Abstract
Rhizobium sp. strain NGR234 possesses a functional type three secretion system (TTSS), through which a number of proteins, called nodulation outer proteins (Nops), are delivered to the outside of the cell. A major constraint to the identification of Nops is their low abundance in the supernatants of NGR234 strains grown in culture. To overcome this limitation, a more sensitive proteomics-based strategy was developed. Secreted proteins from wild-type NGR234 were separated by two-dimensional gel electrophoresis, and the gel was compared to similar gels containing the proteins from a TTSS mutant (NGRΩrhcN). To identify the proteins, spots unique to the NGR234 gels were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry and the data were compared to the sequence of the symbiotic plasmid of NGR234. A nonpolar mutant of one of these proteins was generated called NopB. NopB is required for Nop secretion but inhibits the interaction with Pachyrhizus tuberosus and augments nodulation of Tephrosia vogelii. Flavonoids and a functional TTSS are required for the formation of some surface appendages on NGR234. In situ immunogold labeling and isolation of these pili showed that they contain NopB.
Rhizobia are gram-negative soil inhabitants that induce the formation of highly specialized, nitrogen-fixing organs called nodules on the roots or stems of leguminous plants. Some rhizobial species provoke nodule formation on a limited number of legume genera and are said to have narrow host ranges, e.g., Rhizobium meliloti, which nodulates only three genera of legumes. Other, broad-host-range rhizobia provoke the formation of nodules on many different legumes; e.g., Rhizobium sp. strain NGR234 (hereafter called NGR234) nodulates more than 112 genera of legumes as well as the nonlegume Parasponia andersonii (32, 38). The formation of root nodules is a result of an elaborate developmental program directed by signal exchange between the two partners. These signals include flavonoids, Nod factors, surface polysaccharides, and extracellular proteins (7, 31, 34). After flavonoid induction, NGR234 secretes a number of extracellular proteins called Nops (nodulation outer proteins) via a type III secretion system (TTSS) (24, 41). TTSSs are virulence determinants shared by many diverse gram-negative bacteria that cause disease in plants and animals. The TTSS machinery is highly conserved and encoded by cluster of hrp (hypersensitive response and pathogenesis) or hrc (hrp conserved) genes in phytopathogens (2, 16). Homologues of the hrc genes were found in NGR234 and renamed rhc (Rhizobium conserved) (13, 41). rhcN encodes a protein that shares characteristics of ATPases, which may function as energizers of the secretion process. Mutation of rhcN (strain NGRΩrhcN) abolished the secretion of Nops. The presence or absence of Nops dramatically alters the nodulation capacity of NGR234 in a host-specific manner (41).
Interestingly, the regulation of Nop secretion in NGR234 is flavonoid and NodD1 dependent just like Nod factor synthesis (18, 25, 30, 41). The secretion of Nops requires an additional protein called TtsI, however. TtsI shares characteristics of two-component regulatory systems and is thought to be a transcriptional activator of genes involved in Nop secretion (24, 41). Upon flavonoid induction, TtsI probably binds to a conserved sequence, the tts box that is found upstream of rhizobial genes involved in type III secretion, activating their transcription (20, 26). Pathogenic bacteria use the TTSS to deliver (effector) proteins into the host cell. The involvement of bacterial surface appendages in type three-related interactions with host cells has been studied for both animal and plant pathogens. In Salmonella, so-called “invasome” appendages develop upon contact with cultured epithelial cells (14). Similar structures were also observed on Shiga toxin-producing Escherichia coli cells (11). In plant pathogens, longer appendages (hrp pili) have been reported for Pseudomonas syringae (35), Ralstonia solanacearum (39), and Erwinia amylovora (17).
Of the TTSS-possessing rhizobia, R. fredii strain USDA257 (hereafter called USDA257) makes surface appendages (6 to 8 nm in diameter) when the bacteria are grown in the presence of nod-gene-inducing compounds (21). Similar structures have also been observed on induced cells of NGR234 (W. J. Deakin, C. Marie, M. M. Saad, H. B. Krishnan, and W. J. Broughton, unpublished data). NGR234 secretes at least six Nops via its TTSS; and the functions of these secreted proteins have been characterized by mutagenesis and bioinformatic analyses. Based upon the phenotype of the mutant and its homology to HrpF from Xanthomonas species, NopX is assumed to be a component of the translocon, a pore-like structure formed in the plant cell membrane through which the effector proteins are delivered to the host cell (25). NopL is a putative effector protein of NGR234 that may interfere with plant signal transudation pathways to suppress plant defense responses (5, 6, 25). Recently, a new secreted protein, NopP, was identified which is also believed to be an effector protein (3). NopA is thought to be the major component of the TTSS-dependent surface appendage (pilus) through which the effector proteins are transported to the outside of the cell (25; Deakin et al., unpublished).
