Abstract
Site-directed mutagenesis studies of the signal peptidase of the methanogenic archaeon Methanococcus voltae identified three conserved residues (Ser52, His122, and Asp148) critical for activity. The requirement for one conserved aspartic acid residue distinguishes the archaeal enzyme from both the Escherichia coli and yeast Sec11 enzymes.
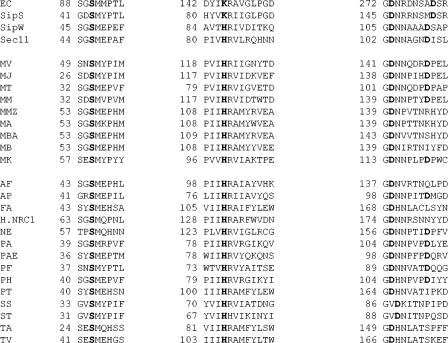
Type I signal peptidases are essential enzymes that are responsible for the removal of N-terminal signal peptide sequences from preproteins during their translocation across various membranes in all three domains of life (1, 5). Analysis of signal peptidases from eukaryotic and prokaryotic cells revealed only weak overall sequence similarity between the bacterial signal peptidases and the eukaryotic Sec11 subunit. Indeed, the Sec11 component has so little sequence homology with the bacterial counterpart that its relatedness was initially missed (5). However, there are five regions of significant sequence homology which are preserved throughout evolution (1), three of which contain conserved amino acids important for catalysis (Fig. 1). In gram-negative and gram-positive bacteria, site-directed mutagenesis studies have been performed to investigate the key amino acid residues involved in catalysis (2, 7). In both cases, a conserved serine and lysine (Ser90 and Lys145 in Escherichia coli) were shown to be critical for activity, supporting the idea of a Ser-Lys dyad catalytic mechanism. No histidines are important for catalysis (5). In addition, the roles of two conserved aspartic acid residues (Asp273 and Asp280 in E. coli) have been investigated. In E. coli, mutation studies indicated that neither of these residues, including the absolutely conserved Asp280, was essential for signal peptidase activity in vivo (2). In contrast, in Bacillus subtilis, similar experiments identified the Asp280 equivalent as essential for activity, in addition to the conserved serine and lysine residues. Mutation of the Asp273 equivalent in B. subtilis also resulted in a decrease in activity, most likely due to disruption of a salt bridge leading to an impairment in structure (7). Recently, mutagenesis work and analysis of the X-ray crystal structure of the E. coli enzyme led to the identification of an additional residue, Ser278, essential for optimal activity (4). This suggested that the Ser-Lys dyad mechanism may be better described as a Ser-Lys-(Ser/Thr) triad (4).
FIG. 1.
Conserved regions of bacterial, eukaryal, and archaeal type I signal peptidases. Boldface type indicates amino acids demonstrated to be important for catalysis in the bacterial and eukaryal enzymes and their conservation in the archaeal enzymes. All available archaeal enzyme sequences are shown. EC, E. coli; SipS, B. subtilis; SipW, B. subtilis; Sec11, yeast; MV, M. voltae; MJ, Methanocaldococcus jannaschii; MM, Methanococcus maripaludis; MT, Methanothermobacter thermautotrophicus; MMZ, Methanosarcina mazei; MA, Methanosarcina acetovorans; MBA, Methanosarcina barkeri; MB, Methanococcoides burtonii; MT, Methanothermus kandlerii; AF, Archaeoglobus fulgidus; AP, Aeropyrum pernix; FA, Ferroplasma acidarmanus; H.NRC1, Halobacterium NRC-1; NE, Nanoarchaeum equitans; PAE, Pyrobaculum aerophilum; PA, Pyrococcus abyssi; PF, Pyrococcus furiosus; PH, Pyrococcus horikoshii; PT, Picrophilus torridus; SS, Sulfolobus solfataricus; ST, Sulfolobus tokodaii; TA, Thermoplasma acidophilum; TV, Thermoplasma volcanium. For Picrophilus torridus, the gene annotated as signal sequence processing protein Sec11 precursor (GenBank accession number YP_024127) was used rather than the gene annotated as signal peptidase I (GenBank accession number YP_023702), as it had a stronger conservation of the conserved regions. Similarly, for F. acidarmanus, two genes are annotated as signal peptidases (GenBank accession numbers ZP_00307053 and ZP_00306921). The former, with stronger hits to other signal peptidases, was used here. For Methanococcoides burtonii, the signal peptidase deposited as GenBank accession number ZP_00148201 is extremely short (79 amino acids) and lacks the universally conserved serine shown to be critical for catalysis in signal peptidases from all three domains. However, examination of the upstream DNA sequence indicates the presence of an in-frame Met codon that, if representing the real translation start site, would result in a protein with conserved regions virtually identical to those of the signal peptidases of Methanosarcina species. This is the start used here.
