Abstract
Bacillus subtilis ATCC 6633 produces the cationic pore-forming lantibiotic subtilin, which preferentially acts on gram-positive microorganisms; self protection of the producer cells is mediated by the four genes spaIFEG. To elucidate the mechanism of subtilin autoimmunity, we transferred different combinations of subtilin immunity genes under the control of an inducible promoter into the genome of subtilin-sensitive host strain B. subtilis MO1099. Recipient cells acquired subtilin tolerance through expression of either spaI or spaFEG, which shows that subtilin immunity is based on two independently acting systems. Cells coordinately expressing all four immunity genes acquired the strongest subtilin protection level. Quantitative in vivo peptide release assays demonstrated that SpaFEG diminished the quantity of cell-associated subtilin, suggesting that SpaFEG transports subtilin molecules from the membrane into the extracellular space. Homology and secondary structure analyses define SpaFEG as a prototype of lantibiotic immunity transporters that fall into the ABC-2 subfamily of multidrug resistance proteins. Membrane localization of the lipoprotein SpaI and specific interaction of SpaI with the cognate lantibiotic subtilin suggest a function of SpaI as a subtilin-intercepting protein. This interpretation was supported by hexahistidine-mediated 0-Å cross-linking between hexahistidine-tagged SpaI and subtilin.
Bacillus subtilis strain ATCC 6633 produces the cationic peptide antibiotic (lantibiotic) subtilin. Lantibiotics contain unusual thioether amino acids, such as meso-lanthionine and 3-methyl-lanthionine (17), which are incorporated into prepeptides through extensive posttranslational modifications (25, 32, 41). The subtilin and the closely related ericin gene clusters (35) encompass genes for posttranslational modification (18), transport (18), immunity (20), and regulation (19). Extracellular B. subtilis serine proteases are involved in the final processing step (7, 37). Subtilin biosynthesis and immunity are under the control of the two-component regulatory system SpaK/SpaR (histidine kinase and response regulator, respectively) and the alternative sigma factor H (36, 38).
Lantibiotics act against a wide range of gram-positive bacteria. The antimicrobial action of nisin produced by Lactococcus lactis, a structurally close relative of subtilin, is based on voltage-dependent pore formation that affects the efflux of small molecules and finally the collapse of the proton motive force (for a review see reference 4). The Bacto prenol-bound peptidoglycan precursor lipid II appears to be both a docking molecule assisting membrane targeting (5) and an integral constituent of the lethal pore itself (14). Gram-positive lantibiotic-producing strains need efficient countermeasures to obviate the lethal action of their own products (31). The nisin self protection (immunity) system is composed of ABC transporter homologue NisFEG and lipoprotein NisI (39).
In the present study we report on the establishment of subtilin immunity in the subtilin-susceptible strain B. subtilis MO1099. Evidence is presented that subtilin immunity is based on two independently acting systems: the lipoprotein SpaI, which interacts with subtilin, and SpaFEG, an ABC transporter homologue that expels subtilin molecules from the membrane into the extracellular medium.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2. B. subtilis strains were grown at 37°C on Difco sporulation or M9 medium (30) supplemented with 50-μg/ml phenylalanine, 20-μg/ml tryptophan, and 0.1% Casamino Acids. For subtilin production B. subtilis ATCC 6633 was grown at 37°C on TY medium (0.8% tryptone, 0.5% yeast extract, 0.5% NaCl). Recombinant plasmids were amplified in Escherichia coli DH5α, TP611, TG1, or RR1. E. coli strains were grown on Luria-Bertani medium (Invitrogen, Karlsruhe, Germany). For selective media 80-μg/ml ampicillin and 5-μg/ml chloramphenicol were used for E. coli and 1-μg/ml erythromycin and 25-μg/ml lincomycin were used for B. subtilis.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 lacU169 [F80lacZM15] hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Gibco BRL |
| TP611 | [F−thi-1 thr-1 leuB6 lacY1 tonA21 supE44 λ−] hsdR hsdM recBC lop11 lig+cya-610 pcnB | |
| TG1 | F′ traD36 lacIq Δ[lacZ]M15 proA+B+/supE Δ[hsdM-mcrB] 5[rK− mK−McrB−]thi Δ[lac-proAB] | Amersham |
| RR1 | F−hsd520 supE44 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | |
| B. subtilis | ||
| MO1099 | Derivative of JH642; MLSr; amyE::erm trpC2 pheA1 | 12 |
| MO1099 Pspac | Derivative of MO1099; Cmr; amyE::cat Pspac | MO1099 × pDR67 DNA |
| MO1099 PspacspaI | Derivative of MO1099; Cmr; amyE::cat Pspac spaI | MO1099 × pHZ33 DNA |
| MO1099 PspacspaIFE | Derivative of MO1099; Cmr; amyE::cat Pspac spaIFE | MO1099 × pHZ50 DNA |
| MO1099 PspacspaFEG | Derivative of MO1099; Cmr; amyE::cat Pspac spaFEG | MO1099 × pHZ44 DNA |
| MO1099 PspacspaIFEG | Derivative of MO1099; Cmr; amyE::cat Pspac spaIFEG | MO1099 × pHZ34 DNA |
| ATCC 6633 | Subtilin biosynthesis gene cluster | ATCCb |
| Plasmids | ||
| pQE9 | Apr; expression vector with 6 histidine codons | QIAGEN |
| pATH1 | Apr; trpE for gene fusions | 21 |
| pDR67 | Apr Cmr; integrative vector with Bacillus Pspac promoter | 16 |
Cmr, chloramphenicol resistance; MLSr, macrolide-lincomycin-streptogramin B resistance; Ap, ampicillin resistance.