Here we report the identification of another secreted protein, NopB, that was isolated by a proteomics-based approach from total extracellular proteins purified from apigenin-induced cultures of NGR234. Secretion of NopB was confirmed with a NopB-specific antibody. Mutational analysis demonstrated that NopB is essential for Nop secretion. To investigate the function of NopB, we visualized the surface appendages of NGR234 by adapting the technique used with P. syringae to visualize hrp pili. Studies done using in situ immunogold labeling support the direct association of NopB with the TTSS-dependent pilus structures produced by NGR234.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. E. coli strains were grown in Luria-Bertani broth at 37°C with shaking at 160 rpm. Rhizobium strains were grown in rhizobia minimal medium (RMS) (8) or yeast extract-mannitol medium (YEM) (40) at 27°C unless otherwise stated. If required, the flavonoid inducer apigenin (Fluka Chemie GmbH, Buchs, Switzerland) was added to a final concentration of 10−6 M. Antibiotics (AppliChem GmbH, Darmstadt, Germany) were used at the following concentrations: ampicillin, 100 μg ml−1, kanamycin, 50 μg ml−1, rifampin, 50 μg ml−1, tetracycline, 25 μg ml−1, spectinamycin, 50 μg ml−1, and gentamicin, 30 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Rhizobium strains | ||
| NGR234 | Rifr derivative of the wild-type isolate of NGR234 | 23 |
| NGRΩrhcN | NGR234 derivative containing an Ω insertion in rhcN, Rifr Spr | 41 |
| NGRΩnopL | NGR234 derivative containing an Ω insertion in nopL, Rifr Kmr | 25 |
| NGRΔnopP | NGR234 derivative in which 0.5 kb of nopP was replaced by an Ω insertion, Rifr Spr | 3 |
| NGRnopB::uidA | NGR234 derivative containing a uidA insertion in nopB, Rifr | This work |
| E. coli DH5α | supE44 ΔlacY169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | BRLa |
| Plasmids | ||
| pJQ200SK | Suicide vector used for directed mutagenesis, Gmr | 33 |
| pRK2013 | Tra+ helper plasmid | 12 |
| PWM3 | pUC1318 derivative containing the uidA2 cassette | 28 |
| PJQB | pJQ200SK derivative carrying a 4,000-bp XhoI-BamHI fragment of pXB110 | This work |
| pMSG2 | pJQB derivative carrying uidA in SmaI site | This work |
| pMSG4 | pLAFR-6 derivative carrying HindIII-XbaI fragment of pXB110, Tetr | This work |
| pLAFR-6 | Broad-host-range vector containing transcriptional terminators flanking cloning sites, Tetr | D. Dahlbeck and B. Staskawicz, unpublished results |
BRL, Bethesda Research Laboratories.
Construction of nopB mutant.
General recombinant DNA and molecular biological techniques were performed according to standard protocols (36). The approximately 4-kb XhoI-BamHI restriction fragment of cosmid pXB110 (29) that carries nopB was subcloned into pJQ200SK (33) to give pJQB. A SmaI-digested uidA2 fragment from pWM3 (28) was inserted into the SmaI site of nopB (pMSG2), and this plasmid was mobilized into NGR234 by triparental mating, using the helper plasmid pRK2013 (12). Marker exchange was selected on RMS plates containing 5% (wt/vol) sucrose. Putative nopB mutants (NGRnopB::uidA) were isolated and confirmed by Southern blotting of restricted genomic DNA according to standard procedures (36). For complementation, a HindIII-XbaI fragment of pXB110 (29) that contains nopB was cloned into pLAFR6 to give pMSG4. The resulting plasmid was mobilized into the nopB mutant by triparental mating, leading to the strain NGRnopB::uidA (pMSG4).
NopB antibody production.
Antibody production was conducted using two synthetic peptides selected from the NopB protein sequences AADSMKNDTASTPVR(C) and (C) DVHRAAPTSPLEDRV; the positions of these peptides are indicated in Fig. 3. Immunization of two rabbits with the coupled peptide mixture was performed according to established protocols (Eurogentec, Herstal, Belgium).
FIG. 3.
Amino acid alignment of NopB homologues. NopB of NGR234, NopB of USDA257, blr1812 of B. japonicum USDA110, and m1r8743 of M. loti MAFF303099 were aligned by using ClustalV. Amino acids common to all the NopB homologues are marked in red. The consensus sequence is shown in bold below the alignment, with the three most conserved domains highlighted in yellow. The positions of the peptides used for antibody production are shown in green above the alignment.
Purification and analysis of secreted proteins and extracellular appendages.
Total extracellular proteins were isolated from various NGR234 derivatives as described by Marie et al. (25). Purification of the extracellular appendages was performed as described previously (21). Proteins were separated by electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels (12 or 15% polyacrylamide) and were stained with silver (4). Concentrations of supernatant proteins were determined by the Bradford assay (Bio-Rad, Hercules, Calif.) with bovine serum albumin as the standard. For immunodetection, separated proteins were transferred to polyvinylidene difluoride (PVDF) Immobilon-P membranes (Millipore Corporation, Bedford, Mass.) and probed with 1:1,000 working dilutions of the NopA, NopB, NopL, NopP, and NopX antibodies or with antiflagellin at 1:5,000 in phosphate-buffered saline with 0.1% Tween 20 (PBS-T) (3, 5, 25). Protein-primary antibody binding was visualized by enhanced chemiluminescence, using horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G as the secondary antibody and the ECL detection system (Amersham Biosciences, Uppsala, Sweden).
2D gel electrophoresis.
Two-dimensional (2D) gel electrophoresis was performed essentially according to the protocols of Amersham Biosciences, but with some modifications. In brief, the protein samples (75 μg) were solubilized in 100 μl of a solution containing 5 M urea, 2 M thiourea, 2% (wt/vol) 3-(3-cholamidopropyl-dimethyl-ammonio)- 1-propanesulfonate (CHAPS), 5 mM tributylphosphine, and 0.5% IPG buffer (pH 3 to 10) (Amersham Biosciences). Isoelectric focusing (IEF) was conducted using Immobiline DryStrip precast gel strips (IPG) (180 mm, linear pH 3 to 10; Amersham Biosciences). The strips were rehydrated for 16 h with the protein samples in a solution containing 8 M urea, 2% (wt/vol) CHAPS, 0.2% (wt/vol) dithioerythritol (DTE), 0.5% IPG buffer (pH 3 to 10), 35 mM Tris, and 0.02% (wt/vol) bromophenol blue. IEF was performed in an IPGphor unit (Amersham Biosciences) for 100 kVh at 18°C. Following IEF, IPG strips were equilibrated for 12 min with shaking in a solution of 0.5 M Tris-HCl (pH 8.5) containing 6 M urea, 30% (wt/vol) glycerol, 2% (wt/vol) SDS, 0.02% (wt/vol) bromophenol blue, and 2% DTE. The strips were then equilibrated for 10 min in the same solution containing 2.5% iodoacetamide instead of DTE. The equilibrated IPG strips were then placed over SDS-12% polyacrylamide gels and covered with 0.5% (wt/vol) hot agarose solution containing 25 mM Tris-HCl, 192 mM glycine, 0.1% (wt/vol) SDS, and a trace amount of bromophenol blue. The gels were run in Bio-Rad Protean II XL cells at 20°C with 35 mA per gel.