The situation in eukaryotic signal peptidases is quite distinct. Mutagenesis work has been performed on the yeast endoplasmic reticulum (ER) signal peptidase complex component Sec11. While Sec11 and other eukaryotic-type signal peptidases contain the conserved Ser90 equivalent, the conserved general base, Lys145, has been replaced with a histidine residue. Both the conserved serine and the histidine, as well as the Asp273 and Asp280 equivalents, were found to be essential for activity in vivo (8). Replacing the conserved histidine with a lysine resulted in an inactive enzyme. Mutagenesis studies revealed that none of the lysines were essential for processing. This led to the suggestion that the type I signal peptidase family was composed of two groups, one with an essential lysine and one without. It was proposed that in the Sec11 enzyme, catalysis was carried out through a Ser-His dyad or a Ser-His-Asp triad. Interestingly, in B. subtilis, where there are multiple signal peptidases, one, SipW, appears to be like the yeast enzyme, with a histidine replacing the conserved lysine. However, unlike in Sec11, replacement of the histidine residue with a lysine results in an active enzyme (6).
In the Archaea, analysis of the signal peptidases indicates a Sec11 type enzyme with a histidine in place of the conserved lysine (1, 3). Archaeal enzymes also contain the conserved Ser residue as well as the absolutely conserved Asp273 equivalent. Furthermore, most but not all of the archaeal signal peptidases contain a second conserved Asp located at positions between 279 and 282 with respect to the E. coli numbering system and thus presumably equivalent to the E. coli Asp280. The only exceptions to this are the enzymes from the euryarchaeote acidophiles Thermoplasma volcanium, Thermoplasma acidophilum, and Ferroplasma acidarmanus, although not Picrophilus torridus (Fig. 1). Alignments indicate that the two conserved aspartic acid residues are separated by 6 amino acids in bacterial-type signal peptidases (including mitochondrial and chloroplast enzymes), while in the ER-type enzymes of eukaryotes, the ER-type enzymes found in a few bacteria (e.g., SipW of B. subtilis), and most archaeal enzymes, the spacing is one amino acid less (Fig. 1) (5).
No studies on the catalytically important residues in the signal peptidases of the Archaea have yet been reported. Recently, we identified the signal peptidase of the methanogenic archaeon Methanococcus voltae and demonstrated its activity when it was expressed in E. coli, using an in vitro assay system with a C-terminally His-tagged version of the M. voltae S-layer protein as a substrate (3). Use of this assay allowed us to investigate the effect of site-directed mutagenesis aimed at the conserved serine, histidine, and aspartic acid residues of the signal peptidase, which is the focus of this report.
