Graphical Abstract
1. INTRODUCTION
1.1. Cancer and Early Detection
Cancer is the second most common cause of death in the United States, trailing only heart disease in incidence. Despite significant worldwide investment in research, cancer remains responsible for 1 in 4 deaths in developed countries.1 Globally, over 14 million cancer diagnoses were reported in 2012, a figure expected to increase to over 22 million cases per annum in the next two decades.2 Estimated to kill over 1/2 million U.S. citizens, and with over 1.6 million new cases predicted to be diagnosed this year,3 cancer continues to present a major, yet unmet challenge to healthcare both globally and in the United States.
Cancer emerges from our own tissues, complicating both detection and treatment methods due to the similarities between the diseased tissue and healthy tissue.4,5 Despite this fact, the mortality rate from cancer is often greatly reduced by early detection of the disease. For example, non-small-cell lung cancer is responsible for the most cancer related deaths worldwide, with patients in the advanced stages of the disease having only 5–15% and <2% 5-year survival rates for stage III and IV patients, respectively.6 In contrast, patients who start therapy in the early stages of the disease (stage I) have markedly improved survival rates, with an 80% overall 5-year survival rate.6 Consequently, early diagnosis is essential to improving cancer patient prognosis.
At present, clinical detection of cancer primarily relies on imaging techniques or the morphological analysis of cells that are suspected to be diseased (cytology) or tissues (histopathology). Imaging techniques applied to cancer detection, including X-ray, mammography, computed tomography (CT), magnetic resonance imaging (MRI), endoscopy, and ultrasound, have low sensitivity and are limited in their ability to differentiate between benign and malignant lesions.7,8 While cytology, such as testing for cervical cancer via a Pap smear or occult blood detection, may be used to distinguish between healthy and diseased cells or tissues, it is not effective at detecting cancer at early stages. Similarly, histopathology, which generally relies on taking a biopsy of a suspected tumor, is typically used to probe the malignancy of tissues that are identified through alternative imaging techniques, such as CT or MRI, and may not be used alone to detect cancer in its early stages. As such, the development of assays and methods for early detection of cancer, before the disease becomes symptomatic, presents a major challenge.
Recent research within the field of nanotechnology has focused on addressing the limitations of the currently available methods for cancer diagnosis. Certain nanoparticle probes possess several unique properties that are advantageous for use in the detection of cancer at the early stages. In this review, we will discuss the advances in the development of nanoparticle-based methods for the detection of cancer by fluorescence spectroscopy. We will divide this topic into three categories: techniques that are designed for (1) the detection of extracellular cancer biomarkers, (2) the detection of cancer cells, and (3) the detection of cancerous tissues in vivo. We will discuss these strategies within the context of the nanoparticle probe used as well as the recognition moieties applied in each approach. Ultimately, the translation of these methods from the laboratory to the clinic may enable earlier detection of cancer and could extend patient survival through the ability to administer therapeutic treatment in the early stages of the disease.
While this review provides a comprehensive overview of the nanoparticle probes that are used to detect cancer in vitro and in vivo through fluorescence, there are several other relevant reviews that may be of interest to our readers, who may refer to the references for more generalized reviews of nanomaterials used for diagnostics and therapy,9–12 or more detailed insight into the specific types of nanoparticle probes (i.e., quantum dots,13 gold nanoparticles,14,15 upconversion nanoparticles,16 polymer dots,17,18 silica nanoparticles,19 polymeric nanoparticles, 20 etc.) for cancer diagnosis.
2. FLUORESCENCE DETECTION
2.1. Background and Theory
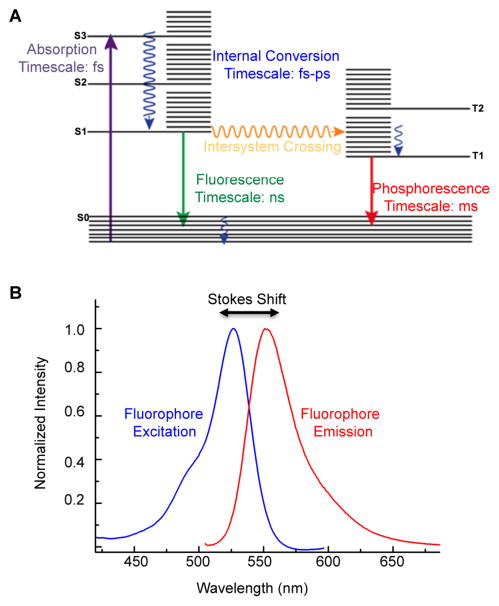
Fluorescence is an optical phenomenon where the absorption of photons at one wavelength results in emission at another, usually longer, wavelength. The loss in energy between the absorbed and emitted photons is the result of vibrational relaxation, and this difference is referred to as a Stokes shift (Figure 1B). A typical Jablonski diagram can be used to describe the process of fluorescence (Figure 1A). In the first phase, known as excitation, absorption of light results in the promotion of an electron from the ground state to the excited state. Once excited, release of the absorbed energy may occur through several photophysical events, including both radiative and nonradiative emission. Vibrational relaxation is often the first route to energy dissipation, and may be followed by internal conversion, intersystem crossing (from a singlet to a triplet state), and subsequent phosphorescence, or fluorescence when the excited electron returns to the ground state and emits energy through the release of a photon.21
Figure 1.
(A) Jablonski diagram including typical time scales of photophysical processes for organic molecules. (B) Molecular fluorescence spectrum illustrating the broadening of the spectral lines due to the presence of vibrational energy levels, and the Stokes shift between the excitation and emission maxima. Adapted from ref 22. Copyright 2010 American Chemical Society.
Fluorescence spectroscopy is a useful technique for the detection of biomolecules, and it is widely used in biological and biomedical applications due to its high spatial and temporal resolution.23,24 Assays utilizing a fluorescent output for detection can employ several methods of analysis, including fluorescence spectroscopy for solution-based assays, microscopy for imaging of cells and arrays used in sandwich assays, flow cytometry for high-throughput imaging of single cells, and in vivo imaging. The use of fluorescence as a detection method depends upon the photophysical properties of the fluorophore used: photostability, quantum yield, Stokes shift, and fluorescence lifetime. These properties, with respect to the advantages that fluorescent nanoparticles offer over organic fluorophores, will be further discussed in the context of the nanoparticle probes that are used in cancer diagnostic applications.
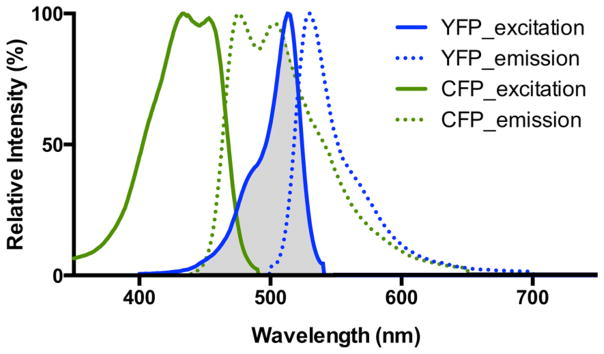
Additionally, Förster resonance energy transfer (FRET), a phenomenon that occurs when an excited donor chromophore transfers energy to an acceptor chromophore via nonradiative dipole–dipole coupling when in close proximity (a few nanometers) to one another, is often used in the design of nanoparticle probes for cancer diagnosis.25,26 The efficiency of energy transfer is dependent upon several factors: (1) spectral overlap between the absorbance spectra of the acceptor fluorophore and the emission spectra of the donor fluorophore (Figure 2), (2) their relative orientations, and (3) their proximity (efficiency is inversely proportional to the distance between the acceptor and the donor to the sixth power).26 The use of FRET probes to detect cancer biomarkers and cells is powerful due to its ability to provide real-time spatial measurements between the donor and acceptor fluorophores, thus allowing the design of more sensitive bioassays for cancer biomarker and cell detection.
Figure 2.
Emission and excitation spectra of CYP and YFP, a commonly used FRET pair, with the spectral overlap shown in gray.
3. NANOPARTICLES FOR FLUORESCENT DETECTION
The application of nanotechnology to cancer diagnosis holds tremendous promise in enhancing the sensitivity and versatility of fluorescence-based methods of detection. In particular, there are several structure-defining traits of nanoparticles that enable the development of novel cancer detection assays: size, shape, high surface area, and unique optical properties. We will review these material-dependent characteristics of nanoparticles with respect to their utility in cancer diagnosis through fluorescence detection, focusing on optical properties and tunable surface functionality.
3.1. Nanoparticle Probe Optical Properties
The optical properties of semiconductor and metallic nanoparticles are highly dependent on nanoparticle size, shape, and composition. In particular, the optical properties most relevant in the design of fluorescence-based biosensors for cancer diagnostics, the intensity and stability of fluorescence emission as well as the effectiveness of fluorescence quenching in “off–on” probes, determine, in part, the sensitivity and dynamic range of a particular assay. These material-dependent optical properties will be further discussed in the context of the nanoparticle probes most widely used in cancer diagnostic applications: quantum dots (QDs), polymer dots (PDs), upconversion nanoparticles (UCNPs), and gold nanoparticles (AuNPs).
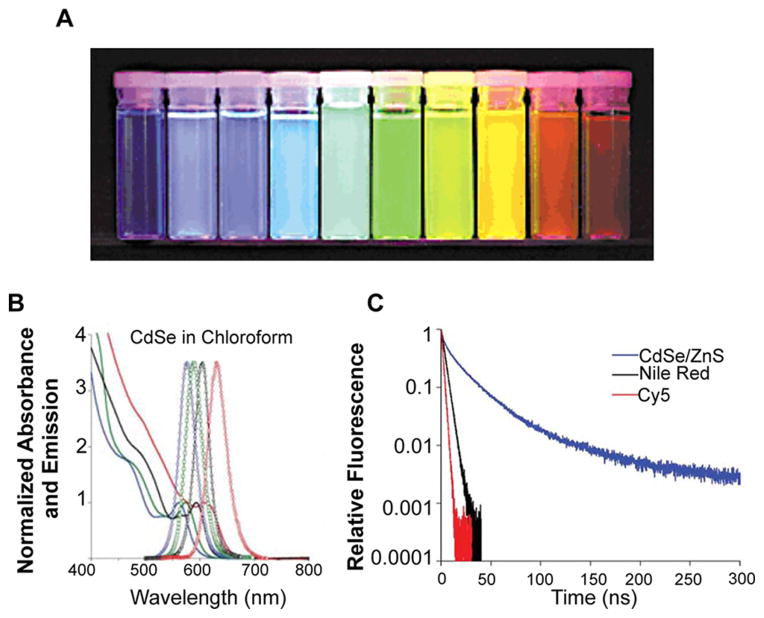
Single crystal semiconductor nanocrystals, or QDs, represent a class of inherently fluorescent nanoparticles with a range of properties that are desirable for biological imaging applications and for the development of novel cancer diagnostics. Semiconducting QDs absorb photons of energy greater than their band gap, resulting in the promotion of electrons from their valence band to their conduction band, generating an electron–hole pair (or exciton).27 Photons are then emitted from discrete bands upon the recombination of the exciton, which generates a narrow emission profile due to their quantum confined properties, which dictate that nanocrystals smaller than the Bohr exciton radius of the material exhibit quantized energy states, with energy levels correlating to QD size.28 This size dependence of QD absorption and emission enables the tunable design of QDs (Figure 3) with a range of imaging applications, especially in multicolor labeling for the simultaneous detection of multiple targets.27,29,30
Figure 3.
(A) Increasing QD size results in a red shift in QD emission: ZnS-capped CdSe QDs of varying size with emission maxima ranging from 443 to 655 nm. Samples were excited with a near-UV lamp. (B) Representative CdSe QD absorption (represented by lines) and emission profiles (represented by circles). QD size increases from left to right, resulting in red-shifted emission. Note the broad absorption. (C) Fluorescence lifetime of CdS/ZnS QD compared to organic dyes Nile Red and Cy5. (A) Reprinted with permission from ref 31. Copyright 2001 Macmillan Publishers Ltd. (B and C) Reprinted with permission from ref 32. Copyright 2008 Macmillan Publishers Ltd.
QD absorption, unlike that of organic dyes, is broad, with large molar absorption coefficients (100 000–1 000 000 M−1 cm−1)33,34 compared to organic dyes (25 000–250 000 M−1 cm−1).32,35–38 QDs exhibit an absorption peak corresponding to the lowest energy level excited state, with absorption increasing at shorter wavelengths due to an increased probability of absorption and the presence of multiple higher energy levels (Figure 3B). This broad absorption allows one to choose the excitation wavelength used, resulting in the ability to excite multiple QD types with a single wavelength of light.
In addition to tunable emission wavelengths, QDs exhibit longer fluorescence lifetimes (>10 ns) compared to organic fluorophores (1–5 ns).39 This property not only enables temporal imaging that is often limited by the short lifetime of organic dyes, but also results in a significant improvement in signal-to-noise ratios in biological applications. Although autofluorescence of tissues and cells often contributes to high background signal, the autofluorescent species present in biological samples have shorter lifetimes. Thus, time-gated fluorescence measurements may be used to image QDs by separating autofluorescence background from positive QD signal.40 This property is especially significant in the application of QDs for enhancing the sensitivity of detecting cancer biomarkers, cells, and tissues, which may be in low abundance at the early stages of the disease.
Polymer dots (PDs) are a class of fluorescent semi-conducting polymer nanoparticles ranging from 5 to 30 nm in size that exhibit broad absorption spectra with narrow emission profiles.17 PDs offer high fluorescence quantum yields (50–60%), which results in bright fluorescence intensity, nearly 3 orders of magnitude higher than that of organic dyes.41–44 In addition, altering the composition of PDs results in tunable emission wavelength,44 which is particularly useful for both in vitro assays45,46 and multiphoton in vivo imaging.47,48 Although PDs have a broader emission spectra than QDs, PDs are brighter in the visible and the near-UV range, are nontoxic, are highly photostable, do not blink, and are therefore utilized in diagnostic and theranostic applications.47
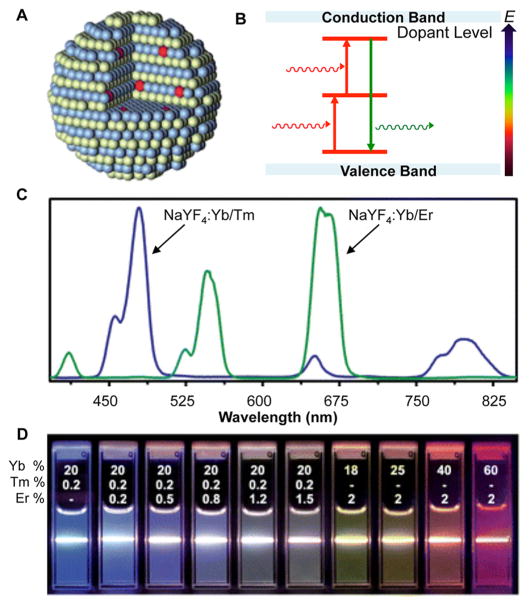
Upconversion nanoparticles (UCNPs) are composed of a rare earth element crystalline host with lanthanide ion (Ln3+) dopants. Most commonly, NaYF4 or NaGdF4 is used as the host lattice, with Yb3+, Tm3+, and Er3+ doped in varying amounts and combinations (Figure 4A).49 In these structures, the Ln3+ ions possess 4fn inner shell electron configurations, which gives rise to fluorescence via intra-4f and 4f–5d electron transitions.50–52 Varying the amounts and types of Ln3+ dopants tunes the emission wavelength53 (Figure 4C,D), which is useful in multicolor imaging applications.49,54
Figure 4.
(A) Schematic representation of rare earth element crystalline host with Ln3+ dopant (red). (B) Two-photon excitation mechanisms common in UCNPs result in the release of a photon of higher energy and an anti-Stokes shift. (C) Emission spectra of NaYF4:Yb/Tm compared to NaYF4:Yb/Er demonstrates the composition-dependent emission profiles of UCNPs. (D) Luminescent photos showing colloidal solutions of UCNPs doped with varying ratios of Yb, Tm, and Er are excited at 980 nm with a 600 mW diode laser. The different colors represent changes in the emission spectra. (A and B) Reprinted with permission from ref 49. Copyright 2010 The Royal Society of Chemistry. (C and D) Adapted from ref 51. Copyright 2008 American Chemical Society.
Unlike organic fluorophores and QDs, UCNPs exhibit an anti-Stokes shift, emitting a photon of higher energy than the absorbed photon. This occurs through multiphoton excitation processes (Figure 4B), which results in the ability to excite UCNPs with near-infrared (NIR) light. This is particularly useful in biological applications due to the minimization of autofluorescence from cells and tissues, as well as enabling deeper tissue penetration through excitation in the tissue-transparent NIR window.49,54
Gold nanoparticles (AuNPs) have also been used in a variety of fluorescent assays for cancer detection. AuNPs exhibit size-dependent absorption in the ultraviolet–visible range due to a size- and shape-related property known as the surface plasmon resonance (SPR).55–58 Incident light on metal atoms in a nanoparticle causes plasmon oscillations in the conduction band of electrons. These collective oscillations result in a strong absorption of light (on the order of 109 M−1 cm−1 for a 40 nm AuNP) and fast electronic relaxation.59,60 Based on their strong absorption, AuNPs are also efficient fluorescence quenchers and thus have been employed in many “off–on” fluorescence probes. Compared to organic quenchers, AuNPs are more efficient due to surface energy transfer processes.55–60
In addition to their use as fluorescence quenchers, AuNPs also have been used as fluorophore labels in imaging applications. Small AuNPs or “Au nanoclusters” (AuNCs) are structures typically less than 3 nm that are composed of a precise number of Au atoms, and unlike larger AuNPs, they do not exhibit SPR absorption in the visible range.61,62 AuNCs do, however, exhibit fluorescence in the visible to near-infrared region with low quantum yields (<1%).62 Despite this, AuNCs have been employed in the design of cancer diagnostic assays, due to their remarkable photostability and resistance to photobleaching.
3.2. Nanoparticle Surface Functionalization and Modes of Targeting
A key advantage to the use of nanoparticles in cancer detection is their large surface area to volume ratio compared to that of bulk materials. In particular, this property enables dense coverage of the nanoparticle surface with moieties that bind and recognize molecules indicative of cancer. Presentation of multiple binding ligands to a cancer cell, for example, often enables multivalent effects that can enhance an assay’s sensitivity. For example, spherical nucleic acid nanoparticles derive their unique properties, including higher binding constants for their complements than free oligonucleotides of the same sequence, from the arrangement of nucleic acids in a dense, highly oriented fashion.63 In addition, since surface atoms contribute more significantly to determining the properties of a nanoparticle, functionalization with a variety of ligands significantly contributes to the collective properties of such structures. Finally, surface curvature can accommodate arrangements of ligands not possible with bulk substrates, leading to unusual and tailorable multivalent effects. Minor variations in nanoparticle surface functionality, ligands, size, and shape can lead to a wealth of properties advantageous for sensing and imaging applications.
A multitude of targeting moieties can also be attached to nanoparticles for use in the diagnosis of cancer, including peptides, antibodies, aptamers, and small molecules, which enable highly specific binding of nanoparticle probes to targets of interest. The targeting moieties relevant to the detection of cancer will be discussed in detail.
Peptides are often used to label cancerous cells based on recognition of their transmembrane proteins. The most commonly used peptide is arginylglycylaspartic acid (RGD), composed of L-arginine, glycine, and L-aspartic acid.64–67 RGD was first isolated from the cell-binding domain of fibronectin, a glycoprotein that binds to integrins, and is involved in cell–cell and cell–extracellular matrix (ECM) attachment and signaling by binding collagen, fibrin, and proteoglycans.64 RGD peptides have the highest affinity for a type of cell surface integrins, αvβ3,68 which are highly expressed in tumoral endothelial cells, but not in normal endothelial cells.69 In addition, the up-regulation of these integrins in breast cancer, glioblastoma, pancreatic tumors, and prostate carcinoma is correlated with increased cell motility and metastasis.70,71 Since RGD peptides are effective in targeting cancer cells (in tissue culture models as well as in mice) through binding and recognition of αvβ3 integrins,68 they serve as promising tools for targeting nanoparticle probes to cancer cells.
One protein that is widely used to detect cancer cells is transferrin, a glycoprotein that binds iron (Fe3+) in the blood with high affinity (10−23 M).72–74 After binding two Fe3+ ions, transferrin is recognized by transferrin receptors, which mediates its uptake into cells via receptor-mediated endocytosis. Transferrin receptors are usually expressed in the basal epidermis, pancreas, hepatocytes, Kupffer cells, testis, and the pituitary gland.75 However, these levels are elevated in various cancers (breast, stomach, colon, kidney, ovarian, lung, pancreas, lymphoma, skin, and bladder) possibly due to the need for increased iron uptake that is usually associated with proliferating cells.73,74 In addition, transferrin is an endogenous protein (2.5 g/mL in normal human serum),76 and is therefore nontoxic and nonimmunogenic when used to bind and detect cancer cells.
Antibodies are widely used in cancer diagnostics in vitro and in vivo since they have been commercialized and are easily procured, have high specificity to their target of interest (both free in solution and on cells), and bind their target with a high affinity (the antigen-binding affinity, Kd, of most antibodies lies in the range 10−6–10−9 M).77–79 In addition, antibodies can be easily conjugated80 to fluorescent dyes81,82 and nanoparticles (AuNPs,83 QDs84), making them perfect candidates for cancer biomarker and cell immunoassays. Antibodies are also widely used for in vivo cancer cell detection due to their relatively low immunogenicity.78 To minimize immunogenicity, smaller fragments consisting of either a single-chain variable fragment85,86 (scFv, a fusion protein of the heavy and light chains) or the fragment-antigen binding87 (Fab) region are used since antibodies containing a nonhuman crystallizable (Fc) region could result in complement system activation. Other types of nonimmunogenic antibodies include chimera antibodies88 (produced by joining a mouse Fab region with a human Fc region) and human antibodies (generated using transgenic mice that have human immunoglobulin genes89,90 or from phage display91). Although antibodies play an important role as targeting moieties for nanoparticles to bind various cancerous biomarkers, cells, or tissues, they also act as effective therapeutics against cancer.78,87,92,93 In fact, there are over 40 FDA-approved antibody drugs available in the United States, and examples of theranostic nanoparticle probes that simultaneously detect and treat cancer in vivo through the use of antibodies will be further discussed in section 6.2.2.
