Abstract
Patients with schizophrenia show decreased processing speed on neuropsychological testing and decreased white matter integrity as measured by diffusion tensor imaging, two traits shown to be both heritable and genetically associated indicating that there may be genes that influence both traits as well as schizophrenia disease risk. The potassium channel gene family is a reasonable candidate to harbor such a gene given the prominent role potassium channels play in the central nervous system in signal transduction, particularly in myelinated axons. We genotyped members of the large potassium channel gene family focusing on putatively functional single nucleotide polymorphisms (SNPs) in a population of 363 controls, 194 patients with schizophrenia spectrum disorder, and 28 patients with affective disorders with psychotic features who completed imaging and neuropsychological testing. We then performed three association analyses using three phenotypes- processing speed, whole-brain white matter fractional anisotropy, and schizophrenia spectrum diagnosis. We extracted SNPs showing an association at a nominal P value of <0.05 with all three phenotypes in the expected direction: decreased processing speed, decreased fractional anisotropy, and increased risk of schizophrenia spectrum disorder. A single SNP, rs8234, in the 3′ untranslated region (UTR) of voltage-gated potassium channel subfamily Q member 1 (KCNQ1) was identified. Rs8234 has been shown to affect KCNQ1 expression levels, and KCNQ1 levels have been shown to affect neuronal action potentials. This exploratory analysis provides preliminary data suggesting that KCNQ1 may contribute to the shared risk for diminished processing speed, diminished white mater integrity, and increased risk of schizophrenia.
Introduction
In the central nervous system, potassium channels serve the fundamental function of supporting action potentials and electrical signal propagation along the neurons and their myelinated axons (Choe 2002). The spread of outward currents of action potentials along the axons are achieved by the opening of the voltage-gated potassium channels at the nodes of Ranvier (Yellen 2002). Indeed, potassium channels are densely expressed in the nodes of Ranvier of myelinated axons where they support saltatory signal propagation, greatly speeding up signal transmissions across brain regions (Buttermore et al 2013). They regulate action potential initiation and conduction (Battefeld et al 2014) and are responsible for the repolarization phase of action potentials, and the speed and selectivity of potassium channels regulate the currents in fast-spiking neurons (Bean 2007, Yellen 2002). There are 92 human genes encoding different potassium channels (HUGO Gene Nomenclature Committee Database www.genenames.org). Some of these channels such as KCNJ10 and KCNJ16 are restricted to oligodendrocytes where they maintain the resting potential of oligodendrocytes necessary for myelin integrity (Brasko et al 2016). Given the neurophysiological roles of potassium channels, genetic variance in potassium channel genes may have a shared common effect on the function of myelinated axons and processing speed.
Cognitive deficits are core symptoms in schizophrenia, and analyses show that processing speed as measured by the digit symbol substitution task (DSST) has one of the largest schizophrenia - control differences among cognitive domains (Dickenson et al 2007; Knowles et al 2010 et al; Henry and Crawford 2005). The large effect size of the DSST compared to other cognitive tasks remains after considering medication effects (Knowles 2010). The heritability estimates for processing speed range between 33% and 68% (Wright et al 2001; Lee et al 2012; Cirulliet al 2010).
White matter impairment has also emerged as one of the most consistent neurobiological deficits identified in schizophrenia (Kochunov et al 2013; Mori et al 2007; Friedman et al 2008; Pérez-Iglesias et al 2010; Davis et al 2003), primarily through measurement of the fractional anisotropy (FA) using diffusion tensor imaging (DTI). White matter abnormalities in patients with schizophrenia have been shown to mediate their processing speed deficit (Karbasforoushan et al 2015; Wright et al 2015). Furthermore, processing speed and white matter FA have been found to be phenotypically and genetically correlated (Kochunov et al 2016a). These findings have led to a search for genes that influence both white matter and processing speed in healthy adults (Giddaluru et al 2016).
As cerebral white matter facilitates the speed of electrical signal processing through myelinated axons (Chang and Rasband 2013), alterations in white matter may affect efficiency of electrical signaling and contribute to the neurocognitive processing speed impairments seen in schizophrenia (Roalf et al 2013). We hypothesized that potassium channel genes may impact white matter microstructures that regulate axonal electric signal processing, which in turn influence processing speed deficits seen in schizophrenia.
In a preliminary exploration of this concept, we used a GWAS (genome wide association study) array to genotype and then select the available genetic variants from potassium channel genes in a modest group of patients and controls. Our eventual goal was to detect potassium channel genetic variants that are associated with the portion of processing speed deficit that is contributed by white matter deficits in schizophrenia. One step in this direction is to test whether there was any variant(s) in these genes that 1) showed schizophrenia - control differences 2) was associated with brain white matter integrity as measured by DTI-FA and 3) was associated with processing speed. As the sample size is small, the above exploratory analyses were focused on putatively functional variants, i.e., single nucleotide polymorphisms (SNPs) that are in 3′ or 5′ untranslated regions (UTRs) or exonic SNPs that are nonsynonymous.
Materials and Methods
Sample
A total of 585 Caucasian individuals participated in this study. Patients were recruited from outpatient clinics of the Maryland Psychiatric Research Center and neighboring outpatient clinics. Community controls were recruited through media advertisements and random digit dialing. The Structured Clinical Interview for DSM-IV (SCID) (First et al 1995) was utilized to obtain DSM-IV diagnoses in all participants, which were based on consensus agreement from two or more research psychiatric clinicians. This resulted in 194 schizophrenia spectrum disorder patients (SSD) and 363 community controls (Table 1). An additional 28 individuals with psychosis had a primary mood disorder and were diagnosed with affective disorders with psychotic features, including 20 with bipolar disorder with psychotic features and 8 with major depressive disorder with psychotic features; these individuals were excluded from diagnosis-based analysis but were included in processing speed and DTI based analyses. Of the 194 schizophrenia spectrum disorder patients, 160 had schizophrenia and 34 had schizoaffective disorder. Community controls had no schizophrenia spectrum diagnosis or psychotic disorders. The majority of schizophrenia patients were on antipsychotic medications. Major medical and neurological illnesses, history of head injury with cognitive sequelae, mental retardation, substance dependence within the past 6 months, or current substance abuse (except nicotine) were exclusionary for all participants. A genomic identity by descent procedure was used to identify and correct for any familial relatedness among individuals in the study (see Statistical Analysis section). For the rs8234 diagnosis-based analysis, four individuals were excluded due to missing genotype. Subjects younger than 16 or older than 65 were excluded for both processing speed and white matter phenotype analyses as both have a strong developmental trajectory and age-specific effects on these phenotypes and their heritability maybe more pronounced in older subjects (Glahn et al 2013; Kochunov et al 2013). All participants gave written informed consent as approved by the University of Maryland IRB.
