Abstract
Analysis of the genome sequence of Enterococcus faecalis allowed the identification of two genes whose protein products showed 33 and 34% identity with those of sigV and yrhM of Bacillus subtilis, respectively. These genes, named sigV and rsiV, are predicted to encode members of the extracytoplasmic function subfamily of eubacterial RNA polymerase sigma and anti-sigma factors, respectively. This group of sigma factors has been shown to regulate gene expression in response to stress conditions. sigV and rsiV were shown to be under the control of the same promoter. The transcriptional start site was determined, and the 1.5-kb mRNA transcript was shown to be overexpressed under glucose and complete starvation, as well as under physicochemical treatments. Three mutants, affected in sigV, rsiV, and both genes, were constructed by double-crossover recombination within the genome of E. faecalis strain JH2-2. Compared with the wild type and the rsiV mutant, the sigV mutants were more susceptible to heat shock, acid, and ethanol treatments and displayed decreased survival during long-term starvation. A nisin-inducible sigV gene construction used in complementation assays restored the wild phenotype of the sigV mutants, confirming the involvement of SigV in the heat shock, ethanol, and acid stress responses. Northern blot analysis carried out with the three mutant strains revealed the inhibition of sigV expression by the related anti-sigma factor gene rsiV. In addition, putative candidates of the sigV regulon determined by computer search for the sigV promoter sequence were analyzed.
Enterococcus faecalis, a gram-positive, nonsporulant, and nonmotile bacterium, is a natural member of the human and animal flora. This ubiquitous microorganism has numerous fields of interest. (i) During its lifetime, E. faecalis can enter the mouth-fecal cycle, as well as colonize various ecological niches (water, food) (2, 6), and is therefore often used as an indicator of fecal contamination. (ii) In the food industry, its abilities to degrade casein and stimulate the growth of certain lactic acid bacteria were exploited for the ripening of some cheeses (cheddar, mozzarella) (2). (iii) In the medical field, although it is a human commensal species, it remains a frightening opportunistic pathogenic bacterium that represents one of the major causes of nosocomial enterococcal infections, affecting mainly children and immunodeficient patients (25, 34, 46).
The responses of E. faecalis to environmental stresses have been well studied during the last decade and have shown its exceptional ability to survive and persist in a variety of adverse environments. Indeed, exponentially growing cells of E. faecalis can resist stresses such as heat, high osmolarity, and the presence of ethanol, detergents, hydrogen peroxide, sodium hypochloride, and heavy metals. Moreover, adaptation by pre-exposure of the culture to sublethal stresses such as 30 min at 50°C or at 37°C in the presence of H2O2 (2.4 mM), sodium dodecyl sulfate (SDS; 0.01%), bile salts (0.08%), cadmium chloride (50 μg ml−1), or pH 4.8 or 10.5 leads to a substantial increase in resistance to the corresponding, usually lethal, stress (10, 27). In addition, starvation promoted by exhaustion of the carbon and energy source glucose or incubation in an oligotrophic microcosm strongly enhances the resistance of E. faecalis to environmental stresses and can be correlated with the increased synthesis of many proteins (13, 14).
Looking at this panel of adaptation and resistances, one can speculate on the presence of transcriptional factors that efficiently control gene expression to ensure that the response will be rapidly induced after exposure to stress. Such a situation was already well characterized for the master regulator of the general stress response σS (RpoS) in Escherichia coli (17) and σB in Bacillus subtilis (15, 16). However, previous investigations and data bank comparisons failed to reveal such general stress sigma factors in E. faecalis. On the other hand, numerous genes encoding putative transcriptional regulators potentially involved in the E. faecalis stress response are present in the genomic sequence of E. faecalis V583 (38). Of these, eight two-component systems (29, 48) and HypR, a transcriptional regulator of the LysR family (51), were clearly shown to be involved in the E. faecalis stress response.
At the beginning of this work, analysis of the unfinished genomic sequence of E. faecalis strain V583 revealed one open reading frame (ORF) whose predicted protein product showed 33% identity with that of the sigV gene of B. subtilis. This gene encodes one of the ECF (extracytoplasmic function) σ factors (47), which are a subfamily of alternative σ factors belonging to the σ70 class (30). Members of this subfamily are involved in regulating bacterial interactions with the extracellular environment, including adaptation to stress and, in some cases, bacterial virulence (32). Indeed, ECF sigma factors control a vast range of processes in different gram-negative and gram-positive bacteria and respond to changes in environmental conditions including temperature upshifts, oxidative stress, acid pH, osmotic shock, and exposure to detergents (9, 18, 20, 40-42, 56). The best-characterized ECF sigma factors of B. subtilis are σW and σX, which are both maximally expressed late in growth and are involved in the response to alkali shock (55) and to high temperature and oxidative stress (20), respectively. Recently, SigM of B. subtilis was reported to be activated in response to cell wall antibiotic, ethanol, heat, acid, and superoxide stress (50). Thus, we were interested in investigating whether sigV of E. faecalis is involved in the exceptional ability of this bacterium to survive and persist in a variety of hostile environments.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. faecalis strain JH2-2 (57), derivatives thereof, and strain V583 (43) were grown at 37°C without shaking in M17 medium (49) supplemented with 0.5% glucose (GM17) or in semisynthetic medium (13). When required, erythromycin (100 μg ml−1) and chloramphenicol (20 μg ml−1) were added. Viability was determined by spreading 0.5 ml of appropriate serial dilutions on brain heart infusion agar. E. coli strains XL1-Blue and EC101 were cultured with vigorous shaking at 37°C in LB medium (44) with ampicillin (100 μg ml−1) or erythromycin (300 μg ml−1) when required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. faecalis | ||
| JH2-2 | Fusr Rifr; plasmid-free wild-type strain | 57 |
| S26 | JH2-2 isogenic derivative sigV mutant | This work |
| AS39 | JH2-2 isogenic derivative rsiV mutant; 75-bp rsiV median deletion | This work |
| SAS | JH2-2 isogenic derivative sigV-rsiV double mutant; 743-bp median deletion within sigV-rsiV operon | This work |
| V583 | Clinical isolate; Vanr | 43 |
| E. coli | ||
| EC101 | KanrsupE thi (lacproAB) (F′ traD36 proAB lacIqZΔM15) repA | 28 |
| XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pORI19.1 | pWV01 derivative; Emr Ori+ RepA−lacZ′ | 28 |
| pG+host3 | pWV01 derivative; Cmr repAts (previously named pVE6007) | 31 |
| pMSP3535 | EmrnisR nisK PnisA (nisin-inducible promoter) | 4 |
Mapping of transcriptional start sites (TSSs).
The 5′ end of the sigV mRNA was mapped from a 5′ RACE (rapid amplification of cDNA ends) PCR product obtained with the 3′/5′ RACE kit (Roche Molecular Biochemicals). For this purpose, we used total RNA extracted from E. faecalis JH2-2 cells harvested 1 h after the onset of stationary phase, corresponding to overexpression of the sigV gene. Briefly, the first-strand cDNA was synthesized from total RNA with sigV-specific primer SP1 (Fig. 1A and Table 2), avian myeloblastosis virus reverse transcriptase, and the deoxynucleotide mixture of the 3′/5′ RACE kit as recommended by the manufacturer. After purification and dA tailing of the cDNA, a PCR with the (dT)-Anchor oligonucleotide primer and the specific sigV SP2 primer (Fig. 1A and Table 2) led to a PCR product of 320 bp revealed by 2% agarose gel electrophoresis. This PCR product was purified with the QIAquick kit (QIAGEN, Santa Clara, Calif.) and sequenced by the dideoxy-chain termination method with the ABI prism sequencing system (PE Biosystems) and primer SP3 (Fig. 1A and Table 2).
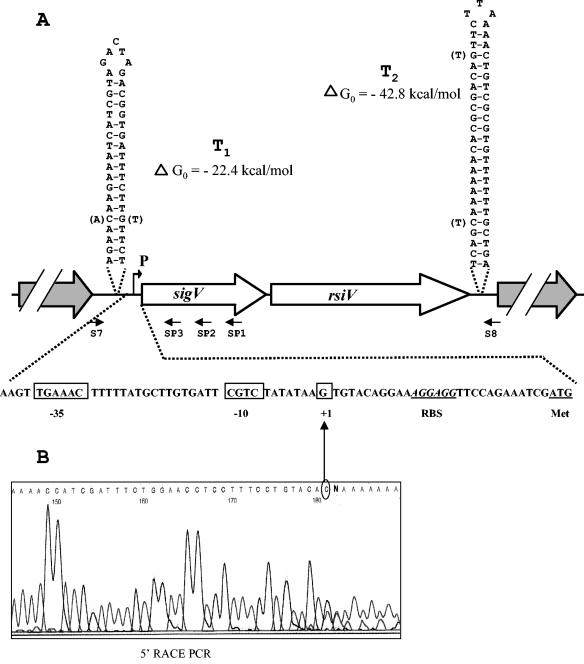
FIG. 1.
