Abstract
In bacteria, whereas disruption methods have been improved recently, the use of plasmid complementation strategies are still subject to limitations, such as cloning difficulties, nonphysiological levels of gene expression, or a requirement for antibiotics as plasmid selection pressure. Moreover, because of the pleiotropic modifications of cell physiology often induced by plasmid-based complementation, these strategies may introduce biases when biological process such as adhesion or biofilm formation are studied. We developed a plasmid-free approach that combines the lambda-red linear DNA recombination method with site-directed insertion of a repression and expression (RExBAD) cassette which places a functional pBAD promoter upstream of a target gene. We showed that this method permits both inactivation and modulation of most Escherichia coli gene expression, including expression of toxin and essential genes. We used this strategy to study adhesion and bacterial biofilms by placing the RExBAD cassette in front of the tra operon, encoding the DNA transfer and pilus genes on the F conjugative plasmid, and in front of flu, the antigen 43 (Ag43) autotransporter adhesin-encoding gene. In silico analysis revealed the existence of 10 genes with homology to the Ag43 gene that were good candidates for genes that encode putative new adhesins in E. coli. We used the RExBAD strategy to study these genes and demonstrated that induction of expression of four of them is associated with adhesion of E. coli to abiotic surfaces. The potential use of the RExBAD approach to study the function of cryptic or uncharacterized genes in large-scale postgenomic functional analyses is discussed.
In spite of the rapid accumulation of prokaryotic sequence information, gene-to-function studies remain relatively slow. As a consequence, only a fraction of the wealth of available microbial genomic information has been explored, and filling the gap between in silico data and the functional characterization of genes is clearly one of the main challenges of the postgenomic era. Recently, significant progress toward large-scale functional analysis has been made with several model prokaryotic organisms. Knockouts of all the genes in a bacterial genome are available for naturally competent bacteria (e.g., Bacillus subtilis), and the development of simple reverse-genetic methods now allows rapid inactivation of a large number of genes in gram-negative bacteria such as Escherichia coli (4, 9, 10, 27, 35, 43, 52). These gene replacement methods, however, do not alleviate the need for genetic complementation in order to validate the phenotype of a gene mutation. More importantly, gene knockout strategies are informative only if the gene of interest is expressed under the experimental conditions used. In addition, workers typically clone and place the candidate genes under the control of an active promoter in order to assess the phenotypic consequences of its expression.
In contrast to gene inactivation, cloning and complementation strategies still require time-consuming steps that limit their use in high-throughput global analysis. Moreover, nonphysiological levels of plasmid-based gene expression may affect the physiological relevance of the complementation phenotype. To circumvent these limitations, several strategies have been developed; these strategies range from the use of low-copy-number plasmids to methods that integrate the complementing allele as a stable, single chromosomal copy. However, most of these methods still involve at least one cloning step. New strategies are required to unravel the functions of genes that have not yet been addressed by traditional microbiology.
Bacterial biofilms are matrix-enclosed communities of bacteria that interact with each other and/or with a surface. Biofilms are thought to require the differential expression of genes that are otherwise cryptic or phenotypically silent under experimental conditions that address the classical planktonic lifestyle. Therefore, physiological gene expression strategies are particularly critical for biofilm studies. Indeed, the pleiotropic modifications of adhesion properties due to the use of antibiotic selection, protein overproduction, or slow growth due to plasmid burden often lead to aberrant results, limiting the utility of plasmid-based complementation strategies.
Here we describe a one-step, combined repression and expression (RExBAD) strategy that allows functional analysis of any chromosomal gene, including toxic and essential genes. We used this approach in the context of biofilm formation and showed that the chromosomal expression of previously uncharacterized large open reading frames (ORFs) that display homology to the gene encoding the E. coli surface autotransporter adhesin antigen 43 (Ag43) are associated with biofilm formation.
Our results validate a new physiologically relevant alternative to conventional inactivation and plasmid-based expression methods. Our approach is applicable to most members of the Enterobacteriaceae, and, considering the high proportion of genes with unassigned or poorly characterized functions in most completed microbial genomes, could be instrumental in high-throughput postgenomic phenotypic analysis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Strains were constructed by transformation and the λ-red linear DNA gene inactivation method (see below), followed by P1vir transduction in a fresh E. coli background.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| TG1 | F′[traD36 proAB+lacIqlacZΔM15] supE hsdΔ5 thi Δ(lac-proAB) | Laboratory collection |
| TG1cat-araC | cat gene inserted downstream of the araC gene, Cmr | This study |
| TG1spec-araC | aadA7 gene inserted downstream of the araC gene, Specr | This study |
| TG1ΔtraA | F′ΔtraA::aphaIII, Kmr | Laboratory collection |
| TG1 F′(catRExBADtraY) | F′tra operon placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| TG1 F′(ΔccdAspecRExBADccdB) | F′ΔccdA, ccdB placed under control of the pBAD promoter of the specRExBAD cassette, Specr | This study |
| TG1catRExBADftsL | ftsL placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655 | l−rph-l | Laboratory collection |
| MG1655ΔlacIZ | ΔlacIZ::aphaIII, Kmr | Laboratory collection |
| MG1655ΔaraBADΔlacIZ | Ara− derivative of MG1655ΔlacIZ, Specr Kmr | This study |
| MG1655Δflu | Δflu::cat, Cmr | Laboratory collection |
| MG1655ΔoxyR | oxyR::aphaIII, Kmr | Laboratory collection |
| MG1655ΔoxyRΔflu | oxyR::aphaIII Δflu::cat, Kmr Cmr | Laboratory collection |
| MG1655catRExBADlacZ | lacZ placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655ΔaraBADcatRExBADlacZ | Ara− derivative of MG1655 catRExBADlacZ, Specr Kmr | This study |
| MG1655catRExBADflu | flu placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655ΔoxyRcatRExBADflu | P1vir transduction of ΔoxyR in MG1655 catRExBADflu, Kmr Cmr | This study |
| MG1655ΔypjA | ΔypjA::aphaIII, Kmr | This study |
| MG1655ΔyfaL | ΔyfaL::aphaIII, Kmr | This study |
| MG1655ΔycgH | ΔycgH::aphaIII, Kmr | This study |
| MG1655ΔydhQ | ΔydhQ::aphaIII, Kmr | This study |
| MG1655ΔycgV | ΔycgV::aphaIII, Kmr | This study |
| MG1655ΔydeKU | ΔydeKU::aphaIII, Kmr | This study |
| MG1655ΔyeeJ | ΔyeeJ::aphaIII, Kmr | This study |
| MG1655catRExBADypjA | ypjA placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655catRExBADyfaL | yfaL placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655catRExBADycgH | ycgH placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655catRExBADydhQ | ydhQ placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655catRExBADycgV | ycgV placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655catRExBADydeKU | ydeKU placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| MG1655catRExBADyeeJ | yeeJ placed under control of the pBAD promoter of the catRExBAD cassette, Cmr | This study |
| Plasmids | ||
| pKOBEG | pSC101 ts, araC arabinose-inducible λ redγβα operon, Cmr | 4 |
| pKOBEGA | Like pKOBEG but Ampr instead of Cmr | 4 |
| pKKccdA | ccdA cloned in the pKK223-3 plasmid, pTac controlled, Ampr | L. Van Melderen |
Growth conditions.