ATCC, American Type Culture Collection.
TABLE 2.
Plasmids constructed in this study
| Plasmid | Description | PCR primers used for amplificationa (5′-3′) |
|---|---|---|
| pHZ24 | 1,370-bp PCR fragment containing spaFE cloned into pQE9 BamHI/SalI fragment | GGTTTTAATAAGGATCCAAAAGGAATAAGCCATCCAAGTCGACAAAGGATCATTG |
| pHZ38 | 422-bp PCR fragment containing 3′ part of spaI cloned into pATH1 EcoRI/XbaI fragment | TTATCTGCTTGTGGATCCTTAACAAAG, CAAACTTTTTTGTAAAGCTTTTGGTTTC |
| pHZ33 | 715-bp PCR fragment containing spaI cloned into pDR67 HindIII/XbaI fragment | CGCCCAAAAGCTTAAAGTTTCCAG, TGCCAAACTAGTTTGTAAGACTTTTGG |
| pHZ34 | 3,013-bp PCR fragment containing spaIFEG cloned into pDR67 HindIII/XbaI fragment | CGCCCAAAAGCTTAAAGTTTCCAG, GTCAACTAGTTGCTGGTCACGGC |
| pHZ44 | 2,434-bp PCR fragment containing spaFEG cloned into pDR67 HindIII/XbaI fragment | TAGTACAAACGAAGCTTCTGATGCC, GTCAACTAGTTGCTGGTCACGGC |
| pHZ50 | 2,182-bp PCR fragment containing spaIFE cloned into pDR67 HindIII/XbaI fragment | CGCCCAAAAGCTTAAAGTTTCCAG, TCTTCTAGAAATCAGCTTGTAAACACC |
Restriction sites within the PCR primers are underlined; altered nucleotides are in boldface.
Molecular biology techniques.
Established protocols for molecular biology techniques were followed (30). DNA was cleaved according to the conditions recommended by the supplier (Roche Molecular Biochemicals, Mannheim, Germany). DNA fragments were eluted from agarose gels by the Gene Clean Kit III (Bio101, Vista, Calif.). The alkaline extraction procedure (2) was used to isolate DNA from E. coli. PCR was carried out according to standard procedures (30) in an Eppendorf Microcycler E. Oligonucleotides were obtained from ARK Scientific GmbH Biosystems (Darmstadt, Germany), and DNA was sequenced by Scientific Research and Development GmbH (Oberursel/Frankfurt, Germany).
The vector pDR67 containing an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Bacillus promoter Pspac and a chloramphenicol resistance cassette (16) was used for integration into the amyE locus of B. subtilis MO1099 (12). This strain harbors a macrolide-lincomycin-streptogramin B resistance marker at the amyE locus, which gets lost after double-homologous recombination via marker exchange. B. subtilis was transformed by the competence method (1, 18). Gene expression was induced with 1 to 2 mM IPTG.
Subtilin isolation, activity, and sensitivity tests.
Subtilin was isolated by reversed-phase chromatography (36). Subtilin sensitivity of B. subtilis cells was determined in agar diffusion tests previously described (39).
Quantitative subtilin transport assay.
A peptide release assay was used as previously described (39). Stationary B. subtilis strains grown overnight in TY with 1% (wt/vol) glucose were harvested and washed with 50 mM Tris-HCl (pH 8). The cell density was adjusted to optical density at 578 nm (OD578) of 10 with 50 mM sodium phosphate (pH 7)-0.5 M NaCl-0.5% (wt/vol) glucose. One-milliliter aliquots of the cell suspension were incubated with subtilin (30 min, 37°C). After centrifugation (10,000 × g, 10 min) quantitative high-performance liquid chromatography (HPLC) analyses of the supernatants were performed on a Beckman Gold HPLC system using an analytical ODS-Hypersil column (Maisch, Ammerbuch, Germany). Subtilin was eluted with a linear gradient from 30 to 40% acetonitrile containing 0.1% (vol/vol/vol) trifluoroacetic acid over 30 column volumes and detected by measuring the absorption at 214 nm. The flow rate (0.4 ml/min) was chosen so that a Gauss distribution of the subtilin absorption peak was obtained; this allows a quantitative determination of the subtilin amount. Subtilin attached to cells was extracted by gently mixing with 20% acetonitrile and 0.1% (vol/vol/vol) trifluoroacetic acid in water at room temperature for 5 min. After centrifugation (10,000 × g, 10 min) the level of subtilin in the supernatant was quantitatively determined.
SDS-PAGE, Western blotting, and antibody isolation.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (24) and Western blot analyses were performed as described previously (39). Molecular weight standards for SDS-PAGE were purchased from Sigma-Aldrich, Munich, Germany. Antiserum against the hexahistidine tag was obtained from QIAGEN, Hilden, Germany. A truncated N-terminal fragment of SpaI (amino acids 28 to 165) was fused to TrpE (vector pATH1 [21]), expressed in E. coli, and purified with preparative SDS. The fusion protein was used for rabbit immunization (Eurogentec, Seraing, Belgium).