Schematic representation of the genetic organization of the sigV-rsiV chromosomal region of E. faecalis JH2-2. (A) Large arrows represent the ORFs, and their orientation shows the transcriptional direction. The nucleotide sequences of the sigV promoter region and of the putative Rho-independent terminators (T1 and T2) are shown. Nucleotides corresponding to the differences observed in the E. faecalis V583 genome sequence are in parentheses. Primers used for amplification of the overall DNA of the sigV locus (S7 and S8) and for mapping of the TSS (SP1 to SP3) are represented. The TSS (+1) and the putative −35 and −10 motifs are boxed. (B) Electropherogram obtained from a 5′ RACE PCR experiment. The sequence in the electropherogram was obtained with primer SP3 and dA-tailed cDNA. The last base (C) upstream of the dA tail corresponds to the first nucleotide transcribed (TSS).
TABLE 2.
Primers used for PCR, mutagenesis, and 5′ RACE PCR experimentsa
| Name | Sequence | Characteristic |
|---|---|---|
| S1 | 5′-CAAATTCtGCaGCGTTTAGAAAC-3′ | (+) PstI |
| S2 | 5′-CTTCGGAGcTCATTCAAACTG-3′ | (−) SacI |
| S3 | 5′-ggatcccgggAAAtGATGAACATGTTTT AGCC-3′ | (+) SmaI |
| S4 | 5′-ggatcccgggTTCaAATATGAACAGCTT CTCG-3′ | (−) SmaI |
| S7 | 5′-CGTCCTGTTGATTGGTCGC-3′ | (+) |
| S8 | 5′-CCATTGCCATTCGCCCTTTC-3′ | (−) |
| S12 | 5′-GTCGATTGGtaCCTGAGATTAC-3′ | (+) KpnI |
| S1B | 5′-GGAAAGGAGGaTCCAGAAATC-3′ | (+) BamHI |
| S2B | 5′-CCATTATTTTTTCGgaTCCTTTC-3′ | (−) BamHI |
| AS1 | 5′-CAACGGATCTAgAtAGCGGAAG-3′ | (+) XbaI |
| AS2 | 5′-CTCCTTGGGGtAcCACAATCAC-3′ | (−) KpnI |
| AS3 | 5′-catgtacccgggtGAACCTTCACAGAAAT TAAAGGC-3′ | (+) SmaI |
| AS4 | 5′-catgtacccgggCtATTGCCCTTTGACTT GTTGA-3′ | (−) SmaI |
| AS6 | 5′-CAGTAAAACCGgAtCcGACTTGG-3′ | (−) BamHI |
| SP1 | 5′-CGTTAATATCCAGGATCAAGGC-3′ | (−) RACE PCR |
| SP2 | 5′-GCTTCTCGTTTCTTTTTCCGCC-3′ | (−) RACE PCR |
| SP3 | 5′-GTCCTTTTTGGATGGCCTTC-3′ | (−) RACE PCR |
| Pgd5 (SP3) | 5′-TGATTCCTTTTCTAACTTGGTC-3′ | (−) RACE PCR |
| Pgd6 (SP2) | 5′-TAGGACTTGTGGGGTTATTTG-3′ | (−) RACE PCR |
| Pgd7 (SP1) | 5′-CCCGTTTGGATATAACGTTGC-3′ | (−) RACE PCR |
| 1934C (SP3) | 5′-CTAGCATACCATTAAAAAATGG-3′ | (−) RACE PCR |
| 1934D (SP2) | 5′-TCTTGATTTGTTGATAATCTGC-3′ | (−) RACE PCR |
| 1934E (SP1) | 5′-GCTTGTTAATTTTCAACATTTCG-3′ | (−) RACE PCR |
| 0159C (SP3) | 5′-GGCACTTAGGATAATTGCCAA-3′ | (−) RACE PCR |
| 0159D (SP2) | 5′-GCCATCAATCGAGCCACTAT-3′ | (−) RACE PCR |
| 0159E (SP1) | 5′-TGGCGTGAATAACGCACCAA-3′ | (−) RACE PCR |
| 0315C (SP3) | 5′-ATGCTCTAGAAAAGGAGCACT-3′ | (−) RACE PCR |
| 0315D (SP2) | 5′-TTCTACTTCTTTCGTCCACTC-3′ | (−) RACE PCR |
| 0315RT2 (SP1) | 5′-GCCAATTCCCGATAAAATCC-3′ | (−) RACE PCR |
| dC-Anchor | 5′-GACCACGCGTATCGATGTCGA C15D*-3′ | RACE PCR |
Nucleotides in lowercase are not complementary to the target sequence. Underlined nucleotides indicate sites inserted within primer sequences and corresponding to the restriction enzymes reported in the right column. (+), primer directed toward the 3′ end of the gene; (−), primer in the opposite direction. The indicated RACE PCR primers were used for first-strand cDNA synthesis (SPI), PCR amplification (SP2), or the sequencing reaction (SP3). *D = A, G or T.
Considering the AT-rich genome of E. faecalis and in order to facilitate determination of the TSSs of the sigV regulon candidates, we replaced the dT-Anchor primer with a dC-Anchor primer and instead of dA tailing, we carried out dG tailing of cDNA by using 2 mM dGTP instead of 2 mM dATP as recommended by the manufacturer. The primers used for this purpose are listed in Table 2.
Analysis of mRNA by Northern and dot blot experiments.
Total RNAs of E. faecalis strain JH2-2 and derivatives thereof were isolated from exponentially growing cells, from cells harvested 1 h after the onset of stationary phase, or from stressed cells with the RNeasy Midi kit (QIAGEN). For Northern blot assays, 10 μg of total RNA was electrophoretically resolved per lane and transferred onto Hybond-N+ membranes (Amersham International, Little Chalfont, United Kingdom) by standard procedures (44). Sizes of transcripts were estimated by comparison with an RNA ladder (0.56 to 9.4 kb; Amersham International). For dot blot assays, 1 or 2 μg of total RNA from cells incubated for 10 min under the different individual stress conditions was spotted onto Hybond-N+ membranes. Membrane-bound nucleic acids were hybridized at 55°C in 0.5 M sodium phosphate buffer (pH 7.0) containing 5% SDS with a single-stranded labeled probe complementary to the sigV mRNA. After hybridization, membranes were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS (10 min) and twice in 0.5× SSC-0.1% SDS (10 min) at 55°C and then exposed to a storage phosphor screen (Packard Instrument Company, Canberra, Australia).
Preparation of the single-stranded labeled probe was done as follows. First, a DNA fragment was amplified by PCR from chromosomal DNA of E. faecalis JH2-2 with primers S1 and AS2 (Fig. 2; Table 2). The probe was then synthesized by elongating the specific oligonucleotide SP1 (Fig. 1; Table 2) with Taq DNA polymerase; 2 μM (each) dCTP, dGTP, and dTTP; 2 μCi of [α-32P]dATP; and 10 ng of the previous PCR DNA fragment as the template. Thirty cycles of 20 s at 94°C, 30 s at 52°C, and 45 s at 72°C were performed.
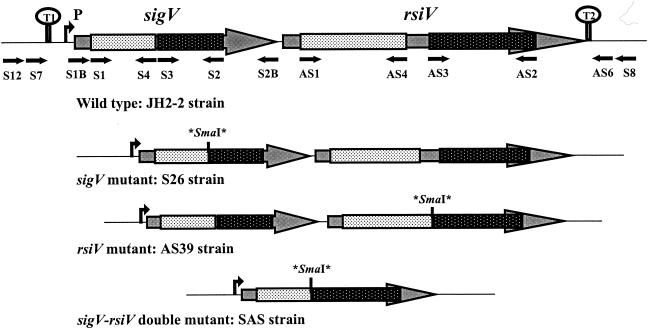
FIG. 2.
Schematic representation of the genetic organization of the sigV locus in the E. faecalis wild-type strain and in its isogenic derivative mutants. Asterisks correspond to stop codons flanking the SmaI site inserted by mutagenesis (see text). The primers used for the mutagenesis experiments, PCR cloning, and sequence verification are indicated by black arrows.
Construction of sigV, rsiV, and sigV-rsiV mutants.
