Abstract
Salmonella enterica serovar Typhimurium has two manganese transport systems, MntH and SitABCD. MntH is a bacterial homolog of the eukaryotic natural resistance-associated macrophage protein 1 (Nramp1), and SitABCD is an ABC-type transporter. Previously we showed that mntH is negatively controlled at the transcriptional level by the trans-acting regulatory factors, MntR and Fur. In this study, we examined the transcriptional regulation of sitABCD and compared it to the transcriptional regulation of mntH by constructing lacZ fusions to the promoter regions with and without mutations in putative MntR and/or Fur binding sites. The presence of Mn caused transcriptional repression of the sitABCD and mntH promoters primarily via MntR, but Fur was also capable of some repression in response to Mn. Likewise, Fe in the medium repressed transcription of both sit and mntH primarily via Fur, although MntR was also involved in this response. Transcriptional control by MntR and Fur was disrupted by site-specific mutations in the putative MntR and Fur binding sites, respectively. Transcription of the sit operon was also affected by the oxygen level and growth phase, but the increased expression observed under high oxygen conditions and higher cell densities is consistent with decreased availability of metals required for repression by the metalloregulatory proteins.
Salmonella enterica serovar Typhimurium has two known manganese transport systems, MntH and SitABCD (20-22). MntH is a bacterial homolog of the eukaryotic natural resistance-associated macrophage protein 1 (Nramp1), a cation transporter (22, 24). Loss of Nramp1 in mice leads to increased susceptibility to intracellular pathogens, including Salmonella (39). Homologs of Nramp1 are found in several bacterial species, including Bacillus subtilis, Escherichia coli, and Salmonella (22, 24, 32). The second manganese transporter of Salmonella is an ABC-type transporter named SitABCD (20). The genes encoding SitABCD are located in the pathogenicity island SPI-1 and were first identified upon sequencing of that island (43). Although both MntH and SitABCD can transport iron, it is believed that manganese is the primary divalent metal transported by these two systems under physiological conditions (20, 22).
Control of metal ion transport is often mediated by metalloregulatory proteins. In response to the presence or absence of metal ions, these proteins activate or repress gene transcription. Several metalloregulatory proteins have been identified and include Fur, DtxR, Mer, and SmtB/AsrR, each being representative of a larger family of proteins (5, 7, 15). Fur was initially discovered in E. coli, and homologs in other bacteria, including Salmonella, were found later (11, 15). In response to iron, Fur regulates genes encoding iron transport systems as well as transport systems for other metals, metabolic enzymes, oxidative stress response functions, and virulence factors (11). Other Fur-like homologs (Zur and PerR) that respond to metals other than iron, namely, Zn and Mn, are known (6, 29). Regulation by Fur and putative Fur binding sites in the promoter regions were reported for mntH of E. coli (30) and Salmonella (19) and for sitABCD of Salmonella (18, 43). DtxR was first identified as an iron-dependent repressor of toxin production in Corynebacterium diphtheriae and later found to control other iron-responsive genes (37). Although there is no sequence similarity between DtxR and Fur (less than 20% identity), there is evidence for structural homology (12). Recently, a manganese-dependent metalloregulatory protein that was similar to MntR of B. subtilis and belonged to the DtxR family of proteins was discovered in E. coli (30). After the protein was purified, it was shown by DNase I footprinting that it bound to an inverted repeat located in the mntH promoter of E. coli. Similar inverted repeats are found in the mntH and sitABCD promoter regions of Salmonella (22), and an MntR ortholog was identified in Salmonella (19).
The importance of manganese and manganese transport in Salmonella pathogenesis is only beginning to be studied (21, 42). Although the genes encoding SitABCD are located in the pathogenicity island SPI-1, responsible for invasion of epithelial cells, the sit operon does not appear to be involved in invasion (18, 41, 43). Consistent with those results, sit appears to be induced in mice after invasion of the intestinal mucosa (18) and after uptake by macrophage cells in culture (41). Similarly, mntH is not involved in the invasion of HeLa cells, and the expression of mntH is induced after phagocytosis by macrophages (22, 41). Virulence studies show a clear role for both sit and mntH during infection (4, 18, 21, 41). A virulence defect is particularly evident when both Mn transporters are mutated (4, 41). The phenotype is also enhanced in Nramp1+/+ mice, suggesting that the host and bacterial transporters could compete for available Mn (41).
In this study, we examine the transcriptional regulation of sitABCD in Salmonella by divalent metals Fe and Mn. Fur and MntR, acting at their cognate binding sites in the promoter regions, control the sitABCD operon and mntH gene. Although Fur responds primarily to iron and MntR responds primarily to manganese, each metalloregulatory protein also represses transcription in the presence of the other metal, albeit at lower efficiency. Transcriptional changes in response to the environment primarily reflect the availability of metals.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All Salmonella strains used in this study (except JS198) are isogenic derivatives of S. enterica serovar Typhimurium strain 14028 (American Type Culture Collection) and are shown in Table 1. M63 medium supplemented with 0.2% glucose (34) or M63 medium supplemented with 0.2% glucose and 0.1% Casamino Acids, followed by a batch treatment with a chelating resin (Chelex 100; Bio-Rad), was used for minimal medium conditions. Luria-Bertani (LB) broth or agar was used as rich medium (34). When required, the following antibiotics were included in the culture medium at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 25 μg/ml. Transcriptional lacZ fusion constructs used in β-galactosidase assays were grown by the following basic procedure. A single colony was used to inoculate LB broth and incubated at 37°C with shaking for 20 h. The culture was washed twice with 0.85% NaCl, and the final pellet was resuspended in an equal volume of 0.85% NaCl. The washed cells were diluted 1:100 in minimal medium with or without added metals and grown to stationary phase by incubating at 37°C with shaking for 20 h.
TABLE 1.
