Abstract
The generalized transducing double-stranded DNA bacteriophage ES18 has an icosahedral head and a long noncontractile tail, and it infects both rough and smooth Salmonella enterica strains. We report here the complete 46,900-bp genome nucleotide sequence and provide an analysis of the sequence. Its 79 genes and their organization clearly show that ES18 is a member of the lambda-like (lambdoid) phage group; however, it contains a novel set of genes that program assembly of the virion head. Most of its integration-excision, immunity, Nin region, and lysis genes are nearly identical to those of the short-tailed Salmonella phage P22, while other early genes are nearly identical to Escherichia coli phages λ and HK97, S. enterica phage ST64T, or a Shigella flexneri prophage. Some of the ES18 late genes are novel, while others are most closely related to phages HK97, lambda, or N15. Thus, the ES18 genome is mosaically related to other lambdoid phages, as is typical for all group members. Analysis of virion DNA showed that it is circularly permuted and about 10% terminally redundant and that initiation of DNA packaging series occurs across an approximately 1-kbp region rather than at a precise location on the genome. This supports a model in which ES18 terminase can move substantial distances along the DNA between recognition and cleavage of DNA destined to be packaged. Bioinformatic analysis of large terminase subunits shows that the different functional classes of phage-encoded terminases can usually be predicted from their amino acid sequence.
Generalized transducing bacteriophages are valuable members of the arsenal of tools for the genetic study of bacteria. ES18 is a temperate, generalized transducing double-stranded DNA (dsDNA) phage that naturally infects Salmonella enterica serovar Typhimurium (48), as well as some serovar Enteriditis, Dublin, Pullorum, Gallinarum, and Paratyphi B strains (52, 76). In addition, it can infect Escherichia coli if it displays the Salmonella surface receptor (47). ES18, which has also been called typing phage A18, was originally isolated in about 1953 after its release from Salmonella sp. strain BA19, in which it apparently resided as a prophage (8, 75). It has an estimated genome size of about 46,000 bp (74). It is of technical interest because, unlike the well-characterized Salmonella transducing phage P22, it can infect rough strains that do not produce full-length O-antigen, for which no other transducing phages have been studied (48). Yamamoto (98) showed that ES18 is able to recombine to form viable hybrid phages with both the short-tailed phage P22 and the long-tailed phage Fels-1, both of which are now considered to be lambdoid phages (34). Thus, ES18 was also thought to be a lambdoid phage. This was tentatively confirmed in studies by Schmieger and colleagues (74), which found that the ES18 prophage repressor and lysis regions are very similar to those of the short-tailed phage P22 in both sequence and genome location. It has been reported (without publication of the data) that ES18 has a long flexible tail (48, 98), and the ES18 head and tail gene DNAs have been shown not to hybridize to the DNAs of phages P22 or λ (74). Because it seemed possible that it harbored a novel set of head assembly genes, and in order to foster further research on this interesting and potentially useful phage, we have sequenced and annotated the complete ES18 genome and analyzed its DNA packaging strategy.
MATERIALS AND METHODS
Bacterial and phage strains and virion production.
ES18 was propagated on S. enterica strain Q1 (98) or on strain DB7000 that was cured of the Fels-2 prophage (7) (this prophage restricts ES18 growth [48]). All bacterial cultures were grown in Luria-Bertani medium at 37°C. ES18 virions were partially purified as follows: cells were shaken at 37°C for 90 min after infection, at which point the culture was shaken with chloroform to complete lysis. Cell debris was removed by centrifugation for 20 min at 8,000 rpm in a Beckman JA-10 rotor at 4°C, and phage particles were pelleted by spinning overnight at 8,000 rpm in the same rotor at 4°C. The pellet was gently resuspended in TM (10 mM Tris-Cl [pH 7.4], 1 mM MgCl2), and insoluble material was removed by low-speed centrifugation. The resulting preparation was pelleted onto a CsCl cushion (density, 1.4 g/ml), dialyzed against TM, and finally sedimented through a 10-to-40% sucrose gradient at 25,000 rpm for 2 h at 4°C in an SW40.1 Beckman rotor; ES18 virions sediment as a visibly opalescent band about three-quarters of the way down the gradient, which was harvested by syringe puncture of the tube. DNA was isolated from ES18 virions by sodium dodecyl sulfate (SDS) release and ethanol precipitation (17), followed by phenol extraction. Virions on carbon-coated Parlodion films supported by 400-mesh copper grids were stained for electron microscopy with 0.5% uranyl formate and examined with a Philips Morgani 268 microscope operating at 80 kV.
DNA and protein methods.
Nucleotide sequence determination was performed by dideoxy chain termination methods as previously described (66). Bulk sequencing of a random plasmid library of sheared DNA was performed until an average of >7-fold coverage was obtained (768 sequencing runs). Twenty-two primers were designed to sequence across weak areas by using virion DNA as template, and PhredPhrap (32) assembled the sequence data into one circularsequence. The DNA sequence analysis software used was DNA Strider (24), GeneMark (5), Staden programs (78), BLAST (2), BPROM (http://www.softberry.com/berry.phtml?topic=gfindb), tRNAscan (57; http://www.genetics.wustl.edu/eddy/tRNAscan-SE), and DNA Master (http://cobamide2.bio.pitt.edu/). Neighbor-joining trees were constructed with CLUSTAL X for the Macintosh (43, 84), and maximum likelihood analysis was performed with PHYLIP (28; http://evolution.genetics.washington. edu/phylip.html). Polyacrylamide gel electrophoresis of proteins in SDS, staining with Coomassie brilliant blue R-250 (Bio-Rad, Richmond, Calif.), and determination of the N-terminal amino acid sequence by the University of Utah Protein Facility were performed as previously described (27). Agarose gel electrophoresis and contour-clamped homogeneous electric field (CHEF) electrophoresis of DNA molecules were performed as previously described, as were Southern (77) analyses of electrophoresis gels (14).
RESULTS AND DISCUSSION
ES18 virions.
ES18 virions are unusual in that they do not band in CsCl equilibrium density gradients at a density near 1.5 g/ml as do nearly all other tailed-phage virions; this is presumed to be due to the inability of Cs+ ions to penetrate the head shell, although other explanations are possible. Heating the virions to 45°C for 10 min or addition of Triton X-100 in the presence of CsCl did not alter the banding density of ES18 virions (data not shown). This property has precluded simple, one-step equilibrium density purification of the virions to near homogeneity. ES18 virions were thus partially purified by differential centrifugation as described in Materials and Methods.
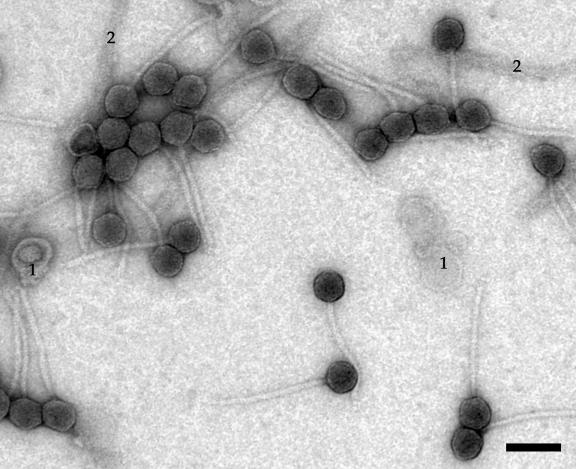
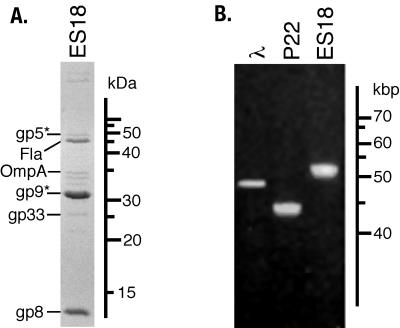
ES18 virions have been reported informally to have long, flexible tails by Kuo and Stocker (48) and Yamamoto (98) (although its morphology has been incorrectly discussed elsewhere as being “P2-like” [60]). Figure 1 shows that ES18 does indeed have a long, flexible, noncontractile tail, as well as a head that is hexagonal in outline and so almost certainly icosahedral. Its head is 56 nm in diameter, and its tail is 210 nm long and 12 nm wide. We observed no obvious side tail fibers or other tail tip structures. Figure 2A shows an SDS-12.5% polyacrylamide electrophoresis gel of the partially purified virion preparation. The genes that encode several of the virion proteins have been identified (discussed below), and two host proteins, flagellin and OmpA, that are present in large contaminating structures (flagella and outer membrane vesicles, respectively) have been identified by their N-terminal amino acid sequences (indicated in Fig. 2A). ES18 virions contain DNA molecules that are 51.5 ± 1 kbp in length as measured by CHEF gel electrophoresis (Fig. 2B). This indicates that the virion chromosome is about 10% terminally redundant, since the sequence of the genome is 46,900 bp in length (below). In addition, like P22 DNA, the width of the ES18 virion DNA electrophoresis band is wider than λ DNA (Fig. 2B). This is a typical feature of headful packaging phages, since size measurement of their packaged chromosome is imprecise (12, 83). Such terminal redundancy and imprecise length measurement is not unexpected, since ES18 is a generalized transducing phage (48) and all characterized naturally occurring generalized transducing phages utilize the headful packaging strategy.
