Abstract
The Staphylococcus aureus lrg and cid operons encode homologous proteins that regulate extracellular murein hydrolase activity and penicillin tolerance in a diametrically opposing manner. Although their specific regulatory functions remain unknown, it has been postulated that the functions of CidA and LrgA are analogous to those of bacteriophage holins and antiholins, respectively, and that these proteins serve as molecular control elements of bacterial programmed cell death. Although these studies demonstrated that cidBC transcription is abundant in σB-proficient strains, cidABC transcription was only minimally expressed under standard growth conditions. In this study, we demonstrate that cidABC and lrgAB transcription in the clinical isolate UAMS-1 is induced by growth in the presence of 35 mM glucose and that this enhances murein hydrolase activity and decreases tolerance to vancomycin and rifampin. The effect of glucose on murein hydrolase activity was not observed in the cidA mutant, indicating that the induction of this activity was dependent on enhanced cidABC expression. Furthermore, we demonstrate that the effects of glucose on cidABC and lrgAB transcription are mediated by the generation of acetic acid produced by the metabolism of this and other carbon sources. These results shed new light on the control of the S. aureus cidABC and lrgAB genes and demonstrate that these operons, as well as murein hydrolase activity and antibiotic tolerance, are responsive to carbohydrate metabolism.
Recent work in our laboratory has identified and characterized the cid and lrg operons of Staphylococcus aureus, whose gene products affect extracellular murein hydrolase activity and penicillin tolerance (11, 22, 35). Disruption of the lrg operon in the laboratory strain RN6390 increased extracellular murein hydrolase activity and decreased penicillin tolerance, whereas disruption of the cid operon decreased extracellular murein hydrolase activity and increased penicillin tolerance (22, 35). The precise mechanism by which the Lrg and Cid proteins regulate these two processes is currently unknown. However, the lrgA and cidA gene products display structural similarities to the bacteriophage holin family of proteins, which are known regulators of murein hydrolase activity and ultimately control the timing and onset of bacteriophage-induced cell lysis (4, 11, 22, 35). Based on these similarities, along with the phenotypic consequences of the cid and lrg mutations, it has been proposed that the cidA and lrgA gene products regulate murein hydrolase activity in a manner analogous to those of holins and anti holins, respectively (4, 34). Furthermore, it has been recently suggested that the lrg and cid gene products are molecular control elements involved in the regulation of programmed cell death (PCD) in S. aureus (3, 34).
To better understand the role of the lrg and cid gene products in modulating murein hydrolase activity and antibiotic-induced killing, recent studies have been performed to examine the environmental factors that influence expression of these genes. It was previously shown that lrgAB transcription is positively regulated by the LytSR two-component regulatory system encoded immediately upstream of lrgAB (10, 11). However, the signal to which this system responds has not been identified. Furthermore, lrgAB expression is positively regulated by the virulence factor regulators Agr and SarA (17). The cidABC operon has been recently shown to be comprised of two overlapping transcripts: a cidABC transcript that is expressed during exponential growth and is detectable only by reverse transcriptase PCR, and a smaller cidBC transcript that is also expressed maximally during exponential growth and is easily detectable by Northern blot analysis (35, 36). These studies also demonstrated that cidBC transcription is dramatically enhanced by σB, whereas lrgAB transcription was downregulated, suggesting that these genes are members of the σB stress regulon (36). Highly propagated S. aureus laboratory isolates such as 8325-4 and RN6390 contain a naturally occurring 11-bp deletion in rsbU (24), whose gene product regulates the activity of σB. Presumably, these laboratory strains do not respond to stress or undergo PCD in the same manner as would low-passage or clinical strains. Based on these findings it was proposed that study of the lrg and cid operons should be carried out in a low-passage clinical isolate in order to better define what role their gene products play in the cell's stress response and/or PCD (34, 36).
Although the factors that affect expression of the full-length S. aureus cidABC transcript have not been reported, a recent transcriptome analysis of carbon catabolite protein A (CcpA)- and glucose-dependent gene expression in Bacillus subtilis revealed that transcription of its cidAB homologues (ywbHG) was enhanced in cultures supplemented with 1% (wt/vol) glucose (30). The data generated by our study presented here reveal that both cidABC and lrgAB expression in strain UAMS-1, a previously characterized low-passage clinical isolate (5, 7, 8, 20), are also stimulated by growth in the presence of glucose. This effect was found to be dependent on the accumulation of acetic acid in the culture supernatant, a consequence of glucose metabolism. Furthermore, murein hydrolase activity and sensitivity to the antibiotics vancomycin and rifampin were greatly enhanced when cells were grown in the presence of glucose. A cidA mutant derivative of UAMS-1 displayed a complete loss of murein hydrolase activity as well as increased antibiotic tolerance, a phenotype similar to that of the previously characterized cidA mutant of RN6390 (35). These results reaffirm the role of the cidABC operon in the regulation of murein hydrolase activity and antibiotic tolerance and demonstrate that expression of both cidABC and lrgAB is responsive to by-products of glucose metabolism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. aureus UAMS-1 strain used throughout this study is a recently isolated osteomyelitis strain (20) that was kindly provided by Mark Smeltzer (Department of Microbiology and Immunology, University of Arkansas for Medical Sciences, Little Rock). S. aureus KB1050 (cidA::Erm; Emr) is a cidA mutant derivative of UAMS-1. All S. aureus strains were grown in either tryptic soy broth (Difco Laboratories, Detroit, Mich.) or NZY broth (3% [wt/vol] N-Z amine A [Sigma Chemical Co., St. Louis, Mo.], 1% [wt/vol] yeast extract [Fisher Scientific, Fair Lawn, N.J.]; adjusted to pH 7.5 unless otherwise indicated), and supplemented as necessary with 1.5% (wt/vol) granulated agar (Difco). Where specified below, NZY cultures were adjusted to pH 5.0 with either HCl or glacial acetic acid and/or were supplemented with one of the following: 35 mM glucose, 30 mM sodium acetate (Na-acetate), or 30 mM sodium chloride (NaCl). All of these supplements were stored as filter-sterilized stock solutions and were diluted to the appropriate concentration in the culture medium at the time of inoculation. All of the cultures were grown in Erlenmeyer flasks at 37°C with shaking at 250 rpm in a volume that equaled 8 to 10% of the flask volume. All antibiotics were purchased from either Sigma Chemical Co. or Fisher Scientific and were used at the following concentrations: erythromycin (Em; 5 μg · ml−1); tetracycline (Tc; 5 μg · ml−1).
Allele replacement of the cidA gene in UAMS-1.
