Abstract
A normality assumption is typically adopted for the random effects in a clustered or longitudinal data analysis using a linear mixed model. However, such an assumption is not always realistic, and it may lead to potential biases of the estimates, especially when variable selection is taken into account. Furthermore, flexibility of nonparametric assumptions (e.g. Dirichlet process) on these random effects may potentially cause centering problems, leading to difficulty of interpretation of fixed effects and variable selection. Motivated by these problems, we proposed a Bayesian method for fixed and random effects selection in nonparametric random effects models. We modeled the regression coefficients via centered latent variables which are distributed as probit stick-breaking (PSB) scale mixtures. By using the mixture priors for centered latent variables along with covariance decomposition, we could avoid the aforementioned problems, and allow efficient selection of fixed and random effects from the model. We demonstrated the advantages of our proposed approach over other competing alternatives through a simulated example, and also via an illustrative application to a dataset from a periodontal disease study.
Keywords: Centered latent variables, nonparametric Bayes, probit stick-breaking process, fixed and random effects selection, stochastic search
1. Introduction
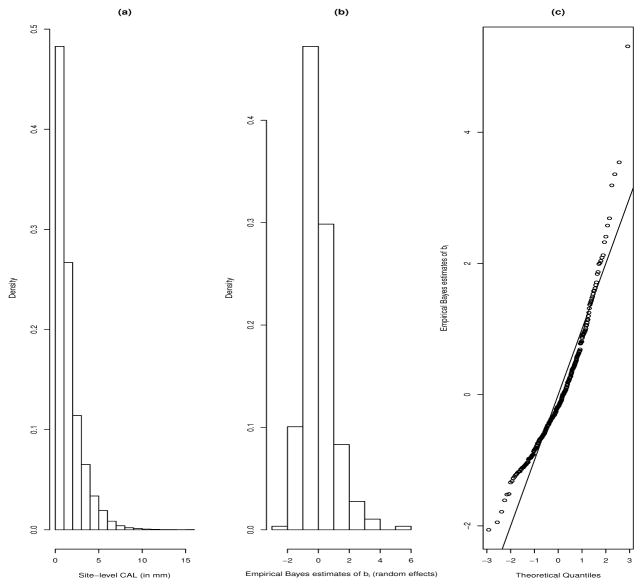
Linear mixed (effects) models are routinely used to analyze clustered and longitudinal data, where a common feature is the fidelity to the ‘Gaussian’ paradigm for the random effects and within subject random errors. Even though normality might be a reasonable model assumption, its violations may potentially impact the underlying estimation, prediction, etc of both the fixed and random effects. For example, consider the motivating data example from a clinical study conducted at the Medical University of South Carolina (MUSC) to determine periodontal health status of Gullah-speaking African American Type-2 diabetic (GAAD) subjects. One of the most important biomarkers to assess periodontal disease (PD), the clinical attachment level (or CAL, in mm), was measured for various pre-specified sites within a mouth/subject, giving rise to a typical clustered data framework. Figure 1 (Panel a) plots the density histogram of site-level CAL for the full data, while Panels b and c display the density histogram and Q-Q plots of the empirical Bayes’ estimates of the subject-level random effects, obtained after fitting a classical linear mixed model (LMM), controlling for some clinical covariables as fixed effects (such as Age, Gender, etc, more details in Section 5). These plots are indicative of the violation of the normality assumptions for the random effects, typically for a LMM analysis.
Figure 1.
Plots of density histogram for site-level CAL (Panel a); density histogram (Panel b) and Q-Q plots (Panel c) of empirical Bayes estimates of the subject-level random effects, obtained from fitting a LMM to the GAAD dataset.
To allow for flexibility of distributions of the random effect, several frequentist considerations are available [1, 2, 3]. Under the Bayesian framework, a vast majority of current research centers around the nonparametric Dirichlet process (DP) priors [4], DP mixtures [5], and other specifications, allowing unknown distributions for random effects [6, 7, 8, 9]. Under a LMM framework, inclusion of a covariate (say, age) only as a fixed effects component would quantify only the ‘average effect’ of age on the mean CAL (response), and leave out important information on how the age effect might vary across subjects. Hence, there is a need to also include a ‘random’ age effect to control for this with the ultimate goal to accommodate uncertainty of predictors and simultaneously achieve parsimony through variable selection and variance-covariance component selections. However, all of the methods described above do not accommodate this predictor uncertainty. One may potentially calculate AIC/BIC for each candidate model, yet this is infeasible unless the number of candidate predictors is modest. There does not exist any general consensus on the penalty for model complexity for a random effects model. Related frequentist propositions include score tests for random effect selection [10, 11, 12], a generalized likelihood ratio test [13], etc. However, these methods can not be directly utilized for the general subset selection problem. In a Bayesian context, a majority of work [14, 15, 16, 17, 18] focuses on variable/model selection in normal variance component models. Relaxing the normality assumption by a DP mixtures for the (unknown) random effects distribution, one may adopt the Basu and Chib [19] approach to compare the resulting semiparametric Bayesian model with the fully parametric linear model that excludes the random effect using marginal likelihoods and Bayes factors for DP mixtures. Such an approach is potentially feasible only when the number of competing random effect models is modest.
Under DP-related models for random effects, there is a difficulty in interpreting posterior inference for fixed effects and variance components of random effects due to the potential bias resulting from the unknown (distributional) specification of random effects. Under normality assumptions in the LMM, Chen and Dunson [17] proposed a Bayesian approach for random effects selection via a stochastic search variable selection algorithm [20] through a special decomposition of the random effects covariance. The Chen and Dunson’s model was extended [21] to fixed effects and random effects selection under linear and logistic mixed models. Unfortunately, it is not straightforward to modify these approaches to allow unknown random effects distributions due to difficulties in incorporating moment constraints. A center-adjusted approach was also proposed [22], however, it is difficult to incorporate random effects selection. The Chen and Dunson’s approach was also extended incorporating unknown distribution for random effects [23] using the centered stick-breaking mixtures [24]. A potential problem still remains because the variance of the latent variables related to random effects is not equal to one, resulting in non-unique decompositions of the covariance matrix for the random effects. Such decompositions may potentially affect the variance component selection and inferences. Cai and Dunson [25] developed a variable selection approach under the nonparametric random effects model with centered latent variables. However, the potential bias might still exist due to the nonparametric specification on the random effects.
In this article, we addressed some of the aforementioned limitations and developed a Bayesian approach for fixed and random effects selection with nonparametric distributions for random effects. By reparameterizing the random coefficients using centered latent variables relating to the fixed and random effects components, the proposed approach avoids the need for moment constraints, and the potential bias in estimation and variable selection. The centered latent variables were modeled by the probit stick-breaking (PSB) scale mixtures [24], allowing latent variables to be centered at the fixed effects. In addition, the centered reparametrization provided a way to incorporate variance-covariance components selection without violating the definition of decomposition of covariance matrix of the random effects. With these characteristics, the proposed method had more appropriate interpretation of the fixed effects and more efficient mixing behavior.