Nucleic acid aptamers, oligonucleotides that bind specific targets of interest (i.e., proteins, small molecules, cells, or other nucleic acids), can also be used as targeting moieties.94 In many cases, aptamers are discovered using a process called “systematic evolution of ligands by exponential enrichment” (SELEX).95,96 In this process, large oligonucleotide libraries (typically 1 × 1014 unique sequences) are created using a single stranded DNA (ssDNA) template consisting of defined 5′ and 3′ ends and a randomized region, typically between 40 and 80 nucleotides in length, within which specific binding motifs can be evolved against proteins and other biomolecules of interest. After generating the library using automated DNA synthesis, a double stranded DNA (dsDNA) library can be generated through polymerase chain reaction (PCR) and used as a starting point for either a DNA or RNA (via transcription with T7 RNA polymerase) in vitro selection process. First the sequences are introduced to a biological target of interest and isolated based on their ability to bind the target through several rounds of in vitro selection. Bound sequences are typically isolated through size exclusion chromatography and other analogous techniques, and then PCR amplified to identify the sequence that binds the target of interest. The bound sequences, along with permutations of new sequences, then go through this cycle 10–15 times, with each cycle yielding more sequences that bind the target of interest with higher specificity. This method generates aptamers that have a high specificity and affinity for their target (Kd of approximately 10−8 M), and may be used to develop aptamers that bind a wide range of targets, including cells (cell-SELEX).97 This makes aptamers ideal candidates for targeting cancer biomarkers and cells for which antibodies may not be available.
4. DETECTION OF EXTRACELLULAR CANCER BIOMARKERS
4.1. Introduction to Biomarkers for Cancer Detection
One promising approach in the early detection of cancer is to identify and detect substances in the blood or other bodily fluids that are correlated with the presence of cancer. These substances, known as biomarkers, may be proteins98–111 (either cell surface glycoproteins or secreted proteins), carbohydrates, 112–114 or nucleic acids115–131 (i.e., genome sequences or RNA transcripts) that are associated with cancerous cells. Measuring the levels of particular cancer biomarkers from a patient’s blood, urine, feces, or saliva could enable the detection of cancer at the early stages of the disease, identification of tumor recurrence, prediction of a patient’s risk to a new or existing cancer, and the ability to monitor a therapy’s efficacy during treatment. However, some primary challenges of early cancer detection include low abundance of biomarkers in plasma at the early stages of the disease,132,133 heterogeneity in the timing and abundance of these biomarkers among patients,110 and difficulties in executing prospective studies (those that are serial in nature, including the collection and storage of prediagnostic samples).134
Research toward the identification of biomarkers that are indicators of cancer has generated thousands of biomarker candidates, but relatively few have been granted FDA clearance (Table 1).113,135 Of the FDA-cleared biomarkers, the majority are utilized for monitoring the progression of cancer, rather than enabling its early detection. However, despite the importance of early detection and the considerable research efforts directed toward it, the use of these assays for early diagnosis of cancer is limited. In fact, no early cancer biomarker assay has been FDA-approved or -cleared, which highlights the challenges of developing sensors for cancer biomarkers.
Table 1.
FDA-Approved or -Cleared Cancer Biomarkersa
| biomarker | cancer type | sample specimen | clinical use |
|---|---|---|---|
| alpha fetoprotein L3% (AFP-L3%) | hepatocellular | serum | risk assessment |
| alpha fetoprotein (AFP) | testicular | serum, plasma | management of cancer |
| cancer antigen 125 (CA 125) | ovarian | serum, plasma | monitoring |
| cancer antigen (CA 15-3) | breast | serum, plasma | monitoring |
| cancer antigen 19-9 (CA 19-9) | pancreatic | serum, plasma | monitoring |
| cancer antigen 27.29 (CA 27.29) | breast | serum | management and prognosis |
| carcinoembryonic antigen (CEA) | not specified | serum | prediction of progression and survival |
| c-Kit | gastrointestinal stromal | FFPE tissue | prognosis, response to therapy |
| estrogen receptor (ER) | breast | FFPE tissue | monitoring |
| fibrin/fibrinogen degradation product (DR-70) | colorectal | serum | discriminate cancer from benign disease |
| human epididymis secretory protein 4 (HE4) | ovarian | serum | assessment for therapy |
| human epidermal growth factor receptor 2 (Her-2/neu) | breast | FFPE tissue | detection of fetal occult blood |
| human hemoglobin (fecal occult blood) | colorectal | feces | diagnosis and monitoring |
| nuclear mitotic apparatus protein (NuMA, NMP22) | bladder | urine | prediction of malignancy |
| ovalbumin (OVA1, multiple proteins) | ovarian | serum | prediction of malignancy |
| p63 protein | prostate | FFPE tissue | aid in differential diagnosis |
| Pro2PSA | prostate | serum | discriminate cancer from benign disease |
| progesterone receptor (PR) | breast | FFPE tissue | prognosis, response to therapy |
| prostate specific antigen (PSA) | prostate | serum | free PSA: monitoring<br1>total PSA: diagnosis and monitoring |
| risk of ovarian malignancy algorithm (ROMA: He4 + CA 125) | ovarian | serum | prediction of malignancy |
| thyroglobulin | thyroid | serum, plasma | monitoring |
FFPE tissue refers to formalin fixed paraffin embedded tissue. Adapted from ref 113.
Although cancer biomarker assay development has faced both fundamental and technical issues, researchers are uniquely positioned to use nanotechnology to address these concerns, as the unique properties of certain classes of nanoparticle probes offer the potential to produce rapid, inexpensive, tailorable, high-throughput assays with high sensitivity and selectivity. Below, we will discuss nanoparticle-mediated techniques for the detection of biomarkers separated by the method of analyte detection. Biomarkers targeting bladder, breast, bone, cervical, colorectal, gastric, hepatocellular, lung, pancreas, prostate, ovarian, and thyroid cancer, will be covered in this section.
4.2. Detection of Cancer Biomarkers Using Quantum Dots
QDs are especially promising for the detection of cancer biomarkers by fluorescence due to their high quantum yields, large molar extinction coefficients, and tunable emission maxima, all of which are advantageous for reducing an assay’s limit of detection.28 Recent examples of QD-based biosensors for cancer biomarker detection are reviewed below.
The most common design motif for the detection of biomarkers is a sandwich-type assay, which consists of several components: substrate, capture antibody, analyte of interest (biomarker), a second capture antibody, and a secondary antibody, usually tagged with a fluorescent probe. With such an assay, the immobilized monoclonal primary antibody binds to the biomarker. Next, a second capture antibody, specific for the biomarker, is introduced and sandwiches the target. A secondary antibody, usually fluorophore-labeled, binds to the second primary antibody, thus generating a fluorescent signal that is detected using microscopy or a fluorescence spectrophotometer. These sandwich-type assays have high specificity due to the high affinity and selectivity a primary antibody has for its analyte, and high sensitivity when QDs are used to fluorophore-label the secondary antibody due to the QD’s-conjugated antibodies’ intense signal.
Sandwich-type assays can occur both on a substrate and free in solution. Suspension assays are advantageous because they exhibit faster kinetics in solution compared to assays performed on a substrate, allowing for faster readouts and higher sample throughput.136 These homologous assays can be easily tailored for detecting various analytes, and do not require optimization for immobilizing antibodies or antigens or extensive wash steps to separate bound versus unbound moieties.
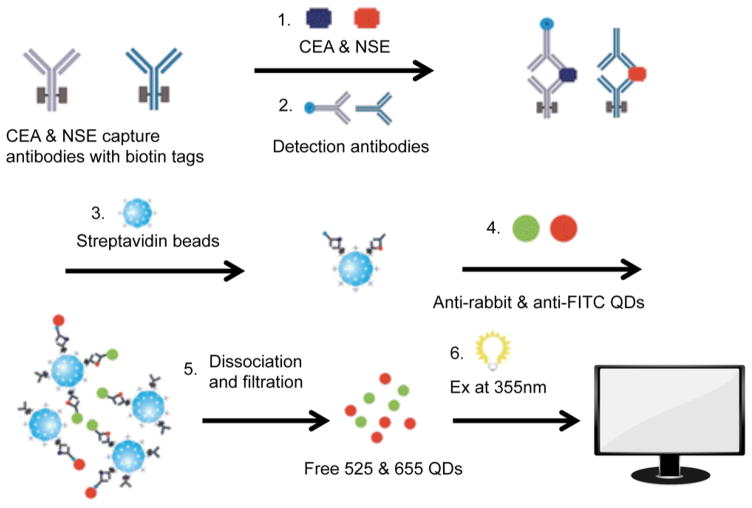
One example of a homogeneous in-solution assay was reported by Li et al., who used QD-conjugated antibodies to detect two biomarkers: carcinoembryonic antigen (CEA, one of the most widely studied cancer biomarkers to monitor anticancer treatments and predict tumor recurrence postsurgical resection in patients with late-stage cancer)137–139 and neuron specific enolase (NSE, an enzyme that catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate140 and is associated with small cell lung carcinoma, carcinoids, islet cell tumors upon secretion at a concentration over 15 ng/mL141–143), each with a limit of detection of 1.0 ng/mL (Figure 5).144 Since the levels of these biomarkers are relatively low (on the order of several nanograms per milliliter), it is important to develop assays with an even lower limit of detection to accurately monitor these biomarkers in patients. Cao and co-workers improved upon this fluoroimmunoassay by immobilizing capture antibodies on polystyrene microspheres instead of streptavidin beads to reduce the limit of detection of each of the two biomarkers to 0.625 ng/mL.145
Figure 5.
Homogenous in-solution sandwich assay for detecting CEA and NSE. (1, 2) Biotinylated capture antibodies and QD-functionalized detection antibodies against each biomarker bind the analyte of interest to form a sandwich. (3) Streptavidin beads are then used to capture the in-solution sandwich constructs. (5, 6) Finally, QDs are freed from the sandwich, and detected using a plate reader (fluorophore excitation at 355 nm). Adapted with permission from ref 144. Copyright 2011 The Royal Society of Chemistry.
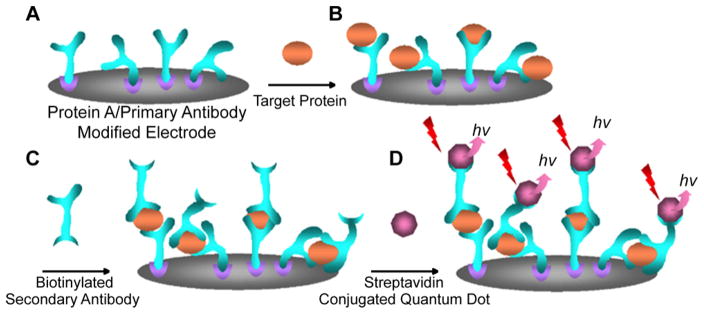
Another type of QD-based immunosensor consists of either an analyte or a capture antibody immobilized onto a surface commonly composed of glass, silicon, or gold.146–152 Unlike homogeneous in-solution immunoassays, these heterogeneous immunosensors require small amounts of patient samples and can provide rapid and high-throughput detection of analytes of interest.146 In one study, Kerman and co-workers developed an immunosensor that could detect total prostate specific antigen (TPSA) in human serum samples with a detection limit of 0.25 ng/mL (Figure 6).150 Prostate specific antigen (PSA), one of the most widely tested biomarkers for prostate cancer, is a glycoprotein produced by the prostate gland that is part of the kallikrein-related peptidase family (serine proteases) and exists in multiple forms in the body (e.g., free and bound to serum proteins).153 Since high PSA concentrations are not always indicative of prostate cancer due to biodiversity in the patient population,98,154–159 PSA velocity, the rate of increase in PSA levels over time, can be monitored since a significant increase is indicative of prostate cancer.160,161 While neither types of PSA monitoring can serve as long-term predictors of prostate cancer, they serve as valuable and definitive indicators in monitoring changes over time to detect cancer recurrence or treatment efficacy.98,153–161
Figure 6.
Scheme for a QD immunosensor utilized to detect TPSA. (A) Protein A and an anti-TPSA antibody are immobilized onto a screen-printed carbon substrate. (B) Upon introduction to the sensor, the analyte binds to the capture antibody and (C) a biotinylated a second antibody. (D) Streptavidin functionalized QDs sandwich the analyte onto the sensor, and produce fluorescence with an emission at 525 nm. Adapted with permission from ref 150. Copyright 2007 Elsevier.
In order to analyze biomarkers in a high-throughput fashion, Gokarna and co-workers developed QD-based protein microand nanoarrays for the detection of PSA.147 The PSA microand nanoarrays were fabricated using dip pen nanolithography (DPN)162–176 and introduced to PEGylated QDs functionalized with an anti-PSA antibody. The presence of PSA (marked by QD fluorescence) was evaluated using a microarray scanner. Since these micro- and nanoarrays allow for rapid screening of many compounds, they may enable high-throughput multiplexed screens for clinical use. In order to improve the sensitivity (to less than 0.5 pM or 0.1 ng/mL) and speed (to 15 min) of biomarker detection, Mukundan et al. used a waveguide based biosensor to detect CEA.151,152 First, anti-CEA monoclonal antibodies were immobilized onto the sensor surface and bound to CEA present in human serum patient samples. Next, CdSe/ZnS core/shell QD-conjugated anti-CEA monoclonal antibodies sandwiched the immobilized biomarker–antibody complex, generating a fluorescent signal as quantified by a fiber optic spectrometer.
Microfluidic devices are often used in clinical assays to reduce the amount of patient sample needed for the detection of cancer biomarkers based upon the ability to precisely control fluids on a small scale.177–179 These “lab-on-a-chip” technologies operate under the premise that large laboratory experiments can be conducted on a small milli- or centimeter size scale,177–179 allowing for cost-effective and faster diagnoses.180–184 For example, Hu et al. used a polydime-thylsiloxane (PDMS) microfluidic device to detect alpha fetoprotein (AFP, a plasma protein found mostly in the fetus that is the most widely researched biomarker for the post-treatment prognosis of hepatocellular carcinoma)185,186 and CEA at a limit of detection of 0.25 nM for each analyte (both individually and in combination).180,183 To do so, capture antibodies for CEA or AFP were immobilized on the device surface, before the analyte was introduced. Next, anti-CEA or anti-AFP primary antibodies and QD-functionalized secondary antibodies bound to the newly immobilized biomarker. Finally, the biochip was imaged using fluorescence microscopy.
To improve upon the detection limit of cancer biomarkers, Jokerst and co-workers developed a microfluidic microporous agarose bead array to detect CEA both individually and in combination with cancer antigen 125 (CA 125, a cell surface glycoprotein that is overexpressed in patients with ovarian cancer and is used as a late-stage prognosis and monitoring tool)139,187–189 in patient blood and saliva samples.181 To do this, capture antibodies are immobilized onto agarose beads. Samples are then introduced, and the target antigen is captured by the immobilized antibodies. Detection antibodies conjugated with QDs then sandwiches the antigen and provides a means for measuring its presence. The fluorescence intensity of photomicrographs of the fluorophore-labeled beads corresponds to the analyte concentration, and the limit of detection of CEA was determined to be 20 pg/mL or 0.11 pM. Hu et al. are able to achieve an even lower limit of detection for CEA (50 fM) in an analogous assay.182
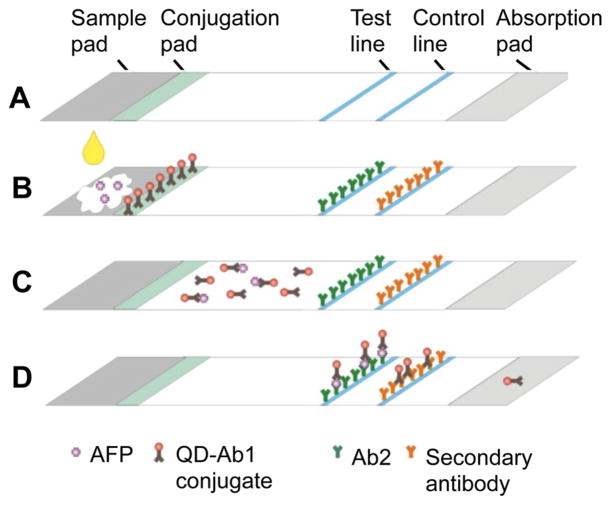
One type of microfluidic device for cancer biomarker detection is an immunochromatography test strip (ICTS), which takes advantage of capillary action to carry sample through the strip.190–192 These test strips are usually composed of either paper or membrane, further reducing the cost of production and analysis, as they do not require extra machinery (i.e., pumps are normally required for microfluidic devices) or expensive imaging equipment (homemade test strip readers are available).193–195 The setup of an ICTS usually includes a sample pad (where sample is loaded), a conjugation pad (antibodies bind analyte of interest), a test line (where fluorophore-tagged antibody–analyte sandwiches are immobilized), and a control line (a positive control to confirm the fluorescence of fluorophore-tagged antibodies). In one study, Yang and co-workers developed an ICTS to detect AFP with a limit of detection of 1 ng/mL (Figure 7).191 Another ICTS assay was developed by Cheng et al. to detect C-reactive protein (CRP, a protein secreted from the liver as a marker of inflammation that is associated with colorectal and lung cancer in addition to diabetes and cardiovascular disease)196 with a limit of detection of 0.63 U/mL.192
Figure 7.
(A) Immunochromatography test strip for AFP detection. (B) AFP-containing sample is loaded onto the sample pad, and binds QD antibodies (QD-Ab1 conjugates) on the conjugation pad. (C) Next, AFP-QD-Ab1 travels to the test line and binds to immobilized anti-AFP antibodies. (D) Unbound QD-Ab1 conjugates bind to a secondary antibody. Fluorescence along the test and control line is quantified using a fluorescence reader. Adapted with permission from ref 191. Copyright 2011 Elsevier.
FRET-based biosensors take advantage of FRET, a phenomenon in which energy is transferred from a donor fluorophore to an acceptor fluorophore (as a result of their spectral overlap, their spatial proximity, and their relative orientation), due to its high sensitivity of detection and high signal-to-noise ratio. There are two types of FRET-based sensors: those that turn “on” in the presence of analyte197–202 (if (1) both chromophores are fluorescent, but when they bind the analyte, FRET occurs, resulting in energy transfer to the acceptor chromophore and a measurable increase in fluorescence at the acceptor’s emission wavelength or if (2) a quencher absorbs the excitation energy of a donor chromophore, but is released upon binding of analyte, resulting in FRET between the donor and acceptor chromophore) and those that turn “off” in the presence of analyte203 (a biomolecule binds and FRET no longer occurs, resulting in a decrease in fluorescence emission by the acceptor). QDs are ideal for FRET due to their tunable emission, broad absorption, and long fluorescence lifetime, such that more than one dye could act as an acceptor fluorophore with one excitation event.204
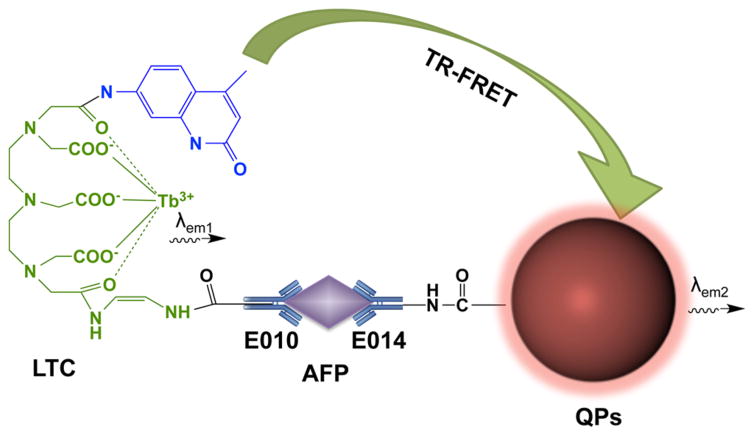
Wei and co-workers developed an in-solution sandwich fluoroimmunoassay to detect estrogen receptor beta (ER-β antigen, a tumor suppressor that is down-regulated in the later stages of various cancers and can be used to monitor treatment efficacy)205,206 utilizing FRET.197 ER-β antigen is incubated with QD-labeled anti-ER-β monoclonal antibody and Alexa Fluor labeled anti-ER-β polyclonal antibody, forming a sandwich. The proximity of the QD 565 (donor) and the Alexa Fluor dye (acceptor) enables FRET, and results in an increase in Alexa Fluor fluorescence, which may be measured using confocal microscopy. This assay is rapid (only a 30 min incubation time), simple, and sensitive (limit of detection was 0.05 nM or 2.65 ng/mL). Another in-solution fluoroimmunoassay was developed by Chen and co-workers to detect AFP with a limit of detection of 0.4 ng/mL using a luminescent terbium chelate (LTC)–QD FRET pair (Figure 8).198 In a similar setup, Wegner et al. designed an immunoassay that also utilized a terbium–QD FRET pair to detect cancer biomarkers. In this case, six different primary antibodies were used to bind a model biomarker (PSA), enabling PSA detection at concentrations as low as 1.6 ng/mL.200
Figure 8.
In-solution FRET fluoroimmunoassay to detect AFP. QDs are incorporated onto the surface of polymeric microparticles (QPs) that are conjugated to an anti-AFP antibody. Luminescent terbium chelates (LTC) are also conjugated to an anti-AFP antibody, and in the presence of AFP, the target is sandwiched, and brings the QDs and LTC in close proximity to each other, initiating FRET. Adapted with permission from ref 198. Copyright 2012 Elsevier.
Kim and co-workers developed a QD-based sandwich immunoassay on a glass substrate consisting of a vertical zinc oxide (ZnO) nanowire array for the detection of CEA.201 The ZnO nanowire substrate provides a large surface area with many binding sites for capture antibodies to bind the analyte of interest. FRET occurs between ZnO nanowires and QD labeled detection antibodies upon sandwich formation due to the presence of CEA. The fluorescence enhancement is quantified using fluorescence microscopy, with a large dynamic range for detecting CEA (from 0.001 to 100ng/mL).
