Abstract
The Ll.LtrB group II intron from the low-G+C gram-positive bacterium Lactococcus lactis was the first bacterial group II intron shown to splice and mobilize in vivo. This retroelement interrupts the relaxase gene (ltrB) of three L. lactis conjugative elements: plasmids pRS01 and pAH90 and the chromosomal sex factor. Conjugative transfer of a plasmid harboring a segment of the pRS01 conjugative plasmid including the Ll.LtrB intron allows dissemination of Ll.LtrB among L. lactis strains and lateral transfer of this retroelement from L. lactis to Enterococcus faecalis. Here we report the dissemination of the Ll.LtrB group II intron among L. lactis strains following conjugative transfer of the native chromosomally embedded L. lactis sex factor. We demonstrated that Ll.LtrB dissemination is highly variable and often more efficient from this integrative and conjugative element than from an engineered conjugative plasmid. Cotransfer among L. lactis strains of both Ll.LtrB-containing elements, the conjugative plasmid and the sex factor, was detected and shown to be synergistic. Moreover, following their concurrent transfer, both mobilizable elements supported the spread of their respective copies of the Ll.LtrB intron. Our findings explain the unusually high efficiency of Ll.LtrB mobility observed following conjugation of intron-containing plasmids.
Group II introns are large ribozymes that splice autocatalytically from their pre-mRNAs (1, 14, 17, 22). Some self-splicing group II introns are also mobile retroelements that invade new DNA sites in a duplicative process using an RNA intermediate, like retrotransposons and retroviruses (1). They can reinsert either in cognate intronless alleles (homing site [HS]) by retrohoming or in nonhomologous sites by retrotransposition (1, 5, 6, 11, 12). Mobile group II introns harbor a multifunctional open reading frame (ORF) that is directly involved in their mobility processes (1). Group II introns are found in eubacteria (7, 15), archaea (8, 25), and eukaryotic organelles derived from bacteria such as fungal and plant mitochondria and plant chloroplasts (1, 14, 17, 22). Horizontal transfer of group II introns between organisms is a well-accepted model of intron dissemination and evolution. This model suggests that group II introns are not only mobile within cells but can also be transferred between species, where they can invade new sites (3, 14).
The Lactococcus lactis LtrB group II intron (Ll.LtrB) is the first bacterial group II intron that was shown to splice and mobilize in vivo (18, 19, 23). L. lactis, an industrially important low-G+C gram-positive bacterium, is extensively used in the dairy industry. Ll.LtrB mobility via the retrohoming (5) and retrotransposition pathways (6, 11, 12) was studied in both L. lactis and Escherichia coli. The Ll.LtrB group II intron (2.5 kb) harbors an ORF called LtrA (599 amino acids) that exhibits reverse transcriptase, endonuclease, and RNA maturase activities (16). These three functions are essential for retrohoming of Ll.LtrB to intronless alleles (5). Following translation, LtrA binds to its harboring intron within the pre-mRNA as a dimer (21). The maturase function of LtrA promotes splicing of Ll.LtrB and concurrent ligation of its flanking exons. Following intron excision, the LtrA dimer remains bound to the intron RNA lariat as a ribonucleoprotein particle (RNP) (intron RNA lariat plus two LtrA proteins). Upon recognition of the homing site by these RNPs, the intron RNA reverse splices into the sense strand of its double-stranded DNA target. The antisense strand is then nicked nine nucleotides downstream from the intron insertion site by the endonuclease domain of LtrA. Using the 3′ end generated by endonuclease cleavage as a primer, LtrA reverse transcribes the intron RNA by a process called target-primed reverse transcription (TPRT). The final steps of the Ll.LtrB retrohoming pathway are thought to be carried out by host DNA repair mechanisms independent of the RecA-dependent homologous recombination pathway (5, 19).
The Ll.LtrB group II intron interrupts the relaxase gene (ltrB) that is present in three L. lactis mobilizable elements: two conjugative plasmids, pRS01 (48.4 kb) (18) and pAH90 (26.5 kb) (20), and the chromosomally embedded sex factor (50 kb) (23). The relaxase enzyme functions by nicking the plasmid at its origin of transfer (oriT) to initiate conjugation; hence, splicing of the Ll.LtrB intron is essential for relaxase production and plasmid transfer (3, 13, 18, 27). The pRS01 plasmid and the chromosomal sex factor are very similar and were probably derived from a common ancestor (4). They are considered integrative and conjugative elements (ICEs) because they excise by site-specific recombination into a circular form, self transfer by conjugation, and integrate into the host genome (4).
We previously demonstrated that transfer of an intron-harboring conjugative plasmid among different L. lactis strains and from L. lactis to Enterococcus faecalis supports intron dissemination and lateral transfer within the recipient cells (3). This plasmid contained the conjugative transfer regions Tra1-2 from pRS01 (7.5 kb) harboring the oriT, ltrE, and ltrB genes; the latter was interrupted by the Ll.LtrB intron. Following its transfer via plasmid-based conjugation, the Ll.LtrB intron was shown to invade either its recognition site (homing site [HS]) harbored on a resident plasmid by retrohoming or different nonhomologous sites present within the chromosome of the recipient cells by retrotransposition (3). While studying intron dissemination by plasmid conjugation, we consistently observed an unusually high efficiency of Ll.LtrB mobility in some analyzed isolates. Moreover, we observed very efficient Ll.LtrB mobility for some intron variants carrying mutations that inactivated any of the three catalytic activities of LtrA (reverse transcriptase, maturase, and endonuclease), although these LtrA functions were demonstrated to be essential for Ll.LtrB retrohoming (5).
Why were there unusually high levels of Ll.LtrB mobility products in some transconjugant isolates following conjugation of intron-carrying plasmids? Using genetic as well as conjugation/retrohoming assays, we describe the dissemination of the Ll.LtrB group II intron following transfer of the chromosomally embedded L. lactis sex factor between L. lactis strains. We show that (i) the chromosomal sex factor can be cotransferred along with a conjugative plasmid, leading to the dissemination of the Ll.LtrB intron it conveys; (ii) cotransfer of a conjugative plasmid and the sex factor is synergistic; (iii) Ll.LtrB dissemination is highly variable and often more efficient from the sex factor than from a vector carrying a portion of the pRS01 conjugative plasmid. This work reveals the nature and origin of the unusually high Ll.LtrB mobility efficiencies observed following conjugation of intron-containing plasmids (3).
MATERIALS AND METHODS
Strains and plasmids.