Salmonella serovar Typhimurium strains used in this study
| Strain | Genotype | Source or referencea |
|---|---|---|
| 14028 | Wild-type | ATCCb |
| JS198 | LT2 metE551 metA22 ilv-452 trpB2 hisC527(Am) galE496 xyl-404 rpsL120 flaA66 hsdL6 hsdSA29 zjg-8103::pir+recA1 | 10 |
| JS210 | 14028 Φ(sitA′-lac+)110 | 10 |
| JS211 | JS210 Δfur-41::cat | 10 |
| JS390 | JS210 ΔmntR51 | |
| JS391 | JS210 ΔmntR51 Δfur-41::cat | |
| JS392 | JS210 fnr-2::Tn10 Δarc | |
| JS401 | 14028 ΔmntR51 | |
| JS402 | 14028 Δfur-41::cat | |
| JS403 | 14028 ΔmntR51 Δfur-41::cat | |
| JS404 | 14028 attλ::pAH125::sitA′-lac+ (wild-type binding sites) | |
| JS405 | 14028 attλ::pAH125::sitA′-lac+ (mutated Fur binding site) | |
| JS406 | 14028 attλ::pAH125::sitA′-lac+ (mutated MntR binding site) | |
| JS407 | 14028 attλ::pAH125::sitA′-lac+ (mutated MntR and Fur binding sites) | |
| JS408 | JS404 ΔmntR51 | |
| JS409 | JS405 ΔmntR51 | |
| JS410 | JS406 ΔmntR51 | |
| JS411 | JS407 ΔmntR51 | |
| JS412 | JS404 Δfur-41::cat | |
| JS413 | JS405 Δfur-41::cat | |
| JS414 | JS406 Δfur-41::cat | |
| JS415 | JS407 Δfur-41::cat | |
| JS416 | JS404 ΔmntR51 Δfur-41::cat | |
| JS417 | JS405 ΔmntR51 Δfur-41::cat | |
| JS418 | JS406 ΔmntR51 Δfur-41::cat | |
| JS419 | JS407 ΔmntR51 Δfur-41::cat | |
| JS420 | 14028 attλ::pAH125::mntH′-lac+ (wild-type binding sites) | |
| JS421 | 14028 attλ::pAH125::mntH′-lac+ (mutated Fur binding site) | |
| JS422 | 14028 attλ::pAH125::mntH′-lac+ (mutated MntR binding site) | |
| JS423 | 14028 attλ::pAH125::mntH′-lac+ (mutated MntR and Fur binding sites) | |
| JS424 | JS420 ΔmntR51 | |
| JS425 | JS421 ΔmntR51 | |
| JS426 | JS422 ΔmntR51 | |
| JS427 | JS423 ΔmntR51 | |
| JS428 | JS420 Δfur-41::cat | |
| JS429 | JS421 Δfur-41::cat | |
| JS430 | JS422 Δfur-41::cat | |
| JS431 | JS423 Δfur-41::cat | |
| JS432 | JS420 ΔmntR51 Δfur-41::cat | |
| JS433 | JS421 ΔmntR51 Δfur-41::cat | |
| JS434 | JS422 ΔmntR51 Δfur-41::cat | |
| JS435 | JS423 ΔmntR51 Δfur-41::cat | |
| JS436 | 14028 Φ(mntR+-lac+)52 | |
| JS437 | JS436 rpoS::tet | |
| JS438 | JS436 Δfur-41::cat |
This study, unless otherwise noted.
ATCC, American Type Culture Collection.
For the growth phase assays, washed cells from the culture grown overnight were inoculated into minimal medium containing 20 μM Mn or into LB broth and then incubated at 37°C with shaking until early exponential phase of growth was reached (optical density at 600 nm [OD600] of 0.2). The culture was then diluted 1/2 with fresh appropriate medium and incubated until it reached an OD600 of 0.25. The cells were then washed with 0.85% NaCl, resuspended in the same medium, diluted 1:100 in the appropriate medium, and incubated until early exponential phase (OD600 of 0.2) or stationary growth phase (20 h incubation) was attained. Aerobically grown cells were incubated with shaking in loosely capped tubes, and anaerobically grown cells were incubated statically in 14.5 ml of broth in tightly capped 15-ml tubes.
Molecular biology and biochemical methods.
Standard molecular biology techniques were used and are briefly described here. Plasmids, chromosomal DNA, and amplified DNA fragments were isolated and purified using commercially available kits (QIAGEN). Amplification of DNA was performed by PCR using Taq polymerase (Promega) or Pfx DNA polymerase (Invitrogen) according to suggested protocols. Primers were synthesized by Integrated DNA Technologies, Inc. DNA sequence analyses were conducted by the W. M. Keck Center for Comparative and Functional Genomics (University of Illinois at Urbana-Champaign). Restriction enzymes were used in accordance with the manufacturer's suggestions (Invitrogen or New England Biolabs). Transformation by electroporation and transduction were performed by the methods of Maloy et al. (25). Mutations in specific genes were created by the Lambda Red recombination method (8, 40) as described by Ellermeier et al. (10). Transcriptional activity was determined by performing β-galactosidase assays in microtiter plates as described previously (35).
Construction of lacZ transcriptional fusions.
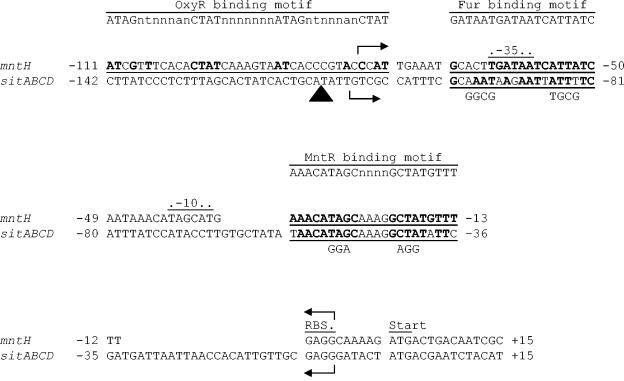
The single-copy lacZ transcriptional fusions to sitA used in some experiments and the fusions to mntR were constructed using pCE37 and pKG136, respectively, as described previously (10). Fusions of the mntH and sit promoter regions, with wild-type or mutant sequences, were constructed in the single-copy lacZ fusion vector pAH125 (14). The promoter region cloned upstream of lacZ, along with bases changed in the Fur and MntR binding sites, is indicated in Fig. 1.
FIG. 1.
Comparison of the sitABCD and mntH promoter sequences. The DNA sequence of the sitABCD promoter region is aligned with the promoter region of mntH. Nucleotide bases are numbered relative to the beginning of the start codon, which is designated +1. The consensus OxyR binding motif (38), Fur binding motif (9, 23), and MntR binding motif (based on the mntH promoter of E. coli) (30) are shown above the corresponding putative sites underlined in the mntH (19) and sitABCD promoter regions. Bases that match between the putative binding sequences and the consensus sequences are shown in bold type. Bases that were changed in the Fur binding and/or MntR binding site mutants are shown below the putative binding sequences. The predicted translational start site (Start), ribosomal binding site (RBS.), −10 site, and −35 site are indicated. The boundary of SPI-1 based on Mills et al. (26) is shown in the sitABCD sequence by a large black triangle. Arrows demarcate the regions cloned in the lacZ fusion constructs.