KB1050, a cidA mutant derivative of UAMS-1, was created by transferring the plasmid pBF650, previously described and used to create a cidA mutant of RN6390 (35), by electroporation (39) into UAMS-1. This was followed by growth at the nonpermissive temperature (44°C) in the presence of 2 μg of erythromycin ml−1 to select for cells in which the plasmid had integrated into the chromosome via homologous recombination. To promote a second recombination event, a single colony was inoculated into antibiotic-free tryptic soy broth medium and grown at 30°C for 5 days, with 1:1,000 dilutions into fresh antibiotic-free medium each day. After the fifth day, dilutions of the culture were spread on tryptic soy agar medium to yield isolated colonies, which were subsequently screened for Emr and Tcs. Verification that 142 bp (nucleotides 2626675 to 2626533 of the 8325 genome; http://www.genome.ou.edu/staph.html.) had been deleted from the 5′ end of the cidA gene was carried out by PCR amplification and Southern blot analyses.
Measurement of growth, pH, and acetate concentrations.
Growth of all S. aureus cultures was monitored by measuring the optical density at 600 nm (OD600) using an UltraSpec 4000 spectrophotometer (Pharmacia-Biotech, Piscataway, N.J.). To harvest culture supernatants, 2 to 3 ml of each culture was centrifuged at 2,000 × g for 15 min, and the culture supernatants were decanted into sterile test tubes. The pH of 1.5-ml aliquots of each supernatant was measured with an Accumet Basic pH meter (Fisher Scientific). One-hundred-microliter aliquots of each supernatant were also stored at −80°C until used for quantifying acetate concentrations using an acetic acid detection kit purchased from R-BioPharm, Inc. (Marshall, Mich.), following the manufacturer's protocols.
RNA isolation and Northern blot analysis.
For all RNA isolations, overnight S. aureus cultures were used to inoculate Erlenmeyer flasks containing an 8 to 10% volume of NZY to an initial OD600 of 0.1. The cultures were then grown for the period of time indicated for each experiment, and cells were harvested for RNA isolation by centrifugation. Total RNA was isolated using the TRIzol reagent (Invitrogen Life Technologies, Carlsbad, Calif.) and the FASTPREP cell disruptor (QBiogene, Inc., Carlsbad, Calif.) as previously described (14, 31, 33). For Northern blot analysis, 5 to 10 μg of each RNA sample as indicated for each experiment was subjected to electrophoresis through a 1% (wt/vol) agarose gel containing 0.66 M formaldehyde and morpholinepropanesulfonic acid (MOPS) running buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA; pH 7.0). The RNA samples were subsequently transferred to nylon membranes (Micron Separations Inc., Westboro, Mass.) by overnight capillary transfer in 20× SSC (0.3 M Na3-citrate, 3.0 M NaCl; pH 7.0) and fixed to the membrane by baking at 80°C for at least 2 h. Hybridization of the immobilized RNA with digoxigenin (DIG)-labeled DNA probes and subsequent washing and detection steps were performed using buffers and reagents of the DIG system (Roche Applied Science, Indianapolis, Ind.), following the manufacturer's recommendations for Northern blot analysis. DIG-labeled DNA probes were synthesized using the PCR-based DIG probe synthesis kit (Roche) and the primer pairs cidA-F-cidA-R, cidB1-F-cidB1-R, and lrgA1-F-lrgA1-R to synthesize cidA-, cidB-, and lrgA-specific probes, respectively (Table 1).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Location (8325 genome) or reference |
|---|---|---|
| cidA1-F | ccccatatgCACAAAGTCCAATTA | 2626127-2626141 |
| cidA1-R | cccctcgagTTCATAAGCGTCTACACC | 2625752-2625769 |
| cidB1-F | TGATTTTGTTGACTGTCGTT | 36 |
| cidB1-R | TCATGTGACACTTCGATACC | 36 |
| lrgA1-F | ccccatatgGTCGTGAAACAACAAAAAGACGC | 36 |
| lrgA1-R | cccctcgagATCATGAGCTTGTGCCTCCTC | 36 |
Lower case letters represent addition of restriction site.
Murein hydrolase assays.
Overnight S. aureus cultures were used to inoculate Erlenmeyer flasks containing an 8% volume of NZY to an initial OD600 of 0.1 and grown for 16 h at 37°C and 250 rpm. The culture supernatant (containing extracellular murein hydrolases) was harvested by centrifugation for 20 min at 1,900 × g and concentrated approximately sixfold in a Centricon-3 concentrator (Millipore, Bedford, Mass.). Protein concentrations of the concentrated extracellular proteins were determined using the Bradford assay (Bio-Rad Laboratories, Hercules, Calif.), according to the manufacturer's recommended protocols. Quantitative cell wall hydrolysis assays were performed as previously described (22), except that 50 μg of concentrated extracellular proteins was used to lyse a suspension of 1 mg · ml−1 Micrococcus luteus cell walls (Sigma), and the turbidity of the samples was determined by measuring the OD580. Zymogram analysis of extracellular murein hydrolase activity was performed as described previously (22).
Antibiotic tolerance assays.
Vancomycin- and rifampin-induced killing of UAMS-1 and KB1050 were assessed by dilution plating based on previously described methods (22, 35), with the following modifications: overnight S. aureus cultures were each diluted to an OD600 of 0.1 in 125-ml Erlenmeyer flasks containing 10 ml of NZY broth, both in the presence and absence of 35 mM glucose. For assessing vancomycin sensitivity, the cultures were then grown for 2 h at 37°C and 250 rpm prior to the addition of 40 μg of vancomycin ml−1. For assessing rifampin sensitivity, the cultures were grown for 4 h at 37°C and 250 rpm prior to the addition of 2 μg of rifampin ml−1. In both types of experiments, cell viability was monitored by dilution plating.
Light microscopy.
Overnight cultures of UAMS-1 and KB1050 were each diluted to an OD600 of 0.1 in 125-ml Erlenmeyer flasks containing 10 ml of NZY broth, both in the presence and absence of glucose. The cultures were grown for 8 h at 37°C and 250 rpm, and samples were harvested for light microscopy analysis. For preparation of microscope slides, a smear of each culture was heat fixed to a standard glass slide and stained with Gram's crystal violet for 1 min, followed by destaining with deionized H2O. Slides of each culture were prepared in duplicate. The slides were then viewed under oil at 1,000× magnification with an Olympus BX41 microscope (Olympus America, Inc., Melville, N.Y.), and images representative of each culture were captured using Magnafire SP (version 2.1B) software.