Construction and isolation of hexahistidine-tagged SpaI.
A spaI copy was amplified by PCR from B. subtilis ATCC 6633 DNA with primers SpAI1, GTTATCTGCTTGTGGATCCTTAACAAAG, and SpaI2, CATGTCAAGCTTTCCCTTATTCC, and inserted into the pQE9 vector (QIAGEN). The E. coli strain M15(pREP4) transformed with the resulting plasmid was grown in Luria-Bertani medium to an optical density at 600 nm of 0.5. After induction with 2 mM IPTG the cells were incubated for 4 h, harvested, suspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]), and disrupted by sonication. After removal of cell debris (centrifugation, 17,000 × g, 30 min, 4°C) the supernatant was incubated with Ni-nitrilotriacetic acid-agarose (QIAGEN) with gentle shaking (1 h, 4°C). The protein was eluted with the same buffer containing 200 mM imidazole and dialyzed against the storage buffer (20 mM Tris-HCl, 10% glycerol, 5 mM dithiothreitol [pH 6.5]).
SpaI-subtilin interaction studies and hexahistidine-mediated cross-linking.
Assays of interactions between SpaI solubilized from B. subtilis membrane vesicles and subtilin were performed as described previously (39), and products were probed with native PAGE gel (e.g., without addition of SDS). Hexahistidine tags were complexed with Ni2+ in the presence of 2 mM Ni-acetate, and complexes were incubated for 10 min at room temperature. The cross-linking reaction with 0.1 to 1 mM magnesium monoperoxyphthalic acid (MMPP) was allowed to proceed for 6 min (10). Reactions were terminated by the addition of SDS sample buffer.
Database research.
Transmembrane regions were predicted with the DAS-Transmembrane Prediction server (8) (http://www.sbc.su.se/∼miklos/DAS/). For homology searches BLAST 2 Sequences at the National Center for Biotechnol-ogy Information (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/bl2.html) wasused, and for multiple sequence alignments ClustalW, version 1.8, from the Baylor College of Medicine (BCM Search Launcher; http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html) was used.
Nucleotide sequence accession number.
The nucleotide sequence of the subtilin immunity gene locus of B. subtilis ATCC 6633, consisting of two overlapping ORFs, spaF and spaE, has been assigned EMBL gene bank accession number U09819.
RESULTS
The subtilin immunity operon.
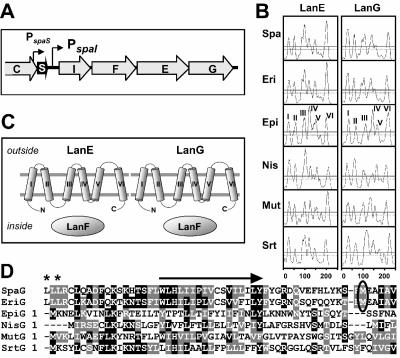
Resequencing the subtilin immunity gene locus of B. subtilis ATCC 6633 revealed that the previously predicted spaF open reading frame (ORF) (20) consists of two overlapping ORFs, spaF and spaE (Fig. 1A). As control, we fused a hexahistidine-encoding sequence to spaF; for the His6-tagged SpaF protein a molecular mass of 30 kDa was observed (not shown), which exactly fits with the calculated value derived from the revised spaF sequence. Thus, the organization of the subtilin immunity system (Fig. 1A) corresponds to closely related LanIFEG systems for ericin (35) and nisin (9, 23) immunity.
FIG. 1.
Subtilin immunity genes. (A) The operon of subtilin immunity genes spaIFEG resides downstream of the subtilin structural gene spaS; encoded proteins are SpaI (165 amino acids [aa], 19.3 kDa), SpaF (247 aa, 27.2 kDa), SpaE (251 aa, 28.9 kDa), and SpaG (254 aa, 28.5 kDa). (B) Hydropathy plots of the hydrophobic lantibiotic immunity LanEG proteins. TMDs predicted with the DAS-Transmembrane Prediction server (8) above the solid line are highly probable. The TMDs of the epidermin immunity proteins EpiEG (roman numerals) are representative. (C) Schematic representation of lantibiotic ABC transporter immunity systems encompassing the two membrane proteins LanEG with six TMDs each (see panel B) and the hydrophilic nucleotide binding protein LanF. (D) Alignment of LanG protein N termini. Previously assigned start methionines of SpaG and EriG (circled) and most probable start methionines of SpaG and EriG (asterisks; encoded by TTG codons) are indicated. Arrow, highly conserved N-terminal TMD I of derived proteins. Black and gray shading represents identical and similar amino acids, respectively. Spa, subtilin, B. subtilis ATCC 6633 (sequence update, EMBL gene bank accession number U09819); Eri, ericin, B. subtilis A1/3 (35); Nis, nisin, L. lactis (34); Epi, epidermin, Staphylococcus epidermidis (33); Mut, mutacin II, Streptococcus mutans (6); Srt, putative lantibiotic streptin, Streptococcus pyogenes (11).