The JH2-2 derivative strain affected in sigV was constructed by introducing two translational stop codons flanking an SmaI site within the coding sequence via a double-crossover (DCO) event by a method based on the conditional replication of the pORI19.1/pG+host3 system previously described (28). A 457-bp sigV internal DNA fragment was amplified by PCR with total DNA of E. faecalis JH2-2 and primers S1 and S2 containing PstI and SacI sites, respectively (Fig. 2; Table 2). The PstI-SacI-digested PCR fragment was then cloned into vector pORI19.1 (Table 1), which had previously been digested by the same restriction enzymes, and then transformed into electrocompetent E. coli EC101cells. The resulting plasmid was used as the DNA template for a second PCR with primers S3 and S4 harboring a nonhomologous tail of 10 nucleotides (nt) containing an SmaI site (Fig. 2; Table 2). The 2.7-kb PCR product obtained was digested with SmaI, purified, ligated, and used to transform EC101 cells. The recombinant plasmids were analyzed by SmaI digestion and sequenced to confirm the mutation within the sigV coding sequence introduced by two translational stop codons. Representative recombinant pORI19.1 plasmids carrying the JH2-2 sigV mutant DNA were used to transform E. faecalis JH2-2 into which plasmid pG+host3 (31), encoding a thermosensitive RepA protein, had previously been introduced. After electroporation, the two plasmids were maintained together at a permissive temperature of 30°C by plating cells on GM17 agar containing erythromycin and chloramphenicol. Clones resistant to both antibiotics were shown to harbor plasmid pG+host3 and recombinant plasmid pORI19-1. One of these clones was grown for 1 h at 30°C in GM17 broth with no antibiotic and then incubated for 3 h at 42°C before being plated on GM17 agar medium containing only erythromycin. After 48 h at 42°C, Emr clones were analyzed by PCR and shown to be single-crossover mutants containing both the wild-type and mutated alleles of the target gene. To set up the second homologous recombination event, the single-crossover mutants were transformed with helper plasmid pG+host3 under the permissive conditions described above. After 100 generations at 30°C on GM17 broth containing chloramphenicol, the resulting transformants were grown for 1 h at 30°C on GM17 with no antibiotic and transferred at 42°C for 3 h before being plated on solid GM17 and incubated at 42°C. Following this step, 25 out of 225 clones tested were found to be Ems. Results of the SmaI digestion of PCR fragments amplified with primers S1 and S2 showed that 21 of the Ems clones kept the wild-type copy while the remaining 4 harbored an SmaI site within the sigV gene and were considered DCO mutants. Total DNA was extracted from these four Ems clones, digested with ClaI or HaeII, and hybridized with a sigV-specific probe. The Southern blot result revealed that the four clones were effectively generated by a second crossover event that resulted in excision of pORI19.1 harboring the sigV wild-type allele (data not shown). The nucleotide sequencing of the sigV gene from these mutants confirmed the presence of the SmaI site flanked by two stop codons leading to a truncated SigV protein in its C-terminal half.
This procedure was also used to construct the rsiV and sigV-rsiV mutants. Primer pairs AS1-AS2 and S12-AS6 (Fig. 2; Table 2) were used to amplify the internal fragment of the rsiV gene and the overall bicistronic structure of the sigV locus, respectively. Following the cloning step in pORI19.1, two other pairs of primers, AS3-AS4 and AS3-S4 (Fig. 2; Table 2), were used to generate the SmaI site. This site was flanked by two translational stop codons and replaces a median deletion of 75 or 743 bp within the rsiV gene or the operon structure, respectively (Fig. 2). The overall procedure described above allowed us to isolate two DCO mutants each for the rsiV gene and the bicistronic operon structure. The JH2-2 isogenic mutant strains selected for further analysis are S26, AS39, and SAS, corresponding to the sigV and rsiV single mutants and the sigV-rsiV double mutant, respectively (Table 1 and Fig. 2).
Challenge treatments and complementation assays.
For determination of survival following different individual physical and chemical challenges, cultures of wild-type E. faecalis JH2-2 and its isogenic sigV locus mutants S26, AS39, and SAS (Table 1) were grown in 10 ml of GM17 broth or semisynthetic medium at 37°C to an optical density at 600 nm of 0.5 (mid-exponential growth phase). Bacteria were harvested by centrifugation, resuspended in 10 ml of fresh medium, and then incubated at 37°C (control culture) or exposed to lethal doses of the following stresses. (i) For high-temperature heat shock, the cultures were transferred to 62°C. (ii) For acid shock, the pH was adjusted to 3.2 with lactic acid. (iii) For ethanol stress, ethanol was added to a final concentration of 22% (vol/vol). (iv) For detergent stress, the final concentration of bile salts or SDS was 0.3 or 0.017% (wt/vol), respectively. (v) For oxidative stress, the medium was supplemented with 45 mM H2O2 or 20 mM t-butyl hydroperoxide (tBOOH). (vi) For salt stress, solid NaCl was added to a final concentration of 28.5% (wt/vol). Sample aliquots of 0.5 ml were removed from each culture for plating at the specified times during the course of the experiment. Tenfold serial dilutions in 0.9% NaCl were plated on solid GM17 at each time point, and colonies were enumerated after 48 h of incubation at 37°C by counting two plates at two different dilutions. Challenges were performed for 2 h, and experiments were carried out at least in triplicate for each stress condition. Percent viability represents the ratio of the number of viable cells after exposure to stress to the number of cells surviving prior to the challenge. The long-term survival of the four strains was followed for 110 days by incubation at 15°C in rich medium (GM17 broth) or oligotrophic medium (seawater).
For the complementation assays, the sigV gene was cloned into plasmid pMSP3535 (Table 1). Briefly, the wild-type sigV gene was amplified by PCR with primers S1B and S2B (Fig. 2; Table 2), both containing BamHI sites, with JH2-2 genomic DNA as the template. The PCR product was then digested with BamHI and inserted into pMSP3535 previously digested with the same restriction enzyme. The recombinant vector (pMSP3535-sigV) was verified for the integrity of the sigV gene by sequencing and used to transform the wild-type JH2-2 strain and its derivative S26 and SAS mutant strains. Attempts to restore the wild-type phenotype were carried out by overexpression of the SigV protein at 0 (control) and 300 ng of nisin ml−1 in the presence of 100 μg of erythromycin ml−1 added to the GM17 broth at the onset of the experiment.
2D protein gel electrophoresis.
Sample preparation and two-dimensional (2D) protein gel electrophoresis were carried out as previously described (13).
General molecular methods.
Restriction endonucleases, T4 polynucleotide kinase, alkaline phosphatase, and T4 DNA ligase were obtained from Roche Molecular Biochemicals, Amersham International, and Eurogentec and used in accordance with the manufacturers' instructions. PCR assays were carried out with 1 μg of chromosomal DNA from E. faecalis JH2-2, 20 pmol of primers, and Taq DNA polymerase (Amersham). When necessary, PCR products were purified with the QIAquick kit (QIAGEN). E. coli and E. faecalis were transformed by electroporation with a Gene Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.) as described by Dower et al. (7) and Holo and Nes (19), respectively. Plasmids were purified with a QIAprep Miniprep kit (QIAGEN). Other standard techniques were carried out as described by Sambrook et al. (44).
DNA sequencing and sequence analysis.
PCR fragments were sequenced by the dideoxy-chain termination method (45) with the ABI Prism sequencing system (PE Biosystems). DNA and amino acid sequences were analyzed with the Mac Vector program (Kodak Scientific Imaging Systems), and database searches were performed with the BLAST program (1).
Nucleotide sequence accession number.
The sequence of the sigV locus of E. faecalis strain JH 2-2 has been deposited in the GenBank database under accession no. AY312057.
RESULTS
Identification and genetic organization of the sigV locus of E. faecalis strain JH2-2
The release of the partial genome sequence of E. faecalis strain V583 by The Institute of Genomic Research (http://www.tigr.org.) allowed us to identify a member of the ECF subfamily of eubacterial RNA polymerase sigma factors that was 33% identical (59% similar) to the ECF-type sigma factor SigV of B. subtilis. Within this sigV locus, an additional ORF was found immediately downstream of sigV and its protein product was 34% identical (54% similar) to the product of yrhM, a putative ECF anti-sigma factor of B. subtilis. In E. faecalis, these two genes were named sigV and rsiV (for regulator of σV) on the basis of their homology to the related genes in B. subtilis (47). On the basis of the sequence of E. faecalis strain V583, oligonucleotides S7 and S8 (Table 2 and Fig. 1) were designed and used to PCR amplify the overall DNA region corresponding to the sigV locus from the genomic DNA of E. faecalis strain JH2-2. The resulting DNA was sequenced and deposited in the GenBank database under accession number AY312057.
Analysis of the nucleotide sequence of the sigV locus revealed that the sigV and rsiV genes encode predicted proteins of 165 and 294 amino acid (aa) residues, respectively. They are oriented in the same direction, and rsiV lies immediately downstream of sigV, separated by only 24 nt (Fig. 1A). No obvious evidence of the presence of a promoter sequence upstream of the rsiV gene was detected. However, putative ribosome binding site sequences AGGAGG and AGGAAG were detected 12 and 10 nt upstream of the initiation codon (ATG) of sigV and rsiV, respectively. The sigV locus is flanked by genes coding for a hypothetical protein upstream of sigV and a peptidase downstream of rsiV. DNA stretches of inverted repeats encoding secondary structures that could correspond to putative strong Rho-independent terminators T1 and T2 were detected in both intergenic regions (Fig. 1A). These characteristics strongly suggested the genetic organization of the sigV locus as a bicistronic operon structure, as suggested for sigV and yrhM of B. subtilis (47).
Comparison of the sigV locus nucleotide sequences of E.faecalis strains V583 and JH2-2 revealed only minor differences, resulting in two and five changes in the amino acid sequences of the protein products of sigV and rsiV, respectively (data not shown). A base pair replacement and two other nucleotide changes, leading to a perfect match, were detected within the sequences of putative terminators T1 and T2, respectively (Fig. 1A).