Briefly, the wild-type promoters were amplified from S. enterica serovar Typhimurium strain 14028 by PCR using primers sitP100 (5′-ACAACTGCAG TCGCCATTTC GCAAATAAGA ATTATTTTCA TT-3′) and sitP160 (5′-CGAATTCCTC GCAACAATGT GGTTAATTAA TCATC-3′) for the sit promoter and primers mntP100 (5′-ACAACTGCAG CCATTGAAAT GCACTTGATA ATCATTATCA AT-3′) and mntP150 (5′-CGAATTCCTC AAAAACATAG CCTTTGCTAT GTTTCA-3′) for the mntH promoter. These primers add PstI (sitP100 and mntP100) or EcoRI (sitP160 and mntP150) restriction sites to the ends of the amplified promoter sequences. Primers used to generate mutations in the putative Fur binding sites (sitP101 for the sit promoter, 5′-ACAACTGCAG TCGCCATTTC GCGGCGAAGA ATTTGCGTCA TT-3′; mntP101 for the mntH promoter, 5′-ACAACTGCAG CCATTGAAAT GCGGCGGATA ATCTGCGTCA AT-3′) and/or MntR binding sites (sitP151 for the sit promoter, 5′-GGTTAATTAA TCATCGAATA CCTCCTTTGT CCTGTTATA-3′; mntP151 for the mntH promoter, 5′-CGAATTCCTC AAAAACACCT CCTTTGTCCT GTTTCA-3′) were substituted for one or more of the wild-type primers in the amplification step. In the case of sit constructs with mutations in the MntR binding site, the fragments were initially amplified using primers sitP100 and sitP151, and the resulting product was used as the template in a PCR using primers sitP100 (or sitP101) and sitP160.
All amplified fragments were purified by gel extraction, digested with PstI and EcoRI, and ligated into the corresponding restriction sites of the conditional replication, integration, and modular (CRIM) reporter plasmid, pAH125 (14). The cloned inserts were verified by DNA sequence analysis. The plasmid constructs were then electroporated into serovar Typhimurium JS198 (r− m+ π+), reisolated, and electroporated into serovar Typhimurium strain 14028 containing the CRIM helper plasmid pINT-ts for integration into the host chromosome at attλ by the method of Haldimann and Wanner (14). Finally, the chromosomally integrated promoter-lacZ fusions were transduced into the following strains using P22Htint: 14028, 14028 ΔmntR (JS401), 14028 Δfur::cat (JS402), and 14028 ΔmntR Δfur::cat (JS403).
RESULTS
Comparison of the sitABCD and mntH promoter sequences.
The two known Mn transport systems in Salmonella belong to the ABC-type or Nramp-type transport system. Although the transporters involved are not functionally similar, the promoter regions of sitABCD and mntH do share common features (Fig. 1). The mntH promoter of Salmonella contains putative sites for binding OxyR, Fur, and MntR, consistent with the regulation of mntH by hydrogen peroxide, iron, and manganese, respectively (19, 22). The sitABCD promoter also contains putative Fur and MntR binding sites. Indeed, it has been shown previously that transcription of sitABCD is repressed by Fur in response to iron (18, 43). The putative Fur binding site in sitABCD (Fig. 1) was identified by comparison with the Fur binding sites of mntH from E. coli (30) and Salmonella (19, 22) and the known consensus Fur binding sequence (9, 23). This site is directly upstream of the Fur binding site suggested by Zhou et al. (43). However, it is likely that the Fur site is larger than the 19-bp consensus sequence extending to position −74 in Fig. 1, showing an F-F-x-R-R architecture as defined by Lavrrar and McIntosh (23).
Genomic analysis suggests four MntR binding sites in the S. enterica serovar Typhimurium LT2 sequence (19). These four sites include sites upstream of mntH, sitABCD, mntR, and yebN, which encodes a putative integral membrane protein of unknown function. Stojiljkovic et al. (36) also identified a Fur binding site in the yebN promoter region. The putative MntR binding site in the sit promoter is very similar to that in the mntH promoter, except for the spacing with respect to the putative Fur binding sites (Fig. 1). In sit, these sites are separated by an additional 8 bp or approximately a 3/4 turn of the helix.
Unlike mntH (19), an OxyR binding motif (38) is not apparent in the sitABCD promoter, and we have shown that activation by hydrogen peroxide does not occur for sitABCD (data not shown). In fact, sitABCD lies at the end of the SPI-1 pathogenicity island (43) such that the boundary of SPI-1 is located immediately upstream of the Fur binding site (26) and within the DNA that corresponds to the OxyR binding site of mntH (19) (Fig. 1). It is interesting to speculate that an OxyR binding site may have existed in sitABCD but it was disrupted during the formation and/or acquisition of SPI-1 in the Salmonella chromosome. Alternatively, MntH could be particularly suited to function under conditions of oxidative stress.
The sitABCD promoter is regulated by MntR and Fur.
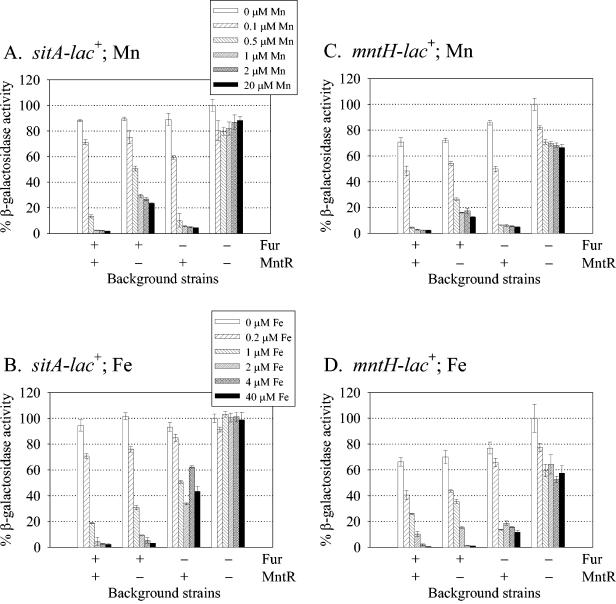
The similarity of the sitABCD promoter with the mntH promoter and the presence of a putative MntR binding site suggested that sitABCD is regulated by MntR in addition to Fur. To directly test this hypothesis, we monitored expression of the sitABCD and mntH promoters in the presence and absence of MntR and/or Fur. Transcriptional lacZ fusions were constructed with the wild-type sit and mntH promoter regions containing the putative Fur and MntR binding sites (Fig. 1). The OxyR binding site of the mntH promoter was not included in the mntH construct for comparison with the sitABCD promoter (which does not contain an OxyR binding site) and to remove any confounding effects of OxyR regulation. The fusion constructs were integrated in single copy into the chromosome at attλ. Thus, the chromosomal sit and mntH loci are wild type in these strains. The fusion strains were grown in defined minimal medium that had been treated with Chelex resin to minimize the amounts of available Mn and Fe. Various concentrations of Mn and Fe were added back to the minimal medium (between 0.1 and 20 μM for Mn and between 0.2 and 40 μM for Fe) to observe any repressive effects on transcription (Fig. 2).