The paper proceeds as follows. Section 2 describes our Bayesian nonparametric specification of the LMM, the reparameterization of random coefficients, and the variable selection strategy. Section 3 outlines the posterior computational strategy, and related sampling procedures. Section 4 evaluates the performance of our method with existing alternatives using simulated data. Section 5 applies the methodology to the motivating PD dataset. Finally, Section 6 concludes, with some discussions.
2. Statistical Model
2.1. Nonparametric priors for random effects
We start with the definition of a typical LMM. Let yij be a response variable for the jth observation (j = 1, …, ni) from subject i (i = 1, …, n), xij and zij be a p × 1 vector and a q × 1 vector of candidate predictors, respectively. The LMM for yi can be written as
| (1) |
where yi = (yi1, …, yini)′, Xi = (xi1, …, xini)′, Zi = (zi1, …, zini)′, β is a p × 1 vector of fixed effects regression coefficients, bi = (bi1, …, biq)′ ~ Nq(0, Σ) is a q × 1 vector of subject-specific random effects with covariance matrix Σ, and εi is a residual error vector, typically assumed to be εi ~ N(0, σ2I).
To allow for flexibility of the distributional assumption for the random effects, if all of the candidate predictors are included, one may choose bi ~ G, where G is the unknown random effects distribution. Following the Bayesian approach [7], a prior distribution for G with support on the space of random probability measures can be chosen. A natural choice would be the DP prior which could be specified as G ~ DP(αG0), where α is a precision parameter and G0 is the base distribution of the DP. Under this specification, for any partition B = (B1, …, Bk)′ of ℜ, we have {G(B1), …, G(Bk)} ~ D(αG0(B1), …, αG0(Bk)), where D(·) denotes the finite Dirichlet density. Under the stickbreaking representation of Sethuraman (1994) [26], we have
| (2) |
with δε denoting the degenerate distribution with all its mass at ε and V0 = 0. Hence, the random distribution G can be represented as an infinite set of point masses at locations generated independently from the base distribution. In addition, we have E(G) = G0, with a natural choice of G0 being the Nq(0,Σ) distribution, so that the prior is centered at the LMM. Under this specification, the expected value of yi (conditional on Xi and Zi) is E(yi |Xi,Zi, β,G) = Xiβ + ZiE(bi). Now, under the nonparametric DP setting, the random effects distribution cannot guarantee that the mean of random effects is centered at 0 as the random mean of G, E(bi) = ∫ bidG(bi), is typically non-zero. This leads to potential complications of interpretation and inference for the fixed effects corresponding to the random effects. In addition, the computational efficiency of Gibbs sampling algorithms for posterior computation in the LMM(and other hierarchical models) tends to depend strongly on the parameterization used [27]. For greater efficiency, one can focus on the centered parameterization: yi = Xiβi + εi, with βi ~ G, G ~ DP(αG0), and G0 = Np(μ,Σ), assuming Xi = Zi so p = q. In this case, the fixed effects can be expressed as β = ∫ βidG(βi) by integrating out the random effects. Li et al. [22]proposed a moment-adjustment procedure for inference on the fixed effects that are paired with the random effects and the variance components of the random effects with hierarchically centered DP prior. However, this approach assumes that all predictors are certainly included in the fixed and random effects components. In addition, it is not straightforward for this method to be extended to the case where fixed and random effects selection is taken into account. The reason is that the fixed and random effects must be paired in order to avoid the non-zero mean of random effect due to the DP setting. Yang [23] proposed a method for fixed and random effects selection with nonparametric distributions for the random effects. To avoid the potential biases caused by the nonparametric DP prior, the random effects are modeled nonparametrically by using the probit stick-breaking (PSB) scale mixtures [24].
The PSB approach [28, 29] was proposed for conditional distribution modeling with variable selection. Briefly, a probability measure G follows a PSB with base measure G0 if it has a representation of the form
| (3) |
where Φ(·) denotes the cumulative standard normal distribution. [24] proposed priors for the residual density based on PSB scale mixtures and symmetrized PSB (sPSB) location-scale mixtures. Note that expression (3) is identical to the stick-breaking representation [26] of the DP, but the DP is obtained by replacing the stick-breaking weight Φ(ηh) with a beta(1, α) distributed random variable. Hence, the PSB process differs from the DP in using probit transformations of Gaussian random variables instead of betas for the stick lengths. In addition, ηh is not necessarily to be predictor-dependent, though it can be generalized as a predictor-dependent parameter. To allow the residuals of the linear regression model to follow an unknown distribution, a normal hierarchical structure was used with the proposed priors. For the scale PSB process mixture of Gaussian, the nonparametric distribution f(·) can be expressed as , where ph’s are defined as in (3), τh ~ 𝒢(aτ, bτ) and 𝒢(a, b) denote a gamma prior with mean of a/b and variance of a/b2. Note that the unknown density f(·) is expressed as a countable mixture of Gaussians centered at zero but with varying variances. Observations will be automatically allocated to clusters, with outlying clusters corresponding to components having large variance. Similarly, the location-scale sPSB mixture of Gaussians can be expressed as , and remains centered at zero while allowing for multimodal densities. The resulting property of centering at zero from the PSB approach provides us a solution to the non-zero centering problem from the conventional DP setting.
2.2. Fixed and random effects selection
In this article, our focus is on selecting the predictors to be included in the fixed effects and random effects components of the model under nonparametric settings. The fixed effects and random effects components have p and q candidate predictors respectively. One of the methods on subset selection for the fixed effects predictors is based on mixture priors for the regression coefficients β [30, 31]. In particular, because βl = 0 corresponds to the lth candidate predictor being effectively excluded from the fixed effects component, a prior that assigns positive probability to both H0l : βl = 0 and H1l : βl ≠ 0, for l = 1, …, p, allows for uncertainty in the subset of predictors to be included. In linear regression models, many choices of mixture priors have been proposed, and a variety of algorithms are available for posterior computation.
Compared to fixed effects selection, selection of random effects components is more challenging. One may intuitively follow the idea of fixed effects selection by inserting a vector of indicator variables, γ = diag(γ1, …, γq) in the LMM specification resulting in Ziγbi. With γl = 1, the lth random effects is included and γl = 0 otherwise. One may then combine it with bi ~ G with G ~ DP(αG0). Although the steps are straightforward, this approach is not immune to flaws such as the uncentered parameterization resulting in potential estimation biases, modeling the covariance random effects indirectly through G0 instead of G, and impossibility of selection of off-diagonal elements in the random effects covariance matrix. Chen and Dunson [17] proposed a modified Cholesky decomposition of Σ in developing a stochastic search variable selection algorithm, but their approach relies on introducing standard normal latent variables underlying the random effects. Here, the LMM takes the form yi = Xiβ + ZiΛΓεi + εi, where the latent variables εi are constrained to follow N(0,1), Λ is a diagonal matrix, and Γ is a lower triangular matrix with 1’s in the diagonal entries. With the reparameterizations and constraints, E(bi) = 0 and Var(bi) = ΛΓΓ′Λ′. This Cholesky decomposition allows for selection of both diagonal elements (variances) and off-diagonal elements (covariances) through mixture priors. In the nonparametric case, one could instead model the latent variables as having an unknown distribution with mean 0 and variance 1. However, such constraints are non-trivial to include in nonparametric models. The approach by Yang [23] (described earlier) assumes the latent variables εi to follow the centered PSB and sPSB mixtures of Gaussians. Integrating out random effects results in the decomposition of the covariance matrix of the random effects, such that Var(bi) = ΛΓΩΓ′Λ′ with Var(εi) = Ω. Selecting elements in Λ regardless of Ω in the approach may lead to potential biases in fixed effects and random effects selection.
