Abstract
An increasing number of cases of herb-induced liver injury (HILI) have been reported, presenting new clinical challenges. In this study, taking Polygonum multiflorum Thunb (PmT) as an example, we proposed a computational systems toxicology approach to explore the molecular mechanisms of HILI. First, the chemical components of PmT were extracted from 3 main TCM databases as well as the literature related to natural products. Then, the known targets were collected through data integration, and the potential compound-target interactions (CTIs) were predicted using our substructure-drug-target network-based inference (SDTNBI) method. After screening for hepatotoxicity-related genes by assessing the symptoms of HILI, a compound-target interaction network was constructed. A scoring function, namely, Ascore, was developed to estimate the toxicity of chemicals in the liver. We conducted network analysis to determine the possible mechanisms of the biphasic effects using the analysis tools, including BiNGO, pathway enrichment, organ distribution analysis and predictions of interactions with CYP450 enzymes. Among the chemical components of PmT, 54 components with good intestinal absorption were used for analysis, and 2939 CTIs were obtained. After analyzing the mRNA expression data in the BioGPS database, 1599 CTIs and 125 targets related to liver diseases were identified. In the top 15 compounds, seven with Ascore values >3000 (emodin, quercetin, apigenin, resveratrol, gallic acid, kaempferol and luteolin) were obviously associated with hepatotoxicity. The results from the pathway enrichment analysis suggest that multiple interactions between apoptosis and metabolism may underlie PmT-induced liver injury. Many of the pathways have been verified in specific compounds, such as glutathione metabolism, cytochrome P450 metabolism, and the p53 pathway, among others. Hepatitis symptoms, the perturbation of nine bile acids and yellow or tawny urine also had corresponding pathways, justifying our method. In conclusion, this computational systems toxicology method reveals possible toxic components and could be very helpful for understanding the mechanisms of HILI. In this way, the method might also facilitate the identification of novel hepatotoxic herbs.
Keywords: computational systems toxicology, TCM, Polygonum multiflorum Thunb, herb-induced liver injury, hepatotoxicity, pathway enrichment
Introduction
Traditional Chinese medicine (TCM) has played an important role in human health for thousands of years1. However, prolonged use (a few weeks or months) of some TCM herbs may adversely affect organs, such as the liver2. In 2011, a detailed report estimated the risk of hospitalization for liver injury associated with the use of TCM products containing Chaihu (Radix bupleuri). This report identified 61 liver injury cases in 639 779 patients with chronic hepatitis B virus infection3. In 2012, Teschke et al summarized cases of herbal hepatotoxicity taken from 185 publications4 in which the administration of 60 different herbs resulted in hepatotoxicity, attracting concern worldwide. The risk of herb-induced liver injury (HILI) associated with several TCM herbs has been reported on the websites of the National Institutes of Health (NIH) and LiverTox. Thus far, a total of 18 classifiable TCM herbal mixtures, a group of unclassifiable TCM herbal mixtures and 39 TCM herbs have been reported to have potential hepatotoxic effects3. TCM often induces liver injury via its chemical components or metabolites. Because the liver is the most important organ for chemical metabolism, it can easily accumulate compounds and toxic metabolites. Therefore, it is imperative to identify hepatotoxicants and the mechanisms of hepatotoxicity early and efficiently for the safe use of TCM.
Polygonum multiflorumThunb (PmT), known as He Shou Wu ( in Chinese) in China, is traditionally used to blacken hair, tonify the liver and kidneys and to slow the effects of aging with low toxicity5. Although PmT is usually used to treat various liver diseases, such as nonalcoholic fatty liver disease, an increasing number of cases of potential PmT-induced hepatotoxicity resulting in severe drug-induced liver injury and even death have been reported6. In 2011, 25 new hepatotoxicity cases were reported. In these cases, hepatotoxicity occurred after one year of consuming the TCM mixture of Shouwu Pian in which PmT is the main ingredient3. Case reports and case series on liver damage associated with PmT include a total of 450 cases from 76 articles. Symptoms of liver damage emerge approximately one month after taking PmT and mainly include jaundice, fatigue, anorexia, and yellow or tawny urine7. High doses of the herb had either an injurious effect on normal rats or a therapeutic effect on rats with chronic liver injury, indicating both harmful and protective effects in the liver8. More than 100 chemical components have been isolated from this herb. The majority of the components are stilbenes, quinones, and flavonoids. Stilbenes have anti-aging effects and hepatoprotective activities9. However, many studies have also demonstrated that some quinones and stilbenes of PmT can lead to hepatotoxicity, especially emodin and rhein10,11. How these compounds produce these biphasic effects (liver injury and liver protection) remains to be determined.
in Chinese) in China, is traditionally used to blacken hair, tonify the liver and kidneys and to slow the effects of aging with low toxicity5. Although PmT is usually used to treat various liver diseases, such as nonalcoholic fatty liver disease, an increasing number of cases of potential PmT-induced hepatotoxicity resulting in severe drug-induced liver injury and even death have been reported6. In 2011, 25 new hepatotoxicity cases were reported. In these cases, hepatotoxicity occurred after one year of consuming the TCM mixture of Shouwu Pian in which PmT is the main ingredient3. Case reports and case series on liver damage associated with PmT include a total of 450 cases from 76 articles. Symptoms of liver damage emerge approximately one month after taking PmT and mainly include jaundice, fatigue, anorexia, and yellow or tawny urine7. High doses of the herb had either an injurious effect on normal rats or a therapeutic effect on rats with chronic liver injury, indicating both harmful and protective effects in the liver8. More than 100 chemical components have been isolated from this herb. The majority of the components are stilbenes, quinones, and flavonoids. Stilbenes have anti-aging effects and hepatoprotective activities9. However, many studies have also demonstrated that some quinones and stilbenes of PmT can lead to hepatotoxicity, especially emodin and rhein10,11. How these compounds produce these biphasic effects (liver injury and liver protection) remains to be determined.
Systems toxicology incorporates experimental and computational approaches to explore the complex mechanisms of the side effects (SEs) of drugs12,13. In systems toxicology, the SEs are often viewed as perturbations of biological pathways, meaning that we must overcome the 'one drug-one target' paradigm used in traditional drug design. Systems toxicology emphasizes system-wide effects that cause a drug to have unforeseen pharmacological effects14,15. Pathway and network analyses can explain the causal nature of correlations, for example, the roles of certain proteins in SE etiology and pathogenesis16. Many studies have successfully demonstrated that systems toxicology (particularly pathway enrichment) is a promising method for determining the mechanisms of toxicity17,18. In our previous study, using a systems pharmacology approach, we demonstrated that FXR antagonism contributes to liver injury induced by non-steroidal anti-inflammatory drugs19. Because TCM herbs are considered as multi-component and multi-target therapeutics, the methodologies of network pharmacology are suitable for understanding the relationship between active interactions and relevant targets, which in turn highlight the mechanisms of action20,21. As with efficacy, toxicity may result from a combination of active compounds rather than a single chemical entity. However, to the best of our knowledge, most of the representative studies have focused on the pharmaceutical action of molecules to understand their mechanisms of action and to evaluate their efficacy but have rarely focused on the adverse effects of these compounds22. In this study, we attempted to understand the mechanisms of HILI by applying systems toxicology.
In this study, we used PmT as an example to propose a pathway-based systems toxicology approach to explore the mechanism of action (MOA) of HILI. Pathway enrichment and network analyses were used to explain the causal nature of correlations, including the roles of proteins in SE etiology and pathogenesis, which may increase our understanding of the biphasic mechanisms and guide the safe use of TCM by implementing herb combinations that reduce toxicity.