FIG. 2.
Effects of various concentrations of Mn or Fe on the transcriptional activities of the sitABCD and mntH promoters. Transcriptional lac fusions with the wild-type sit (A and B) or mntH (C and D) promoters were assayed in wild-type, mntR, fur, and mntR fur background strains. Strains were grown overnight in minimal medium or minimal medium containing various concentrations of Mn (A and C) (0.1 to 20 μM) or Fe (B and D) (0.2 to 40 μM). β-Galactosidase activities were normalized to the activity of the mntR fur background strain grown in minimal medium without added metals. Data are presented as means ± standard deviations (error bars) (n = 4). Strains JS404, JS408, JS412, JS416, JS420, JS424, JS428, and JS432 were used.
The addition of Mn caused at least a 34-fold decrease in sit transcription in a wild-type background (Fig. 2A). Essentially full repression was evident with 1 μM Mn. This repression was mediated primarily by MntR. However, in the absence of MntR, the promoter was still repressed threefold with 1 μM Mn, and this repression was dependent on Fur. Similarly, the sit promoter was repressed at least 21-fold in the presence of 2 μM Fe (Fig. 2B). Most of this repression was dependent on Fur, but MntR could repress the promoter more than twofold in response to Fe. A caveat to this interpretation is that the metals that we are adding could be contaminated. However, on the basis of the manufacturer's chemical analyses, we would expect that the 20 μM Mn would contain less than 0.3 nM contaminating Fe, while 40 μM Fe would contain approximately 0.06 μM contaminating Mn. Thus, contaminating metals are not sufficient to explain the results. These data show that sit transcription is negatively controlled by both MntR and Fur. While MntR primarily responds to the Mn concentration and Fur primarily responds to the Fe concentration, both proteins can use the other metal and repress transcription. In addition, MntR and Fur appear to act independently of one another. For example, at higher Mn concentrations, Fur causes a two- to threefold reduction in sit transcription in both the presence and absence of MntR (Fig. 2A).
Similar results were obtained for the mntH promoter (Fig. 2C and D). As was reported previously (19), the mntH promoter is regulated in response to both Mn and Fe, primarily via MntR and Fur, respectively. However, Fur can respond to Mn, and MntR can respond to Fe to cause repression. This repression is certainly less than when the proteins are responding to their cognate metals but is still significant. However, we noted a reduction in mntH transcription in response to both Mn and Fe, even in the absence of both regulatory proteins (Fig. 2C and D). This is not an inherent property of pAH125-derived lac fusions as evidenced by the absence of an effect on the sit fusion, especially in response to Fe (Fig. 2A and B). Thus, although there are subtle differences, the mntH and sit promoters are regulated similarly by Fur and MntR in response to Fe and Mn.
Effects of mutations in the Fur and MntR binding sites on the sitABCD and mntH promoters.
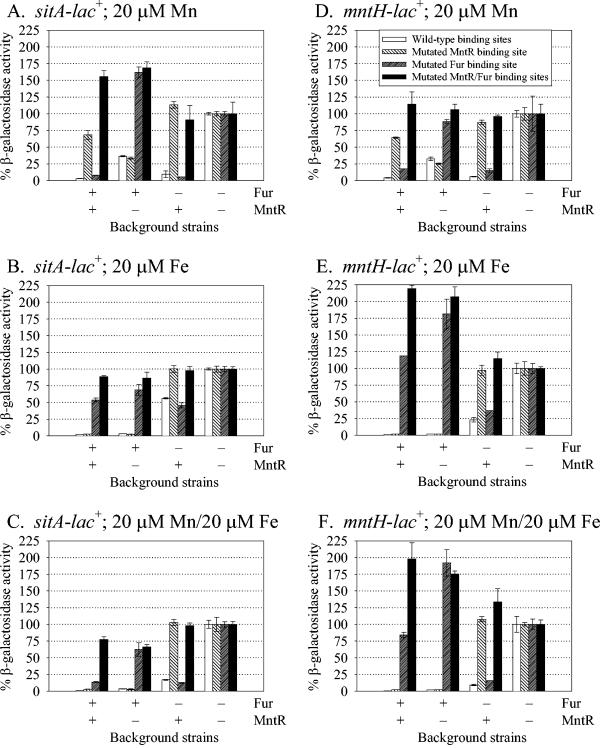
In order to determine the importance of the putative MntR and Fur binding sites to the transcriptional activities of sitABCD and mntH, equivalent lacZ transcriptional fusions with specific mutations in the MntR and/or Fur binding sites were constructed. Nucleotides that were highly conserved in known MntR or Fur (36) binding sites were changed in order to disrupt binding to MntR or Fur, respectively (Fig. 1). Because of the overlap between the binding sites and the promoter, the introduced mutations affected the absolute level of transcription of the fusion constructs. In the absence of regulators, the mutations in the MntR binding site resulted in a 43% decrease in sitABCD transcription, but only a 5% decrease was observed for the mntH promoter. Mutations in the Fur binding site decreased the activity of the sit promoter by 81% and the activity of the mntH promoter by 91%. Mutations in both the MntR and Fur binding sites decreased the activity of the sit promoter by 94% and the activity of the mntH promoter by 82%. These results suggest that changes in the binding sites may have altered the binding of the RNA polymerase at the promoters independent of changes due to altered binding of MntR and/or Fur with their respective binding sites. However, by normalizing the transcriptional activity of each promoter construct, we could determine the relative role of each binding site in transcriptional regulation.
The presence of Mn in the medium greatly reduced transcriptional activity of sitABCD (Fig. 3A) and mntH (Fig. 3D) only when there was both a wild-type MntR and MntR binding site present, suggesting that MntR uses Mn as a cofactor and binds to the MntR binding site. However, when MntR was mutated, repression by Mn was still evident (although there was less repression than with wild-type MntR), and this repression was greatly reduced when Fur or the Fur binding site was also mutated, suggesting that Fur can also use Mn as a cofactor to bind at the Fur binding site. In a fur strain (MntR+) with a mutated MntR binding site and in an mntR strain (Fur+) with a mutated Fur binding site, there was little or no repression, indicating that in response to Mn, neither Fur nor MntR binds to the heterologous binding site.
FIG. 3.
Effects of mutations in putative Fur and/or MntR binding sites on the transcriptional activities of the sitABCD and mntH promoters. Transcriptional lac fusions with the wild-type sit promoter or the sit promoter with site-specific mutations in Fur and/or MntR binding sites were assayed in wild-type, mntR, fur, or mntR fur background strains (A to C). Likewise, transcriptional lac fusions with the wild-type mntH promoter or the mntH promoter with site-specific mutations in Fur and/or MntR binding sites were assayed in wild-type, mntR, fur, or mntR fur background strains (D to F). Strains were grown overnight in minimal medium or minimal medium supplemented with 20 μM Mn (A and D), 20 μM Fe (B and E), or both 20 μM Mn and 20 μM Fe (C and F). β-Galactosidase activities were normalized to the activities of the corresponding mntR fur background strain. Data are presented as means ± standard deviations (error bars) (n = 4). Strains JS404 to JS435 were used.