All experiments were performed in M63B1 minimal medium or in Luria-Bertani (LB) medium at 37°C (45). The following antibiotics were added when required: kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), ampicillin (100 μg/ml), and spectinomycin (50 μg/ml). All media and antibiotics were used as described by Miller (33).
Induction of the RExBAD constructs was achieved with arabinose (Ara) concentrations ranging from 0.004 to 1%. Glucose (Glu) was added to a final concentration of 0.4 or 1%. For studies of the essential gene ftsL, overnight cultures of TG1catRExBADftsL grown in LB medium supplemented with 0.2% arabinose were washed, resuspended in LB medium, and then diluted 1:100 into LB medium or LB medium supplemented with 0.2% glucose or with 0.2% arabinose. After about 2 h, optical microscopy images (magnification, ×40) were captured each 30 min.
Three-step PCR.
In order to interrupt Ag43 homolog-encoding genes, as well as to place chromosomal target genes under control of the RExBAD cassette, we used a three-step PCR procedure described previously (9, 10) and at our website (http://www.pasteur.fr/recherche/unites/Ggb/methodes.ang.html). The primers used to disrupt or insert the cassette upstream of the genes used in this study (lacZ, flu, ftsL, ccdB, tra operon, ypjA, ydhQ, ycgH, yfaL, ydeK-ydeU, ycgV, and yeeJ) are listed in Table 1S in the supplemental material at http://www.pasteur.fr/recherche/unites/Ggb/supmat.html. Chromosomal insertion of the pBAD promoter of the catRExBAD or specRExBAD cassette was done under inducing conditions (medium containing Ara) for the ftsL essential gene and under repressing conditions (medium containing Glu) for the ccdB toxic gene.
Construct verification.
All constructs were checked by PCR performed with specific primers (Table 1S in the supplemental material). The integrity of the cassette was verified by sequencing the junction between the RExBAD cassette and the target gene by using primers described in Table 1S.
β-Galactosidase assays.
To determine the β-galactosidase enzyme activity, MG1655, MG1655ΔlacIZ, and MG1655catRExBADlacZ cultures were grown in LB medium for 8 h. The cultures were diluted 1:100 in LB medium or M63B1-0.4% Glu medium containing various amounts of arabinose or glucose and grown overnight (16 to 18 h). The enzyme activity was assayed in triplicate for each strain as described by Miller (33) and was expressed in arbitrary Miller units.
Aggregation test.
Cells were grown in LB medium for 8 h. The cultures were diluted 1:100 in LB medium or M63B1-0.4% Glu medium containing various amounts of arabinose or glucose and grown overnight (16 to 18 h). The culture optical density at 600 nm (OD600) was adjusted to 2.5 by dilution with LB medium or M63B1 medium, and 3-ml portions of each culture were transferred to 5-ml tubes. These tubes were incubated without agitation at room temperature for 24 h before image capture. The final OD600 of the upper part of each standing tube culture was determined after incubation for 24 h at room temperature.
Ag43 immunodetection.
For each culture the equivalent of 0.2 OD600 unit was analyzed by sodium dodecyl sulfate—10% polyacrylamide gel electrophoresis, followed by immunodetection of Ag43. Equivalent protein loads in the lanes were verified by staining the nitrocellulose membranes with Ponceau S. Immunodetection was performed by using a 1:10,000 dilution of polyclonal rabbit antiserum raised against the α domain of Ag43.
Biofilm formation assay in microtiter plates.
The initial steps of biofilm formation were assayed by determining the ability of cells to adhere to the wells of 96-well polyvinyl chloride (PVC) microtiter dishes (14, 40). M63B1 medium containing 0.4% glucose with or without 0.1% arabinose (100 μl/well) was inoculated with a 1/100 dilution from an overnight M63B1-0.4% glucose medium culture. After inoculation, the plates were incubated at 37°C for 24 h and rinsed, and 125 μl of a 1% solution of crystal violet was added to each well. The plates were incubated at room temperature for 15 min and rinsed, and biofilm formation was tested as follows: the crystal violet was solubilized by addition of 200 μl of ethanol-acetone (80:20), and the OD570 was determined. The results are averages for four replicate wells in three independent experiments.
Biofilm formation assay in microfermentors.
All experiments were performed in triplicate in M63B1 minimal medium containing 0.4% glucose supplemented with 0.004, 0.04, or 0.4% arabinose at 37°C. Sixty-milliliter microfermentors containing a removable glass slide were configured as continuous-flow culture bioreactors with a flow rate of 40 ml · h−1 (3, 15). Bacterial inocula equivalent to an OD600 of 1 from overnight precultures grown in M63B1 medium containing 0.4% glucose supplemented with appropriate antibiotics when required were used to inoculate the microfermentors and were cultivated for 24 h. Images of each removable glass slide were captured at the end of the incubation period, and the OD600 of the biofilm organisms resuspended in 10 ml of M63B1 medium was determined.
RESULTS
Efficient site-directed promoter replacement with the RExBAD cassette.
Recently, Judson et al. developed a procedure using a Mariner-based transposon (TnAraOut) that identified essential genes by replacing their natural promoters with the positively regulated, arabinose-inducible pBAD promoter (18, 26, 37, 46). Their results indicated that random chromosomal replacement of native promoters with the pBAD promoter can lead to a knockout phenotype. Haldimann et al. (19) also showed that the expression of chromosomal pBAD promoter fusions to the phoB and phoR genes at the araCBAD locus could be modulated by catabolite repression with different sugars. Their approach, however, still required cloning of the gene of interest into a suicide plasmid and subsequent selection for two recombination events in a strain in which the gene of interest had been deleted previously.
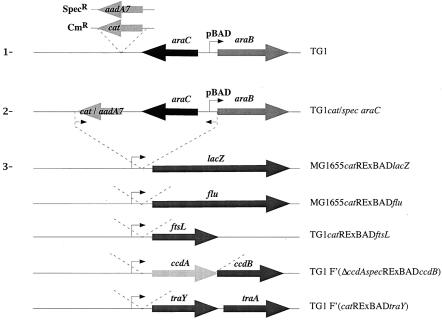
Here, we used λ-red-mediated homologous recombination to place different target genes under the control of the inducible pBAD promoter directly on the chromosome at their native loci. We first generated a selectable pBAD repression/expression cassette, RExBAD, by inserting the chloramphenicol acetyltransferase gene (cat) downstream of the araC gene in the araB region on the chromosome of E. coli K-12 strain TG1 (Fig. 1). The resulting strain, TG1cat-araC, was used as a template to amplify the 2,239-bp catRExBAD cassette carrying both the cat selection marker and the araC-pBAD region. This DNA region encodes a functional arabinose-inducible pBAD promoter, as well as the araB ribosome-binding site. A similar specRExBAD cassette, with the aadA7 gene encoding spectinomycin resistance, was also constructed (Fig. 1).
FIG. 1.