Materials and methods
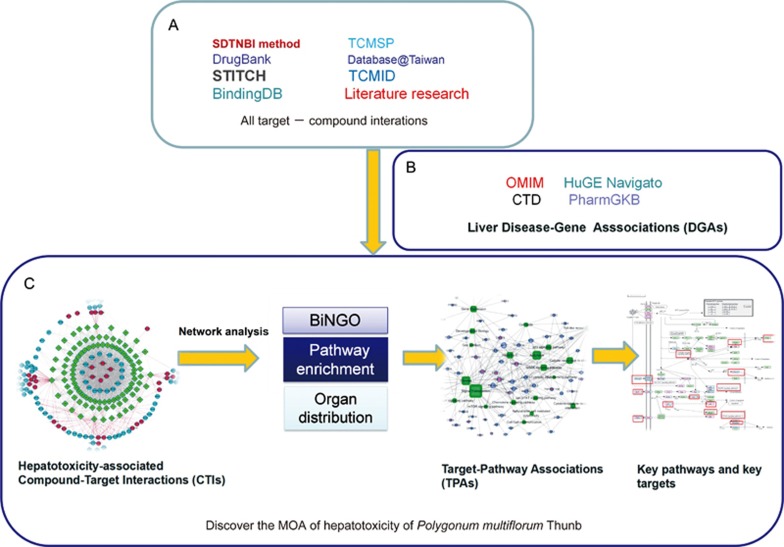
The entire pathway-based systems toxicology approach is illustrated in Figure 1.
Figure 1.
Diagram of the pathway-based systems toxicology approach developed to identify the potential molecular mechanisms of Polygonum multiflorum Thunb (PmT)-induced liver injury. (A) Collections of compound-target interactions: validated targets were from four public databases, putative targets were predicted by the SDTNBI method. The components were obtained from four public databases and the literature. (B) Collection of liver disease-target associations (TDAs). (C) Filtering out the potential proteins involved in PmT-induced liver injury to construct compound-target interactions. To determine its molecular mechanism, many network analysis tools were used, including BiNGO, pathway enrichment and organ distribution analysis. Finally, the hepatotoxicity-associated target-pathway association network was constructed to determine the key pathways and their corresponding targets.
Data collection and preparation
Based on a review of PmT5, we extracted the chemical components of PmT from three typical TCM databases, namely TCMSP23, TCMID24 and TCM Database@Taiwan25. All of the compounds collected were normalized to the canonical SMILES format. Duplicates and compounds without structures were removed. We then calculated the intestinal absorption properties of the compounds using a model integrated within Pipeline Pilot (version 7.5.2, BIOVIA, San Diego, CA, USA)26 to rank the molecules into the following four classes: good (0), moderate (1), poor (2) and very poor (3). The very poor class of compounds was discarded. The remaining compounds were further analyzed.
Subsequently, we collected experimentally determined compound-target pairs from four databases, namely STITCH (version 5.0)27, BindingDB28, PubChem29 and DrugBank30. All of the related targets were normalized to the official gene symbols using the UniProt database (http://www.uniprot.org/uploadlists/). Duplicates from different sources were excluded.
Prediction of putative targets
Putative targets of the components were predicted using our substructure-drug-target network-based inference (SDTNBI) method31, an integrated network and chemoinformatics tool for the systematic prediction of compound-target interactions, particularly new chemical entities. For this purpose, the canonical SMILES format was converted into the substructure fingerprint (FP4) format using PaDEL-Descriptor software32. For each compound, the top 50 predicted targets were stored as putative targets. Next, the targets were normalized to the official gene name using the UniProt database as described above.
Identification of tissue-specific and liver disease-associated targets
To analyze the target distribution within the liver, we examined the mRNA expression data from the BioGPS database33, a centralized gene annotation portal that enables researchers to access distributed gene annotation resources. In 2015, BioGPS34 was expanded by pre-loading approximately 2000 datasets from the NCBI GEO repository and approximately 6000 datasets from the EBI ArrayExpress repository. These datasets were obtained from nine common microarray platforms from humans, mice and rats. We chose the human species and ranked the target expression patterns based on their expression levels in the liver. For example, if the target expression was highest in the liver among the 84 organ patterns, its rank was 84. If the target expression was lowest in the liver, its rank was 1.
Next, we identified the targets related to the PmT liver injury symptoms. The symptoms could be classified into nine types of liver disease. The disease-target pairs of the nine diseases were collected from four public databases, including the Online Mendelian Inheritance in Man (OMIM) database35, HuGE Navigator36, PharmGKB37 and the Comparative Toxicogenomics Database (CTD)38. All liver diseases were annotated using either MeSH or UMLS vocabulary39. The targets were further annotated using the official gene symbols as described above. To ensure the accuracy of our results, we excluded computationally predicted and duplicated target-disease pairs from the different data sources.
Construction of the compound-target-pathway-disease networks
To obtain a global perspective of the interactions between the hepatotoxicity-associated targets and compounds, the targets associated with liver diseases obtained above were selected for further analysis. We constructed the compound-target interaction (CTI) network using Cytoscape (version 3.2.1)40. The BiNGO41 plugin in Cytoscape (version 3.2.1) was used to determine significantly enriched functional annotation gene terms that were able to further support module or complex predictions.
To evaluate the potency of the associations between compounds and hepatotoxic targets, we developed the 'Ascore' scoring function to semi-quantify the action of a compound on liver targets using the following equation:
where Ri represents the rank of the predicted targets of a specific compound in the liver as mentioned above, Re indicates the rank of the experimentally determined targets of the specific compound in the liver and the coefficient k weights the experimentally verified target.
Three factors can influence the Ascore value, i.e., the number of hepatotoxic targets of a compound, the expression rank of all hepatotoxicity-related targets of a compound in the liver, and the source of the compound-target pairs. (i) Each compound has a certain number of hepatotoxicity-related targets. The more hepatotoxicity-related targets a compound has, the higher its Ascore value. (ii) In the same way, each target has a corresponding rank for its distribution in the liver relative to all of the organ patterns in the BioGPS database. Higher target ranks are associated with higher Ascore values. (iii) A weight factor was applied to the experimentally verified targets. The experimentally determined targets of a compound are more reliable than the predicted targets. The equation considers all three factors. Higher Ascores indicate a stronger association between the compound and liver disease.
The experimentally verified targets should be weighted. The weight factor k should be greater than 1. To determine the optimal k value, different weight factors were tested (1, 1.5, 2, 3 and 4) to compare the final results.
To obtain a global perspective of the associations between the targets and the hepatotoxic pathways, we determined the pathway enrichment by mapping the CTI targets to the Gene Set Analyzer (GAzer)42 of CTD. GAzer can generate KEGG and REACT pathways for groups of targets. All of the hepatotoxic pathways were selected to construct the target-pathway association (TPA) network using Cytoscape (version 3.2.1) by linking the targets to the corresponding pathways and different liver diseases. To analyze the important pathways, we visualized the key targets in the entire pathway by mapping them to the KEGG database. We chose the Homo sapiens (human) + disease/drug pathway as the specific organism pathway. The key targets were mapped and their names were highlighted in red.
To obtain a global perspective of the associations between the targets and diseases, we constructed a target-disease association (TDA) network between the CTI targets and nine liver diseases using Cytoscape (version 3.2.1) by linking the targets to their corresponding liver diseases.
Chemical interactions with CYP450 enzymes
To confirm the compounds related to cytochrome P450 (CYP450) enzymes and to further elucidate their specific functions as either inhibitors or substrates, we calculated the chemical ADMET properties using our own web server admetSAR43. AdmetSAR integrated high quality and predictive QSAR models to predict approximately 50 ADMET endpoints, including substrates of CYP1A2, 2C9, 2C19, 2D6 and 3A4, and inhibitors of CYP1A1, 1A2, 2A5, 2A6, 2C9, 2C19, 2D6 and 3A4. The area under the receiver operating characteristic curve (AUC) was used to evaluate the reliability of the predictions of 22 classification models via 5-fold cross validation. In addition, all classification models were assigned a probability output ranging from 0 to 1. Compounds with a probability >0.8 were selected to obtain more accurate results. Only the compounds predicted to be related to CYP450 by both admetSAR and SDTNBI were selected for further analysis.
