Abstract
As demonstrated with alr2835 (hepA) and alr2834 (hepC) mutants, heterocysts of Anabaena sp. strain PCC 7120, a filamentous cyanobacterium, must have an envelope polysaccharide layer (the Hep+ phenotype) to fix dinitrogen in an oxygen-containing milieu (the Fox+ phenotype). Transpositions presumptively responsible for a Fox− phenotype were localized in open reading frames (ORFs) near hepA and hepC. A mutation in each of nine of these ORFs was complemented by a clone bearing only that single, intact ORF. Heterocysts of the nine mutants were found to lack an envelope polysaccharide layer. Complementation of mutations in alr2832 and alr2840 may have resulted from recombination. However, alr2825, alr2827, alr2831, alr2833, alr2837, alr2839, and alr2841, like hepA and hepC, are required for a Hep+ Fox+ phenotype.
When Anabaena spp. and certain other filamentous cyanobacteria are deprived of combined nitrogen in the presence of O2, 5 to 10% of their vegetative cells differentiate into cells called heterocysts. Differentiation takes place at semiregular intervals along the filaments, forming a spacing pattern (34, 38). The process of differentiation involves changes in cellular biochemistry that collectively produce a micro-oxic intracellular milieu in which O2-labile nitrogenase in heterocysts is protected from inactivation by O2 (15, 16; J. Elhai and C. P. Wolk, Abstr. 7th Int. Symp. Photosynthetic Prokaryotes, abstr. 114B, 1991). Protection involves inactivation of the O2-producing photosystem II of vegetative cells (1), deposition of extracellular layers of polysaccharide and glycolipid that greatly decrease entry of O2 (25, 33), increased respiration by which O2 that does enter is reduced to H2O (25), and intercellular interactions that provide heterocysts with the requisite reductant (38).
We shall denote as Fox genes genes that are required specifically for nitrogen fixation in the presence of oxygen. Genes required for the formation of heterocyst envelope polysaccharide are Fox genes and include the following: devRA (alr0442) and hepK (all4496), whose products interact as parts of a two-component regulatory system (42); alr0117 and alr1086 (26; our unpublished results), whose products also resemble elements of such a system; hepB (alr3698) and hepC (alr2834), whose predicted products show greatest similarity to a glycosyl transferase and a UDP-galactose-lipid carrier transferase, respectively (23, 34, 35, 39, 44); and hepA (alr2835), which encodes a member of the family of ATP-binding proteins of ATP-binding cassette transporters and is activated in response to nitrogen deprivation (12, 18, 37). These genes are dispersed at Mb positions 0.12, 0.52, 1.27, 3.45 (hepC and hepA), 4.47, and 5.38 in the 6.41-Mb chromosome of Anabaena sp. strain PCC 7120. Heterocyst differentiation in Anabaena sp. strain PCC 7120 was analyzed with microarrays whose elements contained part or all of one to eight open reading frames (ORFs)(12). In addition to hepA, ORFs close to it in the Anabaena sp. genome (20), including (i) alr2823 and/or alr2824, (ii) alr2826, alr2828, alr2832, alr2833, alr2836, and alr2839, and (iii) one or more of alr2841 through all2843, were found to be up-regulated early in heterocyst differentiation (12, 18). Because up-regulation does not imply essentiality for differentiation or function, we asked whether ORFs other than hepA and hepC in this region are Fox genes, and we identified seven that are.
MATERIALS AND METHODS
Growth conditions for cyanobacteria.
Anabaena sp. strain PCC 7120 was maintained in AA/8 medium (19) at 30°C in the light (ca. 3,500 ergs cm−2 s−1) on a rotary shaker. Liquid cultures to be transposon mutagenized and liquid cultures of derivatives of Anabaena sp. (Table 1) were grown under the same conditions but in AA/8+N medium (19) in the presence of appropriate antibiotics, unless specified otherwise. Tests of complementation (see below) and some preparation for microscopy made use of medium AA or AA+N solidified with 1.2% home-purified (Difco) Bacto agar (19), plus antibiotics as appropriate.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Derivation and/or relevant characteristicsa |
|---|---|
| Strains of Anabaena sp. | |
| PCC 7120 | Wild type, from R. Haselkorn |
| DR1069 | Smr Spr; alr2835 (hepA) mutation resulting from double recombination with pRL1069 (5) |
| FQ57 | Bmr Nmr Smr; alr2827::Tn5-1063 |
| FQ344 | Bmr Nmr Smr; alr2841::Tn5-1063 |
| FQ428 | Bmr Nmr Smr; alr2837::Tn5-1063 |
| FQ470 | Bmr Nmr Smr; alr2833::Tn5-1063 |
| FQ610 | Bmr Nmr Smr; alr2838::Tn5-1063 |
| FQ773 | Bmr Nmr Smr; alr2832::Tn5-1063 |
| FQ794 | Bmr Nmr Smr; alr2839::Tn5-1063 |
| FQ885 | Bmr Nmr Smr; alr2825::Tn5-1063 |
| FQ1630 | Bmr Nmr Smr; alr2840::Tn5-1063 |
| FQ1631 | Bmr Nmr Smr; alr2831::Tn5-1063 |
| Strains of E. coli | |
| DH10B | 17 |
| DH5αMCR | Invitrogen Corp., received courtesy of J. C. Meeks |
| HB101 | 6 |
| Plasmids | |
| anc0293 | Cmr Emr; Anabaena sp. strain PCC 7120 chromosomal DNA from bp 3436903 to 3449816 in the BamHI site of pRL838 |
| anc0626 | Cmr Emr; Anabaena sp. strain PCC 7120 chromosomal DNA from bp 3451665 to 3468470 in the BamHI site of pRL838 |
| anc0901 | Cmr Emr; Anabaena sp. strain PCC 7120 chromosomal DNA from bp 3446424 to 3461510 in the BamHI site of pRL838 |
| anp02206 | Apr; Anabaena sp. strain PCC 7120 chromosomal DNA from bp 3440325 to 3447227 in the BamHI site of pUC18 |
| anp04627 | Apr; Anabaena sp. strain PCC 7120 chromosomal DNA from bp 3444424 to 3452207 in the BamHI site of pUC18 |
| anp08632 | Apr; Anabaena sp. strain PCC 7120 chromosomal DNA from bp 3457063 to 3463731 in the BamHI site of pUC18 |
| pGEM-T Easy | Apr; cloning vector (Promega Corp.) |
| pK18 | Kmr; cloning vector (28) |
| pRL443 | Apr Tcr; conjugative plasmid (13) |
| pRL838 | Cmr Emr; BAC vector (20) (GenBank accession no. AF403425) |
| pRL1124 | Kmr; methylase-encoding plasmid (13) |
| pRL1383a | Smr Spr; RSF1010-based cloning vector (GenBank accession no. AF403428) |
| pRL2665b | Apr Cmr Emr; source of C.CE3-oriT(RK2) cassette to make derivatives of pUC18 and pGEM-T Easy conjugable to, and selectable in, Anabaena sp. strain PCC 7120 (GenBank accession no. AY661563) |
| pRL2683a | Kmr Smr Spr; pRL1124 made mobilizable by the insertion, at its unique PstI site, of an oriT (RK2) fragment (32) in whose SalI site is inserted Smr Spr cassette C.S4 (3, 37) |
| pRL2686 | Kmr; Smr Spr marker removed from pRL2683a by SalI digestion and religation |
| pRL2792 | Apr; internal fragment of alr2833 amplified by PCR using genomic DNA from Anabaena sp. strain PCC 7120 as template and primers IDT145 and IDT146, cloned in pGEM-T Easy |
| pRL2815 | Apr Cmr Emr; PstI fragment containing C.CE3-oriT cassette from pRL2665b transferred to the PstI site of pRL2792 |
| pRL2831a | Smr Spr; glutamine synthetase promoter PglnA on a 0.7-kb EcoRI fragment from pRL559 (14) transferred to the EcoRI site of pRL1383a, oriented toward the BamHI site |
