Abstract
BACKGROUND
To date, no data are available regarding the effects of probiotics on the pathway of tryptophan/serotonin metabolism among human immunodeficiency virus (HIV) 1–infected individuals. Because a condition of dysbiosis might be responsible for the altered use of tryptophan described in this population, the aim of this study was to investigate the link between probiotic supplementation and serotonin levels in combined antiretroviral therapy–treated patients and the subsistence of an interplay with inflammation.
METHODS
We conducted a pilot study that included 8 HIV-positive subjects. We collected blood and fecal samples before and after 6 months of probiotic supplementation, to measure the level of serotonin in serum and tryptophan in stool, the expression of CD38 and HLA-DR on peripheral CD4+ T lymphocytes (as immune activation markers), the expression of indoleamine 2,3-dioxygenase 1 messenger RNA (mRNA) and IFN-γ mRNA (as markers of tryptophan metabolism and systemic inflammation).
RESULTS
After probiotic supplementation, we observed a significant increase in concentration of serum serotonin (P = .008) and a decreased level of tryptophan in plasma. Moreover, a significant reduction in CD38 and HLA-DR expression on the surface of peripheral CD4+ T cells (P = .008) and a reduced expression of indoleamine 2,3-dioxygenase 1 mRNA on peripheral blood mononuclear cells (P = .04) were observed.
CONCLUSIONS
Considering that this probiotic (Vivomixx® in EU; Visbiome® in USA) has an influence on tryptophan metabolism, larger studies on this topic are needed.
Keywords: Probiotics, IDO-1, tryptophan, serotonin, HIV, IFN-γ
Introduction
Chronic human immunodeficiency virus (HIV) infection in humans induces a serious damage in the gut, which leads to the loss of mucosal barrier integrity, microbial translocation, systemic inflammation, and, in the end, to chronic immune activation.1–6 The use of combined antiretroviral therapy (cART) plays a fundamental role in the management of HIV-1 infection given its ability to suppress viremia, thus limiting the seeding of viral reservoir7; furthermore, with the use of cART, HIV-1–positive patients are able to achieve a better health status and prolonged longevity.7 However, antiretrovirals do not fully resolve systemic immune activation and inflammation; thus, HIV-1–infected subjects show an increased risk of developing various disorders and comorbidities, if compared with the general population.8 An intestinal dysbiosis exerting pro-inflammatory effects has been frequently described among HIV-positive individuals9: the damage of the mucosal immune system can result in the modification of the usual gut-resident microbial community with the outgrowth of a “dysbiotic” microbial flora. This dysbiosis can affect the mucosal immune system, actively inducing an inflammatory microenvironment.
Along with the mucosal disequilibrium, alterations of the pathway of tryptophan metabolism, through a modification of the interferon-inducible enzyme indoleamine 2,3-dioxygenase 1 (IDO-1) activity, have been described.10,11
Tryptophan, 1 of the 9 essential amino acids, cannot be synthesized in humans and is usually obtained through dietary intake or released during protein turnover. This amino acid represents a critical component of several metabolic functions and can act as an important determinant of mood, cognition, and behavior.12,13 Tryptophan is used in the synthesis of proteins and in the production of molecules such as the aminergic neurotransmitter serotonin (5-hydroxytryptamine [5-HT]) or nicotinamide adenine dinucleotide (NAD/NADH), via the kynurenine biosynthetic pathway. Kynurenine is the first stable metabolite to be synthesized when tryptophan is oxidized under the influence of l-tryptophan 2,3-dioxygenase, IDO-1, or IDO-2.14 Peripheral tryptophan levels in HIV-infected individuals are predominantly dependent on the activity of IDO-1,14–18 and studies have shown that the accelerated conversion of tryptophan to kynurenine is correlated with an elevation of levels of immune activation.
Probiotics have been proposed as a resource to counterbalance the dysbiosis associated with liver,19 kidney,20 cardiovascular,21 metabolic,22 and neurological disorders.23
We hypothesize that probiotic supplementation might exert an influence on tryptophan metabolism among HIV-1–infected individuals through the correction of HIV-associated dysbiosis, resulting in an increase in serotonin levels. Because no data have been published to date on this topic, we performed the first longitudinal pilot study to evaluate the impact of a high-concentration multistrain probiotic product on fecal levels of tryptophan, serum levels of serotonin, immune activation markers, and IDO-1 messenger RNA (mRNA) expression in peripheral blood mononuclear cells (PBMCs) of HIV-1–infected patients on effective cART.