The SpaFEG proteins are homologous to counterparts of other lantibiotic-producing organisms (Table 3), particularly the hydrophilic putative ATP-binding protein SpaF. But the hydrophobic LanEG proteins also share a number of features with SpaFEG proteins, including (i) a size of about 250 amino acid residues, (ii) hydrophobicity patterns, and (iii) the sizes of and the distance between the predicted transmembrane segments (Fig. 1B); these similarities suggest similar functions. The sequence similarity of SpaG proteins (Fig. 1D) clearly argues against previously assigned translational start ATG codons of spaG (20) and eriG (35); putative TTG start codons, with the preceding appropriate ribosomal binding site GGAGG, are located 150 bp upstream of both the spaG and eriG genes. Remarkably, the derived protein regions are similar to corresponding regions of NisG, EpiG, SrtG, and MutG proteins, in particular, within highly conserved transmembrane domain (TMD) I (Fig. 1B and D).
TABLE 3.
Amino acid identities of LanFEG proteins
| LanFEG protein | % Identical amino acidsa in:
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EriF | EpiF | NisF | MutF | SrtF | SpaE | EriE | EpiE | NisE | MutE | SrtE | EriG | EpiG | NisG | MutG | SrtG | |
| SpaF | 88 | 48 | 44 | 66 | 52 | |||||||||||
| SpaE | 100 | 74 | 25 | 19 | 32 | 23 | ||||||||||
| SpaG | 19 | 20 | 19 | —b | — | 25 | 71 | 17 | — | 28 | 26 | |||||
As determined by BLAST 2 Sequences.
—, less than 15% identical amino acids.
Effect of subtilin immunity genes in B. subtilis strain MO1099.
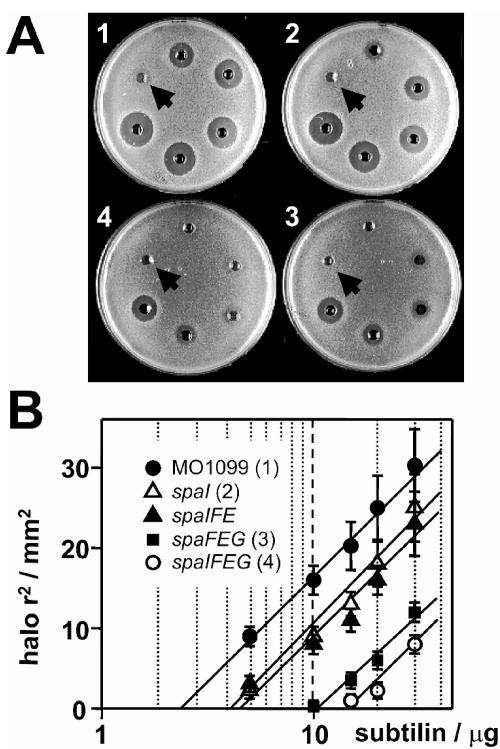
Different combinations of subtilin immunity genes spaI, spaF, spaE, and spaG have been integrated into the genome of B. subtilis strain MO1099 under the control of the IPTG-inducible Pspac promoter. B. subtilis MO1099 is a derivative of B. subtilis JH642 (Table 1) that completely lacks the spa gene cluster and thus is highly sensitive to subtilin. The strongest acquired subtilin tolerance level was obtained for B. subtilis MO1099 cells that coexpressed immunity genes spaIFEG (Fig. 2). B. subtilis cells expressing the complete ABC transporter system SpaFEG (Fig. 2A, plate 3) are less susceptible to subtilin than both wild-type (plate 1) and SpaI-expressing cells (plate 2). In contrast, an incomplete version of the ABC transporter, for example, one without the SpaG subunit, produced no subtilin tolerance (Fig. 2B). These data clearly show establishment of subtilin immunity in the subtilin nonproducer B. subtilis strain MO1099. On the other hand our results show that the lipoprotein SpaI and the ABC transporter homologue SpaFEG can act independently.
FIG. 2.
Functional analysis of subtilin immunity in B. subtilis MO1099. Subtilin sensitivities of B. subtilis MO1099 strains expressing different combinations of subtilin immunity genes were investigated in agar diffusion tests (39). (A) B. subtilis MO1099 transformed with the empty vector plasmid pDR67 (plate 1) or expressing spaI (plate 2), spaFEG (plate 3), or spaIFEG (plate 4). Applied quantities of subtilin clockwise starting from the arrow: 2.5, 5, 10, 15, 20, and 30 μg. (B) According to the second law of diffusion (also referred to as Fick's law), the square of the diffusion distance of a given solute in a liquid is directly proportional to the natural logarithm of its initial concentration. Thus, using standard volumes (60 μl) and sufficient diffusion times, linear dependencies between the square of the halos shown in panel A and the natural logarithm of the applied subtilin amounts were obtained. Results for B. subtilis MO1099 transformed with the empty vector plasmid pDR67 or expressing spaI, spaIFE (not shown in the plate assay), spaFEG, or spaIFEG are shown. Standard errors were <15% for all values (means of three independent assays).
Remarkably, the nisin susceptibility of MO1099 cells expressing subtilin immunity genes was comparable to the susceptibility of wild-type cells (not shown). This clearly shows the high specificity of the subtilin immunity system, which is able to differentiate between the cognate subtilin and the structurally closely related lantibiotic nisin.
Functions of SpaFEG.
