Abstract
The sex pheromone plasmids in Enterococcus faecalis are one of the most efficient conjugative plasmid transfer systems known in bacteria. Plasmid transfer rates can reach or exceed 10−1 transconjugants per donor in vivo and under laboratory conditions. We report the completion of the DNA sequence of plasmid pCF10 and the analysis of the transcription profile of plasmid genes, relative to conjugative transfer ability following pheromone induction. These experiments employed a mini-microarray containing all 57 open reading frames of pCF10 and a set of selected chromosomal genes. A clear peak of transcription activity was observed 30 to 60 min after pheromone addition, with transcription subsiding 2 h after pheromone induction. The transcript activity correlated with the ability of donor cells to transfer pCF10 to recipient cells. Remarkably, aggregation substance (Asc10, encoded by the prgB gene) was present on the cell surface for a long period of time after pheromone-induced transcription of prgB and plasmid transfer ability had ceased. This observation could have relevance for the virulence of E. faecalis.
The advent of microarray technology allows for a comprehensive analysis of gene expression patterns associated with various biological processes, providing insights into complex regulatory networks. One of the most complex processes is the transfer of large portions of genetic material from a donor cell into a recipient cell by means of conjugation. The plasmids of the sex pheromone family in Enterococcus faecalis are among the most efficient bacterial conjugation systems known (16). The family consists of over 20 plasmids and shows extensive sequence homologies (28). E. faecalis strains can host several of these plasmids. This is exemplified by strain V583, the first vancomycin-resistant isolate in the United States (45), chosen for genome sequencing by The Institute for Genomic Research (TIGR; www.tigr.org). V583 contains two sex pheromone plasmids with homology to the well-characterized pAD1 (pTEF1) and pCF10 (pTEF2) plasmids, respectively. The complete sequences for the pheromone plasmids pAD1 and pAM373 became available recently (14, 19). Analysis of the sequences of this group of plasmids allows comparisons and insights into the evolution of these elements.
Although the sex pheromone plasmids can be disseminated among enterococcal populations very efficiently, plasmid transfer is highly regulated and only induced by recipient cells in close proximity to plasmid donors. The recipient cells secret 7- to 8-amino-acid-long hydrophobic sex pheromones that are bound by a plasmid-encoded binding protein (44, 51). The pheromone is then taken up into the cell (32) and releases a transcriptional block of the PrgX/TraA family of repressors (5). One of the early transcripts after induction encodes for the surface protein aggregation substance (AS) (9). Expression of AS results in tight physical contact between donor and recipient, allows for plasmid transfer rates of up to 10−1 transconjugants/donor (16), and is necessary for the characteristic aggregate formation.
Its highly efficient plasmid transfer and its unique regulation sparked interest in this group of plasmids. The plasmids can carry antibiotic resistance markers but also encode virulence factors, like cytolysin on plasmid pAD1 (23) and AS itself, which has been implicated as an adhesin in a variety of model systems (26, 40, 52). The AS of plasmid pCF10 is expressed in human plasma independent of the presence of the inducing pheromone cCF10 (27). These features only increase the concern that rapid spread of antibiotic resistance in enterococci could make these organisms harder to treat. Currently, enterococci are ranked third in nosocomial infections and are associated with considerable mortality (39).
The sequence information thus far available for plasmid pCF10 (25, 29, 41, 44) includes regions for the uptake of the pheromone, regulatory regions, and the AS gene prgB. Studies of the transcriptional response of the plasmid-encoded genes on sex pheromone plasmids have focused on the regions upstream of the respective AS genes. Regulation in the pAD1 system shows some differences, most notably the presence of an apparent in trans regulatory protein (TraE1) that is absent in the pCF10 system (34, 37). In pCF10, the transcriptional start site for the prgB transcript is 5 kb upstream of the gene start (9), in the prgQ locus, which encodes the iCF10 inhibitor peptide and several RNAs involved in regulation of expression of downstream genes. Although the prgQ promoter is very active in both induced and uninduced cells, the prgB transcript is exclusively seen after induction with the pheromone cCF10. The complex regulation of expression of prgB and other genes downstream from prgQ is a complex process that is controlled at both transcriptional and posttranscriptional levels by protein and RNA regulators. These include PrgX, which is the primary regulator of the prgQ promoter, and also the cytoplasmic receptor for pheromone cCF10; these regulatory mechanisms are described in much more detail in several previous publications (2-5, 30). In the present study we were especially interested in comparing the effects of pheromone induction on the transcriptional profile of all pCF10 genes to that of the prgQ-prgB region, which was analyzed previously. No change in transcripts in response to pheromone induction has been noted for the genes prgN, O, P, W, Z, and Y. The pair prgZ/Y forms presumably a transcriptional unit (7). The pcfG gene encoding the relaxase of pCF10 was recently characterized (49), but transcriptional analysis of this region of the plasmid has not been reported.
Here we present the completion of the sequence analysis of the 67.6-kb sex pheromone plasmid pCF10 (including the transposon Tn925). The obtained sequence was used to generate a mini-microarray containing all plasmid genes and three Tn925 genes. In addition, probes for several genomic open reading frames (ORFs) were included in the arrays. We demonstrate the kinetics of gene expression on pCF10 after induction with the pheromone cCF10. Gene expression reached a peak after 30 min to 1 h and subsided thereafter, returning to the uninduced state after 2 h. These results were also mirrored in the donor cells' ability to transfer pCF10 to recipients, which ceased after 4 h. In contrast to the plasmid transfer ability of the donor cells, the AS protein was still detected 8 h after the initial induction.
MATERIALS AND METHODS
DNA sequencing of pCF10.
Sequencing was performed at the Advanced Genetics Analysis Center (University of Minnesota) with automated sequencing using ABI 377 automated fragment analyzers. PCR primers were selected using the Primer3 program (www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) and the TIGR E. faecalis V583 pCF10-like plasmid (pTEF2) as a template when needed. Templates for the sequencing reactions were plasmid subclones of the EcoRI fragments B to H isolated earlier (44), which were purified from Escherichia coli DH5α using QIAGEN plasmid purification kits (QIAGEN, Valencia, Calif.). The Tn925-containing EcoRI fragment A showed instability in cloning attempts, and fragments for sequencing were therefore PCR amplified with standard procedures in an Eppendorf Mastercycler (Eppendorf, Inc., Newton, Conn.). All sequence data presented are the result of double-stranded sequencing with the exception of the Tn925 sequence, which was conducted for the most part as single strand only, because its sequence is nearly identical to that of Tn916 (17). All nucleotide changes in Tn925 relative to Tn916 were confirmed by double-strand sequencing. Sequence assembly was performed with the AssemblyLIGNprogram (Accelrys, San Diego, Calif.) with Tn916 and the pCF10 homologue pTEF2 as matrices. Sequence analysis was performed using the GCG package, and for similarity searches the National Center for Biotechnology Information BLAST engine was used as well as the TIGR comprehensive microbial database (tigrblast.tigr.org/cmr-BLAST/).
List of chosen genomic ORFs.
The chromosome-encoded ORFs included on the microarray are listed in Table 1. TIGR designations are given with the gene symbol if provided. The included genes containing precursors of pheromone peptides have been designated ccfA (1, 11), cpdA, and cobA, respectively. The putative gene function is given where sufficient homology is provided. (Several ORFs with no designated function were chosen; for these, only the TIGR designations are given.)
TABLE 1.