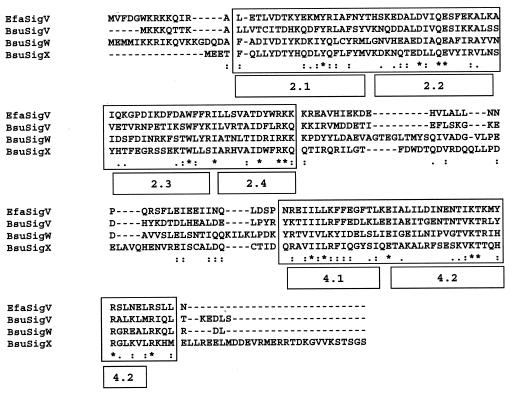
Comparison of the SigV amino acid sequence to the data banks revealed significant homology with more than 100 proteins, all corresponding to ECF-type sigma factors such as SigV and SigW of B. subtilis; SigV, SigW, and SigY of B. halodurans; SigV of C. tetani; SigH of P. putida; Sig24 of C. thermocellum; SigX of C. acetobutylicum; SigR of Streptomyces coelicolor; and AlgU of P. aeruginosa, with identities ranging from 24 to 33%. The E. faecalis SigV amino acid sequence alignment with SigV (best identity) and SigW and SigX (sharing with E. faecalis SigV a highly homologous promoter sequence consensus [see below]) of B. subtilis showed that the most highly conserved regions are region 2.2, an area of overlap at the end of 2.3 and the 2.4 part, and region 4 (Fig. 3). Extended amino acid sequence alignment of E. faecalis SigV with other ECF sigma factors also revealed conserved regions 2 and 4 (data not shown). These areas of conservation are similar to those seen within the ECF family as a whole and are implicated in promoter melting (region 2.3) and −10 (region 2.4) and −35 (region 4.2) promoter recognition (30).
FIG. 3.
Multiple alignment of ECF sigma factors SigV of E. faecalis (EfaSigV) and SigV (BsuSigV), SigW (BsuSigW), and SigX (BsuSigX) of B. subtilis. Highly conserved regions 2 and 4 are boxed, and the subdomains are indicated. Periods, colons, and asterisks indicate weakly similar, strongly similar, and identical amino acids, respectively.
Although a large number of SigV-related proteins exist, only 20 homologous sequences were detected by BLAST searches with RsiV. Among them, only the first hit was described as a putative anti-sigma factor and corresponded to the product of yrhM, which is located immediately downstream of sigV in the B. subtilis genome (47). RsiV is predicted to be a membrane-associated protein since a putative helix transmembrane is detected by the Goldman-Engelman-Steitz hydrophobicity scale (8), spanning amino acid residues 51 to 71 with a certainty score of 1.63. All of these features strongly suggest that SigV and RsiV are RNA polymerase ECF sigma and anti-sigma factors, respectively, and that they are likely organized in a bicistronic operon structure.
Mapping of the TSS of the sigV locus.
In order to locate the TSS of the sigV locus, a 5′ RACE PCR experiment was carried out with total RNA of E. faecalis JH2-2 cells. The sigV 5′ end of the electropherogram sequence is represented in Fig. 1B and shows that the TSS corresponds to the G located 29 nt upstream of the SigV start codon. This TSS is not located downstream of any obvious canonical σ70, σ32, or σE promoters. However, an alignment of the DNA region located upstream of sigV with that of other ECF sigma factors revealed −10 and −35 boxes very conserved with respect to those of the B. subtilis PX and PW promoters (23). The −10 sequence (CGTC) is located 7 nt upstream of the TSS and separated by 17 nt from a −35 motif (TGAAAC). Indeed, the promoters most closely related to that of sigV are PW (TGAAAC N16 CGTA) and PX (TGTAAC N17 CGAC) (23). These observations are in agreement with data in the literature stipulating that the promoter consensus sequences recognized by the members of the ECF subfamily sigma factors are highly conserved mainly in the −35 motif and in the distance between the −35 and −10 boxes (32).
Transcriptional analysis of the sigV locus.
Northern blot analysis with total RNA extracted from exponentially growing cells incubated or not incubated for 3 h in seawater and from cells harvested 1 h after the onset of stationary phase was carried out. For each experimental condition, the hybridization result showed only one RNA band of 1.5 kb (Fig. 4A), in agreement with the combined sizes of sigV and rsiV. A similar transcript size has been obtained with an rsiV-specific probe (data not shown), confirming the expected bicistronic structural organization of the sigV locus. These results also showed that the mRNA transcript was overexpressed 1 h after the entry into stationary phase because of glucose starvation (Fig. 4A, lane 2) and after 3 h of incubation in seawater (complete starvation) (Fig. 4A, lane 3). Similar results were obtained by RNA dot blot hybridization after incubation of exponentially growing cells in tap water (Fig. 4B, spots 3 and 4).
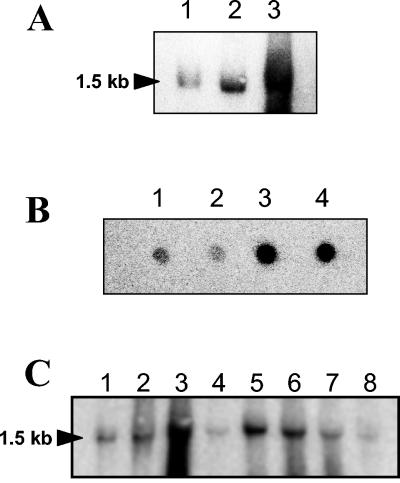
FIG. 4.
(A) Northern blot hybridization of E. faecalis JH2-2 RNA extracted from exponentially growing cells (lane 1) and from cells harvested 1 h after the onset of stationary phase (lane 2) and from exponentially growing cells incubated for 3 h in seawater (lane 3). (B) Dot blot hybridization of total RNA (1 μg) harvested from exponentially growing E. faecalis JH2-2 cells (spot 1) and from exponentially growing cells incubated for 10 min in 50 μM CdCl2 (spot 2) or incubated for 1 and 3 h in tap water (spots 3 and 4, respectively). (C) Northern blot hybridization of E. faecalis JH2-2 RNA extracted from exponentially growing cells (lane 1) and from exponentially growing cells incubated for 10 min in 0.08% bile salts (lane 2), 2 mM tBOOH (lane 3), 2.4 mM H2O2 (lane 4), 50°C (lane 5), 0.01% SDS (lane 6), pH 4.8 (lane 7), or 1 M NaCl (lane 8). Hybridizations were performed with a single-stranded DNA probe which corresponds to the SP1-S1 region. The size of the transcript determined with RNA molecular size markers (Amersham) is indicated on the left.
Northern blot analysis was also carried out with cultures exposed for 10 min to bile salts, tBOOH, H2O2, 50°C, SDS, pH 4.8, and NaCl stress. In comparison to that in unstressed cells harvested during the exponential growth phase, transcription of the sigV operon seems to be influenced by some environmental stresses. As did glucose and complete starvation, tBOOH, heat shock, and exposure to SDS significantly induced expression of the 1.5-kb mRNA transcript whereas exposure to acid pH, bile salts, H2O2, NaCl (Fig. 4C), or CdCl2 (Fig. 4B, spot 2) produced no obvious overexpression of the sigV operon. These combined results suggest the involvement of the sigV locus in the response of E. faecalis JH2-2 cells to environmental stress conditions.
rsiV negatively regulates expression of the sigV operon.
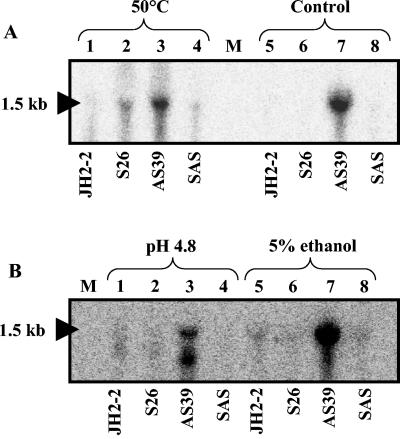
In order to determine whether sigV regulates its own expression, a transcriptional analysis was carried out. Total RNAs were extracted from cells of wild-type strain JH2-2 and its derivative mutants S26, AS39, and SAS growing exponentially at 37°C (standard conditions) and after 10 min of exposure to heat (50°C), acid pH (pH 4.8), or ethanol (5%). Under standard conditions (control), a strong mRNA transcript signal corresponding to the overall structure of the sigV-rsiV operon (1.5 kb) was detected only for mutant AS39, which is affected in the anti-sigma factor (rsiV) gene (Fig. 5A, lane 7). This result is in agreement with reports in the literature that many ECF sigma factors have been found to be cotranscribed with and negatively regulated by their cognate anti-sigma factors (24, 32, 58). Similar results, in which the 1.5-kb mRNA transcript was mainly detected, were obtained for the rsiV mutant (strain AS39) exposed to heat shock (Fig. 5A, lane 3), acid pH (Fig. 5B, lane 3), or ethanol (Fig. 5B, lanes 7). The 1-kb mRNA band detected within the rsiV mutant (Fig. 5B, lane 3) is unexplained and likely resulted from mRNA degradation. These results, showing that the up-regulation of the operon in the rsiV mutant is lost in the sigV-rsiV double mutant, suggest that this operon is autoregulated and that SigV is negatively controlled by RsiV, as recently shown for SigV of B. subtilis (58).
FIG. 5.
Northern blot analysis of E. faecalis strains JH2-2, S26, AS39, and SAS. Total RNAs were extracted from exponentially growing cells under standard conditions (control) (A, lanes 5 to 8) and exposed for 10 min to heat (50°C) (A, lanes 1 to 4), pH 4.8 (B, lanes 1 to 4), or 5% ethanol (B, lanes 5 to 8). Hybridizations were performed with a single-stranded DNA probe corresponding to the SP1-S1 DNA region. The size of the transcript determined with RNA molecular size markers (lane M; Amersham) is indicated on the left.
sigV mutants are more sensitive to environmental stresses.