One multimodal biosensor that utilizes electron transfer to a quencher in the absence of an analyte was developed by Jou et al., who takes advantage of both FRET and chemiluminesence resonance energy transfer (CRET) to enhance the sensitivity of detecting micro-RNA-141 (miR-141),202 a nucleic acid biomarker that is found in the blood and is indicative of prostate, ovarian, and gastric cancers.202,207–210 In this assay, a FRET quencher is covalently bound to nucleic acid functionalized CdSe/ZnS QDs and is released upon miR-141 binding, resulting in an increase in fluorescence that enables the detection of miR-141 with a limit of detection of 1 pM. To further increase the sensitivity of analyte detection, the QDs are introduced to G-quadruplex-forming telomerase and dNTPs. Next, hemin is added to intercalate into the G-quadruplexes on the QD, which catalyzes the oxidation of luminol by H2O2, resulting in CRET and reducing the limit of detection of miR-141 to 0.28 pM.
Ge and co-workers developed a paper-based immunosensor device to detect AFP, CA 125, CA 15-3 (a biomarker also derived from mucins that is used to detect tumor recurrence in breast cancer patients),139,211–213 and CEA with detection limits of 0.3 pg/mL, 6.1 × 10−5 U/mL, 2.9 × 10−4 U/mL, and 1.4 pg/mL, respectively, through the use of a QD-FRET system.199 First, antibodies immobilized onto the paper sensor and copper oxide (CuO) nanoparticle–antibody conjugates sandwich the analyte. Next, dithizone (DZ)-quenched CdTe QDs are added to the sensor surface. Finally, the sensors are treated with HCl, which releases Cu2+ from the CuO NPs in the presence of analyte and results in the recovery of QD fluorescence.
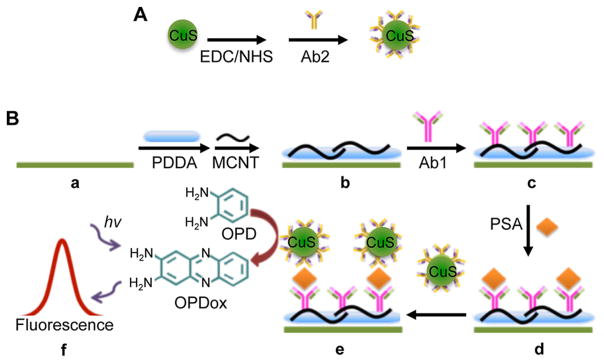
QDs can also be used as an oxidizing agent for the detection of cancer biomarkers using a sandwich assay. In one such study, Zhu et al. developed an immunosensor to detect PSA by using CuS QDs to oxidize a small molecule (o-phenylenediamine, OPD) to generate a fluorescent signal (Figure 9).214 The fluorescence of the oxidized small molecule 2,3-diaminophenazine (OPDox) was measured using fluorescence spectroscopy to detect PSA with a limit of detection of 0.1 pg/mL. High specificity for PSA was obtained even in the presence of 10 ng/mL of interfering substances, such as serum proteins. This CuS-based fluorescent immunosensor compares favorably to nonfluorescence based immunosensors (electrochemical, colorimetric, electrochemiluminescence, and ELISA based).215–218
Figure 9.
Multiwalled carbon nanotube (MCNT) immunosensor for PSA detection. (A) First, CuS QDs are functionalized with an anti-PSA antibody while (B) (a–c) ITO substrates are functionalized with carbon nanotubes and poly(diallyldimethylammonium chloride) (PDDA) before anti-PSA capture antibodies are immobilized to the surface. (B) (d, e) Next, the anti-PSA capture antibodies and the CuS QDs sandwich PSA, resulting in the oxidation of o-phenylenediamine (OPD) to 2,3-diaminophenazine (OPDox), thus producing a fluorescence signal. Adapted with permission from ref 214. Copyright 2014 The Royal Society of Chemistry.
4.3. Detection of Cancer Biomarkers Using Gold Nanoparticles
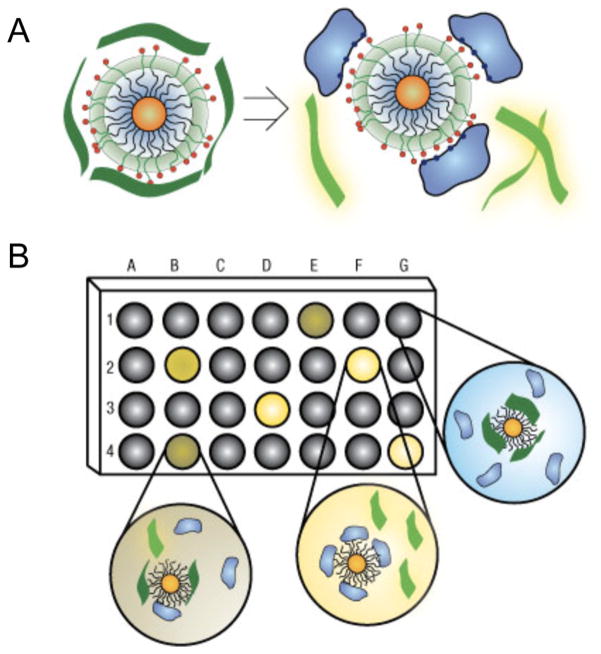
Many biosensors utilize AuNPs as fluorescence quenchers due to their strong absorbance, which can yield sensors with lower background signal than those utilizing organic fluorophore quenchers. For example, You et al. developed a “turn-on” nanoparticle probe for detecting cancer biomarkers based on nonspecific electrostatic adsorption of protein biomarkers to a library of fluorophore-tagged AuNPs. This system was used to detect two biomarkers: acid phosphatase and alkaline phosphatase, both of which are isoenzymes found in various organs that are indicative of cancer when up-regulated. 212,219–221 (Figure 10).222 Though this assay is capable of distinguishing between multiple protein biomarkers, it is not possible to design the system to detect any arbitrary target without screening large AuNP libraries for specificity to the desired target.
Figure 10.
Schematic representation of a nanoparticle probe for the detection of various cancer biomarkers. (A) This in-solution assay uses a library of AuNPs capped with various cationic functional groups that is then coated with an electrostatically associated fluorophore-labeled polymer (poly(p-phenyleneethynylene) or PPE). In the absence of target, the AuNP quenches the fluorescence of the fluorophore, and the sensor is in an “off state.” Binding of various proteins to the AuNP can trigger the dissociation of PPE from the nanoparticle, resulting in a fluorescence enhancement and the transition of the nanoparticle probe to an “on state”. (B) In a typical experiment, one AuNP from a library is contained within each of the wells of a microplate, and a protein sample is added to each well. Due to the differences in the surface charge of various proteins, the biomarkers that are analyzed bind to each of the AuNPs in the library to varying extents, thereby generating a unique “fingerprint” that enables their differentiation through fluorescence spectroscopy. Reprinted with permission from ref 222. Copyright 2007 Macmillan Publishers Ltd.
In order to develop a detection system for platelet derived growth factor (PDGF, a growth factor that is associated with various cancers when up-regulated)223–226 with high specificity, Huang et al. used aptamers in a similar “off–on” solution-based assay.227 In this case, the fluorescence of a small molecule intercalator, N, N′-dimethyl-2,7-diazapyrenium dication (DMDAP), is quenched in the absence of analyte by the AuNP, and it is restored upon introduction of PDGF, which releases DMDAP. The limit of detection for this assay was 8 pM. In a similar study, Cheng and co-workers used the analyte of interest hyaluronidase (HAase, an endoglycosidase that degrades hyaluronic acid, and is indicative of high-grade tumors in bladder cancer patients when elevated228–230) to “turn on” fluorescence from the nanoparticle probe.231 The limit of detection was 0.625 U/mL from urine samples.
Rather than using AuNPs as fluorescence quenchers, Cho and co-workers took advantage of surface enhanced fluorescence between AuNPs and Cyanine 3B (Cy3B), an organic fluorophore, to detect vascular endothelial growth factor (VEGF, another growth factor that is implicated in various cancers when elevated)223,224,232–235 in an “on–off” nanoparticle probe.236 Specifically, Cy3B-labeled aptamers targeting VEGF were electrostatically associated onto 80 nm AuNPs using positively charged poly-L-lysine. Upon VEGF addition, the aptamer dissociated from the nanoparticle surface to bind VEGF, causing a decrease in fluorescence intensity as measured by a fluorescence microscope.
4.4. Detection of Biomarkers via Fluorophore-Labeled Nanoparticles
Fluorophore-labeled nanoparticles can also be used to detect cancer biomarkers. For example, Khazanov et al. fabricated a microarray biosensor to detect alpha 1-antitrypsin precursor (AIAT, a protease inhibitor that is a biomarker for gastric cancer)237 using boradiazaindacene (BODIPY, 660–680) labeled nanoparticles.238 Trypsin was grafted onto a glass surface to capture AIAT. Next, BODIPY NPs conjugated to anti-AIAT antibodies bound to the analyte, resulting in a measurable increase in fluorescence as monitored by confocal laser-scanning microscopy. The limit of detection for this assay was 10 μg/mL.
5. DETECTION OF CANCER CELLS
5.1. Cancer Metastasis and Circulating Tumor Cell Detection
Early detection of cancer enables timelier treatment, and significantly improves patient outcomes. As cancer develops, metastasis may occur, which can drastically diminish cancer patient prognosis. Current clinical methods to diagnose cancer metastasis, such as imaging via computerized tomography, rely on the detection of secondary tumors.239–241 Unfortunately, this presents a significant limitation in the current state of cancer patient care, since treatments administered after metastasis has already occurred are less effective and are associated with poorer survival rates.240–242 As a result, the need to develop novel tools to identify cancer metastasis before the formation of secondary tumors is strong.
Metastatic tumor formation occurs through the spread of cancer cells from the primary tumor site to the bloodstream and lymphatic system, and to new sites at distal organs and tissues to form secondary tumors.243,244 Circulating tumor cells (CTCs) are key in this process, and detecting their presence in the blood early provides a means to predict metastatic potential before the formation of secondary tumors.239–241 Studies have indicated that the presence of CTCs before cancer treatment begins correlates with poorer survival rates in patients with metastatic cancer, and that the presence of CTCs after treatment suggests a higher likelihood of tumor relapse.4 Based on this trend, the ability to isolate and detect cancerous cells in the bloodstream would enable the assessment of metastatic risk and may provide the opportunity to begin treatment prior to the development of a secondary tumor to improve cancer patient outcomes. To achieve this, however, significant effort must be afforded to probe the correlation of CTC presence with cancer patient prognosis. Additionally, quantification of CTCs from individual patients throughout their treatment plan may serve as a means to monitor the patient’s response through a simple blood test.
The identification of cells as cancerous is challenging, since cancer emerges from abnormal growth and differentiation of healthy tissue, and is a highly heterogeneous disease that may be manifested in significant differences in gene expression from cell to cell.245–247 Nonetheless, in general, cancer cells do indeed differ from healthy cells, and there are many known biomarkers that may be used to differentiate between the two populations. Existing technology to detect CTCs relies on the immunomagnetic separation of cells based upon their expression of cell surface antigens, such as epithelial cell adhesion molecule (EpCAM), which is a transmembrane protein involved in cell signaling, migration, and proliferation that is implicated in cancer progression.248,249 While this approach has been FDA-cleared and commercialized through CellSearch technology (Janssen Diagnostics), it is limited by the need to recognize cell surface antigens and can result in the inability to distinguish CTC populations whose protein expression level is not sufficient for isolation, and it does not enable the quantification of gene expression.249
Beyond the recognition of cell-surface markers, monitoring the intracellular expression levels of certain genes may also be used to distinguish cancerous cells from healthy cells. Cancer cells tend to share high expression levels of classes of genes (oncogenes) involved in survival, proliferation, and differentiation. 4,250–252 Over the past several years, researchers have identified several crucial genetic changes that cause cancer cells to become more metastatic in nature. Recent studies have indicated that cancer cells undergo the epithelial to mesenchymal transition (EMT) during metastasis253,254 and that this process results in the loss of epithelial cellular markers and the development of mesenchymal cellular markers.253–256 This process typically occurs in development and in wound healing, but cancer cells use this process to enable their extravasation from tumor tissue into the bloodstream.257,258 Expression of cell adhesion proteins, such as E-cadherin, is suppressed to facilitate the metastatic cancer cell’s dissociation from neighboring tumor cells. Meanwhile, the expression of cell migration proteins such as N-cadherin, vimentin, and fibronectin is up-regulated, causing the cell to invade the bloodstream and begin the metastatic process.254 Based on the known differences in the genetic expression of metastatic cancer cells compared to healthy cells, quantification of the intracellular expression profiles of EMT markers enables the identification of cells that present a risk of cancer metastasis.
Many of the current methods available to study cancer cell populations, such as quantitative real time PCR (qRT-PCR) and the enzyme-linked immunosorbent assay (ELISA), interrogate bulk cell samples, providing information on the population average. Therefore, these techniques are not capable of monitoring sample heterogeneity. This is a serious limitation in the study of cancer cell populations, which inherently vary significantly in gene expression. In particular, a small subset of circulating tumor cell populations, known as cancer stem cells (CSCs), is thought to initiate cancer metastasis and increase the risk of recurrence.259–261 This subset of cells differs in gene expression from the majority of tumor cells, and they may be unidentifiable using conventionally available techniques such as qRT-PCR and ELISA that provide population averages and do not offer single-cell resolution.
Certain classes of nanoparticles are uniquely suited for the design of probes that address the challenges traditional methods face in detecting small populations of cancer cells via the generation of a fluorescent output. For example, QDs and UCNPs conjugated to moieties that bind and recognize cancer cell surface markers offer several important advantages over organic dyes,262–281,243 including high quantum yields, size-dependent emission tunability, and enhanced photostability. 30 In addition, noble metal nanoparticles and AuNPs, in particular, are commonly used in combination with organic fluorophores to design “off–on” probes for intracellular gene expression analysis. These probes exhibit lower background signal than traditional molecular approaches due to the efficient quenching capabilities of AuNPs.282–284
Based upon their fluorescent output, the systems highlighted in this section may utilize a variety of techniques for analysis, including microscopy and flow cytometry. Importantly, flow cytometry provides the unique capability of interrogating single cells, while surveying large samples. This ability is critical in the study of cancer cells, which are quite heterogeneous, and improves upon the currently available technologies that may be unable to distinguish CSCs from a diverse tumor cell population or resolve relatively scarce populations of cancer cells from large populations of healthy cells. In addition, the use of fluorescence activated cell sorting (FACS) enables isolation of cancer cells detected with a fluorescent readout, providing the opportunity for further downstream analysis. Notably, biocompatible nanoparticle probes maintain the health of suspected tumor cells, enabling additional studies that may be used to develop effective treatment plans for individual cancer patients based upon the gene expression profile of the tumor cell population. The recent developments in the use of nanoparticle probes for cancer cell detection significantly expand upon the existing approaches to detect cancer cells, and represent promising advances in the early detection of metastatic cancer with potential impacts for cancer patient prognosis in the clinic.
5.2. Detection through Cell Surface Protein Marker Recognition
Many groups have explored the use of nanoparticles for the detection of cancer cells through cell surface marker recognition using fluorescence. Targeting moieties, such as antibodies,262–276 aptamers,277–281 and small molecules,285–295 can be conjugated to the surface of the nanoparticle probe to induce selective binding to or uptake by cancer cells, fluorophore-labeling them for easy identification over noncancerous cells. In these approaches, a variety of fluorescent nanoparticles ranging from UCNPs271,272,296 and QDs267,274,277,281,293 to silica nanoparticles with fluorescent dyes encapsulated264,270,279,288 are used. We will divide recent advances in this field first by the core composition of the nanoparticle probe and then by the moieties that are used to recognize and bind cancer cells.
5.2.1. Types of Nanoparticles Used To Fluorophore-Label Cancer Cells
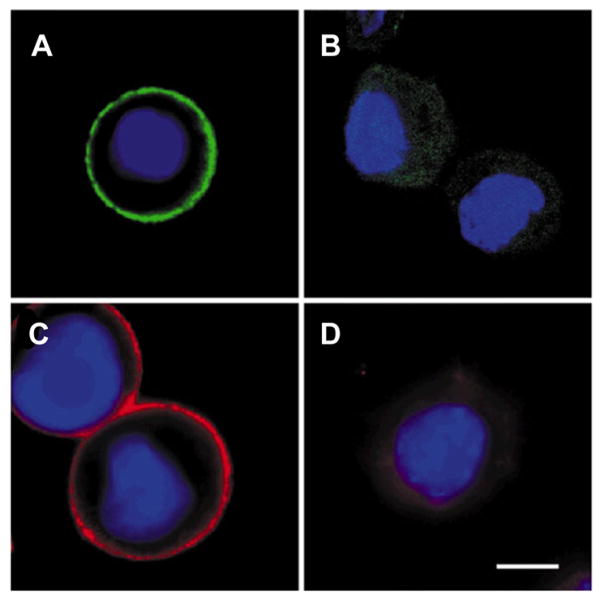
As previously described, QDs exhibit unique optical properties that make them useful for detecting cancer cells. In particular, the detection of cancer cell populations that are present in low abundances greatly benefits from the use of QDs due to their high quantum yields. Wu et al. first reported the use of QDs in the detection of breast cancer cells.274 In their approach, antibody-conjugated QDs were used to label Her2 in fixed SK-BR-3 breast cancer cells (Figure 11). In addition to labeling cancer cells, the QDs were also used to detect Her2-expressing cells in fixed mammary tissue isolated from transgenic mice, demonstrating the potential use of QDs in the pathological and histological diagnosis of cancer. Since this work, other groups have also used QDs in cancer cell detection, expanding the platform to target alternate cancer cell types.267,274,276,277,281,293
Figure 11.
(A, C) Fixed breast cancer SK-BR-3 cells labeled with anti-Her2 535-nm-emitting QDs and anti-Her2 630-nm-emitting QDs. (B, D) SK-BR-3 cells treated with IgG coated 535-nm-emitting QDs and 630-nm-emitting QDs were not specifically labeled. Cell nuclei were stained with Hoechst 33342 (blue), and the scale bar represents 10 μm. Adapted with permission from ref 274. Copyright 2003 Macmillan Publishers Ltd.
Conjugated polymer nanoparticles (CPNs) are sometimes useful for the detection of cancer cells due to their optical properties, which may be tuned by altering the conductive polymer composition, as well as their tailorable surface chemistry, which may be modulated to present various moieties that bind cancer cells.18 However, many water-soluble conductive polymer nanoparticle probes suffer from low fluorescence quantum yields.297,298 To overcome this, the McNeil group developed PDs from a variety of conductive hydrophobic polymers to yield nanoparticles that have high quantum yields, are photostable, and are nontoxic.17 As a result, PDs are great candidates for detecting circulating tumor cells. In one study, Wu et al. demonstrated the use of PDs to target and fluorophore-label MCF-7 breast cancer cells through antibody recognition.45 PD-labeled MCF-7 cells exhibited 25 times higher fluorescence than QD-labeled cells, and 18 times higher fluorescence than Alexa Fluor labeled cells, as analyzed through flow cytometry. In another example, semiconducting fluorescent polymer nanoparticles were encapsulated in PLGA (230–260 nm in size) and functionalized with a Her2 antibody to preferentially identify SK-BR-3 (high Her2 expressing) cells over MCF-7 and NIH/3T3 (low Her2 expressing) cells both on a substrate and free in solution.46
Rare earth element doped fluorescent UCNP probes have also been used for cancer cell detection.271,272,296 In these examples, UCNPs are often chosen for fluorescent labeling based upon the ability to excite UCNPs with near-infrared (NIR) to infrared (IR) light to generate fluorescent emission in the visible region (through multiphoton mechanisms299), resulting in minimization of background noise that is often observed due to cellular autofluorescence.300 This property is especially advantageous in the detection of cancer cells, which often depends upon the ability to distinguish between a small subset of malignant cells among a larger population of healthy cells.
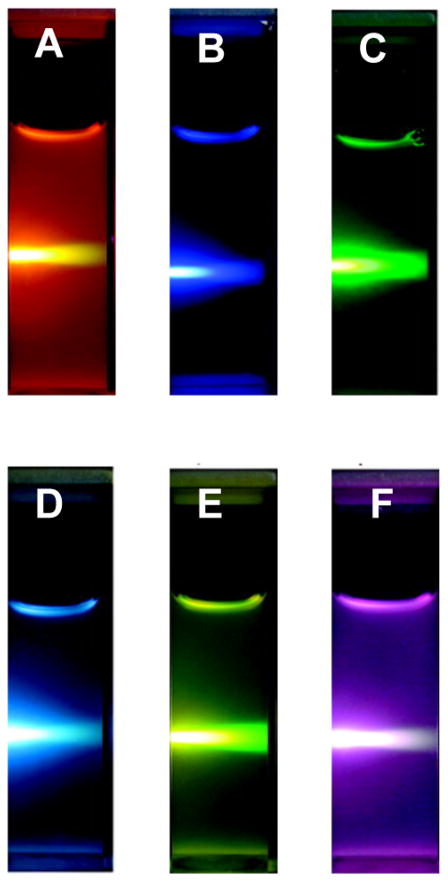
In addition, the tunable emission profiles of UCNPs enable simultaneous multicolor imaging to detect cancer cells. Wang et al. reported tuning NaYbF4 UCNP composition to generate UCNPs that emit orange, yellow, green, cyan, blue, or pink fluorescence upon excitation with 980 nm NIR light (Figure 12).272 These UCNPs were used to label HeLa cervical cancer cells based on CEA antibody recognition. Using the tunable emission profiles of UCNPs, it is possible to simultaneously label several cancer cell surface proteins to detect malignant cancer cells based upon the presence of multiple cancer markers.
Figure 12.
