to B. cereus G9241.
Conclusion.
This study demonstrates the existence of four classes of PlcR-PapR couples, defining four distinct pherotypes in the B. cereus group. This result reveals the polymorphism of the PlcR-PapR quorum-sensing system in B. cereus and suggests a coevolution of PlcR and PapR. In agreement with phylogenetic analyses based on housekeeping genes, the four PlcR-PapR pherotypes emphasize the weakness of the present B. cereus group classification system. Both analyses clearly show that strains belonging to a given species (e.g., B. thuringiensis) may be more related to strains belonging to another species (e.g., B. cereus or B. weihenstephanensis) than to strains from the same species. The pherotypes deduced from the PlcR-PapR specificities might improve our understanding of the relationships between the various members of the B. cereus group. However, it does not appear possible to establish a strict correlation between the four pherotypes that we have defined in this study and the phylogenetic trees based on the analysis of housekeeping genes. This conclusion is in agreement with the absence of a phylogenetic relation between plcR and housekeeping genes, as indicated by a recent MLST study (13). The lack of congruence between pherotypes and phylogenetic classifications has previously been observed for the competence quorum-sensing system in B. subtilis (27) and for the Agr system in Staphylococcus species (3). It is generally suggested that the evolutionary force driving the hypervariability and the coevolution of the quorum-sensing components is the accumulation of point mutations or recombinational exchanges in an element of the genetic system followed by a rigorous positive selection of compensatory mutations in the second element (2-4, 27). The divergence between the pherotype and phylogenetic classifications might be due to the efficient and specific horizontal transfer of the quorum-sensing genes. It is now necessary to characterize and to analyze the precise PlcR regions involved in the specific interaction with PapR to investigate the evolution of this quorum-sensing system.
Nucleotide sequence accession numbers.
The GenBank accession numbers of PlcR sequences from B. weihenstephanensis, Bcc, B. mycoides, B. thuringiensis serotype 3 strain kurstaki HD1, Bt12, Bt44, and Bc90 are AY776139 to AY776145, respectively. Accession numbers for the PapR sequences from B. weihenstephanensis, Bcc, B. thuringiensis serotype 3 strain kurstaki HD1, Bt12, Bt44, and Bc90 are AY776146 to AY776153, respectively.
Acknowledgments
We acknowledge Anne-Brit Kolstø and Ole Andreas Økstad for the gift of B. cereus ATCC 10987, Josette Chaufaux for the gift of B. thuringiensis and B. cereus strains from the INRA collection, and Jean-François Charles for the gift of B. thuringiensis strains from the Institut Pasteur collection. We are grateful to Myriam Gominet for her everyday help. We thank Denis Bourguet, Michel Gohar, and Vincent Sanchis for helpful discussions and critical reading of the manuscript.
This work was supported by the Institut National de la Recherche Agronomique, the Institut Pasteur, and the Centre National de la Recherche Scientifique. L. Slamti received a Ph.D. grant from the Ministère de la Recherche and a Pasteur-Weizman fellowship.