Comparison of the growth patterns of the wild-type JH2-2 strain and its derivative mutants revealed no significant difference in GM17 or semisynthetic medium at 37°C, indicating that the sigV and rsiV genes are dispensable under normal growth conditions (data not shown). To determine a potential role for these genes in the stress response of E. faecalis, the three mutant strains (S26, AS39, and SAS) and wild-type strain JH2-2 were assessed for survival under several distinct stress conditions.
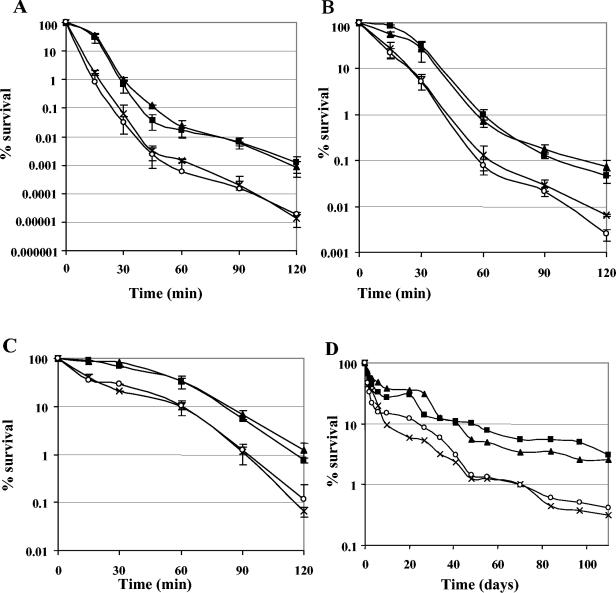
The four strains used were severely impaired by exposure to a temperature of 62°C, and their viability dropped dramatically within 45 min (Fig. 6A). However, striking differences in survival among the four strains were apparent. After the first 15 min of treatment, the sigV mutants (S26 and SAS) were 20- to 50-fold more sensitive than the wild-type strain, respectively (Fig. 6A). Interestingly, the rsiV mutant (AS39) showed survival similar to that of the JH2-2 strain. The survival curves of the latter two strains were characterized by the presence of a shoulder, indicating second-order decay kinetics, whereas those of the sigV mutants showed first-order decay kinetics. At the end of the experiment (2 h), the survival factors differentiating the two groups of strains (JH2-2 and AS39 versus S26 and SAS) were 80- to 100-fold (Fig. 6A).
FIG. 6.
Survival of wild-type E. faecalis strain JH2-2 (squares) and the derivative mutants S26 (cross), AS39 (triangles), and SAS (circles) under different lethal stress conditions. Cells were grown in GM17 to an optical density at 600 nm of 0.5 and submitted to heat challenge at 62°C (A), ethanol stress (22% ethanol) (B), or acid treatment (pH 3.2) (C). The values shown are means ± the standard deviations of three independent experiments. Percent viability represents the ratio of the survival number after exposure to challenge conditions to the survival number prior to the challenge. Panel D shows one representative experiment of the long-term survival of the four strains in seawater (oligotrophic medium).
A comparable result regarding the sensitivity of the sigV mutants and their decay kinetics was also obtained with ethanol stress (Fig. 6B) and acid pH treatment (Fig. 6C), as it was reported that ethanol and acid pH challenges may induce cell damage similar to that caused by heat shock (35). Indeed, the sigV mutants showed an approximately 10- to 20-fold greater drop in cell survival in response to 22% ethanol treatment or exposure to GM17 broth adjusted to pH 3.2, in comparison with the rsiV mutant and wild-type JH2-2 strains. The difference in cell survival between the two groups of strains was less marked than during the heat shock stress but was also observed after 15 min of treatment (Fig. 6). Furthermore, the S26 and SAS strains were also shown to be more sensitive to long-term survival in rich medium (data not shown) or under oligotrophic conditions (Fig. 6D). In addition, no significant differences in survival were observed with the other stresses tested, including exposure to 20 mM tBOOH or 0.017% SDS, despite the fact that these treatments at moderate concentrations significantly induced expression of the mRNA transcript of the sigV operon (Fig. 4).
Complementation assays.
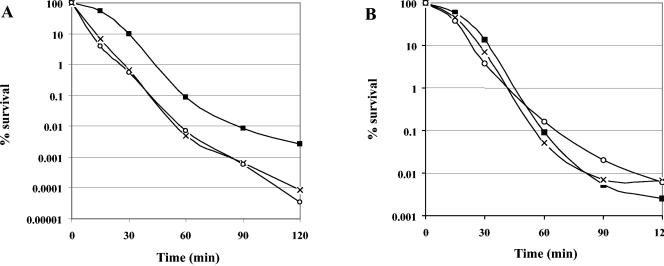
To determine whether the sensitivity to heat, ethanol, and acid pH is the result of disruption of sigV, a wild-type copy of the sigV gene cloned in the pMSP3535 vector under the control of the inducible PnisA promoter was used to transform the JH2-2, S26, and SAS strains. The transformed strains were subjected to previously described heat (62°C), ethanol (22%), and acid pH (pH 3.2) challenges and induced or not induced with nisin. In the absence of nisin, the sigV mutants harboring pMSP3535-sigV showed a marked drop in survival in comparison with their counterpart wild-type JH2-2(pMSP3535-sigV) strain during heat treatment (Fig. 7A). The wild-type JH2-2 strain transformed with the empty pMSP3535 vector showed behavior similar to that of JH2-2 harboring pMSP3535-sigV under the same conditions (data not shown). In addition, the difference in cell survival between the mutant and wild-type strains and the global levels of survival of the three strains are similar to those observed during the physiological assays described above (Fig. 7A versus 6A). In the presence of 300 ng of nisin ml−1, the S26 and SAS mutants harboring pMSP3535-sigV showed a gain in cell survival of approximately 50- to 100-fold, which conferred on them behavior similar to that of JH2-2(pMSP3535-sigV) cells during heat treatment (Fig. 7B). Ethanol and acid treatments carried out under the same conditions showed that the sigV mutants were substantially complemented by introduction of the wild-type sigV gene (data not shown). Note also that overexpression of SigV in a wild-type background did not modify sensitivity to these treatments. These results confirmed that the sigV gene effectively restores the wild phenotype.
FIG. 7.
Effect of SigV complementation on sigV mutant strains S26 and SAS. Survival of exponentially growing cells of wild-type E. faecalis strain JH2-2 (squares) and the derivative mutants S26 (cross) and SAS (circles) transformed with the pMSP3535-sigV recombinant vector and subjected to heat shock (62°C) in the absence of nisin (A) and in the presence of 300 ng of nisin per ml (B). Percent viability represents the ratio of the survival number after exposure to challenge conditions to the survival number prior to the challenge.
Identification of potential candidates of the sigV regulon.
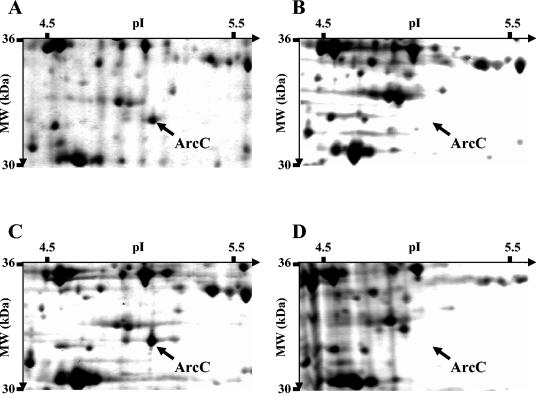
In order to determine whether sigV has a pleiotropic effect on gene expression, we analyzed the protein profile of wild-type strain JH2-2 and its derivative sigV (S26) and rsiV (AS39) mutant strains by 2D gel electrophoresis by extracting proteins under different physiological conditions. Among the slight differences observed in comparison with the proteome of JH2-2 or AS39, S26 cells showed one obvious spot with significantly decreased intensity, suggesting that this polypeptide is encoded by a sigV-regulated gene. This spot has been previously identified by N-terminal sequencing as ArcC, a carbamate kinase protein (13). In comparison with the proteome of the AS39 mutant, which overexpressed the sigV locus mRNA transcript, this polypeptide was undetectable within S26 cells after 3 h of incubation in seawater (Fig. 8B versus A) or 3 h after the onset of stationery phase in GM17 (Fig. 8D versus C). In comparison with the proteome of JH2-2, its intensity decreased progressively within S26 cells since its expression was drastically reduced 1 h after the onset of stationary phase in rich medium and undetectable 2 h later (data not shown).
FIG. 8.
Parts of silver-stained 2D gels of E. faecalis AS39 (A and C) and S26 (B and D) cells harvested after 3 h of incubation in seawater (A and B) and 3 h after the onset of the stationary phase in GM17 (C and D). MW, molecular mass.