Materials and Methods
Samples
Eight HIV-1–positive patients successfully treated with cART were recruited at the Division of Infectious Diseases, Department of Public Health and Infectious Diseases, Hospital of “Sapienza” University of Rome (Italy). The study was approved by the institutional review board (Department of Public Health and Infectious Diseases, “Sapienza” University of Rome and the Ethics Committee of Umberto I General Hospital, Rome). Inclusion criteria for this study were as follows: (1) signature of the written informed consent, (2) men or women at least 18 years old, (3) being on cART, and (4) presenting with HIV-1 RNA <37 copies/mL and CD4+ T counts >400 cells/mm3. Exclusion criteria were as follows: (1) known or suspected allergy or intolerance to study probiotic formulation, (2) use of probiotics or antibiotics during the 3 weeks prior the enrollment, (3) drug addiction, (4) history of or current inflammatory diseases of the small or large intestine, (5) diarrhea, (6) any current or previous systemic malignancy, and (7) pregnancy. Patients received a high concentration of lyophilized multistrain probiotic supplement twice a day for 6 months. The probiotic preparation contained 1.8 × 1012 live bacteria/day (Lactobacillus plantarum DSM24730, Streptococcus thermophilus DSM24731, Bifidobacterium breve DSM24732, Lactobacillus paracasei DSM24733, Lactobacillus delbrueckii subsp bulgaricus DSM24734, Lactobacillus acidophilus DSM 24735, Bifidobacterium longum DSM24736, and Bifidobacterium infantis DSM24737) and is currently sold under the brand Vivomixx in Europe and Visbiome in the United States and Canada. All patients underwent blood and fecal sample collection prior to initiation (T0) and after 6 months (T6) of probiotic supplementation. No adverse event was observed during the follow-up and all subjects maintained undetectable plas-matic viral load before and after probiotic treatment.
Specimen processing
About 20 mL of whole blood was collected by venipuncture in Vacutainer tubes containing EDTA (BD Biosciences, San Jose, CA, USA) at each study visit. Peripheral blood mononuclear cells were separated by Ficoll gradient centrifugation (Lympholyte; Cedarlane Labs, Hornby, ON, Canada) and washed twice in phosphate-buffered saline solution.24 Freshly isolated PBMCs were used immediately for immune phenotyping and activation staining. About 10 mL of whole blood was collected by BD Vacutainer Plus Plastic Serum. After centrifuge, serum was stored in aliquots at −80°C.
Bacterial DNA isolation from fecal samples
Bacterial DNA from fecal samples was extracted using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany). Approximately, 200 mg of feces were cut from frozen samples using sterile disposable scalpel, resuspended in 1.4 mL of ASL lysis buffer from the stool kit, added with glass beads (150–212 μm; Sigma-Aldrich, St. Louis, MO, USA) and homogenized thoroughly. The suspension was incubated at 95°C for 5 minutes, and DNA was purified according to the manufacturer’s instructions. DNA was eluted in 200 μL of AE buffer (provided in the kit) and stored at −20°C.
Real-time polymerase chain reaction assay
Real-time polymerase chain reaction (PCR) was used to quantify bifidobacteria using genus-specific primers and conditions described by Matsuki et al25 and to quantify the expression of IDO-1 and IFN-γ mRNA. Briefly, PCR amplification and detection were performed on optical-grade 96-well plates using the Applied Biosystems 7500 Real-Time PCR instrument (Applied Biosystems Inc., Norwalk, CT, USA).
To quantify bifidobacteria, the reaction mixture (25 μL) was composed of SensiMix SYBR Low-ROX (Bioline, Taunton, MA, USA), 500-nM primers for Bifidobacterium genus and 2.5 μL of template DNA. The fluorescent products were detected at the last step of each of 40 cycles. A melting curve analysis was made after amplification to distinguish the targeted PCR product from the nontargeted PCR products. Standard curves were created using serial 10-fold dilutions of bacterial DNA extracted from B breve. All samples were analyzed in duplicate in 2 independent real-time PCR assays. For IDO-1 and IFN-γ mRNA expression, total RNA was extracted from PBMC, using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. The housekeeping gene β-glucuronidase was used as an internal control. Gene expression values were calculated by the comparative Ct method.26
T-cell phenotyping by flow cytometry
Phenotypes and activation markers were evaluated by Miltenyi Biotec flow cytometer-MACSQuant Analyzer (8 fluorescence channels, 3 lasers) on freshly isolated PBMC. Immune activation was evaluated by multiparameter flow cytofluorimetric analysis by the following antihuman monoclonal antibodies: CD3-PerCP, CD4-APC-Vio770, CD8-FITC, CD45ROPE-Vio770, CD27-VioBlue, CD38-APC, and HLA-DR-PE (Miltenyi Biotec, Bergisch Gladbach, Germany).
Enzyme-linked immunosorbent assay
Serum serotonin levels were detected by enzyme-linked immunosorbent assay (ELISA) kit (Abnova, Taipei, Taiwan) based on an indirect assay with an antigen-antiserum bound and a conjugate antibody using a 3,3′,5, 5′-tetramethylbenzidine (TMB) as a substrate for the detection. For serotonin detection, we used an ELISA assay with an antiserotonin antibody precoated microplate, a serotonin antiserum (rabbit antiserotonin antibody), a conjugate enzyme (goat antirabbit immunoglobulins conjugated with peroxidase), and a substrate for the reaction (containing tetramethylbenzidine, substrate buffer and hydrogen peroxide). The reaction was monitored at 450 nm. Quantification was achieved by comparing their absorbance with a reference curve prepared with known standard concentrations.
Virological analysis
Human immunodeficiency virus 1 RNA copy numbers were evaluated in plasma prepared from blood obtained in EDTA-containing tubes and stored at −80°C. Levels of HIV-RNA were measured with Versant kPCR (Siemens Healthcare Diagnostic Inc., Tarrytown, NY, USA) with a detection limit of 37 copies/mL.