FIG. 1.
Electron micrograph of ES18 virions. Virions were prepared, negatively stained, and examined by electron microscopy as described in Materials and Methods. Membrane vesicles and flagellar fragments (1 and 2 in the figure, respectively) are present in ES18 virion preparations (see text). Bar, 100 nm.
FIG. 2.
ES18 virion proteins and DNA. (A) Separation of ES18 proteins by SDS-polyacrylamide gel electrophoresis. The protein components of purified ES18 virions were displayed by SDS-12.5% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. The experimentally identified proteins are indicated on the left. (B) Display of ES18 virion DNA by agarose gel electrophoresis. Purified whole DNAs from the virions indicated above were subjected to CHEF electrophoresis as described in Materials and Methods.
Mosaic structure of the ES18 genome.
The complete nucleotide sequence of the ES18 genome (GenBank accession no. AY736146) was determined by sequencing random DNA clones of virion DNA fragments as described in Materials and Methods. The sequence assembled unambiguously into a single circular sequence 46,900 bp in length; we assigned bp 1 to the first base pair of the predicted small terminase subunit gene, so its map can be easily compared to the standard phage P22 and λ maps.
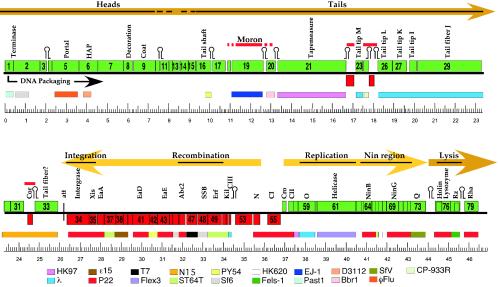
We describe briefly here the predicted ES18 genes from left to right across the genome. Figure 3 shows a map of the 79 predicted ES18 genes, and Table 1 lists salient features of these protein-encoding genes (no tRNA genes were found in the ES18 genome). Most of the morphogenetic portion of the putative ES18 late operon (the bulk of the left half of the virion chromosome) is not closely related to morphogenetic regions of previously characterized phages. Genes 1, 2, 5 and 6 are putative head genes, because their protein products have weak similarities to phage small and large terminase subunits, portal proteins, and phage SPP1 gene 7 head assembly factor (HAF) (81), respectively (Table 1). These functions of the ES18 head genes, as tentatively deduced from their weak sequence matches and the relative abundance of the encoded proteins in virions, fit the stereotypical lambdoid phage morphogenetic gene order, which is 5′-small terminase-large terminase-portal- HAF-decoration-coat-3′ on the mRNA (HAF refers to homologues of the phage SPP1 gene 7 head assembly factor [81]).
FIG. 3.
Map of the ES18 genome. The genome of phage ES18 is shown with its circular sequence arbitrarily opened at the start codon of gene 1. Predicted genes are indicated as rectangular boxes; rightward-transcribed genes are above the central black line and are green; leftward-transcribed genes are below the line and are red. Between the kilobase pair scale and the genome, thinner colored boxes indicate the best matches from the other phages at the bottom of the figure. These matches vary from quite weak to identity; see Table 1 for more details. Flex3 and Past1 are defective prophages in the Shigella flexneri and P. multocida genomes, respectively (9). Above the genome, the names of better-studied, homologous λ or P22 genes or the predicted function is given for some genes, potential late operon morons (see text) are marked by horizontal red lines, and the major predicted early and late lytic transcripts are indicated by the thick light and darker orange arrows, respectively. Stem-loop structures mark the positions of predicted rho-independent transcription terminators.
TABLE 1.
Predicted genes of bacteriophage ES18
| Genea | Start codon | Stop codon | SD (spacer) | AAsc | Predicted function | Best matches (phage/gene) | % Identity/ no. of AAs |
|---|---|---|---|---|---|---|---|
| 1 ↓ | ATG | TGA | AA (4) | 143 | Small subunit terminase | Pasteurella prophage Past1/PM1777 | 49/133 |
| Haemophilus prophage φFlu/HI1411 | 55/81 | ||||||
| 2 ↓ | ATG | TAG | AGGAG (12) | 439 | Large subunit terminase | Photorhabdus prophage/plu2901 | 78/442 |
| Sf6/2 | 33/223 | ||||||
| APSE-1/18 | 31/225 | ||||||
| SPP1/2 | 26/423 | ||||||
| 3 ↓ | TTG | TGA | AGGG (8) | 88 | Unknown | None | |
| 4 ↓ | GTG | TAA | GAGGA (6) | 52 | Unknown | None | |
| 5 ↓ | ATG | TAA | GGA (6) | 449 | Portal protein | Actinobacillus prophage/Aple109001 | 33/441 |
| Haemophilus prophage φFlu/HI1409 | 19/357 | ||||||
| 6 ↓ | ATG | TAA | GGAG (5) | 308 | Procapsid assembly | D3112/28 | 23/152 |
| Mu/30 | 17/161 | ||||||
| 7 ↓ | ATG | TAA | GGAGA (7) | 421 | Unknown | Actinobacillus prophage/Aple109201 | 30/448 |
| 8 ↓ | ATG | TAA | GGAGA (6) | 149 | Decoration? | None | |
| 9 ↓ | ATG | TAA | AAGGA (9) | 358 | Coat? | None | |
| 10 ↓ | ATG | TAG | AAGGA (6) | 62 | Unknown | None | |
| 11 ↓ | ATQ | TAA | AGAGG (5) | 133 | Unknown | None | |
| 12 ↓ | ATG | TGA | GGGGA (6) | 59 | Unknown | None | |
| 13 ↓ | ATG | TGA | GGAGG (6) | 120 | Unknown | Actinobacillus prophage/Aple109701 | 30/120 |
| 14 ↓ | ATG | TGA | GAGG (5) | 131 | Unknown | None | |
| 15 ↓ | ATG | TAA | AAGGA (8) | 128 | Unknown | Sp5/ECs1210 | 36/36 |
| stx2φI/p153 | 36/36 | ||||||
| 16 ↓ | ATG | TAA | GGAGA (7) | 245 | Tail shaft | PY54/13 | 46/88 |
| Felix-01/79 | 50/86 | ||||||
| HK97/10 | 34/94 | ||||||
| 17 ↓ | ATG | TAA | GGGG (4) | 217 | Unknown | Actinobacillus prophage/Aple110101 | 26/219 |
| 18 ↓ | ATG | TAG | GGA (8) | 57 | Unknown | None | |
| 19 ↓ | ATG | TAG | GGGAA (7) | 509 | Unknown; moron | Pasteurella prophage/orf5 | 36/461 |
| EJ-1/03 | 32/485 | ||||||
| Streptococcus prophage φ8382.5/1806 | 40/291 | ||||||
| 20 ↓ | ATG | TAA | GGGA (7) | 151 | Unknown; moron? | Bordetella prophage Bbr1/BB1705 | 32/85 |
| 21 ↓ | ATG | TAG | GGAGA (7) | 1,124 | Tape measure | CP-933P/Z6034 | 27/809 |
| HK97/14 | 28/768 | ||||||
| 22 ↑ | ATG | TGA | AGG (4) | 132 | Unknown; moron? | None | |
| 23 ↓ | ATG | TAA | GGAGG (5) | 115 | Tail tip | Gifsy-1/STM2593 | 38/111 |
| lambda/M | 32/114 | ||||||
| 24 ↓ | ATG | TAA | GGGAGA (4) | 86 | Unknown; moron | E. coli 933R prophage/Z2372 | 33/69 |
| 25 ↑ | ATG | TAA | AGGAG (5) | 95 | Unknown; moron | None | |
| 26 ↓ | GTG | TGA | GGGAG (6) | 234 | Tail tip | Gifsy-2/STM1045 | 47/215 |
| lambda/L | 43/215 | ||||||
| 27 ↓ | ATG | TGA | AGAAGG (7) | 239 | Tail tip | Fels-1/STM0922 | 45/236 |
| lambda/K | 49/198 | ||||||
| 28 ↓ | ATG | TAA | GAGG (8) | 115 | Tail tip | lambda/I | 40/57 |
| Gifsy-2/STM1047 | 34/95 | ||||||
| 29 ↓ | ATG | TGA | AAGG (7) | 1,051 | Tail tip fiber (host range) | lambda/J | 40/847 |
| Gifsy-1/STM2589 | 39/846 | ||||||
| N15/21 | 32/1,069 | ||||||
| 30 ↓ | ATG | TAA | GGGGG (7) | 104 | Unknown | HK022/24 | 43/102 |