Representation of the chromosomal genes of E. faecalis represented on the microarraya
| TIGR designation | Gene symbol | Function |
|---|---|---|
| EF0201 | tufA | Translation elongation factor Tu |
| EF0221 | rplF | Ribosomal protein L6 |
| EF0224 | rpsE | Ribosomal protein S5 |
| EF0775 | ebsG | LTA biosynthesis (Hirt and Dunny, unpublished) |
| EF0782 | rpoN | Sigma 54 family |
| EF0909 | oppB2 | Oligopeptide permease, membrane protein |
| EF0912 | oppF2 | Oligopeptide permease, ATP-binding protein |
| EF0997 | ftsZ | Cell division |
| EF1045 | pfkA | 6-Phosphofructokinase |
| EF1046 | pykA | Pyruvate kinase |
| EF1727 | ebsA | LTA biosynthesis |
| EF1728 | ebsB | LTA biosynthesis |
| EF1730 | ebsC | LTA biosynthesis |
| EF1821 | fsrB | agrB homologue |
| EF1822 | fsrA | agrA homologue |
| EF1891 | LtrA homologue | |
| EF1974 | relA | GTP pyrophosphokinase |
| EF2076 | efaA | Endocarditis-specific antigen |
| EF2326 | LtrA homologue | |
| EF2403 | cpdA | Encodes cPD1 precursor |
| EF2496 | cobA | Encodes cOB1 precursor |
| EF3106 | oppA | Oligopeptide permease, peptide-binding protein |
| EF3108 | oppB1 | Oligopeptide permease, membrane protein |
| EF3109 | oppF1 | Oligopeptide permease, ATP-binding protein |
| EF3331 | ccfA | Encodes cCF10 precursor |
All sequences were obtained from the TIGR database (E. faecalis V583). Included on the array but not shown are the three Tn916 genes int, xis and tet(M) (17). Two oligopeptide permease gene clusters can be found in the V583 genome. Only one (oppB-F1) also includes an oppA determinant.
Construction of the pCF10 microarray.
The sequence of pCF10 served as template for the construction of the array. Genes are represented by PCR products optimized for 500 bp whenever possible. Primer pairs were selected using the program Primer3; sequences of these primers are available at www.mmicab.umn.edu/faculty/Dunny.html. PCR products were purified and subjected to a second round of PCR to reduce contamination with genomic DNA. CsCl-purified pCF10 was used as template, and genomic DNA was isolated according to standard procedures (54). The PCR products were spotted on poly-l-lysine-coated glass slides by using a Microgrid II robot (BioRobotics, Boston, Mass.). Every ORF was represented in five copies on the array.
RNA isolation, cDNA synthesis, and hybridization.
RNA was isolated at the indicated time points using the RNeasy kit (QIAGEN). The generation of cDNA by reverse transcription of purified bacterial RNA employed random hexamers as primers. Amino-allyl UTP was incorporated into cDNA by reverse transcription followed by chemical coupling with the dyes Cy5 and Cy3, and hybridized to the array for 16 h at 63°C. The detailed protocols for the labeling and hybridization procedures used in these studies are provided at the website http://www.microarrays.org/protocols.html.
Detection and data analysis.
Image capture of the hybridized arrays was accomplished with a Scanarray 5000 microarray scanner (GSI Lumonics, Watertown, Mass.). Two independent hybridizations were made for each time point, using independent RNA extractions. Fluorescence intensities were normalized based on the total intensity of fluorescence in the Cy3 and Cy5 channels. Data obtained from hybridizations were subjected to the following exclusions before being considered admissible for data analysis. Spots were checked for regularity and conformity before considered admissible. Spot intensity had to pass a threshold of 1.5 times the background spot intensity. The remaining data points formed an initial average with standard deviation. Data points that exceeded 1.5 times this standard deviation were eliminated, and a final average was determined from the remaining data points. The presence of five copies of the genes on the array assured that at least three data points formed the average in >95% of ORFs. Increases were considered significant if 1.5-fold induction was detected. For cluster analysis of expression profiles, Spotfire software was accessed through the University of Minnesota Supercomputing Institute (http://www.msi.umn.edu/).
Real-time PCR.
Several ORFs were chosen to confirm data obtained from the microarray analysis. Quantitative real-time PCR was performed on cDNA generated as described above and using gene-specific primers with the QuantiTect SYBR-Green PCR kit (QIAGEN) in a Bio-Rad I cycler. The sequences of the primers used for PCR, the amplification conditions, and the detailed results of these experiments can be accessed at the website www.mmicab.umn.edu/faculty/Dunny.html. Experiments were performed in triplicate.
Bacterial strains, media, and experimental conditions.
The E. faecalis strains OG1RF(pCF10) (15) and OG1SSp (16) served as donor and recipient, respectively. All experiments were performed in static cultures at 37°C in Todd-Hewitt broth (Difco, Detroit, Mich.). For the induction experiment, OG1RF(pCF10) cells were inoculated 1:20 in 100 ml of Todd-Hewitt broth and allowed to grow for 30 min. Pheromone cCF10 was then added at 100 pg/ml to the induced culture, and cell samples were taken at the indicated time points from both the induced and uninduced control cultures. The experiment was performed in duplicate. The donor culture was treated identically in the mating experiment; however, at the corresponding time points cells were removed and added to recipient cultures of OG1SSp at a ratio of 1 donor CFU/10 recipient CFU. Matings were allowed to proceed for 10 min, after which the mating mixture was briefly vortexed, serially diluted in 0.9% NaCl, and plated on selective agar plates containing rifampin (200 μg/ml) or streptomycin (1 mg/ml) and tetracycline (10 μg/ml) to select and enumerate donor and transconjugants, respectively. CFU were determined in triplicate. Plasmid transfer was expressed as transconjugants per donor (T/D).
To examine the presence of AS on the cell surface, donor cells were harvested at the indicated time points and sedimented, and cell surface extracts were prepared as described previously (21). Polyacrylamide gel electrophoresis and Western blot analysis followed established procedures (26).
RESULTS
pCF10 sequence.
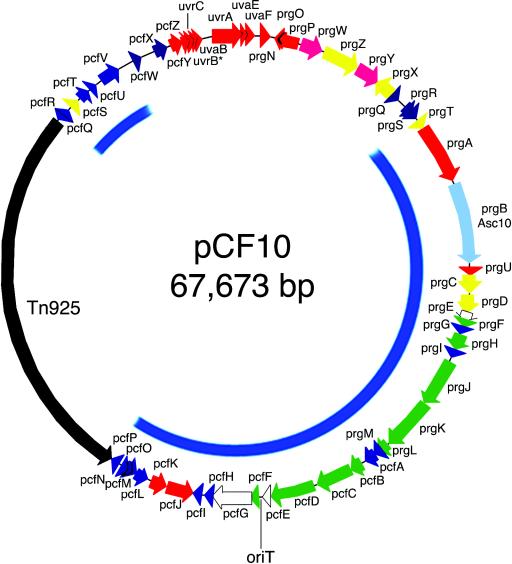
Plasmid pCF10 consists of 67,673 bp, with the transposon Tn925 accounting for 18,032 bp. The G of the unique SalI site was chosen as reference site (bp 1) and is located in the reading frame uvrA. All ORFs listed in Table 2 are numbered in relation to this site. Transposon Tn925 in pCF10 shows very high similarity with Tn916 (17). Sequence changes in Tn925 are most prominent in the tet(M) gene region. Tn925 spans from 40,068 to 58,100 bp and is integrated in counterclockwise orientation. The pCF10 ORFs described in Table 2 do not include Tn925 sequences. The nomenclature for these reading frames follows the previously established pattern of prg (pheromone-responsive gene) designations. Most remaining ORFs are denoted pcf (plasmid pCF10). The region around the SalI site contains ORFs showing homology to UV resistance determinants (42) that are designated uvr and uva. A summary of the homology of predicted pCF10 gene products to other proteins is presented in Fig. 1. Homology searches were updated for the previously sequenced region of pCF10 from prgN to prgC. Deduced amino acid sequences of PrgP and PrgO show the highest homology to products from ORFs of the recently described conjugative plasmid pRE25, a multiple resistance broad-host-range plasmid most similar to pIP501 and not a member of the pheromone plasmid family (47). PrgZ and PrgX show the highest homology in the pheromone family to TraC and TraA on pAM373, although the identity is rather limited due to the pheromone specificity of these two components. PrgT has also its closest homologue on pAM373, which is somewhat surprising, since pAM373 lacks a surface exclusion protein homologue. prgT is transcribed in an operon with prgA (7). The AS on pCF10 is identical to the AS on pTEF2 and shows near identity to an AS in a pathogenicity island that was recently described (48). A small ORF located between the prgB and prgC genes was not mentioned previously. It shows 98% sequence homology on the protein level with orf3 of plasmid pAD1 and 39% with pd125 and EP0040 of plasmids pPD1 and pAM373, respectively (14, 28). It is designated prgU. Interestingly, the degree of homology to gene products of the known pheromone plasmids pAD1, pPD1, and pAM373 diminishes downstream of prgU.