| pRL2831b | Smr Spr; same as pRL2831a, but PglnA oppositely oriented |
| pRL2862 | Smr Spr; alr2831-containing StuI-HindIII fragment of anc0293, subcloned between the Ecl136II and HindIII sites of pRL2831b |
| pRL2863 | Smr Spr; alr2832-containing EcoRV-XhoI fragment of anc0293 transferred between the StuI and XhoI sites of pRL2831a |
| pRL2864 | Smr Spr; alr2827-containing XmnI-XbaI fragment of anc0293, subcloned between the StuI and XbaI sites of pRL2831a |
| pRL2865a | Smr Spr; alr2833-containing HindIII fragment of anp04627 transferred to the HindIII site of pRL2831b |
| pRL2866 | Smr Spr; alr2841-containing HindIII fragment of anc0626, transferred to the HindIII site of pRL2831b |
| pRL2871 | Kmr; alr2825-containing KpnI-XmnI fragment of anp02206 transferred between the KpnI and HincII sites of pK18 |
| pRL2872 | Kmr; alr2839-containing EcoRV fragment of anp08632 inserted in the HincII site of pK18 |
| pRL2873 | Smr Spr; alr2840-containing HincII-XbaI fragment of anp08632, transferred between the StuI and XbaI sites of pRL2831a |
| pRL2875 | Smr Spr; alr2825-containing KpnI-SphI fragment of pRL2871, transferred between the same sites of pRL2831b |
| pRL2876 | Smr Spr; alr2839-containing KpnI-SphI fragment of pRL2872, transferred between the same sites of pRL2831b |
| pRL2877 | Smr Spr; alr2837-containing XbaI-BglII fragment of anp08632 transferred between the XbaI and BamHI sites of pRL2831a |
| pUC18 | Cloning vector (41) |
Sm, streptomycin; Bm, bleomycin; Cm, chloramphenicol; Ap, ampicillin; Tc, tetracycline; Km, kanamycin.
Transposon mutagenesis, identification of mutants, and localization of transposons.
Anabaena sp. cells were mutagenized by transposon Tn5-1063 (36), mutant colonies were selected by growth in the presence of 400 μg of neomycin sulfate (Nm) per ml, and those that were presumptively Fox− were identified by their persistent change of color from blue-green to yellow and lack of protracted growth in response to nitrogen deprivation. Such colonies were grown with nitrate supplemented with Nm for selection, and their DNA was extracted (7). DNA contiguous with the transposon was amplified by inverse PCR (27; Q. Fan et al., unpublished data), the PCR products were sequenced, and the sequences were localized within the genome of Anabaena sp. (20; http://www.kazusa.or.jp/cyano/Anabaena/).
Induction of heterocysts, staining with Alcian blue, and extraction of glycolipids.
To induce heterocyst formation, strains that had been grown in AA/8+N medium were washed three times with nitrogen-free AA/8 medium, and a 5-μl portion of suspension was spotted on agar-solidified AA medium. One week later, the agar bearing a spot was excised, stained with 2 μl of a 1% aqueous solution of Alcian blue for 40 s, washed with distilled water to remove nonadsorbed stain, and examined by microscopy. For extraction of glycolipids, the agar bearing a spot of cells was immersed for 5 min in a solution of chloroform and methanol (2:1, vol/vol) and the adhering organic solution was permitted to evaporate. Photographs were taken with a Nikon CoolPix 4300 digital camera mounted on a Wild microscope.
Construction of plasmids.
RSF1010-based vectors pRL2831a and pRL2831b (plasmids are described in Table 1) were constructed to express single ORFs from the glnA promoter. “a” and “b” refer to two orientations of that promoter, allowing cloning into one end or the other of an extensive polylinker. Plasmid RSF1010 was chosen because it can be mobilized to, and can replicate in, cells of Anabaena sp. (31), and a glnA promoter was chosen because it is expressed both in heterocysts and vegetative cells (14). To facilitate transfer of inserts, the parental plasmids bore the same polylinker as did BAC vector pRL838 (see the next paragraph).
Complementation experiments.
Complementation experiments were carried out initially with mapped, pRL838-based BAC clones from the Anabaena sp. strain PCC 7120 sequencing project (20) to determine whether the mutation that was responsible for the mutant phenotype was localized near the transposon insertion. Conjugative plasmid pRL443 and a mobilizable methylating plasmid, either pRL2683a or pRL2686, were introduced into Escherichia coli strain DH10B bearing a BAC. The resulting strain was mated diparentally with a corresponding mutant with selection for resistance to Nm, conferred by the transposon, and erythromycin (Em), conferred by the BAC. Recombination of the BAC with the genome, permitting replication of the selective marker, resulted routinely in the growth of hundreds of colonies. Three or four colonies from such a mating were grown individually in medium AA/8+N supplemented with 50 μg of Nm per ml and 2 μg of Em per ml and then spotted on petri dishes containing AA agar, AA+N agar, and AA+N agar plus 400 μg of Nm and 10 μg of Em per ml. Liquid cultures of wild-type Anabaena sp. strain PCC 7120 and of the original mutant were also spotted. The mutation was considered complemented (i) if spots from two or more of the cultures and the wild type greened and grew, while the mutant yellowed, remained yellow, and failed to grow, on AA agar; (ii) if all cultures grew on AA+N agar; and (iii) if the presumptive exconjugants grew and only the original mutant and the wild-type strain died on AA+N agar containing the two antibiotics.
In further complementation experiments, RSF1010-based plasmids in E. coli strain DH5αMCR were transferred by triparental matings with E. coli strain DH10B bearing pRL443 and pRL2686. Exconjugants were selected on agar-solidified AA+N medium with 200 μg of Nm per ml, to select for the presence of the transposon, plus 10 μg of spectinomycin dihydrochloride (Sp) per ml to select for a derivative of RSF1010. Subsequent testing of exconjugants was performed as described for BAC-bearing exconjugants, but with Sp replacing Em for selection.
Construction of an alr2833 mutant by insertional mutagenesis.
An internal fragment of alr2833 was generated by PCR using primers IDT145 and IDT146 (Integrated DNA Technologies, Coralville, Iowa) (Table 2) with DNA from wild-type Anabaena sp. as template. The PCR product was cloned into vector pGEM-T Easy (Promega). A cassette (from pRL2665b) permitting mobilization into, and selection in, Anabaena sp. was introduced into the PstI site of the resulting plasmid, yielding pRL2815 in E. coli HB101. Mating with E. coli DH10B containing pRL443 and pRL2686 resulted in coresidence of the three plasmids in E. coli HB101. pRL2815 was introduced into Anabaena sp. by a diparental mating, and the phenotypes of Em-resistant exconjugants were determined. The position of the insertion was verified by recovery (36) in E. coli DH5αMCR of pRL2815 and contiguous DNA, following digestion of genomic DNA with HindIII, and analysis of transformants by restriction, separately, with PstI and XmnI, and by sequencing through the unique HindIII site of the plasmid with primer IDT384.