| N15/22 | 46/102 | ||||||
| 31 ↓ | ATG | TAA | GGAGA (7) | 217 | Unknown | N15/23 | 33/203 |
| HK022/25 | 34/201 | ||||||
| 32 ↓ | ATG | TGA | AGGGA (5) | 80 | Superinfection exclusion (N15 cor) | N15/24 (cor) | 62/74 |
| HK022/26 | 61/78 | ||||||
| 33 ↓ | ATG | TGA | AGGAG (5) | 378 | Tail fiber? | N15/25 | 73/161 |
| φYe03-12/17 | 25/234 | ||||||
| 34 ↑ | ATG | TAA | AGAGGA (5) | 387 | Integrase | P22/int | 96/387 |
| SfV/int | 88/386 | ||||||
| 35 ↑ | ATG | TGA | GGAG (4) | 116 | Excisionase | P22/xis | 96/115 |
| SfV/27 | 85/115 | ||||||
| 36 ↑ | GTG | TAA | GGAGG (6) | 118 | Unknown | P22/eaA | 69/99 |
| Fels-1/STM0895 | 67/49 | ||||||
| 37 ↑ | ATG | TGA | GAGGAA (10) | 186 | Unknown | ST64T/eaa1 | 75/68 |
| φ4795/p04 | 84/51 | ||||||
| 38 ↑ | ATG | TAA | GGAGG (7) | 126 | Unknown | ɛ15/46 | 85/68 |
| 39 ↑ | ATG | TAA | GGGAG (4) | 72 | Unknown | ɛ15/47 | 88/72 |
| 933W/L007 | 50/72 | ||||||
| 40 ↑ | ATG | TGA | GAGGA (6) | 81 | Unknown | None | |
| 41 ↑ | ATG | TAA | AGGAG (8) | 274 | Unknown | P22/eaD | 68/154 |
| ɛ15/45 | 68/154 | ||||||
| 42 ↑ | ATG | TGA | AGGGG (7) | 121 | Unknown | PY54/48 | 54/93 |
| 43 ↑ | ATG | TGA | AGGA (7) | 208 | Unknown | P22/eaE | 93/56 |
| ɛ15/44 | 42/208 | ||||||
| 44 ↑ | ATG | TAA | GAGAG (6) | 56 | Unknown | P22/orf56 | 92/56 |
| Sf6/23 | 4/51 | ||||||
| 45 ↑ | ATG | TGA | GAGGA (6) | 95 | Unknown | Sf6/24 | 66/95 |
| P2/orf83 | 39/91 | ||||||
| 46 ↑ | TTG | TGA | AAG (9) | 105 | Recombination | P22/abc2 | 78/97 |
| HK97/39 | 77/97 | ||||||
| 47 ↑ | ATG | TAA | GG (9) | 165 | Unknown | T7/7.7 | 42/131 |
| 48 ↑ | ATG | TAA | AGG (8) | 156 | Single-strand DNA binding protein | E. coli/ssb | 90/153 |
| Sf6/27 | 89/157 | ||||||
| 49 ↑ | ATG | TAA | GAGG (6) | 235 | Recombination (P22 erf) | ST646T/orf235 | 96/235 |
| Sf6/28 | 91/235 | ||||||
| 50 ↑ | ATG | TGA | GGAGG (5) | 62 | Unknown | ST64T/orf66 | 98/66 |
| HK620/hkaM | 66/62 | ||||||
| 51 ↑ | ATG | TGA | AGGAG (8) | 37 | Inhibition of host septation | ST64T/kil | 100/37 |
| lambda/kil | 58/36 | ||||||
| 52 ↑ | ATG | TGA | AGGAG (6) | 28 | Inhibition of host HflB protease | lambda/cIII | 85/27 |
| HK620/hkaO | 85/27 | ||||||
| 53 ↑ | ATG | TGA | AGGAA (7) | 282 | Unknown | Xp10/44 | 29/149 |
| 54 ↑ | ATG | TAA | GAGGA (7) | 109 | Transcription antitermination | ST64T/24 | 76/108 |
| P22/24 | 85/75 | ||||||
| 55 ↑ | ATG | TAA | GATGA (7) | 216 | Repressor (lambda cI) | P22/c2 | 100/216 |
| 434/cI | 51/207 | ||||||
| 56 ↓ | ATG | TAA | AAAG (12) | 61 | Repressor (lambda cro) | P22/cro | 100/61 |
| 21/cro | 98/61 | ||||||
| 57 ↓ | ATG | TGA | AGGAA (9) | 92 | Transcription activation | P22/cI | 100/92 |
| (lambda cII) | Sf6/41 | 84/92 | |||||
| 58 ↓ | ATG | TAA | AGGAG (5) | 53 | Unknown | HK97/53 | 94/53 |
| Sf6/42 | 94/53 | ||||||
| 59 ↓ | ATG | TGA | AGGAG (6) | 282 | Replication, ori recognition | lambda/O | 93/193 |
| P22/18 | 42/150 | ||||||
| 60 ↓ | ATG | TGA | AGG (6) | 35 | Unknown | None | |
| 61 ↓ | ATG | TGA | GAGG (5) | 626 | Primase-helicase | Flex3/SF0682 | 95/371 |
| T7/4 | 25/355 | ||||||
| 62 ↓ | ATG | TGA | GAG (6) | 92 | Unknown | None | |
| 63 ↓ | ATG | TGA | AGGAG (8) | 26 | Unknown | P22/ninA | 100/25 |
| Sf6/45 | 100/25 | ||||||
| 64 ↓ | ATG | TGA | AGAGG (5) | 145 | Unknown | P22/ninB | 97/145 |
| lambda/ninB | 97/145 | ||||||
| 65 ↓ | ATG | TAA | GGAGA (6) | 57 | Unknown | P22/ninD | 98/57 |
| lambda/ninD | 73/57 | ||||||
| 66 ↓ | ATG | TGA | AGGAGG (4) | 58 | Unknown | ST64T/ninE | 78/55 |
| lambda/ninE | 76/55 | ||||||
| 67 ↓ | ATG | TAG | GAGGAG (6) | 60 | Unknown | HK620/hkbB2 | 52/57 |
| HK97/64 (ninF) | 50/57 | ||||||
| 68 ↓ | ATG | TAA | GGAGG (6) | 72 | Unknown | None | |
| 69 ↓ | ATG | TGA | GGAG (6) | 201 | Unknown | P22/ninG | 94/201 |
| lambda/ninG | 89/203 | ||||||
| 70 ↓ | ATG | TGA | GAGG (5) | 74 | Unknown | P22/ninY | 100/74 |
| 71 ↓ | ATG | TAA | GGGAGG (4) | 67 | Unknown | P22/ninH | 98/67 |
| lambda/ninH | 71/57 | ||||||
| 72 ↓ | ATG | TGA | AGAGG (5) | 59 | Unknown | ST64T/ninZ | 92/57 |
| P22/ninZ | 89/57 | ||||||
| 73 ↓ | ATG | TGA | GGGAGG (9) | 254 | Transcription antitermination | E. coli prophage CP-933M/Z1345 | 45/274 |
| (lambda Q) | SfV/46 | 41/257 | |||||
| 74 ↓ | ATG | TAA | GAGG (6) | 108 | Lysis timing, holin inhibitor | P22/13 | 100/108 |
| lambda/S | 89/106 | ||||||
| 75 ↓ | ATG | TAA | GAA (4) | 105 | Lysis, holin | P22/13′ | 100/106 |
| lambda/S′ | 89/104 | ||||||
| 76 ↓ | ATG | TGA | GAGG (6) | 145 | Lysis, lysozyme | P22/R | 97/146 |
| Gifsy-1/STM2612 | 51/150 | ||||||
| 77 ↓ | GTG | TAA | AGAGCG (11) | 156 | Lysis (lambda Rz) | Fels-1/STM0908 | 85/145 |
| lambda/Rz | 69/155 | ||||||
| 78 ↓ | ATG | TGA | AGGA (8) | 62 | Lysis (lambda Rz1) | lambda/Rz1 | 73/60 |
| 933W/Rz1 | 70/60 | ||||||
| 79 ↓ | ATG | TAA | AGA (8) | 229 | Affects growth in IHFb host | Haemophilus prophage φFlu/HI1412 | 34/95 |
| P22/orf201 (rha) | 24/121 |
Arrow indicates direction of transcription.
IHF defective.
AA, amino acid.
Amino-terminal sequencing of virion proteins isolated from SDS-polyacrylamide electrophoresis gels (Fig. 2A) showed that the products of genes 8 and 9 are major virion components; we determined their N-terminal sequences to be H2N-SRYRRV and H2N-AVGGFTR, respectively. Neither protein has any recognizable homologue in the current sequence database. The gene 8 protein's N-terminal methionine has been removed (common when a penultimate serine is present [30]), and the N-terminal 51 amino acids have been cleaved from the 358-amino-acid primary translation product of the gene 9 protein. The predicted size of the cleaved protein (33,248 Da) agrees well with the measured size of 32,000 Da. Quantitation of the Coomassie brilliant blue staining intensity of the bands in SDS-polyacrylamide electrophoresis gels indicated that the molar ratio of gp8/gp9 was 0.8 ± 0.1 in virions; since staining efficiencies for different proteins may vary, it seems likely that there is one gp8 for each gp9 molecule in the virion. The major components of known phage heads are coat proteins, which build the head shell, and decoration proteins, which stabilize the coat shell by binding to its exterior surface. Proteolytic removal of an N-terminal peptide from phage coat proteins during assembly is common (13), and head decoration proteins are usually rather small and are not known to be cleaved. In addition, decoration protein genes are in some cases immediately 5′ of the coat protein gene. From these observations, we feel it is very likely that gene 8 encodes a decoration protein and gene 9 encodes the coat protein.