RESULTS
Transcription of cidABC and lrgAB is induced by glucose.
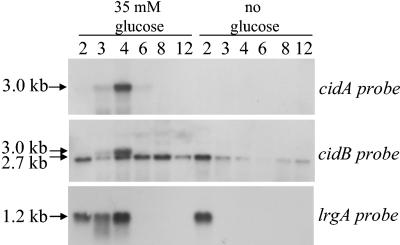
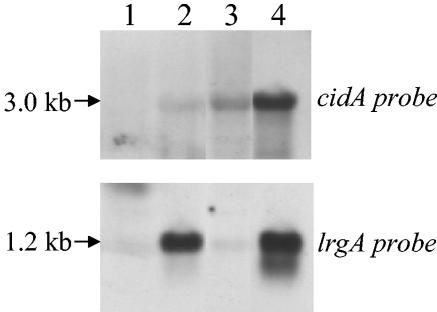
Previous work in our laboratory revealed that the cid operon is comprised of two overlapping transcripts: a 2.7-kb cidBC transcript whose expression is σB dependent, and a full-length cidABC transcript that is expressed at low, nearly undetectable levels during early exponential growth under standard S. aureus culture conditions (36). Since the cidA gene product has been postulated to exert a positive effect on extracellular murein hydrolase activity and antibiotic tolerance by acting in a manner analogous to that of an effector holin (3, 34, 35), we hypothesized that expression of the cidABC transcript is tightly regulated and increased expression may only occur under specific but as-yet-unknown conditions. In this respect, a previously published transcriptome analysis of B. subtilis revealed that transcription of the ywbH and ywbG gene products, which share 34 and 31.9% identity to CidA and CidB, respectively, was induced when B. subtilis was grown in the presence of 1% (wt/vol) glucose (30). To determine if cidABC transcription is also induced by growth in the presence of glucose in S. aureus, a Northern blot analysis was performed on RNA samples from cultures of the clinical isolate UAMS-1 grown in NZY broth in both the presence and absence of 35 mM glucose (Fig. 1). In agreement with previous findings (35, 36), cidABC transcription was not detectable by Northern blot analysis at any of the time points examined in the control (no-glucose) culture. However, in cultures containing glucose, cidABC was dramatically induced at 4 h growth (mid-exponential phase) and declined to undetectable levels by 8 h. The temporal pattern of lrgAB expression was also affected by growth in the presence of glucose: lrgAB transcription in the control culture disappeared after 2 h of growth, whereas expression of lrgAB was decreased in the glucose culture at 2 h of growth but expression persisted at 3 h of growth and was maximal at 4 h of growth. The 2.7-kb cidBC transcript also appeared to be somewhat upregulated (at time points beyond 4 h of growth) when UAMS-1 was grown in the presence of glucose, but this effect was not nearly as dramatic as that observed for cidABC and lrgAB transcription. Further analysis revealed that high-level cidABC and lrgAB transcription occurred in media containing a minimum of 25 mM glucose (unpublished data), indicating that this concentration represents a threshold level that results in high-level cidABC and lrgAB transcription.
FIG. 1.
Northern blot analysis of cid and lrg transcription in S. aureus UAMS-1. Total cellular RNA was isolated from UAMS-1 cells cultured in NZY broth in either the presence of 35 mM glucose or in the absence of glucose at 2, 3, 4, 6, 8, and 12 h postinoculum (as indicated above each lane of the blot). Ten micrograms of each RNA sample was separated through a 1% (wt/vol) agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to cidA-, cidB-, and lrgA-specific DIG-labeled probes. The sizes of each transcript were determined by comparison to an RNA ladder (Invitrogen) run on the same gel. It should be noted that the corresponding culture cell density (as measured by the OD600), pH, and acetate concentration for each time point in this experiment are represented in Fig. 5.
Effect of glucose on extracellular murein hydrolase activity.
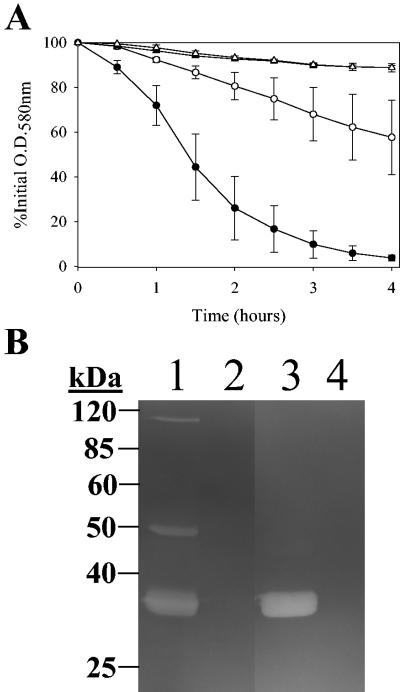
Previous work in our laboratory demonstrated that a cidA mutant derivative of the laboratory strain RN6390 displayed decreased extracellular murein hydrolase activity (35). Given that these findings suggested a role for the cidA gene product as a positive regulator of murein hydrolase activity, and since cidABC expression is increased in cultures containing glucose, we hypothesized that extracellular murein hydrolase activity may also be increased by growth in the presence of glucose. Likewise, we speculated that the phenotype of decreased murein hydrolase activity associated with a cidA mutant would not be affected by growth in the presence of glucose. Therefore, the cidA mutation was transferred to the clinical isolate UAMS-1, and both isogenic strains were subsequently assessed for extracellular murein hydrolase activity when grown in both the presence and absence of glucose (Fig. 2). As predicted, the extracellular murein hydrolase activity of UAMS-1 was dramatically increased when cultured in the presence of glucose, compared to the UAMS-1 no-glucose control (Fig. 2A). By the end of the quantitative cell wall hydrolysis assay (4 h), the no-glucose NZY UAMS-1 sample had solubilized approximately 40% of the M. luteus cell walls, whereas the glucose sample had solubilized nearly 100% of the cell walls. Zymogram analysis revealed alterations in the pattern of murein hydrolases present in the culture supernatants of UAMS-1 grown in the presence and absence of glucose. Specifically, 34-, 50-, and 110-kDa extracellular murein hydrolases were observed in the culture supernatant of UAMS-1 grown in the absence of glucose (Fig. 2B, lane 1). When UAMS-1 was grown in the presence of glucose, the 50- and 110-kDa murein hydrolases disappeared and the 34-kDa murein hydrolase appeared to be more intense (Fig. 2B, lane 3) relative to its counterpart in the no-glucose culture (Fig. 2B, lane 1). These observations suggest that the increase in the amount of the 34-kDa murein hydrolase is responsible for the overall increase in activity of the UAMS-1 glucose culture observed in the quantitative cell wall hydrolysis assay (Fig. 2A). In contrast to UAMS-1, KB1050 (cidA mutant derivative of UAMS-1) displayed a nearly complete loss of extracellular murein hydrolase activity (Fig. 2A), solubilizing less than 10% of the M. luteus cell walls. Furthermore, this loss of activity was unaffected by culturing this strain in the presence of glucose. This loss of murein hydrolase activity was confirmed by zymogram analysis of the culture supernatants of KB1050 grown in the absence and presence of glucose (Fig. 2B, lanes 2 and 4, respectively): in either case, there were no murein hydrolase bands detected. Taken together, these observations suggest that the increased murein hydrolase activity observed in UAMS-1 grown in the presence of glucose was dependent on the presence of an intact cidA gene.