TABLE 2.
Primers used
| Primer | Sequence | Use |
|---|---|---|
| IDT93 | 5′-TAA CCA GAA TCA TGA CCG TTT G-3′ | For PCR amplification of hepA (IDT93 and -94) |
| IDT94 | 5′-CAC CAC AAC CTT ATC TGC TTT G-3′ | |
| IDT145 | 5′-ATT ATA CTT CTG GCG ACC CTG A-3′ | To clone an internal fragment of alr2833 (IDT145 and -146) |
| IDT146 | 5′-ACT GTT TCA TCC GTG GAG AAC T-3′ | |
| IDT249 | 5′-AAT ATA CAG GTG ATT CGA CAA AGG-3′ | P3 for alr2825 |
| IDT250 | 5′-TAA ATC TCC GCC TTG TTG TTG AAT-3′ | P4 for alr2825 |
| IDT251 | 5′-TTG GAA ACT TGC AGG CAA ACA CGC-3′ | P3 for alr2827 |
| IDT252 | 5′-TAA CCA TCC AAT GGC TAG CAC CAA-3′ | P4 for alr2827 |
| IDT253 | 5′-GAA TTT TAT ACC CCA GGA TAT GAA-3′ | P3 for alr2831 |
| IDT254 | 5′-TCT TTG CGT TGG TTG TCA CTA ACA-3′ | P4 for alr2831 |
| IDT255 | 5′-AAA TAC TAT TAT TTA CTA GCG CGC-3′ | P3 for alr2832 |
| IDT256 | 5′-AAT AAA TGA ATG TTG AGT AAT CAG-3′ | P4 for alr2832 |
| IDT259 | 5′-ATA ACT TCG TAT AGC ATA CAT T-3′ | P1 for alr2827, alr2832, alr2837, and alr2840; P2 for alr2825, alr2833, alr2839, and alr2841 |
| IDT260 | 5′-GAT CTT ATT TCA TTA TGG TGA-3′ | P2 for alr2827, alr2832, alr2837, and alr2840; P1 for alr2825, alr2833, alr2839, and alr2841 |
| IDT271 | 5′-GGC GTT GGC GGT TGC AGA CC-3′ | For PCR amplification of rnpB (IDT271 and -272) |
| IDT272 | 5′-AGT TGG TGG TAA GCC GGG TTC-3′ | |
| IDT295 | 5′-GAA AGA GAA AGA TTA CAA GCC-3′ | P3 for alr2833 |
| IDT296 | 5′-TTG AGA ATT ATT TGC TGA ATA-3′ | P4 for alr2833 |
| IDT297 | 5′-ATC GAC GCA CAA AAT ATT ATC-3′ | P3 for alr2837 |
| IDT298 | 5′-AGC AAT TTT CCA AAT CCT CAG-3′ | P4 for alr2837 |
| IDT299 | 5′-ATG GTA AAA GTT CCT TGA TGG-3′ | P3 for alr2839 |
| IDT300 | 5′-TTT CAA TTT GCG CTA ACC AAT-3′ | P4 for alr2839 |
| IDT301 | 5′-TTA CTG CTA GTT CCA CAG GTG-3′ | P3 for alr2840 |
| IDT302 | 5′-AAA TAG ATA AAA AGC AAA GGT-3′ | P4 for alr2840 |
| IDT303 | 5′-GCA GCG ATT TGA TGA AGA CCA-3′ | P3 for alr2841 |
| IDT304 | 5′-TGA TTG GAC TCG GCT TGT TAG-3′ | P4 for alr2841 |
| IDT347 | 5′-GGC AAC CTC ATG TCC TCA TC-3′ | P1 for alr2831 |
| IDT348 | 5′-GCC GCA TAC GAT TTA GGT GA-3′ | P2 for alr2831 |
| IDT384 | 5′-CGC ACA CAT CTT TTT ATT CAG C-3′ | Sequencing primer to test plasmid recovered from Anabaena sp.::pRL2815 |
| IDT390 | 5′-AAT GAT TGG CGC TGG TTT TA-3′ | To amplify an internal fragment of alr2831 by PCR (IDT390 and -391) |
| IDT391 | 5′-TCA CGT TTG AGA CGA CAT CC-3′ | |
| IDT392 | 5′-GGC AGA ACG GGT ATT TGA A-3′ | To amplify an internal fragment of alr2832 by PCR (IDT392 and -393) |
| IDT393 | 5′-AAA GAA AAC TGG CTC AGA AAA A-3′ | |
| IDT394 | 5′-CCT GAT GAC TGC AAG AAC CA-3′ | To amplify an internal fragment of alr2833 by PCR (IDT394 and -395) |
| IDT395 | 5′-GGC TGT AGC TTG AGC GAT TT-3′ | |
| IDT396 | 5′-TGA CAA GCG TAA TAG TTC CAA A-3′ | To amplify an internal fragment of alr2834 by PCR (IDT396 and -397) |
| IDT397 | 5′-TTT TGT AGG CTT GCG TTC CT-3′ |
PCR analysis.
An ORF cloned in a replicating plasmid may complement either in trans or, if the clone recombined into the Anabaena sp. genome, in cis. PCR analysis was used to look for possible single or double recombination. Primers P1 and P2 were chosen that flanked the cloning region of the vector in question; for each gene to be tested, primers P3 and P4 were chosen that were specific to the genome outside of the region of the clone. PCR with Taq polymerase (Invitrogen) was performed by denaturation at 94°C for 3 min; 30 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 2 or 4 min; and then 72°C for 10 min. DNA templates were isolated (7) from the wild-type strain and the mutants and from complemented mutants that were subcultured to liquid AA/8 from spots growing on AA agar.
Reverse transcription-PCR analysis.
Cells of a 50-ml culture of wild-type Anabaena sp. strain PCC 7120 were sedimented after deprivation of fixed nitrogen for 8 h and resuspended in 400 μl of T0.1E buffer (10 mM Tris, 0.1 mM EDTA; pH 8.0), and RNA was extracted at 65°C in the same way as was DNA (6), except that acidic phenol (pH 4.8) was used. RNA was precipitated with a one-fourth volume of 8 M LiCl and 1 volume of isopropanol at −80°C for 15 min. The pellet was suspended in 100 μl of 0.1 M sodium acetate and 5 mM MgSO4 and treated twice at 37°C for 15 min with RNase-free DNase (Roche). RNA was then loaded on an RNeasy column (QIAGEN) and treated again twice with RNase-free DNase (QIAGEN) for 15 min each according to the manufacturer's instructions, and purified RNA was then eluted from the column and tested for the absence of contaminating DNA by PCR amplification with primers for hepA (IDT93 and IDT94) and rnpB (IDT271 and IDT272). cDNA was generated with random primers using 0.5 μg of RNA and Superscript II (Invitrogen) according to the manufacturer's instructions. PCR with Taq polymerase (Invitrogen) was performed by denaturation at 94°C for 3 min; 29 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 4 min; and then 72°C for 10 min.