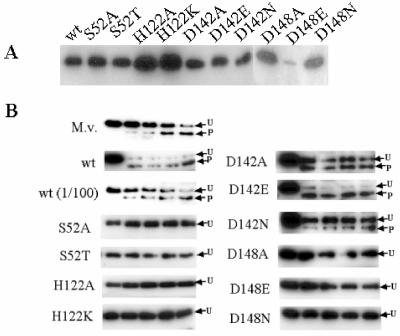
Site-directed mutagenesis was performed with a QuikChange mutagenesis kit from Stratagene (La Jolla, Calif.) and the primers listed in Table 1. Gradient PCR (annealing temperature range from 40 to 60°C) was performed by using pKJ385 (pET23a carrying M. voltae signal peptidase I cloned to create a C-terminal His-tagged protein) as the template (3). All mutant sequences were confirmed by sequencing. His tagging of the signal peptidase variants allowed easy visualization of the proteins expressed in E. coli BL21(DE3)/pLysS cells. Mutant proteins were all expressed to levels similar to that of the wild-type version of the enzyme, with the exception of the D148E variant, which was poorly expressed (Fig. 2A).
TABLE 1.
Primers pairs used to create the site-directed mutations in M. voltae signal peptidase 1
| Primer | Sequence (5′-3′)a | Mutation | Corresponding plasmid |
|---|---|---|---|
| GTTGTTTCCAATAGTATGTATCCAATAATGG | S52wt | ||
| 22866 | GTTGTTTCCAATGCTATGTATCCAATAATGG | S52A | pKJ440 |
| 22867 | CCATTATTGGATACATAGCATTGGAAACAAC | ||
| 22868 | GTTGTTTCCAATACTATGTATCCAATAATGG | S52T | pKJ442 |
| 22869 | CCATTATTGGATACATAGTATTGGAAACAAC | ||
| GGCCTGTAATTCATAGGATTATTGG | H122wt | ||
| 22870 | GGCCTGTAATTGCTAGGATTATTGG | H122A | pKJ455 |
| 22871 | CCAATAATCCTAGCAATTACAGGCC | ||
| 22872 | GGCCTGTAATTAAAAGGATTATTGG | H122K | pKJ458 |
| 22873 | CCAATAATCCTTTTAATTACAGGCC | ||
| CATCAAAGGGGATAATAATCAGGATAGG | D142wt | ||
| 22874 | CATCAAAGGGGCTAATAATCAGGATAGG | D142A | pKJ445 |
| 22875 | CCTATCCTGATTATTAGCCCCTTTGATG | ||
| 22876 | CATCAAAGGGGAAAATAATCAGGATAGG | D142N | pKJ451 |
| 22877 | CCTATCCTGATTATTATTCCCTTTGATG | ||
| 22878 | CATCAAAGGGGAAAATAATCAGGAATGG | D142E | pKJ449 |
| 22879 | CCTATCCTGATTATTTTCCCCTTTGATG | ||
| CAGGATAGGGACCCCGAACTTGTCAAACC | D142wt | ||
| 22880 | CAGGATAGGGCCCCCGAACTTGTCAAACC | D148A | pKJ453 |
| 22881 | GGTTTGACAAGTTCGGGGGCCCTATCCTG | ||
| 22882 | CAGGATAGGAACCCCGAACTTGTCAAACC | D148N | pKJ447 |
| 22883 | GGTTTGACAAGTTCGGGGGTTCTATCCTG | ||
| 22884 | CAGGATAGGGAACCCGAACTTGTCAAACC | D148E | pKJ456 |
| 22885 | GGTTTGACAAGTTCGGGTTCCCTATCCTG |
Underlined nucleotides indicate the wild-type sequence or corresponding changes from the wild type.
FIG. 2.
Expression and activity of wild-type and mutant M. voltae signal peptidases. (A) E. coli BL21(DE3)/pLysS cells, carrying either the wild-type or mutated M. voltae signal peptidase I gene as an NdeI/XhoI fragment in pET23a, were grown in Luria broth at 37°C with shaking to an optical density at 600 nm of approximately 0.8, induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and further incubated for 1.5 h. Induced whole-cell samples were analyzed by Western blotting; blots were developed with anti-His antibodies. Equal loading of all lanes was confirmed in a duplicate gel stained with Coomassie blue (not shown). (B) Signal peptidase activity of wild-type and mutated M. voltae signal peptidases. All assays were identical except for the potential source of enzyme, which consisted of E. coli membranes containing overexpressed wild-type or mutated signal peptidases, as indicated. M.v., M. voltae membranes as the enzyme source. Samples were taken at 0, 5, 10, 15, and 60 min and analyzed by Western blotting utilizing anti-His antibodies. Unprocessed (U) and processed (P) forms of the S-layer substrate are indicated.