2.3. Reparameterization and prior specification
To resolve the aforementioned drawbacks, we reparameterize the random effects with centered nonparametric distributions for the centered latent variables. In practice, it is typically unknown which covariates will be included/excluded in terms of fixed and random effects. To allow for selection of fixed and random effects for all covariates, we let Xi = Zi. Then, model (1) can be expressed as
| (4) |
Instead of modeling βi as the random effects centered at the fixed effects, we model βi as
| (5) |
where denotes a vector of independent latent variables underlying βi, Γ denotes the lower triangular matrix with 1’s in the diagonal entries, and β denotes the fixed effects as in (1). In model (4), the candidate predictors included in the random effects are chosen as the candidate predictors included in the fixed effects, which allows all predictors to possibly vary across subjects. In addition, with the reparameterization of the random coefficients in (5), the proposed model allows for the nonparametric distributions of latent variables with possibility of avoiding the centering and scaling problems.
Let , for l = 1, …, p, where . The measure Q(·) is a random distribution of λl. To incorporate random effects selection into the model, we choose Q(·) as a mixture prior consisting of a point mass at zero (with probability πl0) and a nonparametric PSB component:
| (6) |
where plh is defined as in (3), πl0, al and bl are hyperparameters. Typically, πl0 is taken as 0.5 to reflect an equal preference of inclusion and exclusion. The prior probability that the lth predictor of the p candidate predictors is excluded from the random effects components is then πl0 = Pr(λl = 0). Thus, the latent random coefficients ’s follow
| (7) |
With πl0, the distribution of ’s reduces to a point mass distribution at βl, implying that ’s, for i = 1, …, n, are replaced with βl. In this case, the lth predictor only has no random effects. With 1 − πl0, the resulting nonparametric distribution for implies that the latent variables are expressed as a countable mixture of Gaussians centered at the fixed effects βl, but with varying variances. Under this scenario, there is heterogeneity of the effect of the lth predictor across subjects.
With the centering property of the PSB, it is obvious that the random coefficients βi are centered at the fixed effects β with E(βi) = β. In addition, it can be shown that Var(βi) =ΓΨΨ′Γ′ which is the standard Cholesky decomposition of the covariance matrix with , where . When λl = 0 with probability πl0, ψl = 0 and all the atoms in Gl are effectively generated from a point mass at βl, such that there is no heterogeneity in the βil coefficients among subjects. In this case, the corresponding off-diagonal elements γlr and γsl, for r = 1, …, l − 1, s = l + 1, …, p are removed by setting their values to 0. Note that γlr is only included in the model when both the lth and rth random effects are included, which occurs when ψl > 0 and ψr > 0. This procedure has no effect on the likelihood, but does impact posterior computation. We choose a prior for γψ, the elements of γ that are included in the model. To facilitate posterior computation, we choose a conditionally conjugate N(γψ; Eγψ, Vγψ) prior. In order to allow zero off-diagonal elements in the random effects covariance matrix, this prior can be easily modified to include a mass at zero. The overall prior probability of excluding all the random effects from the model is . When ψl > 0 with 1 − πl0, it is clear that . [32] showed that with a truncated stick-breaking representation, with almost surely. For the choice of the truncation of the mixture, [33] suggested to use a reasonably large value such as 30, or the sample size.
To allow βl to effectively drop out of the model, we choose a mixture prior consisting of a point mass at zero (with probability νl0) and a normal density:
| (8) |
We refer to the prior (8) as a point-mass mixture prior, . The prior probability that the lth predictor of the p candidate predictors is excluded from the fixed effects component is then νl0 = Pr(βl = 0). From the perspective of fixed effects and random effects selections, our specification can drop predictors by choosing mixture priors for the parameters β and Λ without being complicated by the nonparametric characterization. Following standard convention, we choose a conjugate gamma prior for the residual precision of the model, π(σ−2) 𝒢(c, d) with hyperparameters c and d.
3. Posterior computation
We choose priors for the parameters as described in Section 2.3. After initializing values for the parameters, the proposed Markov chain Monte Carlo (MCMC) algorithm proceeds through the following steps:
-
Following [32], we first update the cluster allocation parameter Hil, for i = 1, …, n and l = 1, …, p. The latent variable Hil indicates the cluster that βil belongs to. Let , where Γ* = I − Γ, Γ−l denotes the submatrix of Γ excluding the lth column, and denotes the subvector of with being excluded. Then Hil can be drawn from its full conditional posterior distribution, , where
with , and Γl being the lth column of Γ.
Under the current allocation {Hil = h : h ∈ (1, …, Nl)}, we update latent variable , for i = 1, …, n and l = 1, …, p, from its full conditional posterior distribution given the data and other parameters, .
- To update plh = Φ(ηlh)∏r<h(1 − Φ(ηlr)), for h = 1, …, Nl and l = 1, …, p (following [24]), a latent variable ϕlh is introduced such that ϕlh ~ N(ηlh, 1). Thus, plh = P(ϕlh > 0, ϕlr < 0, for r < h). Then
Updating ηlh, for h = 1, …, Nl and l = 1, …, p, is straightforward from its full conditional posterior distribution, , where and .
-
Following Geweke [30] and Kuo and Mallick [34], we update the variance component ψl, for l = 1, …, p, from the full conditional mixture distribution with point mass at 0. The conditional probability of ψl = 0 is calculated by integrating out λlh, for h = 1, …, Nl,with
where L( , β, Γ, σ−2; y) = N(zi; XiΓl , σ2Ini), = ( ,…, )′, âlh = al + , b̂lh = bl + Σi:Hil = h ( − βl)2, and nlh = ♯{i : Hil = h}. With π̂l, we choose ψl from the degenerate distribution δ0(·), which means that we have ψl = 0 and . Otherwise, we generate λlh from ℐ𝒢(âlh, b̂lh), for h = 1, …, Nl.
-
Similarly, Following [30, 34], we update the parameters related to the fixed effects and random effects selection. The fixed effects βl, for l = 1, …, p, can be sampled from the mixture distribution with the point mass at 0, given bywhere the probability of βl = 0 is calculated by integrating out βl,
where denotes the lth column of Γ*, denotes the submatrix of Γ* excluding the lth column, and β−l denotes the subvector of β with βl excluded. When ψl = 0, .
-
The non-zero lower triangular elements γψ can be generated from
where V̂ψ = (σ−2 )−1, Êψ = V̂{σ−2 (yi − Xi ) + Eψ}, Vi = (Vi1,…,Vini with , and , and denotes Vi removing the elements corresponding to zeroes of ψ.