RESULTS AND DISCUSSION
Localization of transposons within the genome of Anabaena sp.
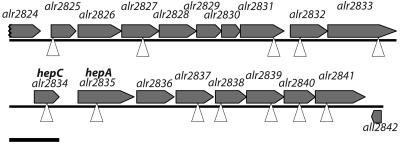
Anabaena sp. strain PCC 7120 was mutagenized with transposon Tn5-1063 and screened for Fox− mutants. The positions of representatives of 63 of the transposition loci thereupon identified and found to be clustered in its chromosome from alr2825 to alr2841 are shown in Fig. 1. The corresponding mutants listed in Tables 1 and 3 were selected for further study. As illustrated by the lack of an insertion in hepC, our transposon mutagenesis achieved only near-saturation (Fan et al., unpublished).
FIG. 1.
Map (20) of genes in an “expression island” (12) of the genome of Anabaena sp. strain PCC 7120. The positions of transposons in mutants that were successfully used for complementation are indicated by triangles. The sole insertion found in alr2834 was reported earlier (44). The marker extends 1.5 kb.
TABLE 3.
ORFs in the hepA region, annotated functions, and complementation
| ORF | No. of transposon mutants | Representative mutant | Complementing BAC | Complementing replicating plasmid | Annotated functiona |
|---|---|---|---|---|---|
| alr2822 | 0 | Similar to hypothetical protein YegL of E. coli | |||
| alr2823 | 0 | Similar to hypothetical protein YegK of E. coli | |||
| alr2824 | 0 | Similar to hypothetical protein YegI of E. coli | |||
| alr2825 | 6 | FQ885 | anc0293 | pRL2875 | Glucose-1-P cytidyltransferaseb |
| alr2826 | 0 | Dehydrogenase | |||
| alr2827 | 1 | FQ57 | anc0293 | pRL2864 | Epimerase/dehydrataseb |
| alr2828 | 0 | Glycosyltransferaseb | |||
| alr2829 | 0 | Hypothetical protein | |||
| alr2830 | 0 | dTDP-4-dehydrorhamnose 3,5-epimerase and related enzymesb | |||
| alr2831 | 12 | FQ1631 | anc0293 | pRL2862 | NAD(P)-dependent oxidoreductaseb |
| alr2832 | 3 | FQ773 | anc0293 | pRL2863 | Glycosyltransferase |
| alr2833 | 7 | FQ470 | anc0901 | pRL2865a | Hypothetical protein (see text)b |
| alr2834 | 0 | hepCb | |||
| alr2835 | 3 | hepAb | |||
| alr2836 | 0 | Glycosyltransferase | |||
| alr2837 | 8 | FQ428 | anc0901 | pRL2877 | Glucosyltransferase (see text) |
| alr2838 | 1 | FQ610c | anc0901 | —c | Glycosyltransferase |
| alr2839 | 6 | FQ794 | anc0901 | pRL2876 | Glycosyltransferase |
| alr2840 | 6 | FQ1630 | anc0626 | pRL2873 | Glycosyltransferase |
| alr2841 | 10 | FQ344 | anc0626 | pRL2866 | Unknownb |
The text discusses possible relationships of these genes to the biosynthesis of an LPS.
Analysis of FQ610, whose mutation was incompletely segregated and not complemented by a single-gene construct (data not shown), is continuing.
Phenotypes of presumptive Fox− mutants.
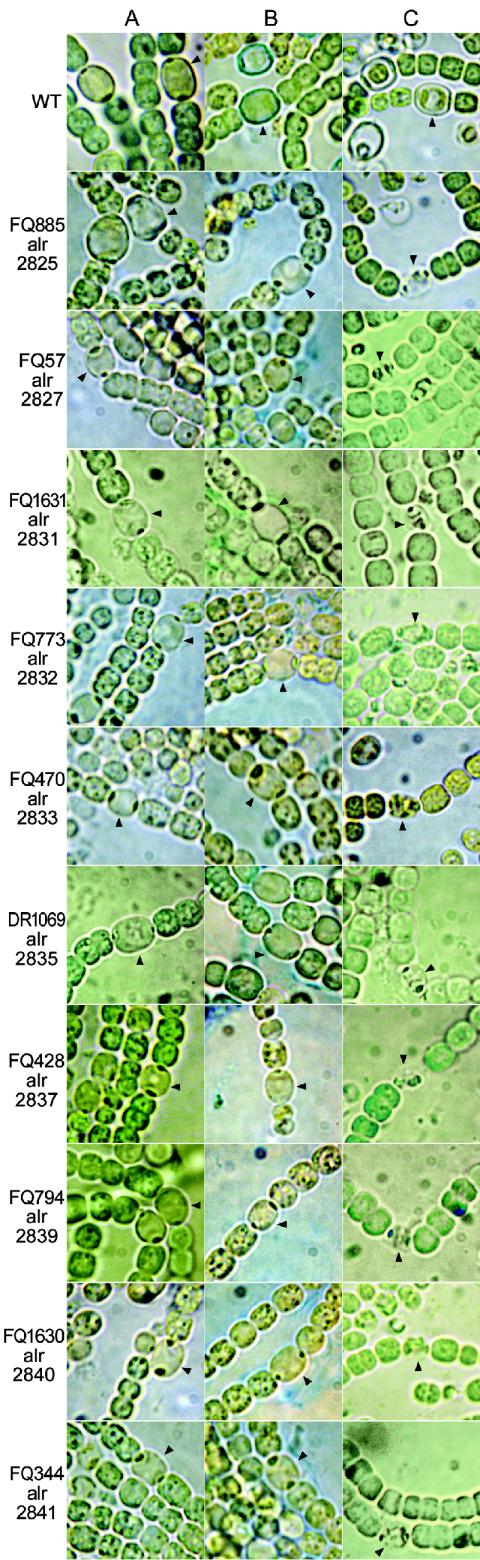
In response to nitrogen deprivation on an agar-solidified medium, each transposon mutant herein described formed heterocysts (Fig. 2A). Solid medium was used because the shear in shaken liquid cultures can disperse a heterocyst envelope glycolipid layer that lacks a protective envelope polysaccharide layer. To determine whether the mutants, like hepA and hepC mutants, lacked heterocyst envelope polysaccharide, we tried to stain them with Alcian blue. This stain colors the envelope of heterocysts of wild-type Anabaena sp. strain PCC 7120 blue (M. Gantar, J. Elhai, J. Jia, and M. Ow, Abstr. 5th Cyanobacterial Mol. Biol. Workshop, p. 25, 1995) (Fig. 2B) but did not stain the heterocyst envelope of hepA mutant DR1069, which lacks a polysaccharide layer (34) (Fig. 2B), or of any other transposon mutant shown in Fig. 2. In response to extraction with organic solvents, the protoplasts of heterocysts shrank. In the wild-type strain, a gap was seen within the residual polysaccharide layer, whereas in those mutants no thick envelope remained (Fig. 2C). The lack of staining with Alcian blue and loss of a pronounced envelope layer upon lipid extraction suggest that transposon insertion in any one of those mutated ORFs is sufficient to block deposition of heterocyst envelope polysaccharide. Alcian blue did stain heterocyst envelopes of incompletely segregated mutant FQ610 (Tables 1 and 3 and data not shown).
FIG. 2.