Results
A total of 105 chemical components of PmT were obtained from the databases and were numbered from 1 to 105 (Table S1). Fifty-four of these compounds exhibited good intestinal absorption and were used for further analysis (Table 1). Two hundred and sixty experimentally determined compound-target pairs were obtained from four databases, including 18 compounds and 146 targets. The other 36 compounds had no known targets, indicating that target prediction for these compounds was a necessity. Therefore, we predicted 2700 putative compound-target pairs with SDTNBI using 54 compounds. In total, we obtained 2939 compound-target pairs among the 54 compounds and 224 targets (Table S1).
Table 1. Fifty-four compounds with good intestinal absorption from PmT.
| Compound id | Compound name | Intestinal absorption level | Prediction for CYP450 | Ascore |
|---|---|---|---|---|
| 6 | 2,3,5,4′-Tetrahydroxystilbene-2-O-(6″-O-acetyl)-β-D-glucopyranoside | 2 | 1879 | |
| 7 | (E)-Resveratrol 3-xyloside | 1 | 1939 | |
| 13 | Resveratrol | 0 | 1A2_Inh/2C19_Inh | 4029 |
| 14 | Polydatin | 1 | 2051 | |
| 15 | (Z)-4′-O-Methylpiceid | 1 | 2202 | |
| 22 | Emodin | 0 | 1A2_Inh | 7773 |
| 23 | Aloe-emodin | 0 | 2392 | |
| 24 | Chrysophanol | 0 | 1A2_Inh | 2479 |
| 25 | Physcion | 0 | 1A2_Inh | 2365 |
| 26 | Rhein | 0 | 1A2_Inh | 2766 |
| 27 | 1,6-Dimethyl ether-emodin | 0 | 2200 | |
| 28 | Emodin-8-methyl ether | 0 | 1A2_Inh | 2200 |
| 29 | Citreorosein | 0 | 2200 | |
| 30 | Citreorosein-8-methyl ether | 0 | 2030 | |
| 31 | Emodin-3-methyl ether | 0 | 1A2_Inh | 1983 |
| 32 | Fallacinol | 0 | 2067 | |
| 33 | Emodin-6,8-dimethylether | 0 | 2105 | |
| 34 | 2-Acetylemodin | 0 | 1A2_Inh | 2176 |
| 42 | 2-Methoxy-6-acethyl-7-methyliuglone | 0 | 1879 | |
| 43 | Tricin | 0 | 1A2_Inh | 1997 |
| 45 | Luteolin | 0 | 1A2_Inh/3A4_Inh | 3069 |
| 46 | Quercetin | 1 | 1A2_Inh/3A4_Inh | 4847 |
| 47 | Kaempferol | 0 | 1A2_Inh/3A4_Inh | 3250 |
| 49 | Apigenin | 0 | 1A2_Inh/3A4_Inh | 4081 |
| 55 | Copaene | 0 | 2051 | |
| 57 | Hexanoic acid | 0 | 2202 | |
| 58 | Hexadecanoic acid methyl ester | 1 | 2176 | |
| 59 | Hexadecanoic acid ethyl ester | 2 | 2131 | |
| 69 | Catechin | 0 | 2295 | |
| 70 | Epicatechin | 0 | 2350 | |
| 75 | Gallic acid | 0 | 3889 | |
| 76 | Methylgallate | 0 | 2217 | |
| 77 | N-trans-feruloyltyramine | 0 | 2C9_Inh | 1920 |
| 78 | N-trans-feruloyl-3-methyldopamine | 0 | 2128 | |
| 79 | Indole-3-(L-α-amino-α-hydroxypropionicacid)methyl ester | 1 | 2004 | |
| 80 | 7-Hydroxy-4-methylcoumarin-5-O-β-D-glucopyranoside | 1 | 1907 | |
| 81 | 7-Hydroxy-3,4-dimethylcoumarin-5-O-β-D-glucopyranoside | 1 | 1836 | |
| 82 | n-Butyl-β-D-fructopyranoside | 0 | 2061 | |
| 83 | 1,3-Dihydroxy-6,7-dimethylxanthone-1-O-β-D-glueopyranoside | 1 | 2061 | |
| 84 | Daucosterol | 2 | 3A4_Sub | 2061 |
| 85 | 4-Hydroxybenzaldehyde | 0 | 2072 | |
| 86 | 5-Carboxymethyl-7-hydroxy-2-methylchromone | 0 | 2C19_Inh | 2061 |
| 87 | 1,2-Propanediol-1-(4-hydroxy-phenyl) | 0 | 1901 | |
| 90 | Torachrysone-8-O-β-D-glucopyranoside | 2 | 1969 | |
| 95 | β-Sitosterol-3-O-β-D-glucoside | 2 | 3A4_Sub | 2034 |
| 96 | 2,5-Dimethyl-7-hydroxychromone | 0 | 1A2_Inh/2C9_Inh | 1859 |
| 97 | Schizandrin | 0 | 2221 | |
| 99 | 2,3-Di-hydro-3,5-dihydroxy-6-methyl-4(H)-pyran-4-one | 0 | 2034 | |
| 100 | Hydroxymaltol | 0 | 2137 | |
| 101 | 5-Hydroxy methyl-furfuran | 0 | 1A2_Inh | 2058 |
| 102 | Butanedioicacid | 0 | 2107 | |
| 103 | 5-Dihydroxy-6-methyl-4(H)-pyran-4-one | 0 | 1836 | |
| 104 | Danthron | 0 | 1A2_Inh | 2487 |
| 105 | Piceid | 1 | 1879 |
The term 'Compound id' ranges from 1 to 105, meaning all the 105 compounds collected at first. But only the 54 compounds with good intestinal absorption were displayed here. Intestinal absorption levels: '0' stands for good, '1' for moderate and '2' for poor. 'Inh' means inhibitor and 'Sub' means substrate.
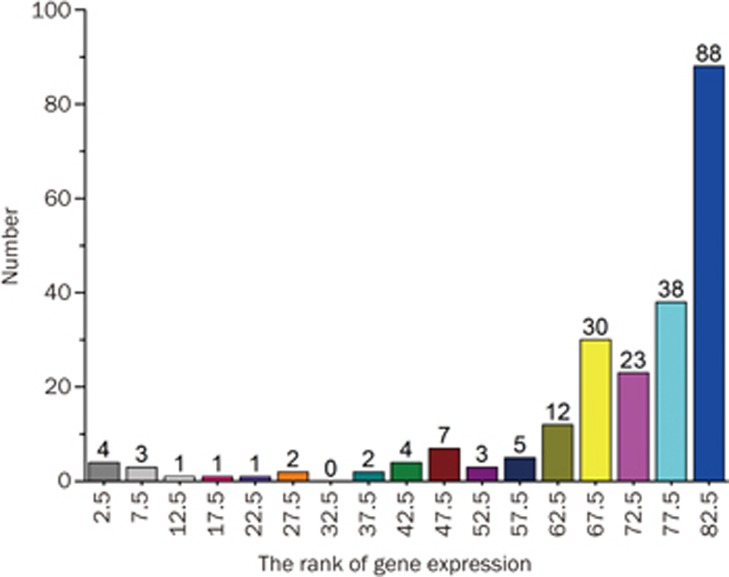
The tissue-specific mRNA expression patterns can provide important clues regarding the target function. The expression rankings in the liver of the 224 targets are shown in Supplementary Table S1. There were 88 targets whose liver expression levels ranked amongst the top 5 of all 84 expression patterns. One hundred and twenty-six targets had liver expression levels that ranked in the top 10 (Figure 2). Therefore, the targets that were highly expressed in the liver may underlie PmT-induced liver damage.
Figure 2.