The predicted gene 5 protein has weak sequence similarity to other phage portal proteins, and it was identified in virions, since its amino-terminal sequence was determined to be H2N-XALNDA, where ALNDA is identical to amino acids 12 through 16 of the predicted ES18 gene 5 protein (Fig. 2A). The unidentified amino acid X in position 1 of the determined sequence is amino acid number 11 (a histidine) in the predicted gene 5 protein sequence, indicating that 10 N-terminal amino acids have been removed by proteolysis from the primary translation product. Removal of a small number of N-terminal portal protein amino acids by proteolysis during head maturation has precedent in phage λ gene B protein's cleavage (35, 90). The staining intensity relative to that of coat protein of the gene 5 protein (Fig. 2A) is consistent with the expected 12 molecules of portal protein per virion (13). The cleavage of the putative coat protein and the stereotyped order of lambdoid head gene clusters suggest that ES18 gene 7 may be the capsid maturation protease, but further experimentation will be required to test this hypothesis.
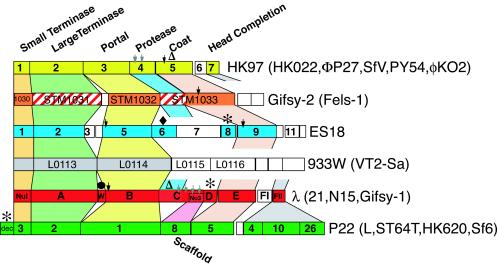
It is noteworthy that the ES18 region of genes 1 through approximately 11 defines a new type of head gene module that has not been previously observed; this module is shown in comparison to the five previously known lambdoid head types in Fig. 4. These six modules encode the formation of heads that are indistinguishable in the electron microscope, and almost certainly all have an icosahedral T=7 structure. This diagram is not meant to imply that additional head types or mosaics of these very different types will not be found. For example, φKO2 (11), PY54 (41), ΦP27 (72), and SFV (1) comprise a rather distantly related subtype within the HK97 group, and Sf6 (17) and HK620 (21) have a large terminase subunit that is more closely related to the ES18 terminase than that of other members of the P22 group (see below). In addition, we point out that although these six groups are very highly diverged, a number of the genes within each functional group are nonetheless homologous (i.e., diverged from a common ancestor). For example, most of these six large terminase and portal protein types can be shown to be members of the extended terminase and portal families after multiple rounds of Ψ-BLAST (2) analysis, and the HK97 and P22 coat proteins are thought to have the same polypeptide fold in spite of no recognizable amino acid sequence similarity (33, 44, 93). On the other hand, the putative proteases of lambda and HK97 are members of the ClpP and procapsid protease families with different folds (20) and so may have arrived at parallel functions in head assembly by convergent evolution.
FIG. 4.
The six known lambdoid phage head types. Physical maps are shown of the head gene clusters of the six different head gene types known in otherwise convincingly lambdoid phages that infect enteric bacteria; rectangles represent genes. Selected gene names are shown for the prototypical member of each phage group as indicated immediately to the right of each head region (the only exception is that the dec gene in the P22 group is present in phages L [Gilcrease and Casjens, unpublished] and ST64T [62] but not in P22, Sf6, or HK620 [27, 67]); the phage names in parentheses at the right are other phages with the same head type. Colored trapezoids vertically connect genes with similar function, as deduced by experimentation or sequence homology. Genes within each such functional column are colored according to sequence similarity; i.e., different gene colors represent sequences that are not brought into the same sequence family by several rounds of Ψ-BLAST analysis (2) of the extant database. Unlike the colored genes, white genes are not recognizably homologous to one another or other known proteins and their functional roles are unknown. Red cross-hatching of Gifsy-2 genes STM1031 and part of STM1033 indicates that these sequences are weak matches (only patches are ≤25% identical) to lambda genes A and C, respectively. Other symbols indicate special gene product features as follows: black arrows mark cleavage sites for assembly-dependent removal of an N-terminal protein fragment; gray arrows mark genes whose products are known to be proteolytically cleaved at multiple sites, resulting in the removal of most or all of the protein from virions; asterisks mark decoration protein genes (these proteins are non- or semiessential and bind to outside of coat protein shells); the black circle indicates the “out-of-place” W head completion gene in lambda; open triangles denote proteins that undergo covalent protein cross-linking—the HK97 (and HK022) coat protein molecules undergo autocatalytic covalent cross-linking to one another (26, 93), and about 10 molecules of lambda C protein are covalently joined to its coat protein molecules (36, 37); the black diamond denotes that ES18 gene 6 is homologous to phage SPP1 gene 7 HAF (81) (no other known lambdoid phage carries a member of this gene family). The Gifsy-2 coat protein (STM1033) sequence (59) predicts that a protease motif near its N terminus might be responsible for self-cleavage from the C-terminal portion which has weak similarities to some other phage coat proteins. The coat protein gene of 933W and its relatives has not been identified.
Genes approximately 12 through 29 include the putative tail genes of ES18. Gene 16 and 21 proteins have weak similarities to the major tail shaft subunits and tail tape measure proteins, respectively, of other lambdoid phages that have long flexible tails, most notably those of phage HK97 (45). Our failure to obtain N-terminal amino acid sequence from the putative tail shaft protein suggests that its N terminus is blocked. Genes 23, 26, 27, 28, and 29 are homologues of phage λ tail tip genes M, L, K, I, and J, respectively (Table 1). The gene J protein is the host range-determining protein in λ virions which binds to the target cell's surface receptor (46, 91), and J protein and its homologues are not known to utilize surface polysaccharides as receptors. This agrees with the fact that ES18 is known to infect both O-antigen-producing (smooth) and O-antigen-defective (rough) strains of Salmonella enterica (48). It adsorbs to the host outer membrane FhuA protein, a TonB-dependent ferrichrome transporter that is also the receptor in E. coli for phages φ80, T1, and T5 (6, 47). However, the ES18 J-like protein is only rather distantly related to receptor binding proteins in these three phages (φ80 [G. Plunkett, personal communication], T1 [73], and T5 [accession no. AY543070]), and it has been shown that φ80 and ES18 bind to different surfaces of the FhuA protein (47), suggesting that they may have evolved independently to utilize the same receptor.
Genes 30, 31, 32, and 33 are homologues of phage N15 (71) and HK022 (45) genes that are located in parallel regions of those genomes. Of the latter four genes, only ES18 genes 32 and 33 have homologues of known function. Gene 32 is similar to the N15 24 and φ80 cor genes, which are lysogenic conversion genes that function to exclude superinfecting phages such as φ80 and T1 (58, 89); however, unlike the N15 and φ80 (and HK022) cor genes, ES18 gene 32 lies in the opposite orientation from the late operon. The N-terminal sequence analysis (H2N-SAGT) of the isolated virion proteins (above) suggests that gp33, which has weak homology to some phage tail fibers, is present in virions; the gp33 band in the gel is substantially smaller than the predicted translation product, suggesting that it might be cleaved (Fig. 2A). The functions of the other two genes in this region, 30 and 31, are not known.
Although the ES18 tail genes show overall similarity and organization to the tail genes of better-studied lambdoid phages with long noncontractile tails, ES18 has four predicted genes, 17 through 20, between the tail shaft subunit (gene 16) and the tape measure (gene 21). Other lambdoid tail gene clusters of this type which have been examined carry only two open reading frames in this interval. It therefore seems likely that one or more of these four genes are “morons,” or non-tail assembly genes that have been recently inserted into this location (38, 45). This idea is supported by the fact that gene 19 homologues are found in E. coli phage ΦP27 (72) and a Pasteurella multicoda prophage (70), where in both cases they are adjacent to phage-carried toxin genes; in the former it is clearly not in the tail cluster and in the latter it lies at a different location but near putative tail genes. The two open reading frames in this interval in phage λ encode a tail assembly chaperone (97), and the downstream one is only expressed through translational frameshifting from the first gene (53); such pairs of overlapping genes related by a translational frameshift are in fact nearly universally present among phages with long tails (97). We are unable to identify the site of the expected frameshift for ES18 with confidence, but we suggest that a plausible possibility, based on patterns seen in other phages, would be a +1 frameshift occurring at the end of gene 17, into the open reading frame that extends just to the beginning of gene 18. Finally, tail region genes 22, 24, and 25 (functions unknown) do not have homologues in the studied lambdoid phages. Genes 3, 18 to 20, 22 to 25, and 32 (cor) correspond to the four portions of the late operon with the lowest G+C contents, suggesting that they have a different evolutionary history from the rest of the late operon, and so are all moron candidates.
ES18's divergent early operons are in a typically lambdoid arrangement and have similar control elements. Between these operons, the immunity region contains genes 55 and 56 (Repressor and Cro) which, as previously pointed out by Schicklmaier and Schmieger (74), encode proteins that are identical to their homologues in P22. This agrees with the observation that ES18 has the same immunity (repressor) specificity as P22 (48, 82). In addition, ES18 gene 57 protein is identical to the P22 c1 gene protein. Transcriptional antitermination should be exerted on the early and late operons by gene 54 protein (85% identical over 75 amino acids to the P22 gene 24 protein) and gene 73 protein (a distantly related λ Q protein homologue), respectively. The former appears to have a target specificity identical to that of the P22 protein, since sequences identical to P22's nut sites are present at the expected locations, but the latter has no known close relatives and so likely has a novel target specificity.