Electron microscopy.
Methods for electron microscopy were essentially those described by Brown et al. (9). Carbon-Formvar-coated grids (300 mesh) were incubated with 20-μl drops of bacterial suspensions grown over night in YEM (with or without apigenin) and then adjusted to an optical density at 600 nm of 0.2. Grids were incubated at 28°C for 40 h and then fixed by the addition of 50 μl of 2% formaldehyde and 0.5% (vol/vol) glutaraldehyde in 50 mM sodium cacodylate buffer (pH 7.2) to the 20-μl droplet. Grids were left to fix for 30 min before being incubated on a fresh drop of fixative at 4°C overnight. Afterwards, the grids were washed with PBS-T by passing them through six 20-μl drops of cacodylate buffer. Then the grids were negatively stained in 1% phosphotungstate (pH 6.5) for 20 s and examined in a Hitachi H-7000 transmission electron microscope (TEM) (Hitachi Ltd., Tokyo, Japan) at an accelerating voltage of 75 kV. For in situ localization experiments, the fixed bacteria on the grids were incubated for 1 h with anti-NopB antibody or preimmune serum (both at a 1:100 dilution) at room temperature. The washing step was repeated, and the grids were then incubated with goat anti-rabbit immunoglobulin G secondary antibodies with 10-nm-diameter gold particles attached (British Biocell International, Cardiff, United Kingdom), diluted 1:50, for 1 h at room temperature. The grids were washed three times with PBS-T and dried for 3 min. The grids were then negatively stained in 1% phosphotungstate for 20 s and observed by TEM as before.
Plant material and assays.
Seed sources are listed in reference 32. Nodulation tests were performed in Magenta jars (Magenta Co., Chicago, Ill.) as described previously (41). Three independent (replica) experiments were performed. Nodules were counted 6 weeks postinoculation. Rhizobia were isolated from nodules following surface sterilization in 70% (vol/vol) ethanol for 10 min and then washed with sterile water three times before being sliced in half with a sterile razor blade. The open face of each nodule was streaked onto RMS agar containing rifampin and then incubated at 27°C for 3 to 5 days. Single colonies were then tested for their antibiotic resistance or sensitivity.
RESULTS
Identification of NopB.
Induced NGR234 cells secrete a number of extracellular proteins into the growth medium. By comparing the extracellular proteins from induced NGR234 and NGRΩrhcN cultures by one-dimensional SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 1A), at least six extra proteins that originate from NGR234 can be detected. Five of these proteins have been identified, NopA, NopC, NopL, NopP and NopX, as indicated by black arrows in Fig. 1A. (3, 25, 41; Deakin et al., unpublished). The sixth, a 16-kDa protein, to which we assigned the code name Nop16, was targeted for further study. To increase the resolution and sensitivity of detection, 2D electrophoresis was carried out (Fig. 1B). Most of the more than 40 TTSS-dependent spots that could be detected after silver staining were located in the acidic part of the gel.
FIG. 1.
Identification of NopB as a TTSS-dependent secreted protein (A) Extracellular proteins of apigenin-induced cultures of Rhizobium sp. strain NGR234 and NGRΩrhcN (a TTSS mutant) were electrophoretically separated by one-dimensional SDS-15% PAGE. Molecular masses of the marker proteins are indicated on the right. The identified Nops are named on the left. A 16-kDa protein labeled as Nop16 was unique to induced wild-type NGR234 and absent in NGRΩrhcN. (B) 2D gel electrophoresis of total extracellular proteins from apigenin-induced cultures of NGR234 (left panel) and NGRΩrhcN (right panel). Each sample contained 75 μg of protein and was IEF for a total of 100,000 Vh. The IPG strips were then placed on top of SDS-12% PAGE gels and after separation were stained with silver nitrate. Black circles indicate the (approximately 16-kDa) protein spots that were subjected to matrix-assisted laser desorption ionization-time of flight mass spectrometry.
Two protein spots originating from NGR234 with estimated molecular masses of 16 kDa (i.e., possibly Nop16) were seen with isoelectric points (pI) of approximately 6.1 and 6.3 (marked with black circles in Fig. 1B). The two spots were excised and digested with trypsin to generate peptide fragments. The peptide mixtures were subjected to matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis. The output masses (peptide mass fingerprints) were matched to the trypsin-digested theoretical peptide libraries generated from the Swiss-Prot and TrEMBL protein sequence databases. A match to the nodulation protein NolB was clearly found (Swiss-Prot entry P55713) with both spots, and the predicted molecular mass (16.8 kDa) and pI (6.27) of NolB are very similar to those of the Nop16 spots. For these reasons, Nop16 (and thus NolB) was renamed as NopB per Marie et al. (24). The reason that NopB appears as two spots is possibly due to posttranslational modification or to a general modification that occurred during sample preparation, e.g., carbamylation or oxidation of methionine residues, leading to change in the total net charges. Thus, two or more isoforms of the protein could occur with very similar molecular weights but different pIs.
Construction of a nonpolar nopB mutant.