Analysis of the target expression ranks of all 84 expression patterns in the liver.
Analysis of compound-target interactions
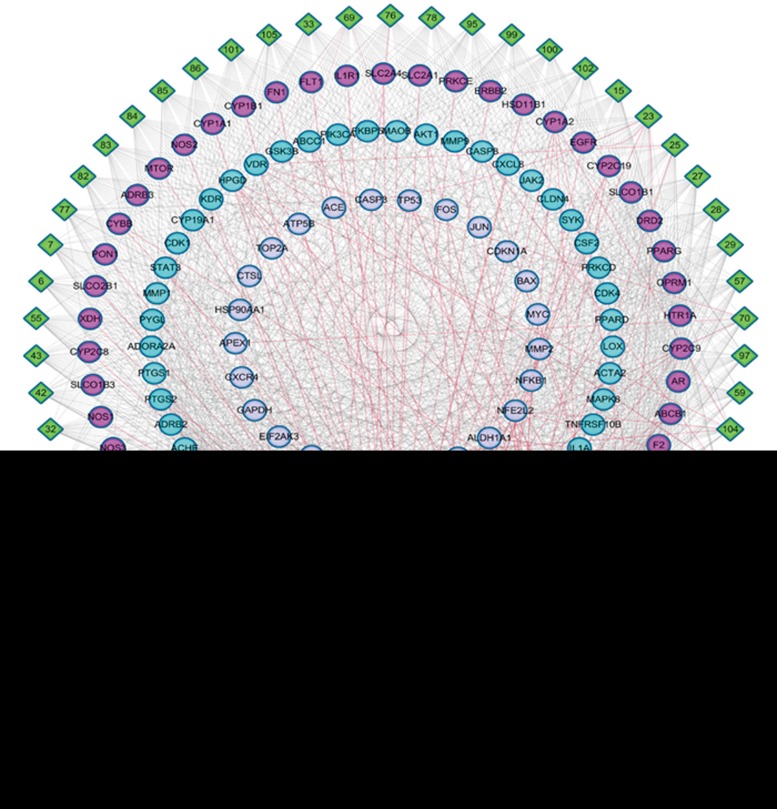
To identify the liver disease-associated targets, 2138 target-disease pairs obtained from 1209 targets and 9 liver diseases were collected from four databases, including OMIM, HuGE, PharmGKB and CTD (Table S2). After mapping the tissue-specific targets, we identified 125 targets related to PmT hepatotoxicity. The 1599 PmT compound-target pairs obtained from the 125 targets (Table S2) were then used to construct the CTIs (Figure 3). The top 5 tissue-specific target expression nodes in the liver are highlighted in red in the CTIs. The top 10 nodes are highlighted in indigo. Using network analysis, we evaluated the degree of the nodes. We obtained an average degree of 29.61 targets per compound and 12.79 compounds per target. Many of the targets were connected to multiple compounds. All of the compounds were connected to multiple targets.
Figure 3.
Target-compound interactions. A compound node and a target node were linked if the target was associated with the compound.
We developed the Ascore scoring function to evaluate the association between compounds and hepatotoxicity targets. To determine the optimal weight factor (k), we assessed a range of different values (1, 1.5, 2, 3 and 4). By comparing the Ascore values of the compounds with different k values, it was easy to see that all of the ranks were the same when k=2, 3 or 4, however, some of the ranking results using k=1 or 1.5 differed from those using k=2. Therefore, we determined that 2 was a reasonable weight factor for this study (Table S3). The Ascore values of these compounds are listed in Table 1. Among the 54 compounds, emodin, an important component of PmT, had the highest degree (56) and Ascore value (7773), greatly exceeding the values determined for the other compounds. The Ascore values of quercetin, apigenin and resveratrol were above 4000. The Ascore values of gallic acid, kaempferol and luteolin exceeded 3000. Most of these components were flavonoids, with the exception of gallic acid. Fourteen compounds had Ascore values higher than 2200, most of which were quinones.
BiNGO41 analysis revealed that the biological processes of the 125 targets could be divided into the following four classes: apoptosis, the regulation of cell death and proliferation, the regulation of metabolic processes, and the regulation of signal transduction (Table S2). These biological processes are closely related to hepatotoxicity. Moreover, these processes were both positively and negatively regulated, indicating that the liver plays a critical role in the metabolism of toxic substances by maintaining the balance between these processes. PmT can influence the metabolism of toxic substances by disturbing the balance between these processes. Of the 125 targets, some were specific detoxifying enzymes, such as MAOB and AChE, while others were non-specific detoxifying enzymes, such as CYP3A4, CYP2C9, CYP1A2, free radical SOD, and free radical COST. Both the non-specific and specific detoxifying enzymes were important for the metabolism of toxic substances and protected the liver from injury. However, some targets represented liver cell death, such as DCD (CASP3) and autophagy mTOR. This finding may explain why PmT has both protective and damaging effects in the liver11.
Analysis of target-pathway and target-disease association networks
The robustness of a phenotype may be understood in terms of alternative compensatory signaling routes within the complex biomolecular networks. To this end, proteins involved in diseases may provide insights into the downstream effects on multiple biological and disease pathways. Some cellular network or signaling pathway databases have systematically collected targets associated with specific diseases reported in the literature44.
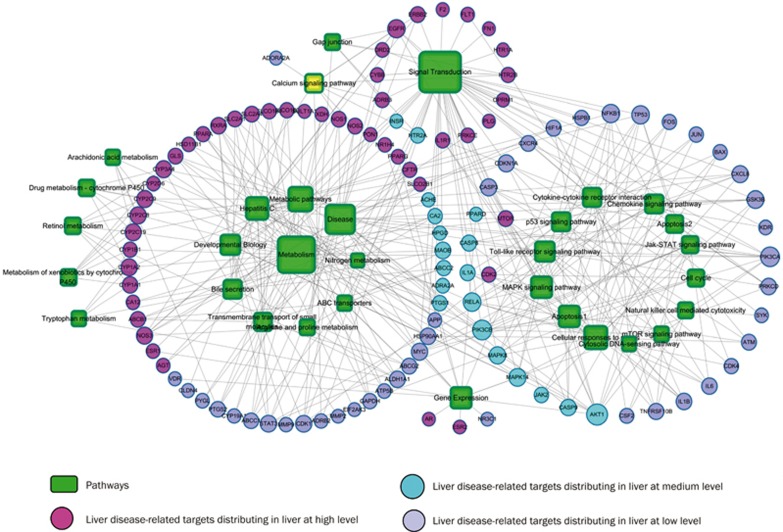
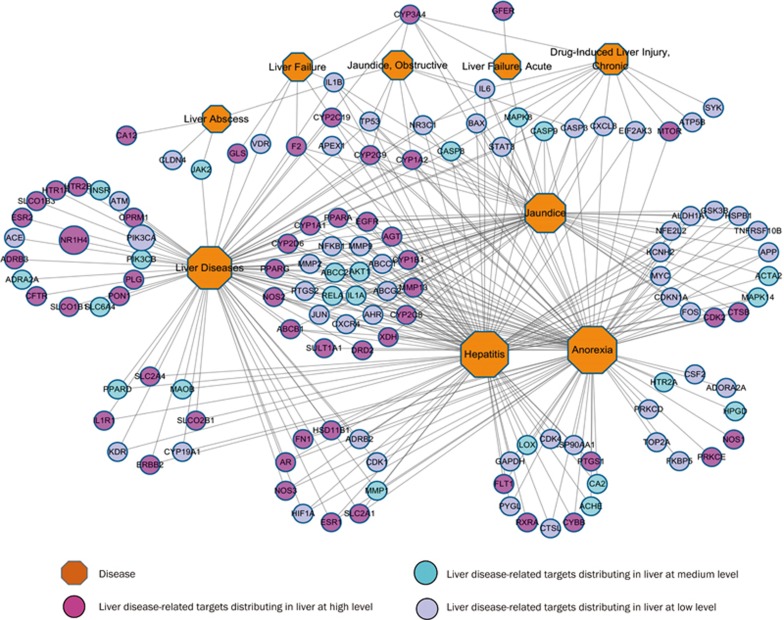
We obtained 75 KEGG pathways and 15 REACT pathways in total (Table S4) by mapping the 125 targets to the GAzer of the CTD databases. The 397 target-pathway pairs obtained from the 32 hepatotoxicity-associated pathways and the 125 targets were used to construct the target-pathway association network (Figure 4). The target-disease association network was constructed from nine liver diseases and 125 targets with 341 target-disease pairs (Figure 5). The multi-targeted compounds can modulate these essential proteins through parallel or similar functional pathways to produce enhanced or decreased effects.