FIG. 2.
Quantitative cell wall hydrolysis assay (A) and zymogram analysis (B) of extracellular murein hydrolase activities of UAMS-1 and KB1050. (A) Aliquots of 50 μg of extracellular proteins isolated from 16-h cultures of UAMS-1 (wild-type; circles) and KB1050 (cidA mutant; triangles) grown in NZY broth in either the presence of 35 mM glucose (closed symbols) or in the absence of glucose (open symbols) were each added to a 1-mg · ml−1 suspension of M. luteus cell walls, and the murein hydrolase activity of each sample was measured as a decrease in turbidity over a 4-h time course experiment. These data represent the average of three independent experiments, and the error bars correspond to the standard errors of the means. (B) Fifteen micrograms of extracellular proteins, isolated from 16-h cultures of UAMS-1 and KB1050 grown in either the presence of 35 mM glucose or in the absence of glucose, was separated in a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel containing 1 mg of M. luteus cell wall · ml−1, followed by an overnight incubation at 37°C in a buffer containing Triton X-100 and staining with methylene blue. This zymogram is representative of results obtained from three independent experiments. The migrations of molecular mass markers (in kilodaltons) are indicated to the left of the gel. Lane 1, UAMS-1 (no glucose); lane 2, KB1050 (no glucose); lane 3, UAMS-1 (35 mM glucose); lane 4, KB1050 (35 mM glucose).
Effect of glucose on antibiotic tolerance.
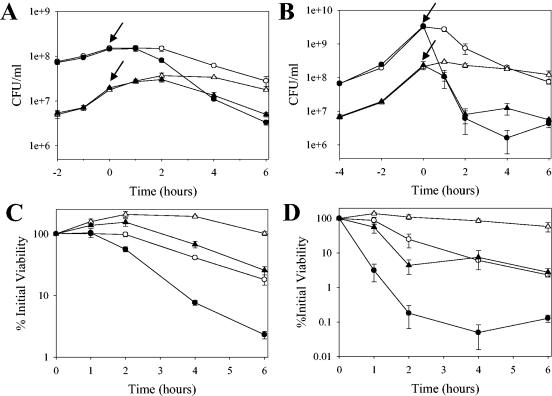
It was also previously found that a cidA mutant derivative of RN6390 displayed increased tolerance to penicillin-induced killing relative to its parental strain (35). Since these studies suggested a role for CidA in antibiotic tolerance, we likewise assessed antibiotic-induced killing of UAMS-1 and KB1050 grown in the presence and absence of glucose (Fig. 3). Although the UAMS-1 strain was found to produce β-lactamase (unpublished data), which precluded the use of penicillin as was performed with the RN6390 laboratory strain, it was found that the cidA mutation had a similar effect on killing induced by the cell wall-active antibiotic vancomycin. As shown in Fig. 3A and C, the cidA mutant (KB1050) displayed increased vancomycin tolerance relative to UAMS-1 when grown in the absence of glucose, yielding almost 1 log unit higher percent viable cell counts compared to UAMS-1 after incubation with this antibiotic. The magnitude of these results was similar to the penicillin tolerance exhibited by the cidA mutant derivative of RN6390 (35). In the presence of glucose, conditions under which cidABC expression was induced, UAMS-1 displayed a decrease in vancomycin tolerance, exhibiting a 2-log decrease in percent viable cell counts compared to just over a 1-log decrease for UAMS-1 grown in the absence of glucose (Fig. 3C). Interestingly, the KB1050 strain grown in the presence of glucose also displayed decreased vancomycin tolerance relative to its growth in the absence of glucose, but it still exhibited increased tolerance relative to UAMS-1 grown in the presence of glucose (Fig. 3C).
FIG. 3.
Comparison of the effect of glucose on antibiotic sensitivity between UAMS-1 and KB1050. Vancomycin (40 μg · ml−1) or rifampin (2 μg · ml−1) was added to cultures of UAMS-1 (wild-type; circles) and KB1050 (cidA mutant; triangles) cells grown in NZY broth in either the presence of 35 mM glucose (closed symbols) or in the absence of glucose (open symbols), and viable cell counts of each culture were determined by dilution plating on tryptic soy agar. These data represent the average of three independent experiments, and error bars correspond to the standard errors of the means. (A and B) Graphs depicting the CFU per milliliter of each culture before and after addition of vancomycin (A) or rifampin (B). The time at which each antibiotic was added to each culture is indicated by an arrow. (C and D) Graphs depicting the percent viability relative to the time of vancomycin (C) or rifampin (D) addition (referred to as the zero time point) for each culture.
Surprisingly, similar results were obtained when rifampin, a transcription inhibitor, was used to challenge cultures of UAMS-1 and KB1050 grown in the presence and absence of glucose (Fig. 3B and D). Specifically, KB1050 had increased rifampin tolerance relative to UAMS-1 when grown in the absence of glucose, displaying greater than 1 log unit higher percent viable cell counts compared to UAMS-1 (Fig. 3D). When grown in the presence of glucose, both UAMS-1 and KB1050 displayed decreased tolerance to rifampin relative to the no-glucose cultures, but the cidA mutant still displayed more tolerance to rifampin relative to UAMS-1 under these conditions (Fig. 3D). Furthermore, UAMS-1 grown in the presence of glucose exhibited a 3-log decrease in percent viable cell counts in response to rifampin challenge compared to just over a 1-log decrease for UAMS-1 grown in the absence of glucose (Fig. 3D). Collectively, these results illustrate that growth in the presence of glucose confers decreased tolerance to both vancomycin and rifampin and that the cidA gene product contributes to this effect.