The early left operon of ES18 is typical of lambdoid phages; it contains 21 genes (34 through 54) that are mosaically related to other known lambdoid phages. From 5′ to 3′ (right to left on the map), the putative early left operon mRNA carries genes that are similar to λ's genes N, cIII, and kil, HK620 gene hkaM, P22 erf, Sf6 gene 27 (a bacterium-type single-stranded DNA binding protein), P22 abc2, HK97 gene 38, P22 eaE and eaD, PY54 gene 46, 933W gene L007, ST64T gene eaa1, ɛ15 gene 46, and P22 eaA (see Table 1 for details). Only two genes in this region, 40 and 53, have no known homologues. Finally, the ES18 integration function integrase and excisionase (encoded by genes 34 and 35, respectively) are 96% identical to those of P22 and its putative attP integration site is identical to that of P22, and so it is virtually certain that ES18 utilizes the P22 attB attachment site for integration into the host chromosome as was suggested by Kuo and Stocker (48).
The ES18 early right operon is also typical of lambdoid phages. It contains 18 genes, 56 through 73, of which only 60, 62, and 68 do not have homologues in parallel locations in other lambdoid phage genomes. The putative ES18 DNA replication initiation gene 59 protein is 93% identical to the λ O gene product (and its putative 85-bp replication origin sequence has only three differences from that of lambda), but its putative partner gp61 is only a very distant relative (if it is a homologue at all) of the λ P protein; gp61 contains primase and helicase homologies, and its closest relatives are in a Shigella defective prophage Flex3 (9) and the phage T7 gene 4 primase protein (85). The ES18 Nin region's genes 63 through 72 are most closely related to those of P22.
At the right end of the genome, near the promoter-proximal end of the ES18 late operon, as Schicklmaier and Schmieger (74) previously reported, the holin and lysozyme lysis genes, 74/75 and 76, respectively, are nearly identical to those of P22, and λ Rz and Rz1 homologues, gene 77 and 78, are present but less closely related. Immediately downstream, gene 79 is homologous to the phage P22 rha (orf201) gene, whose product is detrimental for lytic growth in the absence of host integration host factor function (10, 40).
Thus, ES18 genome is a perfectly “typical” lambdoid phage in that it has a standard lambda-like transcriptional program, its predicted gene functions have the canonical lambdoid arrangement on the genome, and its genome is clearly mosaically related to other characterized lambdoid phage genomes. Perhaps the most notable feature of this mosaicism is that the ES18 head assembly genes are very different from any of the currently known lambdoid phages.
Initiation of DNA packaging by ES18.
Bacteriophage headful nucleases have no strong sequence specificity. This lack of specificity allows the packaging of some host DNA, and all naturally occurring generalized transducing phages utilize a headful packaging strategy. The generalized transducing phages that have been studied in detail, most notably phages P22 and P1, use a DNA recognition-cleavage mechanism that utilizes a pac site for recognition of DNA that is to be packaged (79, 80, 96). Replicated phage concatemeric DNA is recognized at a pac site by the phage terminase, a cut is made in the DNA at or near that point, and a processive series of packaging events proceeds in one direction from the DNA break thus produced. The result is virion DNA that is terminally redundant and partially circularly permuted (4, 42, 86, 87). When such virion DNA is cleaved by a restriction enzyme, a unique fragment, one of whose ends is the packaging series initiation cut, is generated only from the first member of each packaging series. This pac fragment is thus present in submolar amounts relative to the true restriction fragments (usually one-half to one-fourth of that expected if they were present in every DNA molecule). The presence of such a submolar fragment is considered to be diagnostic of this type of headful DNA packaging, as opposed to mechanisms like those of phages λ or T7, which generate ends at precisely the same location in all virion DNA molecules. However, the virion DNAs of some apparently headful packaging phages do not exhibit an obvious pac fragment, e.g., 933W (68), A118 (55), HK620 (21), APSE-1 (88), φHSIC (J. Paul, personal communication), Sf6 (17), φ11 (56), φ42 (64), Aaφ23 (94), and probably T4 (54). We have recently presented evidence that Sf6 packages DNA in a manner that is very similar to that of the phages that do exhibit a pac fragment; however, the DNA cut that initiates the packaging series occurs not at a precise location but at scattered locations within an 1,800-bp region such that the pac fragment electrophoresis band, although present, is so diffuse as to be essentially invisible to staining (17).
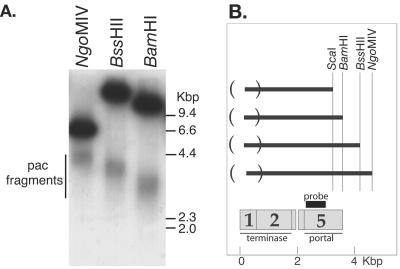
Schicklmaier and Schmieger (74) reported that ES18 virion DNA does not have cohesive ends and does not exhibit a pac fragment in ethidium bromide-stained electrophoresis gels. Our findings above show that its virion chromosome is about 10% terminally redundant and has an imprecise virion DNA length, strongly suggesting that ES18 utilizes a headful packaging mechanism similar to that of Sf6. To determine whether this is true, we searched for the diagnostic diffuse pac fragment. Figure 5A shows that restricted ES18 virion DNA does indeed exhibit a submolar, diffuse DNA band in electrophoresis gels when cleaved by the restriction endonucleases BamHI, BssHII, ScaI, or NgoMIV. This diffuse band is visible upon Southern analysis when using an appropriate probe, but it is not visible after ethidium bromide staining. Analysis of double digests showed that the variable end of each of these fragments is its left end (e.g., double digests of BamHI with BssHII or NgoMIV yields only the BamHI-sized diffuse band [data not shown]), and Fig. 5B shows that the diffuse ends of all of these fragments coincide and are centered on the putative ES18 small terminase subunit encoding gene 1. Each of these diffuse putative pac fragment bands is about 1,000 bp in width.
FIG. 5.
The variable-ended ES18 DNA pac fragment. (A) Southern analysis. DNA from a 1% agarose electrophoresis gel was transferred to a nylon membrane and probed with a 32P-labeled PCR-generated DNA from bp 2207 to 2829 of the ES18 nucleotide sequence. DNA in each lane was cut by the restriction endonuclease indicated above each lane. The upper dark band in each lane is the intact true restriction fragment that spans this region, and the lower diffuse band is the pac fragment (see text); neither band was present in uncut DNA (data not shown). (B) Initiation of ES18 packaging series. Dark horizontal lines represent the average length of the pac fragments aligned on the right according to the restriction enzyme that created them, and parentheses on the left enclose the region within which fragment left ends occur. Below, the locations of the probe used and the ES18 terminase and portal genes are indicated.
These findings support a model for ES18 DNA packaging in which initiation of DNA packaging series occurs at many locations between bp −300 (46,600) and 700 in the genome sequence, and DNA insertion into the procapsid proceeds rightward on the sequence from the site of initiation. This large initiation region is reminiscent of packaging by phage Sf6 (17) and is related to the well-characterized sequential headful packaging by phage P22 (96); however, ES18 initiation occurs within a region that is larger than P22's 120 bp and smaller than Sf6's 1,800 bp. Since the diffuse pac fragment band is submolar compared to true restriction fragments from outside the initiation region and because intact restriction fragments that span this region are present (Fig. 5A), it is almost certain that ES18, like P22, packages additional headfuls of DNA in a sequential manner along the concatemeric product of rolling-circle DNA replication. We note that in P22, Sf6, SPP1, T4, and P1 the region where the packaging series initiates is within or overlaps the gene that encodes the small terminase subunit (19, 54, 80, 96), the protein that carries the specificity for recognition of the DNA to be packaged (15, 16, 19). In P22 this site has been genetically identified, and it lies near the center of the 120-bp region within which P22 initiates DNA packaging (96); similarly, the Sf6 pac site also lies near the center of the 1,800-bp region within which it initiates packaging (E. Gilcrease and S. Casjens, unpublished results). We predict that ES18 will similarly carry a pac DNA packaging recognition site within its 1,000-bp packaging series initiation region, and we note that 8 of 10 bp and 10 of 11 bp at positions 240 to 250 and 462 to 472, respectively, are identical to sections of the pac site region of phage Sf6 (Gilcrease and Casjens, unpublished). It seems possible that one or both are part of the ES18 pac site. If true, its terminase must be able to either move or reach up to 500 bp along the DNA from the recognition site before cleaving to start a packaging series.
Terminases: functional types, phylogenetics, and genetic exchange.
Phage terminases recognize DNA for packaging and have a nuclease activity that is responsible for creating the ends of the virion chromosome (18). The putative gene 2-encoded ES18 large terminase subunit's closest relatives among the characterized phages are those from phages Sf6, HK620, APSE-1, and φHSIC, which are all headful packaging phages that exhibit no obvious, sharp pac fragment band upon electrophoretic separation of their restricted virion DNAs (see above). This observation prompted us to ask if the terminases that create different types of virion DNA ends were members of recognizably separable amino acid sequence groups. The dsDNA, tailed phage virion chromosomes are known to have several different types of end structures, including the (i) 5′- and (ii) 3′-protruding single-stranded cohesive ends with no terminal redundancy or circular permutation (e.g., 5′ phages λ and P2, 3′ HK97), (iii) direct terminal repeats with no circular permutation (e.g., phages T3 and T7), (iv) host DNA attached to both ends (e.g., phage Mu), and (v) the headful packaging phages that have virion DNAs with both terminal redundancy and circular permutation (e.g., phages T4, SPP1, P1, and P22). Although we did not include them in this analysis, we note that the eukaryotic herpesviruses appear to also have terminase proteins which are involved in viral genome maturation and which have sequences that are related to the phage terminases (69). In addition, phage φ29 and its relatives are unique among the tailed dsDNA phages in that they do not cleave the DNA during packaging and have a protein covalently bound to each DNA end. Although arguments can be made that φ29 gene 16 protein and its close relatives have similar (perhaps even homologous?) functions to other large terminase subunits, they have no convincing overall sequence similarity to the other terminases, so they were not included in the following analysis.