L. lactis strains [NZ9800, NZ9800ΔltrB::Tetr, and MMS372(recA)] were grown without shaking in M17 medium supplemented with 0.5% glucose (GM17) at 30°C. The chromosomes of the NZ9800 and NZ9800ΔltrB::Tetr strains contain a copy of the conjugative sex factor, while the MMS372 strain is sex factor free. E. coli strains (DH5α, DH10β), used for cloning and mobility scoring, were grown shaken at 37°C in Luria-Bertani (LB) broth. Milk plates used in conjugation assays were made of 5% dry milk (Carnation), 1% dextrose, and 1.5% agar. pLE12I plasmid consists of the Tra1-2 regions from pRS01 cloned into the pLE1 vector at its unique PstI site, where the td group I intron with portions of its exons was subsequently inserted into the Ll.LtrB intron downstream from LtrA (3, 5). pMNHS plasmid (pMN1343) contains a 271-bp homing site (exon 1, 179 bp; exon 2, 92 bp) (HindIII) inserted at the unique HindIII restriction site in the pDL278 vector (3, 5, 19). pMNHS-CR plasmid contains, in both exons, polymorphic sites that do not interfere with Ll.LtrB mobility (E1, −7 and −30/35; E2, +7 and +25). The modified homing site was isolated from pLHS-CR plasmid (XbaI) (5) and cloned blunt into the pDL278 vector (HindIII). Selective medium contained the following concentrations of antibiotics: chloramphenicol (Cam), 10 μg/ml; spectinomycin (Spc), 300 μg/ml; kanamycin (Kan), 20 μg/ml; tetracycline (Tet), 3 μg/ml.
Conjugation assays.
L. lactis strains (donor and recipient) were diluted (0.4 or 0.8 ml/10 ml) from cultures saturated overnight and were grown for 7 h at 30°C with appropriate antibiotics. Cells were recovered by centrifugation and the pellets were mixed (1:1), spread on milk plates containing DNase I and RNase I (100 U of each/ml) (3, 26), and incubated at 30°C for 12 h. Typical conjugation control experiments used only DNase I, but because the Ll.LtrB intron is a retroelement, we also controlled for possible nonconjugative uptake of active RNP particles (intron RNA lariat plus two LtrA proteins). Cell mixtures were recovered with 1× PBS (1 ml), and serial dilutions were made to score donor (Camr), recipient (Spcr), and transconjugant (Camr/Spcr) cells (3, 18). Conjugation efficiencies (three assays) were calculated as the number of transconjugants (Camr/Spcr)/donor cell (Camr). In sex factor conjugation assays, the identity of the recipient strain [MMS372(recA)] was confirmed by its resistance to Kan and its UV sensitivity.
Mobility assay (colony patch hybridization).
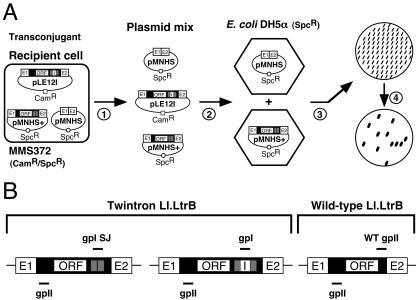
The plasmid mix (donor plasmids, recipient plasmids, and mobility products) from 10 independent transconjugant cells was prepared (same conjugation assay) and retransformed into E. coli (DH5α), and bacteria were plated on LB/Spc plates to select for cells containing recipient plasmids that were or were not interrupted by the intron (Fig. 1A). To calculate the intron mobility efficiency measured by the percentage of recipient plasmids that received the intron (mobility products), 100 isolated colonies (Spcr) were patched for each independent assay (LB/Spc plates). The patches were lifted on nylon membranes and hybridized with the appropriate 5′-end-labeled probe (32P) (Fig. 1B). The group II probe (gpII; 5′-CCGTGCTCTGTTCCCGTATCAGC-3′) (5′ end of the intron) (5) is general and recognizes the three types of mobility events (Fig. 1B). Three other probes (gpI SJ, gpI, and WT gpII) are specific, and each recognizes only one type of mobility event. The td group I intron splice junction probe (gpI SJ; 5′-ATTAAACGGTAGACCCAAGAAAAC-3′) (5) recognizes the two td ligated exons (12 nucleotides each) flanking it and gives a positive signal only when the group I intron is absent in Ll.LtrB mobility products (2, 3, 5, 6). This probe is used as a retromobility indicator. The group I probe (gpI; 5′-GGAGATATAGTCTGCTCTGCA-3′) hybridizes within the td group I intron and reveals mobility events still harboring it. The wild-type group II probe (WT gpII; 5′-AAACACAAGTGAATTTTTACGA-3′) spans the region where the td group I intron is inserted just downstream from the LtrA stop codon (engineered SalI site). We designed this probe to specifically recognize the wild-type Ll.LtrB intron and to not hybridize with either of the two twintron mobility products.
FIG. 1.
Mobility assay and probes. (A) Scoring of intron mobility efficiency after conjugation (colony patch hybridization). Plasmid mixes (donor, recipient, and mobility products) from 10 independent transconjugants were recovered (1) and retransformed in E. coli DH5α cells (LB/Spc) (2). One-hundred colonies were patched on LB/Spc plates (3) and transferred onto a nylon membrane that was then hybridized with an intron-specific probe (4) to calculate the ratio of recipient plasmids that received the intron. (B) Different probes used to identify the two categories of Ll.LtrB mobility products. The mobility products observed were generated either from the plasmid (twintron Ll.LtrB) or the sex factor (wild-type Ll.LtrB). The wild-type intron generates only one kind of mobility product, while the twintron potentially generates two types of mobility products, harboring or lacking the td intron. The group II probe (gpII) recognizes the three types of mobility products. The other three probes (gpI SJ, gpI, and WT gpII) are specific and recognize only one type of mobility product. The td group I intron splice junction probe (gpI SJ) recognizes the two td ligated exons (12 nucleotides for each exon) and gives a positive signal only upon group I intron loss. The group I probe (gpI) hybridizes within the td group I intron and reveals twintron mobility products still harboring it. The wild-type group II probe (WT gpII) recognizes wild-type Ll.LtrB mobility products and does not hybridize with either of the two twintron mobility products. Ll.LtrB group II intron, black; Ll.LtrB exons, E1 and E2; td intron, I; td exons, grey.
RESULTS
Unusually high efficiency of Ll.LtrB mobility among L. lactis strains is consistently observed following conjugation of intron-harboring plasmids. To study the variable efficiencies of Ll.LtrB mobility observed following its transfer by plasmid conjugation, we performed a typical conjugation assay between NZ9800/pLE12I (donor strain) and MMS372(recA)/pMNHS-CR (recipient strain) (Fig. 2A) (3). In this assay, the nonautonomously conjugative donor plasmid (pLE12I; Camr) carried an engineered Ll.LtrB intron harboring the td group I intron from phage T4 flanked by portions of its exons, while the recipient plasmid (pMNHS-CR; Spcr) contained the intron recognition site (HS) (Fig. 2A) (3, 5). Conjugation machinery is provided by the sex factor embedded in the chromosome of NZ9800 (3). After mating, the recipient cells that received the conjugative plasmid (transconjugant; MMS372(recA)/pMNHS-CR/pLE12I) were selected on Camr/Spcr plates, while the donor and recipient strains were isolated on Camr and Spcr plates, respectively. The presence of the td self-splicing intron within Ll.LtrB (twintron) allowed us to determine whether the intron invaded its homing site by using an RNA intermediate. The occurrence of mobility events missing the group I intron confirms that these products are generated using an Ll.LtrB RNA intermediate (3, 5, 6, 11, 12).