Wild-type and mutant strains of Anabaena sp. strain PCC 7120 unstained (A), stained with Alcian blue (B), and lipid extracted with chloroform and methanol (2:1, vol/vol) (C). Only the wild-typestrain retained heterocyst envelope polysaccharide, as assessed by staining with Alcian blue and retention of an envelope layer evident after lipid extraction. Aberrant overall coloration resulting from various intensities of illumination was partially corrected with Photoshop 7.0. Arrowheads point to heterocysts.
High-resolution electron microscopy had earlier shown that heterocysts of hepA (alr2835) and hepC (alr2834) mutants deposit only glycolipids (34, 44). A thicker glycolipid layer appears to be formed in Hep− mutants than in the wild-type strain, a finding reminiscent of the observation that increased pO2 increases formation of heterocyst envelope glycolipid (21); deposition of heterocyst envelope glycolipid may be adjusted in response to permeant O2 (38).
Complementation of presumptive Fox− mutants with BACs and with replicating plasmids.
The phenotype of a transposon-generated mutant may result from a second mutation elsewhere in the genome combined with antibiotic resistances conferred by the transposon. To provide an initial test of which developmental changes were due to insertions of the transposon, we attempted to complement the mutations by recombination with clones containing the wild-type ORFs that had been intercepted in the mutants. The BAC-based plasmids used as bridging clones in the Anabaena sp. sequencing project bore inserts that averaged ca. 18 kb in size. Because of the very low copy number of these F-based plasmids, they are believed to be less liable to deletions than are plasmids of higher copy number. Mutations in the newly mutated genes indicated in Fig. 1 were complemented by recombination with predicted BAC clones.
Although a mutation was complemented by a BAC, the Fox− mutant phenotype might have been due to a mutation in a position near the transposon-mutated ORF and, if the ORF were in an operon but not at the 3′ terminus of the operon, might have resulted from a polar effect of the transposon on the expression of a downstream gene or genes in that operon. Tests of complementation by a single gene in a replicating plasmid were predicated on the following idea. If a gene complemented in trans, the effect could be attributed to no other gene, because no other entire gene would have been present in the complementing DNA. In particular, the mutant phenotype could not be attributed to a polar effect of the mutation on the transcription of a downstream gene because transcription of that gene would not have been affected in trans.
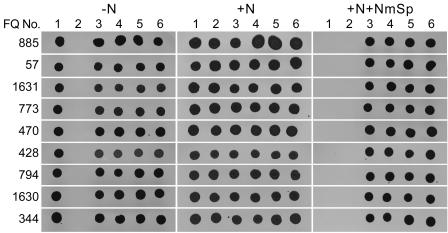
Complementation by a single gene was tested by subcloning the entirety of that gene together with at most a fragment of its neighboring genes into vectors pRL2831a or pRL2831b. Mutations in nine of the BAC-complemented, presumptive, newly identified Fox genes (Fig. 1) were complemented by clones bearing a corresponding wild-type gene (Table 3; Fig. 3). These results were consistent with the interpretation that each mutant phenotype resulted from insertion of the transposon in the corresponding gene. However, complementation might have resulted from recombination between the complementing plasmid and the mutated chromosome. A presumptive recA gene (all3272) has been identified in Anabaena sp. strain PCC 7120 (4; http://www.kazusa.or.jp/cyano/Anabaena/), but no cyanobacterial recA mutant has been isolated despite extensive attempts (24). Therefore, our work was carried out with a recombination-proficient strain. alr2840 overlaps alr2839 and is separated from alr2841 by only 24 bp, suggesting that those genes may be cotranscribed. In turn, complementation of mutations in alr2839 and alr2840 by their respective genes suggested that recombination might have occurred.
FIG. 3.
Tests of complementation by clones bearing, intact, only the gene mutated in the corresponding mutant: FQ885 (alr2825) plus pRL2875, FQ57 (alr2827) plus pRL2864, FQ1631 (alr2831) plus pRL2862, FQ773 (alr2832) plus pRL2863, FQ470 (alr2833) plus pRL2865a, FQ428 (alr2837) plus pRL2877, FQ794 (alr2839) plus pRL2876, FQ1630 (alr2840) plus pRL2873, and FQ344 (alr2841) plus pRL2866. Spots were grown from cells of wild-type Anabaena sp. strain PCC 7120 (lanes 1), a particular FQ mutant (lanes 2), and four independent, exconjugant clones of the particular FQ mutant bearing the corresponding pRL plasmid (lanes 3 to 6). The left, center, and right panels present, respectively, the results observed (for any one mutant, simultaneously) 2 to 3 weeks after spotting cells on agar-solidified media AA, AA+N, and AA+N plus 200 μg of Nm ml−1 and 10 μg of Sp ml−1. Cells of the wild-type strain grew in the presence or absence of nitrate but failed to grow in the presence of antibiotic; cells of each mutant grew in the presence of nitrate unless counterselected by antibiotics, but failed to grow in the absence of nitrate; addition of the corresponding cloned genes permitted the mutants to grow with only N2 as nitrogen source or in the presence of antibiotics.
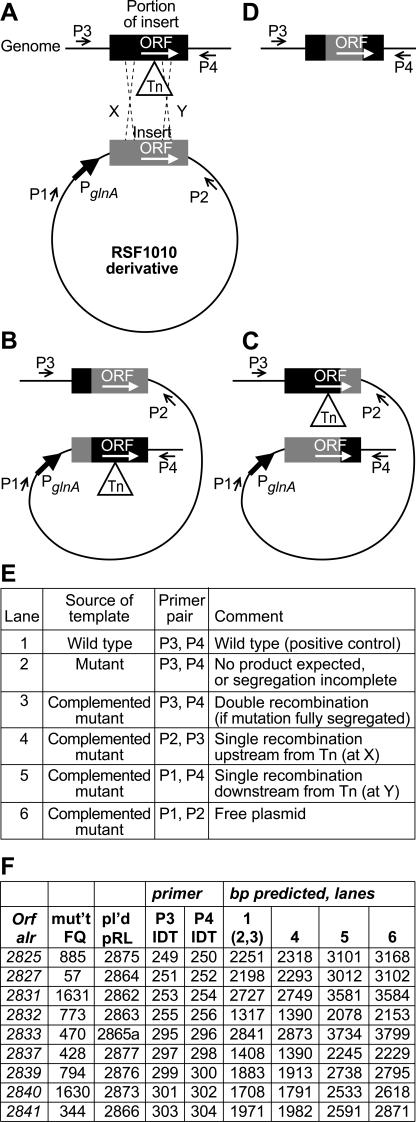
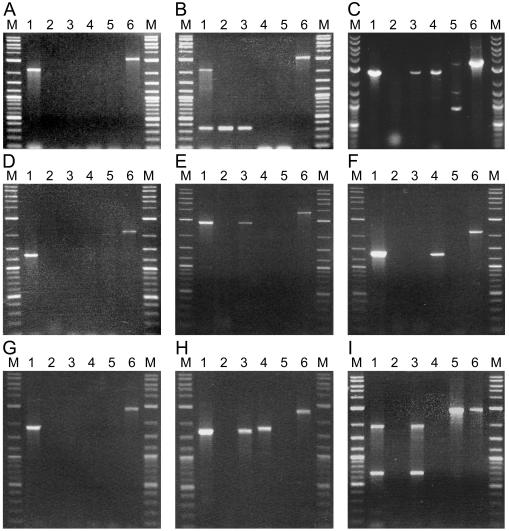
PCR analysis (Fig. 4 and 5) was used to determine whether complementation may have resulted from recombination. Complemented mutations in (i) alr2825, (ii) alr2827, and (iii) alr2839 showed no evidence of recombination (Fig. 5A, B, and G, respectively), implying that those ORFs are Fox genes. (iv) Complemented alr2837 mutant FQ428 showed evidence (Fig. 5F) of a single recombination event in which only that ORF could be expressed from its natural promoter. The clone in complementing plasmid pRL2877 extended 183 bp into alr2836 (and 38 bp 3′ from alr2837, 134 bp 5′ from alr2838). There is a small chance that the mutation conferring the phenotype of FQ428 may lie within alr2836. However, it seems exceedingly unlikely that the phenotype of all eight independent mutations in alr2837 would be similarly attributable. Therefore, we conclude that alr2837 is a Fox gene.