In parallel with the proteomic analysis, we searched the E.faecalis V583 genome for sequences resembling the sigV autoregulatory promoter site as described for defining the sigX and sigW regulons of B. subtilis (5, 21, 23). For this purpose, we determined the ECF-type promoter element upstream of the sigV gene to be TGAAAC N17 CGTC and hypothesized that this sequence might be a sigV-dependent promoter, PsigV. Identification of sigV-dependent promoters by computer-aided genome analysis revealed 15 perfect matches with the 10-bp sequence TGAAAC N16-17 CGTC. Of the 15 matches, 5 (including PsigV) are positioned such that transcription would initiate between 36 and 226 bases upstream of the start codon of the corresponding gene (Table 3). The remaining 10 matches are not positioned upstream of genes and are thus unlikely to be active promoters. The EF0315 5′ untranslated region appeared abnormally long, but extensive analysis revealed the presence of a second sequence (TGAAAC N15 CGTC) located 69 bases upstream the start codon (Table 3), suggesting that this gene may be under the conditional control of one of the two promoters.
TABLE 3.
Alignment of sigV-like consensus elements
| Name (gene) | Candidate promoter region | 5′ UTRa |
|---|---|---|
| −35−10+1 | ||
| EF3180 (sigV) | AAAGTTGAAACTTTTTATGCTTGTGATTCGTCTATATAAGTGTAC | 36 |
| EF1843 (pgdA) | AAAATTGAAACTTTTTGGTCCTCAAATTCGTCTATAACTAATAGG | 81 |
| EF0159 | CTCTTTGAAACAGTTGATGATACAGAAGCGTCCGTTATGGAAAAC | 40 |
| EF1934b | GAAGATGAAACCAATCAATTGAATGA-ACGTCTAGAGGAAATGAA | 89 |
| EF0315 | AAGATTGAAACGTTAAGAGATGCAG-TACGTCACGAAAGTGCAAA | 226 |
| EF0315 | TGTATTGAAACAAAAAGAGC--CTACCTCGTCCGCCAACGAAGTA | 59 |
| Consensus | -----TGAAAC-----------------CGTC---------- |
The length of the 5′ untranslated region (UTR) is measured from the end of the −10 motif to the start codon of the indicated gene. The determined TSSs (+1) are represented by underlined bold letters.
See Fig. 9B for a characterization of the TSS of EF1934.
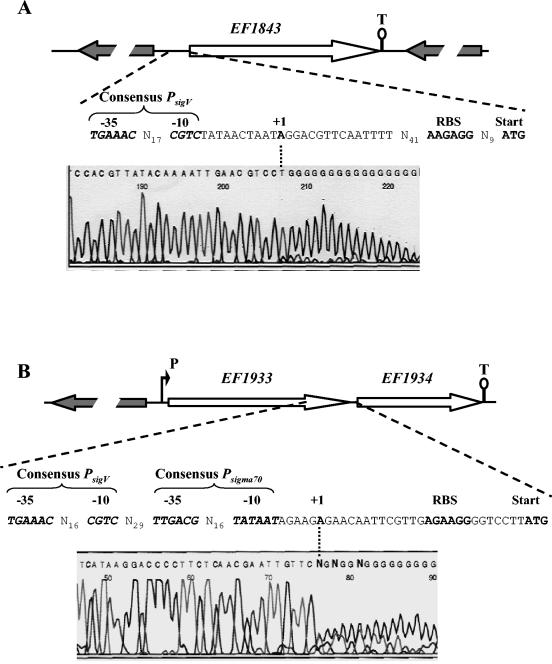
Of the four ORFs, only EF1843 and EF1934 are present within the genome of strain JH2-2. On the basis of the strain V583 genomic sequence, EF1843 appears as a monocistronic gene while EF1934 could be expressed as a monocistronic or bicistronic messenger (Fig. 9). EF1934 codes for a putative hypothetical conserved protein of 53 aa and EF1843 codes for a protein of 301 aa residues showing 44% identity with a putative deacetylase of Streptococcus mutans, 35% identity with the peptidoglycan N-acetylglucosamine deacetylase (PgdA) of Streptococcus pneumoniae, and 34% identity with an endo-1,4-β-xylanase of Clostridium perfringens. Determination of the TSSs of these two genes by the 5′ RACE PCR method was carried out under conditions in which the mRNA sigV locus transcript is overexpressed (from total RNA of JH2-2 and AS39 cells harvested 3 h after the onset of stationary phase) or not overexpressed (from total RNA of exponentially growing cells of JH2-2). EF1843, which exhibited the closest PsigV promoter sequence (66.7% identity), and EF1934 revealed only one and the same TSS under the different conditions tested, corresponding to the A located 71 and 27 nt upstream of the start codon of EF1843 and EF1934, respectively (Fig. 9A and B).
FIG. 9.
Schematic representation of the genetic organization of the EF1843 (A) and EF1934 (B) chromosomal regions of E. faecalis JH2-2 and respective electropherograms obtained from 5′ RACE PCR experiments. Large arrows represent the ORFs, and their orientation shows the transcriptional direction. The putative promoter (P) and Rho-independent terminators (T) are shown. Sequences in the electropherograms were obtained with specific primer SP3 (Table 2) and dG-tailed cDNA. The last base (A) upstream of the dG tail corresponds to the first nucleotide transcribed (TSS), which is in boldface and designated +1. The −10 and −35 motifs of the promoter sequences, the ribosome binding site (RBS), and the start codons are shown.
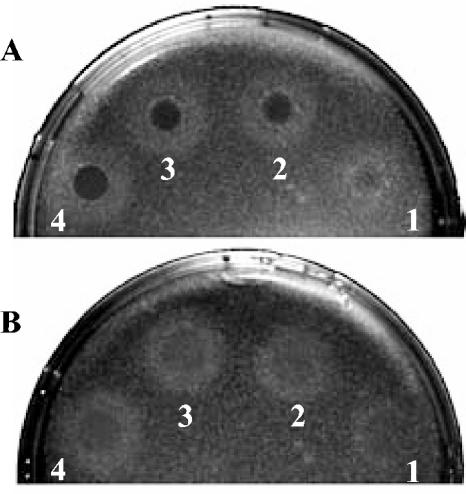
The TSS of EF1843 was ideally positioned relative to the PsigV consensus promoter sequence (Fig. 9A; Table 3), indicating that this gene may be effectively sigV regulated. In the hypothesis that EF1843 really corresponds to a pgdA homologue and is mainly sigV regulated, one would expect that the sigV mutant should be more sensitive to lysozyme treatment than the wild-tye JH2-2 strain, as shown for the pgdA mutant of S.pneumoniae (53, 54). Indeed, the sigV mutant S26 strain is much more sensitive to lysozyme treatment than are the wild-type JH2-2 strain (Fig. 10) and the rsiV mutant AS39 strain (data not shown). This result strongly suggests that SigV effectively regulates the expression of the EF1843 gene.
FIG. 10.
Lysozyme sensitivity of sigV mutant strain S26 (A) compared to that of the wild-type JH2-2 strain (B). A dilution of 106 cells of each strain ml−1 taken at 1 h after the onset of stationary phase was plated on LB agar to give confluent colonies. Immediately after plating, 10 μl of egg white lysozyme at 40 (spot 1), 60 (spot 2), 80 (spot 3), or 100 (spot 4) mg ml−1 was spotted onto the plates. Zones of clearing (dark circles) were photographed after 24 h of incubation at 37°C.
TSS identification showed that EF1934 was rather expressed by a housekeeping Psigma70 promoter (Fig. 9B), but this does not exclude the possibility that the PsigV promoter is active under hitherto unidentified conditions. Following the reverse transcription-PCR assay carried out during TSS identification, we observed the presence of a 600-bp DNA product whose sequence corresponds to the overall structure of EF1933 and EF1934 (data not shown). This result confirmed that the latter gene could effectively be expressed under a monocistronic or bicistronic structure, as suggested by the genetic organization of the EF1934 locus (Fig. 9B). Furthermore, no sequence even distantly related to the PsigV promoter was detected upstream of EF1933. In addition, no evidence for the presence of any TSS immediately upstream of arcC1 (EF0106) was found under the conditions tested (data not shown).
EF0159 and EF0315 (coding for a putative membrane protein of 654 aa and a putative hypothetical conserved protein of 229 aa, respectively), assessed by PCR amplification with genomic DNA of JH2-2 and V583, were found to be present only within the genome of strain V583. This was confirmed by Southern blot hybridization with specific probes (data not shown). Although these two genes are not involved in the stress response of E. faecalis strain JH2-2, we determined their TSSs from total RNA of exponentially growing V583 cells. We detected only one TSS of EF0315, corresponding to the G located 59 nt upstream of the start codon and found no evidence for the presence of any TSS immediately upstream of EF0159 (data not shown). As for EF1843, the TSS of EF0315 was ideally positioned relative to the PsigV consensus promoter sequence (Table 3), strongly suggesting that this gene is effectively sigV regulated.
DISCUSSION
The ECF family of sigma factors constitutes a diverse but distinct subfamily of the σ70 type of sigma factors found in both gram-negative and gram-positive bacteria (30). Features common to members of this subfamily include regulation of ECFs and function as effector molecules responding to extracytoplasmic stimuli (32). Bacterial genome sequencing revealed an exponential increase in the number of new members of this class of σ factors (33). Determining the roles of these numerous regulatory proteins is a formidable challenge. In this context, we identified and characterized the first ECF sigma and anti-sigma factors of lactic acid bacteria, namely, the sigV and rsiV genes of E. faecalis.