Photographs of 1 wt% colloidal solutions of NaYbF4:Er/Tm/Ho upconversion nanoparticles of (A) NaYbF4:2% Er, (B) NaYbF4:2% Tm, (C) NaYbF4:2% Ho, (D) NaYbF4:1% Tm, 1% Ho, (E) NaYbF4:1% Er, 1% Ho, and (F) NaYbF4:1% Er, 1% Tm excited with 980 nm near-infrared light. Despite their differing emission profiles (as seen by the different colors in the photographs), all samples can be excited with the same wavelength of near-infrared light. Adapted from ref 272. Copyright 2009 American Chemical Society.
Biocompatible silica nanoparticle probes have also been used for fluorescence-based detection of cancer cells.264,270,279,288 In most cases, silica nanoparticles are doped with a fluorophore and coated with a targeting moiety to bind a cancer cell specific biomarker. Though these approaches may not offer the enhanced photostability and high quantum yields that QD and UCNP probes do, the surface functionalization of silica nanoparticles enables multivalent interactions with target receptors that tag cancer cells with multiple fluorophores to generate brighter signals. Tao et al. reported the use of either Rhodamine 6G-doped or tris(2, 2′-bipyridyl)-dichlororuthenium(II) (Rubpy)-doped mesoporous silica nanoparticles for the detection of 7721 liver cancer cells based upon CD155 antibody recognition.270 Similarly, Huang et al. reported the detection of ovarian cancer cells (SKOV-3) using tetramethylrhodamine-doped silica nanoparticles through antibody targeting.264
Deng and co-workers expanded upon the use of dye-doped silica nanoparticles for the detection of cancer cells by monitoring the change in fluorescence anisotropy (using fluorescence polarization anisotropy (FPA)) upon binding of the nanoparticle probe to cancer cells.279 In their approach, methylene-blue-encapsulating silica nanoparticles were used to detect T-cell acute lymphoblastic leukemia cells. Since anisotropy changes with respect to the rotational time constant of the fluorophore, binding of the dye-doped silica particles to a comparatively large cancer cell results in a measurable change in anisotropy. Target T-cell acute lymphoblastic leukemia cells were spiked into healthy blood samples, and the technique was shown to have a linear range of detection from 4000 to 70 000 cells/mL of whole blood. Though the use of FPA may enable more sensitive detection, it requires more sophisticated instrumentation than standard fluorescence spectroscopy, limiting its potential for use in clinical cancer diagnosis.
In addition, both single-walled273 and multiwalled290 carbon nanotubes (SCNTs and MCNTs) have been used for detecting cancer cells by fluorescence. Though the quantum yield of SCNTs is low (3–8%),301–303 their NIR fluorescence emission is strong enough to enable selective in vitro labeling and fluorescent imaging of cancer cells. In a report by Welsher and co-workers, antibodies against a cancer cell specific marker, CD20, were conjugated to SCNTs.273 The system was used to distinguish between T cell and B cell lymphoma cells based on the selective recognition of the antibody-labeled SCNTs by B cell lymphoma cells due to their overexpression of the CD20 cell surface marker.
Beyond using nanoparticle probes to fluorophore-label cancer cells, nanoparticles can be used for simultaneous fluorescent and magnetic resonance imaging of cancer cells (refs 244, 262, 268, 269, 275, 280,286, 293, 295, 304–307). Typically, iron oxide core nanoparticles are modified with fluorescent dye molecules either covalently or noncovalently to provide two parallel methods of tracking nanoparticle localization. In one such example, Wang et al. reported a layer-by-layer assembly method to coat Fe3O4 nanoparticles with poly(amidoamine) dendrimers conjugated to fluorescein isothiocyanate (FITC) and folic acid.295 In their system, the iron oxide nanoparticle core was used in parallel with the FITC label to determine their specificity in targeting KB cells (a HeLa contaminant papillary carcinoma cell line that overexpresses folate receptors) through magnetic resonance and fluorescence imaging.
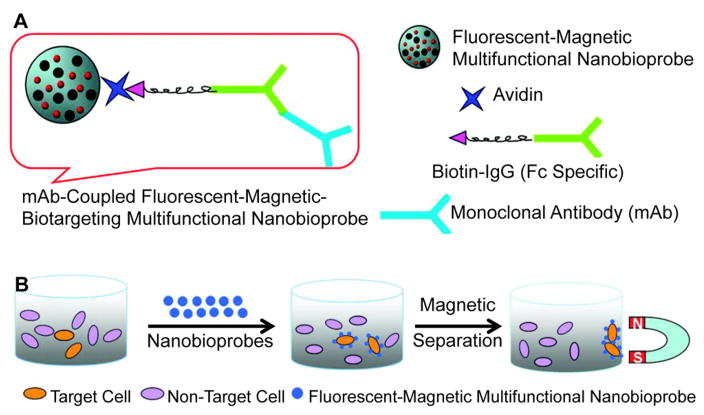
In addition to providing a secondary method of tracking nanoparticle localization, magnetofluorescent nanoparticles enable the simultaneous detection and isolation of cancer cells through magnetic separation.266,268,286,304–306,308 A 2011 study by Song et al. described the use of fluorescent-magneticbiotargeting-multifunctional nanobioprobes (FMBMNs) in detecting and isolating leukemia (Jurkat T) and prostate cancer (LNCap) cells (Figure 13).268 FMBMNs targeting either prostate specific membrane antigen (overexpressed by LNCap cells) or CD3 (overexpressed by Jurkat T cells) were able to detect and isolate target cancer cells through magnetic separation, even with only 0.01% target cells present in a mixture including noncancerous cells.
Figure 13.
Fluorescent magnetic bifunctional nanoparticles (FMBNs) used in the simultaneous fluorescence detection and magnetic isolation of target cancer cells. (A) FMBNs consist of CdSe/ZnS core/shell QDs and Fe2O3 nanoparticles that are encapsulated in a copolymer nanosphere. Biotinylated monoclonal antibodies against a target protein are recognized by the avidin-conjugated FMBNs through avidin–biotin interactions. (B) Upon binding of the FMBNs to target cells, magnetic separation may be performed to isolate the cancer cells that express the cell surface marker of interest. Adapted from ref 268. Copyright 2011 American Chemical Society.
In some cases, nanoparticle probes are used in the design of “off–on” fluorescent sensors for the detection of cancer cells. AuNPs, in particular, are commonly used due to their ability to efficiently quench fluorescence and decrease background signal in the “off state”.263,287,309 Generally, a fluorophore is held in close proximity to the AuNP surface such that the fluorescence is quenched in an “off state”. Upon selective interaction with cancer cell surface moieties, the fluorophore is released, resulting in a measurable fluorescent response. In one such example reported by Lee and co-workers, AuNPs were functionalized with fluorophore-labeled heparin, which is a major component of the extracellular matrix that is degraded by the overexpression of heparanase and heparinase in metastatic cancer cells.287 While conjugated to the AuNPs, the fluorophore label is quenched, but upon degradation by metastatic cancer cells, the heparin fragments are released and generate a fluorescence enhancement. This method was used to selectively detect cancer cell lines that express high levels of heparanase (HeLa cervical cancer cells) over cancer cells with low expression levels (MCF7 breast cancer cells), and noncancerous cells (NIH-3T3 fibroblasts).
In another approach, Bajaj et al. reported an AuNP-based “chemical nose” to differentiate cell types and cancer states using AuNPs that are capped with ligands of varying hydrophobicity and are coated with green fluorescent protein (GFP).285 Based on the differences in chemical structure of the capping ligands used, each AuNP–GFP complex associates with cancer cells to varying extents due to the differences in cell membrane composition. When the AuNP–GFP construct interacts with a cell, the GFP is displaced, generating a fluorescence enhancement. This method required as few as 5000 cells for detection and was able to differentiate between breast cancer cells (MCF7), hepatocellular carcinoma cells (HepG2), cervical cancer cells (HeLa), and testicular cancer cells (NT2). Though this capability may be useful in the study of cancer cell membrane heterogeneity and the identification of cancer cells, it is limited by the inability to rationally design a nanoparticle probe to target a specific cell of interest.
Silver nanoclusters (AgNCs) have also been used for cancer cell detection in “off–on” fluorescent systems. AgNCs consisting of 2–30 Ag atoms may be synthesized using single stranded DNA templates to yield a fluorescent probe that exhibits lower cytotoxicity, exhibits brighter fluorescence, and is more photostable than organic dyes.310,311 In addition, recent research has found that AgNCs may be turned “on” and “off,”312 or tuned in emission wavelength,313 based on their chemical environment. This advantage has been exploited to design fluorescent biosensors for DNA, RNA,312,313 and protein314–316 detection. In 2010, Yeh et al. reported that AgNCs exhibit a 500-fold enhancement in red fluorescence when in close proximity to guanine-rich DNA.312 The extent of fluorescence turn-on was determined to be dependent upon the number of guanine bases in proximity to AgNCs, and it was originally hypothesized that this effect was due to charge transfer between the guanine residues and the AgNCs.317 However, more recent studies have indicated that this may not be the mechanism of AgNC fluorescence enhancement,318 and the underlying cause of guanine-proximity-induced fluorescence of AgNCs is still under investigation.319
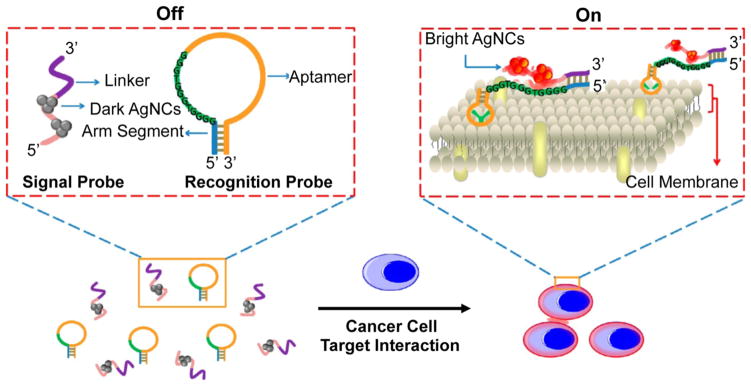
Following the recent advances in the use of AgNCs for biosensing, Yin et al. designed a two-component DNA/AgNC probe to detect as few as 1000 CCRF-CEM acute leukemia cancer cells (Figure 14).320 Unlike many of the other methods reported to fluorophore-label cell surface markers for cancer cell detection, this approach provides a switchable fluorescent output, which may lead to reduced background signal from nonspecific binding of nanoparticle probes to noncancerous cells.
Figure 14.
Schematic representation of silver nanoclusters (AgNCs) used in the detection of CCRF-CEM acute leukemia cells. In the “off state”, the system consists of two DNA strands: the “signal probe” which is tethered to AgNCs that are not fluorescent and a linker region that is complementary to the arm segment of the “recognition probe”. Upon binding of the scg8c aptamer region of the recognition probe to CCRF-CEM cells, the arm segment is exposed, enabling the hybridization of the signal probe and the recognition probe. This brings the AgNCs in close proximity of the G-rich segment of the recognition probe, leading to enhanced AgNC fluorescence in the “on” state. Adapted from ref 320. Copyright 2013 American Chemical Society.
5.2.2. Modes of Cancer Cell Surface Marker Recognition
A range of moieties is used to recognize and bind cell surface markers for nanoparticle-mediated fluorescence detection of cancer cells. In some cases, proteins are conjugated to fluorescent nanoparticles to generate probes that bind to cancer cell surfaces through recognition by cell surface receptors. For example, Mi et al. have conjugated transferrin glycoprotein to UCNPs and achieved fluorescent imaging and detection of HeLa cervical cancer cells based upon recognition by transferrin receptors, which are up-regulated in many types of cancers.296
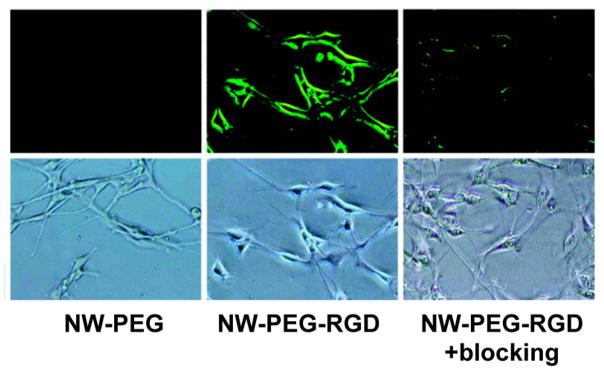
Similarly, short peptides may be conjugated to nanoparticles for targeted cancer cell imaging. In particular, fluorescent nanoparticles labeled with RGD peptide are recognized by integrin αvβ3, a cell surface receptor that is implicated in cancer angiogenesis and metastasis,321,322 and have therefore been used to detect cancer cells.307,323 Hong et al. demonstrated the ability of fluorescent zinc oxide nanowires coated with RGD peptide to selectively label integrin αvβ3 positive human glioblastoma cells (U87MG) over integrin αvβ3 negative human breast cancer cells (MCF-7) by fluorescence microscopy, highlighting the ability to differentiate between cancer types (Figure 15).323
Figure 15.
RGD peptide functionalized ZnO nanowires (NW-PEG-RGD) were used to selectively label integrin αvβ3 in U87MG glioblastoma cells. Cells were treated with either PEGylated ZnO nanowires (NW-PEG) or NW-PEG-RGD. As a control, integrin αvβ3 was also blocked on U87MG cells by pretreating with cyclic RGDYK peptide, resulting in decreased labeling of the U87MG cells. Note that images were taken under 200× magnification. Adapted from ref 323. Copyright 2011 American Chemical Society.
Antibodies are frequently used to recognize cancer cell surface markers based on their ability to specifically bind target cell surface receptors with high affinity. Her2, which is overexpressed in breast cancer, is frequently used for detection through antibodies conjugated to nanoparticle probes for cancer cell detection.264–267,274,324 Additionally, epidermal growth factor receptor (EGFR) is often targeted for selective fluorophore labeling of cancer cells due to its overexpression in a range of cancer cell types.262,263,275
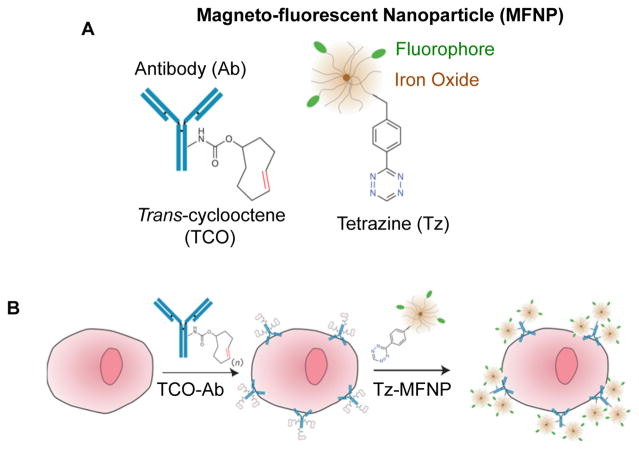
In 2010, Haun et al. expanded upon the use of antibody-coated nanoparticles for detecting cancer cells by developing a bio-orthogonal nanoparticle detection (BOND, Figure 16).262 Cycloaddition of trans-cycloocetene (TCO)-modified antibodies and tetrazine-coated magnetofluorescent nanoparticles occurs in biological media, including cell culture medium and serum. This method was shown to detect cancer cells through either Her2, EGFR, EpCAM, mucin 1, or CD45 receptor recognition, demonstrating the tailorability of the platform to analyze multiple targets in parallel and profile cancer cell populations based on the expression of various surface markers in complex biological media.
Figure 16.
(A) Schematic representation of a tetrazine-labeled magnetofluorescent nanoparticle (MFNP) and a trans-cyclooctene (TCO)-functionalized antibody used in the detection of cancer cells. (B) Cells are first treated with the TCO-Ab and then treated with MFNPs, which react in cell culture conditions to fluorophore-label cells expressing the target of interest. Adapted with permission from ref 262. Copyright 2010 Macmillan Publishers Ltd.
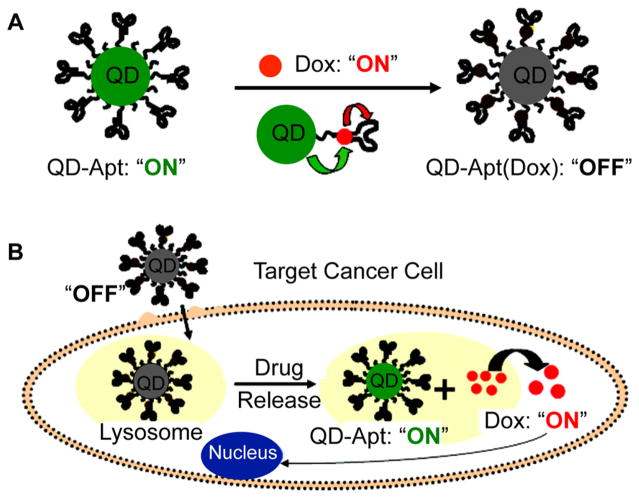
Another method of targeting cancer cells utilizes oligonucleotide aptamers, which may be designed to fold and bind with high selectivity and affinity to any target of interest. Prostate-specific membrane antigen (PSMA) is a common target used for aptamer-based recognition of prostate cancer cells.277,278 Bagalkot and co-workers demonstrated that QDs coated with aptamers against PSMA could be used to selectively deliver doxorubicin to prostate cancer cells (LNCaP) for combined cancer cell detection and treatment (Figure 17).277 Prior to cellular entry, the fluorescence of both the doxorubicin and the QDs is quenched through bi-Förster energy transfer processes. Upon cellular entry, doxorubicin is released, generating an increase in fluorescence from both the doxorubicin and the QD. This method provides an avenue for the design of theranostic nanomaterials to simultaneously detect and treat cancer cells.
Figure 17.
Schematic representation of aptamer-functionalized QDs for combined cancer cell imaging and therapy. (A) Doxorubicin (Dox) is intercalated into DNA aptamers bound to the QD probe. (B) Aptamers recognize cell surface markers of a target cancer cell, and once the construct is internalized, Dox is released. Adapted from ref 277. Copyright 2007 American Chemical Society.
A development in utilizing aptamers for cancer cell targeting is the use of cell based systematic evolution of ligands by exponential enrichment (cell-SELEX) to design aptamers that can recognize complex targets, including whole cells based upon interactions between the aptamer and cell membrane components.97 Aptamers that were designed through cell-SELEX to target cancer cells of interest have been conjugated to nanoparticles to enable their detection through fluorescence. 305,306,308,320,325,326 In one example, an 88-mer DNA aptamer designed to bind acute leukemia CCRF-CEM cells with high affinity (Kd = 5 nM) was appended to tris(2,2′-bipyridyl)dichlororuthenium(II) (Rubpy) dye doped polymeric nanoparticles and utilized for the detection of CCRF-CEM cells through fluorescence.304 Though the specific binding interactions of aptamers designed to target cancer cells are not well understood, cell-SELEX is a generalizable method that may be used to target cell populations for which antibodies are not available.
An alternative approach to detect cancer cells is to use small molecules that bind specific cancer cell receptors. Several groups have demonstrated that folic acid may be conjugated to fluorescent nanoparticles for the detection of cancerous cells, which often overexpress folate receptors.286,288–291,293–295 In one example, Rosenholm and co-workers conjugated folic acid to hybrid mesoporous silica nanoparticles for HeLa cervical cancer cell targeting.288 Human embryonic kidney epithelial cells (Hek-293) with low folate receptor expression exhibited low uptake of the hybrid particles, suggesting their ability to selectively target cancerous cells. An advantage of this approach over others is that some small molecules are relatively inexpensive compared to proteins and antibodies. When small molecules bind cell surface receptors that are not known for a cancer type of interest, large-scale screens of small-molecule-functionalized nanoparticles may be conducted to uncover specific interactions of certain molecules with target cells. Though this approach is not based on rational design, it is a powerful high-throughput approach to uncover cancer cell interactions with small molecules for diagnostic applications. In 2005, Weissleder et al. synthesized a library of nanoparticle constructs conjugated to one of 146 small molecules, and screened their ability to discriminate between cell populations of interest.292 Specifically, the cellular uptake of these constructs into human pancreatic ductal adenocarcinoma cells (PaCa-2) compared to human umbilical vein endothelial cells (HUVEC) and human macrophages (U937) was studied. Of the 146 nanoparticles tested, two exhibited selective uptake (those functionalized with either 5-chloro-isatoic anhydride or isatoic anhydride) into PaCa-2 cells. Though this observed difference in uptake may be used to fluorophore-label and identify pancreatic cancer cells, the underlying mechanism is not known and the application of this approach for the detection of additional cancer types would require additional screens.
5.3. Detection Based on Gene Expression
5.3.1. NanoFlares for Intracellular mRNA Detection
In 2007, the Mirkin group introduced the NanoFlare, a spherical nucleic acid (SNA) AuNP-based platform, which is shown to be useful for detecting and knocking down intracellular mRNA (Figure 18A).327 Based on the highly oriented, dense oligonucleotide coating, NanoFlares enter cells efficiently without the use of cytotoxic transfection agents.328–333 In addition, the density of the oligonucleotides leads to enhanced stability against degradation, making the NanoFlare less susceptible to background fluorescence based upon the degradation of DNA compared to traditional molecular beacon and fluorescence in situ hybridization (FISH) probes.332,334 Unlike FISH probes, which require fixation of cell samples prior to analysis,335 no observable cytotoxicity336,337 is seen following treatment of cells with NanoFlares, which enables one to detect genetic327,338,339 and small molecule content in live cells.340
Figure 18.
(A) NanoFlares are used in the fluorescence-based detection of intracellular mRNA and consist of thiolated “recognition” antisense DNA adsorbed onto the surface of a spherical AuNP. The “reporter flare,” a shorter complementary DNA with a Cyanine 5 (Cy5) fluorophore, is hybridized to the recognition strand, resulting in the quenching of the Cy5 fluorophore. Upon target binding, the reporter flare is released generating a measurable fluorescence signal. (B) NanoFlares have been shown to enable the detection and isolation of circulating tumor cells from a murine model of triple negative breast cancer. Blood samples from mice with xenografted mCherry labeled MDA-MB-231 tumors were treated with vimentin-targeting NanoFlares, and MDA-MB-231 cells were retrieved based on NanoFlare fluorescence. Representative scatter plots (N = 1 mouse per scatter plot) are shown for a mouse that was not injected with mCherry MDA-MB-231 breast cancer cells (Control) and mice that were injected with mCherry MDA-MB-231 breast cancer cells and developed widespread metastases (Experimental 1 and 2). Cancerous cells are shown in red, and noncancerous cells are shown in black (B). Adapted with permission from ref 343. Copyright 2014 National Academy of Sciences of the United States of America.