Table 1.
Demographic and clinical data. SSD: Schizophrenia Spectrum Disorder. Data for age, processing speed, and fractional anisotropy (FA) (mean ± standard deviation) are based on subjects age 16 to 65. BPRS: Brief Psychiatric Rating Scale, is based on all SSD subjects and affective disorder subjects that had BPRS data. Processing speed data is given as raw, age scaled, and normalized. Processing speed and fractional anisotropy are normalized for age, age2, sex, sex×age, and sex×age2. N refers to number of individuals with diagnosis phenotype.
| Controls N=363 | SSD N=194 | Affective Disorder N=28 | Statistics (F or χ2) | P value | |
|---|---|---|---|---|---|
| Age | 40.36±14.08 | 39.47±12.92 | 44.79±13.73 | 1.86 | 0.156 |
| Sex (M:F) | 161:202 | 139:55 | 16:12 | 37.02 | <0.001 |
| Raw Processing Speed | 71.17±16.54 | 51.40±18.33 | 49.41±11.73 | 79.58 | <0.001 |
| Age Scaled Processing Speed | 10.06±2.94 | 6.45±2.57 | 6.54±1.82 | 97.13 | <0.001 |
| Normalized Processing Speed | 0.66±0.76 | −0.30±0.92 | −0.28±0.54 | 79.11 | <0.001 |
| Whole Brain Fractional Anisotropy | 0.13±0.87 | −0.16±1.03 | 0.11±1.10 | 3.05 | 0.049 |
| BPRS | n/a | 38.76±11.76 | 34.00±9.10 | 1.6 | 0.211 |
Processing speed task
We used the digit symbol substitution task (DSST) to measure processing speed (Ashe and Georgopoulos 1994; Lancaster et al 2005; Lutz et al 2005). The task is part of the WAIS-3 and is considered a test for speed of information processing and psychomotor response (Wechsler 1997), although some authors have argued that it also has an executive function component (Knowles et al 2015). The processing speed association analysis included 466 subjects (170 SSD, 268 controls and 28 with affective disorders with psychotic features). The test has 133 digits. The number of correct symbols within the allowed 120 seconds was measured. The WAIS-3 norms were used to report age scaled scores. For the rs8234 processing speed association analysis, three subjects were excluded for missing genotype.
Imaging
Imaging was performed on three generations of 3T Siemens scanners: Allegro 3T with a 6-channel headcoil, Trio 3T with a 12 channel headcoil, and Trio 3T with a 32 channel headcoil. The DTI data acquisition and preprocessing are fully described (Kochunov et al 2016b). All data passed QA control of <3 mm accumulated motion during the scan. There was no significant difference in the head motion parameters between patient and controls. The HARDI data was processed using a tract-based spatial statistics (TBSS) method, distributed as a part of FMRIB Software Library (FSL) package (Smith et al 2006). The population-based DTI cerebral WM tract atlas developed at John Hopkins University and distributed with the FSL package (Wakana et al 2004) was used to calculate average FA values along the spatial course of major WM tracts as described elsewhere (Kochunov et al 2011a, Kochunov et al 2011b, Kochunov et al 2012). Whole brain averaged FA was the primary measure. The DTI association analysis included 313 subjects (96 SSD, 199 controls and 18 with affective disorders with psychotic features). For the rs8234 fractional anisotropy association analysis, two subjects were excluded for missing genotype data.
Data homogenization
The ENIGMA-DTI mega-analysis algorithm was used to combine the DTI and processing speed data from three cohorts into a single population following the publically available ENIGMA regression of nuisance covariates and data homogenization (Kochunov et al 2014; Kochunov et al 2016b). The algorithm uses two normalization steps: regression of covariates per cohort, followed by the per-cohort inverse Gaussian normalization of data. This produced the mega-analytic sample for testing the primary hypothesis. The validity of mega-analytical homogenization in these three cohorts was shown by demonstrating that results obtained using mega-analysis were nearly identical (r>0.99) to those produced using a classical meta-analysis approach (Kochunov et al 2016b). Processing speed and fractional anisotropy were normalized for age, age2, sex, sex×age, and sex×age2. There were no age or sex covariates for the diagnosis phenotype.
Genotyping
Whole blood was collected for DNA extraction and genotyping was performed on the Omni2.5-8 Bead Chip Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s standard protocols. PLINK version 1.9 (Chang et al 2015) was used for genetic analysis. Individual genotype call rate and heterozygosity rate were calculated using PLINK, and outlying samples with heterozygosity rates three standard deviations from the mean were identified as outliers in otherwise Gaussian distributions and removed. To minimize computational complexity of sample relatedness, SNPs were pruned so that no pair of SNPs within a window of 50 SNPs has an r2 value >0.2. We also excluded regions of strong linkage disequilibrium, such as the major histocompatibility complex region. Identity by descent was estimated from the pruned data set. Related individuals (identity by descent> 0.1875) were kept in the analysis to maximize power because mixed model association analysis can correctly account for relatedness. Principal components analysis, implemented in SMARTPCA (Eigenstrat), was used to identify samples of non-European ancestry which were excluded from the current study. Principal components were defined using population samples from the 1000 Genomes Project and then projected into this sample. SNPs with minor allele frequency (MAF) of less than 0.1%, Hardy-Weinberg equilibrium P <0.001, call rate lower than 98%, and those that failed the PLINK non-random differential missing data rate test between cases and controls (P < 1 × 10−5) were excluded.