TABLE 2.
Genes encoded on plasmid pCF10a
| ORF | Gene | pCF10 location | Orientation | Gene/protein size (bp/AA) | Homology | Identity/similarity (%) | Function |
|---|---|---|---|---|---|---|---|
| 1 | uvrA | 66603-258 | cw | 1,392/443 | uvrA (pAD1) | 99 | UV resistance |
| 2 | uvaE | 255-605 | cw | 351/116 | orfB (pAD1) | 96/96 | |
| 3 | uvaF | 562-774 | cw | 213/70 | orfC (pAD1) | 94/97 | |
| 4 | prgN | 1207-1503 | cw | 297/98 | orfE (pAD1) | 91/97 | |
| 5 | prgO | 1935-1606 | ccw | 330/109 | orf57 (pRE25) | 81/90 | |
| 6 | prgP | 2788-1859 | ccw | 930/309 | orf58 (pRE25) | 79/90 | |
| 7 | prgW | 3035-4036 | cw | 1,002/333 | repA (pPD1) | 95/98 | Replication |
| 8 | prgZ | 4195-5832 | cw | 1,638/545 | traC (pAM373) | 38/59 | Pheromone receptor |
| 9 | prgY | 5843-6997 | cw | 1,155/384 | traB (pPD1) | 77/91 | Prevention of self-induction |
| 10 | prgX | 7983-7030 | ccw | 954/317 | traA (pAM373) | 25/45 | Negative regulator |
| 11 | prgQ | 8192-8263 | cw | 72/23 | iCF10 | ||
| 12 | prgR | 8721-9125 | cw | 405/134 | |||
| 13 | prgS | 9125-9397 | cw | 273/90 | |||
| 14 | prgT | 9699-9887 | cw | 189/62 | EP0043 (pAM373) | 69/85 | |
| 15 | prgA* | 10015-12690 | cw | 2,677/891 | sea 1 (pAD1) | 82/90 | Surface exclusion |
| 16 | prgB* | 12883-16800 | cw | 3,918/1,305 | EF0005 (PI, E. faecalis) | 96/97 | AS |
| 17 | prgU* | 16862-17218 | cw | 357/118 | orf3 (pAD1) | 98/100 | |
| 18 | prgC* | 17246-18103 | cw | 859/285 | EP0037 (pAM373) | 63/74 | |
| 19 | prgD* | 18145-19044 | cw | 900/299 | EP0038 (pAM373) | 25/47 | |
| 20 | prgE* | 19064-19498 | cw | 435/144 | BK5-T (L. lactis) | 37/58 | ssb |
| 21 | prgF* | 19545-19772 | cw | 228/75 | gbs1142 | 26/54 | |
| 22 | prgG* | 19786-20082 | cw | 297/98 | EFB0016 (pTEF2) | 100 | |
| 23 | prgH* | 20097-20897 | cw | 801/206 | gbs1362 | 33/55 | |
| gbs140 | 29/52 | ||||||
| 24 | prgI* | 20899-21252 | cw | 354/117 | EFB0018 (pTEF2) | 100 | |
| 25 | prgJ* | 21359-23596 | cw | 2,238/745 | gbs1135 | 41/64 | |
| 26 | prgK* | 23608-26223 | cw | 2,616/871 | gbs1359 | 35/50 | |
| gbs1133 | 31/47 | ||||||
| 27 | prgL* | 26247-26873 | cw | 627/208 | gbs1132 | 41/60 (129 aa) | |
| 28 | prgM* | 26860-27105 | cw | 246/81 | EFB0022 (pTEF2) | 98/100 | |
| 29 | pcfA* | 27089-27697 | cw | 609/202 | EFB0023 (pTEF2) | 97/98 | |
| 30 | pcfB* | 27856-28341 | cw | 487/161 | gbs1129 | 27/55 | |
| 31 | pcfC* | 28341-30170 | cw | 1,830/609 | gbs1128 | 44/64 | TrsK-like |
| 32 | pcfD* | 30221-32380 | cw | 2,160/719 | gbs1126 | 34/54 | |
| ltrC (pMRC01) | 31/49 | ||||||
| 33 | pcfE* | 32413-32685 | cw | 273/90 | ltrD (pRS01) | 36/56 | |
| 34 | pcfF* | 32931-33287 | cw | 357/118 | gbs1122 | 30/61 | |
| 35 | pcfG* | 33288-34973 | cw | 1,686/561 | ltrB (pRS01) | 50/68 | Relaxase |
| gbs1121 | 38/61 | ||||||
| 36 | pcfH* | 35006-35356 | cw | 351/116 | EFB0031 (pTEF2) | 100 (72 aa) | |
| 37 | pcfI* | 35680-35847 | cw | 168/55 | EFB0032 (pTEF2) | 100 | |
| 38 | pcfJ* | 37210-35873 | ccw | 1,338/445 | ORF63 (pAD1) | 81/90 | |
| 39 | pcfK* | 37983-37207 | ccw | 777/258 | ORF62 (pAD1) | 91/94 | |
| 40 | pcfL* | 38674-38126 | ccw | 549/182 | EFB0035 (pTEF2) | 100 | |
| 41 | pcfM* | 38889-38698 | ccw | 192/63 | EFB0036 (pTEF2) | 90/90 | |
| 42 | pcfN* | 39059-38874 | ccw | 186/61 | EFB0037 (TEF2) | 100 | |
| 43 | pcfO* | 39514-39065 | ccw | 450/149 | EFB0038 (pTEF2) | 100/100 | |
| pcfO′ | 39619-39701 | ccw | 84/28 | orf60 (pAD1) | 67** | ||
| 39881-39075 | 195/65 | ||||||
| 44 | pcfP* | 39801-40001 | cw | 201/66 | EFB0040 (pTEF2) | 100 | |
| 45 | pcfQ* | 58372-58142 | ccw | 231/76 | EFB0041 (pTEF2) | 100 | |
| 46 | pcfR* | 58427-58921 | cw | 495/164 | EFB0042 (pTEF2) | 99/100 | |
| 47 | pcfS* | 58987-59439 | cw | 453/150 | EP0029 (pAM373) | 65/76 | ssb |
| 48 | pcfT* | 59617-60195 | cw | 579/192 | EF0045 (pTEF2) | 91/91 | Thermonuclease |
| 49 | pcfU* | 60277-60606 | cw | 330/109 | EFB0046 (pTEF2) | 97/100 | |
| 50 | pcfV* | 60829-61854 | cw | 1,026/341 | EFB0047 (pTEF2) | 100 (pTEF2: 486 aa) | |
| 51 | pcfW | 62776-63048 | cw | 273/90 | |||
| 52 | pcfX | 63710-64369 | cw | 660/219 | BT2225 Bacteroides thetaiotaomicron | 31/54 | |
| 53 | pcfY | 64451-65071 | cw | 621/206 | orf86 (pAD1) | 95/98 | DNA invertase |
| 54 | pcfZ | 65079-65375 | cw | 297/98 | EFA0074 (pTEF1) | 96/97 | |
| 55 | uvrC | 65369-65593 | cw | 225/74 | uvrC (pAD1) | 95/98 | |
| 56 | uvrB | 65654-65863 | cw | 183/60 | uvrB (pAD1) | 94/100** | |
| 57 | uvaB | 65875-66048 | cw | 174/57 | EFA0076 (pTEF1) | 94/100 (truncated) |
Identity/similarity refers to the protein products of the respective ORF. Due to the high similarity with plasmid pTEF2, the highest scoring sequences currently included in the NCBI database other than pTEF2 are given for the location. Reading frames with a closest homologue on pTEF2 are indicated (*). For previously published pCF10 sequences (prgN-prgC), the closest homologue is listed. All plasmids with the exception of pRS01 and pMRC01 (L. lactis) are E. faecalis plasmids. AA, amino acids, gbs, S. agalactiae NEM316; PI, pathogenicity island; ssb, single-strand binding protein; cw, clockwise; ccw, counterclockwise. For findings marked with **, see text for details. For identity/similarity, the number of amino acids (aa) in the homolog is given in parentheses if it differs significantly from the pCF10 open reading frame sequence.