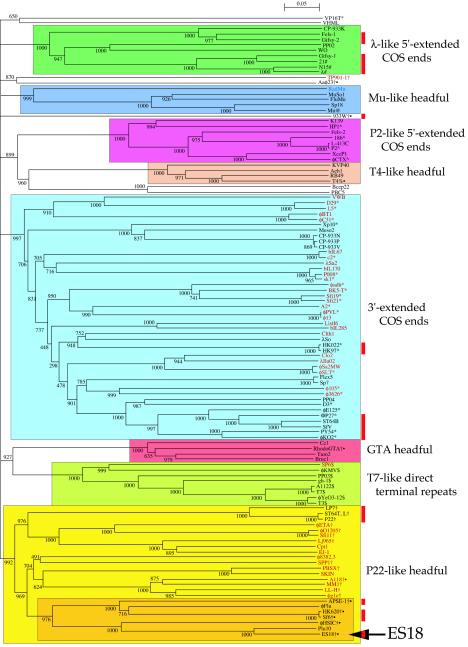
We compared a set of 114 large terminase subunit sequences that includes nearly all those for which both the terminase sequence and virion DNA end structure are known, as well as a number whose end structure is not known. This set includes all of the five types mentioned above. Figure 6shows a neighbor-joining tree of these large terminase subunits. They fall into at least eight robust groups that have bootstrap values of ≥992 out of a possible 1,000, each of which includes only terminases known to create a single type of DNA end (when they have been studied). Several of the major DNA packaging types form single, convincing groups, while others divide into several types. The 5′ cohesive end-forming enzymes split cleanly into two types, exemplified by those of phages λ and P2. The 3′ cohesive end-forming terminases cluster, with one exception (phage VP16T), into a robust single large group, as do the T7-like enzymes. The Mu-like phages—so classified on the grounds that they all appear to utilize a transposition replication mechanism—also form one robust terminase group. The headful packaging terminases fall into several separable groups at this resolution (see below). Maximum likelihood analysis (data not shown) gave nearly identical terminase clustering, further supporting the robustness of these groupings. Previous smaller studies (51, 61) have noticed relationships among some of these terminases but did not include enough terminases to see the complete picture presented here. Clearly, terminases with similar enzymatic end-generating functions usually cluster together.
FIG. 6.
Neighbor-joining tree of large terminase subunit amino acid sequences. The numbers near bifurcations are bootstrap values for 1,000 trials. Short bifurcating branches linking the major groups shown here, all of which had low bootstrap values as determined by CLUSTAL X, were originally present in the analysis. Thus, since this analysis does not robustly indicate the historical relationships among the major groups, these unsupported bifurcations were merged and the major deep branches are all shown as coming from a single source (the location of the root of this tree is unknown). The names of the phages or prophages are shown at the right of each terminal branch (some prophage names are those proposed by Casjens [9]); those in black print infect proteobacteria, those in red infect firmicutes, and blue is a Dienococcus prophage (63, 92). Those phages whose virion DNA termini have been reported are indicated as follows: #, 5′ cohesive ends in lambdoid phages; ^, 5′ cohesive ends in P2-like phages; *, 3′ cohesive ends; $, T7-like phages with direct terminal repeats and no circular permutation; @, Mu-like phages with host DNA at termini; †, headful packaging in non-T4-like phages; %, headful packaging in T4-like phages; •, headful packaging phages for which no obvious pac fragment band has been identified in electrophoresis gels of restricted DNA (see text). Vertical red bars on the right mark those lambdoid phages shown in Fig. 4.
Thus, we believe that the structure of virion DNA ends can be accurately predicted for phages where this is not known experimentally, if their putative terminase amino acid sequence falls convincingly within one of these robust groups. For example, we predict that Gifsy-2 (29, 59), SfV (1), and VWB (3) will be found to have λ type 5′ cohesive ends, 3′ cohesive ends, and headful packaging ends, respectively, in spite of the fact that several of these (e.g., Gifsy-2 and VWB) have terminases that are quite different from any studied terminase. This improved understanding of terminase relationships could also be helpful in searches for generalized transducing phages of particular bacteria, since all known generalized transducing phages utilize headful packaging and many such terminases can be recognized from their sequence.
Although the vast majority of large terminase subunits do fall into the above groups, we hasten to point out that these groups do not encompass all known terminases. In particular, 3′ cohesive end phages TM4, MS1, and r1t and headful packaging phages P1 and φ11, as well as T7-like VpV262, have terminases that are quite different from the majority of such enzymes and were not included in our analysis. TM4, MS1, and r1t, which form a related subgroup, usually fell within the 3′ cohesive end group. The inclusion of the above seven phage terminases sometimes (depending upon which combination was included) tended to lower the overall bootstrap value for their groups by virtue of their relatively weak affinities for those groups.
Several of the major Fig. 6 terminase groups have phage members that infect different classes of bacteria (e.g., proteobacteria and firmicutes in the 3′ cohesive end, P22-like headful, and T7-like groups), and terminases from different groups can be found in phages with very similar molecular lifestyles and that infect closely related host bacteria (e.g., the 3′-extended cohesive end terminases, 5′-extended cohesive end terminases, and headful terminases are found in the clearly otherwise lambdoid phages HK97, λ, and P22, respectively). This is no doubt only a small fraction of such relationships that exist, since the genomes of tailed phages that infect only a few of the many bacterial classes have been studied or sequenced, but it does suggest that either all types of terminases were present before the classes of bacteria diverged or, perhaps more likely, that in the past phages have switched hosts or phages have exchanged terminase genes across vast evolutionary distances (39). Possible examples of relatively recent terminase exchange among the lambdoid phages are the similar terminases of phages ES18 and Sf6, which are otherwise quite different members of this group; similarly, the terminases of Bacillus subtilis (firmicute) phage φ105 and E. coli (proteobacteria) ΦP27 lie within the same rather robust subgroup.
On the other hand, in spite of the accumulating evidence that phages have engaged in much horizontal exchange of genetic material over the eons, terminases do not appear to have been shuffled randomly among the dsDNA phages. For example, among those phages that have been studied in sufficient detail to know, all the terminases within the P2-like, Mu-like, T4-like, and T7-like groups (Fig. 6) are found only in phages that are sufficiently similar in other ways (e.g., transcriptional organization and replication strategies) to form the same groups without reference to their terminases. Such relationships may reflect as-yet-poorly understood functional connections between DNA packaging and other aspects of the lifestyle of these phages.
The headful packaging terminases appear to be more diverse than the other types. Three of the four well-characterized headful terminases fall into two separable groups, typified by T4 and P22/SPP1 (Fig. 6), while the fourth, that of phage P1 (not shown in Fig. 6), does not fall convincingly into any of the observed clusters. In addition, the Mu-like terminases and gene transfer agent (GTA) terminase, although they have not been studied in detail, must have headful properties, since neither packages DNA with sequence-specific ends. The Rhodobacter GTA (49, 50) putative terminase clusters with several others from prophage-like entities in the published Brucella (23), Agrobacterium (31, 95), and Caulobacter (65) genome sequences, and it will be interesting see if the latter three also have gene transfer function. Similarly, phage BBC5 (a Sinorhizobium meliloti phage; GenBank accession no. AF448724) and Bcep22 (a Burkholderia cepacia phage; GenBank accession no. AY349011) terminases cluster fairly robustly near the T4-like group and, although their packaging mechanisms have not been reported, this analysis suggests that they are likely to be headful packagers. Finally, the headful TP901-1 and Aaφ23 terminases form a small, moderately supported group, and those of 933W and φ11 (the latter is not shown in Fig. 6) do not cluster strongly with each other or any other terminase in our analysis set. This larger diversity of headful terminases compared to the other types suggests that they may be the ancestral terminase type.
The ES18 large terminase subunit falls into a headful subgroup with seven members (Fig. 6), five of which, ES18, Sf6, HK620, φHSIC, and APSE-1, have been reported to have virion DNA that has no cohesive end and no obvious pac fragment (the other two members of this subgroup are prophages in bacterial genome sequences whose parental phages have not been studied; both of these, φFlu in Haemophilus influenzae Rd and Plu10 in Photorhabdus luminescens TT01, are probably defective prophages [25, 39]). Headful terminases that do not generate a pac fragment are not, however, limited to this subgroup. For example, phages A118, 933W, φ11, and Aaφ23 are headful packagers that do not exhibit an obvious pac fragment, and their terminases fall outside of this subgroup (and, except for A118, are not robustly within any of the Fig. 6 groups). It may be that the presence of a pac fragment is a somewhat artificial distinction, since the three headful packaging phages that do show a pac fragment and have been studied in detail do not actually initiate packaging at a truly precise site, but the initiation cuts are distributed over a number of base pairs. In phages SPP1, P1, and P22, packaging series initiate with cuts that occur within 9-, 12-, and 120-bp regions, respectively (22, 80, 96). It may well be that no headful packagers initiate at a precise site and that the size of the regions over which different phages initiate packaging series is a continuum rather than falling neatly into “pac fragment” and “no pac fragment” categories. If true, it would not be surprising that the two types do not all break into neatly distinct groups.