Figure 4.
Target and pathway association network. A link was placed between the target and the pathway if the target is on the pathway. The area of the pathway node is proportional to degree. The pathway information was obtained by mapping the target proteins onto the CTD database.
Figure 5.
Target and disease association network. A link was placed between the target and disease if the target was related to the disease. The area of the disease node is proportional to the degree.
To further map the target-pathway interactions, we applied a target-based approach to probe the pathways most likely involved in hepatotoxicity. The results demonstrated that pathways strongly connected to PmT targets were critical for hepatotoxicity, including signal transduction (degree=48), metabolism (degree=40), apoptosis (degree=15) and hepatitis (degree=17). Some of these pathways have been validated.
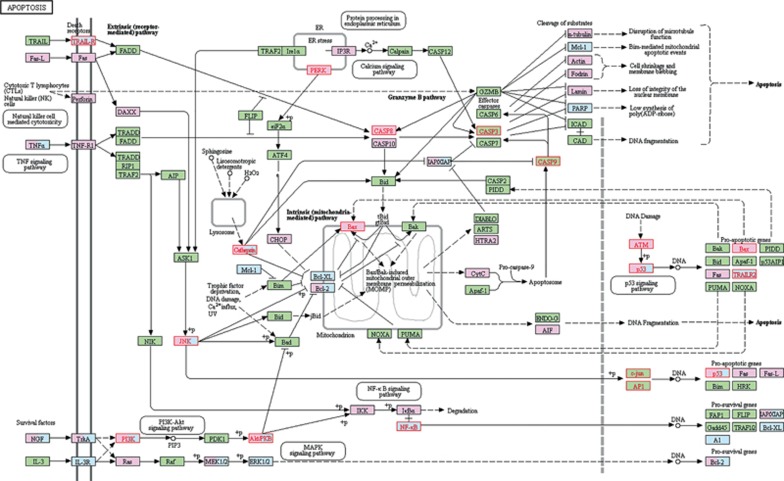
Many pathways can be considered specific mechanisms of cell death, such as the MAPK signaling pathway, signal transduction, cellular responses to stress, cytokine-cytokine receptor interaction, and the p53 signaling pathway, among others (Figure 4). Some of these pathways were demonstrated in many compounds45,46. The apoptosis pathway was analyzed specifically (Figure 6). The pink and blue rectangles represent the disease and drug targets, respectively. Eighteen targets were mapped out; their names are highlighted in red. Apoptosis is a genetically programmed process that eliminates damaged or redundant cells by activating caspases (aspartate-specific cysteine proteases). The balance between the pro-apoptotic and anti-apoptotic signals eventually determined whether cells would undergo apoptosis, survival or proliferation. Both of the death receptors TRAILR and NK (natural killer) were present, which may explain why PmT has both cytotoxic and protective effects47.
Figure 6.
Apoptosis pathway. This figure was generated from KEGG PATHWAY Database. The mapped genes were marked in red by pathway mapping tools embedded in KEGG on the KEGG map 04210. We chose the Homo sapiens (human)+ disease/drug pathway as the specific organism pathway. Pink rectangles indicate disease targets. Blue rectangles indicate drug targets. The names of the 18 mapped targets are highlighted in red.
In addition to apoptosis, hepatitis, bile secretion and the nitrogen metabolism pathways corresponded to symptoms of hepatitis, jaundice and tawny urine. Hepatitis is the most common and obvious side effect of PmT hepatotoxicity48,49. In the current study, the hepatitis pathway was mapped with 18 targets to explain the symptom. We hypothesized that PmT could control the upstream targets of TRAILR, PERK(EIF2AK3) and PI3K (PIK3CA, PIK3CB) to influence the downstream targets such as p53 (TP53), jun (MAPK8) and Akt (AKT1) contributing to apoptosis and other symptoms of hepatitis. The bile secretion pathway was mapped with 11 targets, indicating that PmT may cause bile acid (BA) disruptions. FXR (encoded by target NRIH4) is highly expressed in the liver and other digestive organs. FXR function rapidly expanded from its initial role in controlling the metabolism of bile acids, lipids and glucose, to regulating cell growth, fibrosis, cirrhosis, immunological responses, inflammation and malignance. The role of hepatic FXR in liver-related diseases has captured more attention because increasing evidence suggests that FXR plays a crucial role in liver regeneration and repair, and inflammatory responses. SLCO1B1, ABCB1, ABCC2, ABCG2 and CFTR are important for the transportation of substances during the entire process. The nitrogen metabolism pathway may explain the presence of yellow or tawny urine. CA12, CA2 and GLS can translate CO2 into HCO3, which further influence the carbon amino acid metabolism.
Compounds interacting with CYP450s and the metabolic pathway
The metabolic pathway is a major site for liver injury. Once the liver is injured, its metabolic function is critically affected. If a compound has the potential to disturb substance metabolism in human liver cells, it may produce hepatotoxicity. For example, GSH synthesis plays a central role in cell protection. GSH depletion causes a series of metabolic disorders, including oxidative stress and fatty acid oxidation disorder.
Xenobiotic metabolism by CYP450, arginine and proline metabolism, tryptophan metabolism, retinol metabolism and linoleic acid metabolism have similar effects on human liver cell metabolism and cause hepatotoxicity (Figure 4). Six targets were mapped in the arginine and proline metabolism pathway, namely ALDH1A1, GLS, MAOB, NOS1, NOS2 and NOS3. Arachidonic acid metabolism is associated with PTGS1 and PTGS2. The other metabolic pathways are mainly related to CYP450s, ALDH1A1 and MAOB. In addition, the intake of specific drugs may efficiently induce CYP450s. The levels of CYP1A2 significantly differed among human subjects. Therefore, individuals with high enzymatic activity of CYP1A2 may be more susceptible to potential adverse effects.
The pathway enrichment and organ distribution analyses using BioGPS revealed that CYP450s are important hepatic proteins. It is important to determine which compounds interact with CYP450s as either inhibitors or substrates. Using our web server (admetSAR), we predicted that 16 compounds may act as CYP1A2 inhibitors, 5 as CYP2C19 inhibitors, 4 as CYP3A4 inhibitors and 2 as CYP3A4 substrates (Table 1). Most were quinones and flavonoids, including emodin. The values are given in Table S5.
Discussion
TCM has played an important role in human health for thousands of years. Despite its long history, an increasing number of hepatotoxicity cases have been reported, presenting new clinical challenges. The identification of these toxic compounds and the MOA of hepatotoxicity are critical for applying TCM safely. Studies applying systems pharmacology to TCM have typically focused on pharmaceutical action and drug efficacy, rather than adverse effects. In this study, we used PmT as an example to propose a systems toxicology approach to investigate the molecular mechanisms of hepatotoxicity induced by TCM herbs.