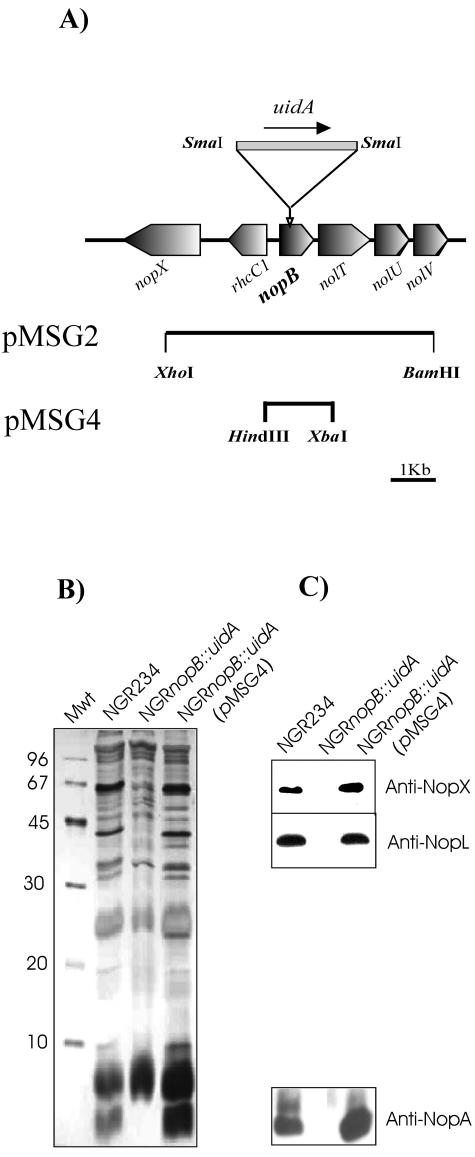
nopB is the first gene of a large operon containing a number of genes (Fig. 2A)that are crucial for the assembly and functioning of the TTSS machinery in NGR234 (41). To examine the role of NopB, a nonpolar mutation was constructed to avoid disturbance to downstream genes by inserting a “GUS gene” (uidA) that lacks transcriptional signals into nopB, creating NGRnopB::uidA, which was tested for Nop secretion (Fig. 2B). Extracellular protein profiles of induced cultures of NGRnopB::uidA were compared those of wild-type strains, and this showed that all TTSS-dependent protein secretion was abolished in the nopB mutant. To verify that protein secretion was abolished in the absence of NopB (not because of unexpected polar effects caused by the insertion), complementation of the mutant was carried out. A DNA fragment of pXB110 containing nopB and lacking genes downstream of nopB was subcloned (pMSG4) into a low-copy-number plasmid (pLAFR6) which was then mobilized into NGRnopB::uidA. Nop secretion was restored in NGRnopB::uidA(pMSG4) (Fig. 2B and C).
FIG. 2.
Construction and analysis of a nopB mutant. (A) Genetic organization of part of the TTSS locus of NGR234 showing the location of nopB. A 4-kb XhoI-BamHI fragment of pXB110 was cloned in pJQ200SK. A nonpolar mutant was constructed by inserting uidA (Gus gene), digested by SmaI, into the SmaI site of nopB (pMSG2). The black arrow shows the direction of the transcription of the Gus gene. pMSG4 is pLAFR6 containing a HindIII-XbaI fragment of pXB110 that was used to complement the nopB mutant. (B) Silver-stained SDS-15% PAGE of proteins isolated from the supernatants of apigenin-induced cultures of NGR234, NGRnopB::uidA, and NGRnopB::uidA (pMSG4). Bacterial cells were grown in RMS for 40 h. Molecular mass markers (Mwt) in kilodaltons are shown at the left. (C) In parallel, the samples were transferred to PVDF membranes, and immunological identification of the Nops was performed by probing the membranes with antibodies raised against NopX, NopL, and NopA.
The presence of different Nops was verified by Western blot analysis (Fig. 2C). Blots were probed with antibodies raised against NopA, NopL, and NopX (5, 25). Nops were detected among purified extracellular proteins of NGR234 and the complemented nopB strain [NGRnopB::uidA (pMSG4)], while the antibodies failed to detect any Nops secreted by the nopB mutant. Thus, it appears that TTSS-dependent protein secretion is abolished in the nopB mutant.
Symbiotic phenotype of the nopB mutant.
Because the nopB mutant blocks TTSS-dependent protein secretion, we suspected that the presence or absence of NopB would affect symbiotic development. Nodulation tests were performed with a number of plants representing different groups of legumes, which have been reported to show different responses to the presence or absence of Nops (Table 2). The NopB mutant exhibited three different symbiotic phenotypes highly similar to those seen with the previously characterized TTSS mutant NGRΩrhcN (41) On Pachyrhizus tuberosus, the absence of NopB (and hence of all the Nops) allows the plant to form a large number of nitrogen-fixing nodules. Conversely, with NGR234, few nodules are formed. On the other hand, the absence of NopB decreases the nodulation efficiency on Tephrosia vogelii to 50% compared to NGR234. Only small differences were observed in the number of nodules formed on Vigna unguiculata by the various strains.
TABLE 2.
The nopB mutant (NGRnopB::uidA) and the complemented strain modulate nodule number on certain legumesa
| Strain inoculated | No. of nodules on:
|
||
|---|---|---|---|
| V. unguiculata | T. vogelii | P. tuberosus | |
| NGR234 | 34.3 (±0.5; 16) | 15.5 (±0.7; 16) | 1.9 (±0.2; 20) |
| NGRΩrhcN | 33.4 (±0.6; 16) | 6.8 (±0.3; 16) | 29.3 (±0.6; 20) |
| NGRnopB::uidA | 27.3 (±0.4; 16) | 6.9 (±0.4; 16) | 28.9 (±0.6; 16) |
| NGRnopB::uidA(pMSG4) | 34.6 (±0.6; 16) | 13.0 (±0.6; 16) | 28.6 (±0.7; 16) |
Plant tests were performed in Magenta jars. For each test, the standard error of the mean and the total number of plants are shown in parentheses. Only nitrogen-fixing (Fix+) nodules were counted.