Analysis of the nucleotide sequence of the sigV locus of E. faecalis revealed the presence of two ORFs flanked by two strong putative rho-independent terminators (Fig. 1A). The sigV gene in E. faecalis encodes a protein with a calculated molecular mass of 19.7 kDa belonging to the ECF sigma factor family as it has in common with the members of this family conserved region 2, containing both the −10 promoter recognition helix and the primary core RNA polymerase binding determinant, and conserved region 4, which is involved in binding to the −35 promoter element. The conservation of these regions possibly indicated their essential functional roles and also showed that they form a distinct group in comparison with the sequences of other housekeeping sigma factors (30, 32).
Immediately downstream of sigV is a second ORF named rsiV that encodes a protein with a calculated molecular mass of 33.8 kDa. Database searches with the predicted amino acid sequence of RsiV revealed only one hit, the anti-sigma factor corresponding to the yrhM product located immediately downstream of sigV within the B. subtilis genome (47). A property common to the ECF anti-sigma factors characterized so far is that they are inner membrane proteins with at least one transmembrane domain (24). Indeed, E. faecalis RsiV exhibited a putative transmembrane domain within its N-terminal part, as was the case of YrhM, which was recently shown to interact through this N-terminal domain with its cognate sigma factor (SigV) within B. subtilis (58). The anti-sigma function of RsiV was further supported by the cotranscription of sigV and rsiV revealed by Northern blot hybridization experiments. This global organization of the sigV locus, where the ECF sigma factor is immediately followed by its related anti-sigma factor, has also been described for some other ECF sigma factors such as sigR and rsrA (26) and sigU and rsuA (12) in S. coelicolor; sigH and orf2 in Mycobacterium smegmatis (9); sigV and yrhM (47), sigX and ypuN (20), and sigW and ybbM (22) in B. subtilis; and rpoE and orf2 in Vibrio angustum (18). On the basis of these results, we suggest that sigV and rsiV are members of an operon, functioning as sigma and anti-sigma factors, respectively. In comparison with the well-studied promoter sequences of the autoregulatory ECF sigma factors SigX and SigW of B. subtilis (21-23, 39), we have determined the PsigV promoter sequence of the E. faecalis sigV locus to be TGAAAC N17 CGTC. These findings further support the hypothesis that SigV may act as an autoregulatory ECF sigma factor in E. faecalis.
Transcriptional analysis of the sigV locus of E. faecalis strain JH2-2 revealed that the sigV-rsiV operon mRNA is differentially expressed following exposure to various stress conditions. It was highly overexpressed under glucose and complete starvation and following exposure to tBOOH. To a lesser extent, this overexpression was also observed under heat shock and SDS treatment (Fig. 4). These results suggest the involvement of the sigV-rsiV operon in the response of E. faecalis JH2-2 cells to environmental stress conditions. This is in agreement with data in the literature reporting that the ECF subfamily of RNA polymerase sigma factors is involved in bacterial stress responses such as survival at high temperature (20, 42) and survival following heat shock and oxidative stress (9, 18, 41) or under conditions of acidic pH and exposure to detergent (56).
To further characterize the role of the sigV and rsiV genes in the regulation of the stress response, mutants affected in each gene individually and in both genes were constructed by DCO events. The ability to generate a gene disruption in E. faecalis indicates that these genes are not essential for viability. However, the sigV gene appears to play a role in stressed cells of E. faecalis, as suggested by the increased sensitivity of sigV mutants (S26 and SAS strains) in comparison to the wild-type strain when exposed to severe heat shock, ethanol, and acid pH treatments (Fig. 6). These results are consistent with reports in the literature that mutants affected in alternative sigma factor σB of B. subtilis (52) or in ECF sigma factors such as SigE of M. smegmatis (56) and SigM of B. subtilis (50) are more sensitive to heat, ethanol, and acid stresses than are their counterpart wild-type strains. Long-term survival experiments carried out with rich medium and under oligotrophic conditions showed that the sigV mutants displayed decreased survival compared with wild-type strain JH2-2 and the rsiV mutant strain. Thus, SigV may contribute to the starvation survival potential of E. faecalis whereas SigB of B. subtilis was shown to have no effect on starvation survival under glucose or phosphate limitation in minimal medium (52). However, it was demonstrated that ECF SigE of V. angustum is involved in carbon starvation-induced cross-protection against oxidative stress (18) and SigB and SigH of B. subtilis reduced stationary-phase viability in either alkaline or acidic media and contributed to the ability to grow in medium containing high ethanol concentrations (11). In addition, complementation assays showed that the presence of a functional copy of SigV is required in E. faecalis for its response to heat shock and exposure to ethanol and acid pH. Together, these results confirmed that the sigV gene is effectively involved in the mediation of the response of E. faecalis to environmental stresses.
Most of the ECF sigma factors examined to date are in operons that also encode negative regulators (3, 12, 20, 26, 36, 37). By using a yeast two-hybrid system, it was recently shown that this is also the case for the bicistronic sigV-yrhM operon of B. subtilis (58), whose gene products exhibited the best identities with SigV and RsiV of E. faecalis. In addition, the fact that the deletion in the rsiV gene led to drastic overexpression of the sigV-rsiV operon mRNA transcript (Fig. 5) suggested that RsiV may directly affect SigV activity as an anti-sigma factor. Thus, these two proteins may function in accordance with the hypothesis stipulating that a rapid increase in the activity of the sigma factor by either inactivation or removal of its cognate anti-sigma factor is a common regulatory strategy (24, 42).
One possible way to identify potential members of the sigV regulon of E. faecalis is direct comparison of the proteomes of wild-type and sigV and rsiV mutant strains. The expected changes in the protein profiles as a result of the loss of SigV function mainly revealed one obvious spot that was shown to be undetectable within S26 cells under glucose or complete starvation (Fig. 8). This spot has been previously identified as the carbamate kinase ArcC1, which was highly induced in glucose-starved cells of E. faecalis (13). This enzyme catalyzes the last reaction of the degradation pathway of arginine. One ATP molecule per carbamoyl phosphate molecule is generated during this reaction, allowing glucose-starved cells to produce energy. It is noteworthy that four paralogues of this gene (arcC1 to arcC4) are present in operon structures within the genome of the V583 strain. A computer search revealed that no putative sigV-dependent promoter was identified upstream of the genes constituting these operon structures. This probably indicates that none of the arcC genes is under direct control of SigV.
Furthermore, as described for the sigX and sigW regulons (5, 21, 23), comparisons of whose promoter sequences have proven to be a valuable approach to defining sigma factor regulons (39), the E. faecalis V583 genome search allowed us to identify, in addition to sigV, four candidates for PsigV-regulated ORFs. Of these, only EF1843 and EF1934 are present within the genome of strain JH2-2. Under conditions in which the sigV locus mRNA is overexpressed or not, EF1934 seems likely to be Psigma70 dependent while EF1843 exhibits only one and the same TSS ideally positioned relative to PsigV, suggesting that it is an effectively PsigV-regulated gene. In addition, this gene, present in a monocistronic organization within the E. faecalis V583 genome, exhibited a gene product 35% identical to that which encodes PgdA of S. pneumoniae, which may contribute to pneumococcal virulence by providing protection against host lysozyme (53, 54). On the other hand, of the two ORFs only present within the genome of the V583 strain, only EF0315, which codes for a putative hypothetical protein, seems to be a PsigV-regulated gene.
In conclusion, our results correspond, to our knowledge, to the first characterization of ECF sigma factors within the important group of lactic acid bacteria. The involvement of SigV in the response to heat shock, ethanol, and acid pH treatments and during long-term survival of E. faecalis strain JH2-2 is clearly demonstrated. The negative control of SigV by the related anti-sigma factor RsiV suggests a mode of action in which SigV may be part of a positive regulatory loop in which increased amounts of SigV will lead to increasing sigV transcription and the negative regulatory element, RsiV, breaks this loop and allows the cell to maintain a steady state of sigV, as described for other members of ECF subfamily of sigma factors (24, 42).
Three additional putative ECF sigma factors were detected within the genome of E. faecalis after the release of the complete annotated sequence of the strain V583 genome. The multiplicity of these E. faecalis ECF sigma factors is in agreement with the hypothesis that it is essential for the bacterial cell to adjust its metabolism to changing environmental conditions. Thus, it seems reasonable to speculate that a species adapted to a constant environment has a smaller repertoire of regulatory mechanisms than a species that is forced to cope with many changing environmental conditions (33). In this way, it will be of great interest to see whether these alternative sigma factors could also be involved in the stress response and/or virulence of E. faecalis.