In initial studies, the NanoFlare was used for the fluorescent detection of the mRNA transcript of the oncogene survivin.327 NanoFlares were used to distinguish between breast cancer cells with high survivin expression (SK-BR-3) and non-cancerous mouse endothelial cells (C166). Importantly, this construct has enabled the profiling of cancer cells based on intracellular genetic content. This is significant, since relying on extracellular protein markers may result in the inability to identify subpopulations of cancerous cells that do not express the surface marker of interest.249 In 2009, the NanoFlare was reported as a single-entity agent for the simultaneous detection and regulation of target mRNA.339 Based on the design of NanoFlares, binding of target mRNA to the recognition stand may also be used in antisense gene regulation. This provides an opportunity to combine targeted mRNA detection and gene regulation in a single construct. As such, the NanoFlare was shown to be useful for detecting and knocking down expression of survivin transcripts.
Additionally, the Mirkin and Tang groups have expanded upon the NanoFlare construct with the development of multiplexed NanoFlares.338,341 In these studies, functionalization of AuNPs with two338 or three341 DNA recognition strands and subsequent hybridization of their short complement reporter strands yielded NanoFlares that are capable of simultaneous intracellular detection of multiple mRNA transcripts. In particular, the use of multiplexed NanoFlares in detecting survivin in addition to actin, a housekeeping gene, was studied as a means to normalize NanoFlare fluorescence to account for differences in cellular uptake and to make the technique comparable with qRT-PCR in quantifying intracellular mRNA, but at the single live cell level.338,341 Notably, the Tan group has developed a “lab-on-a-nanoparticle” system by incorporating multiple DNA recognition stands onto the NanoFlare construct in order to perform intracellular DNA logic gating to simultaneously monitor the presence of multiple small molecules.342
In 2014, the NanoFlare platform was used for the detection and isolation of live circulating tumor cells from whole blood.343 NanoFlares targeting known markers of the EMT were designed, including vimentin and fibronectin. The NanoFlares were used to detect metastatic breast cancer cells (mCherry labeled MDA-MB-231), and were doped into healthy human blood samples. Samples treated with Nano-Flares were analyzed by flow cytometry, and the average recovery of cells was determined to be 68 ± 14%, enabling the detection of as few as 500 cells/mL of blood. In addition, mCherry MDA-MB-231 cells were isolated from the blood of xenografted mice following tumor metastases (Figure 18B). Blood samples were treated with NanoFlares and processed following the same procedure used for human blood experiments. It was noted that 87–90% of the mCherry labeled cells recovered were detected by NanoFlare fluorescence.
Recently, the Mirkin group further expanded upon the NanoFlare platform to enable both intracellular RNA quantification and spatiotemporal localization in living cells.344 Sticky-Flares were designed to target β-actin mRNA and U1 short nuclear RNA such that, upon target binding to a recognition sequence on the Sticky-Flare, a fluorophore is transferred to the RNA transcript, allowing for expression quantification and tagging it for intracellular tracking through fluorescence microscopy. This construct was utilized to visualize the real-time transport of β-actin mRNA in live mouse embryonic fibroblasts using confocal microscopy. Notably, it is difficult, if not impossible, to analyze the dynamics of RNA transport with FISH probes, since they require cell samples to be fixed and permeabilized prior to analysis. Smart-Flares may be applied to the study of gene expression in cancer cells to our further understanding of RNA expression, localization, and transport within cancerous and metastatic cells.
Since their development, NanoFlares have been commercialized by EMD Millipore and sold under the trade name SmartFlare in over 230 countries. At present, there are over 1700 versions of these constructs that can be used to target different mRNA sequences in a variety of flow cytometry and imaging experiments. In addition to providing the only way of measuring genetic content in single live cells, these structures change the paradigm of cell sorting based upon extracellular protein markers to intracellular genetic and small molecule markers.340 Several groups have since utilized the SmartFlare to study a range of biological processes including pluripotency, 345,346 the inflammatory response to infection with DNA viruses,347 and the distribution of mRNA within Purkinje neurons.348
Further, the Hendrix group has utilized SmartFlares to study melanoma tumor cell heterogeneity.349 In their work, Nodal, an embryonic morphogen that is typically silent following early development, was chosen as a target since it is reexpressed in aggressive melanoma cells.350–353 Tumor cell heterogeneity was examined by sorting SmartFlare-treated melanoma cells based upon their expression level of Nodal using fluorescence activated cell sorting. The metastatic potential of the subpopulations was further studied, enabling the correlation of gene expression levels to tumor cell phenotypes. This work highlights the unique capability of the SmartFlare to quantify genomic expression at the single-cell level as well as the immediate implications in expanding our knowledge of cancer and metastasis through the use of SmartFlares. Ultimately, the continued study of cancer cell heterogeneity is essential in the development of more efficacious chemotherapeutics as well as the identification of novel markers for early cancer diagnosis.
5.3.2. Molecular-Beacon-Modified AuNPs for Intracellular mRNA Detection
AuNPs modified with molecular beacon DNA have also been used for the fluorescent detection of cancer cells.354–360 Traditional molecular beacons consist of hairpin DNA that targets mRNA of interest with a quencher and fluorophore pair conjugated to each end of the DNA strand. In the absence of target, the proximity of the quencher to the fluorophore results in low fluorescence, but when target mRNA binds, the hairpin opens, increasing the distance between the quencher and fluorophore to generate fluorescence “turn-on”. In the AuNP-based systems, traditional molecular beacons are improved upon through the enhanced fluorescence quenching efficiency of AuNPs compared to organic dyes. The Tang group has expanded the AuNP–molecular beacon platform by designing systems that can detect two358 to four356 mRNA targets.
In addition, this system has been expanded to enable the simultaneous release of oligonucleotide and the chemotherapeutic drug doxorubicin in a theranostic application.355,359 In these designs, doxorubicin is intercalated into the molecular beacon structures in their “off state,” and is released upon target mRNA binding. This design was shown to detect SKBR-3 cells based upon the expression of target cyclin D1 mRNA. In addition, doxorubicin-induced cell death of SKBR-3 cells was selective, and the viability of MCF-10A cells, which express low levels of cyclin D1, was not affected by treatment with the doxorubicin-loaded molecular beacon–AuNPs.359
6. DETECTION OF TUMOR TISSUE IN VIVO
The detection of cancer, metastasis and tumor growth in particular, is essential for designing effective treatment courses. Beyond the ability to diagnose cancer through ex vivo analysis based upon the detection of cancer biomarkers and cancer cells from patient samples of blood, feces, or urine, the ability to identify cancerous tissues and tumors in the body presents several advantages in the detection and treatment of cancer. The development of nanoparticle probes has enabled opportunities for both prognostics361–392 (understanding the effect of a treatment on tumor growth and metastasis) and treatment393–402 (image guided surgery and theranostics) for improving patient outcomes. Though research in this field has resulted in several nanoparticle probes that may be used in the diagnosis of cancer, several challenges must be overcome before such technologies are viable for use in clinical settings. For example, instruments capable of fluorescence imaging on human patient scale are limited to narrow fields of view that are primarily useful for image-guided surgery. Thus, the development of instrumentation capable of generating fluorescence images of patients is essential for the advancement of these technologies into the clinic. Ultimately, translation of these nanoparticle probes toward clinical use may result in sensitive imaging platforms for the detection of tumors and metastases at the earlier stages of their development, which may be confirmed through histopathological evaluation.
In addition to fluorescence-based imaging, several alternative modalities have been developed for the diagnosis of cancer through in vivo imaging: computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), single photon emission computed tomography (SPECT), ultrasound, and photoacoustic imaging, each with their own strengths and limitations (Table 2). Besides SPECT, which is relatively high-cost and low-throughput, fluorescence is the only other imaging modality available for simultaneous detection of multiple probes.403,404 Notably, this feature is advantageous in the diagnosis of cancer where the ability to distinguish malignant cell populations from benign populations is enhanced by monitoring the presence of multiple cancer markers. QDs and UCNPs are particularly useful in this respect, due to their emission profiles that are easily tuned through changes in composition and size.32,35–38,53
Table 2.
Comparison of in Vivo Imaging Techniquesa
| technique | label | signal measured | cost | throughput | sensitivity (mol of label detectable) | resolution | penetration depth | simultaneous imaging of multiple labels |
|---|---|---|---|---|---|---|---|---|
| CT | none | X-rays | high | low | 10−6 | 50 μm | no limit | no |
| MRI | paramagnetic and superparamagnetic contrast agents | alterations in magnetic fields | high | low | 10−9–10−6 | 50 μm | no limit | no |
| PET | radiolabeled molecules | positrons from radionuclides | high | low | 10−15 | 1–2 mm | no limit | no |
| SPECT | radiolabeled molecules | γ-rays | high | low | 10−14 | 1–2 mm | no limit | yes |
| ultrasound | microbubbles, which can be combined with targeted contrast agents | sound | low | high | 10−8 | 50 μm | several cm | no |
| photoacoustic | probes that absorb light and create sound signals | sound | low | high | 10−12 | 50 μm | <5 cm | no |
| fluorescence | fluorophores | light (e.g., NIR region) | low | high | 10−12 | 1–3 mm | <1 cm to <5 cm | yes |
Though fluorescence imaging is somewhat limited by lower tissue penetration depths and resolution than alternative methods, including MRI, fluorescence offers high sensitivity of detection at a lower cost and is less time-consuming.403–405 Additionally, fluorescence emission may be tuned to optimize tissue penetration depths, based upon the relative transparency of tissue absorbance in the NIR window (Figure 19).406 QDs and UCNPs are especially well-suited to address these challenges due to their tunable NIR emission profiles,32,35–38,53 as well as their large Stokes and anti-Stokes shifts, respectively, which minimize background signal due to tissue autofluorescence. 40,49,54
Figure 19.
NIR window is optimal for in vivo imaging due to minimal light absorption by hemoglobin (Hb), oxyhemoglobin (HbO2), and water in tissues from 650 to 900 nm. Adapted with permission from ref 406. Copyright Macmillan Publishers Ltd.
In this section, we will discuss the advances that have been made toward the use of nanoparticles in vivo, as well as the development of methods to image and, in some cases, treat cancer in animal models. Research within this field primarily focuses on the use of fluorescent nanoparticle probes to image and diagnose cancer in vivo based upon preferential accumulation of nanoparticles in tumor tissue through either passive or active targeting.
6.1. Behavior of Nanoparticles in Vivo and Considerations for their Design
To achieve the goal of diagnosing cancer through the use of nanoparticle probes for fluorescent imaging in patients, significant research in understanding the behavior of nanoparticles in vivo must be done. While in vitro assays are useful for characterizing nanoparticle interactions on the cellular level, these models do not always accurately represent cancer development and metastasis in vivo. As such, in order to design a nanoparticle for biomedical applications, and cancer diagnosis in particular, it is necessary to use animal models of cancer to understand nanoparticle probe behavior in vivo. An ideal nanoparticle probe for cancer tissue detection should have a long circulation time with specificity to the tumor tissue and low toxicity to surrounding healthy tissue. The fundamental design rules for developing such a probe are an active area of ongoing research. We will discuss these recent advances, focusing on the properties that affect nanoparticle circulation times and biodistribution (especially tumor accumulation), and potential toxicity.
6.1.1. Nanoparticle Accumulation in Tumor Tissue
Nanoparticle size, shape, surface charge, and surface modification each play important roles in determining in vivo behavior and contribute to the complex interactions between nanomaterials and biological systems. Here, we will discuss these interactions within the context of nanoparticle association with blood proteins, the uptake and clearance of nanoparticles by the reticuloendothelial system (RES) organs, penetration into solid tumors, and the optimization of active (versus passive) targeting for cancer diagnosis.
Passive targeting refers to the ability of nanoparticles with diameters 10–150 nm to preferentially extravasate from the bloodstream into tumor tissue.407–409 The tumor’s rapid growth initiates local angiogenesis, the process of forming new blood vessels, to supply cancer cells with nutrients.5 As a result, the tight junctions between endothelial cells do not form properly, generating “leaky” blood vessels. In terms of blood supply, oxygen, and nutrient delivery, these newly formed vessels are inefficient, necessitating the formation of additional blood vessels. In vivo delivery of diagnostic and therapeutic nanoparticles can take advantage of the abundance of vasculature at tumor sites in addition to the poor formation of tight junctions (which are larger than the usual 8 nm size in tumor tissue) for preferential accumulation of nanoparticles in tumor tissue.410–413 In addition, large tumors tend to have poor lymphatic drainage, leading to long retention times of extravasated nanoparticles in the tumor tissue. This form of passive nanoparticle entry into the tumor microenvironment is termed the enhanced permeability and retention (EPR) effect (Figure 20),410,412,414 which was observed nearly 30 years ago with the transport of macromolecules into tumor tissue.415,416 Despite the ability to passively target nanoparticles to tumor tissue via the EPR effect, there are several challenges associated with this approach, including heterogeneity within and between tumor types, which may compromise the utility of passive targeting in clinical settings.417,418 Thus, active tumor targeting, in which a moiety that specifically binds a cell surface marker that is associated with cancer, is frequently used to enhance the delivery of nanoparticles to tumor tissue for cancer imaging applications.419
Figure 20.
Diagram depicting the enhanced permeability and retention effect displayed by tumor tissue. Due to the leaky tumor vasculature as a result of poor lymphatic drainage, nanoparticles will escape the blood stream and preferentially localize in tumor tissue, also known as passive targeting. Reprinted with permission from ref 414. Copyright 2007 Macmillan Publishers Ltd.
Though the EPR effect allows nanoparticles to passively extravasate from circulation and accumulate within tumor tissue, it is well understood that the adsorption of serum proteins onto a nanoparticle surface (opsonization) significantly alters in vivo nanoparticle trafficking, uptake, and clearance.420–425 The formation of a nanoparticle protein corona enables the recognition of nanoparticles by cell-surface receptors on macrophages, resulting in rapid clearance rates.420–425 It is widely known that nanoparticle composition, shape, size, and surface charge can dictate the types of proteins that adsorb to the nanoparticle surface and can significantly alter in vivo pharmacokinetics and biodistribution.420,422,424–434 This must be taken into consideration when designing nanoparticle probes for in vivo cancer diagnosis to enhance tumor accumulation and improve signal.
One common method to reduce nonspecific adsorption of serum proteins while also enhancing nanoparticle circulation time is to use poly(ethylene glycol) (PEG) to passivate the surface of nanoparticles.435–440 PEG is widely accepted as a noncytotoxic molecule that imparts decreased sequestration by resident tissue macrophages in the RES organs by minimizing the formation of a protein corona.441–451 Experimentally, PEGylation of a range of nanoparticles, including AuNPs and QDs, results in an enhancement in nanoparticle circulation time and slower accumulation into the liver and spleen when administered by intravenous (IV) injection into the tail vein of mice.452–454 Since circulation time and uptake into various organs can be tuned by altering PEG length453 and packing density455 on the nanoparticle surface, PEGylation provides an additional avenue for designing in vivo nanoparticle probes for cancer diagnosis. It is important to note, however, that the accelerated blood clearance (ABC) phenomenon may result in the rapid clearance of PEGylated nanoparticles following repeat intravenous injections.456–461 Due to this challenge, the design of alternatives to enhance the blood residence time of pharmaceuticals remains an active field of research.462–478
In addition to the EPR effect and the formation of a protein corona, nanoparticle size and shape are also important considerations in the design of nanoparticle probes that exhibit high tumor accumulation. Nanoparticles smaller than 10 nm are rapidly cleared by the renal system, minimizing their ability to localize in tumor tissue.479–481 Additionally, the microenvironment surrounding tumor tissue is complex and extremely heterogeneous: there are areas of hypoxia and necrosis, varying degrees of hyperpermeability in blood vessels, differences in the proliferative capacity between cells at the edge of the tumor and the core, and dense extracellular matrix surrounding the solid tumor.482,483 These heterogeneities can lead to uneven distribution of diagnostic nanoparticle probes within tumor tissue and afford smaller nanoparticles with the ability to permeate tumor tissue more deeply.484,485
Nanoparticle shape also affects in vivo behavior, and must be considered when designing nanoparticle probes for the detection of tumor tissue. Generally speaking, anisotropic particles exhibit longer circulation times in the blood,486,487 which is most likely due to the decreased probability of anisotropic nanoparticles permeating the endothelial gaps found in the fenestrations of the liver that range from hundreds of nanometers to tens of micrometers.488 Due to this, flexible nanoparticles with high aspect ratios may enable accumulation in tissues that are more difficult to access (such as the brain), and carry a larger number of active targeting or drug molecules compared to a spherical nanoparticle of the same diameter. 489,490 Such effects may alter nanoparticle accumulation in tumor tissue, and should be considered in the design of diagnostic nanoparticles for in vivo imaging.
6.1.2. Safety of Systemic Nanoparticle Administration
In order to effectively diagnose and treat cancer in vivo through the use of novel nanoparticle probes, it is necessary to assess any possible toxicity effects caused by their systemic administration. While some nanoparticles, such as those derived from silica and certain polymers, are generally regarded as safe due to their biocompatibility and biodegradability,486,491–500 the long-term safety of systemic injection of metal and semiconductor nanoparticles is still under investigation. Ultimately, the specific toxicity concerns associated with each nanoparticle probe must be thoroughly addressed on a case-by-case basis as the developing technologies are translated to clinical use.
Mesoporous silica nanoparticles are often employed as carriers of drugs and other small molecules into cells. The ability to modify both the surface chemistry and internal pore size of these nanoparticles can increase the loading capacity to greater than the solubility limit of many drugs. This is a feature unique to mesoporous silica nanoparticles, and one that is highly desirable for tumor targeting and treatment. Furthermore, it is generally accepted that mesoporous silica nanoparticles are biocompatible and that any observed toxicity in vivo is due to altered surface chemistry, specifically the surface silanol groups.486,491–496
Polymeric nanoparticles, including micelles, hydrogels, and polymer–drug conjugates, are often useful for in vivo diagnostics and therapeutics due to their biodegradability, biocompatibility, and near complete clearance of degradation products.497–500 Some polymers, such as poly(glycolic acid) and poly(lactic acid), have been approved by the FDA for medical applications,501,502 making the use of such polymers highly attractive for the rapid development of novel nanoparticles for cancer diagnostics in vivo.
The use of AuNPs for biomedical applications, and cancer diagnostics in particular, is rapidly gaining interest due to their ease of surface modification and attractive optical properties. AuNPs have long been considered to be both bioinert and biocompatible, and they have not been shown to cause cytotoxicity in human cell lines in vitro.454 However, these cell culture conditions do not replicate the complexity of in vivo conditions, and in vivo experimentation is necessary to better understand the potential side effects of AuNP accumulation in organs and tissues over time.503–506 Though several studies have aimed to understand the role of AuNP size,454 shape,507–512 dose,513,514 and route of administration515 on biocompatibility, the results are often conflicting and difficult to compare based on variations in study design and nanoparticle synthesis.
The safety of administering UCNPs and QDs is also controversial, due to the concern that toxic metals (Cd, in particular, for QDs) may leach from the nanoparticle core, causing adverse effects in vivo. Though few reports characterize the in vivo toxicity of UCNPs, some studies have used Caenorhabditis elegans (C. elegans) to investigate biocompatibility and have observed minimal toxicity.516–518 Furthermore, a few studies have claimed that the injection of mice with UCNPs does not cause any overt toxic effects.519,520
Meanwhile, QDs have consistently been shown cytotoxicity in vitro,521 but translation of these results to systemic toxicity is not always straightforward. Recent reports have demonstrated contrasting accounts of safety and toxicity of QD administration in various small animal models, which is attributed to differences in administration dosage, QD synthesis strategies, and surface ligands.522–524 Ye et al. reported that when a certain class of QDs was injected into nonhuman primates, no adverse effects on body weight, daily behavior, immune response, kidney and liver function, or blood chemistry were observed for 90 days post injection (Figure 21).525 Though this study, which was the first to use an animal model with high genetic similarity to humans, supports the notion that QDs may be safe for use in humans, QDs have not yet been approved for human application, and many in the community are skeptical about their potential for in vivo clinical use.
Figure 21.
Histological (hematoxylin and eosin staining) analysis of the major organs of nonhuman primates 90 days after injection of phospholipid micelle-encapsulated QDs. Tissues were collected from control (left image) and treated (right image) animals. Tissue analysis shows no significant differences in the (A) brain, (B) heart, (C) liver, (D) spleen, (E) kidneys, or (F) lymph nodes. Reprinted with permission from ref 525. Copyright 2012 Macmillan Publishers Ltd.
6.2. Nanoparticles for Tumor Tissue Detection
Many groups have investigated the use of fluorescent nanoparticle probes for imaging tumor tissue in vivo. Current research in this field utilizes animal models (usually mouse models) to study the accumulation of fluorescent nanoparticle probes in tumor tissue for cancer diagnosis. In these examples, a nanoparticle is typically injected systemically and preferentially localized to tumor tissue either passively via the EPR effect or actively through conjugation of a surface moiety that binds and recognizes the cancer cells found within tumors. We will discuss these approaches within the context of the nanoparticle probe used to generate the fluorescent signal and the recognition moiety used to bind and tag cancer cells within the animal model.