From these, we extracted SNPs that were associated with the 92 potassium channel genes as curated by HGNC (HUGO Gene Nomenclature Committee) (Supplementary Table 1). Human potassium channel genes constitute the largest and most varied family of ion channels (Trimmer 2015). Potassium channels can be grouped into different subfamilies depending on their structure, gating mechanism, and direction of ion flow: for instance constitutively open potassium channels, inwardly rectifying potassium channels, and calcium or sodium activated channels (Trimmer 2015). The largest subfamily is voltage-gated potassium channels with over 40 members, named by letter designation A, B, C, D, F, G, H, Q, S, and V. SNPs were filtered to extract those with MAF ≥ 5% that were in 3′ or 5′ UTRs or nonsynonymous as annotated by Annovar (Wang et al 2010). These filters resulted in 136 3′ UTR, 25 5′ UTR, and 24 nonsynonymous SNPs from potassium channel genes (n=185 genotyped SNPs). Of the nonsynonymous SNPS, six were predicted by PolyPhen-2 (Adzhubei et al, 2010) to be possibly or probably damaging.
Statistical analyses
We performed genome-wide association analysis using solar-eclipse software (www.solar-eclipse-genetics.org) which uses maximum likelihood methods to decompose the variance of a trait into genetic and environmental components by modeling the covariance between individuals as a function of their genetic proximity (Day-Williams et al 2011). To account for any genetic relatedness among the participants (15.9 % of the participants have one or more family members in the study), an empirical kinship matrix was obtained from the genotype data to accommodate for shared common genetic variance among related subjects (Day-Williams et al 2011). Each SNP dosage was coded as 0, 1 or 2 and tested one at a time assuming an additive effect of alleles. The significance of each SNP is adjusted for shared genetic variance among subjects by comparing the null model without the SNP, but with the coefficient of relationship matrix to the model of interest with the SNP (Sprooten et al 2014). To account for population stratification, the first four principal components of the genome-wide SNP data were also added to this model. The fast exact likelihood calculation method was employed for all analyses (Blangero et al 2013).
Our hypothesis is that particular potassium channel SNPs may contribute to decreased FA, decreased PS, and increased risk of schizophrenia spectrum disorder, which would indicate potassium channel variants that may affect processing speed impairments in schizophrenia through reducing white matter FA. To test this hypothesis, we filtered the three association analyses by direction such that one allele of the extracted SNP had the same direction of association with risk of SSD, reduced PS and reduced FA. We used two statistical steps for testing this. First, we identified nominally significant association of a SNP with each of the three phenotypes (each P<0.05). Second, we compared the actual P value from the combined P values to the Bonferroni corrected P value for multiple comparisons of 185 SNPs (<0.05/185=0.00027). To define the actual significant P value, we combined the P values of the association analyses of the three phenotypes by Fisher’s formula (Fisher 1932), which was further divided by 4 for the directions of the associations all aligning under the null hypothesis.
Results
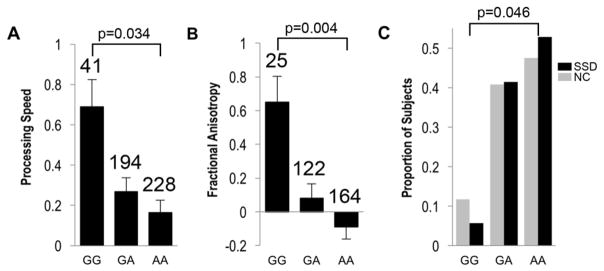
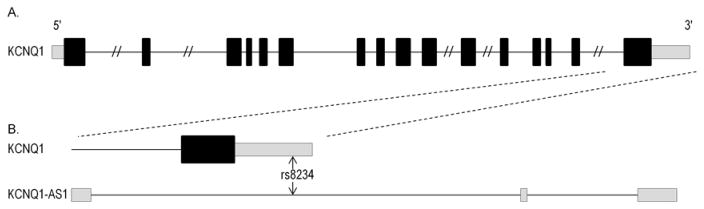
We found one SNP, rs8234, that was nominally significant for schizophrenia spectrum disorder, processing speed, and fractional anisotropy phenotypes in the expected joint directions (P=0.046, β=0.05; P=0.034, β=−0.05; and P=0.004, β=−0.12, respectively) (Figure 1). The combined P value after adjusting for alignment was 0.00013, which was smaller than the Bonferroni corrected threshold of 0.00027. Rs8234 is located in the 3′UTR of voltage-gated potassium channel subfamily Q member 1 (KCNQ1) which also overlaps with the intronic region of the KCNQ1 anti-sense RNA gene (KCNQ1-AS1) (Neyroud 1999) (Figure 2). Rs8234 did not deviate significantly from Harvey-Weinberg equilibrium (P=0.56).
Figure 1.
Rs8234 and joint phenotype analysis. A. Mean normalized processing speed values for rs8234 genotypes. B. Mean normalized whole brain fractional anisotropy values for rs8234. The number above each genotype is the number of individuals with that genotype. Error bars represent standard error. C. Proportion of schizophrenia spectrum disorder patients (N=193) (black color bar) and controls (N=360) (grey color bar) for the three genotypes of rs8234. SSD: Schizophrenia Spectrum Disorder. NC: Community controls.
Figure 2.
Genomic location of rs8234. A. Simplified structure of KCNQ1, which consists of 16 exons spanning over 400 kilobases. B. Magnified view of the 3′UTR of KCNQ1 is shown with the location of rs8234. Below is KCNQ1 anti-sense RNA gene (KCNQ1-AS1) consisting of 3 exons spanning over 20 kilobases. Grey regions indicate untranslated regions of transcripts.
We also used ENIGMA2 imputation protocol (http://enigma.ini.usc.edu/protocols/genetics-protocols/), which was based on the 1000 Genome standard, to impute additional SNPs from the same regions. This resulted in a set of 466 imputed SNPs that had MAF ≥ 5% and were in 3′ or 5′ UTRs or nonsynonymous. There were no imputed SNPs that showed P values <0.05 across all three phenotypes.