As expected, V. unguiculata and T. vogelii show similar phenotypes when inoculated with NGR234 or the complemented nopB mutant NGRnopB::uidA(pMSG4). Surprisingly, P. tuberosus that was inoculated with the complemented strain failed to restore the wild-type phenotype. Presumably, P. tuberosus exerts a strong negative selection pressure for TTSS mutants, thus selecting for the loss of pMSG4 from NGRnopB::uidA. Isolation of bacteria from nodules formed on plants initially inoculated with NGRnopB::uidA(pMSG4) confirmed the absence of pMSG4 (data not shown). There were no large differences in the numbers of nodules formed on plants that were inoculated with the nopB mutant strain carrying pLAFR6 (empty vector without nopB) compared with those inoculated with the nopB mutant (data not shown). Thus, the phenotype seen was due to the nopB mutant itself and not because of any effects from the vector.
Homologues of NopB in other rhizobia and production of antibodies.
BLAST searches of the protein databases revealed a few homologues to NopB: these were aligned using ClustalV (Fig. 3). However, all of them are restricted to rhizobial strains reported to have genes coding for type three secretion machines. In NGR234, NopB has 164 amino acids and shares 98% homology to NolB of USDA257. There is also 63% homology with mlr873 of Mesorhizobium loti MAFF303099 and 43% homology with blr1812 (NolB) of Bradyrhizobium japonicum USDA110. This initial bioinformatics analysis did not give any insight into a possible function of NopB, however, as all the NopB homologues are uncharacterized. Thus, we investigated in more detail the NopB sequence. Although NopB homologues show conservation throughout the protein, certain regions are more conserved (Fig. 3). There are three highly conserved peptides among the rhizobial NopB homologues, and each of these was used in further BLAST searches. Two of the peptides did not reveal any new homologous proteins, but interestingly, the conserved peptide at the carboxy (C) terminus shows significant homology to a flagellar hook-associated protein 1 (HAP1, which has been renamed FlgK) from Rhodopseudomonas palustris. FlgK proteins are associated (as minor components) with the flagella of many gram-negative bacteria, and flagella are constructed by an export and assembly pathway highly similar to that used for TTSSs (1). As is the case for TTSS-possessing phytopathogens, NGR234 also synthesizes surface appendages in a TTSS-dependent manner, predominantly composed of NopA (25; Deakin et al., unpublished). Mutations in genes encoding these external components of the type III secretion machinery generally block TTSS-dependent protein secretion. Thus, based upon the (nonsecreting) phenotype of the nopB mutant and the homology of part of NopB to FlgK, we propose that NopB might be an essential component of the TTSS-dependent surface appendages on NGR234.
To facilitate detection of NopB in NGR234 surface appendages, polyclonal antibodies were raised against synthesized peptides from NopB. The most immunogenic and accessible peptides were chosen for antibody production (the positions of the two peptides are indicated in Fig. 3). Anti-NopB sera were initially tested to detect NopB in purified extracellular proteins from various strains of NGR234 (Fig. 4). NopB was specifically detected as being secreted by NGR234, but not by NGRΩrhcN, confirming that NopB is indeed secreted by the TTSS of NGR234. Furthermore, NopB was not secreted by NGRnopB::uidA, but secretion was restored when the nopB mutant was complemented. Mutations in both nopL and nopP had been shown not to affect secretion of other Nops, and this was also the case with NopB.
FIG. 4.
Detection of NopB in NGR234 and derivatives. Extracellular proteins of induced NGR234, NGRΩrhcN, NGRΩnopL, NGRΔnopP, NGRnopB::uidA, and NGRnopB::uidA (pMSG4) were resolved by SDS-15% PAGE and electrophoretically blotted to PVDF membranes. Immunological detection of NopB was carried out using anti-NopB antibodies at a 1:1,000 dilution.
NopB is associated with surface appendages.
To test our hypothesis that NopB could be part of the TTSS-dependent surface appendages, total surface structures were isolated from apigenin-induced cultures of NGR234 and NGRnopB::uidA and analyzed by SDS-PAGE. The separated proteins were stained with silver nitrate or blotted onto PVDF membranes and then probed with antibodies raised against the Nops. Two different media were used to grow the bacteria, RMS and YEM. YEM was used essentially as a positive control for the extraction procedure, as it is known that flagella are synthesized in this media, whereas flagella are absent when NGR234 is grown in RMS.
The associated proteins with surface appendages isolated from induced NGR234 cells grown in RMS consist of three major proteins (Fig. 5), with molecular masses of 69, 18, and 7 kDa, that are absent from the nopB mutant. These proteins were identified as NopX, NopB, and NopA by using corresponding antibodies in Western blots. Isolation of NopA was expected, as it is thought to be the major component of TTSS pili (25; Deakin et al., unpublished). Similar results were obtained using YEM medium, although an approximately 33-kDa protein was observed in extracts from both the wild-type and the nopB mutant which was not seen when RMS was used (Fig. 5). This protein was identified as flagellin by probing the membrane with antiflagellin antibodies. As a negative control, we used NopL antibodies (NopL is a putative effector protein of NGR234 and thus not thought to be part of the TTSS machinery). NopL antibodies failed to detect NopL in both preparations. NopB was detected in the surface appendage preparations however, and for this reason we tried to directly localize NopB on TTSS-dependent extracellular structures.
FIG. 5.
Analysis of isolated surface appendages of NGR234. Isolation of the proteins associated with the NGR234 surface appendages was performed using two different media: RMS (left panel) and YEM (right panel). The proteins were separated by SDS-15% PAGE, and stained with silver nitrate. In parallel, the separated proteins were transferred to PVDF membranes and probed with antibodies against NopX, NopL, NopB, NopA, and flagellin. Lanes 1, isolated surface filaments from apigenin-induced cultures of NGR234; lanes 2, NGRnopB::uidA.
Visualization of the NGR234 surface appendages.