Construction of RExBAD cassettes. The cassettes were constructed in three steps: (i) introduction of the cat gene, encoding resistance to chloramphenicol, or of the aadA7 gene, encoding resistance to spectinomycin downstream of the araC gene on the chromosome of E. coli K-12 strain TG1 by using a three step PCR (step 1); (ii) amplification of the 2,239-bp catRExBAD or 2,400-bp specRExBAD cassette by using primers annealing to the 3′ end of the antibiotic resistance gene and just upstream of the 5′ end of the araB gene (primers are represented by small arrows) (step 2); and (iii) insertion of the catRExBAD or specRExBAD cassette by a three-step PCR upstream of the target genes lacZ, ftsL, flu, ccdB, and traY (step 3). In the case of ccdB, the specRExBAD cassette replaces ccdA. The primers used to construct the cassettes are described in Table 1S in the supplemental material at http://www.pasteur.fr/recherche/unites/Ggb/supmat.html.
The functionality of the catRExBAD cassette was tested by inserting it upstream of the lacZ gene, between its native promoter and the lacZ start codon in the E. coli K-12 strain MG1655 chromosome (Fig. 1). This event replaced the native lacZ promoter with the arabinose-inducible promoter pBAD (MG1655catRExBADlacZ). This site-directed promoter replacement resulted in a functional transcriptional fusion, as indicated by the blue color of MG1655catRExBADlacZ colonies when they were streaked on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) agar plates supplemented with arabinose, the inducer of the pBAD promoter (data not shown).
These results indicate that the RExBAD cassette can be used to place a target gene under the control of the inducible pBAD promoter directly on the E. coli chromosome.
Chromosomal replacement of native promoters with RExBAD permits down-regulation that is phenotypically equivalent to gene deletion.
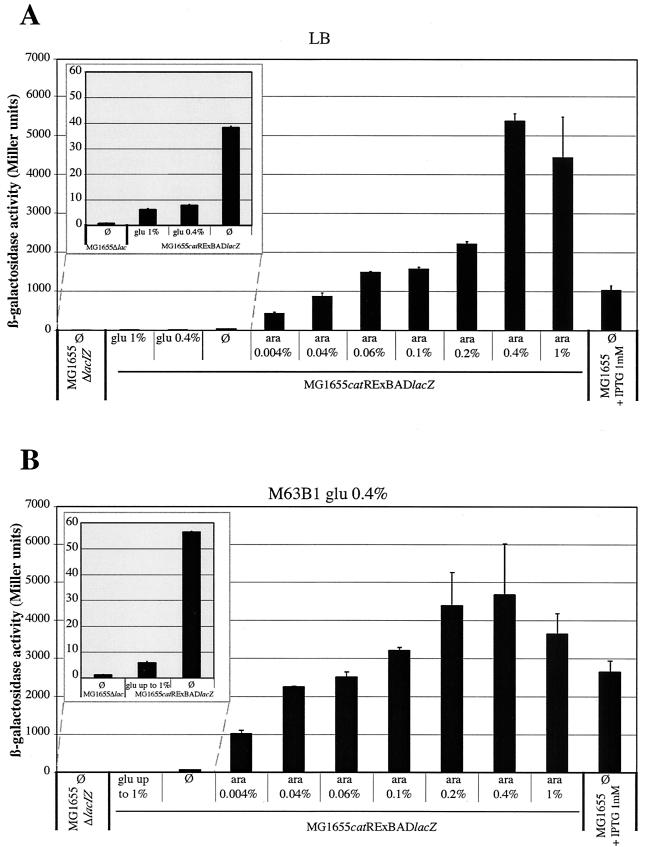
In order to determine the extent of the repression conferred by the RExBAD cassette, we investigated the level of expression of the lacZ gene in strain MG1655catRExBADlacZ grown under repressing conditions and compared it to the level of expression in the lac-minus strain MG1655ΔlacIZ. Whereas MG1655ΔlacIZ had no observable β-galactosidase activity in the absence or presence of any sugar, MG1655catRExBADlacZ cultures displayed low (<10 U) but detectable β-galactosidase activity in LB medium and M63B1 medium supplemented with 0.4 and 1% glucose (Fig. 2, insets). As expected, reducing the concentration of glucose or omitting glucose from the culture medium led to a small increase in the β-galactosidase activity detected in MG1655catRExBADlacZ (up to ca. 40 to 55 U) (Fig. 2, insets). Thus, we tested whether the repression of the lacZ gene in MG1655catRExBADlacZ could mimic a lacZ null mutant phenotype. First, the araBAC operon of strains MG1655ΔlacIZ and MG1655catRExBADlacZ was deleted to avoid the use of arabinose as a carbon source under minimal medium conditions. Like MG1655ΔaraBADΔlacIZ, MG1655ΔaraBADcatRExBADlacZ could not grow on M63B1 minimal medium supplemented with 0.4% lactose as a sole carbon source. However, addition of 0.1% arabinose was enough to restore its growth (data not shown).
FIG. 2.
RExBAD cassette can modulate gene expression. The histograms show the β-galactosidase activities of strain MG1655ΔlacIZ, strain MG1655catRExBADlacZ, and wild-type strain MG1655 grown to the stationary phase in LB medium (A) and in M63B1 medium containing 0.4% glucose (B). The concentration of the added sugar, either glucose (glu) or arabinose (ara), is indicated under each bar. Ø indicates that no sugar was added. The MG1655ΔlacIZ strain corresponded to a lac-minus control strain, whereas the expression of the lac operon by MG1655 grown in presence of 1 mM IPTG was considered wild-type expression. (Insets) Enlargements of the parts of the graphs related to comparisons of a real lac mutant strain and MG1655catRExBADlacZ under repressing conditions. The experiments were performed in triplicate.
This result demonstrated that without arabinose induction and in the presence of glucose, the strict repression induced by the RExBAD cassette at the chromosomal level can lead to phenotypes that are functionally equivalent to a complete gene deletion.
The RExBAD cassette allows plasmid-free modulation of expression of chromosomal target genes.
In order to study whether the RExBAD cassette could be used to modulate and precisely control the expression of chromosomal target genes, we assessed the range of expression of the lacZ gene in cultures supplemented with various concentrations of arabinose. As shown in Fig. 2, the β-galactosidase activity increased gradually with concentrations of arabinose ranging from 0.004 to 0.4%. Wild-type levels of β-galactosidase activity (1 mMisopropyl-β-d-thiogalactopyranoside [IPTG] induction inMG1655) were observed with between 0.04 and 0.06% arabinose in both LB medium and M63B1 medium containing 0.4% glucose. The maximum levels of β-galactosidase activity were observed with 0.4% arabinose in both media. With low levels of arabinose, the activation of lacZ gene expression was stronger in M63B1 medium containing 0.4% glucose than in LB medium. This observation is consistent with a previous report which indicated that pBAD promoter expression is more tightly regulated in LB medium than in minimal medium (17).
These experiments demonstrated that the RExBAD cassette can be used to modulate the expression of a target gene in order to achieve levels of expression ranging from very low levels to levels exceeding the wild-type level.
The RExBAD cassette allows study of essential and toxic genes.
Our results obtained for lacZ gene expression suggested that addition of glucose at concentrations of 0.4 to1% was enough to repress the expression of lacZ in strain MG1655catRExBADlacZ (Fig. 2). In the absence of glucose, a low level of lacZ expression from the RExBAD cassette promoter was detected, although the expression was not sufficient to promote growth on lactose minimal medium (Fig. 2 and data not shown).