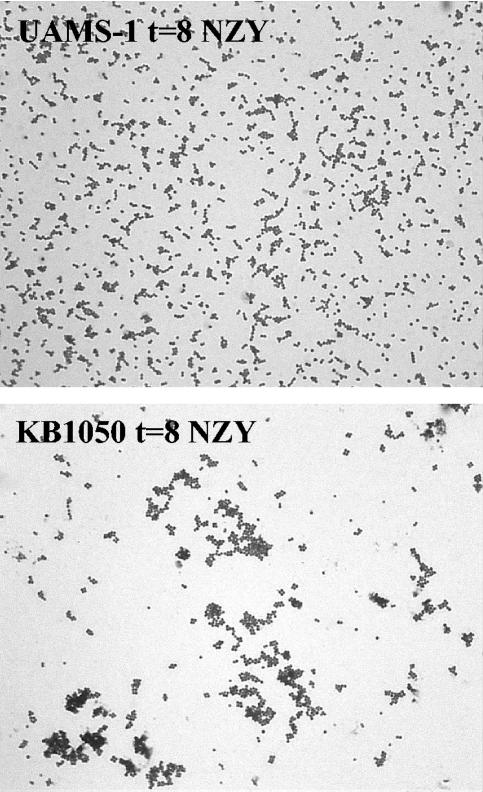
It is important to note that the initial inoculum of both KB1050 cultures appeared to be consistently lower than that of the UAMS-1 cultures, as determined by CFU counts per milliliter (Fig. 3A and B). However, the initial optical densities, as well as the growth rates of UAMS-1 and KB1050, were nearly identical (unpublished data), suggesting that the lower CFU per milliliter values displayed by KB1050 may have been due to cell clumping. To verify that this was the case, light microscopy was performed on both KB1050 and UAMS-1 cultures grown to early stationary phase (Fig. 4). Indeed, KB1050 tended to form large aggregates relative to UAMS-1, and glucose did not appear to have an effect on the degree of clumping in either strain (unpublished data). Interestingly, this clumping phenotype was also found to be associated with disruption of the atl-encoded murein hydrolases of S. aureus (42). Thus, it is likely that the reduction in murein hydrolase activity exhibited by KB1050 is responsible for the clumping phenotype observed with this strain.
FIG. 4.
Light microscopy of UAMS-1 and KB1050. Cultures of UAMS-1 and KB1050 were grown in NZY broth to early stationary phase, and aliquots were heat fixed to glass slides and stained with Gram's crystal violet. Each micrograph is representative of several fields of view. Magnification, ×1,000.
Acetic acid accumulation induces cidABC and lrgAB expression.
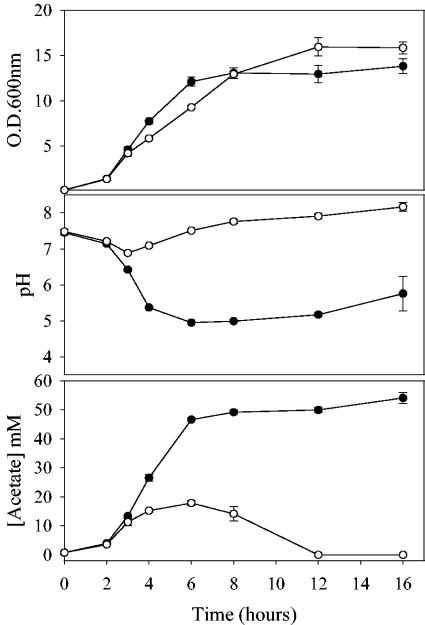
In an effort to elucidate the mechanism by which growth in the presence of glucose increases cidABC and lrgAB expression, the growth yield (measured by optical density), pH, and acetate concentrations of both the glucose-containing cultures and the control (no-glucose) cultures of UAMS-1 were monitored. As shown in Fig. 5, a dramatic drop in the pH (from 7.5 to approximately 5.0) as well as a sharp increase in acetate concentration (from 0 to approximately 25 mM) was observed after 4 h of growth (mid-exponential phase) in the glucose cultures. In comparison, cells grown in the absence of glucose displayed a change in pH from 7.5 to approximately 7.0 and acetate accumulated to approximately 15 mM at 4 h of growth. Therefore, we hypothesized that low pH and/or accumulation of acetate in cultures containing glucose may be the signal that induces high-level expression of cidABC and lrgAB. To test this, a Northern blot analysis was performed on RNA isolated from UAMS-1 cells grown for 4 h in either neutral (pH 7.5) or acidic (pH 5.0) glucose-free NZY medium (Fig. 6). As expected, growth of UAMS-1 in neutral NZY broth did not increase the expression of either cidABC or lrgAB (Fig. 6, lane 1), whereas growth in NZY broth that was supplemented with glucose resulted in increased expression of both transcripts (Fig. 6, lane 2). Growth of UAMS-1 in NZY broth adjusted to pH 5.0 with HCl elevated the expression of cidABC relative to growth in neutral media, whereas an effect on lrgAB expression was negligible (Fig. 6, lane 3). However, growth in NZY broth adjusted to pH 5.0 with acetic acid had a pronounced positive effect on both cidABC and lrgAB expression (Fig. 6, lane 4).
FIG. 5.
Comparison of the growth rate, pH, and acetate concentration in UAMS-1 cultures grown in the presence and absence of glucose. The growth rate, as determined by the OD600 (top graph), pH (middle graph), and acetate concentration (bottom graph), was measured in UAMS-1 cultures grown in NZY broth in either the presence of 35 mM glucose (closed circles) or in the absence of glucose (open circles). Each parameter represents the average of three independent experiments, and error bars correspond to the standard errors of the means.
FIG. 6.
Transcription of cidABC and lrgAB is increased by the presence of acetic acid in S. aureus UAMS-1. Total cellular RNA was isolated from UAMS-1 grown to late exponential growth phase (4 h postinoculum) in either NZY pH 7.5 (lane 1), NZY pH 7.5 plus 35 mM glucose (lane 2), NZY pH 5.0 (HCl) (lane 3), or NZY pH 5.0 (acetic acid) (lane 4), and 10 μg of each sample was separated through a 1% (wt/vol) agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to cidA- and lrgA-specific DIG-labeled probes. The sizes of each transcript were determined by comparison to an RNA ladder (Invitrogen) run on the same gel.