Similar results were observed with Fe, Fur, and the Fur binding site (Fig. 3B and E). Fe in the medium greatly repressed transcription of sit and mntH only when both wild-type Fur and the Fur binding site were present. When Fur was mutated, repression by Fe was still evident, but this repression was lost when MntR or the MntR binding site was also mutated. These results suggest that both Fur and MntR can use Fe as a cofactor to bind to the Fur binding site and MntR binding site, respectively. In a fur strain (MntR+) with a mutated MntR binding site, there was no repression by Fe, indicating that MntR does not bind to the Fur binding site. In an mntR strain (Fur+) with a mutated Fur binding site, there was some repression of sitABCD, suggesting that the mutations in the Fur site might not completely disrupt binding (Fig. 3B). This minor repression was not significantly affected by the addition of the MntR binding site mutation. This Fur-mediated repression was not very strong, nor was it observed for the mntH promoter (Fig. 3E). Transcriptional activities of sitABCD and mntH were also examined in the presence of both Mn and Fe (Fig. 3C and F). In general, the activity of each strain reflected the additive effects of Fur- and MntR-mediated repression. Note that in some instances transcriptional activity of the various fusions was increased up to twofold the normalized value (set at 100%) obtained in a fur mntR double mutant background. These effects are seen even in fusions lacking both MntR and Fur binding sites and are primarily observed when isogenic fur+ and fur mutant strains are compared. Given the pleiotropic nature of fur mutations, this could reflect subtle changes in general physiology. We do not believe that these twofold or less effects impinge on our overall interpretation of the data.
These results suggest that repression of sitABCD and mntH is mediated by MntR binding to the MntR binding site or Fur binding to the Fur binding site, and each repressor can use either Mn or Fe as a cofactor. These studies also confirm the involvement of the proposed MntR and Fur binding sites in the sitABCD and mntH promoters. The relative repression by MntR acting at the MntR binding site or Fur acting at the Fur binding site was independent of mutations in the other binding site (Fig. 3), again suggesting that these regulatory proteins act independently of one another to control transcription at these promoters.
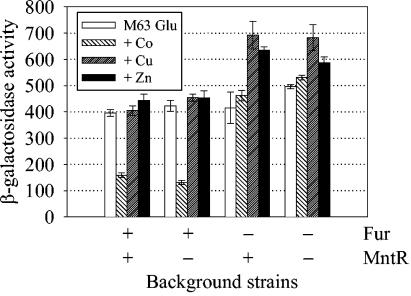
Fur, but not MntR, can use Co as a cofactor to repress sit transcription.
To determine whether Fur or MntR can use divalent metals other than Mn or Fe as a cofactor, we examined the transcription of sit in various background strains (mntR, fur, or mntR fur) in response to 10 μM Co, Cu, or Zn (Fig. 4). We found that the activity of the sitA::lac fusion in the wild-type and mntR background strains were reduced by Co, but not Cu or Zn. In fur and mntR fur background strains, there was no reduction in activity by Co. Therefore, these results suggest that Fur, but not MntR, can use Co as a cofactor to repress sit transcription.
FIG. 4.
Effects of divalent metals other than Mn and Fe on the transcriptional activity of the sitABCD promoter. β-Galactosidase activities of a sitA::lac fusion in wild-type, mntR, fur, and mntR fur background strains were measured after the bacteria were allowed to grow overnight in minimal medium (M63 medium with 0.2% glucose) supplemented with 10 μM (final concentration) of Co, Cu, or Zn. Units are defined as follows: (micromoles of ONP formed per minute × 106)/(OD600 × milliliter of cell suspension). Data are presented as means ± standard deviations (error bars) (n = 4). Strains JS210, JS211, JS390, and JS391 were used.
Effects of oxygen on sit transcription.
The importance of Mn in the biology of prokaryotic cell function is only beginning to be appreciated (21). Mn is required for several enzymes involved in general metabolism but is also required for oxidative stress response proteins, including the superoxide dismutase SodA and the nonheme catalase KatN (33). Indeed, SodA is predicted to be the predominant enzyme containing Mn in the cell. Regulation of SodA production is complex but includes repression by Fnr, ArcA, and Fur, such that expression is repressed under anaerobic conditions (16). Fnr and ArcA are global transcriptional regulators that control several genes in response to oxygen availability.
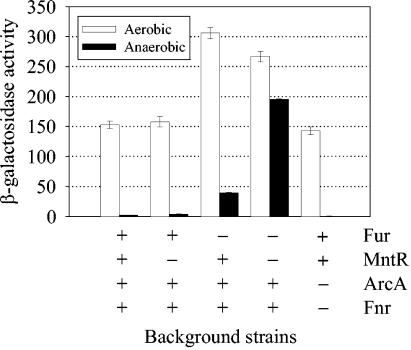
Given the potential role of Mn in protection against oxidative stress, we monitored expression of sitABCD under aerobic versus anaerobic conditions. When grown in LB medium, the activity of a sitA::lac fusion in the wild-type background strain was greatly reduced (by 98%) under low oxygen conditions compared to that under high oxygen conditions (Fig. 5). Transcriptional regulators ArcA and Fnr were not involved, since regulation was normal in an arcA fnr background. Regulation was also normal in an mntR background strain. In a fur background, however, the transcriptional activity was higher under aerobic conditions and the level of repression under anaerobic conditions was significantly affected but still evident (62-fold repression in the wild type; 8-fold repression in the fur mutant). Loss of both MntR and Fur allowed high-level expression under both conditions, although there was still a 27% decrease in expression when the cells were grown anaerobically for reasons that are not clear. These results suggest that Fur and MntR are largely responsible for the increased repression of sit under anaerobic conditions compared to aerobic conditions. Furthermore, repression could be due to the increased availability under anaerobic conditions of reduced metals that act as cofactors with the metalloregulatory proteins.
FIG. 5.
Effects of aerobic and anaerobic growth on the transcriptional activity of the sitABCD promoter. β-Galactosidase activities of a sitA::lac fusion in wild-type, mntR, fur, mntR fur, and arcA fnr background strains were measured after the bacteria were allowed to grow overnight in LB medium under aerobic and anaerobic conditions. Units are defined as follows: (micromoles of ONP formed per minute × 106)/(OD600 × milliliter of cell suspension). Data presented as means ± standard deviations (error bars) (n = 2 or 3). Strains JS210, JS211, JS390, JS391, and JS392 were used.