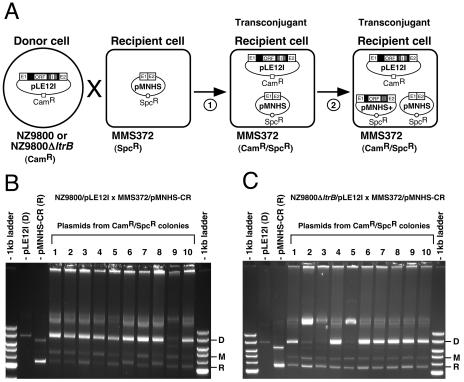
FIG. 2.
Conjugation/retrohoming assays. (A) Schematic of the assays. The first step (1) represents the transfer of pLE12I by conjugation from donor (NZ9800 or NZ9800ΔltrB) to recipient cell (MMS372). The second step (2) shows the invasion (retrohoming) of some homing sites (E1-E2) present on the recipient plasmid (pMNHS) by the Ll.LtrB intron expressed from donor plasmid (pLE12I). Intron-harboring recipient plasmids (pMNHS+) are represented in the transconjugant cell. Recipient plasmid used in these assays contained polymorphic sites in both exons (E1 and E2) (pMNHS-CR). Mobility products harboring the td intron are not depicted. Ll.LtrB group II intron, black; Ll.LtrB exons, E1 and E2; td intron, I; td exons, grey. (B and C) Agarose (0.5%) gels containing undigested plasmid mixes recovered from the progeny of 10 transconjugant cells (Spcr/Camr). Bands corresponding to donor plasmid (D), recipient plasmid (R), and mobility product (M) are highlighted.
Conjugation efficiency of the pLE12I plasmid was 5.2 × 10−3 ± 0.4 × 10−3 transconjugant per donor cell (three independent assays), as previously observed (4.8 × 10−3 ± 1.1 × 10−3) (3). To determine the mobility efficiency of the Ll.LtrB intron following plasmid conjugation, we isolated the plasmids (donor plasmids, recipient plasmids, and mobility products) from 10 independent transconjugant colonies (Fig. 2A). Analyses by agarose gel electrophoresis (Fig. 2B) showed, as previously observed (3), some variability in Ll.LtrB mobility efficiency among the different isolates. Indeed, intensity of bands corresponding to uninterrupted recipient plasmid (R; pMNHS-CR) and mobility product (M; pMNHS-CR plus intron) varied from lane to lane (Fig. 2B).
Efficiency of Ll.LtrB mobility was determined by colony patch hybridization assays for each of the 10 plasmid mixes isolated (Fig. 1A) (3). In these assays, E. coli DH5α cells were transformed with the plasmid mixes recovered from L. lactis transconjugant colonies and were plated on LB/Spc plates to select for cells containing recipient plasmids harboring or not harboring the intron. For each transconjugant event, 100 colonies were patched onto LB/Spc plates, transferred to nylon membranes, and hybridized with intron-specific 32P-labeled probe (gpII; Fig. 1B). Efficiency of Ll.LtrB mobility, defined as the ratio of Ll.LtrB-interrupted recipient plasmids to total recipient plasmids, varied from 13 to 70% for the 10 studied transconjugant events (Table 1, NZ/pLE12I × MMS/pMNHS-CR, gpII).
TABLE 1.
Efficiency of LI.LtrB mobility following its transfer by conjugationa
| Isolate no. | Mobility efficiency (%) after conjugation:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NZ/pLE12I × MMS/pMNHS-CR
|
NZΔltrB/pLE12I × MMS/pMNHS-CRb
|
NZ/pLE12I × MMS/pMNHS
|
NZ/pLE12I × MMS/pMNHSc
|
|||||||||||||
| gpII | gpI SJ | gpI SJ/gpII | gpI | WT gpII | gpII | gpI SJ | gpI SJ/gpII | gp I | gpII | WT gpII | gpII-WT gpII | gpII | gpI SJ | gpI SJ/gpII | WT gpII | |
| 1 | 61 | 25 | 41 | 0 | 36 | 13 | 13 | 100 | 0 | 95 | 91 | 4 | 63 | 16 | 25 | 47 |
| 2 | 51 | 17 | 33 | 0 | 34 | 12 | 12 | 100 | 0 | 88 | 79 | 9 | 34 | 34 | 100 | 0 |
| 3 | 21 | 21 | 100 | 0 | 0 | 12 | 12 | 100 | 0 | 98 | 98 | 0 | ||||
| 4 | 70 | 9 | 13 | 0 | 61 | 19 | 19 | 100 | 0 | 71 | 50 | 21 | ||||
| 5 | 25 | 25 | 100 | 0 | 0 | 24 | 24 | 100 | 0 | 98 | 59 | 39 | ||||
| 6 | 13 | 13 | 100 | 0 | 0 | 13 | 13 | 100 | 0 | 92 | 87 | 5 | ||||
| 7 | 51 | 51 | 100 | 0 | 0 | 14 | 14 | 100 | 0 | 93 | 85 | 8 | ||||
| 8 | 13 | 13 | 100 | 0 | 0 | 11 | 10 | 91 | 1 | 43 | 19 | 24 | ||||
| 9 | 43 | 43 | 100 | 0 | 0 | 6 | 6 | 100 | 0 | 98 | 98 | 0 | 86 | 29 | 34 | 57 |
| 10 | 19 | 18 | 95 | 1 | 0 | 16 | 16 | 100 | 0 | 98 | 96 | 2 | ||||
| Avg ± SEM | 26.4 ± 5.6 | 23.5 ± 4.3 | 14.0 ± 1.5 | 13.9 ± 1.6 | 87.4 ± 5.6 | 76.2 ± 8.2 | 11.2 ± 4.1 | 26.3 ± 5.4 | ||||||||
The mobility efficiencies (%) of the LI.LtrB intron were calculated by colony patch hybridization assays (Materials and Methods and Fig. 1A) analyzing 100 colonies for each of the 10 independent transconjugants (1 to 10). Unexpectedly high LI.LtrB mobility efficiencies are italicized and not included in the means. NZ9800 (NZ) and MMS372 (MMS) are L. lactis strains; NZΔltrB is NZ9800ΔltrB::Tetr (11). gpII, number of colonies revealed with the group II intron probe (5′ end, Fig. 1B); gpI SJ, number of colonies revealed with the td group I intron splice junction probe (ligated exons, Fig. 1B); gpI SJ/gpII, % of LI.LtrB mobility events missing the td group I intron; gpI, number of colonies revealed with the td group I intron probe (Fig. 1B); WT gpII, number of colonies revealed with the wild-type group II intron probe (3′ end, Fig. 1B). The recipient strain in our previous study (3) contained the pMNHS plasmid.