The sources and methods used for the analyses are reliable
First, the chemical components of PmT should be identified. Because oral administration is the most desirable route for TCM, only compounds with good oral absorption are distributed in the liver and have the potential to cause hepatotoxicity. Therefore, we applied an in silico model embedded in the Pipeline Pilot to predict the intestinal absorption of the PmT chemical components. This model26 utilized robust outlier detection to analyze the well-absorbed compounds. Extensive validation of the model with hundreds of known orally delivered drugs demonstrated high predictive accuracy (74%-92%, depending on the dataset and criterion used). Finally, the number of compounds was reduced from 105 to 54, which facilitated the understanding of the molecular mechanisms of liver injury induced by PmT chemical components.
The target expression data were used to explain the organ distribution of PmT, which can indirectly clarify the specific organ toxicity. If a TCM herb acts on many targets that are extensively expressed in the liver, it is more likely to impact the liver and either treat the disease or cause hepatotoxicity. In 2015, BioGPS was updated to provide broader dataset coverage. BioGPS has been applied extensively even in the GeneCards database. To verify our gene expression data, we used another tissue-specific gene/protein database, TiGER50, which was developed by the Bioinformatics Lab at the Wilmer Eye Institute at Johns Hopkins University. After the Ascore was calculated for each compound, the compounds with the top 15 Ascore values determined using the TiGER database were compared to the top 15 compounds using the BioGPS database (Table S3). Twelve of the top 15 compounds overlapped. The top five compounds were identical. All 7 compounds associated with either hepatotoxicity or hepatoprotection were in the top 15. This comparison suggested that BioGPS and our results were reliable.
Our newly developed SDTNBI method was used for target prediction. It was systematically evaluated using 10-fold cross validation, leave-one-out cross validation, and external validation in our previous publication31. Recently, we further improved this method by introducing three tunable parameters that were successfully validated with biological assays. Twenty-seven of the 56 commercially available compounds were experimentally confirmed to have binding affinities for estrogen receptor α with IC50 or EC50 values ≤10 μmol/L51.
Ascore accurately ranked the compounds
To determine the active components for specific TCM pharmacological functions, most previous studies assumed that the more important a compound is as a node in the network, the stronger its disease-initiating effects. As a fundamental topological parameter of a network, the degree of a compound indicates the number of targets it can affect, which may indicate the more influential compounds21. Some studies applied new network-based evaluation methods to identify active ingredients in the network for a given herbal formula. For example, Li et al successfully predicted the active and synergistic ingredients of the anti-rheumatoid arthritis formula of Qing Luo Yin using a new evaluation method52.
To estimate the toxicity of the chemical components in the liver, we developed a scoring function (Ascore) to semi-qualify the action of the compounds on the liver targets. The Ascore considers the target distribution in the liver. If the targets are barely distributed in the liver, the Ascore value will remain small, regardless of how many targets it has. The purpose of target prediction is to consider more potential targets. The known targets themselves provided important clues for clarifying the underlying mechanism. As a result, the Ascore provided a more reasonable measure than the degree for evaluating the liver toxicity in our network analysis. Each compound had a corresponding Ascore value. A higher Ascore value indicates a stronger target action.
Among the 54 compounds, emodin had the highest degree and the highest Ascore, which is consistent with the fact that emodin is the most important component in PmT. Because emodin interacts extensively with hepatotoxicity-association targets, it may be the most important compound for hepatotoxicity, which has been suggested by many studies47. The Ascore for resveratrol was in the top 2 and has been shown to attenuate hepatotoxicity in rats exposed to arsenic trioxide. Gallic acid plays a hepatoprotective role in sodium fluoride-induced liver injury in rats. Quercetin has hepatoprotective effects against acrylonitrile-induced hepatotoxicity in rats. The protective and preventive effects of kaempferol were demonstrated in CCl4-induced hepatotoxicity. Rhein and physcion were shown to cause hepatotoxicity. Based on the Ascore values, 7 of the top 15 compounds were associated with either hepatotoxicity or hepatoprotection. Thus, the Ascore value of a compound reflects the degree of the hepatotoxicity-associated targets it can influence and furthers the possibility of hepatotoxicity. The investigation of other compounds with high Ascore values could provide important insights.
The superiority of pathway enrichment for determining the MOA of toxicity
Apoptosis is a well-known cause of PmT-induced HILI. In fact, many other pathways are considered as specific mechanisms of cell death. Some pathways have been confirmed for many compounds45,46, providing the rationale for our method. The highest concentration of drugs and metabolites after oral administration is usually found in the liver. Most hepatotoxic compounds are metabolized by CYP450 enzymes, which are primarily involved in the generation of reactive metabolites. Drugs can also interact with CYP450 enzymes as either inhibitors or substrates53,54. Our web server admetSAR was used to predict compounds that could interact with CYP450s as either inhibitors or substrates. In addition to the prediction of CYP450 substrates or inhibitors, a total of 22 highly predictive qualitative classification models were implemented. The area under AUC ranged from 0.638 to 0.956 for the 22 classification models using 5-fold cross validation. As described in our previous study, all classification models were assigned a probability output instead of a simple binary output 43. Thus, the 20 PmT components can be regarded as candidate PmT compounds that cause HILI via CYP450s. All of these metabolic disorders consequently contribute to liver cell death. Many studies have reported the influence of metabolism in PmT-induced HILI55,56.
In addition to apoptosis and metabolism, 3 of the 9 phenotypic symptoms were confirmed by the corresponding pathways, which also provided the rationale for our method. Hepatitis is the most common and obvious side effect of PmT-induced hepatotoxicity48,49. Bile is a vital secretion, essential for the digestion and absorption of fats and fat-soluble vitamins in the small intestine. Moreover, bile is an important route of elimination for excess cholesterol and many waste products, bilirubin, drugs and toxic compounds. Bile secretion depends on the function of the membrane transport systems in hepatocytes and cholangiocytes and on the structural and functional integrity of the biliary tree. Bile acids are also important signaling molecules. Coincidentally, Wang et al reported that the perturbation of nine bile acids (BAs) were associated with PmT-induced liver injury in 2015. However, the mechanism was not clear57. This result is consistent with our findings. In addition, 11 targets could explain these mechanisms. The nitrogen metabolism pathway could explain the presence of yellow or tawny urine.
The Toll-like receptor-signaling pathway was mapped with 13 targets. Coincidentally, Wang et al verified this result with animal experiments. After determining the organ-to-body ratio for each rat, histopathological observations of liver samples and mTLR-4 detection using RT-PCR revealed that the Toll-like receptor 4 (TLR4) may be associated with PmT-induced HILI58. Interestingly, most of the 13 targets were downstream targets and TLR4 was not affected, indicating that pathway enrichment could expand our current understanding and help us to view the system network as a whole.
Pathway information can be directly mapped onto each compound. If a compound interacts with a protein that is included in a pathway, the corresponding compound-pathway interaction is considered. However, such an approach does not consider the role of a target in a given pathway. Pathway enrichment was used to determine the possible MOA of hepatotoxicity.
There is a need for realistic evaluation of the toxicity of herbal drugs as they are prepared traditionally and used clinically. Special attention must be paid to toxicities that are not easily detected when used at reasonable doses, including genotoxicity, carcinogenicity, developmental toxicity and organ toxicity, particularly hepatotoxicity. Because a large number of components have variable potencies and affinities for various targets, the complexity of herbal medicine is a major issue for safety assessment. As with efficacy, toxicity may result from a combination of active compounds rather than a single chemical entity; therefore, different methods are required to replace or supplement those routinely used in classical drug risk assessment. Systems toxicology as an in silico tool is generally low-cost, efficient, and easy to handle. Pathway enrichment analysis is very important for systems toxicology. If an effect is caused by the action of a compound on an appropriate target and if this target belongs to a certain pathway, it suggests that any influence on this pathway is related to the observed effect. In this case, we may expect that if a new compound acts on any of the targets of this pathway, it may cause the same effect. This effect is quite possible when using plant extracts because of the presence of many phytocomponents59. We also screened the hepatotoxicity-related targets using the HILI symptoms, which focused our network analysis on hepatotoxicity. The robustness of a phenotype may be understood in terms of alternative compensatory signaling routes within complex biomolecular networks. Furthermore, intestinal absorption prediction, target expression data of organ distribution in the liver, Ascore function and the CYP450 prediction are innovations of the current study that can be used to clarify specific organ toxicity, such as hepatotoxicity.