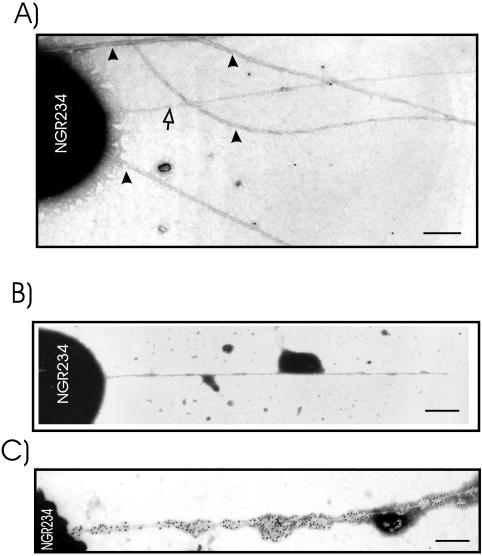
Recently, hrp pili were observed in the phytopathogenic bacterium P. syringae by using a novel electron microscopic technique (9). We applied this technique to NGR234, which was seen to produce a number of filamentous structures in the presence or absence of apigenin (Fig. 6). To identify the TTSS-dependent structures among these filaments, NGRΩrhcN was used as a negative control. As YEM was used, a number of flagella (indicated by arrowheads in Fig. 6) with a thickness of around 14 to 18 nm in diameter were detected in noninduced NGR234 (Fig. 6A), induced NGR234 (Fig. 6B), NGRΩrhcN (Fig. 6C), and NGRnopB::uidA (Fig. 6D). The two mutants appeared to produce more flagella than the wild type, however. In addition, induced NGR234 produces thin filaments with a thickness of around 6 to 8 nm, hereafter called TTSS pili, as they were not seen in either the rhcN or the nopB mutant. Some of them were short and attached to the cell (indicated by white arrows in Fig. 6B), whereas others were unattached to bacterial cells. A number of thin filaments were often observed binding together as a long bundle in NGR234, and in some cases in both NGRΩrhcN and NGRnopB::uidA. Such filamentous structures were not TTSS or flavonoid dependent, and thus NGR234 must have other genes encoding surface structures. DNA sequencing projects have revealed the presence of a conjugal transfer (tra) system on pNGR234a (13) as well as a region encoding a type four pilus on pNGR234b (37), which might be responsible for the extra surface appendages.
FIG. 6.
Electron micrographs of the NGR234 surface appendages. Transmission electron micrographs showing flagella, pili, and other filaments of negatively stained cells of noninduced cultures of NGR234 (A) and apigenin-induced cultures of NGR234 (B), NGRΩrhcN (C), and NGRnopB::uidA (D). The flagella are marked with arrowheads, while the pilus-like structures are marked with white arrows. The bacterial cells were grown in YEM on carbon-Formvar-coated gold grids at 28°C and stained with 1% phosphotungstate. Scale bar, 1 μm.
Immunolocalization of NopB on TTSS pili.
To unambiguously identify TTSS pili and to detect NopB within them, immunoelectron microscopy was performed, using methods developed by Brown et al. (9). NGR234 and derivatives attached to the electron microscopic grids were incubated with either preimmune serum or anti-NopB serum (both diluted 1:100) followed by gold-labeled secondary antibodies. A negligible number of gold particles scattered around some of the filamentous structures (non-TTSS pili and flagella) (Fig. 7A and B), whereas an extensive number of gold particles were seen around TTSS pili when incubation was with anti-NopB (Fig. 7C). The gold particles were randomly distributed along the pilus structures. Labeling of external structures of NGRΩrhcN or NGRnopB::uidA mutants was not observed (data not shown), implying that the gold labeling is specific to the TTSS pili. Thus, NopB is associated with TTSS pili and is a part of these structures.
FIG. 7.
Immunogold localization of NopB. TTSS pili were marked using NopB antibodies and secondary antibodies tagged with 10-nm gold particles. The NGR234 cells were grown in YEM containing apigenin on carbon-coated electron microscope gold grids for 40 h at 28°C. After this, they were incubated with preimmune serum (A and B) or anti-NopB antibodies (C). The grids were then incubated with goat anti-rabbit antibodies labeled with 10-nm gold particles (black particles) and then stained with 1% phosphotungstate. The flagella are marked with arrowheads, while the pilus-like structure is marked with a white arrow. Scale bar, 0.25 μm.
DISCUSSION
By using a proteomics-based approach, we have identified NopB as a flavonoid-inducible protein secreted by the TTSS of NGR234. This was confirmed by raising an antibody to peptides designed from the NopB sequence. NopB has no homology to any proteins of known function in sequence databases. All NopB homologues in rhizobia possess TTSSs. The USDA257 homologue, NolB, has 98% similarity to NopB from NGR234. A polar mutation in nolB was reported to abolish signal response (i.e., TTSS-dependent) protein secretion and to affect nodulation of certain soybean cultivars (22, 27). This secretion block could be due to polar effects on genes downstream of nolB as it located in an operon containing other genes shown to be required for protein secretion (19). As in USDA257, nopB of NGR234 is the first gene of a large operon that contains a number of genes essential for the TTSS machinery (24, 41). Thus, to investigate the role of NopB in the NGR234 TTSS, a nonpolar insertional mutation was made to avoid any disturbance to other genes. TTSS-dependent protein secretion was abolished in the nopB mutant, which was successfully complemented for protein secretion by mobilizing into it a plasmid containing nopB (and no downstream genes) under the control of its own promoter. Thus, NopB is required for the secretion of the NGR234 Nops. The symbiotic phenotype of the nopB mutant was assessed and found to be very similar to that of the previously characterized TTSS mutant NGRΩrhcN (41). Nodulation efficiency was reduced on T. vogelii and improved on P. tuberosus. It is possible that P. tuberosus perceives the Nops as being potentially pathogenic and thus generates a defense response against their source (NGR234) that blocks nodule formation. Conversely, on T. vogelii the Nops may function to suppress an induced plant defense reaction in a manner analogous to that of the effector proteins of phytopathogens (24, 41).