6.2.1. In Vivo Tumor Imaging
Many groups have utilized the EPR effect to passively target fluorescent nanoparticles to tumor tissue for in vivo imaging and diagnostics. Due to their unusual properties, including remarkable photostability, tunable emission, and high quantum yield, QDs have been used in the fluorescent imaging of tumor tissue by passive accumulation via the EPR effect.372,375,382 In 2012, Hong et al. reported the use of silver sulfide (Ag2S) QDs for in vivo imaging of tumor tissues in a xenograft mouse model of 4T1 metastatic breast cancer cells.526 In another example, Popovic and colleagues reported the use of QDs coated with a silica shell of varying thickness to probe the role of nanoparticle size in determining tumor tissue accumulation for cancer diagnostic applications.382 By varying the composition of the QD cores, the researchers found that each of the resulting nanoparticles, with diameters ranging from 12 to 120 nm, exhibited distinct emission colors. A mixture of 12, 60, and 120 nm silica-coated QDs was injected intravenously into a xenografted Mu89 human melanoma mouse model. Due to the unique emission wavelength of each QD used, real-time fluorescence imaging of QD extravasation and tumor penetration was gathered simultaneously for each nanoparticle size. The results indicated that the 12 nm QDs penetrated the tumor tissue with minimal hindrance, while the 60 nm QDs extravasated but remained within 10 μm from the blood vessels. In contrast, the 120 nm QDs did not extravasate to an appreciable extent. This data supports the use of nanoparticles for the detection of cancer in vivo through fluorescence based upon passive accumulation in tumor tissue, and highlights the notion that a nanoparticle probe’s size must be appropriately designed to enhance tumor targeting and penetration.
Additionally, there have been recent advances in the use of QDs for in vivo cancer diagnosis based upon sentinel lymph node imaging. Sentinel lymph nodes are the first lymph nodes that a cancer will metastasize to, and evidence of metastasis in the sentinel lymph nodes serves as an important prognostic for the progression of cancer.362,389 The ability to locate and surgically remove the sentinel lymph node provides an avenue to understanding the development of cancer: the absence of cancerous cells in the sentinel lymph node indicates that the cancer is likely unable to have advanced through the formation of secondary tumors, while the presence of cancerous cells suggests that the cancer has begun the metastatic process.362,389
Kim et al. first reported the use of near-infrared QDs for in vivo mapping of sentinel lymph nodes in both a mouse and pig model.527 To demonstrate the utility of QDs in performing diagnostic surgeries to excise sentinel lymph nodes, QDs were injected into pigs. Localization of the QDs in the sentinel lymph nodes highlighted their location, and the fluorescence signal was used to direct surgery to remove them with penetration depths on the centimeter length scale. The use of QDs enabled deeper penetration depths compared to organic dyes used for sentinel lymph node mapping.527 In 2007, Ballou et al. expanded upon the use of QDs to map sentinel lymph nodes in tumor-bearing animals.364 Mice with subcutaneous M21 human melanoma tumors were injected with PEGylated QDs intratumorally. The QDs rapidly transferred from the tumor tissue to the adjacent lymph nodes, and the fluorescence emission was visible through the skin of the animal almost immediately.
Conducting polymer nanoparticles (CPNs) have also been used in vivo for sentinel lymph node imaging. Kim et al. reported the use of cyanovinylene-backbone CPNs (60 nm) to map sentinel lymph nodes in real time in the NIR wavelength range.47 CPNs were injected intradermally in the paws of mice, and could be tracked by the naked eye (due to their high fluorescence, at 365 nm) as they drained to the lymph nodes.
UCNPs are also used to detect tumor tissue in vivo through fluorescence.365,378,386,391 Cheng and co-workers demonstrated the tunability of UCNPs for in vivo multicolor imaging applications.365 NaYF4 nanocrystals doped with either Er3+/Yb3+ or Er3+/Tm3+ were synthesized and modified by adsorbing one of three organic fluorophores on the nanoparticle surface: rhodamine B, rhodamine 6G, and Tide Quencher 1. Five sets of UCNPs were synthesized, each exhibiting distinct emission profiles. Each UCNP was subcutaneously injected into the backs of nude mice and imaged to demonstrate the potential use of UCNPs in imaging and diagnostic applications (Figure 22).
Figure 22.
Upconversion nanoparticles with varying emission profiles for multicolor in vivo imaging. (A) NaYF4:Yb,Er, (B) NaYF4:Yb,Er–rhodium B, (C) NaYF4:Yb,Er–rhodium 6G, (D) NaYF4:Yb,Er–Tide Quencher 1, and (E) NaYF4:ErTm upconversion nanoparticles subcutaneously injected into the back of a nude mouse. (F) Fluorescence merge of the upconversion nanoparticles and (G) white light image. Adapted from ref 365. Copyright 2011 American Chemical Society.
Beyond multicolor imaging, UCNPs have been used in the design of probes to diagnose cancer through the ability to detect hypoxia in vivo,378 since tumor tissues are often low in oxygen due to their abnormal vasculature.388 Liu et al. designed composite nanoparticles consisting of a UCNP core and a mesoporous silica shell containing tris(4,7-diphenyl-1,10-phenanthroline) ruthenium(II) dichloride ([Ru(dpp)3]2+Cl2). In the presence of oxygen, the [Ru(dpp)3]2+ is quenched due to reversible photochemical oxidation processes.380 In the absence of oxygen, the [Ru(dpp)3]2+ may be excited through energy transfer from the UCNPs, providing a fluorescent output to detect hypoxia in tumor tissues. This system was tested in a zebrafish embryo model of hypoxia (Figure 23), and could be expanded upon for the detection of hypoxic cancerous tissues in vivo.
Figure 23.
Zebrafish embryo induced with cerebral hypoxia imaged using an UCNP sensor for low oxygen detection. Zebrafish embryos were injected with UCNP sensors via intracerebral microinjection prior to treatment with 2,3-butanedione monoxime (BDM) to induce cerebral hypoxia. The increase in fluorescence intensity from the cranium 0–7 min post BDM treatment indicates the decrease in oxygen. Adapted from ref 378. Copyright 2014 American Chemical Society.
AuNCs have also been used in fluorescent in vivo tumor imaging applications based on passive targeting.367,370,390,392 In one report, Wu et al. described the use of AuNCs, about 2.7 nm in diameter, for fluorescent tumor imaging in vivo in cervical cancer (HeLa) and breast cancer (MDA-MB-45) xenograft mouse models.390 As previously described, small AuNCs of this size exhibit inherent fluorescence. The ultrasmall AuNCs were injected systemically into BALB/c nude mice with subcutaneous HeLa or MDA-MB-45 tumors. Within 6 h post injection, fluorescence signal from the tumors increased, demonstrating the potential use of AuNPs for in vivo cancer diagnostics.
For in vivo fluorescence tumor imaging using larger AuNPs, organic fluorophores are typically conjugated to the nanoparticle surface. For example, Chou and Chan used fluorophore-labeled AuNPs of varying sizes to study the effect of nanoparticle size on tumor accumulation for diagnostic and therapeutic applications.367 AuNPs from 15 to 100 nm were synthesized and functionalized with Alexa Fluor 750. Smaller, 15 nm AuNPs dispersed faster into the tumor tissue to provide fluorescence contrast of the cancerous lesions. This approach may be used for tumor tissue detection through passive accumulation, but also aids in our understanding of the size dependence of the EPR effect.
Perrault and Chan later expanded upon the use of AuNPs in cancer tissue imaging through the use of AuNPs as an “anchor” in a xenograft breast cancer (MDA-MD-435) mouse model (Figure 24).381 This approach combines passive targeting of the anchor AuNPs to tumor tissue with active targeting of a fluorophore to label the AuNPs in vivo. This method resulted in a 200 times faster rate of fluorophore accumulation in tumor tissue compared to fluorophore-labeled AuNPs of the same size, based on the active targeting of the fluorophore to the biotinylated AuNPs. This method’s use of actively targeting biotinylated AuNPs within the tumor tissue may be beneficial for imaging cancer tissues when the cell surface markers unique to the cancer cells are not well understood.
Figure 24.
Schematic representation of AuNP “anchors” for in vivo tumor imaging. Biotinylated AuNPs are injected into mice and passively accumulate in target tumor tissue before streptavidin-fluorophore is injected to fluorophore-label the AuNPs in vivo. Reprinted with permission from ref 381. Copyright 2010 National Academy of Sciences of the United States of America.
In 2011, von Maltzahn et al. expanded upon this approach by utilizing photothermal nanoparticles to passively target tumor tissue and locally induce a coagulation cascade that was then actively targeted with fluorescent nanoparticles.387 Gold nanorods (AuNRs) were intravenously injected into MDA-MB-435 breast cancer tumor-bearing mice. Mice were irradiated to locally heat tumor tissue based on passive AuNR accumulation, resulting in local blood vessel disruption and coagulation. In a labeling step, magnetofluorescent iron oxide nanoworms coated with a peptide substrate for the coagulation transglutaminase FXIII were administered intravenously to the mice for the detection of tumor tissue through fluorescence.
In addition to metal and semiconducting nanoparticles, polymeric nanoparticles encapsulating organic fluorophores have also been used for in vivo tumor imaging.371,383,384 While these approaches do not utilize the unique optical properties of inorganic nanoparticles, such as QDs and UCNPs, the nanoparticles’ size enables passive tumor targeting via the EPR effect. In one such report, Fortin et al. describe the use of liposomal nanoparticles loaded with a near-infrared organic fluorophore, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide (DiR), to image brain tumors in a mouse model by fluorescence.371 In a similar study, Schadlich et al. demonstrated the use of polymeric nanoparticles for tumor imaging in a xenograft mouse model of colorectal cancer (DsRed2 labeled colorectal adenocarcinoma HT29 cells).383 Tumor accumulation of the nanoparticles was evaluated in vivo and ex vivo following excision of the tumor tissues (Figure 25). Colocalization of the DsRed2 fluorescence from the tumor tissue and the DiR dye from the PEG–PLA nanoparticles increased from 10 min to 24 h post injection as a result of enhanced retention of the nanoparticles over time.
Figure 25.
(A) Image of xenografted mouse 10 min post injection with DiR-encapsulating PEG–PLA nanoparticles (red) illustrates location of DsRed2 labeled HT29 tumors (green). (B) Time course of DiR fluorescence from 10 min to 48 h post injection of PEG–PLA nanoparticles. (C) Ex vivo imaging of excised tumor demonstrates colocalization of DsRed2 labeled HT29 tumors and DiR from PEG–PLA nanoparticles. Adapted from ref 383. Copyright 2011 American Chemical Society.
In addition to tumor imaging based upon nanoparticle accumulation through passive targeting via the EPR effect, extensive work in the use of nanoparticles that actively target tumor tissue by recognizing cell surface receptors on cancer cells has been studied. Often, these approaches enhance the sensitivity of in vivo tumor detection by increasing the amount of nanoparticles that are delivered to tumor tissue per unit time. We will discuss these approaches, dividing them by the recognition moiety used to target cancer cells in vivo.
Peptides are frequently used to actively target cancerous tissues in vivo. In particular, the RGD peptide, which is recognized by a cell surface receptor that is implicated in cancer angiogenesis and metastasis (integrin αvβ3),321,322 has been used to target tumor tissue in vivo for imaging and diagnostic applications.528–531 In some cases, polymeric nanoparticles conjugated to organic fluorophores are modified with RGD to target tumors.529,530 More commonly, inherently fluorescent nanoparticles, such as UCNPs, are used to target integrin αvβ3 through presentation of the RGD peptide for in vivo imaging of tumors. In one such example, Xiong et al. used PEGylated UCNPs conjugated to RGD to actively target tumors in U87MG glioma xenografted mice.531 To evaluate the effect of RGD active targeting, mice were injected with two xenograft tumors: U87MG glioma, which has high integrin αvβ3 expression, and MCF7 breast cancer, which has low integrin αvβ3 expression. The RGD-conjugated UCNPs were injected intravenously, and the fluorescence intensity of the two tumors was compared (Figure 26). Ex vivo analysis of the UCNP content in organs by inductively coupled plasma atomic emission spectroscopy (based on Y3+ content) revealed that UCNP content in the U87MG tumor was roughly 30 times higher than that of the MCF7 tumor. In another example, Wu and co-workers utilized polymer dots (PDs) modified with chlorotoxin, a tumor targeting peptide, to target medullo-blastoma tumors in ND2:SmoA1 mice.48 High fluorescence intensity of the PDs was detected in the brain tumor regions (2.3-fold increase over wild type mice, compared to a 1.2-fold increase in nontargeting PDs) following intravenous injection via the tail vein. These works highlight the benefit of active tumor targeting compared to passive targeting via the EPR effect for cancer diagnostics, when high contrast enables the detection of tumors at earlier stages.
Figure 26.
Nude mouse with subcutaneous U87MG xenograft tumor (left hind leg, short arrow) and MCF7 xenograft tumor (right hind leg, long arrow) imaged 1 (top) and 4 (bottom) h post intravenous injection with RGD labeled UCNPs. UCNP accumulation is higher in the U87MG tumor as compared to the MCF7 tumor, due to active targeting of integrin αvβ3. Adapted from ref 531. Copyright 2009 American Chemical Society.
Conjugation of antibodies that bind cancer cell surface receptors to fluorescent nanoparticles has also been used for in vivo tumor imaging through active nanoparticle probe targeting.532–536 Kolitz-Domb and co-workers used polymeric fluorescent nanoparticles for in vivo imaging of colon cancer through antibody recognition of carcinoembryonic antigen (CEA).534 Indocyanine green, a near-infrared fluorophore, was encapsulated into proteinoid-poly(lactic acid) nanoparticles, which were used to image colon cancer tumors in a LS174t colorectal cancer orthotopic mouse model. The nanoparticles were administered to mice through the anus, and the animals were sacrificed 4 h postdelivery for ex vivo imaging of nanoparticle tumor accumulation in the colon. Based on the overexpression of CEA by LS174t cells, the nanoparticles localized to tumor tissue and generated fluorescent signal at the tumor sites. In comparison, control nanoparticles without anti-CEA antibody produced no appreciable fluorescence signal, indicating the enhancement of tumor targeting achieved through the use of antibodies.
Aptamers have also been used in the development of nanoparticle probes for in vivo fluorescent imaging of tumor tissue through active targeting.537,538 Tong et al. reported the use of polymeric nanoparticles incorporating Cy5 to target PSMA using a DNA aptamer conjugated to the nanoparticle surface.538 In vitro experiments demonstrated that nanoparticle labeling of prostate cancer cells was specific to LNCaP cells, which have high PSMA expression compared to PC3 cells. Though the nanoparticles were not used to image prostate cancer in vivo, they exhibited no observable adverse effects in mice. In 2015, Ding and co-workers expanded upon this and used an aptamer designed to bind SCG7901 gastric cancer cells through cell-SELEX for in vivo imaging of tumors in a mouse model.537
In addition to the use of peptides, antibodies, and aptamers, small molecules also are used to actively target nanoparticle probes to tumor tissue for cancer diagnostics. In particular, folate is frequently attached to fluorescent nanoparticles to target folate receptors, which are overexpressed by many cancer cells.419,539–543 In one such example, Ma et al. synthesized polymeric nanoparticles loaded with indocyanine green and conjugated to folate for in vivo imaging of xenograft MDA-MB-231 breast cancer tumors in a mouse model.541 Similarly, Xiong and co-workers reported the conjugation of folate to NaYF4:Yb,Er UCNPs for active tumor targeting in cancer diagnostic imaging.543 Folate-tagged UCNPs resulted in an observable fluorescence signal from cervical cancer HeLa tumors in mice 24 h following intravenous injection compared to no observable signal from HeLa tumors in mice injected with control UCNPs lacking the folate tag. To quantify the difference in uptake, inductively coupled plasma atomic emission spectroscopy was used to measure UCNP accumulation in the tumor tissue based on Y3+ content. Approximately 6 times more folate-receptor-targeting UCNPs per gram of tumor tissue than UCNPs without folate was observed, indicating the fold enhancement in nanoparticle accumulation that may be achieved by actively targeting tumor tissue. Such enhancements can also improve fluorescence signal from smaller tumors and enable the early detection of tumors.
Considerable effort has been devoted to the development of nanoparticle probes for simultaneous imaging through both fluorescence and additional techniques, such as MRI, computed tomography (CT), and X-ray imaging. These approaches typically seek to combine the benefits of fluorescence imaging with a complementary imaging modality to expand upon the use of nanoparticle probes for the detection of cancerous tissues in vivo. While the design of multimodal nanoparticle probes has been an active area of research, the infrastructure required to support the use of such technologies, and the availability of instrumentation to conduct fluorescence imaging while simultaneously imaging via an alternative modality, has yet to be developed.
In one such example, Yi et al. reported the design of UCNPs for in vivo imaging through simultaneous upconversion fluorescence and X-ray imaging.391 Similarly, Shen et al. described the synthesis of a UCNP sandwiched structure containing a NaGdF4:Yb/Tm core, a NaLuF4:Yb/Tm middle layer, and a NaYF4 top layer for simultaneous in vivo upconversion fluorescence and CT imaging.386 Zhang et al. described the encapsulation of AuNPs and the organic fluorophore, bis(4-(N-(2-naphthyl)phenylamino)phenyl)-fumaronitrile (NPAPF), in PEGylated micelles for in vivo imaging of colon carcinoma (CT26) tumors through both fluorescence and through CT.392 Combining fluorescence imaging capabilities with either CT or X-ray imaging may be especially useful in the translation of nanoparticle probes from animal models to clinical patient use since the relatively deeper tissue penetration afforded by these methods may be used in parallel with the high sensitivity of fluorescence imaging.
Some approaches to image tumor tissue in vivo focus on the use of magnetofluorescent nanoparticles for simultaneous fluorescent and magnetic resonance imaging. One common approach in the design of magentofluorescent nanoparticles is to synthesize self-assembling fluorophore-labeled polymeric nanoparticles that encapsulate smaller magnetic nanoparticles. In some cases, chitosan nanoparticles containing magnetic and fluorescent components are used for in vivo imaging of tumor tissue through passive targeting by the EPR effect.374,376,379 Since chitosan is a biodegradable polysaccharide, it is regarded as highly biocompatible with low toxicity.361,373 In 2010, Nam et al. used self-assembling chitosan nanoparticles encapsulating a gadolinium (Gd) MRI contrast agent and labeled with the near-infrared dye Cyanine 5.5 (Cy5.5) for squamous carcinoma tumor detection in mice.379 Similarly, Lee et al. reported the use of chitosan nanoparticles for multimodal fluorescence and MRI imaging of tumor tissue in vivo.376 Chitosan nanoparticles encapsulating superparamagnetic iron oxide nanoparticles (SPIONs) with a Cy5.5 fluorophore label were synthesized and intravenously injected into mice bearing glioma U87MG tumors (Figure 27).
Figure 27.
(A) Schematic representation of the synthesis of superparamagnetic iron oxide nanoparticle (SPION) encapsulated in chitosan nanoparticles labeled with Cy5.5. (B) U87MG tumor-bearing mice (B) preinjection, and (C) 1, (D) 3, and (E) 5 h post injection of SPION-loaded Cy5.5-labeled chitosan nanoparticles. Adapted from ref 376. Copyright 2011 American Chemical Society.
Others have utilized antibodies in the development of multimodal imaging platforms for in vivo cancer diagnostics through active tumor targeting. For example, Liu et al. described the use of magnetic, Gd-containing UCNPs for multimodal MRI and fluorescence imaging of small tumors in vivo.535 In another example reported by Chen et al., mesoporous silica nanoparticles were used to actively target tumor tissue for multimodal fluorescence and positron emission tomography (PET) imaging.532 Mesoporous silica nanoparticles were labeled with 64Cu and IR Dye 800, and conjugated to an anti-CD105 TRC105 antibody (TRACON Pharmaceuticals Inc.), which targets a glycoprotein receptor that is overexpressed on tumor vessel endothelial cells and is implicated in angiogenesis and tumor cell proliferation. Following intravenous administration, the 64Cu PET-active label was used to quantify the percentage of the injected dose localized to tumor tissue in vivo. While fluorescence alone may not provide this capability, multimodal imaging systems impart many advantages that may expand upon the capabilities of fluorescence imaging to provide complementary information in cancer diagnosis.
Finally, folate is also used in actively targeting multifunctional nanoparticles to tumor tissue through recognition of folate by folate receptors, as reported by Rolfe et al. in 2014.542 Specifically, a hyperbranched polymer scaffold labeled with Rhodamine B and 19F was assembled into nanoparticles and conjugated to folate to generate magnetofluorescent nanoparticles for multimodal imaging of B16 melanoma tumors in vivo. MRI and fluorescence imaging indicated an enhanced tumor accumulation of magnetofluorescent nanoparticles tagged with folate compared to nanoparticles without folate by combining the high resolution of MRI with the sensitivity of fluorescence imaging within a single nanoparticle construct.
6.2.2. Theranostic Nanoparticle Probes
Theranostic nanoparticle-based applications take advantage of the preferential accumulation of nanoparticles in tumor tissue to simultaneously detect and treat cancer, where simply incorporating a chemotherapeutic drug into a fluorescent nanoparticle used for tumor imaging may also enable therapy. Additionally, many reports describe the use of plasmonically active nanoparticles, such as nanoshells, for tumor detection and light-triggered local heating of cancerous cells, causing death via photothermal therapy. These methods will be discussed in further detail below.
Many reports describe the incorporation of chemotherapeutics into fluorescent nanoparticles for use in theranostic applications.393,394,397–400 These nanoparticles may be passively or actively targeted to the tumor tissue, and are used to treat cancer based upon preferential nanoparticle accumulation in the tumor. A variety of nanoparticles (i.e., polymeric and inorganic) are used in these approaches. The Kwon group, for example, has reported the use of dye-labeled chitosan nanoparticles for the simultaneous detection and treatment of cancer through delivery of paclitaxel to tumor tissue through passive nanoparticle accumulation.398,544 Dye-doped polymeric nanoparticles have also been used to deliver camptothecin402 and irinotecan394 chemotherapeutics for theranostic applications. Additionally, Chen et al. reported the use of magnetofluorescent UCNPs to deliver doxorubicin to tumor tissue via passive targeting for simultaneous cancer imaging and therapy.393
In order to improve nanoparticle’s selectivity for cancer cells, active nanoparticle targeting is often used to deliver theranostic nanoparticle probes to tumor tissue. In some cases, delivery of the targeting moiety to tumor tissue alone may result in a therapeutic response.395,396 Corsi et al. reported the suppression of Her2, a cell surface receptor that promotes cancer cell growth, through active targeting via antibody conjugation to magnetofluorescent UCNPs for cancer imaging applications.395 Clinical treatment of Her2-positive cancers typically utilizes the monoclonal antibody trastuzumab to target Her2 and subsequently inhibit cancer cell proliferation. In their work, Corsi and co-workers demonstrated that conjugation of trastuzumab to UCNPs for in vivo tumor imaging in a breast cancer xenograft mouse model (MCF7) through active targeting also results in a significant decrease in Her2 expression on the tumor cell surface for up to 1 week following intravenous administration (Figure 28). This may be further developed into a theranostic system that can both diagnose and treat Her2-positive cancers.