Signal peptidase assays were carried out as described previously (3). Briefly, crude membrane fractions of E. coli BL21(DE3)/pLysS expressing wild-type or mutated forms of the M. voltae signal peptidase I were used as the enzyme source. Heat-treated E. coli membranes containing truncated, overexpressed S-layer protein with a C-terminal His tag were used as the substrate. Enzyme and substrate were incubated together under conditions (125 mM HEPES [pH 8.5], 1 M NaCl, 1.25% Triton X-100; 37°C) previously determined for maximal processing by the archaeal signal peptidase while inhibiting the E. coli signal peptidase (3). The processed form of the His-tagged S-layer protein was detected as a faster-migrating cross-reacting band in immunoblots developed with anti-His antibodies, due to the removal of the 28-amino-acid leader peptide.
Essential nature of Ser52 and His122 in M. voltae signal peptidase.
Under the standard conditions of the assay (3), with M. voltae membranes as the source of the signal peptidase there is a gradual conversion of the unprocessed form of the S-layer substrate into the processed form (Fig. 2B). With overexpressed wild-type M. voltae signal peptidase as the enzyme source, the conversion to the processed form is much faster, while a 100-fold dilution of the enzyme source again results in a gradual processing. The conserved serine residue (Ser52 in M. voltae) found to be essential for signal peptidase activity in enzymes from gram-positive and gram-negative bacteria, as well as in Sec11, was also found to be essential for activity of the archaeal enzyme. Overexpressed enzyme in which the Ser52 was mutated to either alanine or threonine was inactive (Fig. 2B). The histidine residue (His122), which corresponds to the essential histidine of Sec11, was also necessary for activity. Mutation of this residue to either alanine or lysine resulted in an inactive enzyme (Fig. 2B). In the case of signal peptidase mutated at either the serine or histidine residues, no processing of the substrate could be detected by using undiluted membrane preparations under conditions in which membrane preparations containing wild-type signal peptidase could be diluted 100-fold with easily detectable activity. The failure of the histidine-to-lysine change to lead to enzymatic activity is consistent with eukaryal Sec11 mutagenesis results (7) and distinct from the result obtained with the prokaryotic SipW of B. subtilis, where such a change did result in active enzyme (6).
Role of the conserved aspartic acid residues.
Archaeal signal peptidases, as well as Sec11, contain a conserved aspartic acid residue located immediately following the universally conserved glycine residue, corresponding to position 272 in the E. coli signal peptidase. The sole exceptions are the enzymes from Sulfolobus species, where the sequence is slightly varied in having a glycine-valine-aspartic acid sequence rather than glycine-aspartic acid (Fig. 1). Signal peptidase carrying a D142A or D142N change retains partial activity, while enzyme with the D142E mutation, which retains the carboxylic acid group, shows near-wild-type processing levels (Fig. 2B). These results indicate that the Asp142 residue, in spite of its universal conservation, is not required for activity for the archaeal signal peptidase. Very similar results were obtained with the equivalent aspartic acid residue (Asp146) of SipS in B. subtilis: enzyme with a D146E change had no loss of activity, while the D146N variant was severely reduced in activity (7). Mutation of the second conserved aspartic acid residue in the M. voltae enzyme, Asp148 (equivalent to E. coli Asp280), had a severe negative effect on the signal peptidase activity. Signal peptidase containing either a D148A or D148N mutation was inactive in the in vitro assay (Fig. 2B). No activity could be detected with the D148E change either, although the expression level of this mutant enzyme was considerably lower than those of all other mutants tested and it is not possible to rule out slight residual activity in this protein. These results are again similar to those obtained with SipS, where either a D153E or D153N change resulted in nearly complete loss of enzymatic activity (7). In the ER-type SipW of B. subtilis, the first aspartic acid residue, Asp106 was important but not essential for activity, while the second aspartic acid, Asp112, was not important (6). In E. coli, neither conserved aspartic acid is absolutely required for activity, while in Sec11 both conserved aspartic acids are absolutely essential for activity (5). In having a requirement for one conserved aspartic acid for activity, the archaeal enzyme is clearly distinguished from both the E. coli and yeast paradigms.