The mobility efficiency detected with the WT gpII probe was zero for all transconjugants.
From reference 3. The mobility efficiency detected with the gpI probe was zero for all transconjugants.
Because pLE12I carried the twintron variant of Ll.LtrB, we also assessed which mobility products had lost the group I intron by probing them with the group I splice junction probe (Fig. 1B, gpI SJ). This probe recognizes the group I intron ligated exons and produces a positive signal only if the td group I intron is missing from group II intron mobility products. As previously observed (3), the majority of the transconjugant cells (7 out of 10) harbored mobility products showing nearly 100% loss of the retromobility indicator (Table 1, NZ/pLE12I × MMS/pMNHS-CR, gpI SJ, gpI SJ/gpII, nos. 3 and 5 to 10). These results demonstrate that the majority of mobility products resulted from Ll.LtrB invading its homing site through the retrohoming pathway (5). In sharp contrast, in transconjugant cells that displayed unusually high levels of Ll.LtrB mobility, only a fraction of mobility products appear to have lost the group I intron (Table 1, NZ/pLE12I × MMS/pMNHS-CR, gpI SJ/gpII, nos. 1, 2, and 4). Levels of bona fide retromobility products (gpII and gpI SJ positive) observed in these three transconjugant cells are in the same range as those observed for the other seven transconjugant cells (Table 1, NZ/pLE12I × MMS/pMNHS-CR, compare gpI SJ and gpII mobility averages). Similar unusually high frequencies of Ll.LtrB mobility were previously observed in various crosses among different L. lactis strains where transferred plasmids were carrying different intron derivatives (3). Our findings confirm that unusually high efficiencies of Ll.LtrB mobility following plasmid conjugation are a general and reproducible phenomenon.
Some Ll.LtrB mobility products in transconjugant cells are not generated from pLE12I plasmid.
Does a significant portion of mobility products present in the three isolates showing unusually high mobility efficiency of Ll.LtrB still harbor the td group I intron (Table 1, NZ/pLE12I × MMS/pMNHS-CR, gpI SJ/gpII, no. 1, 2, and 4)? The presence of Ll.LtrB mobility products still harboring the td intron would explain why the group I splice junction probe did not hybridize to this subset of mobility products. However, using a group I intron-specific probe (Fig. 1B, gpI), we found that almost none of these mobility products contained the td group I intron (Table 1, NZ/pLE12I × MMS/pMNHS-CR, gpI). This suggests that these inserted Ll.LtrB introns are not derived from donor pLE12I plasmid, because mobility products coming from this plasmid should harbor either the td splice junction or the unspliced td intron. Only one mobility product still contained the td group I intron (Table 1, NZ/pLE12I × MMS/pMNHS-CR, gpI, no. 10). This mobility product is most likely a retrohoming event in which the td group I intron was unable to splice before the Ll.LtrB intron RNA was reverse transcribed (5).
To demonstrate that these mobility products were not derived from the pLE12I plasmid and that they were probably generated from a wild-type copy of the intron, we designed a probe specific for wild-type Ll.LtrB (Fig. 1B, WT gpII) that would not hybridize to mobility products generated from donor plasmid, harboring or lacking the td group I intron (Fig. 1B). This DNA oligonucleotide probe spans the region where the td group I intron is inserted in Ll.LtrB just downstream of LtrA (engineered SalI site) (5). We confirmed that mobility products that are negative for both group I and group I splice junction probes contained wild-type Ll.LtrB introns not expressed from pLE12I plasmid (Table 1, NZ/pLE12I × MMS/pMNHS-CR, WT gpII). The WT gpII probe gave positive signals only for the three transconjugant isolates where unusual Ll.LtrB mobility efficiency was observed and where group I intron loss from mobility products seemed inefficient (Table 1, NZ/pLE12I × MMS/pMNHS-CR, WT gpII, gpII, and gpI SJ/gpII, no. 1, 2, and 4). Moreover, mobility products harboring the wild-type Ll.LtrB intron corresponded in all three cases to gpII-positive, gpI SJ-negative mobility products (Table 1, NZ/pLE12I × MMS/pMNHS-CR; WT gpII plus gpI SJ = gpII). This implies that mobility products derived from pLE12I plasmid efficiently lost the td intron and therefore were generated by retrohoming.
To determine whether the wild-type Ll.LtrB mobility products were generated through a DNA-based mobility pathway or through the retrohoming pathway, we performed a coconversion analysis of flanking markers around the intron homing site (5). DNA-dependent Ll.LtrB mobility predicts that genetic information located within flanking exons could be exchanged between donor and recipient alleles, while precise insertion of the intron through complete reverse splicing (retrohoming) does not promote genetic exchange between homologous exons (5). The recipient plasmid used in the conjugation assay (Fig. 2A, pMNHS-CR) contained polymorphic sites in both exons that do not interfere with Ll.LtrB mobility (E1, −7 and −30/35; E2, +7 and +25) (5). Sequencing analyses of intron-exon junctions (5′ and 3′) of 10 wild-type Ll.LtrB mobility products (gpII+/gpI SJ−/gpI−/WT gpII+) (Table 1, NZ/pLE12I × MMS/pMNHS-CR, no. 1) were performed. All 10 mobility products had retained their polymorphic nucleotides on both sides of the intron insertion site, showing that no marker coconversion occurred during Ll.LtrB insertion. This suggests that wild-type Ll.LtrB intron invades its recognition site on the recipient plasmid by the RNA-based retrohoming pathway.
Using the same four probes (gpII, gpI SJ, gpI, and WT gpII) (Fig. 1B), we analyzed two previously isolated transconjugant events (no. 1 and 9) along with a control (no. 2) from the same cross that showed an unusual pattern of Ll.LtrB mobility (3). Similarly, in isolates 1 and 9, all Ll.LtrB mobility products that are gpII positive and gpI and gpI SJ negative were recognized by the WT gpII probe, showing that they were also not derived from pLE12I donor plasmid (Table 1, NZ/pLE12I × MMS/pMNHS, isolates from a previous study [3]). Taken together, these results suggest that in cases of unusually high efficiency of Ll.LtrB mobility following plasmid conjugation, two different Ll.LtrB dissemination pathways are operating concurrently.
Wild-type Ll.LtrB mobility products originated from the chromosome of the donor strain.