- Finally, the variance of the error terms σ−2 can be updated straightforwardly from the Gamma distribution
Relying on the above algorithm, we conduct a stochastic search through the fixed effects and random effects model spaces. For updating a single parameter from the non-conjugate distribution, we use adaptive rejection Metropolis sampling [35]. The algorithm was implemented in Matlab [36], with the respective MCMC iteration and burn-in sizes for simulation studies and real data application presented in Sections 4 and 5. After convergence of the samples for the parameters and latent variables, the posterior densities of the parameters and posterior probabilities for each of the different submodels can be straightforwardly calculated. Convergence of model parameters for both simulations and real data analysis were tested using the Geweke’s diagnostics [37] and Gelman-Rubin diagnostics [38], and good mixing behavior was observed.
4. A simulated study
A simulated data example was used to evaluate the performance of the proposed approach. In the simulation design, we combined the following scenarios: 1) the outcome only depends on some of the predictors in terms of fixed and random effects, which allows for selection of fixed and random effects based on the models; 2) the random effects were designed to follow various distributions, including the normal distribution, degenerate distribution, and the multi-modal distribution; 3) the covariance matrix of random effects reflects the varying correlations among the random effects. We generated 100 data sets, each with 200 subjects and 10 repeated measurements for each subject. Ten covariates, xij = (xij1, xij2, xij3, xij4, xij5, xij6, xij7, xij8, xij9, xij10)′, were included, where xij1 = 1 corresponding to the intercept, and the rest are generated from a standard uniform distribution. We chose , and , implying β = (2, 2, 1, 0, 0, 0, 0, 0, 0, 0)′. We chose the off-diagonal elements of Γ as γ = (γ21, γ31, γ32, γ41, γ42, γ43, γ51, …, γ10,9)′ = (0, 0.9, 0, 0.5, 0, 0.6, 0, …, 0)′. Then, the designed covariance matrix for the first four random coefficients (the rest elements are zeros), , is
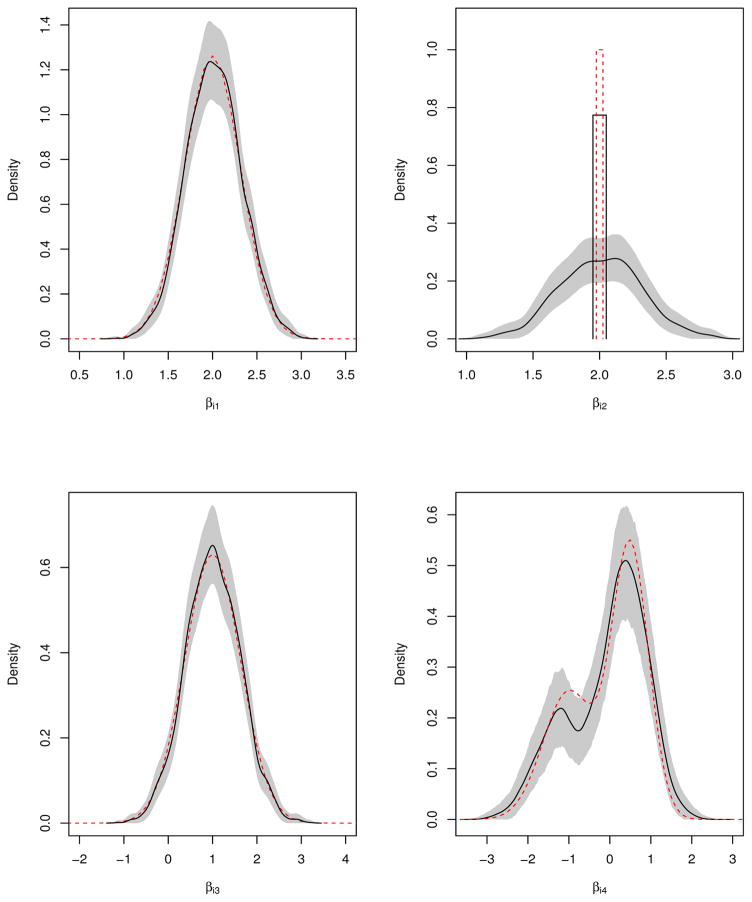
We choose σ2 = 0.4. The response variable yij is sampled from (1). The true distributions of the first four elements of are displayed by the dashed lines in Figure 2.
Figure 2.
Posterior densities (solid lines) and true densities (dashed lines) of the first four parameters βi in the simulated example. The shaded area indicates the 95% credible interval band. The vertical bars denote the probability of the point mass at 2.
Following the priors described in Section 2.3, we chose the prior distribution for σ−2 as 𝒢(0.05, 0.05). The prior distributions for the elements of β were chosen as Nδ0(0, 10). The prior probability of inclusion of a predictor was chosen to be 0.5 to reflect equal weights on inclusion and exclusion. The elements of γψ were chosen to follow independent N(0, 1). The prior for λlh was chosen as ℐ𝒢(1, 1). We ran the Gibbs sampling algorithm described in Section 3 for 10,000 iterations, after a burn-in of 1,000. Geweke’s convergence diagnostic [37] was conducted for the coefficients by calculating Z-scores and the corresponding p-values. The p-values were all larger than 0.35, implying the good mixing and convergence. In addition, the Gelman-Rubin convergence diagnostic test [38] was also applied based on multiple chains with over-dispersed starting values. The range of shrinkage reduction factors is between 1.01 and 1.08, indicating the good convergence. Posterior summaries, such as posterior probabilities of the selected submodels, estimated posterior means, and 95% credible intervals for each of the parameters were obtained based on the post burn-in samples. Sensitivity of the results to the prior specification was assessed by repeating the analyses with different hyperparameters. Although we do not show details, inferences for all models are robust to the prior specification. We noticed that different choices of hyperparameters in the prior for λlh could lead to the results with some variations in parameter estimates and probabilities of selected models. These variations were shown in Table 1 and Table 3. This is not unexpected, given the small sample size and the relatively large number of predictors. In terms of selection of hyperparameters, the bottom line is not to use too small values close to zero to obtain a diffuse prior, which typically yields an improper posterior density [39]. We suggest choosing the prior ℐ𝒢(al, al) for λlh with al value between 0.5 and 10.
Table 1.