Figure 28.
Theranostic trastuzumab-conjugated UCNPs are targeted to tumor tissue via active Her2 recognition. The magnetofluorescence of the UCNPs allows for diagnostic imaging, while trastuzumab results in therapeutic down-regulation of Her2. Immunohistochemical analysis of Her2 expression in tumor sections from MCF7 xenografted mice (A) prior to intravenous injection with trastuzumab-conjugated UCNPs, (B) 5 h post injection, (C) 24 h post injection, and (D) 1 week post injection. Note that brown staining corresponds to Her2 while blue represents cell nuclei, and that the images were taken under 40× magnification. Adapted from ref 395. Copyright 2011 American Chemical Society.
In another example of active tumor targeting for theranostic applications, Santra et al. reported the synthesis of dendrimer polymeric nanoparticles encapsulating cytochrome c, a protein known to initiate apoptosis, and the fluorophore indocyanine green.400 The nanoparticles were tagged with folate for targeted theranostic detection and treatment of A549 lung cancer cells, a cell line that overexpresses folate receptors. Selectivity in cellular uptake and death for A549 cells over MCF7 cells, which have low folate receptor expression, was demonstrated in vitro; this approach may later be applied to in vivo theranostic applications.
In addition to chemotherapeutics, photoactive materials may also be delivered to tumor tissue for theranostic imaging and treatment. In these approaches, materials with strong absorbance in the near-infrared region are used to convert optical energy into heat to locally kill cancer cells.545–565 This method, termed photothermal therapy, typically utilizes NIR light, which does not cause damage to cells, to activate photothermal nanoparticles localized in tumor tissue throughout the body. One concern with this approach, however, is that nanoparticles accumulated in the liver and spleen may damage healthy tissue upon irradiation. This disadvantage hinders the applicability of photothermal therapies to treat cancer once metastasis has occurred, and limits its potential use to local treatment of tumors.
Nanoparticles that allow for both imaging and photothermal therapy have demonstrated passive targeting to tumor tissue via the EPR effect. In some cases, silica or polymeric nanoparticles are designed to encapsulate photothermal-sensitizing dyes, such as protoporphyrin IX566 or naphthalocyanine.567 In one such example, Chen et al. reported the use of a conductive polymer-based nanoparticle for photothermal therapy of 4T1 breast cancer tumors in mice.568 Poly (3, 4-ethylenedioxythiophene):poly(4-styrenesulfonte) (PEDOT: PSS) nanoparticles were synthesized via a layer-by-layer assembly process and labeled with Cy5 fluorophores. The resulting nanoparticles were intravenously administered to tumor bearing mice for fluorescent detection of tumors in vivo. At 48 h post injection, mice were irradiated with a 808 nm laser for 5 min, raising the temperature of the tumor tissue to about 50 °C (Figure 29). One day following laser treatment, the tumors were eliminated, while control groups (no treatment, nanoparticle only, or laser only), showed no change in tumor growth.
Figure 29.
Cy5 labeled PEDOT:PSS passively target 4T1 tumors. (A) Fluorescence images 1–48 h post intravenous administration. (B) Tissue temperature with 10 s to 5 min of laser irradiation. Tumor location is indicated by the arrow. Adapted from ref 568. Copyright 2012 American Chemical Society.
Active targeting of nanoparticles for combined imaging and photothermal therapy can enhance the selectivity of nanoparticle accumulation in tumor tissue.569–573 In some examples, polymeric nanoparticles are used to actively target tumor tissue for theranostic applications. The Cai group encapsulated two photosensitizing dyes, indocyanine green and IR-780571 in polymer nanoparticles to target folate receptor in a breast cancer mouse model. Forty-eight hours following intravenous injection with folate-coated indocyanine green encapsulating nanoparticles, mice were irradiated with a near-infrared laser for 5 min, resulting in an increase in intratumoral temperature to 50 °C. This localized heating resulted in significant cancer cell death, as indicated by histological analysis, and complete tumor ablation.
Bardhan et al. reported the use of Her2 antibody-conjugated nanoshells for combined imaging and therapy of breast cancer.569 Indocyanine green and anti-Her2 antibody were conjugated to gold nanoshells and used to selectively image BT474AZ breast cancer tumors with high Her2 expression over MDA-MB-231 breast cancer tumors with low Her2 expression in mouse models. Within 4 h following intravenous injection, strong fluorescence was observed from the tumors of BT474AZ xenografted mice, which was about 2 times the intensity of tumors in MDA-MB-231 xenografted mice. Though the photothermal treatment of tumors was not examined in vivo, nanoshells have been reported for effective cancer cell ablation,574–576 and the anti-Her2 nanoshells may be further explored for theranostics in vivo.
7. SUMMARY AND OUTLOOK
We have reviewed many of the recently reported nanoparticle-mediated methods for the detection of cancer biomarkers, cells, and tissues through fluorescence. Nanotechnology-based assays for cancer detection are an increasingly relevant alternative to traditional techniques. The characteristics of certain nanoparticle probes, such as high surface-area-to-volume ratio and unique optical properties, allow them to overcome some of the limitations of currently available methods of cancer detection.
In particular, the four main nanoparticle types used in the design of probes for the detection of cancer biomarkers, cells, and tissues through fluorescence are (1) QDs, which exhibit tunable absorption and high quantum yields; (2) PDs, which have high fluorescence quantum yields and show no cytotoxicity; (3) UCNPs, which exhibit anti-Stokes shifts and are excitable in the tissue-transparent NIR window; (4) AuNPs, which are excellent fluorescence quenchers and are thus commonly used in “off–on” probes; and (5) fluorophore-encapsulating polymeric or mesoporous silica nanoparticles, which are useful in theranostic applications that simultaneously deliver chemotherapeutics or photosensitizing agents to image and treat cancer cells.
Beyond the nanoparticle probe’s core composition, surface functionalization with a range of moieties (i.e., antibodies, oligonucleotides, aptamers, peptides, and small molecules) can enable the detection of cancer biomarkers, cells, and tissues by imparting high binding affinity and specificity to a target. The high surface-area-to-volume ratio of nanoparticles can enable dense functionalization, and provides tailorable and sometimes multivalent binding of nanoparticle probes to target proteins, cell surface markers, and oligonucleotides that may be indicative of cancer. This property allows one to detect cancer markers with low LODs and high specificity.
As discussed in this review, the combination of the optical properties of certain classes of nanoparticles (QDs, UCNPs, and AuNPs in particular) with a variety of tailorable surface chemistries has been utilized in the design of nanoparticle probes for the detection of cancer. The desirable properties for each nanoparticle probe are often dependent upon the application. Most notably, the design considerations for nanoparticle probes used in the detection of secreted biomarkers do not require the same cellular and systemic biocompatibility that is required for the detection of cancerous tissues in vivo. For example, although though QDs and UCNP exhibit enhanced photostability and are efficiently excited in the tissue-penetrating NIR window, their long-term safety and toxicity have not yet been fully evaluated. Thus, these types of nanoparticles may be more rapidly translated to clinical use for ex vivo cancer biomarker detection after further in-depth studies regarding the safety of systemic QD and UCNP administration are completed.
While a variety of nanoparticle probes have been designed for the detection of cancer biomarkers through fluorescence, no specific assay has been selected as the “gold standard” for clinical use. Perhaps this is due to difficulties in comparing results across reports with differing experimental conditions. Notably, there is a significant difference between LODs that are obtained by measuring pure target compared to those obtained measuring target in the presence of complex mixtures that more accurately represent patient-derived samples, including serum, for example. In general, however, sandwich-type immunoassays which utilize QDs to generate fluorescence in the presence of a target biomarker have continued to demonstrate low LODs, which are attainable due to the high quantum yield of QDs.
There are two main classes of nanoparticle-based fluorescent assays for the detection of cancer cells: those that rely of the binding of nanoparticle probes to cancer cell surface markers, and those that enter cells and detect genetic content. Most of the nanoparticle probes reported operate solely through binding to cell surface markers. These approaches often improve upon the currently available system for detection of CTCs (i.e., CellSearch), due to the enhanced fluorescence properties (i.e., higher photostability, longer fluorescence lifetimes, and high quantum yields) of certain nanoparticles including QDs and UCNPs. However, they are limited in their ability to detect malignant cells that may not express sufficient quantities of the surface marker. The NanoFlare or SmartFlare (commercialized by EMD Millipore), however, overcomes this challenge by providing a method to detect and quantify intracellular genetic content in live cell samples. This approach has been successfully used to detect, isolate, and culture metastatic breast cancer cells from a murine model as well as from human blood samples, and represents a paradigm shift in the ability to detect cancer cell populations based upon genetic markers. Additionally, multiplexed NanoFlares provide the opportunity to distinguish truly malignant cancer cells based upon the expression levels of multiple oncogenes.
In addition to the detection of cancer biomarkers and cells, nanoparticle probes for the detection of cancerous tissues in vivo have also been discussed in this review. One remarkable property of certain nanoparticles, which enables their utility in cancer tissue detection, is their in vivo behavior. Specifically, the preferential uptake of fluorophore-labeled nanoparticle probes into tumor tissues via the EPR effect or through active tumor targeting offers a powerful tool for tumor detection, image-guided therapy, and theranostics. Methods to detect tumor tissue in vivo are enhanced through active targeting based upon the recognition of cancer cell surface markers by providing higher image contrast in less time. This effect is due to the selective and enhanced accumulation of nanoparticles in cancerous tissue.
Compared to alternative imaging modalities, fluorescence offers relatively high sensitivity and tailorable excitation and emission maxima to optimize tissue penetration depths for in vivo imaging. QDs, PDs, and UCNPs, in particular, are ideal for in vivo use due to their tunable NIR profiles and large Stokes and anti-Stokes shifts, respectively, which minimize background signal caused by tissue autofluorescence to enhance image contrast. Though initial studies on the safety of QDs, PDs, and UCNPs in vivo are promising, showing little to no toxicity in mouse and nonhuman primate models, translation of the use of such nanoparticle probes to the clinic will require significant research on the long-term safety of QD and UCNP administration.
Overall, significant effort has been afforded toward the design of nanoparticle probes for the detection of cancer biomarkers, cells, and tissues through fluorescence. A wide range of assays have been developed, and many of them improve upon the currently available detection methods either through enhanced sensitivity and selectivity, or by offering entirely new and unique capabilities that are not attainable with conventional methods.
In addition to their potential use in cancer diagnosis and prognosis, these technologies may also be used to further study the progression of cancer and may aid in our understanding of the disease. Ultimately, translation of nanoparticle probes to clinical cancer diagnosis will require further knowledge of the correlation between levels of cancer biomarkers, cells, or tissues present in a patient with the stage of their disease to enhance prognostic capabilities. This ability will improve cancer patient care by enabling early detection to improve survival outcomes and by providing a means for monitoring the progress of the disease in response to treatment. The latter will lead to the design of better treatment strategies for an individual patient. With an understanding of the strides made in using nanoparticle probes to detect cancer biomarkers, cells, and tissues, as well as the challenges associated with this type of research, researchers in this field are poised to move nanoparticle probes for cancer diagnosis, prognosis, and therapeutics rapidly into the clinic.
Acknowledgments
This work was supported by the following awards: the Center for Cancer Nanotechnology Excellence (CCNE) initiative of the National Institutes of Health (NIH) Award Number U54 CA151880, and the Defense Advanced Research Projects Agency Award Number HR0011-13-2-0018. A.B.C. and C.M.G. were both supported by the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) Program. J.R.F. was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers T32GM105538 and 5R25GM79300. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. S.N.B. acknowledges a National Science Foundation Graduate Research Fellowship and a P.E.O. Scholar Award. T.J.M. acknowledges an IBNAM-Baxter Early Career Development Award in Bioengineering and support from the International Institute for Nanotechnology of Northwestern University.
ABBREVIATIONS
- [Ru(dpp)3]2+Cl2
tris(4,7-diphenyl-1,10-phenanthroline) ruthenium(II) dichloride
- Ab
antibody
- AFP
alpha fetoprotein
- AgNC
silver nanocluster
- AIAT
alpha 1-antitrypsin precursor
- Apt
aptamer
- AuNC
gold nanocluster
- AuNP
gold nanoparticle
- AuNR
gold nanorod
- BDM
2,3-butanedione
- BODIPY
boradiazaindacene
- BOND
bio-orthogonal nanoparticle detection
- C. elegans
Caenorhabditis elegans
- CA 125
cancer antigen 125
- CA 15-3
cancer antigen 15-3
- CEA
carcinoembryonic antigen
- CPN
conjugated polymer nanoparticle
- CRET
chemiluminescence resonance energy transfer
- CRP
C-reactive protein
- CSC
cancer stem cell
- CT
computed tomography
- CTC
circulating tumor cell
- Cy3B
Cyanine 3B
- Cy5
Cyanine 5
- Cy5.5
Cyanine 5.5
- DiR
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotri-carbocyanine iodide
- DMDAP
N, N′-dimethyl-2,7-diazapyrenium dication
- DNA
deoxyribonucleic acid
- dNTPs
deoxynucleotide triphosphates
- Dox
doxorubicin
- DPN
dip pen nanolithography
- dsDNA
double stranded DNA
- DZ
dithizone
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- ELISA
enzyme-linked immunosorbent assay
- EMT
epithelial to mesenchymal transition
- EpCAM
epithelial cell adhesion molecule
- EPR
enhanced permeability and retention
- ER-β
estrogen receptor beta
- Fab
fragment-antigen binding
- Fc
nonhuman crystallizable
- FDA
Food and Drug Administration
- FISH
fluorescence in situ hybridization
- FITC
fluorescein isothiocyanate
- FMBMNs
fluorescent-magnetic biotargeting-multifunctional nanobioprobes
- FPA
fluorescence polarization anisotropy
- FRET
Förster resonance energy transfer
- GFP
green fluorescent protein
- HAase
hyaluronidase
- Hb
hemoglobin
- HbO2
oxyhemoglobin
- Her2
human epidermal growth factor receptor 2
- HUVEC
human umbilical vein endothelial cells
- ICTS
immunochromatography test strip
- IgG
immunoglobulin G
- IR
infrared
- IV
intravenous
- LTC
luminescent terbium chelate
- LOD
limit of detection
- mAb
monoclonal antibody
- MCNT
multiwalled carbon nanotube
- MFNP
magnetofluorescent nanoparticle
- MRI
magnetic resonance imaging
- mRNA
messenger RNA
- NIR
near-infrared
- NPAPF
bis(4-(N-(2-naphthyl)phenylamino)phenyl)-fumaronitrile
- NSE
neuron specific enolase
- NW
nanowire
- OPD
o-phenylenediamine
- OPDox
2,3-diaminophenazine
- PCR
polymerase chain reaction
- PD
polymer dot
- PDDA
poly(diallyldimethylammonium chloride)
- PDGF
platelet derived growth factor
- PEDOT:PSS
poly(3,4-ethylenedioxythiophene):poly(4-styrenesulfonte)
- PEG
poly(ethylene glycol)
- PET
positron emission tomography
- PLA
poly(lactic acid)
- PPE
poly(p-phenyleneethynylene)
- PSA
prostate specific antigen
- PSMA
prostate-specific membrane antigen
- QD
quantum dot
- qRT-PCR
quantitative real time PCR
- RES
reticuloendothelial system
- RGD
arginylglycylaspartic acid
- RNA
ribonucleic acid
- Rubpy
tris(2,2′-bipyridyl)dichlororuthenium(II)
- scFv
single-chain variable fragment
- SELEX
systematic evolution of ligands by exponential enrichment
- SNA
spherical nucleic acid
- SPECT
single photon emission computed tomography
- SPION
superparamagnetic iron oxide nanoparticle
- ssDNA
single stranded DNA
- SCNT
single-walled carbon nanotube
- TCO
trans-cyclooctene
- TPSA
total prostate specific antigen
- Tz
tetrazine
- UCNP
upconversion nanoparticle
- VEGF
vascular endothelial growth factor
Biographies
Alyssa B. Chinen earned her B.S. in chemistry with a minor in biomedical engineering and departmental honors at Carnegie Mellon University in Pittsburgh, Pennsylvania, in 2011. As an undergraduate, she conducted research under the guidance of Prof. Bruce Armitage and Prof. Catalina Achim to design fluorescent biosensors using protein, DNA, and PNA scaffolds. Currently, she is a Ph.D. candidate in the Department of Chemistry at Northwestern University and is a National Defense Science and Engineering Graduate Fellow. Under the mentorship of Prof. Chad Mirkin, her current research focuses on the use of NanoFlares in cancer diagnostics, as well as understanding the interactions of spherical nucleic acids with serum proteins to enable the design of more efficacious therapeutic nanoparticles.
Chenxia M. Guan received her dual B.S. in biology and chemical-biological engineering from the Massachusetts Institute for Technology in 2011. There, she worked in Prof. Robert Langer’s laboratory to develop delivery vehicles for siRNA as anticancer therapeutics. She is currently a Ph.D. candidate advised by Prof. Chad Mirkin at Northwestern in the Department of Chemical and Biological Engineering, and is also a National Defense Science and Engineering Graduate Fellowship Awardee. Her research is focused on understanding how spherical nucleic acids behave in vivo and utilizing them for cancer immunotherapy applications.
Jennifer R. Ferrer received her B.S. in molecular and cellular biology from the University of Illinois at Urbana–Champaign in 2007. She received her M.S. in biotechnology from Rush University in 2010. She is currently a Ph.D. candidate in the Interdisciplinary Biological Sciences Program, advised by Prof. Chad Mirkin and Prof. Jason Wertheim at Northwestern. Her current research focuses on the application of Toll-like receptor modulating liposomal SNAs for liver disease and cancer immunotherapy.
Stacey N. Barnaby received a B.S. in chemistry from Fordham University in 2011. While at Fordham, she studied the self-assembly and biomineralization of metal nanoparticles using plant phytohormones under the guidance of Prof. Ipsita A. Banerjee. She is currently a Ph.D. candidate in chemistry, a National Science Foundation Graduate Research Fellow, and a P.E.O. Scholar in Prof. Chad Mirkin’s group at Northwestern University. Her current research focuses on investigating the interaction of enzymes with oligonucleotides immobilized on nanoparticle surfaces. She is exploring these materials for applications in medicine, biology, and catalysis.
Timothy J. Merkel earned his B.A. degree in chemistry at Swarthmore College in 1999 and received his M.S. in chemistry from Villanova University in 2006. He earned a Ph.D. in polymer chemistry from the University of North Carolina at Chapel Hill in 2011, where he studied the design, synthesis, in vivo interactions, and biological applications of biomimetic microgel particles under the guidance of Prof. Joseph M. DeSimone. He joined the Mirkin lab at Northwestern University as a postdoctoral fellow in 2011, where his research has focused on the design and application of spherical nucleic acids as therapeutic and diagnostic tools for cancer and other diseases.
Chad A. Mirkin is the Director of the International Institute for Nanotechnology, the George B. Rathmann Professor of Chemistry, Professor of Chemical and Biological Engineering, Professor of Biomedical Engineering, Professor of Materials Science and Engineering, and Professor of Medicine at Northwestern University. He is a world-renowned nanoscience expert, who has authored over 600 manuscripts and over 900 patent applications worldwide (259 issued). He is the founder of multiple companies, including Nanosphere, AuraSense, and AuraSense Therapeutics, which are commercializing nanotechnology applications in the life science industry. Prof. Mirkin has been recognized for his accomplishments with over 100 national and international awards, is the only chemist to be elected to all three U.S. National Academies, and is a member of the President’s Council of Advisors on Science & Technology (Obama Administration). Prof. Mirkin holds a B.S. degree from Dickinson College (1986, elected into Phi Beta Kappa) and a Ph.D. degree in chemistry from Pennsylvania State University (1989). He was a National Science Foundation Postdoctoral Fellow at the Massachusetts Institute for Technology prior to becoming a professor at Northwestern University in 1991.
Footnotes
Author Contributions
A.B.C. and C.M.G. contributed equally.
The authors declare the following competing financial interest(s): C.A.M. is a cofounder of AuraSense, LLC, a startup biotechnology company that licenses the NanoFlare technology from Northwestern University.
References
- 1.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer Treatment and Survivorship Statistics, 2014. Ca-Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2014 www.who.int.