Because the recipient strain does not contain a chromosomal copy of the Ll.LtrB intron, we hypothesized that the detected wild-type intron originated from the chromosome of the donor strain. To test our hypothesis, we performed a conjugation assay similar to the one described above but where the wild-type chromosomal copy of the Ll.LtrB intron was deleted from the donor strain (NZ9800ΔltrB/pLE12I × MMS372/pMNHS-CR) (Fig. 2A). Chromosomal deletion of Ll.LtrB removed portions of the flanking exons, most probably destroying the ltrB gene and preventing production of the relaxase enzyme (LtrB) that is essential to initiate plasmid conjugation (3, 13, 18, 27). However, in our conjugation system, this defect is complemented by expression of the relaxase enzyme from the pLE12I plasmid (3). As previously observed (3), because the relaxase is produced only from the conjugative plasmid, conjugation efficiency was lower than that when the relaxase is also expressed from the chromosome (∼100-fold; 7.5 × 10−5 ± 2.7 × 10−5 versus 5.2 × 10−3 ± 0.4 × 10−3). In this particular case, the presence of the group I intron within Ll.LtrB hinders its splicing efficiency and lowers relaxase expression. This explains the more dramatic reduction of pLE12I conjugation efficiency (∼100-fold) in the NZ9800ΔltrB background compared to that previously observed for pLE12, where the intron is wild type (∼10-fold; 2.2 × 10−4 ± 0.6 × 10−4 versus 2.3 × 10−3 ± 0.6 × 10−3) (3).
Following conjugation of pLE12I, the plasmid mix from progeny of 10 independent transconjugant colonies were studied as described above by using the same four probes (gpII, gpI SJ, gpI, and WT gpII) (Fig. 1B). The gpII probe is general and reveals all types of mobility products, while the other probes are specific for three different subsets of Ll.LtrB mobility products (Fig. 1B). In contrast to what was observed in the first cross (Fig. 2B), all plasmid mixes contained the same ratio of mobility products (M) over uninterrupted plasmids (R), suggesting similar efficiencies of Ll.LtrB mobility (Fig. 2C, compare M and R). This observation was confirmed by colony patch hybridization assays; no mobility products harboring a wild-type copy of Ll.LtrB were found (Table 1, NZΔltrB/pLE12I × MMS/pMNHS-CR, WT gpII) and no unusually high Ll.LtrB mobility was seen (Table 1, NZΔltrB/pLE12I × MMS/pMNHS-CR). Finally, all mobility products hybridized either with the gpI or gpI SJ probe, confirming that all originated from pLE12I donor plasmid. Mobility products harboring a wild-type copy of Ll.LtrB observed following plasmid conjugation most probably acquired their introns from the chromosome of the donor strain.
Conjugative transfer of the chromosomally located sex factor induces dissemination of the Ll.LtrB group II intron.
Can the Ll.LtrB-carrying sex factor embedded within the chromosome of the NZ9800 strain be efficiently transferred, and can conjugation disseminate its copy of Ll.LtrB to other L. lactis strains? We performed a conjugation assay in which the donor strain was plasmid free (Fig. 3A, NZ9800 × MMS372/pMNHS). Because the sex factor and the Ll.LtrB intron are wild type and do not carry a selective marker, we directly probed, via colony patch hybridization, 1,000 colonies of recipient cells (MMS372/pMNHS) for the presence of the group II intron (Fig. 1B, gpII probe). We found 9 positive patches potentially carrying Ll.LtrB mobility products (9 out of 1,000). Recipient plasmids showed extra bands of variable intensity from lane to lane, corresponding to hypothetical Ll.LtrB mobility products (Fig. 3C, M). Colony patch hybridization assays (Fig. 1A) confirmed that all nine isolates contained Ll.LtrB mobility products and that the percentage of interrupted plasmid was highly variable (6 to 96%) (Fig. 3C, % Mob). Because the recipient strain [MMS372(recA); Kanr] is kanamycin resistant and UV sensitive (recA), we could rule out the possibility that the recipient plasmid had been transferred from the recipient to donor cell [NZ9800(recA+); Kans]. Indeed, we observed that, like the MMS372 recipient control strain, all nine isolates were kanamycin resistant and UV sensitive, while the NZ9800 donor control strain was kanamycin sensitive and UV resistant (data not shown).
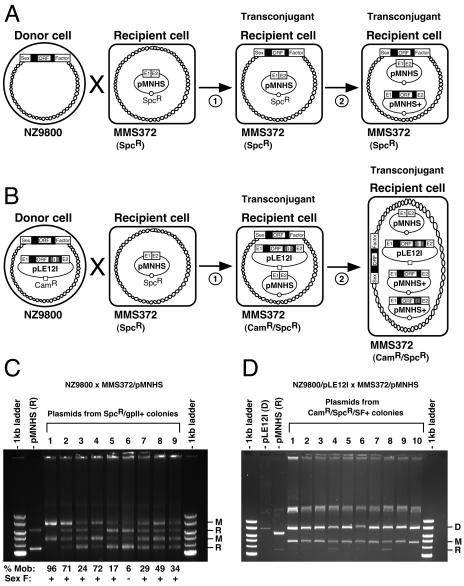
FIG. 3.
Conjugation/retrohoming assays. (A and B) Schematic of the assays. The first step (1) represents the transfer of sex factor (A) or both sex factor and pLE12I plasmid (B) by conjugation from donor (NZ9800) to recipient cell (MMS372). The second step (2) shows invasion (retrohoming) of some homing sites (E1-E2) present on the recipient plasmid (pMNHS) by Ll.LtrB introns expressed either from pLE12I or sex factor. Intron-harboring recipient plasmids (pMNHS+) are also represented in the transconjugant cell. Chromosomes are depicted as circular double-stranded helices. Ll.LtrB group II intron, black; Ll.LtrB exons, E1 and E2. (C and D) Agarose (0.5%) gels containing undigested plasmid mixes recovered from transconjugants containing sex factor (C) or both sex factor and pLE12I plasmid (D). Bands corresponding to donor plasmid (D), recipient plasmid (R), and mobility product (M) are highlighted. Ll.LtrB mobility efficiency, % Mob; +, presence of sex factor (Sex F); −, absence of sex factor.
To determine if the sex factor was present in the nine MMS372 recipient cells carrying Ll.LtrB mobility products, we performed Southern blot analyses on genomic DNA using two sex factor-specific probes that recognize both extremities of the element (10). We confirmed that the sex factor was present in eight of the nine isolated recipient strains (Fig. 3C, Sex F) but was absent from the genomic DNA of the control MMS372 strain (data not shown).
To confirm the frequency at which the sex factor is transferred, we twice repeated the cross described above. Recipient cells were first analyzed with a sex factor-specific probe (10), where we twice found that 7 recipient cells had received the sex factor (7 out of 1,000). The patches were then rehybridized with gpII probe; no additional signals appeared, showing that no recipient cells carried Ll.LtrB without harboring the sex factor. If we exclude isolate 6 from the first cross, because it is not clear whether it harbored the sex factor, the sex factor transfer frequency is high at 7.3 × 10−3 ± 0.3 × 10−3 (three assays) and correlates well with that previously observed (23) and with the transfer frequency of the engineered plasmid pLE12 carrying wild-type Ll.LtrB intron (2.3 × 10−3 ± 0.5 × 10−3) (3).
Taken together, these results demonstrate that the L. lactis sex factor can be efficiently transferred by conjugation from the chromosome of strain NZ9800 to strain MMS372 and can support Ll.LtrB dissemination.
Cotransfer of pLE12I plasmid and sex factor.