Comparison of the estimates of the parameters for the first four covariates in the simulated example.
| Parameter | True value | LMM | CD | Yang | NPME |
|---|---|---|---|---|---|
| β1 | 2 | 2.03(1.96,2.12) | 2.03(1.97,2.13) | 2.02(1.96,2.09) | 2.01(1.98,2.07) |
| β2 | 2 | 2.02(1.93,2.11) | 2.02(1.93,2.09) | 1.98(1.94,2.05) | 2.01(1.96,2.05) |
| β3 | 1 | 1.10(0.96,1.23) | 1.08(0.95,1.19) | 1.04(0.96,1.16) | 1.03(0.96,1.11) |
| β4 | 0 | 0.03(−0.12,0.16) | 0.05(−0.10,0.19) | 0.02(−0.09,0.12) | 0.01(−0.07,0.10) |
| σ11 | 0.1 | 0.12(0.07,0.16) | 0.12(0.07,0.17) | 0.08(0.03,0.12) | 0.09(0.04,0.13) |
| σ21 | 0 | 0.01(−0.05,0.06) | 0.003(−0.02,0.03) | 0.003(−0.02,0.03) | 0.002(−0.01,0.02) |
| σ22 | 0 | 0.30(0.25,0.38) | 0.01(0.00,0.05) | 0.004(0.00,0.05) | 0.003(0.00,0.03) |
| σ31 | 0.09 | 0.07(0.03,0.15) | 0.12(0.06,0.17) | 0.08(0.03,0.14) | 0.09(0.04,0.12) |
| σ32 | 0 | −0.01(−0.07,0.06) | 0.003(−0.03,0.04) | −0.00(−0.03,0.03) | 0.00(−0.02,0.03) |
| σ33 | 0.48 | 0.43(0.33,0.54) | 0.55(0.40,0.69) | 0.44(0.35,0.52) | 0.44(0.36,0.52) |
| σ41 | 0.05 | 0.02(−0.04,0.09) | 0.02(−0.04,0.08) | 0.03(−0.04,0.10) | 0.03(−0.04,0.09) |
| σ42 | 0 | 0.002(−0.06,0.05) | 0.002(−0.04,0.04) | 0.002(−0.04,0.05) | 0.001(−0.04,0.05) |
| σ43 | 0.29 | 0.32(0.26,0.34) | 0.27(0.23,0.33) | 0.27(0.23,0.32) | 0.30(0.26,0.36) |
| σ44 | 0.99 | 1.08(0.97,1.18) | 0.95(0.88,1.03) | 0.96(0.89,1.03) | 0.97(0.90,1.03) |
| σ2 | 0.4 | 0.38(0.34,0.43) | 0.41(0.33,0.49) | 0.43(0.36,0.49) | 0.41(0.36,0.47) |
Table 3.
Estimated model posterior probabilities of top five models in the simulated example. Methods used: LMM = R package MCMCglmm of Hadfield [40]; CD = Chen and Dunson (2003)[17]; Yang = Yang (2013)[23]; NPME = the proposed nonparametric mixed effects model.
| Model | CD | Yang | NPME | DIC | BPIC | |
|---|---|---|---|---|---|---|
| xij1, xij2, xij3, zij1, zij3, |
|
0.736(0.677,0.775) | 0.761(0.720,0.809) | 4137.36 | 4584.62 | |
| xij1, xij2, xij3, zij1, zij2, zij3, zij4 | 0.125(0.076,0.140) | 0.100(0.078,0.136) | 0.111(0.088,0.130) | 4215.06 | 4843.14 | |
| xij1, xij2, xij3, zij1, zij3, zij4, zij9 | 0.030(0.012,0.040) | 0.025(0.016,0.041) | 0.029(0.019,0.035) | 4241.91 | 4775.53 | |
| xij1, xij2, xij3, zij1, zij3, zij4, zij8 | 0.026(0.014,0.044) | 0.021(0.009,0.040) | 0.013(0.008,0.033) | 4247.80 | 4783.78 | |
| xij1, xij2, xij4, zij1, zij3, zij4, zij7 | 0.011(0.001,0.019) | 0.009(0.000,0.017) | 0.008(0.000,0.015) | 4240.06 | 4790.50 |
True model
Posterior probability
Range
To compare the results from our proposed nonparametric mixed effects (NPME) model with other alternatives, we fitted a Bayesian LMM using the R package MCMCglmm [40], Chen and Dunson’s (CD) model [17] with modifications by adding the fixed effects selection, and Yang’s (Yang) [23] method with unimodal distribution. Table 1 presents the true values, posterior estimates, and 95% credible intervals of the parameters corresponding to the first four covariates from the four competing methods. It is shown that the NPME estimates are closer to the true values than those from the other models. The estimates of the parameters for the rest of the covariates are pretty close to zeros from all the methods, which are not shown due to the space limit. Table 2 shows the comparison of the results from the four methods over 100 simulated data sets. In Table 2, we calculated the average of the estimated standard errors (ESE), the sample standard deviation (SSD) of the 100 point estimates and the mean squared errors (MSEs). Although all of the MSEs are small for the estimates of β’s from the four methods, it is clear that the estimates from the proposed method have relatively smaller ESEs, SSDs and MSEs.
Table 2.
Comparison of the estimated standard errors (ESE), the sample standard deviation (SSD) and the mean squared errors (MSEs) of the estimates of β’s based on 100 simulated data sets. Methods used: LMM = R package MCMCglmm of Hadfield [40]; CD = Chen and Dunson (2003)[17]; Yang = Yang (2013)[23]; NPME = the proposed nonparametric mixed effects model.
| Method | β1 | β2 | β3 | β4 | β5 | β6 | β7 | β8 | β9 | β10 |
|---|---|---|---|---|---|---|---|---|---|---|
| LMM | ||||||||||
| ESE | 0.0073 | 0.0274 | 0.0068 | 0.0068 | 0.0073 | 0.0052 | 0.0080 | 0.0070 | 0.0060 | 0.0066 |
| SSD | 0.0075 | 0.0244 | 0.0076 | 0.0072 | 0.0069 | 0.0060 | 0.0073 | 0.0075 | 0.0063 | 0.0066 |
| MSE | 0.0021 | 0.0100 | 0.0075 | 0.0049 | 0.0061 | 0.0055 | 0.0066 | 0.0030 | 0.0049 | 0.0032 |
| CD | ||||||||||
| ESE | 0.0069 | 0.0039 | 0.0080 | 0.0071 | 0.0069 | 0.0058 | 0.0076 | 0.0065 | 0.0069 | 0.0061 |
| SSD | 0.0092 | 0.0043 | 0.0072 | 0.0054 | 0.0072 | 0.0064 | 0.0060 | 0.0060 | 0.0061 | 0.0065 |
| MSE | 0.0007 | 0.0005 | 0.0072 | 0.0068 | 0.0064 | 0.0050 | 0.0061 | 0.0041 | 0.0053 | 0.0032 |
| Yang | ||||||||||
| ESE | 0.0061 | 0.0043 | 0.0059 | 0.0030 | 0.0068 | 0.0056 | 0.0065 | 0.0057 | 0.0068 | 0.0061 |
| SSD | 0.0065 | 0.0042 | 0.0054 | 0.0038 | 0.0067 | 0.0063 | 0.0057 | 0.0055 | 0.0063 | 0.0063 |
| MSE | 0.0009 | 0.0012 | 0.0020 | 0.0012 | 0.0038 | 0.0042 | 0.0040 | 0.0017 | 0.0041 | 0.0025 |
| NPME | ||||||||||
| ESE | 0.0043 | 0.0026 | 0.0034 | 0.0031 | 0.0043 | 0.0039 | 0.0047 | 0.0025 | 0.0031 | 0.0037 |
| SSD | 0.0034 | 0.0039 | 0.0041 | 0.0032 | 0.0040 | 0.0035 | 0.0046 | 0.0030 | 0.0035 | 0.0034 |
| MSE | 0.0008 | 0.0008 | 0.0011 | 0.0010 | 0.0010 | 0.0008 | 0.0012 | 0.0007 | 0.0021 | 0.0008 |
Table 3 presents the posterior probabilities of top five models selected by the proposed mixed effect method and the other two Bayesian methods. We also calculated corresponding deviance information criterion (DIC) [41], obtained after running separate LMM analyses for each model in the list. Although each method chooses the true model as the best model, our NPME approach selected the true model with the higher posterior probability than the CD and the Yang methods. The DICs confirmed the selection based on the posterior probabilities. To avoid the potential overfitting problem when using DIC, we also considered the Bayesian predictive information criterion (BPIC) [42]. Due to the complexity of the proposed semiparametric model, computing the score required for BPIC could be complicated. Instead, we calculated the BPIC based on [43], where we included double of the model complexity in the criterion which provides more accurate penalty in the criterion. The BPICs in Table 3 confirmed the top selected model but there was some disparity among DIC, BPIC and the selection methods for the other models. As pointed out by [41, 43], the penalty term based on the model complexity in DIC and BPIC are not invariant to reparameterization, which may cause this problem. Figure 2 depicts the posterior densities of the random coefficient parameters βi based on our NPME model, and the corresponding true densities. It appears that the proposed NPME successfully captured the right densities of βi.