Since even limited promoter leakage could be an issue in some experimental contexts, this prompted us to investigate whether we could use the RExBAD cassette in rather extreme conditions (for example, with an essential gene when leakage might be sufficient for viability and with a toxic gene when leakage might be lethal).
We first inserted the catRExBAD cassette in front of ftsL, an essential cell division gene present at a level of 25 to 50 copies per cell whose inactivation leads to cell filamentation and death through cell division arrest (16, 17). The construct could be obtained only in the presence of arabinose. As previously shown with the TnAraOut transposon for other essential genes (26), expression of ftsL from the pBAD promoter in catRExBAD under inducing conditions is sufficient for viability. When an exponential-phase culture of TG1catRExBADftsL grown in LB medium containing arabinose was transferred to LB medium containing glucose, repressing ftsL expression, cells rapidly displayed cell division arrest, which led to the characteristic filamentation phenotype and cell death of an ftsL mutant (Fig. 3). Moreover, when strain TG1catRExBADftsL was grown in LB medium alone, the repression of ftL gene expression still led to filamentation and cell death (data not shown).
FIG. 3.
Use of the RExBAD approach to study the essential cell division gene ftsL. Overnight cultures of E. coli TG1 and TG1catRExBADftsL grown in LB medium supplemented with 0.2% arabinose were washed in LB medium twice and diluted 1:100 into LB medium or LB medium supplemented with 0.2% glucose or with 0.2% arabinose, and then they were grown at 37°C for 6 h. Optical light microscopy images were captured with a ×40 objective. (Top panels) E. coli TG1. (Bottom panels) E. coli TG1catRExBADftsL. Cell filamentation was observed 4 h after subculturing in LB medium or LB medium supplemented with glucose for strain TG1catRExBADftsL but not for strain TG1. Filamentation led to cell death and lysis after 6 h in LB medium or LB medium supplemented with glucose.
This indicated that ftsL depletion was achieved through tight repression of ftsL expression in strain TG1catRExBADftsL. Therefore, the leakage of transcription from the catRExBAD cassette was not sufficient to allow the low-abundance cell division protein FtsL to accumulate to levels that supported viability.
We also validated the tight regulation of expression from the RExBAD cassette by placing this cassette in front of the toxic gene ccdB. The ccd module is composed of two genes, ccdA and ccdB, that encode two small proteins, the CcdB toxin that targets DNA gyrase and the CcdA antidote (5). The ccd locus is located on the F plasmid and is involved in the postdivisional killing of the plasmid-free segregants (25, 48). Therefore, the ccdB gene cannot be cloned for expression without its antidote, CcdA (1). In a single recombination step we replaced the ccdA gene with the specRExBAD cassette upstream of ccdB on the F′ plasmid carried by E. coli strain TG1. The resulting strain, TG1 F′(ΔccdAspecRExBADccdB), was viable under repressing conditions (LB medium with or without 0.4% glucose), demonstrating that the cassette pBAD promoter-driven expression was sufficiently repressed to allow viability of the ccdA mutant strain. Whereas the toxic effect of the RExBAD-mediated expression of ccdB was not observed in the absence of arabinose, strain TG1 F′(ΔccdAspecRExBADccdB) was unable to grow when it was streaked on plates supplemented with arabinose (Fig. 4). Consistent with these observations, introduction of the pKKccdA plasmid into strain TG1 F′(ΔccdAspecRExBADccdB) restored the expression in trans of the CcdB antidote, CcdA, and allowed the growth of this strain under all conditions (Fig. 4). Some rare colonies arose from TG1(ΔccdAspecRExBADccdB) grown on arabinose. These colonies proved to be CcdB-resistant bacteria that acquired a spontaneous mutation(s) during the time of the experiment.
FIG. 4.
Use of the RExBAD approach to study the toxic gene ccdB. Strains TG1 (region 1), TG1 F′(ΔccdARExBADccdB) (region 2), and TG1 F′(ΔccdARExBADccdB)/pKKccdA (region 3) were streaked on LB medium, LB medium containing 0.4% glucose, and LB medium containing 0.4% arabinose, and the plates were incubated at 37°C for 16 h.
The data from this set of experiments demonstrated that the RExBAD cassette can be efficiently used to study essential and toxic genes under either inducing or repressing culture conditions.
Application of the RExBAD inactivation and expression method to the study of biofilm formation: induction of the F conjugative pilus.
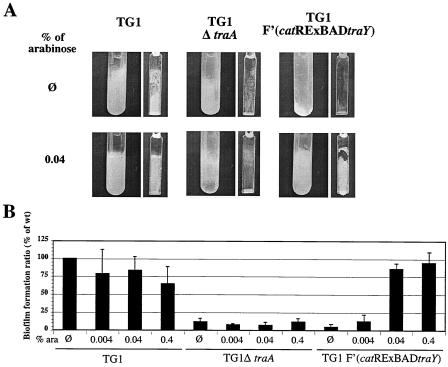
The RExBAD inactivation and expression method was applied to the study of biofilm formation by inserting the catRExBAD cassette into an E. coli strain upstream of the traY gene. traY is the first gene of the main F plasmid conjugative operon (33 kb) that encodes, among other conjugative functions, the F conjugative pilus. The capacity to produce F conjugative pili strongly induces biofilm formation (15). F pili are also necessary for the entry of several phages into bacterial cells (13, 31). The sensitivity of three E. coli strains, TG1, TG1 F′(catRExBADtraY), and TG1ΔtraA (the TraA pilin is the building block of the F conjugative pilus [20, 32]), to M13 phage was first studied in order to test the functionality of the F pili. Whereas the knockout TG1ΔtraA strain was resistant to M13 phage under any conditions, the TG1 F′(catRExBADtraY) strain was resistant to M13 phage under repressing conditions but became as sensitive as the TG1 wild-type strain in the presence of 0.2% arabinose (data not shown).
We then compared the biofilm phenotypes of these three strains grown in microfermentors with increasing concentrations of arabinose. The use of microfermentors allows continuous-flow cultures in which biofilm formation can be reproducibly monitored and controlled under conditions under which a high imposed flow rate is incompatible with planktonic growth. Most of the observed growth occurs on the available surfaces inside the microfermentors, and biofilm formation (1- to 2-mm-thick biofilms form in 24 to 72 h) can be monitored on removable slides (15).
As shown in Fig. 5, neither TG1Δtra or TG1 F′(catRExBADtraY) cultivated in M63B1 medium with glucose was able to form a strong biofilm. By contrast, when strain TG1 F′(catRExBADtraY) was grown in M63B1 medium containing glucose supplemented with increasing concentrations of arabinose, a wild-type biofilm phenotype was progressively restored. Therefore, as previously shown, induction of the production of the F pilus clearly promoted biofilm formation (15). Interestingly, when the growth medium was subsequently switched from M63B1 medium containing glucose and arabinose to M63B1 medium containing only glucose, the biofilm dispersed within 24 h (data not shown). This indicates that induction followed by repression of the production of the F pilus appeared to disrupt associations between bacteria within the F pilus-formed biofilm.