Acknowledgments
This work was supported by grant MCB-990526 to S.C. from the National Science Foundation and by grant GM51975 from the National Institutes of Health to R.H. and G.H.
We thank John Paul and Guy Plunkett for information on the φHSIC terminase and φ80 J gene sequences, respectively, prior to their publication. We thank Jon Seger for help with PHYLIP analysis.
REFERENCES
- 1.Allison, G. E., D. Angeles, N. Tran-Dinh, and N. K. Verma. 2002. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J. Bacteriol. 184:1974-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anne, J., P. Fiten, M. L. Van, B. Joris, G. Opdenakker, and H. Eyssen. 1995. Analysis of the open reading frames of the main capsid proteins of actinophage VWB. Arch. Virol. 140:1033-1047. [DOI] [PubMed] [Google Scholar]
- 4.Bachi, B., and W. Arber. 1977. Physical mapping of BglII, BamHI, EcoRI, HindIII and PstI restriction fragments of bacteriophage P1 DNA. Mol. Gen. Genet. 153:311-324. [DOI] [PubMed] [Google Scholar]
- 5.Borodovsky, M., and J. McInish. 1993. GeneMark: parallel gene recognition for both strands. Comput. Chem. 17:123-133. [Google Scholar]
- 6.Braun, M., F. Endriss, H. Killmann, and V. Braun. 2003. In vivo reconstitution of the FhuA transport protein of Escherichia coli K-12. J. Bacteriol. 185:5508-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunny, K., J. Liu, and J. Roth. 2002. Phenotypes of lexA mutations in Salmonella enterica: evidence for a lethal lexA null phenotype due to the Fels-2 prophage. J. Bacteriol. 184:6235-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Callow, B. R. 1959. A new phage-typing scheme for Salmonella typhimurium. J. Hyg. 57:346-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens, S. 2003. Prophages in bacterial genomics: what have we learned so far? Mol. Microbiol. 249:277-300. [DOI] [PubMed] [Google Scholar]
- 10.Casjens, S., K. Eppler, R. Parr, and A. R. Poteete. 1989. Nucleotide sequence of the bacteriophage P22 gene 19 to 3 region: identification of a new gene required for lysis. Virology 171:588-598. [DOI] [PubMed] [Google Scholar]
- 11.Casjens, S., E. B. Gilcrease, W. M. Huang, K. L. Bunny, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186:1818-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casjens, S., and M. Hayden. 1988. Analysis in vivo of the bacteriophage P22 headful nuclease. J. Mol. Biol. 199:467-474. [DOI] [PubMed] [Google Scholar]
- 13.Casjens, S., and R. Hendrix. 1988. Control mechanisms in dsDNA bacteriophage assembly, p. 15-91. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y.
- 14.Casjens, S., and W. M. Huang. 1993. Linear chromosomal physical and genetic map of Borrelia burgdorferi, the Lyme disease agent. Mol. Microbiol. 8:967-980. [DOI] [PubMed] [Google Scholar]
- 15.Casjens, S., W. M. Huang, M. Hayden, and R. Parr. 1987. Initiation of bacteriophage P22 DNA packaging series. Analysis of a mutant that alters the DNA target specificity of the packaging apparatus. J. Mol. Biol. 194:411-422. [DOI] [PubMed] [Google Scholar]
- 16.Casjens, S., L. Sampson, S. Randall, K. Eppler, H. Wu, J. B. Petri, and H. Schmieger. 1992. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 227:1086-1099. [DOI] [PubMed] [Google Scholar]
- 17.Casjens, S., D. Winn-Stapley, E. Gilcrease, R. Moreno, C. Kühlewein, J. E. Chua, P. A. Manning, W. Inwood, and A. J. Clark. 2004. The chromosome of Shigella flexneri bacteriophage Sf6: complete nucleotide sequence, genetic mosaicism, and DNA packaging. J. Mol. Biol. 339:379-394. [DOI] [PubMed] [Google Scholar]
- 18.Catalano, C. E., D. Cue, and M. Feiss. 1995. Virus DNA packaging: the strategy used by phage lambda. Mol. Microbiol. 16:1075-1086. [DOI] [PubMed] [Google Scholar]
- 19.Chai, S., R. Lurz, and J. C. Alonso. 1995. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J. Mol. Biol. 252:386-398. [DOI] [PubMed] [Google Scholar]
- 20.Cheng, H., N. Shen, J. Pei, and N. V. Grishin. 2004. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 13:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark, A. J., W. Inwood, T. Cloutier, and T. S. Dillon. 2001. Nucleotide sequence of coliphage HK620 and the evolution of lambdoid phages. J. Bacteriol. 311:657-679. [DOI] [PubMed] [Google Scholar]
- 22.Deichelbohrer, I., J. C. Alonso, G. Luder, and T. A. Trautner. 1985. Plasmid transduction by Bacillus subtilis bacteriophage SPP1: effects of DNA homology between plasmid and bacteriophage. J. Bacteriol. 162:1238-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas, S. E. 1994. DNA Strider. A Macintosh program for handling protein and nucleic acid sequences. Methods Mol. Biol. 25:181-194. [DOI] [PubMed] [Google Scholar]
- 25.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 26.Duda, R. L. 1998. Protein chainmail: catenated protein in viral capsids. Cell 94:55-60. [DOI] [PubMed] [Google Scholar]
- 27.Eppler, K., E. Wyckoff, J. Goates, R. Parr, and S. Casjens. 1991. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology 183:519-538. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein, J. 1989. PHYLIP—Phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 29.Figueroa-Bossi, N., E. Coissac, P. Netter, and L. Bossi. 1997. Unsuspected prophage-like elements in Salmonella typhimurium. Mol. Microbiol. 25:161-173. [DOI] [PubMed] [Google Scholar]
- 30.Flinta, C., B. Persson, H. Jornvall, and G. von Heijne. 1986. Sequence determinants of cytosolic N-terminal protein processing. Eur. J. Biochem. 154:193-196. [DOI] [PubMed] [Google Scholar]
- 31.Goodner, B., G. Hinkle, S. Gattung, N. Miller, M. Blanchard, B. Qurollo, B. S. Goldman, Y. Cao, M. Askenazi, C. Halling, L. Mullin, K. Houmiel, J. Gordon, M. Vaudin, O. Iartchouk, A. Epp, F. Liu, C. Wollam, M. Allinger, D. Doughty, C. Scott, C. Lappas, B. Markelz, C. Flanagan, C. Crowell, J. Gurson, C. Lomo, C. Sear, G. Strub, C. Cielo, and S. Slater. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323-2328. [DOI] [PubMed] [Google Scholar]
- 32.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 33.Helgstrand, C., W. R. Wikoff, R. L. Duda, R. W. Hendrix, J. E. Johnson, and L. Liljas. 2003. The refined structure of a protein catenane: the HK97 bacteriophage capsid at 3.44 Å resolution. J. Mol. Biol. 334:885-899. [DOI] [PubMed] [Google Scholar]
- 34.Hendrix, R., and S. Casjens. Bacteriophage λ and its genetic neighborhood. In R. Calendar (ed.), The bacteriophages, 2nd ed., in press. Oxford Press, New York, N.Y.