5. Application: Periodontal Data
We illustrate our approach through analysis of the motivating GAAD dataset (see Section 1) generated from a clinical study at the Medical University of South Carolina [44]. The relationship between PD and diabetes level has been previously studied in the dental literature [45, 46], and the objective of this analysis is to quantify the disease status of this interesting population, and to study the associations between PD status and diabetes level (determined by the popular marker HbA1c, or ‘glycosylated hemoglobin’) in the Type-2 diabetic African-American adults residing in the coastal sea-islands of South Carolina.
Our analysis focused on identifying predictors of one of the most popular bio-markers of PD, the clinical attachment level (CAL). CAL is the distance down a tooth’s root that is no longer attached to the surrounding bone by the periodontal ligament. During a full periodontal exam, CAL is usually measured at six pre-specified sites [47] for each tooth (excluding the third molars, i.e., the wisdom teeth). For a subject with no missing teeth, there are S = 168 measurements for CAL. The CAL measures for each subject are clustered and highly correlated. The subject-level covariates include age (in year), body mass index (BMI) (in kg/m2), gender (1=female, 0=male), HbA1c (1=high, 0=controled) and smoking status (1=smoker, 0=non-smoker). In addition, the total number of available teeth (cluster size) within each mouth/subject is varying, and we included the log(cluster size) for each subject as a predictor as it is highly associated with dental health [48].
In risk assessment studies involving PD, a linear relationship was considered between the response CAL and the associated risk factors in this data set [49, 50, 51]. We followed the same linearity assumption, and proceed by fitting our nonparametric LMM. Predictors included in the fixed effects component have an average effect on the mean CAL, while predictors included in the random effect component vary in their effects across subjects. Let xij = (xij1, xij2, xij3, xij4, xij5, xij6, xij7)′ denote the vector of candidate predictors with xij1 = 1, xij2=age, xij3 =BMI, xij4 =gender, xij5 =HbA1c, xij6 =smoking status, and xij7 =log(cluster size). We included 288 out of 360 patients, consisting of patients with at least one tooth (i.e. 6 measures) and complete covariate information. The cluster size (per subject) varied between 18 and 168.
The prior distributions for the elements of β are chosen as Nδ0 (0, 10) with νl0 = 0.5. The prior distributions for the free elements of γ are independent N(0, 1). The mixture prior distributions of the elements of λlh are chosen as independent ℐ𝒢(1, 1). We also chose 𝒢(0.05, 0.05) as the prior for σ−2. We ran the MCMC algorithm described in Section 3 for 20,000 iterations, with a burn-in size of 10,000. The Geweke’s diagnostic tests [37] for regression coefficients based on Z-scores and the Gelman-Rubin diagnostic [38] demonstrated good mixing. Posterior probabilities for the possible submodels, estimated posterior means, and 95% credible intervals for each of the parameters are calculated thereafter. Sensitivity of the results to the prior specifications were assessed by repeating the analyses with varying choices of hyperparameters, similar to those in the simulated example. The results appeared stable.
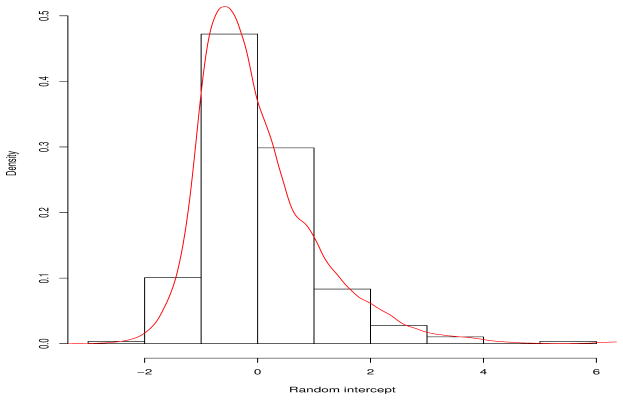
Table 4 presents the posterior means and 95% credible intervals for fixed effects, and the marginal posterior probability of inclusion of predictors in terms of fixed effects and random effects. From the proposed approach, we observe a significant negative effect of gender on CAL, indicating males more likely to be prone to PD than females. A significantly positive effect of HbA1c implies patients with uncontrolled glycemic level are more likely to experience PD. This result is consistent with previous findings [48]. In addition, the log of cluster size of teeth sites confirmed a significantly negative impact on the CAL, which is intuitive given that patients with larger number of available teeth (i.e., higher log cluster size) are expected to have a lower degree of PD. From the Bayesian LMM, only the log of cluster size is significantly and negatively affecting CAL. For comparison purpose, the means and 95% credible intervals for β1, …, β7 from the Bayesian LMM are also presented in Table 4. Although the estimates of the fixed effects from the two approaches are similar, the 95% credible intervals of estimates from the Bayesian LMM are wider than those based on our proposed approach. From the marginal posterior probabilities of inclusion, it is clear that for the fixed effects components, the predictors including gender, HbA1C and log(cluster size) are important in predicting CAL. On the other hand, our nonparametric random effects method suggests to include the random intercept and effects for the smoking status, implying heterogeneity of these effects across subjects, while the effects of the other predictors do not vary substantially. The proposed method selected the top model with the posterior probability of 0.35, including all predictors in fixed effects, and all predictors except age and log(cluster size) in random effects. In contrast, Chen and Dunson’s method and Yang’s method chose the same model with the posterior probability of 0.29 and 0.28, respectively. Based on the marginal probability, the Bayes factor can be calculated [52] as inclusion criterion for a single predictor in terms of fixed and random effects. Kass and Raftery [52] suggest the cutoff points for positive, strong and very strong evidence for a Bayes factor as 3, 20 and 150, respectively. Given the same prior probability for inclusion and exclusion (i.e. 0.5), the marginal posterior probability over 0.96 corresponds to the Bayes factor being over 20, indicating a strong evidence of inclusion. Figure 3 shows the histogram of empirical Bayes estimates of random intercept based on the LMM and the density curve of posterior estimates of the random intercept from the proposed method. In terms of model fits to the data, we calculated the DIC and BPIC for both the models. The DICs for the Bayesian LMM and our proposed model are 54,403.15 and 54,205.68, respectively, and the BPICs for the Bayesian LMM and our proposed model are 54,545.39 and 54,386.00, respectively, implying that the proposed model has a better fit to the data.