FIG. 5.
Modulation of biofilm formation via RExBAD-mediated control of F′ factor conjugative pilus synthesis. The abilities of the different strains to form biofilms in microfermentors were determined by using M63B1 medium containing 0.4% glucose with or without increasing concentrations of arabinose. After 24 h, microfermentor and spatula images were captured (A), and the biofilms formed on the glass spatulas were resuspended in 10 ml of M63B1 medium to obtain OD600 measurements (B). In panel A the results of a representative experiment in which no arabinose or 0.04% arabinose was added are shown. In panel B, the histograms show the results based on averages for three independent experiments in which the OD600 of wild-type strain TG1 grown in LB medium without added sugar was defined as 100%. Ø indicates that no arabinose was added.
These results showed that the RExBAD inactivation and expression method can also be used to efficiently appreciate the role of adhesins in biofilm formation.
Application of the RExBAD inactivation and expression method to the study of cell-cell interactions.
To further confirm the usefulness of the RExBAD approach in the characterization of genes encoding adhesins, we introduced the catRExBAD cassette in front of flu (3.120 kb), the gene encoding the autotransporter (AT) self-recognizing adhesin Ag43 (11, 41). AT proteins are modular proteins composed of an N-terminal passenger domain that is thought to be translocated through a pore in the outer membrane. The pore is created by insertion of a β-barrel formed by the C-terminal part of the protein, called the β AT domain (23, 39). Ag43 promotes cell-cell interactions leading to bacterial aggregation, which can be monitored in culture tubes, as shown in Fig. 6. Ag43 has also been shown to play a role in single or mixed biofilm formation and structure (8, 28). The expression of Ag43 is phase variable, and the shift from Ag43+ (ON) to Ag43− (OFF) cells results from combined regulation of the flu gene by the Dam methylase and the transcriptional regulator OxyR (21, 22, 42, 50). In strain MG1655ΔoxyR, flu expression is not repressed, and most of the cells are in an ON situation, leading to Ag43 expression and Ag43-mediated bacterial aggregation (Fig. 6A and B). We compared the bacterial aggregation properties of MG1655ΔoxyR (aggregation positive) and MG1655ΔoxyRΔflu (aggregation negative) with those of MG1655catRExBADflu cultures grown in LB medium with or without glucose. Figures 6A and B show that MG1655catRExBADflu did not aggregate under these conditions, similar to a ΔoxyR Δflu mutant. The same results were obtained in M63B1 medium containing 0.4% glucose (data not shown). This suggests that Ag43 synthesis is repressed in strain MG1655catRExBADflu cultures when they are grown in the absence of arabinose and that MG1655catRExBADflu is phenotypically identical to the MG1655Δflu strain (Fig. 6A and B).
FIG. 6.
Modulation of bacterial autoaggregation via controlled production of the Ag43 protein. The different strains were grown overnight in LB medium with either 0, 0.4, and 1% glucose (glu) or concentrations of arabinose (ara) ranging from 0.004 to 1%. (A and B) The aggregation of MG1655catRExBADflu increased with the arabinose concentration. After growth cells were diluted to an OD600 of 2.5 in a 3-ml culture, and the autoaggregation of each strain was assessed after incubation for 24 h at room temperature by capturing images of the tubes (A) and determining the OD600 of the upper part of the culture in each standing tube (dashed line) (B). The aggregation tests were performed in triplicate. (C) Ag43 production by MG1655catRExBADflu can be modulated by increasing arabinose concentrations. For each culture the equivalent of 0.2 OD600 unit was loaded on a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gel. Immunodetection was performed by using a polyclonal rabbit antiserum raised against the α domain of Ag43. Ø indicates that no sugar was added.
In order to study the modulation of expression of the flu adhesin-encoding gene from the RExBAD cassette, we tested the capacity of MG1655catRExBADflu to aggregate in the presence of increasing concentrations of arabinose. In contrast to an oxyR mutant, wild-type strain MG1655 does not aggregate (21, 22, 42, 50). As shown in Fig. 6A and B, an increase in the concentration of arabinose from 0.004 to 1% was correlated with increasing bacterial aggregation in tubes. With 0.4% arabinose, the aggregation of MG1655catRExBADflu was equivalent to the aggregation observed in the fully aggregative MG1655ΔoxyR strain.
We checked whether the increased-aggregation phenotype did correspond to an increase in Ag43 synthesis by performing an immunodetection analysis of Ag43 in LB medium (Fig. 6C). Whereas addition of glucose or arabinose to the culture had no effect on Ag43 production in MG1655 wild-type cells (data not shown), no Ag43 was detected in cell extracts of strain MG1655catRExBADflu in the absence of arabinose, mimicking the MG1655Δflu strain results. This was also the case with extracts of cells grown in M63B1 minimal medium containing 0.4% glucose (data not shown). This further demonstrated that expression of the flu gene from the RExBAD cassette in the absence of arabinose resulted in a lack of Ag43 production. Moreover, a progressive increase in the arabinose concentration led to a progressive increase in Ag43 production. Maximal Ag43 production was obtained with 0.4% arabinose, and the levels corresponded to levels obtained with the MG1655ΔoxyR flu-derepressed strain.
These results demonstrated that the RExBAD cassette can be used to fine-tune the expression of a large chromosomal adhesin-encoding gene, such as the gene encoding the autotransporter Ag43, facilitating complementation studies for large genes that are often difficult to handle.
Identification of new putative adhesin-encoding genes in the E. coli K-12 genome.
Autotransporters represent one of the largest groups of secreted proteins in gram-negative bacteria and have different functions, including adhesin, protease, and hydrolytic enzyme functions. We hypothesized that some of the putative E. coli autotransporters are new adhesins that participate in bacterium-surface or bacterium-bacterium interactions that take place during biofilm formation. In order to identify such potential adhesins, we first searched the E. coli K-12 genome sequence for genes with unknown functions that are also homologs of Ag43 genes. Using BLAST comparisons with two different algorithms, we identified 10 genes showing homology to the flu gene (Table 2) (ypjA, yejO, ydhQ, ydbA, ycgH, yfaL, ydeK, ydeU, yaiT, and ycgV). In a phylogenetic study of outer membrane translocated proteins, Yen et al. identified 120 putative members of the AT family (51). ydhQ, ydbA, and ycgV were not identified in that study as recognized homologues of the AT family, and ypjA was misspelled as yejA. yejO, ydbA, and yaiT are described in the Colibri database (genome of E. coli K-12 strain MG1655) as being interrupted by the insertion (IS) elements IS5K, hybrid IS2D-IS30C, and IS3B, respectively (http://genolist.pasteur.fr/Colibri/). In yejO, IS5K is inserted close to the beginning of the protein (between codons 21 and 22), which probably eliminates translation of the protein. In ydbA and yaiT insertion of the IS elements created separation between the passenger and autotransporter domains. This leaves the possibility that the two domains are encoded by separate ORFs but are still functional (51). Nevertheless, no experimental evidence supports this hypothesis. Consequently, we did not investigate yejO, ydbA, and yaiT. In addition, we considered ycgH (b1169-b1170) and ydeK-ydeU (b1509-b1510) as functional units, although the published sequence of ycgH contains a stop within the ORF that separates the two domains, which have been described as two ORFs (b1169 is Orf6 Eco and b1170 is Orf3 Eco in the study of Yen et al. [51]). Finally, ydeU is proposed to contain the β autotransporter domain that translocates the passenger domain of ydeK (51), and ydeK-ydeU is probably organized as a unique transcription unit.