The concentration of acetate in NZY broth adjusted to pH 5.0 with acetic acid (approximately 26 mM) was similar to that found at mid-exponential growth when UAMS-1 was cultured in the presence of 35 mM glucose (Fig. 5). Given that the pKa of acetate is 4.76 (25), these observations suggest that cidABC and lrgAB expression is induced in glucose cultures by the acidic acid that accumulates as a by-product of glucose metabolism as the culture approaches mid-exponential growth (4 h postinoculum). In this respect, UAMS-1 cultures grown in the presence of other utilizable carbohydrate sources, such as sucrose or fructose, induced high-level expression of cidABC and lrgAB at the mid-exponential growth phase and also displayed a concurrent drop in pH and increase in acetate similar to that observed in Fig. 5 (unpublished data). In contrast, growth of UAMS-1 in the presence of xylose, a nonutilizable carbohydrate source, did not upregulate expression of cidABC and lrgAB and likewise did not display a drop in pH and increase in acetate concentration (unpublished data). Furthermore, the moderately increased expression of cidABC observed in NZY broth adjusted to pH 5.0 with HCl (Fig. 6, lane 3) may have been a result of the low levels of acetate that are normally found in NZY at this time point (Fig. 5).
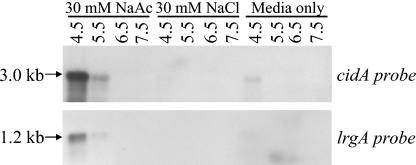
To confirm that acetic acid is the signal responsible for stimulating cidABC and lrgAB expression, the ability of 30 mM acetate to induce cidABC and lrgAB expression was assessed by Northern blot analysis on cultures grown for 4 h at a wide range of pH values (4.5 to 7.5) (Fig. 7). In agreement with our hypothesis, expression of both cidABC and lrgAB dramatically increased in the low-pH medium containing 30 mM Na-acetate (pH 4.5 to 5.5) (Fig. 7), whereas expression of both of these transcripts was undetectable in the high-pH medium containing 30 mM Na-acetate (pH 6.5 to 7.5). By comparison, cidABC and lrgAB expression was barely detectable at any of the pH values tested in medium containing 30 mM NaCl or in medium alone (Fig. 7). Furthermore, when several weak acids were tested for their ability to stimulate cidABC expression, it was found that upregulation of cidABC expression was limited to only acetic acid and lactic acid (unpublished data), suggesting that this effect is specific to weak acids produced as by-products of glucose metabolism.
FIG. 7.
Effect of pH on the ability of acetate to increase transcription of cidABC and lrgAB in S. aureus UAMS-1. Total cellular RNA was isolated from UAMS-1 cells grown to the mid-exponential growth phase (4 h postinoculum) in NZY at increasing pH values (from 4.5 to 7.5, as indicated above each lane of the blot), containing either 30 mM sodium acetate or 30 mM sodium chloride as indicated. Five micrograms of each sample was separated through a 1% (wt/vol) agarose-formaldehyde gel, transferred to a nylon membrane, and hybridized to cidA- and lrgA-specific DIG-labeled probes. The sizes of each transcript were determined by comparison to an RNA ladder (Invitrogen) run on the same gel.
DISCUSSION
This study demonstrates that carbohydrate metabolism has a dramatic impact on murein hydrolase activity and antibiotic tolerance and that these effects can be attributed, at least in part, to changes in the expression of the cid and lrg operons. Metabolism of glucose and generation of acetic acid resulted in increased expression of both the cidABC and lrgAB transcripts during mid-exponential growth. Furthermore, it was found that murein hydrolase activity was increased and tolerance to both vancomycin and rifampin was decreased in these cultures relative to growth in the absence of glucose. Collectively, these observations support the hypothesis that the cid operon positively regulates murein hydrolase activity and tolerance to antibiotics (34, 35). The fact that lrgAB expression was also increased under these conditions seems counterintuitive, given that this operon has previously been shown to have an opposing effect on murein hydrolase activity and penicillin tolerance (22). However, similar to the S107 inhibitor holin produced by bacteriophage lambda, LrgA may be able to act as an effector holin under certain physiological conditions. For example, in the absence of the S105 effector holin of bacteriophage λ, the S107 inhibitor holin was shown to be able to induce cell lysis under conditions that depolarized the cytoplasmic membrane (21). Our results reported here have also demonstrated that the effect of glucose on cidABC and lrgAB transcription is due to, at least in part, the accumulation of acetic acid in the culture supernatant as a consequence of aerobic glucose metabolism (9), since the effects of glucose on expression of these two operons could be mimicked by the addition of 30 mM acetate to low-pH cultures that did not contain glucose. Interestingly, cidABC expression could also be increased when grown in low-pH (5.0) medium, but the effect was less dramatic than that in medium containing acetic acid. One explanation accounting for this is that the levels of acetate produced by growth in glucose-free NZY medium (Fig. 5) were converted to its acid form when the medium was adjusted to pH 5.0, whereas growth in standard NZY medium does not cause this drop in pH.
In addition to the ability of glucose-containing cultures to increase cidABC transcription, increase murein hydrolase activity, and decrease tolerance to antibiotics, a more definitive role for the cidA gene product as a positive regulator of murein hydrolase activity and antibiotic tolerance was also demonstrated in this study. Although we cannot rule out the possibility that the cidA mutant phenotype is due to a polar effect on expression of the cidB and/or cidC genes, two unpublished experiments suggest that this is unlikely: first, Northern blot analysis of KB1050 (cidA mutant) has shown that expression of the cidBC transcript is comparable to the level of expression found in UAMS-1. Second, three different UAMS-1 mutants defective in the cidB and/or cidC genes produced increased murein hydrolase activity compared to the decreased activity observed with the cidA mutant. Unfortunately, attempts at complementation of the UAMS-1 cidA mutant by supplying cidA, cidAB, or cidABC in trans from a plasmid were unsuccessful. However, this was not surprising considering that a similar strategy was previously ineffective in complementing the penicillin tolerance phenotype of KB350, the cidA mutant derivative of RN6390 (35). Collectively, these observations suggest that the phenotype of KB1050 is likely caused by a defect in CidA itself rather than an effect on the downstream cid genes.
Previously, a cidA mutant of the laboratory strain RN6390 displayed a decrease in extracellular murein hydrolase activity and increased penicillin tolerance (35). However, the phenotype of an isogenic cidA mutant of the clinical isolate UAMS-1 was more dramatic, as it displayed a complete loss of extracellular murein hydrolase activity. This loss of murein hydrolase activity displayed by the cidA mutant was also unaffected when this strain was grown in the presence of glucose, suggesting that the increased extracellular murein hydrolase activity observed in UAMS-1 when grown in the presence of glucose may be a result of increased expression of the cidABC transcript. The cidA mutant displayed increased tolerance to the inhibitory effects of vancomycin and rifampin relative to UAMS-1, which is also reminiscent of the increased tolerance previously observed for the RN6390 cidA mutant to penicillin (35). The similar growth rates of the cidA mutant and parental strains, as well as of cells grown in the presence or absence of glucose (Fig. 3A and B), rule out the possibility that differences in antibiotic tolerance are attributable to growth rate effects. Furthermore, these results illustrate that the cid operon affects tolerance to antibiotics with two distinct cellular targets, supporting the previously proposed model that one or more of the cid gene products mediates a common cell death mechanism in response to antibiotic stress (34).