Fecal metabolome
Fecal samples were prepared for nuclear magnetic resonance (NMR) analysis by vortex mixing for 5 minutes 80 mg of stool with 1 mL of deionized water, followed by centrifugation for 15 minutes at 18 000g and 4°C. About 700 mL of supernatant was added to 100 μL of a D2O solution of 3-(trimethylsilyl)-propionic-2,2,3,3-d4 acid sodium salt, 10 mM, set at pH 7.00 with 1-M phosphate buffer. Before analysis, the samples were again centrifuged.
Proton NMR (1H-NMR) spectra were recorded at 298 K with an AVANCE III spectrometer (Bruker, Milan, Italy) operating at a frequency of 600.13 MHz. The Hydrogen Deuterium Oxide (HOD) residual signal was suppressed by presaturation, whereas broad signals from slowly tumbling molecules were removed by including a Carr-Purcell-Meiboom-Gill filter27 to a free induction decay sequence. The filter was made up by a train of 400 echoes separated by 800 μs, for a total time of 328 ms. Each spectrum was acquired by summing up 256 transients using 32 K data points over a 7211.54-Hz spectra (for an acquisition time of 2.27 seconds). The recycle delay was set to 8 seconds, keeping into consideration the longitudinal relaxation time of the protons under investigation. The so-obtained spectra were adjusted for baseline irregularities as explained elsewhere.28
The signals of tryptophan were assigned by comparing their chemical shift and multiplicity with Chenomx software data bank (ver. 8.1; Chenomx Inc., Edmonton, AB, Canada). The doublet signal at 7.747 ppm was finally used for molecular quantification because it was clear from interferences due to signals from different molecules.
Statistical analysis
Statistical analyses were performed using SPSS software, version 22.00 (IBM, Somers, NY, USA) and R environment (version 3.2.2; the R Foundation for Statistical Computing, Vienna, Austria) on data obtained from peripheral samples of HIV-1–positive patients before and after probiotic supplementation. Data obtained at T0 and T6 analysis in peripheral blood were compared by Wilcoxon test for paired samples.29,30 Results, unless differently stated, were given as medians, ranges, and percentages. P values <.05 were considered statistically significant. All graphs were generated using GraphPad Prism (version 5.00; GraphPad Software, Inc., La Jolla, CA, USA).
Results
Demographic and clinical characteristics of HIV-1–positive patients
All study participants were HIV-1–positive white men with a median age of 42 (interquartile range [IQR]: 31-50) years old. Participants started cART during chronic HIV-1 infection and have been treated for a median of 6 years (IQR: 1.75-15.75). Probiotics were taken as a supplement to cART. The median of CD4+ cell count pre-cART was 255 cells/mm3 (IQR: 47.5-360), and HIV-1 RNA median value was 5.0 log/mL (IQR: 4.8-5.55). All subjects have been virologically suppressed (<37 HIV-1 RNA copies/mL) for at least 1 year prior to enrollment; their median CD4+ cell count was 674.5 cells/mm3 before (IQR: 570.5-752.3) and 683 cells/mm3 (IQR: 587-796) after probiotic supplementation of cART.
Peripheral immune activation
We compared the immune activation of PBMCs measuring the levels of CD4 T cells expressing CD38 and HLA-DR markers, before (T0) and after probiotic supplementation (T6). Our results showed a statistically significant reduction in the percentage of CD4+CD38+HLA−DR+ T cells at T6 (median: 0.18 [IQR: 0.04-0.32]) compared with T0 (median: 0.80 [IQR: 0.37-2.16]); P = .008 (Table 1).
Table 1.
Effect of probiotic supplementation in CD4+ T-cell immune activation.
IFN-γ mRNA
To assess systemic immune activation, we measured the levels of expression of IFN-γ mRNA in PBMCs, which is one of the main cytokines produced following CD4+ T-cell activation.
IFN-γ mRNA expression showed a reduction after 6 months of probiotic supplementation, without reaching statistical significance (median: 8.60 [IQR: 2.06-23.80] vs median: 1.06 [IQR: 0.32-2.97]) (P = .09) (Figure 1).
Figure 1.
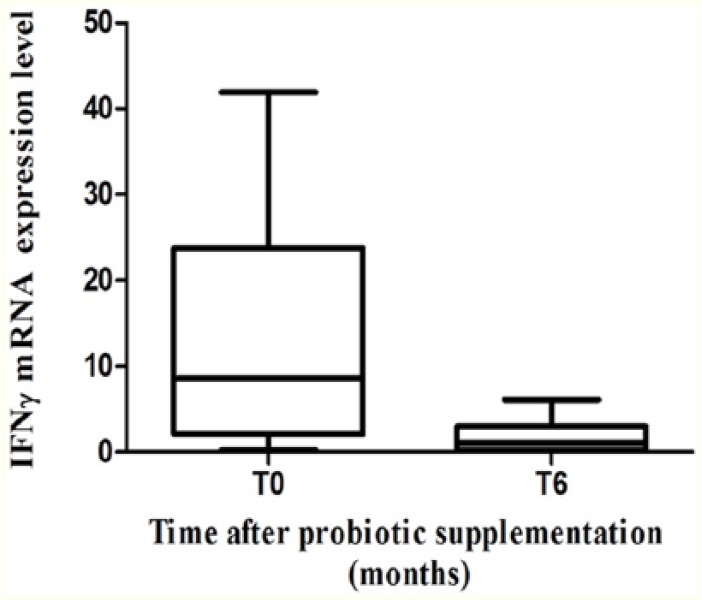
IFN-γ mRNA expression in PBMC. IFN-γ mRNA expression before and after 6 months of probiotic supplementation in PBMC of combined antiretroviral therapy–treated HIV-1–positive patients with suppressed HIV-1 viremia (n = 6) P > .05. HIV-1 indicates human immunodeficiency virus 1; mRNA, messenger RNA; PBMC, peripheral blood mononuclear cell.