TABLE 2.
Putative new adhesins in E. coli K-12 strain MG1655 displaying homology to Ag43
| Gene | Blattner designation | Protein size (amino acids) | % Similarity with flu (ClustaIW)a |
|---|---|---|---|
| flu | b2000 | 1,039 | 100 (1,039) |
| ypjA | b2647 | 1,526 | 46 (1,254) |
| yejOb | b2190 | 842 ± 21 | 52 (1,052) |
| ydhQ | b1664 | 418 | 27 (1,039) |
| ydbAb | b1401-b1405 (b4492) | 852 ± 1,106 | 35 (2,003) |
| ycgH | b1169-b1170 (b4491) | 506 ± 337 | 48 (1,039) |
| yfaL | b2233 | 1,250 | 46 (1,254) |
| ydeK | b1510 | 1,325 | 46 (1,330) |
| ydeU | b1509 | 466 | 29 (1,039) |
| yaiTb | b0371-b0374 | 486 ± 466 | 53 (1,045) |
| ycgV | b1202 | 955 | 54 (1,056) |
| yeeJ | b1978 | 2,358 | 28 (2,358) |
The values in parentheses are the sizes of the ClustalW alignments (in amino acids).
The protein is interrupted by an IS element. The size of the protein corresponds to the size of the reconstituted protein.
The putative YeeJ protein (2,358 amino acids) also exhibits homology with the N-terminal part of the Ag43 protein. Based on the limited sequence conservation of the C-terminal part of the proteins, YeeJ is probably not an autotransported protein. However, this protein might be exported to the surface of the bacteria. YeeJ does possess a signal sequence that is followed by a peptidoglycan-binding domain (LysM) that is often found in cell wall degradation enzymes. In addition, YeeJ also contains 13 bacterial immunoglobulin-like group 1 (Big_1) domains that are typically found in a variety of bacterial and phage surface proteins, such as intimins, which mediate the interaction with host cells. Finally, YeeJ has four polycystic kidney disease protein PKD1 domains that contain an immunoglobulin-like fold which might be involved in protein-protein and protein-carbohydrate interactions.
Functional analysis of the putative adhesins: yfaL, yeeJ, ypjA, and ycgV expression lead to biofilm formation in E. coli.
In order to check whether any of the uncharacterized genes could play a role in the adhesion of E. coli K-12 strain MG1655 during biofilm formation, ypjA, yfaL, ycgH, ydhQ, ycgV, ydeKU, and yeeJ were deleted individually, and the seven MG1655 derivative strains were tested for initial adhesion in microtiter plate assays, as well as for mature biofilm formation in microfermentors. No significant modification of adhesion or biofilm formation was observed in the mutants compared to the parental MG1655 strain (data not shown). This suggests that none of the proteins encoded by these genes plays a role in the adhesion of MG1655 on abiotic surfaces under the conditions examined.
These genes could have been phenotypically silent because of our experimental conditions. We assessed whether their overexpression could lead to detectable adhesion phenotypes. We introduced the catRExBAD cassette in front of ypjA, yfaL, ycgH, ydhQ, ycgV, ydeKU, and yeeJ and compared the adhesion phenotypes of strains MG1655, MG1655ΔoxyR (which constitutively overexpresses Ag43, MG1655catRExBADflu, MG1655catRExBADypjA, MG1655catRExBADyfaL, MG1655catRExBADycgH, MG1655catRExBADydhQ, MG1655catRExBADycgV, MG1655catRExBADydeKU, and MG1655catRExBADyeeJ grown at 37°C in M63B1 medium with 0.4% glucose in the presence or absence of 0.1% arabinose. As shown in Fig. 7, in the absence of arabinose induction, no differences were observed between wild-type strain MG1655 and the different catRExBAD constructs. This is consistent with the results of the knockout experiments. Strain MG1655ΔoxyR, by virtue of its constitutive Ag43 overproduction, displayed a twofold increase in its adhesion properties. Accordingly, expression of the flu gene due to the use of arabinose also increased the adhesion of strain MG1655catRExBADflu. No effect on MG1655 adhesion was seen when ycgH, ydhQ, and ydeKU were induced. This suggests that the three corresponding proteins have no detectable adhesin activity under the conditions tested. However, arabinose induction of yfaL and yeeJ expression and, to a lesser extent, ypjA and ycgV expression increased the adhesion properties of MG1655 when they were measured by a microtiter PVC plate assay (Fig. 7). Since no growth difference was observed between MG1655 and the different constructs in the presence of 0.1% arabinose, our data suggest that these four genes encode proteins with potential adhesins functions. Interestingly, overexpression of these four potential adhesins promoted bacterial adhesion to PVC at a level that was greater than the level obtained with Ag43 overexpression. This suggests that these adhesins are better adapted or more efficient than Ag43 for adhesion to PVC. In microfermentors, in agreement with the results obtained with microtiter plates, no effect was detected when ycgH, ydhQ, and ydeKU were induced, and ypjA induction had no effect on mature biofilm formation, indicating that the contribution of the protein encoded by this gene may be important only in the early stages of adhesion. However, induction of yeeJ, ycgV, and yfaL increased mature biofilm formation by MG1655 (data not shown).
FIG. 7.
Induction of yfaL, yeeJ, ypjA, and ycgV for increases in adhesion to solid surfaces. The adhesion properties of the RExBAD constructs were assessed at 37°C in M63B1 medium containing 0.4% glucose with no arabinose added (open bars) or with 0.1% arabinose added (solid bars) by using a microtiter PVC plate assay. The adhesion capacities were expressed as percentages of the capacity of wild-type strain MG1655 in the absence of arabinose. Cells were grown for 48 h before crystal violet staining and measurement of the optical density at 570 nm. The averages for three independent experiments are shown.
These results demonstrated that YcgV, YeeJ, and especially YfaL are potential adhesins in adhesion to two types of abiotic surfaces (PVC and glass) in either static or flow condition cultures.
DISCUSSION
In this study, we developed a simple and cloning-free approach that fulfills both gene inactivation and gene expression needs and allows workers to study gene function at nearly physiological levels.
RExBAD cassette permits expression knockout.
Insertion of the RExBAD cassette in front of the lacZ gene resulted in residual β-galactosidase activity that could be detected even under repressing conditions, suggesting that the repression of lacZ expression in this construct is not strictly equivalent to the repression achieved by deletion of lacZ. However, the residual lacZ expression in the MG1655catRExBADlacZ strain was unable to promote growth on lactose as a sole carbon source. In addition, when the RExBAD cassette was placed in front of a toxin gene (ccdB), an essential gene (ftsL), or genes involved in adhesion processes (flu, F plasmid tra operon), the repressed expression levels of the target genes were phenotypically equivalent to the effects observed after gene deletion, including the effects on viability (RExBADccdB), cell division arrest (RExBADftsL), the absence of aggregation (RExBADflu), and the low-adhesion and biofilm phenotype (RExBADtraY). Therefore, despite a basal level of expression due to limited leakage of the pBAD promoter, especially in minimal medium (17), our data clearly demonstrated that the RExBAD cassette can be used to create a phenotypic knockout construct that is functionally equivalent to gene deletion for a wide range of cell functions.