NopB could block secretion in two ways. The first would be by functioning as an activator of TTSS-related genes. This was discounted, as NopB shows no homology to any known transcriptional activator. The secretion of NopB itself also suggests that it does not play an intracellular role. Another possibility is that NopB could be part of the external component of the TTSS machinery, like the Hrp pili produced by the TTSS-possessing phytopathogens (15). When mutated, the genes encoding the main structural components of such pili also block TTSS-dependent protein secretion, as was seen with the nopB mutant. Further support for NopB being part of a TTSS-dependent external structure came from specific homology searches with the most conserved region of NopB. These showed that NopB has significant homology to FlgK, a protein associated with the bacterial flagellum, an external structure synthesized by a machine analogous to that of the TTSS of gram-negative pathogens.
It has been shown that two rhizobia, USDA257 and NGR234, produce in a TTSS-dependent manner, extracellular appendages (pili) when grown in the presence of flavonoids (21; Deakin et al., unpublished). The purified pili have been visualized in USDA257 by TEM (21). The genetic and biochemical analyses that were done on the surface appendages made by NGR234 suggest that the main pilus component is NopA (25; Deakin et al., unpublished). Polyacrylamide gel analysis of isolated surface appendages from genistein-induced USDA257 cells showed several abundant proteins, including NopX, Nop38, 25- and 18-kDa proteins, and Nop7. These were copurified or associated with the pilus structures and were not seen in similarly prepared extracts from TTSS mutants of USDA257. The presence of numerous proteins (one of which had approximately the same size as NopB) in the surface appendage preparations of USDA257 led us to investigate whether NopB might be a component of the TTSS pili produced by NGR234. By isolating the surface structures from induced NGR234 and the rhcN and nopB mutants, we were able to show that the TTSS pili are mainly composed of three different proteins: NopX, NopB, and NopA. The presence of NopA was expected from earlier studies. In NGR234, as in USDA257, NopX is also attached to surface appendages. In NGR234, NopX is thought to be a translocated protein and thus also an external component of the TTSS (25), which could explain its appearance in the extractions. The detection of NopB confirmed our hypothesis that it might be a component of the TTSS pili, and thus we attempted to visualize NopB in pili on the surface of NGR234 cells.
Initially, the surface appendages produced by NGR234 and its derivatives were detected in the presence or absence of flavonoid inducers. Bacteria were grown on electron microscopic grids to preserve the surface appendages (9). Mixtures of filamentous structures were observed on induced cells of NGR234. One of these was a pilus-like structure with a diameter of 6 to 8 nm that was absent from the nopB and rhcN mutants. All the strains, however, produce flagella and long thin filaments, even in the absence of apigenin (these could be type IV pili or even conjugative pili). It has been reported that NGR234 carries the genes encoding a type IV pilus on pNGR234b (36) and that pNGR234a has a conjugal transfer (tra) system (13). Type IV pili are structures on the bacterial surface that are found in many gram-negative bacteria, where they play important roles in adhesion to host cells, conjugation, and biofilm formation (10). Combining the techniques of growing the rhizobia on electron microscopic grids with immunogold labeling localized NopB to the TTSS pili. Labeling did not occur when preimmune serum was used. Perhaps NopB interacts with the main component of pili, NopA, to help form or even stabilize the pili. We will continue to focus on whether the effector Nops are secreted through these TTSS pili into leguminous root cells.
Acknowledgments
We thank Christian Staehelin (University of Geneva) for helpful comments and advice. We also thank Birgit Scharf (University of Regensberg, Regensberg, Germany) for her gift of the antiflagellin antibody. We are grateful for Yin -Yin Aung for her help with plant tests and Dora Gerber for her general support.
Financial assistance was provided by the Université de Genève and the Fonds National Suisse de la Recherche Scientifique (projects 31-63893.00 and 3100AO-104097/1).
REFERENCES
- 1.Aizawa, S. I. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157-164. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 1997. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179:5655-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausmees, N., H. Kobayashi, W. J. Deakin, C. Marie, H. B. Krishnan, W. J. Broughton, and X. Perret. 2004. Characterization of NopP, a type III secreted effector of Rhizobium sp. strain NGR234. J. Bacteriol. 186:4774-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, and J. Smith. 1991. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Bartsev, A. V., N. M. Boukli, W. J. Deakin, C. Staehelin, and W. Broughton. 2003. Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 554:271-274. [DOI] [PubMed] [Google Scholar]
- 6.Bartsev, A. V., W. J. Deakin, N. M. Boukli, C. B. McAlvin, G. Stacey, P. Malnoë, W. J. Broughton, and C. Staehelin. 2004. NopL, an effector protein of Rhizobium sp. NGR234, thwarts activation of plant defense reactions. Plant Physiol. 134:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broughton, W. J., S. Jabbouri, and X. Perret. 2000. Keys to symbiotic harmony. J. Bacteriol. 182:5641-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broughton, W. J., C.-H. Wong, A. Lewin, U. Samrey, H. Myint, H. Meyer z.A., D. N. Dowling, and R. Simon. 1986. Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J. Cell Biol. 102:1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, I. R., J. W. Mansfield, S. Taira, E. Roine, and M. Romantschuk. 2001. Immunocytochemical localization of HrpA and HrpZ supports a role for the Hrp pilus in the transfer of effector proteins from Pseudomonas syringae pv. tomato across the host plant cell wall. Mol. Plant-Microbe Interact. 14:394-404. [DOI] [PubMed] [Google Scholar]