Knowing that our donor strain contains two proficient mobilizable elements, we hypothesized that the conjugative plasmid and the sex factor can be simultaneously transferred from donor cell to recipient cell during conjugation and that both support the dissemination of their Ll.LtrB copy. To test our hypotheses, we repeated the NZ9800/pLE12I × MMS372/pMNHS cross (Fig. 3B). Following conjugation, we probed 100 transconjugant cells (MMS372/pMNHS/pLE12I) with a sex factor-specific probe (10) and found that approximately one-third (37.6% ± 2.9% [three assays]) of donor plasmid-containing recipient cells had also acquired the sex factor from the chromosome of the donor strain. Cotransfer frequency of these two conjugative elements is thus in the 10−3 range. Taking into account the transfer frequency of both conjugative elements (10−3 range each), and assuming that they are independently transferred, we expected the number of recipient cells containing both elements to be much lower (10−5 range, 7.3 × 10−3 multiplied by 5.2 × 10−3). This result shows that the conjugative transfer of either pLE12I plasmid or sex factor can happen independently but that cotransfer of both elements occurs quite frequently. We thus consider that pLE12I and sex factor cotransfer is synergistic, suggesting that when the conjugative pores are created to allow mobilization of the chromosomal sex factor between donor and recipient cells, transfer of the conjugative plasmid, present in approximately 25 copies (12) in the cytoplasm of the donor strain, is facilitated.
To analyze Ll.LtrB dissemination from these two elements, we randomly picked 10 colonies corresponding to recipient cells that had acquired both pLE12I plasmid and sex factor. Plasmid mixes (Fig. 3D) were retransformed in DH5α, and patches were first hybridized with WT gpII probe (Fig. 1B) that specifically identified the mobility products generated from the sex factor (Table 1, NZ/pLE12I × MMS/pMNHS, WT gpII). The general gpII probe (Fig. 1B) was then used to reveal all Ll.LtrB mobility products generated from both elements (Table 1, NZ/pLE12I × MMS/pMNHS, gpII). The number of Ll.LtrB mobility products that originated from pLE12I plasmid was calculated by subtracting the products generated by sex factor (WT gpII) from the total amount of Ll.LtrB mobility products (gpII) (Table 1, NZ/pLE12I × MMS/pMNHS, gpII minus WT gpII). We found that for the majority of isolates (8 of 10), both conjugative elements generated Ll.LtrB mobility products. As previously observed (Table 1, NZ/pLE12I × MMS/pMNHS-CR and NZΔltrB/pLE12I × MMS/pMNHS-CR), the efficiency of Ll.LtrB mobility from the sex factor is broadly variable (19 to 98%) (Table 1, NZ/pLE12I × MMS/pMNHS). However, in this assay, average mobility efficiency of the Ll.LtrB intron expressed from the sex factor is significatly higher (Fig. 3D) (76.2% ± 8.2% versus 44.2% ± 10.0%). On the other hand, Ll.LtrB dissemination from pLE12I showed more variability (0 to 39%) than previously observed (Table 1, NZ/pLE12I × MMS/pMNHS) (3). These results suggest that plasmid and sex factor transfer almost invariably stimulates Ll.LtrB dissemination, leading to the generation of mobility products upon their transfer to a recipient cell. In the two isolates showing no Ll.LtrB mobility products generated from pLE12I, 98% of recipient plasmids are interrupted by Ll.LtrB intron produced from the sex factor. This suggests that the sex factor was first transferred to the recipient cell, where its copy of Ll.LtrB invaded almost all available homing sites on recipient plasmids before pLE12I was acquired. This scenario would prevent establishment of Ll.LtrB mobility products generated from pLE12I plasmid. Nevertheless, it is difficult to compare this experiment with the previous crosses presented here and earlier (3) because of the extra steps involved in identifying recipient cells that had acquired both elements before scoring Ll.LtrB mobility efficiencies.
DISCUSSION
Horizontal transfer of group II introns between organisms, a well-accepted model of intron dissemination and evolution (14, 28), was only recently experimentally corroborated (3). This model was proposed to explain the presence of closely related introns at different locations (genes or species) and to rationalize why, in specific genes, introns are more conserved than their flanking exons (14, 28). Interestingly, the great majority of bacterial group II introns is found associated with other mobile elements, such as transposons, insertion sequence (IS) elements, conjugative plasmids, pathogenicity islands, and virulence plasmids (7, 15). Furthermore, a phylogenetic study using numerous bacterial intron-encoded proteins suggests that mobile group II introns originated in the bacterial kingdom and that these prokaryotic introns seem to be subject to a high level of horizontal transfer (28). Taking these facts into consideration, we hypothesized that these various mobile elements carrying group II introns promote dissemination of their hitchhikers to new sites following their transfer between cells (3).
As a first initiative, using plasmid conjugation assays, we showed that the Ll.LtrB group II intron, present within the L. lactis pRS01 conjugative plasmid, can be disseminated and laterally transferred following its conjugative transfer (3). The engineered plasmids used in that study contained a small segment (Tra1-2; 7.5 kb) of the pRS01 conjugative plasmid (48.4 kb) that harbored the relaxase gene interrupted by the Ll.LtrB group II intron. Remaining functions of the conjugation machinery were provided by the integrative and conjugative sex factor embedded in the chromosome of the donor strain (3). Using that system, Ll.LtrB dissemination was shown among L. lactis strains (retrohoming and retrotransposition) and from L. lactis to E. faecalis (retrohoming) (3).
Here, we demonstrated that a fraction of Ll.LtrB mobility products, present in transconjugant cells showing an unusually high level of mobility products after plasmid acquisition (3), was wild type and, thus, not produced from plasmid. We showed that wild-type Ll.LtrB mobility products originated from the chromosome of the donor strain and were generated by the RNA-mediated retrohoming pathway. We then demonstrated that the sex factor present within the chromosome of NZ9800 strain can be efficiently transferred by conjugation to the MMS372 strain and can support Ll.LtrB dissemination at highly variable rates ranging from 6 to 96%. Even if pLE12I contains an Ll.LtrB variant that is approximately 33% less proficient than wild-type Ll.LtrB in transconjugant MMS372 cells (3), frequencies of wild-type Ll.LtrB mobility from the sex factor can be much higher (almost 100%) than ever observed in any of our plasmid conjugation assays (∼20%) (Table 1) (3) or when these two plasmids (pLE12I and pMNHS) are simply cotransformed in the same strain (11%) (5). Furthermore, taking into consideration that the sex factor is maintained at 1 copy per cell while pLE-based plasmids are at 25 copies per cell, efficiency of mobility of the Ll.LtrB intron is significantly higher from the sex factor. These results suggest that regulation of production of active Ll.LtrB RNP particles is different between the whole conjugative element and the simplified context of the engineered conjugative plasmids. This finding is biologically relevant, because the Ll.LtrB group II intron can be spread among L. lactis strains, using the native chromosomal sex factor (50 kb) as a carrier. It also suggests that spreading of Ll.LtrB among L. lactis strains by sex factor conjugative transfer may have previously occurred and could still occur naturally, because these two mobile elements are active in the NZ9800 L. lactis laboratory strain.