Modulation of gene expression from the RExBAD cassette.
Gene function studies require depletion of a gene product via gene deletion, as well as restoration of the function through gene complementation. Here we show that use of the RExBAD cassette permits inducible expression of target genes. The levels of expression of the target genes tested in this study increased over the range from 0.004% arabinose to the threshold of 0.4% arabinose, indicating that the level of gene expression can be adjusted with the arabinose concentration like the level of gene expression from a pBAD plasmid (Fig. 2 and 6), although the induction in synthetic minimal medium is more efficient than the induction in rich medium.
Our results also indicate that the expression of the target gene can be modulated only up to a certain extent. We showed that for lacZ and flu (Ag43) expression driven from the chromosomal RExBAD promoter, gene expression reaches a plateau in the presence of 0.4% of arabinose; beyond this, an increase in the inducer concentration does not result in an increased amount of β-galactosidase activity or protein but rather results in decreased expression (Fig 2 and 6C). These results demonstrate that the RExBAD cassette can be use to study gene expression ranging from low to overexpression levels. However, the use of engineered strains such as the Δara lacI A177C E. coli strains that perform facilitated diffusion of arabinose may prevent the high inducer-low expression effect due to all-or-nothing induction of the arabinose regulon (34). This approach may be difficult to apply to other gram-negative bacteria in which the ara metabolism system is less well characterized, yet it may provide the optimal way to fine-tune the target gene expression level.
Study of essential or toxic gene function in cellular processes.
Microbial genomes contain essential genes, as well as genes whose unregulated expression is lethal. The study of these genes is necessary in order to understand basic cellular processes. Several methods have been developed to study essential genes, and these methods generally rely on identification of conditional mutations that allow expression of the essential genes only under permissive conditions (2, 38, 44). However, the study of such genes often presents considerable technical difficulties. Here we provide evidence that the use of RExBAD cassettes is a robust and rapid alternative to the generation of random or directed conditional mutants to study essential genes. Similarly, an RExBAD cassette can also be used to study toxic genes. Whereas this approach may not be useful for catalytic toxins, which are toxic at very low levels, the use of RExBAD cassettes allows low and titrable modulation of gene expression that may prevent the selection of compensatory mutations under these conditions.
Study of gene expression in antibiotic- and plasmid-free physiological conditions.
The requirement for antibiotic selection to maintain plasmids is another drawback of conventional plasmid-borne gene function studies. In some instances, the use of an antibiotic may alter bacterial physiology and prevent clear interpretation of the complementation experiment. This is the case for the study of bacterial adhesion and biofilm formation, which are highly dependent on the growth rate, cell density, and culture conditions. We observed that both the introduction of plasmids into cells and the use of antibiotics affect the rate of biofilm formation (C. Beloin, P. Latour-Lambert, and J.-M. Ghigo, unpublished observations). By avoiding the use of antibiotics in the depletion and complementation experiments, we found that the use of the stable chromosomal RExBAD cassette is ideal for biofilm formation studies.
Amplification and sequence fidelity can also be a limiting step in plasmid complementation strategies. The RExBAD cassette is a simple alternative for facilitating the study of large adhesins which are often involved in biofilm or adhesion processes (12, 29).
New adhesins identified in E. coli K-12.
Studies of the genes required for biofilm communal growth indicated the importance and the diversity of surface organelles, such as flagella, type I fimbriae, curli, and conjugative pili, and also of membrane adhesins, such as Ag43 (for a review see reference 30). Ag43, a major surface adhesin that promotes cell-cell aggregation and biofilm formation, belongs to the autotransporter protein family, which consists of modular proteins composed of three domains (the signal sequence, the passenger functional domain, and the translocation unit) that contain all the information required to localize to the outer membrane.
In this study we identified several genes encoding putative adhesins exhibiting homology with Ag43 (ypjA, yejO, ydhQ, ydbA, ycgH, yfaL, ydeK, ydeU, yaiT, ycgV, and yeeJ). We investigated their function in adhesion and biofilm formation using the RExBAD approach to demonstrate the roles of these previously uncharacterized E. coli genes in bacterial adhesion. The physiological conditions required for expression of these genes are not known. However, phenotypic analyses of strains from which these putative adhesin genes were deleted suggested that they do not contribute to the adhesion of the wild-type MG1655 strain under laboratory conditions. Nevertheless, single-copy chromosome-based expression of the genes by using the RExBAD strategy demonstrated that expression of four of these genes (yfaL, yeeJ, ypjA, and ycgV) leads to a clear adhesion and biofilm phenotype. Interestingly, in addition to containing several putative domains clearly linked to host cell and bacterial cell-cell interactions, the large YeeJ protein (2,358 amino acids) exhibits 54% similarity with the biofilm-associated protein (Bap) of Staphylococcus aureus. The Bap protein is a 2,276-amino-acid surface protein that promotes biofilm formation on abiotic surfaces by a mechanism that is not yet clear and increases the ability of S. aureus to colonize and persist in the bovine mammary gland (6, 7). Reports of such large proteins implicated in adhesion and biofilm formation are not unique; such proteins include the LapA proteins of Pseudomonas putida and Pseudomonas fluorescens (12, 24) and the Esp protein of Enterococcus faecalis (49).
Our results illustrate the diversity of potential adhesins that can be expressed by E. coli K-12 and suggest that E. coli probably possesses a large arsenal of surface adhesins with different binding specificities, as well as different patterns of expression, possibly in response to different environmental conditions. We cannot exclude the possibility that some of the phenotypically silent putative adhesins (encoded by yejO, ydhQ, ydbA, ycgH, ydeK, ydeU, and yaiT) have adhesion properties that were not revealed by our phenotypic analysis. This matter is currently under investigation. The issue of the physiological expression conditions of these potentially cryptic genes needs to be addressed for each of these newly identified adhesions factors.
Postgenomic applications.
Analysis of the complete prokaryotic genome sequences revealed that genes with unassigned or poorly characterized functions account for a high proportion of most microbial genomes. Because of the rapidity of the RExBAD approach, as well as the wide range of bacteria in which this λ-red recombination system can be used, including E. coli K-12 (3, 9), pathogenic E. coli (36; S. Da Re, personal communication), Yersinia species (10), Shigella flexneri (3), Klebsiella pneumoniae (D. Balestrino, personal communication), and Salmonella enteritidis (47), we assume that this method could be used for a wide range of gram-negative bacteria in which the RExBAD approach could be instrumental for studying these unknown genes. This method permits inactivation and modulation of the expression of target genes. It was developed to study both single genes and full operons and could be a very efficient method for investigating the functions of cryptic genes by studying their phenotypes over a wide range of expression levels.
Acknowledgments
We are grateful to P. Owen for the gift of the Ag43 antiserum and to L. Van Melderen for the gift of plasmid pKKccdA. We thank Jennifer Leeds, Sandra Da Re, Didier Mazel, Cécile Wandersman, and Evelyne Richet for their helpful comments and suggestions on the manuscript and Damien Balestrino for providing results before publication.