- 35.Hendrix, R. W., and S. R. Casjens. 1975. Assembly of bacteriophage lambda heads: protein processing and its genetic control in petit lambda assembly. J. Mol. Biol. 91:187-199. [DOI] [PubMed] [Google Scholar]
- 36.Hendrix, R. W., and S. R. Casjens. 1974. Protein fusion during the assembly of phage lambda heads. J. Supramol. Struct. 2:329-336. [DOI] [PubMed] [Google Scholar]
- 37.Hendrix, R. W., and S. R. Casjens. 1974. Protein fusion: a novel reaction in bacteriophage lambda head assembly. Proc. Natl. Acad. Sci. USA 71:1451-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrix, R. W., J. G. Lawrence, G. F. Hatfull, and S. Casjens. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504-508. [DOI] [PubMed] [Google Scholar]
- 39.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henthorn, K. S., and D. I. Friedman. 1995. Identification of related genes in phages φ80 and P22 whose products are inhibitory for phage growth in Escherichia coli IHF mutants. J. Bacteriol. 177:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hertwig, S., I. Klein, V. Schmidt, S. Beck, J. A. Hammerl, and B. Appel. 2003. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 331:605-622. [DOI] [PubMed] [Google Scholar]
- 42.Jackson, E. N., D. A. Jackson, and R. J. Deans. 1978. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 118:365-388. [DOI] [PubMed] [Google Scholar]
- 43.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 44.Jiang, W., Z. Li, Z. Zhang, M. L. Baker, P. E. Prevelige, Jr., and W. Chiu. 2003. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 10:131-135. [DOI] [PubMed] [Google Scholar]
- 45.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 46.Katsura, I. 1983. Tail assembly and injection, p. 331-346. In R. Hendrix, J. Roberts, F. W. Stahl, and R. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Killmann, H., M. Braun, C. Herrmann, and V. Braun. 2001. FhuA barrel-cork hybrids are active transporters and receptors. J. Bacteriol. 183:3476-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuo, T. T., and B. A. Stocker. 1970. ES18, a general transducing phage for smooth and nonsmooth Salmonella typhimurium. Virology 42:621-632. [DOI] [PubMed] [Google Scholar]
- 49.Lang, A. S., and J. T. Beatty. 2001. The gene transfer agent of Rhodobacter capsulatus and “constitutive transduction” in prokaryotes. Arch. Microbiol. 175:241-249. [DOI] [PubMed] [Google Scholar]
- 50.Lang, A. S., J. T. Beatty, H. LeBlanc, G. Towers, J. Harris, G. Lang, M. K. Collins, D. S. Latchman, K. Orchard, M. Collins, and D. Latchman. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. USA 97:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Marrec, C., D. van Sinderen, L. Walsh, E. Stanley, E. Vlegels, S. Moineau, P. Heinze, G. Fitzgerald, and B. Fayard. 1997. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 63:3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Minor, L., and A. M. Chalon. 1975. Sensitivity to bacteriophage ES18 of strains of “S. dublin,” “S. enteritidis” and “S. blegdam” and related serotypes. Ann. Microbiol. (Paris) 126:327-331. [PubMed] [Google Scholar]
- 53.Levin, M. E., R. W. Hendrix, and S. R. Casjens. 1993. A programmed translational frameshift is required for the synthesis of a bacteriophage λ tail assembly protein. J. Mol. Biol. 234:124-139. [DOI] [PubMed] [Google Scholar]
- 54.Lin, H., and L. W. Black. 1998. DNA requirements in vivo for phage T4 packaging. Virology 242:118-127. [DOI] [PubMed] [Google Scholar]
- 55.Loessner, M. J., R. B. Inman, P. Lauer, and R. Calendar. 2000. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35:324-340. [DOI] [PubMed] [Google Scholar]
- 56.Lofdahl, S., J. Zabielski, and L. Philipson. 1981. Structure and restriction enzyme maps of the circularly permuted DNA of staphylococcal bacteriophage φ11. J. Virol. 37:784-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsumoto, M., N. Ichikawa, S. Tanaka, T. Morita, and A. Matsushiro. 1985. Molecular cloning of the φ80 adsorption-inhibiting cor gene. Jpn. J. Genet. 60:475-483. [Google Scholar]
- 59.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 60.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9845-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell, M. S., S. Matsuzaki, S. Imai, and V. B. Rao. 2002. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 30:4009-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mmolawa, P. T., H. Schmieger, C. P. Tucker, and M. W. Heuzenroeder. 2003. Genomic structure of the Salmonella enterica serovar Typhimurium DT 64 bacteriophage ST64T: evidence for modular genetic architecture. J. Bacteriol. 185:3473-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morgan, G., G. Hatfull, S. Casjens, and R. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317:337-359. [DOI] [PubMed] [Google Scholar]
- 64.Moynet, D. J., and F. M. DeFilippes. 1982. Characterization of bacteriophage φ42 DNA. Virology 117:475-484. [DOI] [PubMed] [Google Scholar]
- 65.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, C. M. Fraser, and J. Eisen. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 67.Pedulla, M. L., M. E. Ford, T. Karthikeyan, J. M. Houtz, R. W. Hendrix, G. F. Hatfull, A. R. Poteete, E. B. Gilcrease, D. A. Winn-Stapley, and S. R. Casjens. 2003. Corrected sequence of the bacteriophage P22 genome. J. Bacteriol. 185:1475-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Przech, A. J., D. Yu, and S. K. Weller. 2003. Point mutations in exon I of the herpes simplex virus putative terminase subunit, UL15, indicate that the most conserved residues are essential for cleavage and packaging. J. Virol. 77:9613-9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pullinger, G. D., T. Bevir, and A. J. Lax. 2004. The Pasteurella multocida toxin is encoded within a lysogenic bacteriophage. Mol. Microbiol. 51:255-269. [DOI] [PubMed] [Google Scholar]
- 71.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 72.Recktenwald, J., and H. Schmidt. 2002. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage ΦP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect. Immun. 70:1896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roberts, M. D., N. L. Martin, and A. M. Kropinski. 2004. The genome and proteome of coliphage T1. Virology 318:245-266. [DOI] [PubMed] [Google Scholar]
- 74.Schicklmaier, P., and H. Schmieger. 1997. Sequence comparison of the genes for immunity, DNA replication, and cell lysis of the P22-related Salmonella phages ES18 and L. Gene 195:93-100. [DOI] [PubMed] [Google Scholar]
- 75.Schmieger, H. 1999. Molecular survey of the Salmonella phage typing system of Anderson. J. Bacteriol. 181:1630-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Selander, R. K., P. Beltran, N. H. Smith, R. M. Barker, P. B. Crichton, D. C. Old, J. M. Musser, and T. S. Whittam. 1990. Genetic population structure, clonal phylogeny, and pathogenicity of Salmonella paratyphi B. Infect. Immun. 58:1891-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Southern, E. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 78.Staden, R. 1986. The current status and portability of our sequence handling software. Nucleic Acids Res. 14:217-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sternberg, N., and J. Coulby. 1987. Recognition and cleavage of the bacteriophage P1 packaging site (pac). I. Differential processing of the cleaved ends in vivo. J. Mol. Biol. 194:453-468. [DOI] [PubMed] [Google Scholar]
- 80.Sternberg, N., and J. Coulby. 1987. Recognition and cleavage of the bacteriophage P1 packaging site (pac). II. Functional limits of pac and location of pac cleavage termini. J. Mol. Biol. 194:469-479. [DOI] [PubMed] [Google Scholar]
- 81.Stiege, A. C., A. Isidro, A. Droge, and P. Tavares. 2003. Specific targeting of a DNA-binding protein to the SPP1 procapsid by interaction with the portal oligomer. Mol. Microbiol. 49:1201-1212. [DOI] [PubMed] [Google Scholar]
- 82.Susskind, M. M., and D. Botstein. 1978. Molecular genetics of bacteriophage P22. Microbiol. Rev. 42:385-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tavares, P., R. Lurz, A. Stiege, B. Ruckert, and T. A. Trautner. 1996. Sequential headful packaging and fate of the cleaved DNA ends in bacteriophage SPP1. J. Mol. Biol. 264:954-967. [DOI] [PubMed] [Google Scholar]
- 84.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toth, E. A., Y. Li, M. R. Sawaya, Y. Cheng, and T. Ellenberger. 2003. The crystal structure of the bifunctional primase-helicase of bacteriophage T7. Mol. Cell 12:1113-1123. [DOI] [PubMed] [Google Scholar]
- 86.Tye, B. K., R. K. Chan, and D. Botstein. 1974. Packaging of an oversize transducing genome by Salmonella phage P22. J. Mol. Biol. 85:485-500. [DOI] [PubMed] [Google Scholar]
- 87.Tye, B. K., J. A. Huberman, and D. Botstein. 1974. Non-random circular permutation of phage P22 DNA. J. Mol. Biol. 85:501-528. [DOI] [PubMed] [Google Scholar]
- 88.van der Wilk, F., A. M. Dullemans, M. Verbeek, and J. F. van den Heuvel. 1999. Isolation and characterization of APSE-1, a bacteriophage infecting the secondary endosymbiont of Acyrthosiphon pisum. Virology 262:104-113. [DOI] [PubMed] [Google Scholar]
- 89.Vostrov, A., O. Vostrukhina, A. Svarchevsky, and V. Rybchin. 1996. Proteins responsible for lysogenic conversion caused by coliphages N15 and φ80 are highly homologous. J. Bacteriol. 178:1484-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Walker, J. E., A. D. Auffret, A. Carne, A. Gurnett, P. Hanisch, D. Hill, and M. Saraste. 1982. Solid-phase sequence analysis of polypeptides eluted from polyacrylamide gels. An aid to interpretation of DNA sequences exemplified by the Escherichia coli unc operon and bacteriophage lambda. Eur. J. Biochem. 123:253-260. [DOI] [PubMed] [Google Scholar]
- 91.Wang, J., M. Hofnung, and A. Charbit. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, C. M. Fraser, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wikoff, W. R., L. Liljas, R. L. Duda, H. Tsuruta, R. W. Hendrix, and J. E. Johnson. 2000. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289:2129-2133. [DOI] [PubMed] [Google Scholar]
- 94.Willi, K., and J. Meyer. 1998. DNA analysis of temperate bacteriophage Aaφ23 isolated from Actinobacillus actinomycetemcomitans. Mol. Gen. Genet. 258:323-325. [DOI] [PubMed] [Google Scholar]
- 95.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr., P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 96.Wu, H., L. Sampson, R. Parr, and S. Casjens. 2002. The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol. Microbiol. 45:1631-1646. [DOI] [PubMed] [Google Scholar]
- 97.Xu, J., R. W. Hendrix, and R. L. Duda. 2004.. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11-21 [DOI] [PubMed] [Google Scholar]
- 98.Yamamoto, N. 1978. A generalized transducing Salmonella phage ES18 can recombine with a serologically unrelated phage Fels 1. J. Gen. Virol. 38:263-272. [DOI] [PubMed] [Google Scholar]