Discussion
We hypothesized that potassium channel genes may contribute to the part of the processing speed impairment in schizophrenia that is due to abnormal white matter related signal transmission deficits in schizophrenia. These exploratory analyses identified a 3′ UTR SNP in the voltage-gated KCNQ1 gene that was nominally but simultaneously significant in association with reduced processing speed, reduced whole brain FA, and schizophrenia spectrum disorder.
There are several lines of evidence supporting the potential relevance of the finding. Potassium channels have been linked to psychiatric illnesses such as bipolar disorder, schizophrenia, and autism (Askland et al 2009; Imbrici et al 2013; Gargus 2006; Griswold et al 2015; Strauss et al 2014; Kuo et al 2014; Heide et al 2012; Belengeanu et al 2014). One potassium channel gene, KCNH2, which is well-known as a cause of long QT syndrome (Curran et al 1995) and antipsychotic related prolonged QT interval (Kongsamut et al 2002), was found to be associated with processing speed and schizophrenia (Huffaker et al 2009; Atalar et al 2010). Genome wide association studies in schizophrenia have identified significant loci containing potassium channel genes, for instance KCNB1 (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014) and KCNJ3 (Yamada et al 2012). Expression studies have also identified a role of potassium channels in schizophrenia with KCNS3 (Georgiev et al 2014) and KCNC1 (Yanagi et al 2014) showing abnormal expression in the cortex of patients with schizophrenia. Another two genes KCNF1 and KCNH7 showed increase expression in brains of patients with schizophrenia (Roussos et al 2012).
Our study differs from previous studies by attempting to restrict the search of genetic variants to those potassium channel gene variants that may be related to the processing speed deficit in schizophrenia through their role in myelinated axons, using a combined related-endophenotype and disease-phenotype approach. Our finding of a KCNQ1 association with this exploratory joint phenotype analysis maybe of interest as members of the KCNQ family are renowned for harboring mutations that cause human diseases (Jentsch 2000). KCNQ1 has been linked to long QT syndrome (Wang et al 1996) as well as deafness (Neyroud et al 1997), and epilepsy (Goldman et al 2009, Tiron et al 2015). The KCNQ-mediated potassium current, also called M current (Wang et al 1998), is crucial in regulating central nervous system neuronal excitability such that a 25% reduction can cause epilepsy (Schroeder et al 1998). Converging evidence supports the potential role of the KCNQ gene family in bipolar disorder (Judy and Zandi 2015; Kaminsky et al 2014; Judy et al 2013). Interestingly, KCNQ pharmacologic modulators have shown psychotropic effects in animal models (Grunnet et al 2013), and there is also evidence that a pharmacologic modulator of KCNQ channels improved learning and memory in animal models (Fontana et al 1994, Zaczek et al 1997).
Though earlier reviews of the KCNQ family (Delmas and Brown 2005; Jentsch 2000) describe KCNQ1 channels as non-neuronal channels, modern gene expression studies show clear evidence of KCNQ1 expression in the central nervous system; Luo et al (2008) showed KCNQ1 expression in human brains, and Goldman et al (2009) showed KCNQ1 expression in frontal, temporal, parietal, and occipital lobes as well as hippocampus and spinal cord including in glia cells of white matter tracts.
The rs8234 variant in the 3′UTR of KCNQ1 is of interest also because there is evidence that it is a functional SNP. Rs8234 has been shown experimentally to influence the expression level of KCNQ1 with the A allele being associated with higher expression than the G allele by luciferase reporter assays as well as by using the clinical phenotype of QT length (Amin et al 2012) though the association with QT has not been replicated (Crotti et al 2016). In our sample, the A allele was jointly associated with reduced processing speed, reduced white matter FA, and higher risk for schizophrenia. Furthermore, increased expression of KCNQ1 in neurons causes decreased action potential frequency, and it is partly through this mechanism that Transcription Factor 4 (TCF4), a strong candidate gene for schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014), alters the intrinsic excitability of prefrontal neurons and this excitability deficit is reversed by KCNQ1 antagonists (Rannals et al 2014).
There were several limitations in our approach. A main limitation of our study is the relatively small sample size though it is within the range of other recent genetic studies using DTI data. Our P values did not reach genome wide significance, and without replication our results can only be viewed as exploratory and should be interpreted with caution. A second limitation is that by covarying out age, sex, age2 and their interactive effects as nuisance variables, we limited the ability to study their potential effects on genotypes and groups. However, the sample did not have the power to comprehensively examine all the possible combinations of these effects. A third limitation is that the processing speed measure was based on a single DSST task. Information processing speed is a complex cognitive construct involving also attention, motivation and other cognitive functions, and many cognitive batteries use a combination of DSST and other tasks such as Trail Making Test Part A and Stroop Color-Word Interference Test to represent processing speed which may be a better approach. However DSST does capture a substantial variance in processing speed likely because it represents a very general constraint on cognitive processing (Salthouse,1996), and DSST is the most robust cognitive task for separating healthy controls and schizophrenia patients (Dickinson et al 2007). Another important limitation of using DSST alone without other cognitive tests is that we did not test for specificity of the findings, which limits the interpretation of whether the findings were specific to information processing speed or more general cognitive deficits.
In conclusion, the results of this preliminary analysis of the potassium channel gene group suggest a potential mechanism by which variants in KCNQ1 may alter white matter electrical signal transmission and thus contribute to cognitive deficits such as slower processing speed in schizophrenia.
Supplementary Material
Acknowledgments
Support was received from NIH grants U01MH108148, R01EB015611, R01DA027680, R01MH085646, P50MH103222, T32MH067533 and a Pfizer research grant.