The biphasic effects of PmT in the liver
Three factors could explain the biphasic effects of liver protection and damage induced by PmT. In terms of target function, the liver plays an important role in the metabolism of toxic substances by maintaining the balance between four main biological processes. Detoxifying enzymes and targets representing cell death were both present. With regard to metabolic function, 16 compounds were predicted to interact with CYP1A2. The level of CYP1A2 significantly varied amongst the individuals. People with high enzymatic activity may be more susceptible to the potential adverse effects of these compounds. In terms of apoptosis, the death receptors and ligands activating anti-apoptotic or cell-survival signals were both present. This result can also explain why PmT had both cytotoxic and protective effects. PmT compounds can interact with each other through pathways to influence hepatotoxicity. In fact, some herbs exhibit delayed or cumulative toxicity that may not be immediately obvious because the toxicity may be associated with the dose. Preclinical and clinical toxicology research on herbal medicines is in most cases inadequate or insufficient to fulfill official drug registration requirements. Toxicological and pharmacokinetic studies of individual constituents of herbal materials must be compared to the qualitative and quantitative profiling data for the total extract.
In this study, a computational systems toxicology approach was proposed to systematically understand the mechanisms of the effects of PmT on particular hepatotoxicity-associated pathways that could result in HILI. Apoptosis and metabolism are two major mechanisms of PmT-induced liver injury. Many of the pathways obtained from pathway enrichment have been verified in certain compounds. The Ascore developed in the current study is a helpful scoring tool that reflects the relationship between a compound and hepatotoxicity-associated targets, and the hepatotoxicity potential of a compound. Metabolic disorders will contribute to liver cell death. The biphasic effects of both liver protection and liver damage are also explained by the target function, metabolism and apoptosis. The methods employed in the current study could be used to identify novel hepatotoxic herbs.
Abbreviations
HILI, herb induced liver injury; PmT, Polygonum multiflorum Thunb.; CTIs, compound-target interactions; MOA, mechanism of action; TCM, traditional Chinese medicine; NIH, National Institutes of Health; SEs, side effects; SDTNBI, substructure-drug-target network-based inference; AUC, the area under the receiver operating characteristic curve; MeSH, medical subject headings; UMLS, Unified Medical Language System; target-pathway association network, TPAs; target-disease association network, TDAs; AChE, acetylcholinesterase; SOD, superoxide dismutase; DCD, dermcidin; NK, natural killer; TRAILR (TNFRSF10B): Tumor necrosis factor receptor superfamily member 10B; PIK3CA, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform; PIK3CB, phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit beta isoform; EIF2AK3, eukaryotic translation initiation factor 2-alpha kinase 3; CDKN1A, cyclin-dependent kinase inhibitor 1; MAPK14, mitogen-activated protein kinase 14; MAPK8, mitogen-activated protein kinase 8; STAT3, signal transducer and activator of transcription 3; BAs, bile acids; FXR(NRIH4), bile acid receptor; ABCB1, ABC transporter B family member 1; ABCC2, ABC transporter C family member 2; ABCG2, ATP-binding cassette sub-family G member 2; CFTR, cystic fibrosis transmembrane conductance regulator; CA12, carbonic anhydrase 12; CA2, carbonic anhydrase 2; NOS1, nitric oxide synthase brain; NOS2, nitric oxide synthase inducible; NOS3, nitric oxide synthase endothelial; PTGS1, prostaglandin G/H synthase 1; PTGS2, prostaglandin G/H synthase 2; TLR4, Toll-like receptor 4; RT-PCR, reverse transcription-polymerase chain reaction.
Author contribution
Yun TANG, Gui-xia LIU, Wei-hua LI, Bo ZHANG, Yin-yin WANG, and Jie LI designed the experiments; Yin-yin WANG performed the research; Yun TANG, Zeng-rui WU, Bo ZHANG, and Hong-bin YANG contributed to the new analytic tools; Yin-yin WANG, Qin WANG, and Ying-chun CAI analyzed the data; Yun TANG, Yin-yin WANG, Jie LI, and Hong-bin YANG wrote the paper.
Acknowledgments
This work was supported by the National Key Research and Development Program (No 2016YFA0502304), the National Natural Science Foundation of China (No 81373329 and 81673356), and the 111 Project (No B07023).
Footnotes
Supplementary information is available on the Acta Pharmacologica Sinica website.
Supplementary Information
Information of 105 compounds
BiNGO result
k-try
All the 90 pathways
ADMET properties predicted by admetSAR
References
- Normile D. Asian medicine. The new face of traditional Chinese medicine. Science 2003; 299: 188–90. [DOI] [PubMed] [Google Scholar]
- Licata A, Macaluso FS, Craxì A. Herbal hepatotoxicity: a hidden epidemic. Pharm Des 2013; 8: 13–22. [DOI] [PubMed] [Google Scholar]
- Teschke R, Eickhoff A. Herbal hepatotoxicity in traditional and modern medicine: actual key issues and new encouraging steps. Front Pharmacol 2015; 6: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschke R, Wolff A, Frenzel C, Schulze J, Eickhoff A. Herbal hepatotoxicity: a tabular compilation of reported cases. Liver Int 2012; 32: 1543–56. [DOI] [PubMed] [Google Scholar]
- Lin L, Ni B, Lin H, Zhang M, Li X, Yin X, et al. Traditional usages, botany, phytochemistry, pharmacology and toxicology of Polygonum multiflorum Thunb: a review. J Ethnopharmacol 2015; 159: 158–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi KQ, Fan YC, Liu WY, Li LF, Chen YP, Zheng MH. Traditional Chinese medicines benefit to nonalcoholic fatty liver disease: a systematic review and meta-analysis. Mol Biol Rep 2012; 39: 9715–22. [DOI] [PubMed] [Google Scholar]
- Lei X, Chen J, Ren J, Li Y, Zhai J, Mu W, et al. Liver damage associated with Polygonum multiflorum Thunb: a systematic review of case reports and case series. Evid Based Complement Alternat Med: eCAM 2015: 459749. [DOI] [PMC free article] [PubMed]
- Pang J, Bai Z, Niu M, Tu C, Ma Z, Zhao Y, et al. The toxic and protective effects of Polygonum multiflorum on normal and liver injured rats based on the symptom-based prescription theory. Yao Xue Xue Bao 2015; 50: 973–9. [PubMed] [Google Scholar]
- Huang CH, Horng LY, Chen CF, Wu RT. Chinese herb radix polygoni multiflori as a therapeutic drug for liver cirrhosis in mice. J Ethnopharmacol 2007; 114: 199–206. [DOI] [PubMed] [Google Scholar]
- Lin L, Lin H, Zhang M, Ni B, Yin X, Qu C, et al. A novel method to analyze hepatotoxic components in Polygonum multiflorum using ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry. J Hazard Mater 2015; 299: 249–59. [DOI] [PubMed] [Google Scholar]
- Lv G, Meng L, Han D, Li H, Zhao J, Li S. Effect of sample preparation on components and liver toxicity of Polygonum multiflorum. J Pharm Biomed Anal 2015; 109: 105–11. [DOI] [PubMed] [Google Scholar]
- Sturla SJ, Boobis AR, FitzGerald RE, Hoeng J, Kavlock RJ, Schirmer K, et al. Systems toxicology: from basic research to risk assessment. Chem Res Toxicol 2014; 27: 314–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MD, Fostel JM. Toxicogenomics and systems toxicology: aims and prospects. Nat Rev Genet 2004; 5: 936–48. [DOI] [PubMed] [Google Scholar]
- Chen M, Vijay V, Shi Q, Liu Z, Fang H, Tong W. FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov Today 2011; 16: 697–703. [DOI] [PubMed] [Google Scholar]