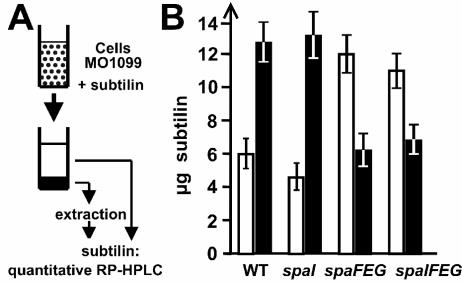
The functions of subtilin immunity proteins were analyzed in quantitative peptide release assays (Fig. 3A). When B. subtilis MO1099 or MO1099 spaI cells were preincubated with subtilin, about two-thirds of the applied subtilin was found to be cell attached and one-third was found to be in the culture supernatant (Fig. 3B). The situation was reversed for cells expressing spaFEG or spaIFEG: only one-third of the applied subtilin was cell attached and two-thirds remained in the supernatant. When only two subunits of the SpaFEG system were expressed (i.e., for the spaIFE strain), the quantity of cell-associated subtilin was similar to that for the MO1099 wild-type strain without any immunity proteins (not shown). Remarkably, in all experiments >90% of the applied subtilin could be recovered, even after longer incubation times in our peptide release assay (30 to 60 min), which argues against a degradation activity involved in subtilin immunity. Our data provide evidence that the contribution of SpaFEG to subtilin immunity is the expulsion of subtilin molecules, most likely from the cytoplasmic membrane into the extracellular medium.
FIG. 3.
Functional analysis of SpaFEG by quantitative subtilin transport assay. (A) B. subtilis cells grown to stationary phase were incubated with different amounts of subtilin. After centrifugation, the quantities of supernatant and cell-associated (after extraction; see Materials and Methods) subtilin were determined by quantitative reversed-phase HPLC (RP-HPLC). (B) Quantities of subtilin determined by the transport assay with 20 μg of applied subtilin. White bars, supernatant subtilin; black bars, subtilin extracted from cells. The values represent the means of three independent assays, for which all determinations were performed twice. Standard errors of less than 20% were obtained for all values.
SpaI: cellular localization and isolation of hexahistidine-tagged species.
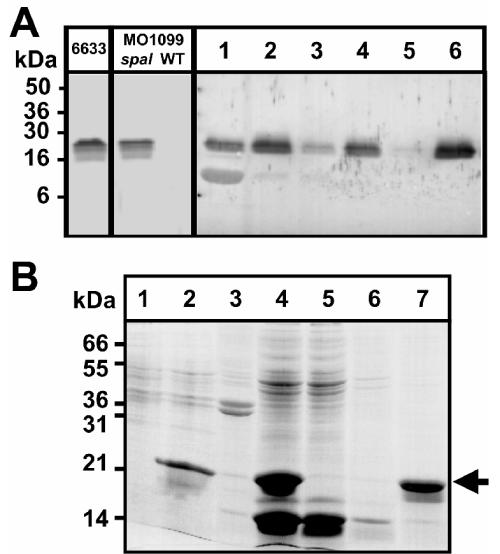
The predominant fraction of SpaI (17.2 kDa, calculated without the signal sequence, residues 1 to 22) expressed in B. subtilis MO1099 was localized in the membrane fraction (Fig. 4A), and thus it appears to be correctly anchored in the cytoplasmic membrane. SpaI possesses an N-terminal signal sequence with the typical lipobox motif LSAC (40), suggesting that SpaI becomes a peripheral membrane protein after (i) lipid modification of the lipobox Cys, (ii) processing, and (iii) transport across the cytoplasmic membrane. The lipoprotein signal sequence, including the anchoring Cys residue (amino acids 1 to 22 of SpaI), was replaced by the sequence MRSGSHHHHHH. The resulting His6-SpaI protein (17.9 kDa) was heterologously produced in E. coli (Fig. 4B, lanes 1 and 2) and could be purified from the soluble protein extract (lanes 3 and 4) by Ni-agarose affinity chromatography (lanes 5 to 7). His6-SpaI was used for interaction studies with subtilin and immunoaffinity purification of the SpaI antibody. The purified antibody showed no cross-reactivity with components of an SDS-PAGE-separated B. subtilis total-cell extract (Fig. 4A), demonstrating its high selectivity.
FIG. 4.
Expression of SpaI in heterologous hosts. (A) SpaI immunoblot of SDS-PAGE-separated proteins. Left three lanes, cell extracts of B. subtilis 6633, and MO1099 (spaI expressing and wild type [WT]). MO1099 spaI cells were disrupted by sonication. Lane 1, supernatant; lane 2, pellet suspended within a comparable volume of lysis buffer after centrifugation (1,000 × g, 60 min). The pellet was washed two times, and after centrifugation (48,000 × g, 30 min) aliquots of each supernatant (lanes 3 and 5) and the pellet corresponding to the membrane fraction (lanes 4 and 6) were analyzed. (B) Bromphenol blue stain of SDS-PAGE-separated extracts of E. coli DH5α producing N-terminal His6-tagged SpaI without a membrane anchor. Lanes 1 and 2, results before and after, respectively, 3-h IPTG induction. After lysis and centrifugation (17,000 × g, 30 min) the predominant fraction of His6-SpaI was found in the supernatant (lane 4) and not in the membrane pellet (lane 3). His6-SpaI was adsorbed to nickel-agarose (lane 5, supernatant; lane 6, supernatant after washing with 10 mM imidazole) and eluted with 200 mM imidazole (lane 7). Arrow, position of His6-SpaI.