FIG. 1.
Physical map of plasmid pCF10. Putative ORFs and their orientations are shown. The colors indicate the closest homology of the protein products to E. faecalis plasmids pAD1 (red), pAM373 (yellow), pPD1 (magenta), pRE25 (orange), and pTEF2 (blue). ORFs found only on pCF10 are purple. Regions with homology to chromosomal regions of S. agalactiae NEM316 are indicated in green, and similarities to L. lactis are shown in white. The blue inner circle indicates the region of 95 to 97% homology on the nucleotide level between the plasmids pCF10 and pTEF2.
The plasmid pTEF2 found in the vancomycin-resistant clinical isolate V583 shows sequence homology to pCF10 in the region from bp 8,288 to 40,063 and bp 58,158 to 62,489, with a similarity on the DNA sequence level between 95 and 97%. Consequently, ORFs in those regions scored the highest homologies in comparison to pCF10. Most notable is the fact that despite near identity in the region encoding the two surface proteins PrgA and PrgB, the proteins in the regulatory region of the plasmids pCF10 and pTEF2 are significantly different. The location of the PrgZ (65% identity) and PrgY (52% identity) homologues in pTEF2 is reversed and resembles the arrangement in plasmids pAD1 and pPD1 (28, 55). A putative regulatory protein and PrgX analogue is located in counterclockwise orientation relative to the other regulatory elements and is consistent with the arrangement in all other pheromone-responsive plasmids. This protein, however, shows only an identity of 28% to PrgX. The limited identity of the pTEF2-encoded PrgZ and PrgX homologues—the two major pheromone-interacting proteins—explains why we found that cCF10 does not trigger a clumping response in E. faecalis V583 (data not shown). Due to the near identity of pCF10 with pTEF2 in the AS downstream region, the listed similarity of pCF10 gene products in Table 2 is given to the next-closest-related gene product other than those encoded by pTEF2. While PrgC protein shares 63 and 62% identity with the products from pAM373 and pAD1, respectively (14, 19), PrgD shows only 29 and 28% homology with its counterpart from pAM373 and pAD1, respectively. Previous DNA hybridization studies had already shown that there is a considerable difference in the regions downstream of AS in pAD1 and pCF10; in fact, pAM373 appeared to show closer homology to pAD1 (28). This was confirmed by the pCF10 sequence determination. Surprisingly, the region following prgD, including the ORFs prgE to pcfH (total of 17 ORFs, ∼16.3 kb) shows homologies between 26% (prgF) and 44% (pcfC) to the two regions gbs1142 to gbs1121 and gbs1360 to gbs1338, respectively, in the genome of Streptococcus agalactiae NEM316, which are presumed to represent plasmids that have integrated into the chromosome (24).
The products of the reading frames pcfD, -E, and -G show homology to LtrC, -D, and -B, respectively (33). PcfG shows 51% homology to LtrB, the relaxase of the Lactococcus lactis conjugative plasmid pRS01. The presence of this type of relaxase on pCF10 differs remarkably from the plasmids pAD1 and pAM373, where no such enzyme was found (14, 19). Instead, an ORF not present on pCF10 has been found to encode the pAD1-specific relaxase (18). The noncoding pCF10 sequence between pcfE and pcfF contains inverted and direct repeat motifs characteristic of functional oriT regions of other mobile plasmids (J. H. Staddon and G. M. Dunny, unpublished data). Our genetic analysis of this region, which will be described in a separate publication (Staddon and Dunny, unpublished), indicates that this region does contain the functional pCF10 oriT.
The reading frames pcfO to pcfI encoded in counterclockwise orientation on the left flank of the Tn925 insertion show homologies of between 71 and 91% to the ORFs 60 to 63 on pAD1. The reading frame prgO is present on both pCF10 and pTEF2; however, the surrounding sequences framing prgO show homology to the reading frame orf60 on plasmid pAD1. The sequence analysis suggests that, in a plasmid that was an ancestor of pCF10, a frameshift mutation in orf60 split this gene into pcfO and pcfO′, which are probably nonfunctional. Tn925 is inserted between pcfP and pcfQ. No reading frame was disrupted by the transposon insertion. The reading frames downstream of the transposon insertion show some homologies to known proteins. PcfS shows a 68% homology to ORF48 of pAD1, a putative single-strand DNA binding protein. PcfT shows the highest homology to EF0031, a reading frame described on a chromosomal pathogenicity island in E. faecalis (48) with putative nuclease activity. Regions surrounding the SalI site show high homology to pAD1 and the ORFs 86, 87 uvrC, uvrA, ORFB, and -C, with homologies at the protein level of >94% (19, 42). The reading frame uvrB is eliminated in pCF10 by the insertion of an adenine on position 4. The insertion creates a new potential reading frame encoding a hypothetical protein of 82 amino acids, present only on the sister plasmid pTEF2.
Phylogenetic analysis.