Footnotes
Financial Disclosures
All authors report no potential conflicts of interest.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nature methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel J, Rio M, de Pontual L, Redon R, Malan V, Boddaert N, Plouin P, Carter NP, Lyonnet S, Munnich A, Colleaux L. Mutations in TCF4, encoding a class I basic helix-loop-helix transcription factor, are responsible for Pitt-Hopkins syndrome, a severe epileptic encephalopathy associated with autonomic dysfunction. American Journal of Human Genetics. 2007;80:988–993. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin AS, Giudicessi JR, Tijsen AJ, Spanjaart AM, Reckman YJ, Klemens CA, Tanck MW, Kapplinger JD, Hofman N, Sinner MF, Muller M, Wijnen WJ, Tan HL, Bezzina CR, Creemers EE, Wilde AA, Ackerman MJ, Pinto YM. Variants in the 3′ untranslated region of the KCNQ1-encoded Kv7.1 potassium channel modify disease severity in patients with type 1 long QT syndrome in an allele-specific manner. European heart journal. 2012;33:714–723. doi: 10.1093/eurheartj/ehr473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe J, Georgopoulos AP. Movement parameters and neural activity in motor cortex and area 5. Cerebral cortex (New York, NY: 1991) 1994;4:590–600. doi: 10.1093/cercor/4.6.590. [DOI] [PubMed] [Google Scholar]
- Askland K, Read C, Moore J. Pathways-based analyses of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Human genetics. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- Atalar F, Acuner TT, Cine N, Oncu F, Yesilbursa D, Ozbek U, Turkcan S. Two four-marker haplotypes on 7q36.1 region indicate that the potassium channel gene HERG1 (KCNH2, Kv11.1) is related to schizophrenia: a case control study. Behavioral and brain functions: BBF. 2010;6 doi: 10.1186/1744-9081-6-27. 27-9081-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battefeld A, Tran BT, Gavrilis J, Cooper EC, Kole MH. Heteromeric Kv7.2/7.3 channels differentially regulate action potential initiation and conduction in neocortical myelinated axons. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2014;34:3719–3732. doi: 10.1523/JNEUROSCI.4206-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean BP. The action potential in mammalian central neurons. Nature reviews. Neuroscience. 2007;8:451–465. doi: 10.1038/nrn2148. [DOI] [PubMed] [Google Scholar]
- Belengeanu V, Gamage TH, Farcas S, Stoian M, Andreescu N, Belengeanu A, Frengen E, Misceo D. A de novo 2.3 Mb deletion in 2q24.2q24.3 in a 20-month-old developmentally delayed girl. Gene. 2014;539:168–172. doi: 10.1016/j.gene.2014.01.060. [DOI] [PubMed] [Google Scholar]
- Blangero J, Diego VP, Dyer TD, Almeida M, Peralta J, Kent JW, Jr, Williams JT, Almasy L, Goring HH. A kernel of truth: statistical advances in polygenic variance component models for complex human pedigrees. Advances in Genetics. 2013;81:1–31. doi: 10.1016/B978-0-12-407677-8.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasko C, Hawkins V, De La Rocha IC, Butt AM. Expression of Kir4.1 and Kir5.1 inwardly rectifying potassium channels in oligodendrocytes, the myelinating cells of the CNS. Brain structure & function. 2016 doi: 10.1007/s00429-016-1199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttermore ED, Thaxton CL, Bhat MA. Organization and maintenance of molecular domains in myelinated axons. Journal of neuroscience research. 2013;91:603–622. doi: 10.1002/jnr.23197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4 doi: 10.1186/s13742-015-0047-8. 7-015-0047-8 eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KJ, Rasband MN. Excitable domains of myelinated nerves: axon initial segments and nodes of Ranvier. Current topics in membranes. 2013;72:159–192. doi: 10.1016/B978-0-12-417027-8.00005-2. [DOI] [PubMed] [Google Scholar]
- Choe S. Potassium channel structures. Nature reviews. Neuroscience. 2002;3:115–121. doi: 10.1038/nrn727. [DOI] [PubMed] [Google Scholar]
- Cirulli ET, Kasperaviciute D, Attix DK, Need AC, Ge D, Gibson G, Goldstein DB. Common genetic variation and performance on standardized cognitive tests. European journal of human genetics: EJHG. 2010;18:815–820. doi: 10.1038/ejhg.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti L, Lahtinen AM, Spazzolini C, Mastantuono E, Monti MC, Morassutto C, Parati G, Heradien M, Goosen A, Lichtner P, Meitinger T, Brink PA, Kontula K, Swan H, Schwartz PJ. Genetic Modifiers for the Long-QT Syndrome: How Important Is the Role of Variants in the 3′ Untranslated Region of KCNQ1? Circulation. Cardiovascular genetics. 2016;9:330–339. doi: 10.1161/CIRCGENETICS.116.001419. [DOI] [PubMed] [Google Scholar]
- Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Archives of General Psychiatry. 2003;60:443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- Day-Williams AG, Blangero J, Dyer TD, Lange K, Sobel EM. Linkage analysis without defined pedigrees. Genetic epidemiology. 2011;35:360–370. doi: 10.1002/gepi.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P, Brown DA. Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nature reviews. Neuroscience. 2005;6:850–862. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- Dickinson D, Ramsey ME, Gold JM. Overlooking the obvious: a meta-analytic comparison of digit symbol coding tasks and other cognitive measures in schizophrenia. Archives of General Psychiatry. 2007;64:532–542. doi: 10.1001/archpsyc.64.5.532. [DOI] [PubMed] [Google Scholar]
- First M, Spitzer R, Williams J, Gibbon M. Structured clinical interview for DSM-IV-patient edition (SCID-P) Biometric Research; New York: 1995. [Google Scholar]
- Fisher R. Statistical Methods for Research Workers. Oliver and Boyd; Edinburgh: 1932. [Google Scholar]