Specific interaction between SpaI and subtilin.
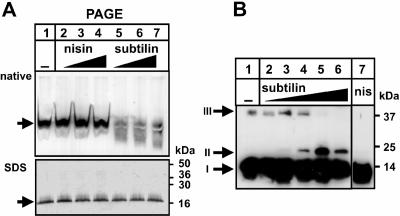
To unravel the function of SpaI, a possible interaction between SpaI and the lantibiotic subtilin or nisin was investigated. SpaI was solubilized from B. subtilis membrane vesicles with the detergents lauryl-maltoside and 6-aminocaproic acid and subsequently incubated with different amounts of subtilin and nisin. Preincubation of SpaI with subtilin led to a depletion of the SpaI immunoblot signal after native PAGE (Fig. 5A, lanes 5 to 7), which was not observed if nisin was used (Fig. 5A, lanes 2 to 4). A probable explanation for this observation is a masking of the SpaI protein by closely associated subtilin molecules, which could produce a weaker recognition by the SpaI antiserum. However, regardless of former incubation with subtilin or nisin, under denaturing conditions all samples revealed comparable SpaI signal intensities (Fig. 5A, all lanes). This argues against a covalent linkage between SpaI and subtilin and suggests an interaction of the lipoprotein SpaI specifically with subtilin.
FIG. 5.
Functional analysis of SpaI: interaction with the cognate lantibiotic subtilin. (A) SpaI-expressing B. subtilis MO1099 membrane fractions were solubilized with 0.17 volumes of 3% dodecyl-β-d-maltoside and 0.17 volumes of 2 M 6-aminocaproic acid as previously described (39). Aliquots of 60 μg of solubilized protein were incubated with different amounts of nisin or subtilin. After the aliquots were split into two parts (70:30), the larger part was separated by native PAGE (top) and the minor part was separated under denaturing conditions by SDS-PAGE (bottom). The lanes are identical for both SpaI immunoblots. Solubilized membrane proteins were incubated with a culture supernatant of a subtilin-negative mutant (lane 1); with 2, 3, and 4 μg of nisin (lanes 2 to 4), or with 2, 4, and 6 μg of subtilin (lanes 5 to 7). Arrows, positions of SpaI. (B) SpaI immunoblots of hexahistidine-mediated 0- Å cross-linking (10) between His6-SpaI and subtilin. Fifty picomoles of His6-SpaI (0.9 μg) was incubated with nickel acetate, MMPP, and 0, 29 (0.1 μg), 290 (1 μg), 860 (3 μg), 2,900 (10 μg), or 8,600 (30 μg) pmol of subtilin (lanes 1 to 6, respectively) or 2.9 nmol (10 μg) of nisin (lane 7). Arrows: I, His6-SpaI; II, putative heterodimer of His6-SpaI and subtilin; III, homodimer of His6-SpaI.
Furthermore, hexahistidine-mediated cross-linking was used to probe for direct interactions of SpaI with subtilin (10). His6-SpaI was complexed with Ni(II), followed by incubation with the peracid MMPP, which oxidized the complexed Ni(II) to Ni(III), resulting in the formation of highly reactive radicals that led to fast and efficient 0-Å cross-linking (i.e., direct cross-linking of amino acid side chains, without a linker in between). Cross-linking of His6-SpaI with subtilin results in an additional signal at 21 kDa (Fig. 5B, II) detected by the SpaI antiserum, which can be interpreted as a heterodimeric complex between His6-SpaI (17.9 kDa) and subtilin (3.3 kDa). Between ratios of subtilin to His6-SpaI of 6:1 and 60:1 (lanes 3 to 5) the signal strength of this complex increases. At a subtilin-to-His6-SpaI ratio of 180:1, the complex signal strength appeared to decrease (lane 6). The absence of a corresponding band when His6-SpaI was incubated with nisin (lane 7) argues for a preferred interaction between SpaI and its cognate lantibiotic subtilin and thus for a preferred His6-SpaI-subtilin 0-Å cross-linking reaction.
DISCUSSION
Gram-positive lantibiotic-producing organisms need effective self protection systems to protect themselves against the lethal activities of their own products. To study subtilin self protection independently from the endogenous production of subtilin, different combinations of spaIFEG were integrated into the genome of the subtilin-sensitive B. subtilis strain MO1099. The establishment of subtilin immunity in B. subtilis MO1099 was highly efficient: the subtilin tolerance level of cells that express spaIFEG was increased more than fourfold compared to that of cells without immunity genes (Fig. 2A, plates 1 and 4), and the level seems to be comparable to that for the subtilin producer B. subtilis ATCC 6633 (not shown). Completely different regulators of immunity gene expression in both strains, via an autregulatory system in the wild type and via an IPTG-inducible Pspac promoter in MO1099, make it difficult to compare the quantities of components of the subtilin immunity system in the two strains. However, the levels of the SpaI lipoprotein in the MO1099 spaI strain and the subtilin producer ATCC 6633 seem to be comparable (Fig. 4A). Our results suggest that SpaI and SpaFEG can act independently, although we cannot rule out cooperation between both systems. Though SpaI solely affected subtilin tolerance in B. subtilis, its contribution is modest compared to the contribution of NisI to nisin tolerance (39). The peptide release assay suggests an export mechanism for SpaFEG which seems to be comparable to LanFEG systems of strains producing nisin (39), mersacidin (13), epidermin (26), and lacticin 481 (28). Nevertheless, the mersacidin, epidermin, and lacticin systems are based only on exporter proteins LanFEG, whereas additional LanI lipoproteins are involved in subtilin and nisin immunity.