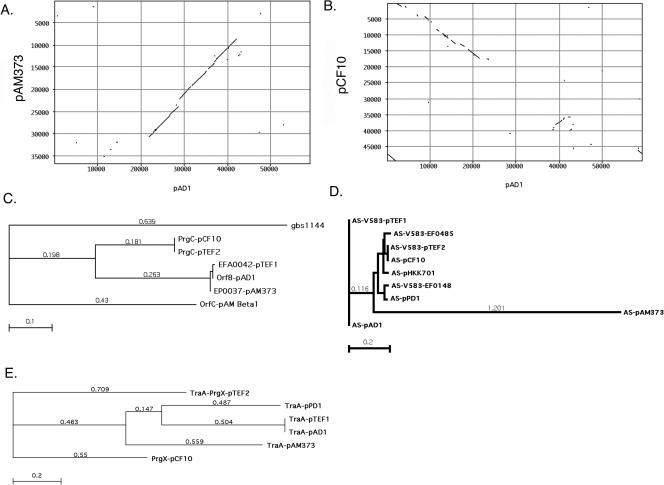
The completed sequence of pCF10 allows a closer look at the phylogenetic relationship among the five pheromone-responsive plasmids sequenced thus far. As described above, pCF10 and pTEF2 are closely related, with identical AS and PrgC (see below) proteins and extensive sequence homologies downstream of AS (Fig. 2C and D and 1). The plasmids pAD1 and pTEF1 share identical AS, TraA, and iAD1 components (Fig. 2D and E) and stretches of close similarity comparable to the similarity between pCF10 and pTEF2. However, pTEF1 has undergone a major rearrangement, and SE, AS, and downstream regions are inverted and separated from the regulatory region containing traA and iad. Somewhat surprisingly, pAM373 shows a closer relationship to pAD1 than pCF10 (Fig. 2A and B), despite the lack of a surface exclusion protein and a different class of AS (Fig. 2D). This closer relationship of pAM373 to pAD1 is exemplified by the protein homologues of TraA (Fig. 2E) and PrgC on pCF10 that are both more closely related to pAD1 than to their pCF10 counterpart. PrgC is a putative surface protein potentially involved in conjugative plasmid transfer with a homologue also present on the conjugative nonpheromone plasmid pAMβ1 (Fig. 2C). The TraA components in pAD1 and pTEF1 are identical, as indicated above, but the TraA homologue PrgX on pCF10 is only distantly related to its homologue on pTEF2. The pTEF2 version of this negative regulator is, however, still the closest relative to PrgX. (Fig. 2E). The AS proteins of the different pheromone plasmids and AS homologues on the chromosome of V583 show a closer phylogenetic relationship (Fig. 2D) than the TraA/PrgX and PrgC proteins, respectively.
FIG. 2.
Phylogenetic analysis of pheromone plasmids and selected plasmid-encoded proteins. (A and B) Pustell DNA matrix comparing pAD1 with pAM373 (A) or pCF10 (B). pAM373 is more closely related to pAD1 than pCF10, despite the lack of a surface exclusion protein and a nonhomologous AS. Tn925 was omitted from pCF10 for the purpose of this comparison. (C) Comparison of the prgC gene product and its homologues situated downstream of AS. (D) Comparison of described AS on pheromone plasmids and the chromosome of E. faecalis V583. The AS of pCF10/pTEF2 and pAD1/pTEF1 are identical. (E) Comparison of the pheromone plasmid negative regulator family TraA/PrgX. TraA of the plasmids pAD1/pTEF1 is identical, in contrast to PrgX on pCF10 and pTEF2. Methods used include neighbor joining and best tree; distance was determined by Poisson correction, and gaps are distributed proportionally.
Effect of pheromone induction on the transcription pattern of pCF10 genes.
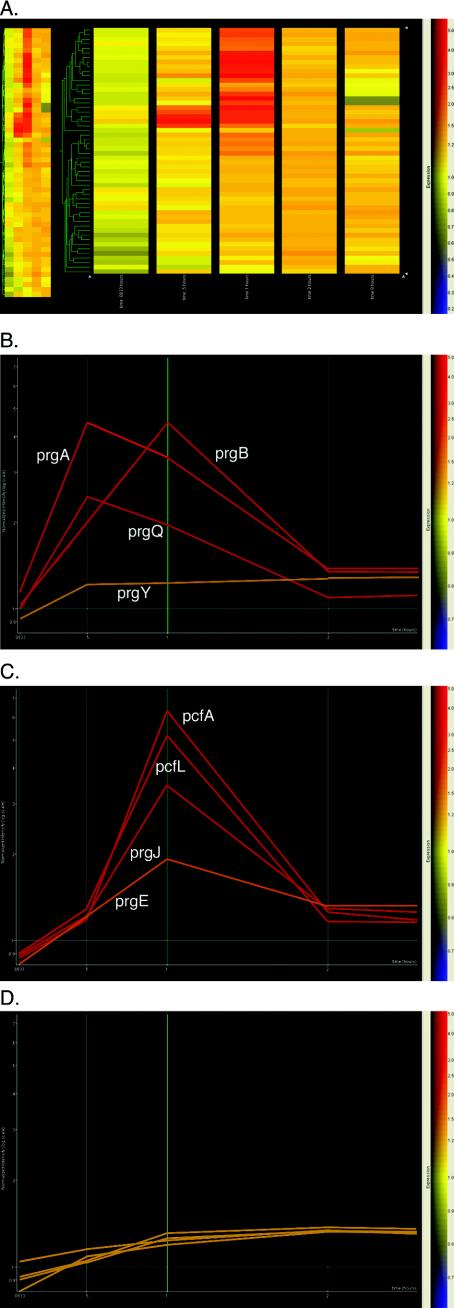
The completion of the pCF10 sequence enabled us to construct a miniarray containing all pCF10 ORFs (excluding Tn925) and to characterize for the first time all transcriptional events that occur after cCF10 induction. We chose an inducing concentration of cCF10 that lies about 10-fold over the concentration of cCF10 found in the supernatant of a recipient culture (38). Cluster analysis of the overall expression profile is shown in Fig. 3A,while graphical depictions of expression profiles of three classes of genes are shown in Fig. 3B to D. The temporal analysis of the data showed a gradual increase in transcripts from genes behind the prgQ promoter, with the highest level of induction seen in prgA and prgR 30 min after induction, with 4.5- and 3-fold over background levels, respectively. The level of transcription in that region then dropped off, and prgA was only 2.5-fold induced over the background level after 1 h. The prgB transcript showed a higher induction than prgA at this time point (Fig. 3B). At the 1-h time point after induction, transcript abundance had shifted to another region of pCF10, and the reading frames pcfA and prgL showed the highest level of induction (Fig. 3C), with about threefold induction over background. Two hours after induction, all transcription activity returned to background levels (Fig. 3A).
FIG.3.
Spotfire analysis of cCF10 induction of plasmid pCF10 in E. faecalis. (A) Gene expression at 15, 30, 60, 120, and 480 min is shown in comparison to that in the uninduced control after induction of strain OG1RF(pCF10) with 100 pg of cCF10/ml. The color bar on the right indicates expression of background. An increase of 1.5-fold is considered significant. (B) Expression profile of the reading frames prgY, prgQ, prgA, and prgB. prgQ and prgA showed their highest expression 30 min after induction and prgB peaked at the 60-min time point, while prgY as a gene in the negative regulatory region remained unaltered during the time course of cCF10 induction. (C) Expression profile of reading frames prgE, prgJ, prgL, and pcfA. The prgB downstream reading frames showed no significant expression at the 30-min time point and reached their peak at 60 min, with prgL and pcfA at higher expression levels than prgB at that time point. The expression of all selected reading frames returned to uninduced levels at the 120-min time point. (D) No significant induction of transcription can be seen in the Tn925 upstream region and the reading frames pcfS, pcfT, pcfY, and pcfZ.
Not all regions of the plasmid showed increased transcriptional activity during the time course of the experiment. Previously obtained data indicated that there is no change in the transcripts for prgN, O, P, W, Z, and Y during pheromone induction (7). These results were confirmed with our analysis (Fig. 3B, prgY). As expected, no activity was detected upon pheromone induction in the region encoding UV resistance determinants. Three genes from Tn925, xis, int, and tet(M), showed no transcriptional activity during pheromone induction. Notably, the regions on pCF10 downstream of the Tn925 insertion did not show significant induction (Fig. 3D). The pcfO to pcfJ genes showed low-level induction. pcfL and pcfJ reached the highest levels of expression of the genes in this region, although if the significant threshold of 1.5-fold induction is considered, no induction was observed. The results obtained by microarray analysis were confirmed by real-time quantitative reverse transcription-PCR with primers for prgX, prgQ, prgA to -C, prgL, pcfA, -C, -D, and -G, and pcfV to -X and for the chromosomal genes ebsG and rplF. Data obtained from this analysis confirmed the conclusions from the microarray analysis (data not shown).