Effects of growth phase on sit transcription.
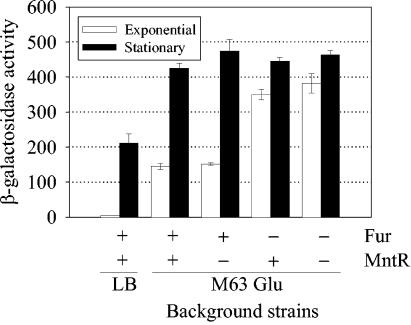
We noted that the sit operon was preferentially expressed in stationary phase. Figure 6 shows the expression of the sitA::lac fusion in various background strains grown to exponential and stationary phase. When the wild-type strain was grown in LB medium, transcriptional activity was approximately 40-fold higher during stationary phase than exponential phase. Growth of the wild-type strain in minimal medium resulted in greater activity during both exponential and stationary phases. Many genes induced in stationary phase are controlled by the alternative sigma factor, RpoS. However, sit does not appear to be controlled by RpoS; there was very little difference in activity in an rpoS background strain compared to a wild-type strain during growth in stationary phase in minimal medium (data not shown). Although there was no difference in activity between an mntR strain and a wild-type strain grown in minimal medium, in a fur background strain (with or without MntR), activity was high during the exponential phase of growth. These results suggest that repression of sit in the wild-type background strain during exponential growth was due mainly to Fur. Furthermore, the increased expression of sit during stationary phase was not due to RpoS but was probably due to the decreased availability of Fe.
FIG. 6.
Effect of growth phase on the transcriptional activity of the sitABCD promoter. β-Galactosidase activities of a sitA::lac fusion in wild-type, mntR, fur, and mntR fur background strains were measured after growth in LB medium or minimal medium (M63 medium with 0.2% glucose) to exponential or stationary growth phase. Units are defined as follows: (micromoles of ONP formed per minute × 106)/(OD600 × milliliters of cell suspension). Data are presented as means ± standard deviations (error bars) (n = 2). Strains JS210, JS211, JS390, and JS391 were used.
Transcriptional regulation of mntR.
We have shown that MntR is the main transcriptional regulator of the sit operon in response to Mn. Therefore, we examined more thoroughly the regulation of mntR transcription. Upstream of mntR we identified a consensus MntR binding box (16 of 18 bases match the consensus sequence), which suggests that MntR may be autoregulated. To directly address the regulation of mntR, we constructed a lacZ transcriptional fusion such that mntR itself was not disrupted. Rather, the fusion joint was immediately downstream of the mntR stop codon and 4 bases into the overlapping coding sequence of an open reading frame called b0818, which apparently is in an operon with mntR. This disruption of b0818 did not affect the repression of the sit operon by MntR in response to Mn (data not shown), suggesting that our lacZ fusion did not affect the expression and/or function of MntR. To address the issue of autoregulation, we monitored the activity of the mntR+::lac fusion in minimal medium with or without added Mn. We did not detect any Mn-dependent changes in mntR transcriptional activity during the exponential or stationary growth phase (Table 2). These results suggest that MntR does not repress its own transcription in response to Mn.
TABLE 2.
Transcriptional activity of the mntR promoter
| Straina | β-Galactosidase activityb
|
|||||
|---|---|---|---|---|---|---|
| Minimal mediumc
|
LB
|
|||||
| Without Mn
|
With Mn
|
|||||
| Exponential | Stationary | Exponential | Stationary | Exponential | Stationary | |
| 14028 mntR+-lac | 70.46 ± 6.17 | 85.16 ± 5.89 | 88.02 ± 10.31 | 95.10 ± 6.06 | 32.41 ± 2.27 | 85.17 ± 5.93 |
| 14028 mntR+-lac rpoS::tet | 69.56 ± 6.62 | 95.96 ± 4.15 | 62.24 ± 3.02 | 97.60 ± 4.78 | 29.59 ± 0.66 | 149.65 ± 8.55 |
Strains JS436 and JS437 were used.
Data presented as means ± standard deviations (n = 6). Units are defined as follows: (micromoles of ONP formed per minute × 106)/(OD600 × milliliters of cell suspension).
M63 medium supplemented with Casamino Acids treated with Chelex resin with or without the addition of 20 μM Mn.
The mntR+::lac fusion appeared to be induced in the stationary growth phase when the bacteria were grown in LB but not in minimal medium (Table 2). The increased activity during the stationary growth phase in LB was not due to rpoS, because activity did not decrease in an rpoS background strain. Activity of the strain grown in minimal medium was also not affected by a mutation in rpoS. Since sit transcriptional activity increased during stationary phase in an RpoS-independent Fur-dependent manner, we examined whether Fur was also involved in mntR expression. Search for a Fur binding site in the mntR promoter region revealed a sequence that matched 12 of 19 bases in the Fur binding consensus sequence. However, several highly conserved bases in the consensus sequence did not match the bases in the sequence found in the mntR promoter. Furthermore, the transcriptional activity of the mntR fusion during exponential growth in LB medium remained unchanged with the introduction of a fur mutation (33.12 ± 1.73 units of activity in the fur background strain JS438 compared to 35.52 ± 1.49 units of activity in the wild-type background strain). Unlike results for sitABCD, neither Fur nor MntR appears to be involved in mntR transcriptional regulation.
DISCUSSION
Initial studies of the sit operon of S. enterica serovar Typhimurium addressed the roles of Fe and Fur on transcription (18, 43), but it has since been discovered that the major metal transported by SitABCD is Mn instead of Fe (20). Studies on the other known Mn transport system of Salmonella, MntH, showed that Fur and MntR are important in regulation in response to Fe and Mn, and this regulation involved Fur and MntR binding sites in the promoter region of mntH (19). Therefore, we examined the transcriptional regulation of sitABCD in response to Fe and Mn in more detail. Our studies indicate that both Fur and MntR regulate sit transcription in response to Fe and Mn, respectively, although both regulators can use the alternative metal to partially repress transcription. Site-specific mutations in putative Fur and MntR binding sites disrupted regulation by Fur and MntR, confirming that those sites are involved in transcriptional regulation. Moreover, Fur and MntR act independently of one another at both the sit and mntH promoters. This is consistent with the relative positions of the sites: the Fur and MntR sites are on approximately the same face of the helix in sit but are three quarters of a turn closer in the mntH promoter.