FIG.4.
Use of PCR to analyze possible recombination products in complemented mutants. (A) A plasmid (RSF1010 derivative) bears a fragment (in gray) that contains a single, intact (wild-type) copy of an ORF (shown as a white arrow) that has not recombined with the transposon (Tn)-interrupted copy of the same ORF in the genome. The ORF in the plasmid may be expressed from the glnA promoter or, perhaps, from an intervening, native promoter. PCR primers P3 and P4 are genomic sequences, up- and downstream from the cloned region, respectively, and primers P1 and P2 are vector sequences present, respectively, up- and downstream from the cloning region of the plasmid (P1 is also upstream from the glnA promoter). (B) Single, homologous recombination upstream from the transposon (at X in diagram A) would give rise to structure B, in which the uninterrupted ORF is positioned downstream from its natural upstream sequence. PCR would be expected to yield a product of predictable size with primers P2 and P3. (C) Single, homologous recombination downstream from the transposon (at Y in diagram A) would give rise to structure C, in which the uninterrupted ORF and any 3′ cotranscribed sequences would be placed under the influence of the glnA promoter and/or an intervening native promoter. PCR would be expected to yield a product of predictable size with primers P1 and P4. (D) Double, homologous recombination at both sides of the transposon would give rise to structure D, identical with that of the wild-type strain. With DNA from the wild-type strain or a double recombinant as template, PCR would be expected to yield a product of predictable size with primers P3 and P4. Such a band should be obtained with DNA from the mutant strain as template only if segregation of the mutation were incomplete, because the extension period was too short for the polymerase to traverse the transposon. If DNA from the mutant yielded no band, a band of the same size as from the wild-type strain, using DNA from the complemented mutant as template, would indicate that a double recombination event had cured the transposon from one or more copies of the genome. Lane numbers in inset tables E and F refer to lane numbers in Fig. 5, below. Inset table F shows PCR band sizes predicted (lanes 1 and 6) or predicted conditionally upon incomplete segregation (lane 2) or single (lanes 4 and 5) or double (lane 3) recombination.
FIG. 5.
PCR analyses of N2-grown cultures derived from complemented mutants. Panels correspond to analysis of total DNA, as template, from wild-type Anabaena sp. strain PCC 7120 (lanes 1), or (in lanes 2) the following mutants: FQ885 (alr2825) (A); FQ57 (alr2827) (B); FQ1631 (alr2831) (C); FQ773 (alr2832) (D); FQ470 (alr2833) (E); FQ428 (alr2837) (F); FQ794 (alr2839) (G); FQ1630 (alr2840) (H); FQ344 (alr2841) (I). Lanes 3 to 6: complemented strains FQ885 plus pRL2875, FQ57 plus pRL2864, FQ1631 plus pRL2862, FQ773 plus pRL2863, FQ470 plus pRL2865a, FQ428 plus pRL2877, FQ794 plus pRL2876, FQ1630 plus pRL2873, and FQ344 plus pRL2866. Lanes M are New England BioLabs 2-log DNA size-standard markers (10, 8, 6, 5, 4, and 3 kb [bright]; 2, 1.5, 1.2, and 1.0 kb [bright]; 0.9, 0.8, 0.7, 0.6, and 0.5 kb [bright]; and 0.4, 0.3, 0.2, and 0.1 kb). Primers were ORF-specific P3 and P4 (lanes 1 to 3), P2 plus P3 (lanes 4), P1 plus P4 (lanes 5), and P1 and P2 (lanes 6), specific for the ends of the cloning region of pRL1383a and its derivatives pRL2831a and pRL2831b. In the panels (see inset table F of Fig. 4), the band in lane 1 shows the size expected for wild-type DNA, indicating the efficacy of P3 and P4 to prime the expected PCR with genomic DNA; lanes 2 lack any band other than from primer dimerization, indicating that segregation of the mutations was complete; and lanes 3 show in several instances (panels C, E, H, and I) the presence of a band in the complemented mutant, indicative of double recombination having taken place in these recombination-proficient cells. Single recombination shown by bands in lanes 4 and 5 is discussed in the text. The band in each lane 6 shows the size expected for the plasmid added to the corresponding strain, indicating the efficacy of P1 and P2 to prime the expected PCR with the plasmid present in total DNA from the complemented strain.
(v) Despite showing no single recombination, alr2833 showed evidence of double recombination (Fig. 5E). An alr2833 mutation was constructed by insertional mutagenesis with pRL2815, and the locus of insertion was confirmed by restriction and sequencing as described in Materials and Methods. Like alr2833 mutant FQ470, the resulting mutant was Fox− and Hep− (data not shown). Thus, the phenotype of FQ470 may be due to a transposition affecting alr2833, a 3′ gene, or both. Its 3′ neighbor, alr2834, is 776 bp distant. The intergenic region has 13 or more stop codons in each reading frame, probably blocking further translation. PCR with primer pairs IDT394-IDT395 and IDT396-IDT397 produced products of the predicted size with, as template, DNA from wild-type cells and (separately) cDNA from cells deprived of fixed nitrogen for 8 h. Therefore, alr2833 and alr2834 were transcribed. Primers IDT394 and IDT397 produced, with the same sample of DNA as template, a predicted band of 3.63 kb and, with the same sample of cDNA as template, no band (data not shown). Therefore, (a) no transcript bridged the two ORFs, so (b) alr2834 has its own promoter, implying (c) that the phenotypes of the insertional mutant with pRL2815 and, by extension, the seven alr2833 transposon mutants are due not to a polar effect but to the effect on alr2833, and so (d) alr2833 is a Fox gene. (vi) Because in complemented FQ773, Fox gene alr2833 may have been expressed from a promoter in pRL2863 (see the faint band in Fig. 5D, lane 5), the transposon in alr2832 may have had a polar effect on alr2833. Therefore, alr2832 may not be a Fox gene.