J.-M.G. and C.B. were supported by the Institut Pasteur, Paris, France, and by CNRS URA2172 grants. A.R. was supported by a Ministère Français de l'Éducation Nationale, de l'Enseignement Supérieur et de la Recherche fellowship.
REFERENCES
- 1.Afif, H., N. Allali, M. Couturier, and L. Van Melderen. 2001. The ratio between CcdA and CcdB modulates the transcriptional repression of the ccd poison-antidote system. Mol. Microbiol. 41:73-82. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, K. A., and R. K. Herman. 1976. Method for the isolation of Escherichia coli K-12 mutants deficient in essential genes. J. Bacteriol. 126:38-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 4.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Critchlow, S. E., M. H. O'Dea, A. J. Howells, M. Couturier, M. Gellert, and A. Maxwell. 1997. The interaction of the F plasmid killer protein, CcdB, with DNA gyrase: induction of DNA cleavage and blocking of transcription. J. Mol. Biol. 273:826-839. [DOI] [PubMed] [Google Scholar]
- 6.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cucarella, C., M. A. Tormo, C. Ubeda, M. P. Trotonda, M. Monzon, C. Peris, B. Amorena, I. Lasa, and J. R. Penades. 2004. Role of biofilm-associated protein Bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 72:2177-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derbise, A., B. Lesic, D. Dacheux, J. M. Ghigo, and E. Carniel. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 38:113-116. [DOI] [PubMed] [Google Scholar]
- 11.Diderichsen, B. 1980. flu, a metastable gene controlling surface properties of Escherichia coli. J. Bacteriol. 141:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Espinosa-Urgel, M., A. Salido, and J. L. Ramos. 2000. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J. Bacteriol. 182:2363-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost, L. S., and W. Paranchych. 1988. DNA sequence analysis of point mutations in traA, the F pilin gene, reveal two domains involved in F-specific bacteriophage attachment. Mol. Gen. Genet. 213:134-139. [DOI] [PubMed] [Google Scholar]
- 14.Genevaux, P., S. Muller, and P. Bauda. 1996. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 142:27-30. [DOI] [PubMed] [Google Scholar]
- 15.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo, J. M., and J. Beckwith. 2000. Cell division in Escherichia coli: role of FtsL domains in septal localization, function, and oligomerization. J. Bacteriol. 182:116-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzman, L. M., J. J. Barondess, and J. Beckwith. 1992. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J. Bacteriol. 174:7716-7728. [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haldimann, A., L. L. Daniels, and B. L. Wanner. 1998. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J. Bacteriol. 180:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, R. L., K. A. Sholl, M. N. Conrad, M. E. Dresser, and P. M. Silverman. 1999. Interaction between the F plasmid TraA (F-pilin) and TraQ proteins. Mol. Microbiol. 34:780-791. [DOI] [PubMed] [Google Scholar]
- 21.Henderson, I. R., M. Meehan, and P. Owen. 1997. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol. Lett. 149:115-120. [DOI] [PubMed] [Google Scholar]
- 22.Henderson, I. R., M. Meehan, and P. Owen. 1997. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli. Adv. Exp. Med. Biol. 412:349-355. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 24.Hinsa, S. M., M. Espinosa-Urgel, J. L. Ramos, and G. A. O'Toole. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol 49:905-918. [DOI] [PubMed] [Google Scholar]
- 25.Hiraga, S., A. Jaffe, T. Ogura, H. Mori, and H. Takahashi. 1986. F plasmid ccd mechanism in Escherichia coli. J. Bacteriol. 166:100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Judson, N., and J. J. Mekalanos. 2000. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18:740-745. [DOI] [PubMed] [Google Scholar]
- 27.Kang, Y., T. Durfee, J. D. Glasner, Y. Qiu, D. Frisch, K. M. Winterberg, and F. R. Blattner. 2004. Systematic mutagenesis of the Escherichia coli genome. J. Bacteriol. 186:4921-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 29.Klemm, P., L. Hjerrild, M. Gjermansen, and M. A. Schembri. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol. Microbiol. 51:283-296. [DOI] [PubMed] [Google Scholar]
- 30.Lejeune, P. 2003. Contamination of abiotic surfaces: what a colonizing bacterium sees and how to blur it. Trends Microbiol. 11:179-184. [DOI] [PubMed] [Google Scholar]
- 31.Manchak, J., K. G. Anthony, and L. S. Frost. 2002. Mutational analysis of F-pilin reveals domains for pilus assembly, phage infection and DNA transfer. Mol. Microbiol. 43:195-205. [DOI] [PubMed] [Google Scholar]
- 32.Maneewannakul, K., S. Maneewannakul, and K. Ippen-Ihler. 1993. Synthesis of F pilin. J. Bacteriol. 175:1384-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, J. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 34.Morgan-Kiss, R. M., C. Wadler, and J. E. Cronan, Jr. 2002. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proc. Natl. Acad. Sci. USA 99:7373-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, K. C. 1998. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newman, J. R., and C. Fuqua. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197-203. [DOI] [PubMed] [Google Scholar]
- 38.Oliver, D. B. 1985. Identification of five new essential genes involved in the synthesis of a secreted protein in Escherichia coli. J. Bacteriol. 161:285-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oomen, C. J., P. Van Ulsen, P. Van Gelder, M. Feijen, J. Tommassen, and P. Gros. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23:1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 41.Owen, P. 1983. Antigens of the Escherichia coli cell envelope, p. 347-373. In O. J. Bjerrum (ed.), Electroimmunochemical analysis of membrane proteins. Elsevier Science Publishing, Amsterdam, The Netherlands.
- 42.Owen, P., M. Meehan, H. de Loughry-Doherty, and I. Henderson. 1996. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 16:63-76. [DOI] [PubMed] [Google Scholar]
- 43.Posfai, G., V. Kolisnychenko, Z. Bereczki, and F. R. Blattner. 1999. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res. 27:4409-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salmond, G. P., and S. Plakidou. 1984. Genetic analysis of essential genes in the ftsE region of the Escherichia coli genetic map and identification of a new cell division gene, ftsS. Mol. Gen. Genet. 197:304-308. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Siegele, D. A., and J. C. Hu. 1997. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc. Natl. Acad. Sci. USA 94:8168-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 48.Tam, J. E., and B. C. Kline. 1989. Control of the ccd operon in plasmid F. J. Bacteriol. 171:2353-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toledo-Arana, A., J. Valle, C. Solano, M. J. Arrizubieta, C. Cucarella, M. Lamata, B. Amorena, J. Leiva, J. R. Penades, and I. Lasa. 2001. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67:4538-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallecha, A., J. Correnti, V. Munster, and M. van der Woude. 2003. Phase variation of Ag43 is independent of the oxidation state of OxyR. J. Bacteriol. 185:2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yen, M. R., C. R. Peabody, S. M. Partovi, Y. Zhai, Y. H. Tseng, and M. H. Saier. 2002. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta 1562:6-31. [DOI] [PubMed] [Google Scholar]
- 52.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]