KB1050, the cidA mutant of UAMS-1, also formed aggregates of cells when growing in broth culture relative to growth of UAMS-1 (Fig. 4), and this cell clumping was reflected by the fact that the CFU of KB1050 per milliliter was consistently lower than that of the parental strain (Fig. 3A and B), despite the observation that the OD600 values of both of these strains were comparable (unpublished data). In light of these observations, it is possible that the increased antibiotic tolerance observed in KB1050 may be attributable to the clumping phenotype of this strain. However, the fact that the cidA mutant derivative of RN6390 does not display cell clumping (unpublished data) yet is still more tolerant to penicillin-induced killing relative to its parental strain (35) supports the idea that the antibiotic tolerance displayed by KB1050 is a consequence, at least in part, of loss of the cidA gene. Since the cidA mutant derivative of the standard laboratory strain, RN6390, did not display cell clumping when grown in either the presence or absence of glucose (unpublished data), it is likely that other, as-yet-unidentified strain-dependent differences account for the ability of the cidA mutation to confer a clumping phenotype in UAMS-1 and not in RN6390. These differences are likely attributable to the observation that the RN6390 cidA mutant only displayed a modest decrease in extracellular murein hydrolase activity (35), whereas the UAMS-1 cidA mutant displayed a nearly complete loss of extracellular murein hydrolase activity. Previously, an S. aureus atl mutant was shown to form large clusters of cells (42), and inactivation of specific murein hydrolases in other bacteria has led to incomplete cell separation, such as long chain formation in a lytB mutant of Streptococcus pneumoniae (15, 18) and long-chain formation in B. subtilis containing multiple murein hydrolase mutations (6). Therefore, the loss of one or more murein hydrolase activities in the cidA mutant of UAMS-1 may be responsible for its cell clumping phenotype. Collectively, these results demonstrate that the cidA gene product regulates extracellular murein hydrolase activity and antibiotic tolerance in a similar manner in two distinct genetic backgrounds.
A significant finding of this study was that acetic acid, a product of glucose metabolism, was capable of inducing transcription of cidABC and lrgAB. The exact mechanism by which glucose metabolism and acetic acid promote high-level expression of these transcripts is unknown. It has been postulated that the undissociated forms of certain weak organic acids are capable of permeating the cell membrane and subsequently dissipating the proton motive force (2). Therefore, one possibility is that the effect of acetic acid on expression of the cid and lrg operons is simply due to general stress induced by weak acids. However, unpublished data from our laboratory have shown that among several weak acids tested, only acetic acid and lactic acid were capable of stimulating cidABC expression, suggesting that this response is specific for acids produced as by-products of glucose metabolism. Another possibility stems from the observation that the CcpA protein of B. subtilis has been shown by microarray analysis to regulate the expression of ywbHG (homologues of the S. aureus cidAB genes) when grown in the presence of 1% (wt/vol) glucose (30). Thus, one prospect is that S. aureus CcpA may also influence expression of cidABC and/or lrgAB when grown in glucose. However, it is important to consider the possibility that the results of the microarray performed by Moreno et al. (30) may also have been attributable, in part, to an indirect effect that deletion of ccpA had on the levels of acetic acid production. The S. aureus ccpA gene was recognized by transposon mutagenesis as contributing to methicillin resistance in strain COL (16), and various purine biosynthesis and glycolysis genes were identified by microarray analysis as being induced in response to vancomycin in vancomycin intermediate-resistant S. aureus strains (29), suggesting that glucose metabolism may be involved in resistance to this antibiotic. A second possible mode of regulation stems from the fact that the cidC gene encodes a putative pyruvate oxidase (36). The PoxB pyruvate oxidase of E. coli produces acetate and carbon dioxide as by-products of the oxidative decarboxylation of pyruvate in stationary phase (1, 12, 13, 19). Whether or not acetate production by a cidC-encoded pyruvate oxidase, in combination with the drop in pH that occurs as a consequence of metabolism of glucose, could represent an autoregulatory mechanism that induces high-level expression of cidABC and/or lrgAB is currently being investigated in our laboratory.
The ability of acetic acid to stimulate high-level expression of cidABC and lrgAB is intriguing, since these gene products have been recently postulated to be involved in bacterial programmed cell death (34, 3). Weak organic acids such as acetic acid and lactic acid have long been used as preservatives in food preparation. However, the exact mechanism by which they exert their antibacterial properties is still unknown (38, 37, 41). Could they function by inducing programmed cell death in bacteria by inducing expression of cidABC and lrgAB? Although more evidence is required to support this idea, a recently published study by Somerville et al. (40) has demonstrated that inactivation of the tricarboxylic acid cycle in S. aureus prevented catabolism of acetate in late exponential growth phase and enhanced survival in stationary phase. Based on these results, they proposed that acetate is required as an energy source for promoting cell death in stationary phase (40). Acetic acid has also been shown to elicit cellular changes similar to mammalian apoptosis in the yeast species Saccharomyces cerevisiae (26, 28), Zygosaccharomyces bailii (27), and Candida albicans (32). These changes included Annexin-V staining, DNA fragmentation, and production of reactive oxygen species. Additionally, programmed cell death in response to treatment of S. cerevisiae with acetic acid has been shown to be dependent on mitochondrial function (26), an exciting observation given that the LrgA and CidA proteins have been proposed to perform functions analogous to the Bax and Bcl protein families that facilitate induction of apoptosis via the mitochondrial pathway (3). As well, the short-chain fatty acids propionate and acetate, produced by two Propionibacterium species, were identified as cytotoxic components found in the bacterial culture supernatants that were capable of inducing apoptosis via the mitochondrial pathway of two colorectal carcinoma cell lines (23). Specifically, the culture supernatants as well as the purified short-chain fatty acids were able to cause several apoptotic phenotypes, such as loss of mitochondrial membrane potential, generation of reactive oxygen species, and nuclear chromatin condensation (23). Involvement of the mitochondrial pathway was illustrated by the finding that induced Bcl-2 expression was able to protect the cells against the effects of acetate and butyrate (23). Whether or not these systems are functionally homologous to the cid/lrg regulatory system described here is the subject of future investigations.