- Fontana DJ, Inouye GT, Johnson RM. Linopirdine (DuP 996) improves performance in several tests of learning and memory by modulation of cholinergic neurotransmission. Pharmacology, biochemistry, and behavior. 1994;49:1075–1082. doi: 10.1016/0091-3057(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Friedman JI, Tang C, Carpenter D, Buchsbaum M, Schmeidler J, Flanagan L, Golembo S, Kanellopoulou I, Ng J, Hof PR, Harvey PD, Tsopelas ND, Stewart D, Davis KL. Diffusion tensor imaging findings in first-episode and chronic schizophrenia patients. The American Journal of Psychiatry. 2008;165:1024–1032. doi: 10.1176/appi.ajp.2008.07101640. [DOI] [PubMed] [Google Scholar]
- Gargus JJ. Ion channel functional candidate genes in multigenic neuropsychiatric disease. Biological psychiatry. 2006;60:177–185. doi: 10.1016/j.biopsych.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Georgiev D, Arion D, Enwright JF, Kikuchi M, Minabe Y, Corradi JP, Lewis DA, Hashimoto T. Lower gene expression for KCNS3 potassium channel subunit in parvalbumin-containing neurons in the prefrontal cortex in schizophrenia. The American Journal of Psychiatry. 2014;171:62–71. doi: 10.1176/appi.ajp.2013.13040468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddaluru S, Espeseth T, Salami A, Westlye LT, Lundquist A, Christoforou A, Cichon S, Adolfsson R, Steen VM, Reinvang I, Nilsson LG, Le Hellard S, Nyberg L. Genetics of structural connectivity and information processing in the brain. Brain structure & function. 2016 doi: 10.1007/s00429-016-1194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Kent JW, Jr, Sprooten E, Diego VP, Winkler AM, Curran JE, McKay DR, Knowles EE, Carless MA, Goring HH, Dyer TD, Olvera RL, Fox PT, Almasy L, Charlesworth J, Kochunov P, Duggirala R, Blangero J. Genetic basis of neurocognitive decline and reduced white-matter integrity in normal human brain aging. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19006–19011. doi: 10.1073/pnas.1313735110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman AM, Glasscock E, Yoo J, Chen TT, Klassen TL, Noebels JL. Arrhythmia in heart and brain: KCNQ1 mutations link epilepsy and sudden unexplained death. Science translational medicine. 2009;1:2ra6. doi: 10.1126/scitranslmed.3000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold AJ, Dueker ND, Van Booven D, Rantus JA, Jaworski JM, Slifer SH, Schmidt MA, Hulme W, Konidari I, Whitehead PL, Cuccaro ML, Martin ER, Haines JL, Gilbert JR, Hussman JP, Pericak-Vance MA. Targeted massively parallel sequencing of autism spectrum disorder-associated genes in a case control cohort reveals rare loss-of-function risk variants. Molecular autism. 2015;6 doi: 10.1186/s13229-015-0034-z. 43-015–0034-z eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunnet M, Strobaek D, Hougaard C, Christophersen P. Kv7 channels as targets for anti-epileptic and psychiatric drug-development. European journal of pharmacology. 2014;726:133–137. doi: 10.1016/j.ejphar.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Heide J, Mann SA, Vandenberg JI. The schizophrenia-associated Kv11.1-3.1 isoform results in reduced current accumulation during repetitive brief depolarizations. PloS one. 2012;7:e45624. doi: 10.1371/journal.pone.0045624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry JD, Crawford JR. A meta-analytic review of verbal fluency deficits in schizophrenia relative to other neurocognitive deficits. Cognitive neuropsychiatry. 2005;10:1–33. doi: 10.1080/13546800344000309. [DOI] [PubMed] [Google Scholar]
- Huffaker SJ, Chen J, Nicodemus KK, Sambataro F, Yang F, Mattay V, Lipska BK, Hyde TM, Song J, Rujescu D, Giegling I, Mayilyan K, Proust MJ, Soghoyan A, Caforio G, Callicott JH, Bertolino A, Meyer-Lindenberg A, Chang J, Ji Y, Egan MF, Goldberg TE, Kleinman JE, Lu B, Weinberger DR. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nature medicine. 2009;15:509–518. doi: 10.1038/nm.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbrici P, Camerino DC, Tricarico D. Major channels involved in neuropsychiatric disorders and therapeutic perspectives. Frontiers in genetics. 2013;4:76. doi: 10.3389/fgene.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch TJ. Neuronal KCNQ potassium channels: physiology and role in disease. Nature reviews. Neuroscience. 2000;1:21–30. doi: 10.1038/35036198. [DOI] [PubMed] [Google Scholar]
- Judy JT, Seifuddin F, Pirooznia M, Mahon PB, Jancic D, Goes FS, Schulze T, Cichon S, Noethen M, Rietschel M, Depaulo JR, Jr, Potash JB, Zandi PP Bipolar Genome Study Consortium. Converging Evidence for Epistasis between ANK3 and Potassium Channel Gene KCNQ2 in Bipolar Disorder. Frontiers in genetics. 2013;4:87. doi: 10.3389/fgene.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judy JT, Zandi PP. A review of potassium channels in bipolar disorder. Frontiers in genetics. 2013;4:105. doi: 10.3389/fgene.2013.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky Z, Jones I, Verma R, Saleh L, Trivedi H, Guintivano J, Akman R, Zandi P, Lee RS, Potash JB. DNA methylation and expression of KCNQ3 in bipolar disorder. Bipolar disorders. 2015;17:150–159. doi: 10.1111/bdi.12230. [DOI] [PubMed] [Google Scholar]
- Karbasforoushan H, Duffy B, Blackford JU, Woodward ND. Processing speed impairment in schizophrenia is mediated by white matter integrity. Psychological medicine. 2015;45:109–120. doi: 10.1017/S0033291714001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles EE, David AS, Reichenberg A. Processing speed deficits in schizophrenia: reexamining the evidence. The American Journal of Psychiatry. 2010;167:828–835. doi: 10.1176/appi.ajp.2010.09070937. [DOI] [PubMed] [Google Scholar]