IDO-1 mRNA expression
We than analyzed whether 6 months of probiotic supplementation and the relative reduction in IFN-γ mRNA levels could affect the expression of IDO-1 mRNA in PBMCs. A significant reduction in IDO-1 mRNA in PBMCs was observed at T6 (median: 0.0034 [IQR: 0.001-0.006]) compared with T0 (median: 0.052 [IQR: 0.0024-0.18]) (P = .04) (Figure 2).
Figure 2.
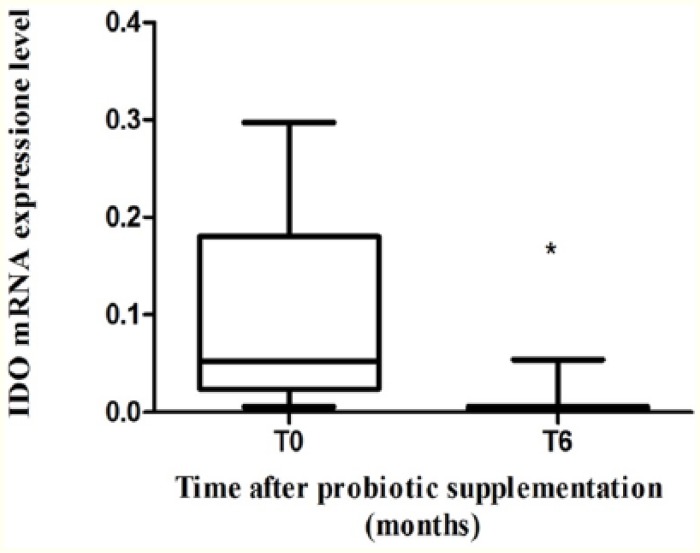
IDO-1 mRNA expression in PBMC. IDO-1 mRNA expression before and after 6 months of probiotic supplementation in PBMC of combined antiretroviral therapy–treated HIV-1–positive patients with suppressed HIV-1 viremia (n = 8) *P = .04. HIV-1 indicates human immunodeficiency virus 1; IDO-1, indoleamine 2,3-dioxygenase 1; mRNA, messenger RNA; PBMC, peripheral blood mononuclear cell.
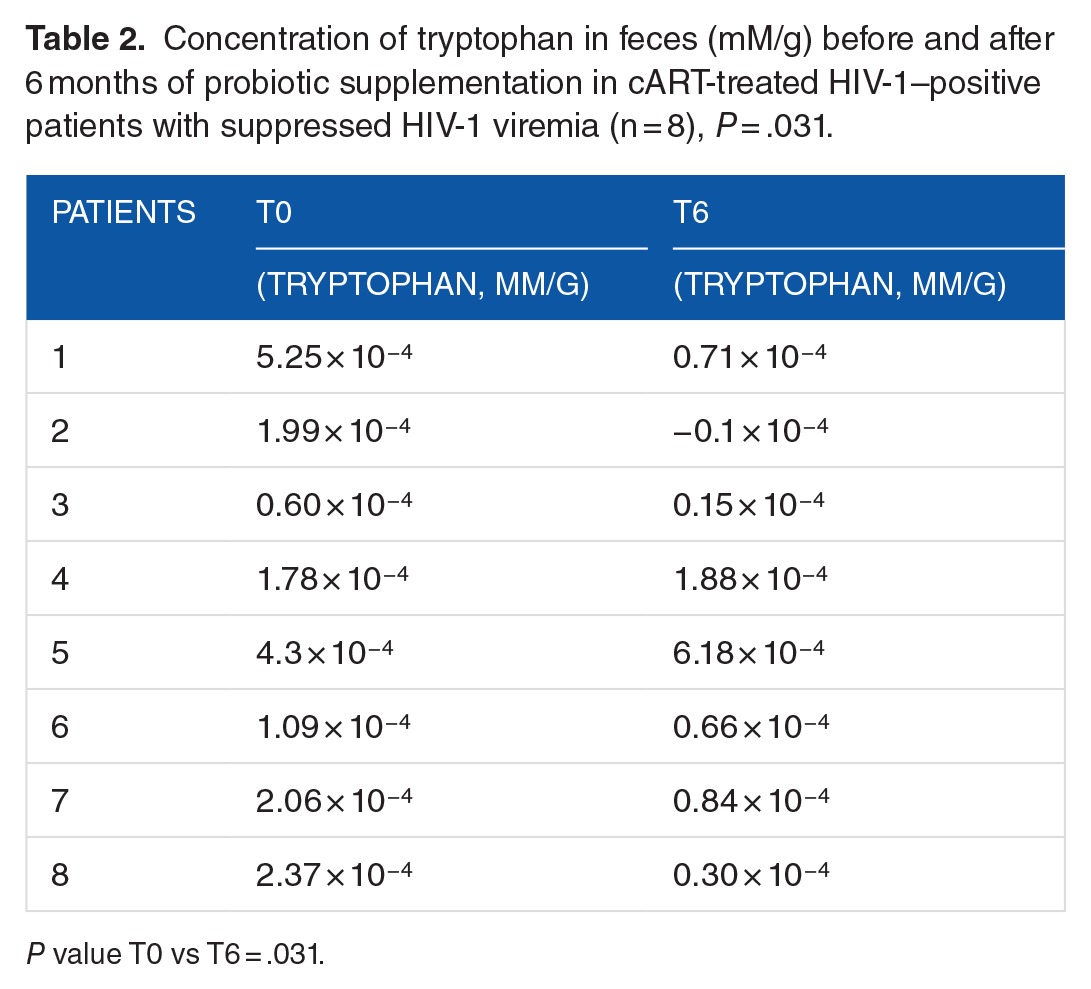
Tryptophan quantification
To assess whether the reduction in IDO-1 mRNA observed in PBMCs could affect tryptophan metabolism, we quantified tryptophan levels in fecal samples through metabolomic analysis by 1H-NMR. A paired comparison revealed a significant decrease in tryptophan level between T0 and T6, as detailed in Table 2 (P = .031).