REFERENCES
- 1.Agaisse, H., M. Gominet, O. A. Økstad, A. B. Kolstø, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 2.Ansaldi, M., and D. Dubnau. 2004. Diversifying selection at the Bacillus quorum-sensing locus and determinants of modification specificity during synthesis of the ComX pheromone. J. Bacteriol. 186:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dufour, P., S. Jarraud, F. Vandenesch, T. Greenland, R. P. Novick, M. Bes, J. Etienne, and G. Lina. 2002. High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 184:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Håvarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helgason, E., N. J. Tourasse, R. Meisal, D. A. Caugant, and A. B. Kolstø. 2004. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl. Environ. Microbiol. 70:191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill, K. K., L. O. Ticknor, R. T. Okinaka, M. Asay, H. Blair, K. A. Bliss, M. Laker, P. E. Pardington, A. P. Richardson, M. Tonks, D. J. Beecher, J. D. Kemp, A. B. Kolstø, A. C. Wong, P. Keim, and P. J. Jackson. 2004. Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 70:1068-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. Maiden, F. G. Priest, M. Barker, L. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. D. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 10.Jackson, P. J., K. K. Hill, M. T. Laker, L. O. Ticknor, and P. Keim. 1999. Genetic comparison of Bacillus anthracis and its close relatives using amplified fragment length polymorphism and polymerase chain reaction analysis. J. Appl. Microbiol. 87:263-269. [DOI] [PubMed] [Google Scholar]
- 11.Ji, G., R. C. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 12.Keim, P., A. Kalif, J. Schupp, K. Hill, S. E. Travis, K. Richmond, D. M. Adair, M. Hugh-Jones, C. R. Kuske, and P. Jackson. 1997. Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J. Bacteriol. 179:818-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko, K. S., J. W. Kim, J. M. Kim, W. Kim, S. I. Chung, I. J. Kim, and Y. H. Kook. 2004. Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR gene. Infect. Immun. 72:5253-5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecadet, M. M., E. Frachon, V. C. Dumanoir, H. Ripouteau, S. Hamon, P. Laurent, and I. Thiery. 1999. Updating the H-antigen classification of Bacillus thuringiensis. J. Appl. Microbiol. 86:660-672. [DOI] [PubMed] [Google Scholar]
- 15.Lechner, S., R. Mayr, K. P. Francis, B. M. Pruss, T. Kaplan, E. Wiessner-Gunkel, G. S. Stewart, and S. Scherer. 1998. Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int. J. Syst. Bacteriol. 48:1373-1382. [DOI] [PubMed] [Google Scholar]
- 16.Lereclus, D., H. Agaisse, M. Gominet, S. Salamitou, and V. Sanchis. 1996. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 178:2749-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mignot, T., M. Mock, D. Robichon, A. Landier, D. Lereclus, and A. Fouet. 2001. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol. Microbiol. 42:1189-1198. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura, L. K. 1994. DNA relatedness among Bacillus thuringiensis serovars. Int. J. Syst. Bacteriol. 44:125-129. [DOI] [PubMed] [Google Scholar]
- 19.Økstad, O. A., M. Gominet, B. Purnelle, M. Rose, D. Lereclus, and A.-B. Kolstø. 1999. Sequence analysis of three Bacillus cereus loci under PlcR virulence gene regulator control. Microbiology 145:3129-3138. [DOI] [PubMed] [Google Scholar]
- 20.Pomerantsev, A. P., K. V. Kalnin, M. Osorio, and S. H. Leppla. 2003. Phosphatidylcholine-specific phospholipase C and sphingomyelinase activities in bacteria of the Bacillus cereus group. Infect. Immun. 71:6591-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Priest, F. G., D. A. Kaji, Y. B. Rosato, and V. P. Canhos. 1994. Characterization of Bacillus thuringiensis and related bacteria by ribosomal RNA gene restriction fragment length polymorphisms. Microbiology 140:1015-1022. [DOI] [PubMed] [Google Scholar]
- 22.Rasko, D. A., J. Ravel, O. A. Økstad, E. Helgason, R. Z. Cer, L. Jiang, K. A. Shores, D. E. Fouts, N. J. Tourasse, S. V. Angiuoli, J. Kolonay, W. C. Nelson, A.-B. Kolstø, C. M. Fraser, and T. D. Read. 2004. The genome sequence of Bacillus cereus ATCC 10987 reveals metabolic adaptations and a large plasmid related to Bacillus anthracis pXO1. Nucleic Acids Res. 32:977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Økstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolstø, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 24.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slamti, L., S. Perchat, M. Gominet, G. Vilas-Bôas, A. Fouet, M. Mock, V. Sanchis, J. Chaufaux, M. Gohar, and D. Lereclus. 2004. Distinct mutations in PlcR explain why some strains of the Bacillus cereus group are nonhemolytic. J. Bacteriol. 186:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortosa, P., L. Logsdon, B. Kraigher, Y. Itoh, I. Mandic-Mulec, and D. Dubnau. 2001. Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J. Bacteriol. 183:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilas-Boas, G., V. Sanchis, D. Lereclus, M. V. Lemos, and D. Bourguet. 2002. Genetic differentiation between sympatric populations of Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 68:1414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]