- Knowles EE, Weiser M, David AS, Glahn DC, Davidson M, Reichenberg A. The puzzle of processing speed, memory, and executive function impairments in schizophrenia: fitting the pieces together. Biological psychiatry. 2015;78:786–793. doi: 10.1016/j.biopsych.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Ganjgahi H, Winkler A, Kelly S, Shukla DK, Du X, Jahanshad N, Rowland L, Sampath H, Patel B, O’Donnell P, Xie Z, Paciga SA, Schubert CR, Chen J, Zhang G, Thompson PM, Nichols TE, Hong LE. Heterochronicity of white matter development and aging explains regional patient control differences in schizophrenia. Human brain mapping. 2016 doi: 10.1002/hbm.23336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Lancaster J, Thompson PM, Kochunov V, Rogers B, Fox P, Blangero J, Williamson DE. Fractional anisotropy of cerebral white matter and thickness of cortical gray matter across the lifespan. NeuroImage. 2011;58:41–49. doi: 10.1016/j.neuroimage.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Nichols TE, Winkler AM, Hong EL, Holcomb HH, Stein JL, Thompson PM, Curran JE, Carless MA, Olvera RL, Johnson MP, Cole SA, Kochunov V, Kent J, Blangero J. Genetic analysis of cortical thickness and fractional anisotropy of water diffusion in the brain. Frontiers in neuroscience. 2011;5:120. doi: 10.3389/fnins.2011.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Rowland LM, Olvera RL, Winkler A, Yang YH, Sampath H, Carpenter WT, Duggirala R, Curran J, Blangero J, Hong LE. Testing the hypothesis of accelerated cerebral white matter aging in schizophrenia and major depression. Biological psychiatry. 2013;73:482–491. doi: 10.1016/j.biopsych.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Jahanshad N, Sprooten E, Nichols TE, Mandl RC, Almasy L, Booth T, Brouwer RM, Curran JE, de Zubicaray GI, Dimitrova R, Duggirala R, Fox PT, Hong LE, Landman BA, Lemaitre H, Lopez LM, Martin NG, McMahon KL, Mitchell BD, Olvera RL, Peterson CP, Starr JM, Sussmann JE, Toga AW, Wardlaw JM, Wright MJ, Wright SN, Bastin ME, McIntosh AM, Boomsma DI, Kahn RS, den Braber A, de Geus EJ, Deary IJ, Hulshoff Pol HE, Williamson DE, Blangero J, van ‘t Ent D, Thompson PM, Glahn DC. Multi-site study of additive genetic effects on fractional anisotropy of cerebral white matter: Comparing meta and megaanalytical approaches for data pooling. NeuroImage. 2014;95:136–150. doi: 10.1016/j.neuroimage.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Thompson PM, Winkler A, Morrissey M, Fu M, Coyle TR, Du X, Muellerklein F, Savransky A, Gaudiot C, Sampath H, Eskandar G, Jahanshad N, Patel B, Rowland L, Nichols TE, O’Connell JR, Shuldiner AR, Mitchell BD, Hong LE. The common genetic influence over processing speed and white matter microstructure: Evidence from the Old Order Amish and Human Connectome Projects. NeuroImage. 2016;125:189–197. doi: 10.1016/j.neuroimage.2015.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Williamson DE, Lancaster J, Fox P, Cornell J, Blangero J, Glahn DC. Fractional anisotropy of water diffusion in cerebral white matter across the lifespan. Neurobiology of aging. 2012;33:9–20. doi: 10.1016/j.neurobiolaging.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsamut S, Kang J, Chen XL, Roehr J, Rampe D. A comparison of the receptor binding and HERG channel affinities for a series of antipsychotic drugs. European journal of pharmacology. 2002;450:37–41. doi: 10.1016/s0014-2999(02)02074-5. [DOI] [PubMed] [Google Scholar]
- Kuo PH, Chuang LC, Liu JR, Liu CM, Huang MC, Lin SK, Sunny Sun H, Hsieh MH, Hung H, Lu RB. Identification of novel loci for bipolar I disorder in a multi-stage genome-wide association study. Progress in neuro-psychopharmacology & biological psychiatry. 2014;51:58–64. doi: 10.1016/j.pnpbp.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Lancaster JL, Cody JD, Andrews T, Hardies LJ, Hale DE, Fox PT. Myelination in children with partial deletions of chromosome 18q. AJNR. American journal of neuroradiology. 2005;26:447–454. [PMC free article] [PubMed] [Google Scholar]
- Lee T, Mosing MA, Henry JD, Trollor JN, Lammel A, Ames D, Martin NG, Wright MJ, Sachdev PS. Genetic influences on five measures of processing speed and their covariation with general cognitive ability in the elderly: the older Australian twins study. Behavior genetics. 2012;42:96–106. doi: 10.1007/s10519-011-9474-1. [DOI] [PubMed] [Google Scholar]
- Luo X, Xiao J, Lin H, Lu Y, Yang B, Wang Z. Genomic structure, transcriptional control, and tissue distribution of HERG1 and KCNQ1 genes. American journal of physiology. Heart and circulatory physiology. 2008;294:H1371–80. doi: 10.1152/ajpheart.01026.2007. [DOI] [PubMed] [Google Scholar]
- Lutz K, Koeneke S, Wustenberg T, Jancke L. Asymmetry of cortical activation during maximum and convenient tapping speed. Neuroscience letters. 2005;373:61–66. doi: 10.1016/j.neulet.2004.09.058. [DOI] [PubMed] [Google Scholar]
- Mori T, Ohnishi T, Hashimoto R, Nemoto K, Moriguchi Y, Noguchi H, Nakabayashi T, Hori H, Harada S, Saitoh O, Matsuda H, Kunugi H. Progressive changes of white matter integrity in schizophrenia revealed by diffusion tensor imaging. Psychiatry research. 2007;154:133–145. doi: 10.1016/j.pscychresns.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Neyroud N, Richard P, Vignier N, Donger C, Denjoy I, Demay L, Shkolnikova M, Pesce R, Chevalier P, Hainque B, Coumel P, Schwartz K, Guicheney P. Genomic organization of the KCNQ1 K+ channel gene and identification of C-terminal mutations in the long-QT syndrome. Circulation research. 1999;84:290–297. doi: 10.1161/01.res.84.3.290. [DOI] [PubMed] [Google Scholar]