Table 2.
Concentration of tryptophan in feces (mM/g) before and after 6 months of probiotic supplementation in cART-treated HIV-1–positive patients with suppressed HIV-1 viremia (n = 8), P = .031.
Levels of serotonin
To better understand the ongoing interrelation between probiotic supplementation, tryptophan metabolism, and neurocognitive functions, we evaluated the modification of serotonin level in serum before and after probiotic intake. Our results showed a significant increase in the levels of serotonin at the end of supplementation (median: 185.3 ng/mL [IQR: 104.4-292]) compared with baseline (median: 80.04 ng/mL [IQR: 45.11-148.3]) (P = .008) (Figure 3).
Figure 3.
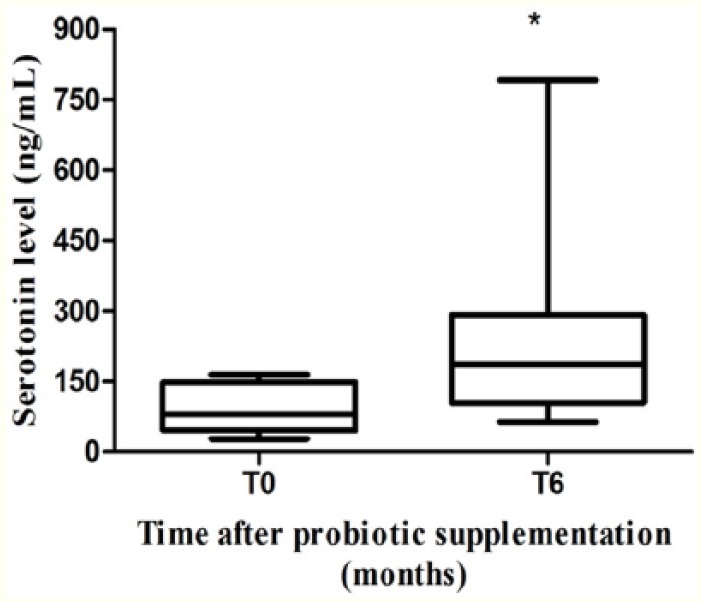
Serum serotonin levels. Serum serotonin levels before and after 6 months of probiotic supplementation in combined antiretroviral therapy–treated HIV-1–positive patients with suppressed HIV-1 viremia (n = 8). *P = .008. HIV-1 indicates human immunodeficiency virus 1.
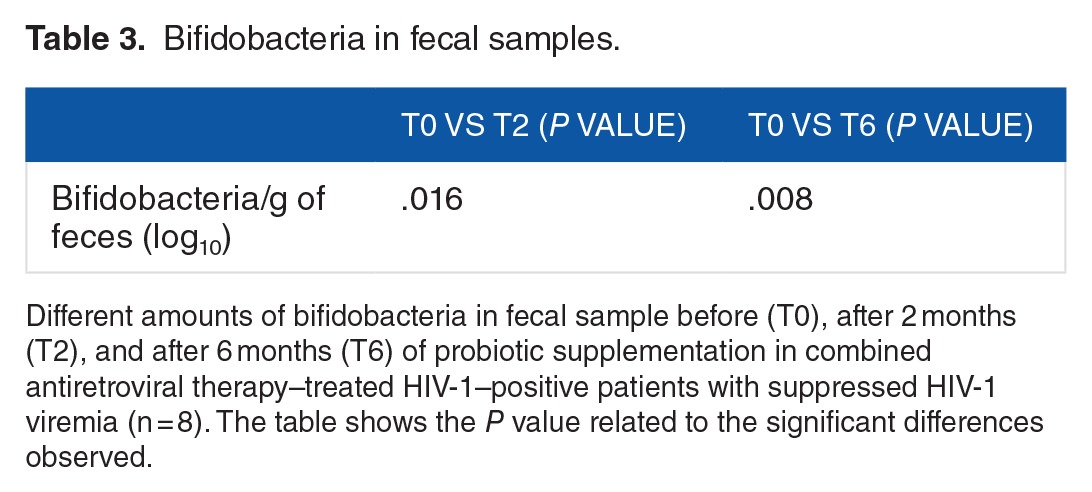
Levels of bifidobacteria in stool
We assessed the amount of bifidobacteria in fecal samples at enrollment (T0), after 2 months (T2), and after 6 months (T6) of probiotic supplementation. We observed a significant increase in this specific bacterial strain at T2 (P = .016) and T6 (P = .008) in comparison with T0 (Table 3). Bifidobacteria were included in the provided product and this was assumed as an evidence of patient’s compliance with probiotic supplementation.