Pheromone induction had no pronounced effect on expression of the chromosomal genes analyzed.
Effect of induction on plasmid transfer and AS.
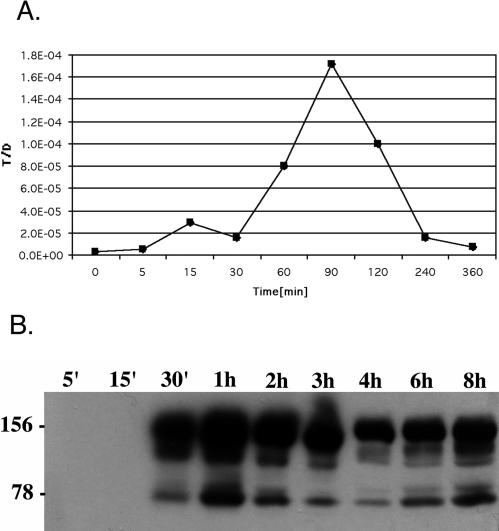
Most previously published pCF10 transfer experiments have been conducted with a 1,000-fold excess of cCF10 (10 ng/ml) compared to recipient supernatant concentration and resulted in T/D rates between 10−2 and 10−1. We decided to examine expression profiles in conjunction with plasmid transfer frequency following induction with a lower level of cCF10 that more closely resembles the pheromone concentrations excreted into the growth medium of recipient cultures. The ability to transfer pCF10 followed the pattern observed in the transcriptional analysis, with a clear peak activity 1.5 h after induction. The plasmid transfer activity diminished thereafter and showed only minimal and nonsignificant levels over background 4 h after induction. Six hours after the induction no significant plasmid transfer activity was detected. Transfer frequencies reached 10−4 at the peak (Fig. 4A).
FIG. 4.
Phenotypic effects of pheromone induction. (A) Plasmid transfer. Donor OG1RF(pCF10) was induced with 100 pg of cCF10/ml. At the indicated time points, donor cells were added to the recipient OG1SSp. After a 10-min mating, donor and transconjugants were enumerated as described in Materials and Methods. The graph represents the T/D frequency. (B) AS expression. OG1RF(pCF10) was induced with 100 pg of cCF10/ml. Surface extracts were prepared at the indicated time points. Protein extracts were separated on an 8% polyacrylamide gel electrophoresis gel, and AS was detected by Western blot analysis with AS-specific antibodies. Equivalent amounts of protein were loaded in each lane. The 156- and 78-kDa markers represent the full-length and amino-terminal segment of AS.
In contrast to the diminishing ability of the donors to transfer pCF10 to recipients, AS was readily detected on the cell surface even 8 h after the initial induction with cCF10. Although the amount of protein isolated from the cells reached a peak 1 h after induction and decreased slightly, a plateau was reached after 4 h and the amount of AS stayed essentially constant (Fig. 4B). The stability of the AS protein is therefore distinct from the components of the plasmid transfer apparatus that appear to be relatively quickly degraded, based on the loss of transfer ability. The characteristic clumping response seen after induction with higher pheromone concentrations was not observed in these cultures, consistent with previous results suggesting higher levels of pheromone are required to produce enough AS to mediate visible clumping.
DISCUSSION
The sequence of the pheromone plasmid pCF10 is the fifth completed pheromone-inducible plasmid sequence available. Plasmid pAM373 claims a unique place in the sex pheromone family, most notably by the lack of a surface exclusion function (13) and with an AS that has no homology to the AS of the rest of the family (13, 14, 35). Its cognitive pheromone is also the only one known thus far to be produced by another bacterial species, Staphylococcus aureus, which is of considerable concern (12, 36). The homologies between the pAD1, pCF10, and pPD1 regulatory regions and AS genes have been described elsewhere (28).
Previous data obtained by DNA hybridizations from pAD1 AS downstream regions had suggested that the plasmids pAD1 and pCF10 might not be as closely related, despite 90% DNA sequence homology for surface exclusion and AS genes. In fact, pAM373 had more ORFs in common with pAD1 than with pCF10 (28). A comparison of the sequence of the plasmids confirmed this earlier observation. A large region of pCF10 downstream of the prgB gene interestingly shows homology to a region of the genome of S. agalactiae strain NEM316. This region is hypothesized to originate from an integration event of a conjugative plasmid into the chromosome (24). A second region on the S. agalactiae chromosome shows a similar degree of homology. Only prgK in that pCF10 region showed homology to a reading frame on pAD1, ORF41 (38%), previously designated orf9 and -10, of which the probe for orf10 showed hybridization with pCF10 (28). Confirming the closer-than-expected similarity between pAD1 and pAM373, a gene encoding a relaxase of the type found on other conjugative plasmids was not identified on the pAD1 and pAM373 plasmids. Instead, pAD1 and pAM373 appear to encode a new class of relaxase (12). DNA relaxases cleave at a specific nic site in the origin of transfer and are therefore necessary for the initiation of the DNA strand transfer. In contrast, pCF10 encodes a common class relaxase in the form of pcfG, with the highest homology to LtrB, the group II intron-interrupted relaxase of the L. lactis conjugative plasmid pRS01 (33). No intron is found in pcfG; however, it was shown recently that the lactococcal intron in LtrB can target pcfG of pCF10 and insert in a conserved target site in pcfG (49). The remaining regions of the plasmid show high homology to some ORFs on pAD1 with putative functions that include single-strand binding proteins, a nuclease, and a DNA invertase (pcfS, pcfT, and pcfY, respectively). The putative nuclease is not found on pAD1 but on an E. faecalis pathogenicity island (48). The region encoding the UV resistance determinants is basically identical between pCF10 and pAD1, with the exception of the uvrB reading frame. An insertion at the fourth base of the reading frame creates a new protein, a situation also found on plasmid pTEF2. Exposure to UV light did not show increased sensitivity of OG1RF:pCF10 in comparison to OG1RF:pAD1 (data not shown).
The regions of homology illustrate the different origins of the plasmid transfer genes. There was obviously a common ancestor of the regulatory regions of the pheromone plasmids, as the homologies between pCF10, pAD1, pPD1, and even pAM373 show. Also very well conserved among the pheromone plasmids (with the notable exception of pAM373) is the cell contact function exemplified by the short evolutionary distance between the AS of the different plasmids. However, the diversity in response to different pheromones apparently led to significant variation in the PrgX/TraA negative regulators. The regions encoding putative DNA transfer functions are also arranged differently, perhaps most apparent in the different classes of relaxases employed by the plasmids. Large regions of pCF10 presumably involved in plasmid transfer are more closely related to plasmids from other gram-positive species. Most intriguing is the homology to portions of the S. agalactiae NEM316 genome that presumably derived from integrated plasmids (24). This represents a fine illustration of the importance and widespread occurrence of horizontal gene transfer within gram-positive bacteria. Taken together, our comparative analysis of the sequences of pheromone plasmids suggests a highly modular structure where gene clusters encoding pheromone-inducible aggregation appear to have assorted DNA processing and DNA transfer modules from a variety of evolutionary sources.
Induction of the pheromone response confirmed data obtained in a conventional manner previously (2, 6, 9, 10). The expression of prgQ increased steadily until the 1-h time point. The amount of prgA and prgB transcript increased over time, reached a peak, and declined to uninduced levels after 2 h. All ORFs downstream of prgQ in clockwise direction up to pcfG showed significant transcriptional activity compared to noninduced cells. It remains to be determined if this activity is due to read-through from the prgQ promoter and one long transcript or if several transcriptional units exist downstream of the prgB gene. The two ORFs pcfL and -J counterclockwise and adjacent to this region also showed minor induction, which could indicate the presence of a trans-acting factor(s). The induction of pcfL and pcfJ was below the threshold that normally is considered significant in microarray experiments (log2 of 0.37 and 0.48, respectively), yet several genes that later showed significant induction showed similar values 5 or 15 min following cCF10 induction. The evaluation of significant induction of genes has proven to be difficult due to the snapshot nature of the microarray experiment without taking phenotypic or biochemical changes into account.