(vii) Plasmid pRL2862 showed single and double recombination with alr2831 mutant FQ1631 (Fig. 5C). A transcript from alr2831 to alr2832 would start with a highly extended stem-loop structure (AAUUGAUUGUUUGGUAGGGUGCGUCAGUAUGAAGAUUUCUGAGUAUAGUUAGGUUCUAUCGCACUGACGCACCCUACUGGAUAGUCUAUU) (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi [45]) and would have at least nine stop codons in each reading frame, presumably blocking further translation. As described for alr2833 and alr2834, PCR and reverse transcription-PCR with primer pairs IDT390 through IDT393 indicated that alr2831 and alr2832 were transcribed, but the two ORFs were not bridged by a transcript (data not shown). The mutations in alr2831 therefore lacked a polar effect. Although the complementing sequence in pRL2862 extends 139 bp into alr2830, where a spontaneous mutation could have been corrected by recombination, it is implausible that the phenotype of all 12 transposon mutations in alr2831 (Table 3) were attributable to such an upstream mutation. Therefore, we conclude that alr2831 is a Fox gene.
(viii) Plasmid pRL2866 showed evidence of single and double recombination with alr2841 mutant FQ344 (Fig. 5I), implying that complementation may have been due to recombinant correction of a spontaneous mutation in alr2840 or all2842, into which the insert of pRL2866 extends 268 and 8 bp, respectively (for comparison, alr2841 and its 5′ and 3′ intergenic regions total 1,608 bp). However, it is hardly credible that such a mutation was parent to the phenotype of all 10 independent mutations observed in alr2841 (Table 3). Moreover, the effect of the alr2841 mutations that we identified cannot be attributed to a polar effect, because all2842, 3′ from alr2841, is oppositely oriented. Therefore, we conclude that alr2841 is a Fox gene.
(ix) Plasmid pRL2876 bearing alr2839 showed no evidence of recombination with mutant FQ794, implying that in complemented FQ794, alr2840 was not expressed from a promoter upstream from alr2839. Therefore, if alr2840 is a Fox gene, a sequence within the 3′ end of alr2839 can promote it. Although six transposon insertions were observed in alr2840 (Table 3), the effects of each of these may be attributable to a polar effect of the mutation on transcription of alr2841 and, because a PCR product attributable to double recombination of pRL2873 with mutant FQ1630 was observed (Fig. 5H), we cannot now conclude that alr2840 is itself a Fox gene.
Evidence of recombination does not imply a need of recombination for complementation. For unknown reasons, perhaps that protein synthesis is more sensitive in heterocysts than in vegetative cells to certain antibiotics, Anabaena sp. shows greater sensitivity to those antibiotics when growing on N2 than on fixed nitrogen. Not wanting to chance counterselecting recombinants, we grew complemented strains without antibiotics. Therefore, it may be more noteworthy that some strains lacked, than that others showed, recombination.
We note the utility of sequencing clones as a source of material for constructions and other purposes. Because their ends have been sequenced, their entire sequences are presumed known once a genomic sequence is finalized, and they are far less subject to point mutations (and, therefore, for a need for confirmatory sequencing) than would be PCR-amplified fragments. If generated from appropriate vectors, e.g., pRL838, they could provide a minimally redundant library of known completeness for complementing mutations of unknown genomic position.
Relationships between syntheses of LPS and heterocyst envelope polysaccharide.
Mutations in all4829 and all4830, respective orthologs of lipopolysaccharide (LPS) biosynthetic genes rfbP and rfbZ, affect synthesis of vegetative cell LPS. The mutants are Fox−; the rfbP mutant synthesizes at least some heterocyst envelope polysaccharide (40). An all4828 (rfbD) mutant is Fox+ (40). Like rfbP and rfbZ, autolysin-encoding hcwA (43) and a gene that encodes a putative penicillin-binding protein (22) are Fox genes. LPS is a constituent of the walls of vegetative cells, HcwA degrades those walls, and penicillin blocks synthesis of those walls. Therefore, the refashioning of cell wall components of the differentiating vegetative cell may be a prerequisite for correct transport and deposition of components of the heterocyst envelope.
Lipid A is the portion of LPS that roots it in the outer cell membrane (29), and HepA itself (Alr2835), used in a BLAST (2) search against Pseudomonas aeruginosa strain PAO1 and E. coli strain CFT073, shows greatest similarity to the lipid A export ATP-binding-permease protein MsbA (in PAO1, 282 bits [Expect = 9e-77] versus 214 bits [Expect = 4e-56] for the next closest match; and in CFT073, 270 bits [Expect = 2e-73] versus 227 bits [Expect = 2e-60] for the next closest match). Conversely, MsbA from E. coli K-12 (GenBank P60752; in E. coli, an msbA knockout mutation is lethal [29]) shows greatest similarity, among the more than 80 proteins annotated as ATP-binding transporters in Anabaena sp. strain PCC 7120, to Alr2835 (313 bits [Expect = 3e-86]) and Alr5199 (316 bits [Expect = 4e-87]).
Other observations also hint that the hepA region may contain genes related to synthesis of LPS in addition to genes required for synthesis of heterocyst envelope polysaccharide. (i) Alr2833 bears a motif (http://www.ebi.ac.uk/interpro/DisplayIproEntry?ac = IPR003856) of a chain length determinant protein (or wzz protein) involved in LPS biosynthesis. (ii) The structure of the heterocyst envelope polysaccharide has been defined in detail for Anabaena cylindrica (9, 10), and in lesser detail for Anabaena variabilis strain ATCC 29413, in which its synthesis has been studied (8, 11), and Cylindrospermum licheniforme (11). Because A. variabilis and Anabaena sp. strain PCC 7120 are genetically highly similar (http://genome.jgi-psf.org/draft_microbes/anava/anava.home.html; http://www.kazusa.or.jp/cyano/Anabaena/), their heterocyst envelope polysaccharides and corresponding biosynthetic pathways are probably very similar. A. variabilis has a cluster of genes that corresponds with the cluster alr2823-alr2841 in Anabaena sp. strain PCC 7120 (we thank Jeff Elhai for assistance with this analysis). Neither rhamnose nor any other deoxy sugar was found in the heterocyst envelope polysaccharide of A. variabilis (11). The predicted product of alr2830 (its ortholog in A. variabilis shows 98% amino acid identity) is similar (as is also Alr4489) to dTDP-4-dehydrorhamnose 3,5-epimerase, a diagnostic gene (rfbC in E. coli) in the synthesis of activated rhamnose en route to LPS synthesis in gram-negative bacteria. The biosynthesis of dTDP-rhamnose also requires a glucose-1-phosphate nucleotide transferase, an epimerase, and a reductase, to which the products of ORFs alr2825, alr2827, and alr2831 show similarity. (iii) Alr2841 (annotated as an unknown protein) shows sequence similarity to O-antigen polymerases that are involved in LPS and exopolysaccharide synthesis in different bacteria (InterPro family PD416824). (iv) Alr2828 shows similarity to glycosyl transferases that are involved in biosynthesis of the O-antigen portion of LPS. Finally, (v) Alr2834 (HepC) shows overall similarity (Expect = 7e-42) to COG2148, WcaJ, sugar transferases involved in LPS synthesis (30). Whether mutations in the hepA region affect LPS and whether ORFs in that region (excluding hepC) in which we found no transposon insertion are Fox genes remain untested.
If genes for synthesis of an LPS and of heterocyst envelope polysaccharide are both present in the hepA region, future studies should clarify whether they are related or merely interspersed. A possibility not hitherto considered is that heterocyst envelope polysaccharide may itself be an LPS that differs from, but may share some biosynthetic genes with, vegetative cell LPS. The possible localization of rhamnose (see above) in the single-copy core region of such an LPS (29), coupled at one end to lipid A and at the other to the highly repeated subunits of heterocyst envelope polysaccharide (10), could account for that sugar not having been observed in the envelope polysaccharide of A. variabilis heterocysts (11).