Table 3.
Bifidobacteria in fecal samples.
Discussion
The aim of this study was to evaluate the existence of a link between probiotic supplementation and the modification of serotonin levels in cART-treated patients and to clarify the role exerted by inflammation on this interplay. Moreover, through probiotic supplementation, we aimed to achieve among study population a reduction in systemic immune activation and its related inflammatory markers. To do so, we selected a high-concentration multistrain probiotic preparation with a known ability in modulating the expression of pro-inflammatory cytokines such as tumor necrosis factor α, interleukin 12, and IFN-γ in patients with inflammatory bowel diseases,31 inhibiting microbial pathogens growth by competing with them for epithelial colonization and in increasing tight junctions expression between enterocytes.32
In this study, the frequency of CD4+CD38+HLA−DR+ T cells resulted significantly decreased after 6 months of probiotic supplementation. The reduction in T-cell activation led to a decrease in the expression of the pro-inflammatory cytokine IFN-γ, although not reaching a statistical significance. Our results are in agreement with Ortiz et al who reported that in simian immunodeficiency virus–infected macaques, the same probiotic formulation, in combination with interleukin 21, reduced the expression of several markers of CD4+ T-cell immune activation, such as lipopolysaccharide-binding protein, antigen Ki-67, and soluble CD14 in PBMCs, bronchoalveolar lavage, jejunum (Jej), lymph nodes, and rectal biopsies.33 Similar results were published by Brenchley et al34; in their study, these authors reported that CD4+ T-cell immune activation from colon samples resulted decreased after supplementation with high-concentration multistrain probiotic. The main hypothesis to explain these evidence is that the probiotic acts on toll-like receptors (TLR) signaling downmodulating TLR-induced immune activation.35
Because we observed a reduction in the production of IFN-γ and a beneficial effect on helper T 17 (TH17) cells induced by this probiotic, we measured the level of IDO-1 expression in PBMCs. According to literature, the activity of IDO-1 is influenced by IFN-γ and is associated with an imbalance in the TH17/regulatory T cells’ ratio36; moreover, its activity can exert divergent effects in immunosuppression or immunostimulation in numerous conditions.37–39 Our results showed a reduction in the expression of IDO-1 mRNA in PBMCs after continuous probiotic supplementation for a period of 6 months, thus corroborating the hypothesis that IDO-1 represents the final player in the complex interaction between gut-resident flora, tryptophan metabolism, TH17 cells, and IFN-γ.
Finally, we observed a concurrent decrease in the levels of fecal tryptophan and an increase in serum serotonin levels after 6 months of probiotic supplementation, although less than 1% of tryptophan is involved in the conversion to serotonin (5-HT) inside the brain.
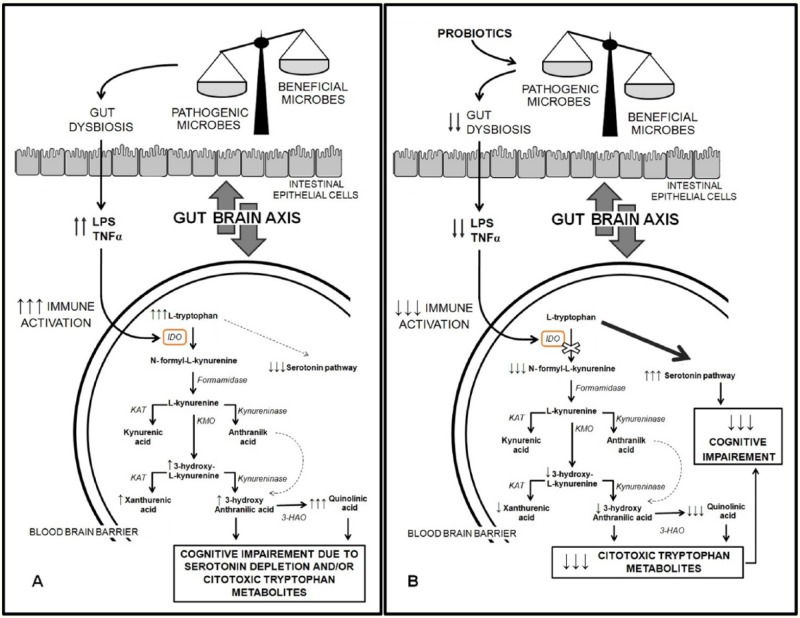
In conclusion, in this study, we highlighted the evidence of a complex relationship between probiotic supplementation, inflammation, and serotonin levels in HIV-1–infected subjects treated with cART together with the safety of this high-concentration multistrain probiotic in this population. This study presents several limitations such as the small size of the included sample, the lack of some tryptophan metabolites’ measurements (eg, plasmatic tryptophan and kynurenine), and the lack of an evaluation of the amount of IDO-1 protein and IDO-1 activity, the latter measured as kynurenine/tryptophan ratio (we only evaluated IDO-1 mRNA expression). If confirmed in subsequent studies, these data could strengthen the hypothesis that this specific probiotic formulation might influence tryptophan metabolism (Figure 4) and suggest novel treatment strategies to resolve or at least reduce some of the common disorders and symptoms observed among HIV-1–infected patients effectively treated with cART.