In this study, we found that Fur, acting at the Fur binding site, can repress mntH and sitABCD in response to Fe or Mn. Consistent with independent action of the two regulators, this Fur-mediated regulation was unaffected by mutations in the MntR binding site. Previously, we suggested that Fur can repress mntH in response to Mn, but the exact mechanism was unclear (19). The complexity arose from the observation that, whereas mutational loss of both MntR and the MntR binding site left only residual regulation of mntH in response to Mn, significant regulation was observed in the mntR strain where the binding site was intact. Mutational loss of both MntR and the Fur binding site also relieved repression. These results suggested that the residual Mn-dependent regulation that was mediated by Fur required both the Fur and MntR binding sites. Our results here suggest a simpler model where Fur acts only at the Fur binding site. Discrepancies between the two studies might be due to differences in the construction of the transcriptional lacZ fusions. In the present study, the promoter regions encompassing the MntR and Fur binding sites were cloned upstream of lacZ and integrated into the chromosome in a single copy. Furthermore, specific base changes were made within the binding motifs only (eight base changes for the Fur binding site and six base changes for the MntR binding site). In our original study, a larger region of the mntH promoter that included the MntR binding site, Fur binding site, and OxyR binding site was cloned upstream of lacZ in a single- or low-copy-number plasmid. Several specific base changes in the motifs were made (12 base changes for the Fur binding site and 16 base changes for the MntR binding site) plus base changes between the motifs were made when restriction sites were incorporated into the region. Perhaps the restriction site introduced immediately downstream of the Fur binding site, along with the mutations in the MntR binding site, affected binding of Fur in response to Mn.
Optimal expression of sitABCD occurs under aerobic conditions in stationary phase. We had previously noted that mntH expression was also induced in stationary phase (19). This initially suggested that additional regulatory circuits might be involved in the transcriptional control. However, our results suggest that oxygen and growth phase primarily affect the availability of metals. Anaerobic repression of sit was unaffected by loss of the oxygen-sensing global regulator ArcA or Fnr. However, this repression was greatly relieved by mutations in both mntR and fur, with single mutations in fur producing a greater effect than mutations in mntR. Under low oxygen conditions, the soluble reduced form of iron (Fe2+) would become available for transport, leading to repression of the Fur regulon. The fact that Mn remains in the reduced state independent of oxygen conditions may explain why MntR had much less effect on anaerobic repression than Fur did. Likewise, stationary-phase induction of the sit promoter was unaffected by loss of the regulator, RpoS. In contrast, loss of both Fur and MntR led to essentially constitutive expression in all phases of growth. The simplest explanation is that available iron and manganese become limited as the cell population increases, leading to induction of the Fur and MntR regulons. Although metal availability is certainly important, additional data (D. G. Kehres and M. E. Maguire, unpublished data) suggest that stationary-phase induction in some media is more complex, and further studies are required to completely understand sit and mntH regulation in response to growth phase.
MntR, acting at its cognate binding site, can repress sitABCD and mntH in response to Fe (shown here and in reference 19). This Fe-mediated repression by MntR is independent of Fur. In a fur background, the Fe concentration is presumably increased, due to constitutive expression of iron transporters (13), but the response of MntR to physiological levels of iron is also seen in a fur+ background where the Fur binding site has been mutated (e.g., Fig. 3E). MntR belongs to the DtxR family of metalloregulatory proteins that can be divided into two groups on the basis of their primary responsiveness to Fe or Mn (13). These proteins share structural similarities, including two residues directly involved in metal coordination that are conserved within each group but differ between groups and have been shown to be important in metal ion selectivity (13). However, selectivity is apparently more complicated and subtle than this would suggest. The E. coli MntR protein reportedly does not respond to Fe (30). The E. coli and Salmonella proteins differ by 12 of 157 amino acids; the metal coordinating amino acids are conserved. This fundamental difference between the two orthologs needs to be confirmed by direct comparison but could be useful in defining metal specificity in this important class of proteins.
Likewise, we found that the Fur protein was able to use both Fe and Mn as a cofactor. In addition, Fur could repress the sitABCD operon with Co. We previously showed that mntH is also repressed by Co, although a higher concentration was required (19). The importance of Co in controlling Mn transport under physiological conditions is unclear, especially since the level of Co in E. coli cells was shown to be extremely low when the cells were grown in LB or minimal medium (27). In in vitro experiments, Mn and Co have been shown to bind to Fur (1), and Fur has been shown to use Mn and Co as a corepressor in DNA binding studies (3). Privalle and Fridovich (31) demonstrated that the Fur protein in E. coli has different metal specificities that affect its function. Whereas the E. coli aerobactin operon was repressed by Fur using either Fe or Mn as a corepressor, sodA was repressed by Fur using Fe but not Mn. Similar to results of the mntH study (19), we found that Cu and Zn do not repress sit transcription. Therefore, whereas Zn is a potent competitive inhibitor of Mn transport by sitABCD (20), the metal apparently cannot be used by Fur or MntR as a corepressor. Nevertheless, the Fur protein of E. coli does bind Zn tightly in a different metal binding site than that of the cofactors (2, 17). The exact mechanism of how the binding of divalent metals into the two metal binding sites influences Fur to bind to the Fur binding site to repress transcription is not known (28). Clearly, the interaction of Fur with different metals is complex and may allow additional discriminatory control over the expression of different genes.
There are apparently four MntR binding sites in the chromosome of Salmonella. One of those sites is located in the promoter of mntR, suggesting autoregulation. The presence of an intact MntR box in the promoter sequence but the absence of any Mn-dependent regulation of this locus presents a quandary. The expression of mntR might be more strictly regulated, and perhaps the binding of MntR to this site might require other factors or different conditions than those tested here.
The mutations in the Fur and MntR binding sites decreased the absolute transcriptional activities of sitABCD and mntH fusions with or without the regulatory proteins present, suggesting that these mutations directly affected the promoters. Mutations in the Fur binding sites decreased transcription 80 to 90%, likely due to changes in the −35 sequence. Interestingly, mutations in the MntR binding site had much less effect on the activity of the mntH promoter (5% decrease) than the equivalent mutations had on the sit promoter (43% decrease). This likely reflects differences in spacing between the MntR binding sites and their respective −10 sites and perhaps Fur binding sites. The absolute levels of activity produced by the lac fusions containing wild-type binding sites in the absence of regulators were approximately 3,200 U for sit and 400 U for mntH, suggesting that the sit promoter is the stronger of the two. Most importantly, repression by Fur could still be observed for promoters mutated in the MntR binding site, and repression by MntR could still be observed for promoters mutated in the Fur binding site, suggesting that the effects of mutations on or differences in inherent promoter strength did not interfere with our ability to determine the relative roles of Fur and MntR in regulation.