Following sex factor conjugative transfer, we also found Ll.LtrB mobility products within a recipient cell that did not harbor the sex factor (Fig. 3C, isolate 6). This implies that the sex factor can transiently express active Ll.LtrB RNP particles and promote intron dissemination even if it is not permanently retained within recipient cells following conjugative transfer. Indeed, it has been shown that sex factor can be lost from its host strains (9). Isolate 6 showed the lowest efficiency of Ll.LtrB mobility among the nine isolates examined in this experiment, perhaps a result of the transient presence of sex factor in the recipient cell. Furthermore, uptake of Ll.LtrB RNP particles directly from medium is an unlikely means of transmission of this mobile element, because RNase was spread on the mating plates.
Unusually high levels of Ll.LtrB mobility products following plasmid conjugation.
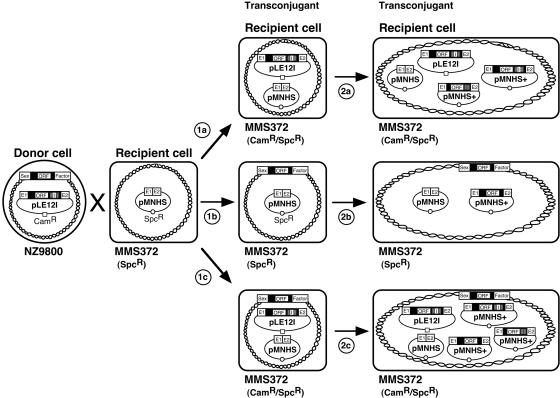
Figure 4 shows different potential outcomes of a typical cross between NZ9800/pLE12I and MMS372/pMNHS. Because the donor strain contains both pLE12I conjugative plasmid and a proficient copy of the integrative and conjugative sex factor embedded within its chromosome, three different types of transconjugant cells can be produced. Acquisition of the intron by recipient cells can occur by conjugation of pLE12I plasmid (1a), sex factor (1b), or both mobilizable elements (1c). We demonstrated that acquisition of pLE12I plasmid by the recipient cell supports Ll.LtrB mobility to relatively low levels (∼20%), potentially creating two types of mobility products that do or do not harbor the td group I intron (step 2a) (Table 1, NZ/pLE12I × MMS/pMNHS-CR and NZΔltrB/pLE12I × MMS/pMNHS-CR) (3). On the other hand, acquisition of sex factor promotes spreading of wild-type Ll.LtrB intron at higher and more variable efficiencies (6 to 96%) and creates only one type of mobility product (step 2b) (Table 1, NZ/pLE12I × MMS/pMNHS-CR and NZ/pLE12I × MMS/pMNHS). Finally, when pLE12I plasmid and sex factor are cotransferred into the same recipient cell, they can both independently promote mobility of their intron variant with the potential of creating three types of mobility products (step 2c) (Table 1, NZ/pLE12I × MMS/pMNHS-CR and NZ/pLE12I × MMS/pMNHS).
FIG. 4.
Plasmid- and sex factor-based Ll.LtrB dissemination between L. lactis strains. A conjugation assay between the donor NZ9800/pLE12I and recipient MMS372/pMNHS strains can generate three types of transconjugant cells. In the first step (1), the recipient cell acquires pLE12I plasmid (1a), sex factor (1b), or both mobilizable elements (1c) by conjugation. The second step (2) represents mobility of the Ll.LtrB intron from its carrying element(s) (pLE12I, sex factor) to the homing site on the recipient plasmid (pMNHS). The percentage and nature of Ll.LtrB mobility products (pMNHS+) depends on which Ll.LtrB-harboring element(s) is present within the recipient cell. Ll.LtrB group II intron, black; Ll.LtrB exons, E1 and E2; td intron, I; td exons, grey.
Using different probes that are specific for each of the three types of Ll.LtrB mobility products, plus a general probe that recognizes all of them (Fig. 1B), we demonstrated that the unusually high level of mobility products consists of those generated from both pLE12I plasmid and sex factor (Fig. 4, step 1c). In our standard conjugation/retrohoming assays, we isolated 10 independent transconjugant cells (Camr/Spcr) that had received the donor plasmid, thus selecting for only two (Fig. 4, 1a and 1c) of the three possible outcomes of the cross (Table 1, NZ/pLE12I × MMS/pMNHS-CR) (3). We typically observed two or three isolates showing unusually high efficiency of Ll.LtrB mobility and reduced td loss (Table 1, NZ/pLE12I × MMS/pMNHS-CR) (3). Taking into consideration that we studied the plasmid mix from only 10 transconjugant cells, a relatively small sample, this ratio (20 to 30%) correlates with pLE12I and sex factor cotransfer frequency (37.6% ± 2.9%).
Group II intron dissemination through conjugation.
Our study demonstrates that L. lactis sex factor can efficiently disseminate the Ll.LtrB mobile group II intron between L. lactis strains. Our findings support the theory that bacterial group II introns can be spread from cell to cell following mobilization of their host elements (3). Furthermore, the association of group II introns with various mobile elements in bacteria may have been a means of survival for these introns and could explain why a great majority of them are found within other mobile elements (3).
This intron dissemination theory is further strengthened by recent studies. It was demonstrated that the Ll.LtrB group II intron has a marked preference to retrotranspose into plasmids rather than chromosomal target sites (12). It was proposed that this bias for plasmid invasion is linked to the nature of the retrotranposition pathway and to target accessibility (12). This propensity to invade plasmids may explain the unexpectedly high representation of bacterial group II introns within plasmids. Moreover, this bias seems advantageous for these mobile elements because invasion of plasmids limits host damage and could also promote their lateral transfer by conjugation if the invaded plasmid is mobilizable. This inclination of group II introns to invade plasmids may ultimately lead to their better chance of survival.
It was recently shown that other relaxase genes present within the enterococcal conjugative plasmid pCF10 and the streptococcal conjugative transposon Tn5252 can also be invaded by the Ll.LtrB intron from L. lactis (24). The site invaded by Ll.LtrB lies within a conserved functional domain of the relaxase enzymes (24). If Ll.LtrB jumps into these available target sites in nature, newly inserted copies of the intron could be spread between cells by these new conjugative carriers, potentially increasing its dispersal. Indeed, the enterococcal plasmid pCF10, like pRS01 plasmid and sex factor from L. lactis, retained conjugative function following Ll.LtrB acquisition (24). This means that splicing of Ll.LtrB from its newly invaded site is efficient enough to produce sufficient amounts of relaxase enzyme to support plasmid transfer. However, it also implies that active RNP particles are produced upon splicing and that Ll.LtrB could invade new sites following conjugative transfer of pCF10.