Acknowledgments
We thank Mark Smeltzer (University of Arkansas Medical Sciences Center, Little Rock) for kindly providing UAMS-1 and Linda Liou for providing technical support.
This work was funded by NIH grant no. R01AI038901, NIH-NRRI grant no. P20RR15587, and DOD grant no. DAAD 19-03-1-0191.
REFERENCES
- 1.Abdel-Hamid, A. M., M. M. Attwood, and J. R. Guest. 2001. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147:1483-1498. [DOI] [PubMed] [Google Scholar]
- 2.Axe, D. D., and J. E. Bailey. 1995. Transport of lactate and acetate through the energized cytoplasmic membrane of Escherichia coli. Biotechnol. Bioeng. 47:8-19. [DOI] [PubMed] [Google Scholar]
- 3.Bayles, K. W. 2003. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 11:306-311. [DOI] [PubMed] [Google Scholar]
- 4.Bayles, K. W. 2000. The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8:274-278. [DOI] [PubMed] [Google Scholar]
- 5.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackman, S. A., T. J. Smith, and S. J. Foster. 1998. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiology 144:73-82. [DOI] [PubMed] [Google Scholar]
- 7.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blevins, J. S., M. O. Elasri, S. D. Allmendinger, K. E. Beenken, R. A. Skinner, J. R. Thomas, and M. S. Smeltzer. 2003. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect. Immun. 71:516-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumenthal, H. J. 1972. Glucose catabolism in staphylococci, p. 111-115. In J. O. Cohen (ed.), The staphylococci. Wiley Interscience, New York, N.Y.
- 10.Brunskill, E. W., and K. W. Bayles. 1996. Identification and characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, Y. Y., and J. E. Cronan, Jr. 1983. Genetic and biochemical analyses of Escherichia coli strains having a mutation in the structural gene (poxB) for pyruvate oxidase. J. Bacteriol. 154:756-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, Y. Y., A. Y. Wang, and J. E. Cronan, Jr. 1994. Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS(katF) gene. Mol. Microbiol. 11:1019-1028. [DOI] [PubMed] [Google Scholar]
- 14.Cheung, A. L., K. J. Eberhardt, and V. A. Fischetti. 1994. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal. Biochem. 222:511-514. [DOI] [PubMed] [Google Scholar]
- 15.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-beta-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 184:4988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lencastre, H., S. W. Wu, M. G. Pinho, A. M. Ludovice, S. Filipe, S. Gardete, R. Sobral, S. Gill, M. Chung, and A. Tomasz. 1999. Antibiotic resistance as a stress response: complete sequencing of a large number of chromosomal loci in Staphylococcus aureus strain COL that impact on the expression of resistance to methicillin. Microb. Drug Resist. 5:163-175. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto, D. F., E. W. Brunskill, and K. W. Bayles. 2000. Analysis of genetic elements controlling Staphylococcus aureus lrgAB expression: potential role of DNA topology in SarA regulation. J. Bacteriol. 182:4822-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia, P., M. P. Gonzalez, E. Garcia, R. Lopez, and J. L. Garcia. 1999. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 31:1275-1277. [DOI] [PubMed] [Google Scholar]
- 19.Gennis, R. B., and L. P. Hager. 1976. Pyruvate oxidase, p. 493-504. In A. Martonosi (ed.), The enzymes of biological membranes, vol. 2. Plenum, New York, N.Y.
- 20.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graschopf, A., and U. Blasi. 1999. Molecular function of the dual-start motif in the lambda S holin. Mol. Microbiol. 33:569-582. [DOI] [PubMed] [Google Scholar]
- 22.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jan, G., A.-S. Belzacq, D. Haouzi, A. Rouault, D. Metivier, G. Kroemer, and C. Brenner. 2002. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ. 9:179-188. [DOI] [PubMed] [Google Scholar]
- 24.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehninger, A. L., D. L. Nelson, and M. M. Cox. 1993. Principles of biochemistry, 2nd ed. Worth Publishers, New York, N.Y.
- 26.Ludovico, P., F. Rodrigues, A. Almeida, M. T. Silva, A. Barrientos, and M. Corte-Real. 2002. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol. Biol. Cell 13:2598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludovico, P., F. Sansonetty, M. T. Silva, and M. Corte-Real. 2003. Acetic acid induces a programmed cell death process in the food spoilage yeast Zygosaccharomyces bailii. FEMS Yeast Res. 3:91-96. [DOI] [PubMed] [Google Scholar]
- 28.Ludovico, P., M. J. Sousa, M. T. Silva, C. Leao, and M. Corte-Real. 2001. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology 147:2409-2415. [DOI] [PubMed] [Google Scholar]
- 29.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 31.Papakyriacou, H., D. Vaz, A. Simor, M. Louie, and M. J. McGavin. 2000. Molecular analysis of the accessory gene regulator (agr) locus and balance of virulence factor expression in epidemic methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 181:990-1000. [DOI] [PubMed] [Google Scholar]
- 32.Phillips, A. J., I. Sudbery, and M. Ramsdale. 2003. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. USA 100:14327-14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, K. C., and K. W. Bayles. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50:729-738. [DOI] [PubMed] [Google Scholar]
- 35.Rice, K. C., B. A. Firek, J. B. Nelson, S.-J. Yang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in the regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rice, K. C., T. G. Patton, S.-J. Yang, A. Dumoulin, M. Bischoff, and K. W. Bayles. 2004. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by the alternative sigma factor B (σB). J. Bacteriol. 186:3029-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Russell, J. B. 1992. Another explanation for the toxicity of fermentation acids at low pH: anion accumulation versus uncoupling. J. Appl. Bacteriol. 73:363-370. [Google Scholar]
- 38.Russell, J. B., and F. Diez-Gonzalez. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 39.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 40.Somerville, G. A., M. S. Chaussee, C. I. Morgan, J. R. Fitzgerald, D. W. Dorward, L. J. Reitzer, and J. M. Musser. 2002. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70:6373-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stratford, M., and P. A. Anslow. 1996. Comparison of the inhibitory action on Saccharomyces cerevisiae of weak-acid preservatives, uncouplers, and medium-chain fatty acids. FEMS Microbiol. Lett. 142:53-58. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi, J., H. Komatsuzawa, S. Yamada, T. Nishida, H. Labischinski, T. Fujiwara, M. Ohara, J. Yamagishi, and M. Sugai. 2002. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol. Immunol. 46:601-612. [DOI] [PubMed] [Google Scholar]