Acknowledgments
We are grateful to C. J. Leenhouts (Department of Genetics, University of Groningen, Groningen, The Netherlands), E. Maguin (INRA, Jouy-en-Josas, France), and M. S. Gilmore (University of Oklahoma) for providing plasmid pORI19.1 and E. coli strain EC101, the pG+host3 plasmid, and E. faecalis strain V583, respectively. We are also grateful to G. M. Dunny (Department of Microbiology, Medical School, University of Minnesota, Minneapolis) for providing plasmid pMSP3535, which was constructed by a system originally developed by researchers at The Netherlands Dairy Research Institute (NIZO). The expert technical assistance of A. Blandin and B. Gillot is greatly appreciated.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Batish, V. K., and B. Ranganathan. 1984. Occurrence of enterococci in milk and milk products. J. Dairy Sci. Technol. 19:189-195. [Google Scholar]
- 3.Brutsche, S., and V. Braun. 1997. SigX of Bacillus subtilis replaces the ECF sigma factor FecI of Escherichia coli and is inhibited by RsiX. Mol. Gen. Genet. 256:416-425. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 5.Cao, M., P. A. Kobel, M. M. Morshedi, M. F. W. Wu, C. Paddon, and J. D. Helmann. 2002. Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, runoff transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316:443-457. [DOI] [PubMed] [Google Scholar]
- 6.Devriese, L. A., L. Laurier, P. De Herdt, and F. Haesebrouck. 1992. Enterococcal and streptococcal species isolated from faeces of calves, young cattle and dairy cows. J. Appl. Bacteriol. 72:29-31. [DOI] [PubMed] [Google Scholar]
- 7.Dower, W. J., J. F. Miller, and C. W. Ragsdal. 1988. High efficiency transformation of Escherichia coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman, D. M., T. A. Steitz, and A. Goldman. 1986. Identifying nonpolar transmembrane helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Chem. 15:321-353. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes, N. D., Q. L. Wu, D. Kong, X. Puyang, S. Garg, and R. N. Husson. 1999. A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181:4266-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flahaut, S., A. Hartke, J. C. Giard, A. Benachour, P. Boutibonnes, and Y. Auffray. 1996. Relationship between stress response towards bile salts, acid and heat treatment in Enterococcus faecalis. FEMS Microbiol. Lett. 138:49-54. [DOI] [PubMed] [Google Scholar]
- 11.Gaidenko, T. A., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gehring, A. M., N. J. Yoo, and R. Losick. 2001. RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor. J. Bacteriol. 183:5991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giard, J. C., A. Hartke, S. Flahaut, P. Boutibonnes, and Y. Auffray. 1997. Glucose starvation response in Enterococcus faecalis JH2-2: survival and protein analysis. Res. Microbiol. 148:27-35. [DOI] [PubMed] [Google Scholar]
- 14.Hartke, A., J. C. Giard, J. M. Laplace, and Y. Auffray. 1998. Survival of Enterococcus faecalis in an oligotrophic microcosm: changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl. Environ. Microbiol. 64:4238-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecker, M., and U. Völker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 16.Hecker, M., and U. Völker. 2001. The general stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 17.Hengge-Aronis, R. 2000. The general stress response in Escherichia coli, p. 101-178. In G. Stortz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 18.Hild, E., K. Takayama, R.-M. Olsson, and S. Kjelleberg. 2000. Evidence for a role of rpoE in stressed and unstressed cells of marine Vibrio angustum strain S14. J. Bacteriol. 182:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, X., A. Decatur, A. Sorokin, and J. D. Helmann. 1997. The Bacillus subtilis σX protein is an extracytoplasmic function σ factor contributing to survival at high temperature. J. Bacteriol. 179:2915-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, X., and J. D. Helmann. 1998. Identification of target promoters for the Bacillus subtilis σX factor using a consensus-directed search. J. Mol. Biol. 279:165-173. [DOI] [PubMed] [Google Scholar]
- 22.Huang, X., K. L. Fredrick, and J. D. Helmann. 1998. Promoter recognition by Bacillus subtilis σW: autoregulation and partial overlap with the σX regulon. J. Bacteriol. 180:3765-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, X., A. Gaballa, M. Cao, and J. D. Helmann. 1999. Identification of target promoters for the Bacillus subtilis extracytoplasmic function σ factor, σW. Mol. Microbiol. 31:361-371. [DOI] [PubMed] [Google Scholar]
- 24.Hughes, K. T., and K. Mathee. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52:231-286. [DOI] [PubMed] [Google Scholar]
- 25.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, J.-G., M. S. B. Paget, Y.-J. Seok, M.-Y. Hahn, J.-B. Bae, J.-S. Hahn, C. Kleanthous, M. J. Buttner, and J.-H. Roe. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 18:4292-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laplace, J. M., A. Hartke, J. C. Giard, and Y. Auffray. 2000. Cloning, characterization and expression of an Enterococcus faecalis gene responsive to heavy metals. Appl. Microbiol. Biotechnol. 53:685-689. [DOI] [PubMed] [Google Scholar]
- 28.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis allows fast analysis of targeted genes. J. Bacteriol. 177:7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Breton, Y., G. Boël, A. Benachour, H. Prévost, Y. Auffray, and A. Rincé. 2003. Molecular characterization of Enterococcus faecalis two-component signal transduction pathways related to environmental stresses. Environ. Microbiol. 5:329-337. [DOI] [PubMed] [Google Scholar]
- 30.Lonetto, M. A., K. L. Brown, K. E. Rudd, and M. J. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase σ factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maguin, E., P. Duwat, T. Hege, and D. Ehrlich. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Missiakas, D., and S. Raina. 1998. The extracytoplasmic function sigma factors: role and regulation. Mol. Microbiol. 28:1059-1066. [DOI] [PubMed] [Google Scholar]
- 33.Mittenhuber, G. 2002. An inventory of genes encoding RNA polymerase sigma factors in 31 completely sequenced eubacterial genomes. J. Mol. Microbiol. Biotechnol. 4:77-91. [PubMed] [Google Scholar]
- 34.Moellering, R. C. 1992. Emergence of Enterococcus as a significant nosocomial pathogen. Clin. Infect. Dis. 14:1173-1178. [DOI] [PubMed] [Google Scholar]
- 35.Mogk, A., A. Völker, S. Engelmann, M. Hecker, W. Schumann, and U. Völker. 1998. Nonnative proteins signal induction of the Bacillus subtilis CIRCE regulon. J. Bacteriol. 180:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman, J. D., J. R. Anthony, and T. J. Donohue. 2001. The importance of zinc-binding to the function of Rhodobacter sphaeroides ChrR as an anti-sigma factor. J. Mol. Biol. 313:485-499. [DOI] [PubMed] [Google Scholar]
- 37.Paget, M. S. B., J.-B. Bae, M.-Y. Hahn, W. Li, C. Kleanthous, J.-H. Roe, and M. J. Buttner. 2001. Mutational analysis of RsrA, a zinc-binding anti-sigma factor with a thiol-disulfide redox switch. Mol. Microbiol. 39:1036-1047. [DOI] [PubMed] [Google Scholar]
- 38.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 39.Qiu, J., and J. D. Helmann. 2001. The −10 region is a key promoter specificity determinant for the Bacillus subtilis extracytoplasmic function σX and σW. J. Bacteriol. 183:1921-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raina, S., D. Missiakas, and C. Georgopoulos. 1995. The rpoE gene encoding the σE (σ24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raman, S., T. Song, X. Puyang, S. Bardarov, W. R. Jacobs, Jr., and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rouvière, P. E., A. De Las Penas, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat shock sigma factor, σE, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaberg, D. R., D. H. Culver, and R. P. Gaynes. 1991. Major trends in microbial etiology of nosocomial infection. Am. J. Med. Suppl. 3B:725-758. [DOI] [PubMed] [Google Scholar]
- 47.Sorokin, A., A. Bolotin, B. Purnelle, H. Hilbert, J. Lauber, A. Düsterhöft, and S. D. Ehrlich. 1997. Sequence of the Bacillus subtilis genome region in the vicinity of the lev operon reveals two new extracytoplasmic function RNA polymerase factors, SigV and SigZ. Microbiology 143:2939-2943. [DOI] [PubMed] [Google Scholar]
- 48.Teng, F., L. Wang, K. V. Singh, B. E. Murray, and G. M. Weinstock. 2002. Involvement of PhoP-PhoS homologs in Enterococcus faecalis virulence. Infect. Immun. 70:1991-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Terzaghi, B. E., and W. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thackray, P. D., and A. Moir. 2003. SigM, an extracytoplasmic function sigma factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verneuil, N., M. Sanguinetti, Y. Le Breton, B. Posteraro, G. Fadda, Y. Auffray, A. Hartke, and J.-C. Giard. 2004. Effects of Enterococcus faecalis hypR gene encoding a new transcriptional regulator on oxidative stress response and intracellular survival within macrophages. Infect. Immun. 72:4424-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vollmer, W., and A. Thomasz. 2000. The pgdA gene encodes for a peptidoglycan N-acetylglucosamine deacetylase in Streptococcus pneumoniae. J. Biol. Chem. 275:20496-20501. [DOI] [PubMed] [Google Scholar]
- 54.Vollmer, W., and A. Thomasz. 2002. Peptidoglycan N-acetylglucosamine deacetylase, a putative virulence factor in Streptococcus pneumoniae. Infect. Immun. 70:7176-7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiegert, T., G. Homuth, S. Versteeg, and W. Schumann. 2001. Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 41:59-71. [DOI] [PubMed] [Google Scholar]
- 56.Wu, Q. L., D. Kong, K. Lam, and R. N. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yagi, Y., and D. B. Clewell. 1980. Recombination-deficient mutant of Streptococcus faecalis. J. Bacteriol. 143:966-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yoshimura, M., K. Asai, Y. Sadaie, and H. Yoshikawa. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150:591-599. [DOI] [PubMed] [Google Scholar]