- 10.Dorr, J., T. Hurek, and B. Reinhold-Hurek. 1998. Type IV pili are involved in plant-microbe and fungus-microbe interactions. Mol. Microbiol. 30:7-17. [DOI] [PubMed] [Google Scholar]
- 11.Ebel, F., T. Podzadel, M. Rohde, A. U. Kresse, S. Kramer, C. Deibel, C. A. Guzman, and T. Chakraborty. 1998. Initial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol. 30:147-161. [DOI] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freiberg, C., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 14.Ginocchio, C. C., S. B. Olmsted, C. L. Wells, and J. E. Galan. 1994. Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell 76:717-724. [DOI] [PubMed] [Google Scholar]
- 15.He, S. Y., and Q. Jin. 2003. The Hrp pilus: learning from flagella. Curr. Opin. Microbiol. 6:15-19. [DOI] [PubMed] [Google Scholar]
- 16.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, Q., W. Hu, I. Brown, G. McGhee, P. Hart, A. L. Jones, and S. Y. He. 2001. Visualization of secreted Hrp and Avr proteins along the Hrp pilus during type III secretion in Erwinia amylovora and Pseudomonas syringae. Mol. Microbiol. 40:1129-1139. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, H., Y. Naciri-Graven, W. J. Broughton, and X. Perret. 2004. Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Mol. Microbiol. 51:335-347. [DOI] [PubMed] [Google Scholar]
- 19.Kovacs, L. G., P. A. Balatti, H. B. Krishnan, and S. G. Pueppke. 1995. Transcriptional organization and expression of noIXWBTUV, a locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 17:923-933. [DOI] [PubMed] [Google Scholar]
- 20.Krause, A., A. Doerfel, and M. Gottfert. 2002. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 15:1228-1235. [DOI] [PubMed] [Google Scholar]
- 21.Krishnan, H. B., J. Lorio, W. S. Kim, G. Jiang, K. Y. Kim, M. DeBoer, and S. G. Pueppke. 2003. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant-Microbe Interact. 16:617-625. [DOI] [PubMed] [Google Scholar]
- 22.Krishnan, H. B., and S. G. Pueppke. 1993. Flavonoid inducers of nodulation genes stimulate Rhizobium fredii USDA257 to export proteins into the environment. Mol. Plant-Microbe Interact. 6:107-113. [DOI] [PubMed] [Google Scholar]
- 23.Lewin, A., E. Cervantes, C.-H. Wong, and W. J. Broughton. 1990. nodSU, two new nod genes of the broad host range Rhizobium strain NGR234 encode host-specific nodulation of the tropical tree Leucaena leucocephala. Mol. Plant-Microbe Interact. 3:317-326. [DOI] [PubMed] [Google Scholar]
- 24.Marie, C., W. J. Broughton, and W. J. Deakin. 2001. Rhizobium type III secretion systems: legume charmers or alarmers? Curr. Opin. Plant Biol. 4:336-342. [DOI] [PubMed] [Google Scholar]
- 25.Marie, C., W. J. Deakin, V. Viprey, J. Kopcinska, W. Golinowski, H. B. Krishnan, X. Perret, and W. J. Broughton. 2003. Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant-Microbe Interact. 16:743-751. [DOI] [PubMed] [Google Scholar]
- 26.Marie, C., W. J. Deakin, T. O. Reuhs, E. Diallo, B. Reuhs, W. Broughton, and X. Perret. 2004. TtsI, a key regulator of Rhizobium species NGR234, is required for type III-dependent protein secretion and synthesis of rhamnose-rich polysaccharides. Mol. Plant-Microbe Interact. 17:958-966. [DOI] [PubMed] [Google Scholar]
- 27.Meinhardt, L. W., H. B. Krishnan, P. A. Balatti, and S. G. Pueppke. 1993. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol. Microbiol. 9:17-29. [DOI] [PubMed] [Google Scholar]
- 28.Metcalf, W. W., and B. L. Wanner. 1993. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene 129:17-25. [DOI] [PubMed] [Google Scholar]
- 29.Perret, X., W. J. Broughton, and S. Brenner. 1991. Canonical ordered cosmid library of the symbiotic plasmid of Rhizobium species NGR234. Proc. Natl. Acad. Sci. USA 88:1923-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perret, X., C. Freiberg, A. Rosenthal, W. J. Broughton, and R. Fellay. 1999. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 32:415-425. [DOI] [PubMed] [Google Scholar]
- 31.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pueppke, S. G., and W. J. Broughton. 1999. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant-Microbe Interact. 12:293-318. [DOI] [PubMed] [Google Scholar]
- 33.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors, which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 34.Rélić, B., X. Perret, M. T. Estrada-Garcia, J. Kopcinska, W. Golinowski, H. B. Krishnan, S. G. Pueppke, and W. J. Broughton. 1994. Nod-factors of Rhizobium are a key to the legume door. Mol. Microbiol. 13:171-178. [DOI] [PubMed] [Google Scholar]
- 35.Roine, E., W. Wei, J. Yuan, E. L. Nurmiaho-Lassila, N. Kalkkinen, M. Romantschuk, and S. Y. He. 1997. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 94:3459-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Streit, W. R., R. A. Schmitz, X. Perret, C. Staehelin, W. J. Deakin, C. Raasch, H. Liesegang, and W. J. Broughton. 2004. An evolutionary hot spot: the pNGR234b replicon of Rhizobium sp. strain NGR234. J. Bacteriol. 186:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trinick, M. J. 1980. Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa spp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J. Appl. Bacteriol. 49:39-53. [Google Scholar]
- 39.Van Gijsegem, F., J. Vasse, J. C. Camus, M. Marenda, and C. Boucher. 2000. Ralstonia solanacearum produces hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol. Microbiol. 36:249-260. [DOI] [PubMed] [Google Scholar]
- 40.Vincent, J. M. 1970. A manual for the practical study of root-Nodule bacteria. Blackwell Scientific, Oxford, United Kingdom.
- 41.Viprey, V., A. Del Greco, W. Golinowski, W. J. Broughton, and X. Perret. 1998. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 28:1381-1389. [DOI] [PubMed] [Google Scholar]