Acknowledgments
We thank Haixia He and Yi Li for skilled technical assistance.
This work was supported by National Science Foundation grant MCB0090232 and United States Department of Energy grant DOE-FG02-91ER20021.
REFERENCES
- 1.Almon, H., and H. Böhme. 1980. Components and activity of the photosynthetic electron transport system of intact heterocysts isolated from the blue-green alga Nostoc muscorum. Biochim. Biophys. Acta 592:113-120. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bancroft, I., and C. P. Wolk. 1989. Characterization of an insertion sequence (IS891) of novel structure from the cyanobacterium Anabaena sp. strain M-131. J. Bacteriol. 171:5949-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bancroft, I., C. P. Wolk, and E. V. Oren. 1989. Physical and genetic maps of the genome of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 171:5940-5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 6.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 7.Cai, Y., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardemil, L., and C. P. Wolk. 1981. Isolated heterocysts of Anabaena variabilis synthesize envelope polysaccharide. Biochim. Biophys. Acta 674:265-276. [DOI] [PubMed] [Google Scholar]
- 9.Cardemil, L., and C. P. Wolk. 1976. The polysaccharides from heterocyst and spore envelopes of a blue-green alga. Methylation analysis and structure of the backbones. J. Biol. Chem. 251:2967-2975. [PubMed] [Google Scholar]
- 10.Cardemil, L., and C. P. Wolk. 1979. The polysaccharides from heterocyst and spore envelopes of a blue-green alga. Structure of the basic repeating unit. J. Biol. Chem. 254:736-741. [PubMed] [Google Scholar]
- 11.Cardemil, L., and C. P. Wolk. 1981. Polysaccharides from the envelopes of heterocysts and spores of the blue-green algae Anabaena variabilis and Cylindrospermum licheniforme. J. Phycol. 17:234-240. [Google Scholar]
- 12.Ehira, S., M. Ohmori, and N. Sato. 2003. Genome-wide expression analysis of the responses to nitrogen deprivation in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 10:97-113. [DOI] [PubMed] [Google Scholar]
- 13.Elhai, J., A. Vepritskiy, A. M. Muro-Pastor, E. Flores, and C. P. Wolk. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elhai, J., and C. P. Wolk. 1990. Developmental regulation and spatial pattern of expression of the structural genes for nitrogenase in the cyanobacterium Anabaena. EMBO J. 9:3379-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fay, P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56:340-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallon, J. R. 1992. Reconciling the incompatible: N2 fixation and O2. New Phytol. 122:571-609. [Google Scholar]
- 17.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 18.Holland, D., and C. P. Wolk. 1990. Identification and characterization of hetA, a gene that acts early in the process of morphological differentiation of heterocysts. J. Bacteriol. 172:3131-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, N. T., T. Thiel, T. H. Giddings, and C. P. Wolk. 1981. New Anabaena and Nostoc cyanophages from sewage settling ponds. Virology 114:236-246. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 21.Kangatharalingam, N., J. C. Priscu, and H. W. Paerl. 1992. Heterocyst envelope thickness, heterocyst frequency and nitrogenase activity in Anabaena flos-aquae: influence of exogenous oxygen tension. J. Gen. Microbiol. 138:2673-2678. [Google Scholar]
- 22.Lázaro, S., F. Fernández-Piñas, E. Fernández-Valiente, A. Blanco-Rivero, and F. Leganés. 2001. pbpB, a gene coding for a putative penicillin-binding protein, is required for aerobic nitrogen fixation in the cyanobacterium Anabaena sp. strain PCC7120. J. Bacteriol. 183:628-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maldener, I., S. Hannus, and M. Kammerer. 2003. Description of five mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst differentiation and identification of the transposon-tagged genes. FEMS Microbiol. Lett. 224:205-213. [DOI] [PubMed] [Google Scholar]
- 24.Murphy, R. C., G. E. Gasparich, D. A. Bryant, and R. D. Porter. 1990. Nucleotide sequence and further characterization of the Synechococcus sp. strain PCC 7002 recA gene: complementation of a cyanobacterial recA mutation by the Escherichia coli recA gene. J. Bacteriol. 172:967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murry, M. A., and C. P. Wolk. 1989. Evidence that the barrier to the penetration of oxygen into heterocysts depends upon two layers of the cell envelope. Arch. Microbiol. 151:469-474. [Google Scholar]
- 26.Ning, D., and X. Xu. 2004. alr0117, a two-component histidine kinase gene, is involved in heterocyst development in Anabaena sp. PCC 7120. Microbiology 150:447-453. [DOI] [PubMed] [Google Scholar]
- 27.Ochman, H., A. S. Gerber, and D. L. Hartl. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120:621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 29.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiel, T. 1994. Genetic analysis of cyanobacteria, p. 581-611. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 32.Trieu-Cuot, P., C. Carlier, P. Martin, and P. Courvalin. 1987. Plasmid transfer by conjugation from Escherichia coli to gram-positive bacteria. FEMS Microbiol. Lett. 48:289-294. [Google Scholar]
- 33.Walsby, A. E. 1985. The permeability of heterocysts to the gases nitrogen and oxygen. Proc. R. Soc. London B 226:345-366. [Google Scholar]
- 34.Wolk, C. P. 2000. Heterocyst formation in Anabaena, p. 83-104. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 35.Wolk, C. P., Y. Cai, L. Cardemil, E. Flores, B. Hohn, M. Murry, G. Schmetterer, B. Schrautemeier, and R. Wilson. 1988. Isolation and complementation of mutants of Anabaena sp. strain PCC 7120 unable to grow aerobically on dinitrogen. J. Bacteriol. 170:1239-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolk, C. P., Y. Cai, and J. M. Panoff. 1991. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc. Natl. Acad. Sci. USA 88:5355-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolk, C. P., J. Elhai, T. Kuritz, and D. Holland. 1993. Amplified expression of a transcriptional pattern formed during development of Anabaena. Mol. Microbiol. 7:441-445. [DOI] [PubMed] [Google Scholar]
- 38.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 39.Wolk, C. P., J. Zhu, and R. Kong. 1999. Genetic analysis of heterocyst formation, p. 509-515. In G. A. Peschek, W. Loeffelhardt, and G. Schmetterer (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 40.Xu, X., I. Khudyakov, and C. P. Wolk. 1997. Lipopolysaccharide dependence of cyanophage sensitivity and aerobic nitrogen fixation in Anabaena sp. strain PCC 7120. J. Bacteriol. 179:2884-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, R., and C. P. Wolk. 2003. A two-component system mediates developmental regulation of biosynthesis of a heterocyst polysaccharide. J. Biol. Chem. 278:19939-19946. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, J., K. Jäger, T. Black, K. Zarka, O. Koksharova, and C. P. Wolk. 2001. HcwA, an autolysin, is required for heterocyst maturation in Anabaena sp. strain PCC 7120. J. Bacteriol. 183:6841-6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, J., R. Kong, and C. P. Wolk. 1998. Regulation of hepA of Anabaena sp. strain PCC 7120 by elements 5′ from the gene and by hepK. J. Bacteriol. 180:4233-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]