Figure 4.
Possible modulation of tryptophan/serotonin pathway by probiotic supplementation.
Footnotes
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 709 words, excluding any confidential comments to the academic editor.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Sapienza University of Rome (Ricerche Universitarie, anno 2015, C26A15WTF4).
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions
Gd’E and VV conceived and designed the experiments. GCS, SNF, IS, NG, SS, CP, MS, LL, AM, and GDG analyzed the data. GCS, SNF, and Gd’E wrote the first draft of the manuscript and agree with manuscript results and conclusions. Gd’E and GC contributed to the writing of the manuscript. GCS and SNF jointly developed the structure and arguments for the paper. Gd’E made critical revisions and approved final version. All authors reviewed and approved the final manuscript.
Disclosures and Ethics
The researchers declare compliance with ethical practices on submission of a manuscript and that clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the institutional review board (Department of Public Health and Infectious Diseases, Sapienza University of Rome and the Ethics Committee (Sapienza University of Rome/Policlinico Umberto I) on January 16, 2014. All study participants signed written informed consent. Furthermore, the authors confirmed that all ongoing and related trials for this drug/intervention are registered and declared that the Department of Public Health and Infectious Disease where patients were recruited is fully equipped for the planned interventions.
REFERENCES
- 1.MacDonald TT Spencer J. Evidence that activated mucosal T cells play a role in the pathogenesis of enteropathy in human small intestine. J Exp Med. 1988;167:1341–1349. doi: 10.1084/jem.167.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chahroudi A, Meeker T, Lawson B, Ratcliffe S, Else J, Silvestri G. Mother-to-infant transmission of simian immunodeficiency virus is rare in sooty mangabeys and is associated with low viremia. J Virol. 2011;85:5757–5763. doi: 10.1128/JVI.02690-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Wang X, Veazey RS. Mucosal immunology of HIV infection. Immunol Rev. 2013;254:10–33. doi: 10.1111/imr.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salas JT, Chang TL. Microbiome in human immunodeficiency virus infection. Clin Lab Med. 2014;34:733–745. doi: 10.1016/j.cll.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tenorio AR, Zheng Y, Bosch RJ, et al. Soluble markers of inflammation and coagulation but not T-cell activation predict non-AIDS-defining morbid events during suppressive antiretroviral treatment. J Infect Dis. 2014;210:1248–1259. doi: 10.1093/infdis/jiu254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunt PW, Sinclair E, Rodriguez B, et al. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis. 2014;210:1228–1238. doi: 10.1093/infdis/jiu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolton DL, Pegu A, Wang K, et al. Human immunodeficiency virus type 1 monoclonal antibodies suppress acute simian-human immunodeficiency virus viremia and limit seeding of cell-associated viral reservoirs. J Immunol Res. 2015;90:1321–1332. doi: 10.1128/JVI.02454-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anzinger JJ, Butterfield TR, Angelovich TA, Crowe SM, Palmer CS. Monocytes as regulators of inflammation and HIV-related comorbidities during cART. J Immunol Res. 2014;2014:569819. doi: 10.1155/2014/569819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mudd JC, Brenchley JM. Gut mucosal barrier dysfunction, microbial dysbiosis, and their role in HIV-1 disease progression. J Infect Dis. 2016;214:S58–S66. doi: 10.1093/infdis/jiw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood KJ, Sawitzki B. Interferon gamma: a crucial role in the function of induced regulatory T cells in vivo. Trends Immunol. 2006;27:183–187. doi: 10.1016/j.it.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 12.Brown RR, Ozaki Y, Datta SP, Borden EC, Sondel PM, Malone DG. Implications of interferon-induced tryptophan catabolism in cancer, auto-immune diseases and AIDS. Adv Exp Med Biol. 1991;294:425–435. doi: 10.1007/978-1-4684-5952-4_39. [DOI] [PubMed] [Google Scholar]
- 13.Bipath P, Levay PF, Viljoen M. Tryptophan depletion in context of the inflammatory and general nutritional status of a low-income South African HIV-infected population. J Health Popul Nutr. 2016;35:5. doi: 10.1186/s41043-016-0042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard DM, Dawes MA, Mathias CW, Acheson A, Hill-Kapturczak N, Dougherty DM. L-Tryptophan: basic metabolic functions, behavioral research and therapeutic indications. Int J Tryptophan Res. 2009;2:45–60. doi: 10.4137/ijtr.s2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs D, Hausen A, Reibnegger G, et al. Interferon-gamma concentrations are increased in sera from individuals infected with human immunodeficiency virus type 1. J Acquir Immune Defic Syndr. 1989;2:158–162. [PubMed] [Google Scholar]