Another L. lactis mobilizable plasmid (pAH90) was shown to harbor a copy of the Ll.LtrB intron that interrupts its relaxase gene at the same position (20). This Ll.LtrB-invaded plasmid can probably also support the dissemination of its stowaway because, like the other Ll.LtrB-harboring elements, it retained its conjugative function (20). By targeting a conserved region within relaxase genes that code for essential enzymes involved in initiating the transfer of their conjugative elements, Ll.LtrB ensures its efficient dissemination and survival. This may explain why identical proficient copies of the Ll.LtrB intron, which are probably recent acquisitions, have been found in three different natural conjugative elements of L. lactis: chromosomal sex factor, pRS01, and pAH90. Conjugation, the most efficient way to transfer genetic information between bacterial cells and even across phyla (14), may have been and probably still is an efficient mode of dissemination, not only for Ll.LtrB but also for other group II introns present on various conjugative elements.
Acknowledgments
We thank M. Belfort for kindly providing NZ9800ΔltrB::Tetr strain and pLHS-CR plasmid. We also thank N. H. Acheson, J. W. Coulton, K. Fiola, R. A. Lease, and H. Le Moual for providing comments and J. Kashul for editing the manuscript.
B.C. is a CIHR New Investigator Scholar and a McGill University William Dawson Scholar. Research in the B.C. laboratory is supported by grants from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Belfort, M., V. Derbyshire, M. M. Parker, B. Cousineau, and A. M. Lambowitz. 2002. Mobile introns: pathways and proteins, p. 761-783. In N. L. Craig, R. Craigie, M. Gellert, and A. M Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 2.Belfort, M., K. Ehrenman, and P. S. Chandry. 1990. Genetic and molecular analysis of RNA splicing in Escherichia coli. Methods Enzymol. 181:521-539. [DOI] [PubMed] [Google Scholar]
- 3.Belhocine, K., I. Plante, and B. Cousineau. 2004. Conjugation mediates transfer of the Ll.LtrB group II intron between different bacterial species. Mol. Microbiol. 51:1459-1469. [DOI] [PubMed] [Google Scholar]
- 4.Burrus, V., G. Pavlovic, B. Decaris, and G. Guédon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 5.Cousineau, B., D. Smith, S. Lawrence-Cavanagh, J. E. Mueller, J. Yang, D. Mills, D. Manias, G. Dunny, A. M. Lambowitz, and M. Belfort. 1998. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94:451-462. [DOI] [PubMed] [Google Scholar]
- 6.Cousineau, B., S. Lawrence, D. Smith, and M. Belfort. 2000. Retrotransposition of a bacterial group II intron. Nature 404:1018-1021. [DOI] [PubMed] [Google Scholar]
- 7.Dai, L., and S. Zimmerly. 2002. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 30:1091-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, L., and S. Zimmerly. 2003. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA 9:14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gasson, M. J., S. Swindell, S. Maeda, and H. M. Dodd. 1992. Molecular rearrangements of lactose plasmid DNA associated with high-frequency transfer and cell aggregation in Lactococcus lactis 712. Mol. Microbiol. 6:3213-3223. [DOI] [PubMed] [Google Scholar]
- 10.Godon, J. J., C. J. Pillidge, C. A. Shearman, and M. J. Gasson. 1995. Molecular analysis of the Lactococcus lactis sex factor. Dev. Biol. Stand. 85:423-430. [PubMed] [Google Scholar]
- 11.Ichiyanagi, K., A. Beauregard, S. Lawrence, D. Smith, B. Cousineau, and M. Belfort. 2002. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol. 46:1259-1272. [DOI] [PubMed] [Google Scholar]
- 12.Ichiyanagi, K., A. Beauregard, and M. Belfort. 2003. A bacterial group II intron favors retrotransposition into plasmid targets. Proc. Natl. Acad. Sci. USA 100:15742-15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein, J. R., Y. Chen, D. A. Manias, J. Zhuo, L. Zhou, C. L. Peebles, and G. M. Dunny. 2004. A conjugation-based system for genetic analysis of group II intron splicing in Lactococcus lactis. J. Bacteriol. 186:1991-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambowitz, A. M., and M. Belfort. 1993. Introns as mobile genetic elements. Annu. Rev. Biochem. 62:587-622. [DOI] [PubMed] [Google Scholar]
- 15.Martinez-Abarca, F., and N. Toro. 2000. Group II introns in the bacterial world. Mol. Microbiol. 38:917-926. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura, M., R. Saldanha, H. Ma, H. Wank, J. Yang, G. Mohr, S. Cavanagh, G. M. Dunny, M. Belfort, and A. M. Lambowitz. 1997. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 11:2910-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michel, F., and J.-L. Ferat. 1995. Structure and activities of group II introns. Annu. Rev. Biochem. 64:435-461. [DOI] [PubMed] [Google Scholar]
- 18.Mills, D. A., L. L. McKay, and G. M. Dunny. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 178:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills, D. A., D. A. Manias, L. L. McKay, and G. M. Dunny. 1997. Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J. Bacteriol. 179:6107-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Sullivan, D. O., R. P. Ross, D. P. Twomey, G. F. Fitzgerald, C. Hill, and A. Coffey. 2001. Naturally occurring lactococcal plasmid pAH90 links bacteriophage resistance and mobility function to a food-grade selectable marker. Appl. Environ. Microbiol. 67:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rambo, R. P., and J. A. Doudna. 2004. Assembly of an active group II intron-maturase complex by protein dimerization. Biochemistry 43:6486-6497. [DOI] [PubMed] [Google Scholar]
- 22.Saldanha, R., G. Mohr, M. Belfort, and A. M. Lambowitz. 1993. Group I and group II introns. FASEB J. 7:15-24. [DOI] [PubMed] [Google Scholar]
- 23.Shearman, C., J. J. Godon, and M. Gasson. 1996. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol. Microbiol. 21:45-53. [DOI] [PubMed] [Google Scholar]
- 24.Staddon, J. H., E. M. Bryan, D. A. Manias, and G. M. Dunny. 2004. Conserved target for group II intron insertion in relaxase genes of conjugative elements of gram-positive bacteria. J. Bacteriol. 186:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toro, N. 2003. Bacteria and archaea group II introns: additional mobile genetic elements in the environment. Environ. Microbiol. 5:143-151. [DOI] [PubMed] [Google Scholar]
- 26.Trieu-Cuot, P., C. Carlier, and P. Courvalin. 1998. Conjugative plasmid transfer from Enterococcus faecalis to Escherichia coli. J. Bacteriol. 170:4388-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou, L., D. A. Manias, and G. M. Dunny. 2000. Regulation of intron function: efficient splicing in vivo of a bacterial group II intron requires a functional promoter within the intron. Mol. Microbiol. 37:639-651. [DOI] [PubMed] [Google Scholar]
- 28.Zimmerly, S., G. Hausner, and X. Wu. 2001. Phylogenetic relationship among group II intron ORFs. Nucleic Acids Res. 29:1238-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]