The ABC family of microbial multidrug resistance proteins expel a broad range of toxic molecules from the cell (29, 43). They consist of six TMDs and conserved cytoplasmic nucleotide-binding domains (NBDs) in various structural themes, from fused domains in a single multifunctional polypeptide to “half-size” transporters (42). ABC-2 transporters contain TMDs and NBDs on two separate polypeptides, exhibiting a (TMD)2(NBD)2 configuration, with two equal or two dissimilar TMDs (27). Based on homology and secondary-structure analyses we propose that the B. subtilis SpaFEG immunity system is a prototype of lantibiotic immunity transporters LanFEG, which fall into the ABC-2 subfamily of multidrug resistance proteins. Whereas LanF represents the separate NBD, the LanEG proteins represent two dissimilar transmembrane proteins (TMD-TMD′), each consisting of six TMDs (Fig. 1B and C). The conservation of the LanEG membrane topology (Table 1; Fig. 1B and C) implies that these TMDs were formed from a common ancestor. An export function of LanFEG proteins would protect the cells by diminishing the quantity of lantibiotic molecules that have already entered the cytoplasmic membrane before or during the formation of lethal pores. A hydrophobic vacuum cleaner model for the removal of the hydrophobic lantibiotic from the inner leaflet of the cytoplasmic membrane, similar to that proposed for multidrug transporters (3), fits with both the structural organization of LanFEG proteins and their function in lantibiotic extrusion.
We provided evidence that the lipoprotein SpaI interacts specifically with subtilin and not the structurally closely related nisin. Several roles for SpaI in subtilin immunity are conceivable. Attached to the outside of the cytoplasmic membrane, it can protect the membrane from subtilin by sequestering subtilin and thus prevent a high local density of subtilin molecules and subsequent pore formation. In L. lactis it has been estimated that nisin molecules dominate the cell-associated lipoprotein NisI by factors of 6 to 20 (22). A similar molar ratio between subtilin and SpaI can be assumed. Strikingly, hexahistidine-mediated cross-linking was efficient for molar excesses of subtilin over SpaI in this range. One important question that arises is how so few LanI proteins can tackle the excess of lantibiotic molecules. For nisin the peptidoglycan precursor lipid II appears to be both the docking molecule assisting membrane targeting (5) and an integral constituent of the pore itself (14). It is tempting to speculate that lipid II plays a similar role for the subtilin pore. One role for SpaI lipoproteins might be interaction with subtilin exhibiting a chaperone-like function, which can circumvent membrane insertion and/or oligomerization of subtilin prior to pore formation. Another mode of SpaI action might be competition with subtilin-lipid II pore formation. Similar mechanisms may be involved in nisin (39) and Pep5 immunity (15); however, different sizes and unrelated sequences of LanI lipoproteins may also reflect different activity modes.
Acknowledgments
This work was supported by a grant from the Stipendien-Fonds des Verbandes der Chemischen Industrie (to S.H.).
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolhuis, H., H. W. van Veen, B. Poolman, A. J. Driessen, and W. N. Konings. 1997. Mechanisms of multidrug transporters. FEMS Microbiol. Rev. 21:55-84. [DOI] [PubMed] [Google Scholar]
- 4.Breukink, E., and B. de Kruijff. 1999. The lantibiotic nisin, a special case or not? Biochim. Biophys. Acta 1462:223-234. [DOI] [PubMed] [Google Scholar]
- 5.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 6.Chen, P., F. Qi, J. Novak, and P. W. Caufield. 1999. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl. Environ. Microbiol. 65:1356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corvey, C., T. Stein, S. Düsterhus, M. Karas, and K. D. Entian. 2003. Activation of subtilin precursors by Bacillus subtilis extracellular serine proteases subtilisin (AprE), WprA, and Vpr. Biochem. Biophys. Res. Commun. 304:48-54. [DOI] [PubMed] [Google Scholar]
- 8.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G. G. A., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fancy, D. A., K. Melcher, S. A. Johnston, and T. Kodadek. 1996. New chemistry for the study of multiprotein complexes: the six-histidine tag as a receptor for a protein crosslinking reagent. Chem. Biol. 3:551-559. [DOI] [PubMed] [Google Scholar]
- 11.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzy-Treboul, G., C. Karmazyn-Campelli, and P. Stragier. 1992. Developmental regulation of transcription of the Bacillus subtilis ftsAZ operon. J. Mol. Biol. 224:967-979. [DOI] [PubMed] [Google Scholar]
- 13.Guder, A., T. Schmitter, I. Wiedemann, H. G. Sahl, and G. Bierbaum. 2002. Role of the single regulator MrsR1 and the two-component system MrsR2/K2 in the regulation of mersacidin production and immunity. Appl. Environ. Microbiol. 68:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasper, H. E., B. De Kruijff, and E. Breukink. 2004. Assembly and stability of nisin-lipid II pores. Biochemistry 43:11567-11575. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann, A., T. Schneider, U. Pag, and H. G. Sahl. 2004. Localization and functional analysis of PepI, the immunity peptide of Pep5-producing Staphylococcus epidermidis strain 5. Appl. Environ. Microbiol. 70:3263-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ireton, K., D. Z. Rudner, K. J. Siranosian, and A. D. Grossman. 1993. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 7:283-294. [DOI] [PubMed] [Google Scholar]
- 17.Jung, G. 1991. Lantibiotics: a survey, p. 1-34. In G. Jung and H.-G. Sahl (ed.), Nisin and novel lantibiotics. ESCOM Science Publishers, Leiden, The Netherlands.