- 16.Fuchs D, Jager H, Popescu M, et al. Immune activation markers to predict AIDS and survival in HIV-1 seropositives. Immunol Lett. 1990;26:75–79. doi: 10.1016/0165-2478(90)90178-s. [DOI] [PubMed] [Google Scholar]
- 17.Wirleitner B, Reider D, Ebner S, et al. Monocyte-derived dendritic cells release neopterin. J Leukoc Biol. 2002;72:1148–1153. [PubMed] [Google Scholar]
- 18.Gostner JM, Becker K, Kurz K, Fuchs D. Disturbed amino acid metabolism in HIV: association with neuropsychiatric symptoms. Front Psychiatry. 2015;6 doi: 10.3389/fpsyt.2015.00097. Article 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung H, Kim SW, Hong M, Suk KT. Microbiota-based treatments in alcoholic liver disease. World J Gastroenterol. 2016;22:6673–6682. doi: 10.3748/wjg.v22.i29.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koppe L, Mafra D, Fouque D. Probiotics and chronic kidney disease. Kidney Int. 2015;88:958–966. doi: 10.1038/ki.2015.255. [DOI] [PubMed] [Google Scholar]
- 21.Thushara RM, Gangadaran S, Solati Z, Moghadasian MH. Cardiovascular benefits of probiotics: a review of experimental and clinical studies. Food Funct. 2016;7:632–642. doi: 10.1039/c5fo01190f. [DOI] [PubMed] [Google Scholar]
- 22.Le Barz M, Anhê FF, Varin TV, et al. Probiotics as complementary treatment for metabolic disorders. Diabetes Metab J. 2015;39:291–303. doi: 10.4093/dmj.2015.39.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert JA, Krajmalnik-Brown R, Porazinska DL, Weiss SJ, Knight R. Toward effective probiotics for autism and other neurodevelopmental disorders. Cell. 2013;155:1446–1448. doi: 10.1016/j.cell.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuss IJ, Kanof ME, Smith PD, Zola H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol. 2009 doi: 10.1002/0471142735.im0701s85. Chapter 7:Unit7.1. [DOI] [PubMed] [Google Scholar]
- 25.Matsuki T, Watanabe K, Fujimoto J, et al. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl Environ Microbiol. 2004;70:167–173. doi: 10.1128/AEM.70.1.167-173.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scagnolari C, Monteleone K, Selvaggi C, et al. ISG15 expression correlates with HIV-1 viral load and with factors regulating T cell response. Immunobiology. 2016;221:282–290. doi: 10.1016/j.imbio.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Meiboom S, Gill D. Modified spin-echo method for measuring nuclear relaxation times. Rev Sci Instrum. 1958;29:688–691. [Google Scholar]
- 28.Barbara G, Scaioli E, Barbaro MR, et al. Gut microbiota, metabolome and immune signatures in patients with uncomplicated diverticular disease [published online ahead of print September 12, 2016] Gut. doi: 10.1136/gutjnl-2016-312377. [DOI] [PubMed] [Google Scholar]
- 29.Datta S, Satten GA. A signed-rank test for clustered data. Biometrics. 2008;64:501–507. doi: 10.1111/j.1541-0420.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 30.Rosner B, Glynn RJ, Lee ML. The Wilcoxon signed rank test for paired comparisons of clustered data. Biometrics. 2006;62:185–192. doi: 10.1111/j.1541-0420.2005.00389.x. [DOI] [PubMed] [Google Scholar]
- 31.Dotan I, Rachmilewitz D. Probiotics in inflammatory bowel disease: possible mechanisms of action. Curr Opin Gastroenterol. 2005;21:426–430. [PubMed] [Google Scholar]
- 32.Lyle S, Erickson T, Walton KLW. Effects of probiotic bacteria on cytokine expression in cultured intestinal epithelial cells. FASEB J. 2016;30(1) Supplement 1259.3. [Google Scholar]
- 33.Manuzak JA, Hensley-McBain T, Zevin AS, et al. Enhancement of microbiota in healthy macaques results in beneficial modulation of mucosal and systemic immune function. J Immunol. 2016;196:2401–2409. doi: 10.4049/jimmunol.1502470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt N, Liu Y, Bentebibel SE, et al. The cytokine TGF-β co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol. 2014;15:856–865. doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenabian MA, Patel M, Kema I, et al. Distinct tryptophan catabolism and Th17/Treg balance in HIV progressors and elite controllers. PLoS ONE. 2013;8:e78146. doi: 10.1371/journal.pone.0078146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mancuso R, Hernis A, Agostini S, et al. Indoleamine 2,3 dioxygenase (IDO) expression and activity in relapsing-remitting multiple sclerosis. PLoS ONE. 2015;10:e0130715. doi: 10.1371/journal.pone.0130715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curti A, Trabanelli S, Salvestrini V, Baccarani M, Lemoli RM. The role of indoleamine 2,3-dioxygenase in the induction of immune tolerance: focus on hematology. Blood. 2009;113:2394–2401. doi: 10.1182/blood-2008-07-144485. [DOI] [PubMed] [Google Scholar]
- 39.Campbell BM, Charych E, Lee AW, Möller T. Kynurenines in CNS disease: regulation by inflammatory cytokines. Front Neurosci. 2014;8:12. doi: 10.3389/fnins.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]