The pcfQ through pcfZ region downstream of the Tn925 insertion showed no activity during pheromone induction. The same held true for the three selected Tn925 genes xis, int, and tet(M), suggesting that the transposon is not activated during the plasmid transfer. This finding is in contrast with findings on pAD1, where resident Tn916 showed increased transposition during plasmid transfer (20, 22). None of the selected chromosomal genes showed an increase in activity during the observed period; rather, a slight but not significant decrease in activity could be seen. The effect of plasmid transfer on genes on the E. faecalis chromosome needs to be determined with a whole-genome array, which could also identify potential host factors involved in the transfer process. Perhaps most surprising is the transcriptional inactivity of the pCF10 region encoding pcfP to pcfZ, considering the fact that it contains reading frames with high homology to pAD1 reading frames. This is especially the case for pcfS (encoding a putative ssb protein) and pcfY (encoding a DNA invertase), gene products thought to be necessary for plasmid transfer. A possible explanation for the inactivity of that region could be that ORFs expressed downstream of prgB in pCF10 are providing the necessary functions. The prgE gene product could represent the active ssb protein, suggested by sufficient similarity (37%) to an identified ssb protein in a lactococcal phage to support this notion. The upregulation of transcription in the pcfS-pcfY region might be only short and transient. The chosen time points for sampling could have prevented the detection of any significant transcriptional activity.
The biological read-out of the transcript analysis is the transfer of pCF10 from the induced donor to the recipient. The T/D frequency showed a rapid increase that peaked around 90 min after cCF10 addition. The maximum T/D frequency achieved was 1.8 × 10−4. This frequency appears to be small in comparison to published data; however, it has to be taken into consideration that those data were accomplished with pheromone concentrations of 10 to 40 ng per ml of culture, 100- to 400-fold higher than those used in the experiments described here. Under the conditions chosen for the time of induction (1:20 dilution of overnight culture representing ∼107 cells/ml), the amount of cCF10 translates to still approximately 1,500 molecules of cCF10 per cell at the time of induction, hardly a limiting condition, since 4 to 6 molecules per cell are thought to be sufficient for pheromone induction (38). The peak in plasmid transfer activity was followed by a steady decline of the transfer ability of the donor and returned to the baseline 4 h after the initial induction. The shut down of transfer activity demonstrates the high degree of regulation and economy of the plasmid transfer process in pCF10; interestingly, the first ORF contained in the prgQ transcript encodes the iCF10 inhibitor peptide, providing a potential mechanism to limit the pheromone response. The other focus of pheromone plasmid research, AS, demonstrates another feature that has to be accounted for in microarray experiments: protein stability. While the expression of prgB returns to background levels 2 h postinduction like all the other plasmid genes, Asc10 (PrgB) remains detectable on the cell surface in almost-unaltered amounts. This could have far reaching consequences for the virulence of E. faecalis carrying pCF10. AS has been shown to contribute to enterococcal virulence in several model systems (8, 27). Recent data provided evidence for AS expression in vivo with the pheromone-sensing system involved in the induction (27). The stability of Asc10 on the cell surface opens the possibility that only a brief induction event—resulting from a recipient cell or an environmental cue—is necessary to present AS on the surface for a considerable amount of time. This could allow pCF10-containing cells to adhere to different host cell types (31, 46, 53), to withstand the attack of phagocytic cells (43, 50), or to bind to potential enterococcal recipient cells that might only be transiently passing through a particular ecological niche, such as the intestinal lumen. This tethering of potential recipients in close proximity could facilitate a pheromone response by the donor cells, ultimately increasing plasmid transfer.
Acknowledgments
This work was supported by National Institutes of Health grants HL51987 and GM49530. This publication was made possible by National Institutes of Health grant number P20 RR16443-03 from the COBRE Program of the National Center for Research Resources.
We thank Wayne Xu of the University of Minnesota Supercomputing Institute for excellent instruction in the use of the software employed for analysis of the microarray results.
Footnotes
Contribution no. 05-68-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Antiporta, M. H., and G. M. Dunny. 2002. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 184:1155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, T., S. Clerc-Bardin, and G. M. Dunny. 2000. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J. Mol. Biol. 297:861-875. [DOI] [PubMed] [Google Scholar]
- 3.Bae, T., and G. M. Dunny. 2001. Dominant-negative mutants of prgX: evidence for a role for PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol. Microbiol. 39:1307-1320. [DOI] [PubMed] [Google Scholar]
- 4.Bae, T., B. Kozlowicz, and G. M. Dunny. 2002. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J. Mol. Biol. 315:995-1007. [DOI] [PubMed] [Google Scholar]
- 5.Bae, T., B. K. Kozlowicz, and G. M. Dunny. 2004. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol. Microbiol. 51:271-281. [DOI] [PubMed] [Google Scholar]
- 6.Bensing, B. A., D. A. Manias, and G. M. Dunny. 1997. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol. Microbiol. 24:285-294. [DOI] [PubMed] [Google Scholar]
- 7.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 8.Chow, J. W., L. A. Thal, M. B. Perri, J. A. Vazquez, S. M. Donabedian, D. B. Clewell, and M. J. Zervos. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 37:2474-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, J. W., and G. M. Dunny. 1992. cis-acting, orientation-dependent, positive control system activates pheromone-inducible conjugation functions at distances greater than 10 kilobases upstream from its target in Enterococcus faecalis. Proc. Natl. Acad. Sci. USA 89:9020-9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, J. W., and G. M. Dunny. 1995. Transcriptional analysis of a region of the Enterococcus faecalis plasmid pCF10 involved in positive regulation of conjugative transfer functions. J. Bacteriol. 177:2118-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clewell, D. B., F. Y. An, S. E. Flannagan, M. Antiporta, and G. M. Dunny. 2000. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol. Microbiol. 35:246-247. [DOI] [PubMed] [Google Scholar]
- 12.Clewell, D. B., M. V. Francia, S. E. Flannagan, and F. Y. An. 2002. Enterococcal plasmid transfer: sex pheromones, transfer origins, relaxases, and the Staphylococcus aureus issue. Plasmid 48:193-201. [DOI] [PubMed] [Google Scholar]
- 13.De Boever, E. H., and D. B. Clewell. 2001. The Enterococcus faecalis pheromone-responsive plasmid pAM373 does not encode an entry exclusion function. Plasmid 45:57-60. [DOI] [PubMed] [Google Scholar]
- 14.De Boever, E. H., D. B. Clewell, and C. M. Fraser. 2000. Enterococcus faecalis conjugative plasmid pAM373: complete nucleotide sequence and genetic analyses of sex pheromone response. Mol. Microbiol. 37:1327-1341. [DOI] [PubMed] [Google Scholar]
- 15.Dunny, G., C. Funk, and J. Adsit. 1981. Direct stimulation of the transfer of antibiotic resistance by sex pheromones in Streptococcus faecalis. Plasmid 6:270-278. [DOI] [PubMed] [Google Scholar]