Table 4.
Estimates of fixed effects, 95% credible intervals, and marginal posterior inclusion probabilities of predictors in the fixed effects and random effects components in the GAAD dataset. Methods compared: LMM = R package MCMCglmm [40]; CD = Chen and Dunson’s method [17]; Yang = Yang’s method [23]; NPME = our proposed nonparametric mixed effects model.
| Predictor | Fixed Effect Estimate (95% CI)
|
NPME Marginal Probability of Inclusion |
||||
|---|---|---|---|---|---|---|
| LMM | CD | Yang | NPME | Fixed Effect | Random Effect | |
| Intercept | 6.43(5.00,8.06) | 6.61(4.88,8.45) | 6.56(5.10,8.38) | 6.21(5.01,7.34) | 1.00 | 0.95 |
| Age | −0.01(−0.21,0.24) | −0.02(−0.11,0.12) | −0.03(−0.08,0.14) | −0.02(−0.12,0.08) | 0.80 | 0.66 |
| BMI | −0.11(−0.33,0.12) | −0.10(−0.45,0.21) | −0.09(−0.28,0.14) | −0.12(−0.23,0.003) | 0.72 | 0.75 |
| Gender | −0.41(−0.89,0.09) | −0.43(−0.81,0.02) | −0.45(−0.80,0.03) | −0.44(−0.70,−0.18) | 1.00 | 0.76 |
| HbA1c | 0.30(−0.06,0.68) | 0.33(−0.01,0.64) | 0.33(−0.03,0.76) | 0.29(0.08,0.48) | 0.98 | 0.79 |
| Smoking Status | 0.16(−0.26,0.60) | 0.13(−0.23,0.51) | 0.14(−0.30,0.56) | 0.16(−0.10,0.40) | 0.81 | 0.97 |
| log(cluster size) | −1.03(−1.42,−0.56) | −1.00(−1.38,−0.61) | −0.99(−1.32,−0.61) | −0.92(−1.16,−0.63) | 1.00 | 0.001 |
Figure 3.
Histogram of empirical Bayes estimates and the density of posterior estimates of the random intercept.
6. Conclusions
We develop a Bayesian approach to the problem of nonparametric random effects models where both the predictors to be included and distributions of their random effects are unknown. Relying on reparameterization of the random coefficients and the centered nonparametric distributions, our proposed approach avoids the potential biases in estimation, which may lead to difficulty in interpretation. Incorporating centered independent latent variables with the decomposition of the dependency of random coefficients allows the approach to be efficient and straightforward to implement. By using latent random coefficients which are centered at fixed effects, the proposed reparameterization allows for the random effects not necessarily being the subset of the fixed effects, resulting in the independent selection of the fixed and random effects. The simulation study shows that the performance of the proposed method is better than the other competing methods available. It is straightforward to extend the method to allow categorical outcomes by using data augmentation as in probit models. Although motivated by the random effects selection problem, the proposed approach provides a general strategy for dependency modeling in related unknown distributions. Future research may focus on analyzing multivariate responses with spatial information observed in datasets from dental epidemiology. In addition, it might be really interesting to analyze non-continuous responses, with variable selection under the similar nonparametric framework. Such methods are less developed and challenging, and will be pursued elsewhere.
Acknowledgments
The authors thank the Center for Oral Health Research at MUSC for providing the motivating data. They also thank the Editor, the Associate Editor, and three anonymous referees, whose constructive comments led to a substantially improved version of this manuscript. This work was supported by NIH/NIDCR grants R03DE020114, R03DE023372 and R01DE024984. The authors would like to thank Dr. Mingan Yang for providing the Fortran code for fitting his method in the simulated example.
References
- 1.Chen J, Zhang D, Davidian M. A Monte Carlo EM algorithm for generalized linear mixed models with flexible random effects distribution. Biostatistics. 2002;3(3):347–360. doi: 10.1093/biostatistics/3.3.347. [DOI] [PubMed] [Google Scholar]
- 2.Lai TL, Shih MC. Nonparametric estimation in nonlinear mixed effects models. Biometrika. 2003;90(1):1–13. [Google Scholar]
- 3.Ghidey W, Lesaffre E, Eilers P. Smooth random effects distribution in a linear mixed model. Biometrics. 2004;60(4):945–953. doi: 10.1111/j.0006-341X.2004.00250.x. [DOI] [PubMed] [Google Scholar]
- 4.Ferguson TS. A Bayesian analysis of some nonparametric problems. The Annals of Statistics. 1973;1:209–230. [Google Scholar]
- 5.Antoniak CE. Mixtures of Dirichlet processes with applications to Bayesian nonparametric problems. The Annals of Statistics. 1974:1152–1174. [Google Scholar]
- 6.Bush CA, MacEachern SN. A semiparametric Bayesian model for randomised block designs. Biometrika. 1996;83(2):275–285. [Google Scholar]
- 7.Kleinman KP, Ibrahim JG. A semiparametric bayesian approach to the random effects model. Biometrics. 1998;54(3):921–938. [PubMed] [Google Scholar]
- 8.Ishwaran H, Takahara G. Independent and identically distributed Monte Carlo algorithms for semiparametric linear mixed models. Journal of the American Statistical Association. 2002;97(460):1154–1166. [Google Scholar]
- 9.Müller P, Rosner GL, Iorio MD, MacEachern S. A nonparametric Bayesian model for inference in related longitudinal studies. Journal of the Royal Statistical Society: Series C (Applied Statistics) 2005;54(3):611–626. [Google Scholar]
- 10.Lin X. Variance component testing in generalised linear models with random effects. Biometrika. 1997;84(2):309–326. [Google Scholar]
- 11.Verbeke G, Molenberghs G. The use of score tests for inference on variance components. Biometrics. 2003;59(2):254–262. doi: 10.1111/1541-0420.00032. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Z, Fung WK. Variance component testing in semiparametric mixed models. Journal of Multivariate Analysis. 2004;91(1):107–118. [Google Scholar]
- 13.Crainiceanu CM, Ruppert D. Restricted likelihood ratio tests in nonparametric longitudinal models. Statistica Sinica. 2004;14:713–729. [Google Scholar]
- 14.Albert J, Chib S. Bayesian tests and model diagnostics in conditionally independent hierarchical models. Journal of the American Statistical Association. 1997;92(439):916–925. [Google Scholar]
- 15.Pauler DK, Wakefield JC, Kass RE. Bayes factors and approximations for variance component models. Journal of the American Statistical Association. 1999;94(448):1242–1253. [Google Scholar]
- 16.Sinharay S, Stern H. Proceedings I, editor. Bayesian Methods with Applications to Science, Policy and Official Statistics. 2001. Bayes factors for variance component testing in generalized linear mixed models; pp. 507–516. [Google Scholar]
- 17.Chen Z, Dunson DB. Random effects selection in linear mixed models. Biometrics. 2003;59(4):762–769. doi: 10.1111/j.0006-341x.2003.00089.x. [DOI] [PubMed] [Google Scholar]