The results of this study present, for the first time, an evaluation of amino acids necessary for the activity of archaeal type I signal peptidases. The mechanism likely involves a Ser-His-Asp triad, as proposed for eukaryotic ER-type signal peptidases, like Sec11. One difference between the archaeal and Sec11 enzymes is that the archaeal enzyme does not have the strict requirement for both conserved aspartic acid residues that Sec11 does. In the case of the M. voltae signal peptidase, the critical amino acids are Ser52, His122 and Asp148. It seems likely that corresponding amino acids will also be found to be critical for the activity of signal peptidases in most other archaea which have aspartic acid residues in the positions corresponding to positions 147 to 151 in the M. voltae enzyme. However, this appears not to be the case in acidophiles of the genera Thermoplasma and Ferroplasma, since an Asp148 equivalent is not found. It is interesting that in these three organisms, a serine is found at the position equivalent to M. voltae position 148 or 149 (E. coli position 279 or 280). In E. coli, a conserved serine located at position 278 is proposed to be part of the Ser-Lys-Ser triad mechanism of catalysis (2).
Acknowledgments
This research was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) to K.F.J. S.L.B. and S.Y.M.N. were supported by postgraduate fellowships from NSERC.
REFERENCES
- 1.Eichler, J. 2002. Archaeal signal peptidases from the genus Thermoplasma: structural and mechanistic hybrids of the bacterial and eukaryal enzymes. J. Mol. Evol. 54:411-415. [DOI] [PubMed] [Google Scholar]
- 2.Klenotic, P. A., J. L. Carlos, J. C. Samuelson, T. A. Schuenemann, W. R. Tschantz, M. Paetzel, N. C. J. Strynadka, and R. E. Dalbey. 2000. The role of the conserved box E residues in the active site of Escherichia coli type I signal peptidase. J. Biol. Chem. 275:6490-6498. [DOI] [PubMed] [Google Scholar]
- 3.Ng, S. Y. M., and K. F. Jarrell. 2003. Cloning and characterization of archaeal type I signal peptidase from Methanococcus voltae. J. Bacteriol. 185:5936-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paetzel, M., R. E. Dalbey, and N. C. J. Strynadka. 2002. Crystal structure of a bacterial signal peptidase apoenzyme: implications for signal peptide binding and the Ser-Thr dyad mechanism. J. Biol. Chem. 277:9512-9519. [DOI] [PubMed] [Google Scholar]
- 5.Paetzel, M., A. Karla, N. C. J. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4579. [DOI] [PubMed] [Google Scholar]
- 6.Tjalsma, H., A. G. Stover, A. Driks, G. Venema, S. Bron, and J. M. van Dijl. 2000. Conserved serine and histidine residues are critical for activity of the ER-type signal peptidase SipW of Bacillus subtilis. J. Biol. Chem. 275:25102-25108. [DOI] [PubMed] [Google Scholar]
- 7.van Dijl, J. M., A. de Jong, G. Venema, and S. Bron. 1995. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis. Structural and functional similarities with LexA-like proteases. J. Biol. Chem. 270:3611-3618. [DOI] [PubMed] [Google Scholar]
- 8.van Valkenbough, C., X. Chen, C. Mullins, H. Fang, and N. Green. 1999. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. J. Biol. Chem. 274:11519-11525. [DOI] [PubMed] [Google Scholar]