- Persson M, Løye AF, Mow T, Hornberg JJ. A high content screening assay to predict human drug-induced liver injury during drug discovery. J Pharmacol Toxicol Methods 2013; 68: 302–13. [DOI] [PubMed] [Google Scholar]
- Ivanov SM, Lagunin AA, Poroikov VV. In silico assessment of adverse drug reactions and associated mechanisms. Drug Discovery Today 2016; 21: 58–71. [DOI] [PubMed] [Google Scholar]
- Desai K, Brott D, Hu X, Christianson A. A systems biology approach for detecting toxicity-related hotspots inside protein interaction networks. J Bioinform Comput Biol 2011; 9: 647–62. [DOI] [PubMed] [Google Scholar]
- Huang LC, Wu X, Chen JY. Predicting adverse side effects of drugs. BMC Genomics 2011; 12 Suppl 5: S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Cheng F, Jiang J, Zhang C, Deng X, Xu Z, et al. FXR antagonism of NSAIDs contributes to drug-induced liver injury identified by systems pharmacology approach. Sci Rep 2015; 5: 08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Zheng C, Li Y, Wang Y, Lu A, Yang L. Systems pharmacology in drug discovery and therapeutic insight for herbal medicines. Brief Bioinform 2014; 15: 710–33. [DOI] [PubMed] [Google Scholar]
- Kibble M, Saarinen N, Tang J, Wennerberg K, Mäkelä S, Aittokallio T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat Prod Rep 2015; 32: 1249–66. [DOI] [PubMed] [Google Scholar]
- Koeberle A, Werz O. Multi-target approach for natural products in inflammation. Drug Discovery Today 2014; 19: 1871–82. [DOI] [PubMed] [Google Scholar]
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014; 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R, Fang Z, Zhang M, Yi Z, Wen C, Shi T. TCMID: traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res 2013; 41 (Database issue): D1089–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CYC. TCM database@ Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS One 2011; 6: e15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan WJ, Merz KM, Baldwin JJ. Prediction of drug absorption using multivariate statistics. J Med Chem 2000; 43: 3867–77. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43: D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res 2007; 35: D198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res 2009; 37: W623–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res 2006; 34: D668–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Cheng F, Li J, Li W, Liu G, Tang Y. SDTNBI: an integrated network and chemoinformatics tool for systematic prediction of drug-target interactions and drug repositioning. Brief Bioinform 2017. doi: 10.1093/bib/bbw012 [DOI] [PubMed]
- Yap CW. PaDEL — descriptor: An open source software to calculate molecular descriptors and fingerprints. J Comput Chem 2011; 32: 1466–74. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 2009; 10: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Jin X, Tsueng G, Afrasiabi C, Su AI. BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res 2016; 44: D313–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 2005; 33: D514–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet 2008; 40: 124–25. [DOI] [PubMed] [Google Scholar]
- Hernandez-Boussard T, Whirl-Carrillo M, Hebert JM, Gong L, Owen R, Gong M, et al. The pharmacogenetics and pharmacogenomics knowledge base: accentuating the knowledge. Nucleic Acids Res 2008; 36: D913–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, King BL, Mockus S, Murphy CG, Saraceni-Richards C, Rosenstein M, et al. The comparative toxicogenomics database: update 2011. Nucleic Acids Res 2011; 39: D1067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res 2007; 35: D5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005; 21: 3448–9. [DOI] [PubMed] [Google Scholar]
- Kim S-B, Yang S, Kim SK, Kim SC, Woo HG, Volsky DJ, et al. GAzer: gene set analyzer. Bioinformatics 2007; 23: 1697–9. [DOI] [PubMed] [Google Scholar]
- Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 2012; 52: 3099–105. [DOI] [PubMed] [Google Scholar]
- Zhao J, Jiang P, Zhang W. Molecular networks for the study of TCM pharmacology. Brief Bioinform 2010; 11: 417–30. [DOI] [PubMed] [Google Scholar]
- Hsu CM, Hsu YA, Tsai Y, Shieh FK, Huang SH, Wan L, et al. Emodin inhibits the growth of hepatoma cells: finding the common anti-cancer pathway using Huh7, Hep3B, and HepG2 cells. Biochem Biophys Res Commun 2010; 392: 473–8. [DOI] [PubMed] [Google Scholar]
- Ni CH, Chen PY, Lu HF, Yang JS, Huang HY, Wu SH, et al. Chrysophanol-induced necrotic-like cell death through an impaired mitochondrial ATP synthesis in Hep3B human liver cancer cells. Arch Pharm Res 2012; 35: 887–95. [DOI] [PubMed] [Google Scholar]
- Kuo TC, Yang JS, Lin MW, Hsu SC, Lin JJ, Lin HJ, et al. Emodin has cytotoxic and protective effects in rat C6 glioma cells: roles of Mdr1a and nuclear factor κB in cell survival. J Pharmacol Exp Ther 2009; 330: 736–44. [DOI] [PubMed] [Google Scholar]
- Laird AR, Ramchandani N, Avula B, Khan IA, Gesundheit N. Acute hepatitis associated with the use of an herbal supplement (Polygonum multiflorum) mimicking iron-overload syndrome. J Clin Gastroenterol 2008; 42: 861–2. [DOI] [PubMed] [Google Scholar]
- Jung KA, Min HJ, Yoo SS, Kim HJ, Choi SN, Ha CY, et al. Drug-induced liver injury: twenty five cases of acute hepatitis following ingestion of Polygonum multiflorum Thunb. Gut Liver 2011; 5: 493–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yu X, Zack DJ, Zhu H, Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics 2008; 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Lu W, Wu D, Luo A, Bian H, Li J, et al. In silico prediction of chemical mechanism-of-action via an improved network-based inference method. Br J Pharmacol 2016; 173: 3372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang X, Li S. An integrative platform of TCM network pharmacology and its application on a herbal formula, Qing-Luo-Yin. Evid Based Complement Alternat Med 2013; Article ID 456747. [DOI] [PMC free article] [PubMed]
- Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012; 13: 6964–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Geng X, Chen M, Zhang J, Wang B, Ilic K, et al. High daily dose and being a substrate of cytochrome P450 enzymes are two important predictors of drug-induced liver injury. Drug Metab Dispos 2014; 42: 744–50. [DOI] [PubMed] [Google Scholar]
- Wang HW, Chen TL, Yang PC, Ueng TH. Induction of cytochromes P450 1A1 and 1B1 by emodin in human lung adenocarcinoma cell line CL5. Drug Metab Dispos 2001; 29: 1229–35. [PubMed] [Google Scholar]
- Yu K, Geng X, Chen M, Zhang J, Wang B, Ilic K, et al. High daily dose and being a substrate of cytochrome P450 enzymes are two important predictors of drug-induced liver injury. Drug Metab Dispos 2014; 42: 744–50. [DOI] [PubMed] [Google Scholar]
- Dong Q, Li N, Li Q, Zhang C, Feng W, Li G, et al. Screening for biomarkers of liver injury induced by Polygonum multiflorum: a targeted metabolomic study. Front Pharmacol 2015; 6: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Wang J, Xie L, Dong Y, Han G, Hu D, et al. A new animal model for Polygonum multiflorum Thunb-induced liver injury in rats and its potential mechanisms. Toxicol Res 2015; 4: 1085–97. [Google Scholar]
- Lagunin AA, Goel RK, Gawande DY, Pahwa P, Gloriozova TA, Dmitriev AV, et al. Chemo-and bioinformatics resources for in silico drug discovery from medicinal plants beyond their traditional use: a critical review. Nat Prod Rep 2014; 31: 1585–611. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information of 105 compounds
BiNGO result
k-try
All the 90 pathways
ADMET properties predicted by admetSAR