What remains unclear is why Salmonella would require both Fur and MntR to alter gene transcription of the manganese transport systems. One explanation is that these transport systems also transport Fe, and therefore, control by Fur would be appropriate. However, under physiological conditions, Mn is the predominant metal transported by MntH and SitABCD, and the transport of Fe is likely not significant (20, 22). Our results indicate that in response to low levels of Mn, MntR is more efficient than Fur at repressing sitABCD and mntH. Likewise, in response to low levels of Fe, Fur can repress sitABCD and mntH better than MntR. Fur has been well studied and is known to control many other genes, and this may be a way to coordinate Mn transport with other cellular functions. As we have shown, sitABCD is controlled by oxygen level and growth phase via the Fur regulatory protein. Currently, MntR is known only to control Mn transport systems, but other cellular functions may be discovered to be regulated by MntR as well. These functions may thus be coordinated with Mn transport in response to Mn levels, whereas Fur-controlled genes may be coordinated with Mn transport in response to Fe levels.
In summary, transcriptional control of sitABCD is similar to mntH in that both involve the metalloregulators, MntR and Fur. MntR represses transcription by acting at the MntR binding site. MntR is most efficient when complexed with Mn but can also use Fe. Fur-mediated repression requires the Fur binding site and is most efficient with Fe, but Fur also responds to Mn (or Co). The regulation of sitABCD is complex and controlled by multiple regulators that may coordinate Mn transport with other cellular functions in response to different environmental conditions.
Acknowledgments
This work was supported in part by grant 00-25 from the Roy J. Carver Charitable Trust (J.M.S.) and NIH grant GM61748 (M.E.M.).
REFERENCES
- 1.Adrait, A., L. Jacquamet, L. Le Pape, A. Gonzalez de Peredo, D. Aberdam, J.-L. Hazemann, J.-M. Latour, and I. Michaud-Soret. 1999. Spectroscopic and saturation magnetization properties of the manganese- and cobalt-substituted Fur (ferric uptake regulation) protein from Escherichia coli. Biochemistry 38:6248-6260. [DOI] [PubMed] [Google Scholar]
- 2.Althaus, E. W., C. E. Outten, K. E. Olson, H. Cao, and T. V. O'Halloran. 1999. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry 38:6559-6569. [DOI] [PubMed] [Google Scholar]
- 3.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, E., I. Bergevin, D. Malo, P. Gros, and M. F. M. Cellier. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 6.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 7.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using λ Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 11.Escolar, L., J. Pérez-Martín, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez de Peredo, A., C. Saint-Pierre, J.-M. Latour, I. Michaud-Soret, and E. Forest. 2001. Conformational changes of the ferric uptake regulation protein upon metal activation and DNA binding: first evidence of structural homologies with the diphtheria toxin repressor. J. Mol. Biol. 310:83-91. [DOI] [PubMed] [Google Scholar]
- 13.Guedon, E., and J. D. Helmann. 2003. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol. Microbiol. 48:495-506. [DOI] [PubMed] [Google Scholar]
- 14.Haldimann, A., and B. L. Wanner. 2001. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183:6384-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 16.Hassan, H. M., and H.-C. H. Sun. 1992. Regulatory roles of Fnr, Fur, and Arc in expression of manganese-containing superoxide dismutase in Escherichia coli. Proc. Natl. Acad. Sci. USA 89:3217-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquamet, L., D. Aberdam, A. Adrait, J.-L. Hazemann, J.-M. Latour, and I. Michaud-Soret. 1998. X-ray absorption spectroscopy of a new zinc site in the Fur protein from Escherichia coli. Biochemistry 37:2564-2571. [DOI] [PubMed] [Google Scholar]
- 18.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 19.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J. Bacteriol. 184:3151-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kehres, D. G., and M. E. Maguire. 2003. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 27:263-290. [DOI] [PubMed] [Google Scholar]
- 22.Kehres, D. G., M. L. Zaharik, B. B. Finlay, and M. E. Maguire. 2000. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol. Microbiol. 36:1085-1100. [DOI] [PubMed] [Google Scholar]
- 23.Lavrrar, J. L., and M. A. McIntosh. 2003. Architecture of a Fur binding site: a comparative analysis. J. Bacteriol. 185:2194-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makui, H., E. Roig, S. T. Cole, J. D. Helmann, P. Gros, and M. F. M. Cellier. 2000. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol. Microbiol. 35:1065-1078. [DOI] [PubMed] [Google Scholar]
- 25.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 26.Mills, D. M., V. Bajaj, and C. A. Lee. 1995. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol. Microbiol. 15:749-759. [DOI] [PubMed] [Google Scholar]
- 27.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 28.Outten, F. W., C. E. Outten, and T. V. O'Halloran. 2000. Metalloregulatory systems at the interface between bacterial metal homeostasis and resistance, p. 145-157. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 29.Patzer, S. I., and K. Hantke. 1998. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28:1199-1210. [DOI] [PubMed] [Google Scholar]
- 30.Patzer, S. I., and K. Hantke. 2001. Dual repression by Fe2+-Fur and Mn2+-MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J. Bacteriol. 183:4806-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Privalle, C. T., and I. Fridovich. 1993. Iron specificity of the Fur-dependent regulation of the biosynthesis of the manganese-containing superoxide dismutase in Escherichia coli. J. Biol. Chem. 268:5178-5181. [PubMed] [Google Scholar]
- 32.Que, Q., and J. D. Helmann. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454-1468. [DOI] [PubMed] [Google Scholar]
- 33.Robbe-Saule, V., C. Coynault, M. Ibanez-Ruiz, D. Hermant, and F. Norel. 2001. Identification of a non-haem catalase in Salmonella and its regulation by RpoS (σs). Mol. Microbiol. 39:1533-1545. [DOI] [PubMed] [Google Scholar]
- 34.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Slauch, J. M., and T. J. Silhavy. 1991. cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J. Bacteriol. 173:4039-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stojiljkovic, I., A. J. Bäumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria: identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 37.Tao, X., N. Schiering, H.-Y. Zeng, D. Ringe, and J. R. Murphy. 1994. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol. Microbiol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 38.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 39.Vidal, S. M., E. Pinner, P. Lepage, S. Gauthier, and P. Gros. 1996. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1D169) mouse strains. J. Immunol. 157:3559-3568. [PubMed] [Google Scholar]
- 40.Yu, D., H. M. Ellis, E.-C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaharik, M. L., V. L. Cullen, A. M. Fung, S. J. Libby, S. L. Kujat Choy, B. Coburn, D. G. Kehres, M. E. Maguire, F. C. Fang, and B. B. Finlay. 2004. The Salmonella enterica serovar Typhimurium divalent cation transport systems MntH and SitABCD are essential for virulence in an Nramp1G169 murine typhoid model. Infect. Immun. 72:5522-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaharik, M. L., and B. B. Finlay. 2004. Mn2+ and bacterial pathogenesis. Front. Biosci. 9:1035-1042. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, D., W.-D. Hardt, and J. E. Galán. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]