- Neyroud N, Tesson F, Denjoy I, Leibovici M, Donger C, Barhanin J, Faure S, Gary F, Coumel P, Petit C, Schwartz K, Guicheney P. A novel mutation in the potassium channel gene KVLQT1 causes the Jervell and Lange-Nielsen cardioauditory syndrome. Nature genetics. 1997;15:186–189. doi: 10.1038/ng0297-186. [DOI] [PubMed] [Google Scholar]
- Perez-Iglesias R, Tordesillas-Gutierrez D, McGuire PK, Barker GJ, Roiz-Santianez R, Mata I, de Lucas EM, Rodriguez-Sanchez JM, Ayesa-Arriola R, Vazquez-Barquero JL, Crespo-Facorro B. White matter integrity and cognitive impairment in first-episode psychosis. The American Journal of Psychiatry. 2010;167:451–458. doi: 10.1176/appi.ajp.2009.09050716. [DOI] [PubMed] [Google Scholar]
- Rannals MD, Hamersky GR, Page SC, Campbell MN, Briley A, Gallo RA, Phan BN, Hyde TM, Kleinman JE, Shin JH, Jaffe AE, Weinberger DR, Maher BJ. Psychiatric Risk Gene Transcription Factor 4 Regulates Intrinsic Excitability of Prefrontal Neurons via Repression of SCN10a and KCNQ1. Neuron. 2016;90:43–55. doi: 10.1016/j.neuron.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roalf DR, Ruparel K, Verma R, Elliott MA, Gur RE, Gur RC. White matter organization and neurocognitive performance variability in schizophrenia. Schizophrenia research. 2013;143:172–178. doi: 10.1016/j.schres.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Katsel P, Davis KL, Bitsios P, Giakoumaki SG, Jogia J, Rozsnyai K, Collier D, Frangou S, Siever LJ, Haroutunian V. Molecular and genetic evidence for abnormalities in the nodes of Ranvier in schizophrenia. Archives of General Psychiatry. 2012;69:7–15. doi: 10.1001/archgenpsychiatry.2011.110. [DOI] [PubMed] [Google Scholar]
- Salthouse TA. The processing-speed theory of adult age differences in cognition. Psychological review. 1996;103:403–428. doi: 10.1037/0033-295x.103.3.403. [DOI] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BC, Kubisch C, Stein V, Jentsch TJ. Moderate loss of function of cyclic-AMP-modulated KCNQ2/KCNQ3 K+ channels causes epilepsy. Nature. 1998;396:687–690. doi: 10.1038/25367. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Sprooten E, Knowles EE, McKay DR, Goring HH, Curran JE, Kent JW, Jr, Carless MA, Dyer TD, Drigalenko EI, Olvera RL, Fox PT, Almasy L, Duggirala R, Kochunov P, Blangero J, Glahn DC. Common genetic variants and gene expression associated with white matter microstructure in the human brain. NeuroImage. 2014;97:252–261. doi: 10.1016/j.neuroimage.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KA, Markx S, Georgi B, Paul SM, Jinks RN, Hoshi T, McDonald A, First MB, Liu W, Benkert AR, Heaps AD, Tian Y, Chakravarti A, Bucan M, Puffenberger EG. A population-based study of KCNH7 p.Arg394His and bipolar spectrum disorder. Human molecular genetics. 2014;23:6395–6406. doi: 10.1093/hmg/ddu335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiron C, Campuzano O, Perez-Serra A, Mademont I, Coll M, Allegue C, Iglesias A, Partemi S, Striano P, Oliva A, Brugada R. Further evidence of the association between LQT syndrome and epilepsy in a family with KCNQ1 pathogenic variant. Seizure. 2015;25:65–67. doi: 10.1016/j.seizure.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Trimmer JS. Subcellular localization of K+ channels in mammalian brain neurons: remarkable precision in the midst of extraordinary complexity. Neuron. 2015;85:238–256. doi: 10.1016/j.neuron.2014.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana S, Jiang H, Nagae-Poetscher LM, van Zijl PC, Mori S. Fiber tract-based atlas of human white matter anatomy. Radiology. 2004;230:77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- Wang HS, Pan Z, Shi W, Brown BS, Wymore RS, Cohen IS, Dixon JE, McKinnon D. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science (New York, NY) 1998;282:1890–1893. doi: 10.1126/science.282.5395.1890. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nature genetics. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- Weschler D. Wechsler Adult Intelligence Scale. Psychological Corporation; San Antonio, TX: 1997. [Google Scholar]
- Wright M, De Geus E, Ando J, Luciano M, Posthuma D, Ono Y, Hansell N, Van Baal C, Hiraishi K, Hasegawa T, Smith G, Geffen G, Geffen L, Kanba S, Miyake A, Martin N, Boomsma D. Genetics of cognition: outline of a collaborative twin study. Twin research: the official journal of the International Society for Twin Studies. 2001;4:48–56. doi: 10.1375/1369052012146. [DOI] [PubMed] [Google Scholar]
- Wright SN, Hong LE, Winkler AM, Chiappelli J, Nugent K, Muellerklein F, Du X, Rowland LM, Wang DJ, Kochunov P. Perfusion shift from white to gray matter may account for processing speed deficits in schizophrenia. Human brain mapping. 2015;36:3793–3804. doi: 10.1002/hbm.22878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Iwayama Y, Toyota T, Ohnishi T, Ohba H, Maekawa M, Yoshikawa T. Association study of the KCNJ3 gene as a susceptibility candidate for schizophrenia in the Chinese population. Human genetics. 2012;131:443–451. doi: 10.1007/s00439-011-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi M, Joho RH, Southcott SA, Shukla AA, Ghose S, Tamminga CA. Kv3.1-containing K(+) channels are reduced in untreated schizophrenia and normalized with antipsychotic drugs. Molecular psychiatry. 2014;19:573–579. doi: 10.1038/mp.2013.49. [DOI] [PubMed] [Google Scholar]
- Yellen G. The voltage-gated potassium channels and their relatives. Nature. 2002;419:35–42. doi: 10.1038/nature00978. [DOI] [PubMed] [Google Scholar]
- Zaczek Robert, Chorvat RJ, Brown BS. Linopirdine: Pharmacology of a Neurotransmitter Release Enhancer. CNS Drug Reviews. 1997;3:103–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.