- 18.Cai B, Dunson D. Bayesian covariance selection in generalized linear mixed models. Biometrics. 2006;62(2):446–457. doi: 10.1111/j.1541-0420.2005.00499.x. [DOI] [PubMed] [Google Scholar]
- 19.Basu S, Chib S. Marginal likelihood and Bayes factors for Dirichlet process mixture models. Journal of the American Statistical Association. 2003;98(461):224–235. [Google Scholar]
- 20.George EI, McCulloch RE. Variable selectionvia Gibbs sampling. Journal of the American Statistical Association. 1993;88:881–889. [Google Scholar]
- 21.Kinney SK, Dunson DB. Fixed and random effects selection in linear and logistic models. Biometrics. 2007;63(3):690–698. doi: 10.1111/j.1541-0420.2007.00771.x. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Müller P, Lin X. Center-adjusted inference for a nonparametric Bayesian random effect distribution. Statistica Sinica. 2011;21(3):1201–1223. doi: 10.5705/ss.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M. Bayesian nonparametric centered random effects models with variable selection. Biometrical Journal. 2013;55(2):217–230. doi: 10.1002/bimj.201100149. [DOI] [PubMed] [Google Scholar]
- 24.Pati D, Dunson DB. Bayesian nonparametric regression with varying residual density. Annals of the Institute of Statistical Mathematics. 2014;66(1):1–31. doi: 10.1007/s10463-013-0415-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai B, Dunson D. Technical Report. Duke University; 2010. Variable selection in nonparametric random effects models. [Google Scholar]
- 26.Sethuraman J. A constructive definition of Dirichlet priors. Statistica Sinica. 1994;4:639–650. [Google Scholar]
- 27.Gelfand AE, Sahu SK, Carlin BP. Efficient parametrisations for normal linear mixed models. Biometrika. 1995;82(3):479–488. [Google Scholar]
- 28.Chung Y, Dunson DB. Nonparametric Bayes conditional distribution modeling with variable selection. Journal of the American Statistical Association. 2009;104:1646–1660. doi: 10.1198/jasa.2009.tm08302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez A, Dunson DB. Nonparametric Bayesian models through probit stick-breaking processes. Bayesian analysis. 2011;6(1):145–177. doi: 10.1214/11-BA605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geweke J. Variable selection and model comparison in regression. In: Berger JO, Bernardo JM, Dawid AP, Smith AFM, editors. Bayesian Statistics 5. Vol. 5. Oxford University Press; 1996. pp. 609–620. [Google Scholar]
- 31.George EI, McCulloch RE. Approaches for Bayesian variable selection. Statistica Sinica. 1997;7:339–373. [Google Scholar]
- 32.Ishwaran H, James LF. Gibbs sampling methods for stick-breaking priors. Journal of the American Statistical Association. 2001;96:161–173. [Google Scholar]
- 33.Ishwaran H, Zarepour M. Markov chain Monte Carlo in approximate Dirichlet and beta two-parameter process hierarchical models. Biometrika. 2000;87(2):371–390. [Google Scholar]
- 34.Kuo L, Mallick B. Variable selection for regression models. Sankhya B. 1998;60(1):65–81. [Google Scholar]
- 35.Gilks WR, Best N, Tan K. Adaptive rejection Metropolis sampling within Gibbs sampling. Journal of the Royal Statistical Society Series C (Applied Statistics) 1995;44(4):455–472. [Google Scholar]
- 36.MATLAB. version 7.10.0 (R2010a) The MathWorks Inc; Natick, Massachusetts: 2010. [Google Scholar]
- 37.Geweke J. Evaluating the accuracy of sampling-based approaches to the calculation of posterior moments. In: Berger JO, Bernardo JM, Dawid AP, Smith AFM, editors. Bayesian Statistics 4. Vol. 4. Oxford University Press; 1992. pp. 169–193. [Google Scholar]
- 38.Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Statistical Science. 2006;1(3):515–533. [Google Scholar]
- 39.Gelman A. Prior distributions for variance parameters in hierarchical models. Bayesian Analysis. 1992;7(4):457–511. [Google Scholar]
- 40.Hadfield JD, et al. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. Journal of Statistical Software. 2010;33(2):1–22. [Google Scholar]
- 41.Spiegelhalter DJ, Best NG, Carlin BP, Van Der Linde A. Bayesian measures of model complexity and fit. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64(4):583–639. [Google Scholar]
- 42.Ando T. Bayesian predictive information criterion for the evaluation of hierarchical bayesian and empirical bayes models. Biometrika. 2007;94(2):443–458. [Google Scholar]
- 43.Ando T. Predictive bayesian model selection. American Journal of Mathematical and Management Sciences. 2011;31(1):13–38. [Google Scholar]
- 44.Bandyopadhyay D, Marlow NM, Fernandes JK, Leite RS. Periodontal disease progression and glycaemic control among Gullah African Americans with Type-2 diabetes. Journal of Clinical Periodontology. 2010;37(6):501–509. doi: 10.1111/j.1600-051X.2010.01564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faria-Almeida R, Navarro A, Bascones A. Clinical and metabolic changes after conventional treatment of type-2 diabetic patients with chronic periodontitis. Journal of Periodontology. 2006;77(4):591–598. doi: 10.1902/jop.2006.050084. [DOI] [PubMed] [Google Scholar]
- 46.Taylor GW, Borgnakke W. Periodontal disease: Associations with diabetes, glycemic control and complications. Oral Diseases. 2008;14(3):191–203. doi: 10.1111/j.1601-0825.2008.01442.x. [DOI] [PubMed] [Google Scholar]
- 47.Darby ML, Walsh M. Dental Hygiene: Theory and Practice. 1. W. B. Saunders; Philadelphia, PA: 1995. [Google Scholar]
- 48.Botero JE, Yepes FL, Roldán N, Castrillón CA, Hincapie JP, Ochoa SP, Ospina CA, Becerra MA, Jaramillo A, Gutierrez SJ, et al. Tooth and periodontal clinical attachment loss are associated with hyperglycemia in patients with diabetes. Journal of Periodontology. 2012;83(10):1245–1250. doi: 10.1902/jop.2012.110681. [DOI] [PubMed] [Google Scholar]
- 49.Bandyopadhyay D, Lachos VH, Abanto-Valle CA, Ghosh P. Linear mixed models for skew-normal/independent bivariate responses with an application to periodontal disease. Statistics in Medicine. 2010;29(25):2643–2655. doi: 10.1002/sim.4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reich BJ, Bandyopadhyay D. A latent factor model for spatial data with informative missingness. The Annals of Applied Statistics. 2010;4(1):439–459. doi: 10.1214/09-AOAS278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reich BJ, Bandyopadhyay D, Bondell HD. A nonparametric spatial model for periodontal data with non-random missingness. Journal of the American Statistical Association. 2013;108(503):820–831. doi: 10.1080/01621459.2013.795487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kass RE, Raftery A. Bayes factors. Journal of the American Statistical Association. 1995;90(430):773–795. [Google Scholar]