- 16.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flannagan, S. E., L. A. Zitzow, Y. A. Su, and D. B. Clewell. 1994. Nucleotide sequence of the 18-kb conjugative transposon Tn916 from Enterococcus faecalis. Plasmid 32:350-354. [DOI] [PubMed] [Google Scholar]
- 18.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45:375-395. [DOI] [PubMed] [Google Scholar]
- 19.Francia, M. V., W. Haas, R. Wirth, E. Samberger, A. Muscholl-Silberhorn, M. S. Gilmore, Y. Ike, K. E. Weaver, F. Y. An, and D. B. Clewell. 2001. Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid 46:117-127. [DOI] [PubMed] [Google Scholar]
- 20.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galli, D., F. Lottspeich, and R. Wirth. 1990. Sequence analysis of Enterococcus faecalis aggregation substance encoded by the sex pheromone plasmid pAD1. Mol. Microbiol. 4:895-904. [DOI] [PubMed] [Google Scholar]
- 22.Gawron-Burke, C., and D. B. Clewell. 1982. A transposon in Streptococcus faecalis with fertility properties. Nature 300:281-284. [DOI] [PubMed] [Google Scholar]
- 23.Gilmore, M. S., R. A. Segarra, M. C. Booth, C. P. Bogie, L. R. Hall, and D. B. Clewell. 1994. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 176:7335-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 25.Hedberg, P. J., B. A. Leonard, R. E. Ruhfel, and G. M. Dunny. 1996. Identification and characterization of the genes of Enterococcus faecalis plasmid pCF10 involved in replication and in negative control of pheromone-inducible conjugation. Plasmid 35:46-57. [DOI] [PubMed] [Google Scholar]
- 26.Hirt, H., S. L. Erlandsen, and G. M. Dunny. 2000. Heterologous inducible expression of Enterococcus faecalis pCF10 aggregation substance Asc10 in Lactococcus lactis and Streptococcus gordonii contributes to cell hydrophobicity and adhesion to fibrin. J. Bacteriol. 182:2299-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirt, H., P. M. Schlievert, and G. M. Dunny. 2002. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect. Immun. 70:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirt, H., R. Wirth, and A. Muscholl. 1996. Comparative analysis of 18 sex pheromone plasmids from Enterococcus faecalis: detection of a new insertion element on pPD1 and implications for the evolution of this plasmid family. Mol. Gen. Genet. 252:640-647. [DOI] [PubMed] [Google Scholar]
- 29.Kao, S. M., S. B. Olmsted, A. S. Viksnins, J. C. Gallo, and G. M. Dunny. 1991. Molecular and genetic analysis of a region of plasmid pCF10 containing positive control genes and structural genes encoding surface proteins involved in pheromone-inducible conjugation in Enterococcus faecalis. J. Bacteriol. 173:7650-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlowicz, B. K., T. Bae, and G. M. Dunny. 2004. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol. Microbiol. 54:520-532. [DOI] [PubMed] [Google Scholar]
- 31.Kreft, B., R. Marre, U. Schramm, and R. Wirth. 1992. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect. Immun. 60:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leonard, B. A., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills, D. A., L. L. McKay, and G. M. Dunny. 1996. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J. Bacteriol. 178:3531-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muscholl, A., D. Galli, G. Wanner, and R. Wirth. 1993. Sex pheromone plasmid pAD1-encoded aggregation substance of Enterococcus faecalis is positively regulated in trans by traE1. Eur. J. Biochem. 214:333-338. [DOI] [PubMed] [Google Scholar]
- 35.Muscholl-Silberhorn, A. 1999. Cloning and functional analysis of Asa373, a novel adhesin unrelated to the other sex pheromone plasmid-encoded aggregation substances of Enterococcus faecalis. Mol. Microbiol. 34:620-630. [DOI] [PubMed] [Google Scholar]
- 36.Muscholl-Silberhorn, A., E. Samberger, and R. Wirth. 1997. Why does Staphylococcus aureus secrete an Enterococcus faecalis-specific pheromone? FEMS Microbiol. Lett. 157:261-266. [DOI] [PubMed] [Google Scholar]
- 37.Muscholl-Silberhorn, A. B. 2000. Pheromone-regulated expression of sex pheromone plasmid pAD1-encoded aggregation substance depends on at least six upstream genes and a cis-acting, orientation-dependent factor. J. Bacteriol. 182:3816-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama, J., G. M. Dunny, D. B. Clewell, and A. Suzuki. 1995. Quantitative analysis for pheromone inhibitor and pheromone shutdown in Enterococcus faecalis. Dev. Biol. Stand. 85:35-38. [PubMed] [Google Scholar]
- 39.National Nosocomial Infections Surveillance System. 1999. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 40.Olmsted, S. B., G. M. Dunny, S. L. Erlandsen, and C. L. Wells. 1994. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J. Infect. Dis. 170:1549-1556. [DOI] [PubMed] [Google Scholar]
- 41.Olmsted, S. B., S. M. Kao, L. J. van Putte, J. C. Gallo, and G. M. Dunny. 1991. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J. Bacteriol. 173:7665-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozawa, Y., K. Tanimoto, S. Fujimoto, H. Tomita, and Y. Ike. 1997. Cloning and genetic analysis of the UV resistance determinant (uvr) encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pAD1. J. Bacteriol. 179:7468-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rakita, R. M., N. N. Vanek, K. Jacques-Palaz, M. Mee, M. M. Mariscalco, G. M. Dunny, M. Snuggs, W. B. Van Winkle, and S. I. Simon. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect. Immun. 67:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruhfel, R. E., D. A. Manias, and G. M. Dunny. 1993. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J. Bacteriol. 175:5253-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sartingen, S., E. Rozdzinski, A. Muscholl-Silberhorn, and R. Marre. 2000. Aggregation substance increases adherence and internalization, but not translocation, of Enterococcus faecalis through different intestinal epithelial cells in vitro. Infect. Immun. 68:6044-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwarz, F. V., V. Perreten, and M. Teuber. 2001. Sequence of the 50-kb conjugative multiresistance plasmid pRE25 from Enterococcus faecalis RE25. Plasmid 46:170-187. [DOI] [PubMed] [Google Scholar]
- 48.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 49.Staddon, J. H., E. M. Bryan, D. A. Manias, and G. M. Dunny. 2004. Conserved target for group II intron insertion in relaxase genes of conjugative elements of gram-positive bacteria. J. Bacteriol. 186:2393-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Süssmuth, S. D., A. Muscholl-Silberhorn, R. Wirth, M. Susa, R. Marre, and E. Rozdzinski. 2000. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect. Immun. 68:4900-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanimoto, K., F. Y. An, and D. B. Clewell. 1993. Characterization of the traC determinant of the Enterococcus faecalis hemolysin-bacteriocin plasmid pAD1: binding of sex pheromone. J. Bacteriol. 175:5260-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanek, N. N., S. I. Simon, K. Jacques-Palaz, M. M. Mariscalco, G. M. Dunny, and R. M. Rakita. 1999. Enterococcus faecalis aggregation substance promotes opsonin-independent binding to human neutrophils via a complement receptor type 3-mediated mechanism. FEMS Immunol. Med. Microbiol. 26:49-60. [DOI] [PubMed] [Google Scholar]
- 53.Wells, C. L., E. A. Moore, J. A. Hoag, H. Hirt, G. M. Dunny, and S. L. Erlandsen. 2000. Inducible expression of Enterococcus faecalis aggregation substance surface protein facilitates bacterial internalization by cultured enterocytes. Infect. Immun. 68:7190-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson, K. 2002. Preparation of genomic DNA from bacteria, vol. 1. John Wiley and Sons, New York, N.Y.
- 55.Wirth, R. 1994. The sex pheromone system of Enterococcus faecalis. More than just a plasmid-collection mechanism? Eur. J. Biochem. 222:235-246. [DOI] [PubMed] [Google Scholar]