- 18.Klein, C., C. Kaletta, N. Schnell, and K. D. Entian. 1992. Analysis of genes involved in biosynthesis of the lantibiotic subtilin. Appl. Environ. Microbiol. 58:132-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein, C., C. Kaletta, and K. D. Entian. 1993. Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl. Environ. Microbiol. 59:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein, C., and K. D. Entian. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koerner, T. J., J. E. Hill, A. M. Myers, and A. Tzagoloff. 1991. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 194:477-490. [DOI] [PubMed] [Google Scholar]
- 22.Koponen, O., T. M. Takala, U. Saarela, M. Qiao, and P. E. Saris. 2004. Distribution of the NisI immunity protein and enhancement of nisin activity by the lipid-free NisI. FEMS Microbiol. Lett. 231:85-90. [DOI] [PubMed] [Google Scholar]
- 23.Kuipers, O. P., M. M. Beerthuyzen, P. G. de Ruyter, E. J. Luesink, and W. M. de Vos. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J. Biol. Chem. 270:27299-27304. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 26.Otto, M., A. Peschel, and F. Götz. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tu3298. FEMS Microbiol. Lett. 166:203-211. [DOI] [PubMed] [Google Scholar]
- 27.Reizer, J., A. Reizer, and M. H. Saier, Jr. 1992. A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci. 1:1326-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rincé, A., A. Dufour, P. Uguen, J. P. Le Pennec, and D. Haras. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 63:4252-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saier, M. H., Jr., I. T. Paulsen, M. K. Sliwinski, S. S. Pao, R. A. Skurray, and H. Nikaido. 1998. Evolutionary origins of multidrug and drug-specific efflux pumps in bacteria. FASEB J. 12:265-274. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Saris, P. E., T. Immonen, M. Reis, and H. G. Sahl. 1996. Immunity to lantibiotics. Antonie Leeuwenhoek. 69:151-159. [DOI] [PubMed] [Google Scholar]
- 32.Schnell, N., K.-D. Entian, U. Schneider, F. Götz, H. Zähner, R. Kellner, and G. Jung. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide rings. Nature 333:276-278. [DOI] [PubMed] [Google Scholar]
- 33.Schnell, N., G. Engelke, J. Augustin, R. Rosenstein, V. Ungermann, F. Götz, and K.-D. Entian. 1992. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur. J. Biochem. 204:57-68. [DOI] [PubMed] [Google Scholar]
- 34.Siegers, K., and K.-D. Entian. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61:1082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein, T., S. Borchert, B. Conrad, J. Feesche, B. Hofemeister, J. Hofemeister, and K.-D. Entian. 2002. Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J. Bacteriol. 184:1703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein, T., S. Borchert, P. Kiesau, S. Heinzmann, S. Kloss, C. Klein, M. Helfrich, and K.-D. Entian. 2002. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 44:403-416. [DOI] [PubMed] [Google Scholar]
- 37.Stein, T., and K.-D. Entian. 2002. Maturation of the lantibiotic subtilin: matrix-assisted laser desorption/ionization time-of-flight mass spectrometry to monitor precursors and their proteolytic processing in crude bacterial cultures. Rapid Commun. Mass Spectrom. 16:103-110. [DOI] [PubMed] [Google Scholar]
- 38.Stein, T., S. Heinzmann, P. Kiesau, B. Himmel, and K.-D. Entian. 2003. The spa-box for transcriptional activation of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 47:1627-1636. [DOI] [PubMed] [Google Scholar]
- 39.Stein, T., S. Heinzmann, I. Solovieva, and K.-D. Entian. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278:89-94. [DOI] [PubMed] [Google Scholar]
- 40.Tjalsma, H., V. P. Kontinen, Z. Pragai, H. Wu, R. Meima, G. Venema, S. Bron, M. Sarvas, and J. M. van Dijl. 1999. The role of lipoprotein processing by signal peptidase II in the gram-positive eubacterium Bacillus subtilis. Signal peptidase II is required for the efficient secretion of alpha-amylase, a non-lipoprotein. J. Biol. Chem. 274:1698-1707. [DOI] [PubMed] [Google Scholar]
- 41.Twomey, D., R. P. Ross, M. Ryan, B. Meaney, and C. Hill. 2002. Lantibiotics produced by lactic acid bacteria: structure, function and applications. Antonie Leeuwenhoek. 82:165-185. [PubMed] [Google Scholar]
- 42.van Veen, H. W., and W. N. Konings. 1998. The ABC family of multidrug transporters in microorganisms. Biochim. Biophys. Acta 1365:31-36. [DOI] [PubMed] [Google Scholar]
- 43.van Veen, H. W. 2001. Towards the molecular mechanism of prokaryotic and eukaryotic multidrug transporters. Semin. Cell Dev. Biol. 12:239-245. [DOI] [PubMed] [Google Scholar]