- 3.Siegel R, Ma J, Zou Z, Jemal A. Cancer Statistics, 2014. Ca-Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Higgins MJ, Ettinger DS. Chemotherapy for Lung Cancer: The State of the Art in 2009. Expert Rev Anticancer Ther. 2009;9:1365–1378. doi: 10.1586/era.09.115. [DOI] [PubMed] [Google Scholar]
- 7.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early Detection of Squamous Cell Lung Cancer in Sputum by a Panel of Microrna Markers. Mod Pathol. 2010;23:1157–1164. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 8.Choi YE, Kwak JW, Park JW. Nanotechnology for Early Cancer Detection. Sensors. 2010;10:428–455. doi: 10.3390/s100100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari M. Cancer Nanotechnology: Opportunities and Challenges. Nat Rev Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- 10.Nie S, Xing Y, Kim GJ, Simons JW. Nanotechnology Applications in Cancer. Annu Rev Biomed Eng. 2007;9:257–288. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 11.Yao J, Yang M, Duan Y. Chemistry, Biology, and Medicine of Fluorescent Nanomaterials and Related Systems: New Insights into Biosensing, Bioimaging, Genomics, Diagnostics, and Therapy. Chem Rev. 2014;114:6130–6178. doi: 10.1021/cr200359p. [DOI] [PubMed] [Google Scholar]
- 12.Wolfbeis OS. An Overview of Nanoparticles Commonly Used in Fluorescent Bioimaging. Chem Soc Rev. 2015;44:4743. doi: 10.1039/c4cs00392f. [DOI] [PubMed] [Google Scholar]
- 13.Smith AM, Duan H, Mohs AM, Nie S. Bioconjugated Quantum Dots for in Vivo Molecular and Cellular Imaging. Adv Drug Delivery Rev. 2008;60:1226–1240. doi: 10.1016/j.addr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang X, Jain PK, El-Sayed IH, El-Sayed MA. Gold Nanoparticles: Interesting Optical Properties and Recent Applications in Cancer Diagnostics and Therapy. Nanomedicine (London, U K) 2007;2:681–693. doi: 10.2217/17435889.2.5.681. [DOI] [PubMed] [Google Scholar]
- 15.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Gold Nanoparticles for Biology and Medicine. Angew Chem, Int Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng L, Wang C, Liu Z. Upconversion Nanoparticles and Their Composite Nanostructures for Biomedical Imaging and Cancer Therapy. Nanoscale. 2013;5:23–37. doi: 10.1039/c2nr32311g. [DOI] [PubMed] [Google Scholar]
- 17.Wu C, Chiu DT. Highly Fluorescent Semiconducting Polymer Dots for Biology and Medicine. Angew Chem, Int Ed. 2013;52:3086–3109. doi: 10.1002/anie.201205133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuncel D, Demir HV. Conjugated Polymer Nanoparticles. Nanoscale. 2010;2:484–494. doi: 10.1039/b9nr00374f. [DOI] [PubMed] [Google Scholar]
- 19.Rosenholm JM, Sahlgren C, Linden M. Multifunctional Mesoporous Silica Nanoparticles for Combined Therapeutic, Diagnostic and Targeted Action in Cancer Treatment. Curr Drug Targets. 2011;12:1166–1186. doi: 10.2174/138945011795906624. [DOI] [PubMed] [Google Scholar]
- 20.Pridgen EM, Langer R, Farokhzad OC. Biodegradable, Polymeric Nanoparticle Delivery Systems for Cancer Therapy. Nanomedicine (London, U K) 2007;2:669–680. doi: 10.2217/17435889.2.5.669. [DOI] [PubMed] [Google Scholar]
- 21.Berezin MY, Achilefu S. Fluorescence Lifetime Measurements and Biological Imaging. Chem Rev. 2010;110:2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berezin MY, Achilefu S. Fluorescence Lifetime Measurements and Biological Imaging. Chem Rev. 2010;110:2641–2684. doi: 10.1021/cr900343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akers WJ, Berezin MY, Lee H, Guo K, Almutairi A, Frechet JMJ, Fischer GM, Daltrozzo E, Achilefu S. Biological Applications of Fluorescence Lifetime Imaging Beyond Microscopy. Proc SPIE. 2010;7576:757612. [Google Scholar]
- 24.Yao J, Yang M, Duan YX. Chemistry, Biology, and Medicine of Fluorescent Nanomaterials and Related Systems: New Insights into Biosensing, Bioimaging, Genomics, Diagnostics, and Therapy. Chem Rev. 2014;114:6130–6178. doi: 10.1021/cr200359p. [DOI] [PubMed] [Google Scholar]
- 25.Förster T. Zwischenmolekulare Energiewanderung Und Fluoreszenz. Ann Phys (Berlin, Ger) 1948;437:55–75. [Google Scholar]
- 26.Piston DW, Kremers GJ. Fluorescent Protein Fret: The Good, the Bad and the Ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Chan WCW, Maxwell DJ, Gao XH, Bailey RE, Han MY, Nie SM. Luminescent Quantum Dots for Multiplexed Biological Detection and Imaging. Curr Opin Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 28.Alivisatos AP. Semiconductor Clusters, Nanocrystals, and Quantum Dots. Science. 1996;271:933–937. [Google Scholar]
- 29.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 30.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum Dots for Live Cells, in Vivo Imaging, and Diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han MY, Gao XH, Su JZ, Nie S. Quantum-Dot-Tagged Microbeads for Multiplexed Optical Coding of Biomolecules. Nat Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 32.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R, Nann T. Quantum Dots Versus Organic Dyes as Fluorescent Labels. Nat Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 33.Yu WW, Qu LH, Guo WZ, Peng XG. Experimental Determination of the Extinction Coefficient of Cdte, Cdse, and Cds Nanocrystals. Chem Mater. 2003;15:2854–2860. [Google Scholar]
- 34.Kucur E, Boldt FM, Cavaliere-Jaricot S, Ziegler J, Nann T. Quantitative Analysis of Cadmium Selenide Nanocrystal Concentration by Comparative Techniques. Anal Chem. 2007;79:8987–8993. doi: 10.1021/ac0715064. [DOI] [PubMed] [Google Scholar]
- 35.Sackett DL, Wolff J. Nile Red as a Polarity-Sensitive Fluorescent-Probe of Hydrophobic Protein Surfaces. Anal Biochem. 1987;167:228–234. doi: 10.1016/0003-2697(87)90157-6. [DOI] [PubMed] [Google Scholar]
- 36.Seybold PG, Gouterman M, Callis J. Calorimetric Photometric and Lifetime Determinations of Fluorescence Yields of Fluorescein Dyes. Photochem Photobiol. 1969;9:229. doi: 10.1111/j.1751-1097.1969.tb07287.x. [DOI] [PubMed] [Google Scholar]
- 37.Mujumdar RB, Ernst LA, Mujumdar SR, Lewis CJ, Waggoner AS. Cyanine Dye Labeling Reagents - Sulfoindocyanine Succinimidyl Esters. Bioconjugate Chem. 1993;4:105–111. doi: 10.1021/bc00020a001. [DOI] [PubMed] [Google Scholar]
- 38.Soper SA, Mattingly QL. Steady-State and Picosecond Laser Fluorescence Studies of Nonradiative Pathways in Tricarbocyanine Dyes - Implications to the Design of near-Ir Fluorochromes with High Fluorescence Efficiencies. J Am Chem Soc. 1994;116:3744–3752. [Google Scholar]
- 39.Efros AL, Rosen M. The Electronic Structure of Semiconductor Nanocrystals. Annu Rev Mater Sci. 2000;30:475–521. [Google Scholar]
- 40.Dahan M, Laurence T, Pinaud F, Chemla DS, Alivisatos AP, Sauer M, Weiss S. Time-Gated Biological Imaging by Use of Colloidal Quantum Dots. Opt Lett. 2001;26:825–827. doi: 10.1364/ol.26.000825. [DOI] [PubMed] [Google Scholar]
- 41.Szymanski C, Wu C, Hooper J, Salazar MA, Perdomo A, Dukes A, McNeill J. Single Molecule Nanoparticles of the Conjugated Polymer Meh-Ppv, Preparation and Characterization by near-Field Scanning Optical Microscopy. J Phys Chem B. 2005;109:8543–8546. doi: 10.1021/jp051062k. [DOI] [PubMed] [Google Scholar]
- 42.Wu C, Peng H, Jiang Y, McNeill J. Energy Transfer Mediated Fluorescence from Blended Conjugated Polymer Nanoparticles. J Phys Chem B. 2006;110:14148–14154. doi: 10.1021/jp0618126. [DOI] [PubMed] [Google Scholar]
- 43.Wu C, Szymanski C, McNeill J. Preparation and Encapsulation of Highly Fluorescent Conjugated Polymer Nanoparticles. Langmuir. 2006;22:2956–2960. doi: 10.1021/la060188l. [DOI] [PubMed] [Google Scholar]
- 44.Wu C, Bull B, Szymanski C, Christensen K, McNeill J. Multicolor Conjugated Polymer Dots for Biological Fluorescence Imaging. ACS Nano. 2008;2:2415–2423. doi: 10.1021/nn800590n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C, Schneider T, Zeigler M, Yu J, Schiro PG, Burnham DR, McNeill JD, Chiu DT. Bioconjugation of Ultrabright Semiconducting Polymer Dots for Specific Cellular Targeting. J Am Chem Soc. 2010;132:15410–15417. doi: 10.1021/ja107196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li K, Liu Y, Pu KY, Feng SS, Zhan R, Liu B. Polyhedral Oligomeric Silsesquioxanes-Containing Conjugated Polymer Loaded PLGA Nanoparticles with Trastuzumab (Herceptin) Functionalization for HER2-Positive Cancer Cell Detection. Adv Funct Mater. 2011;21:287–294. [Google Scholar]
- 47.Kim S, Lim CK, Na J, Lee YD, Kim K, Choi K, Leary JF, Kwon IC. Conjugated Polymer Nanoparticles for Biomedical in Vivo Imaging. Chem Commun (Cambridge, U K) 2010;46:1617–1619. doi: 10.1039/b923309a. [DOI] [PubMed] [Google Scholar]
- 48.Wu C, Hansen SJ, Hou Q, Yu J, Zeigler M, Jin Y, Burnham DR, McNeill JD, Olson JM, Chiu DT. Design of Highly Emissive Polymer Dot Bioconjugates for in Vivo Tumor Targeting. Angew Chem, Int Ed. 2011;50:3430–3434. doi: 10.1002/anie.201007461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang F, Banerjee D, Liu YS, Chen XY, Liu XG. Upconversion Nanoparticles in Biological Labeling, Imaging, and Therapy. Analyst. 2010;135:1839–1854. doi: 10.1039/c0an00144a. [DOI] [PubMed] [Google Scholar]
- 50.Suyver JF, Aebischer A, Biner D, Gerner P, Grimm J, Heer S, Kramer KW, Reinhard C, Gudel HU. Novel Materials Doped with Trivalent Lanthanides and Transition Metal Ions Showing near-Infrared to Visible Photon Upconversion. Opt Mater. 2005;27:1111–1130. [Google Scholar]
- 51.Wang F, Liu XG. Upconversion Multicolor Fine-Tuning: Visible to near-Infrared Emission from Lanthanide-Doped Nayf4 Nanoparticles. J Am Chem Soc. 2008;130:5642. doi: 10.1021/ja800868a. [DOI] [PubMed] [Google Scholar]
- 52.Wang F, Liu XG. Recent Advances in the Chemistry of Lanthanide-Doped Upconversion Nanocrystals. Chem Soc Rev. 2009;38:976–989. doi: 10.1039/b809132n. [DOI] [PubMed] [Google Scholar]
- 53.Chen G, Qju H, Prasad PN, Chen X. Upconversion Nanoparticles: Design, Nanochemistry, and Applications in Theranostics. Chem Rev. 2014;114:5161–5214. doi: 10.1021/cr400425h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang M, Abbineni G, Clevenger A, Mao CB, Xu SK. Upconversion Nanoparticles: Synthesis, Surface Modification and Biological Applications. Nanomedicine. 2011;7:710–729. doi: 10.1016/j.nano.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with Plasmonic Nanosensors. Nat Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- 56.Jain PK, Huang XH, El-Sayed IH, El-Sayed MA. Noble Metals on the Nanoscale: Optical and Photothermal Properties and Some Applications in Imaging, Sensing, Biology, and Medicine. Acc Chem Res. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 57.Homola J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 58.Hutter E, Fendler JH. Exploitation of Localized Surface Plasmon Resonance. Adv Mater. 2004;16:1685–1706. [Google Scholar]
- 59.Kelly KL, Coronado E, Zhao LL, Schatz GC. The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. J Phys Chem B. 2003;107:668–677. [Google Scholar]
- 60.Willets KA, Van Duyne RP. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 61.Jin RC. Quantum Sized, Thiolate-Protected Gold Nanoclusters. Nanoscale. 2010;2:343–362. doi: 10.1039/b9nr00160c. [DOI] [PubMed] [Google Scholar]
- 62.Qian HF, Zhu MZ, Wu ZK, Jin RC. Quantum Sized Gold Nanoclusters with Atomic Precision. Acc Chem Res. 2012;45:1470–1479. doi: 10.1021/ar200331z. [DOI] [PubMed] [Google Scholar]
- 63.Lytton-Jean AK, Mirkin CA. A Thermodynamic Investigation into the Binding Properties of DNA Functionalized Gold Nanoparticle Probes and Molecular Fluorophore Probes. J Am Chem Soc. 2005;127:12754–12755. doi: 10.1021/ja052255o. [DOI] [PubMed] [Google Scholar]
- 64.Pierschbacher M, Hayman EG, Ruoslahti E. Synthetic Peptide with Cell Attachment Activity of Fibronectin. Proc Natl Acad Sci U S A. 1983;80:1224–1227. doi: 10.1073/pnas.80.5.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pierschbacher MD, Ruoslahti E. Cell Attachment Activity of Fibronectin Can Be Duplicated by Small Synthetic Fragments of the Molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 66.Yamada KM, Kennedy DW. Dualistic Nature of Adhesive Protein Function: Fibronectin and Its Biologically Active Peptide Fragments Can Autoinhibit Fibronectin Function. J Cell Biol. 1984;99:29–36. doi: 10.1083/jcb.99.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruoslahti E, Pierschbacher MD. Arg-Gly-Asp: A Versatile Cell Recognition Signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 68.Zitzmann S, Ehemann V, Schwab M. Arginine-Glycine-Aspartic Acid (Rgd)-Peptide Binds to Both Tumor and Tumor-Endothelial Cells in Vivo. Cancer Res. 2002;62:5139–5143. [PubMed] [Google Scholar]
- 69.Danhier F, Le Breton A, Preat V. Rgd-Based Strategies to Target Alpha(V) Beta(3) Integrin in Cancer Therapy and Diagnosis. Mol Pharmaceutics. 2012;9:2961–2973. doi: 10.1021/mp3002733. [DOI] [PubMed] [Google Scholar]
- 70.Desgrosellier JS, Cheresh DA. Integrins in Cancer: Biological Implications and Therapeutic Opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seguin L, Desgrosellier JS, Weis SM, Cheresh DA. Integrins and Cancer: Regulators of Cancer Stemness, Metastasis, and Drug Resistance. Trends Cell Biol. 2015;25:234–240. doi: 10.1016/j.tcb.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fletcher J. The Plasma Clearance and Liver Uptake of Iron from Transferrin of Low and High Iron Saturation. Clin Sci. 1971;41:395–402. doi: 10.1042/cs0410395. [DOI] [PubMed] [Google Scholar]
- 73.Daniels TR, Delgado T, Helguera G, Penichet ML. The Transferrin Receptor Part Ii: Targeted Delivery of Therapeutic Agents into Cancer Cells. Clin Immunol. 2006;121:159–176. doi: 10.1016/j.clim.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 74.Daniels TR, Delgado T, Rodriguez JA, Helguera G, Penichet ML. The Transferrin Receptor Part I: Biology and Targeting with Cytotoxic Antibodies for the Treatment of Cancer. Clin Immunol. 2006;121:144–158. doi: 10.1016/j.clim.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 75.Gatter KC, Brown G, Trowbridge IS, Woolston RE, Mason DY. Transferrin Receptors in Human Tissues: Their Distribution and Possible Clinical Relevance. J Clin Pathol. 1983;36:539–545. doi: 10.1136/jcp.36.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Esterly NB, Brammer SR, Crounse RG. The Relationship of Transferrin and Iron to Serum Inhibition of Candida Albicans. J Invest Dermatol. 1967;49:437–442. [PubMed] [Google Scholar]
- 77.Kohler G, Milstein C. Continuous Cultures of Fused Cells Secreting Antibody of Predefined Specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 78.Carter PJ. Potent Antibody Therapeutics by Design. Nat Rev Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 79.Byrne JD, Betancourt T, Brannon-Peppas L. Active Targeting Schemes for Nanoparticle Systems in Cancer Therapeutics. Adv Drug Delivery Rev. 2008;60:1615–1626. doi: 10.1016/j.addr.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 80.Shen BQ, Xu K, Liu L, Raab H, Bhakta S, Kenrick M, Parsons-Reponte KL, Tien J, Yu SF, Mai E, et al. Conjugation Site Modulates the in Vivo Stability and Therapeutic Activity of Antibody-Drug Conjugates. Nat Biotechnol. 2012;30:184–189. doi: 10.1038/nbt.2108. [DOI] [PubMed] [Google Scholar]
- 81.Brinkley M. A Brief Survey of Methods for Preparing Protein Conjugates with Dyes, Haptens, and Cross-Linking Reagents. Bioconjugate Chem. 1992;3:2–13. doi: 10.1021/bc00013a001. [DOI] [PubMed] [Google Scholar]
- 82.Panchuk-Voloshina N, Haugland RP, Bishop-Stewart J, Bhalgat MK, Millard PJ, Mao F, Leung WY, Haugland RP. Alexa Dyes, a Series of New Fluorescent Dyes That Yield Exceptionally Bright, Photostable Conjugates. J Histochem Cytochem. 1999;47:1179–1188. doi: 10.1177/002215549904700910. [DOI] [PubMed] [Google Scholar]
- 83.Kumar S, Aaron J, Sokolov K. Directional Conjugation of Antibodies to Nanoparticles for Synthesis of Multiplexed Optical Contrast Agents with Both Delivery and Targeting Moieties. Nat Protoc. 2008;3:314–320. doi: 10.1038/nprot.2008.1. [DOI] [PubMed] [Google Scholar]
- 84.Pathak S, Davidson MC, Silva GA. Characterization of the Functional Binding Properties of Antibody Conjugated Quantum Dots. Nano Lett. 2007;7:1839–1845. doi: 10.1021/nl062706i. [DOI] [PubMed] [Google Scholar]
- 85.Begent RH, Verhaar MJ, Chester KA, Casey JL, Green AJ, Napier MP, Hope-Stone LD, Cushen N, Keep PA, Johnson CJ, et al. Clinical Evidence of Efficient Tumor Targeting Based on Single-Chain Fv Antibody Selected from a Combinatorial Library. Nat Med. 1996;2:979–984. doi: 10.1038/nm0996-979. [DOI] [PubMed] [Google Scholar]
- 86.Mayer A, Tsiompanou E, O’Malley D, Boxer GM, Bhatia J, Flynn AA, Chester KA, Davidson BR, Lewis AA, Winslet MC, et al. Radioimmunoguided Surgery in Colorectal Cancer Using a Genetically Engineered Anti-Cea Single-Chain Fv Antibody. Clin Cancer Res. 2000;6:1711–1719. [PubMed] [Google Scholar]
- 87.Chapman AP, Antoniw P, Spitali M, West S, Stephens S, King DJ. Therapeutic Antibody Fragments with Prolonged in Vivo Half-Lives. Nat Biotechnol. 1999;17:780–783. doi: 10.1038/11717. [DOI] [PubMed] [Google Scholar]
- 88.Hwang WY, Foote J. Immunogenicity of Engineered Antibodies. Methods. 2005;36:3–10. doi: 10.1016/j.ymeth.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 89.Green LL, Hardy MC, Maynard-Currie CE, Tsuda H, Louie DM, Mendez MJ, Abderrahim H, Noguchi M, Smith DH, Zeng Y, et al. Antigen-Specific Human Monoclonal Antibodies from Mice Engineered with Human Ig Heavy and Light Chain Yacs. Nat Genet. 1994;7:13–21. doi: 10.1038/ng0594-13. [DOI] [PubMed] [Google Scholar]
- 90.Lonberg N, Taylor LD, Harding FA, Trounstine M, Higgins KM, Schramm SR, Kuo CC, Mashayekh R, Wymore K, McCabe JG, et al. Antigen-Specific Human Antibodies from Mice Comprising Four Distinct Genetic Modifications. Nature. 1994;368:856–859. doi: 10.1038/368856a0. [DOI] [PubMed] [Google Scholar]
- 91.Goletz S, Christensen PA, Kristensen P, Blohm D, Tomlinson I, Winter G, Karsten U. Selection of Large Diversities of Antiidiotypic Antibody Fragments by Phage Display. J Mol Biol. 2002;315:1087–1097. doi: 10.1006/jmbi.2001.5314. [DOI] [PubMed] [Google Scholar]
- 92.Brannon-Peppas L, Blanchette JO. Nanoparticle and Targeted Systems for Cancer Therapy. Adv Drug Delivery Rev. 2004;56:1649–1659. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 93.Wu AM, Senter PD. Arming Antibodies: Prospects and Challenges for Immunoconjugates. Nat Biotechnol. 2005;23:1137–1146. doi: 10.1038/nbt1141. [DOI] [PubMed] [Google Scholar]
- 94.Xiang D, Shigdar S, Qiao G, Wang T, Kouzani AZ, Zhou SF, Kong L, Li Y, Pu C, Duan W. Nucleic Acid Aptamer-Guided Cancer Therapeutics and Diagnostics: The Next Generation of Cancer Medicine. Theranostics. 2015;5:23–42. doi: 10.7150/thno.10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ellington AD, Szostak JW. In Vitro Selection of Rna Molecules That Bind Specific Ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 96.Tuerk C, Gold L. Systematic Evolution of Ligands by Exponential Enrichment: Rna Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 97.Shamah SM, Healy JM, Cload ST. Complex Target Selex. Acc Chem Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 98.Nam JM, Thaxton CS, Mirkin CA. Nanoparticle-Based Bio-Bar Codes for the Ultrasensitive Detection of Proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 99.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, Klein WL, Mirkin CA. Nanoparticle-Based Detection in Cerebral Spinal Fluid of a Soluble Pathogenic Biomarker for Alzheimer’s Disease. Proc Natl Acad Sci U S A. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goluch ED, Nam JM, Georganopoulou DG, Chiesl TN, Shaikh KA, Ryu KS, Barron AE, Mirkin CA, Liu C. A Bio-Barcode Assay for on-Chip Attomolar-Sensitivity Protein Detection. Lab Chip. 2006;6:1293–1299. doi: 10.1039/b606294f. [DOI] [PubMed] [Google Scholar]
- 101.Hill HD, Mirkin CA. The Bio-Barcode Assay for the Detection of Protein and Nucleic Acid Targets Using Dtt-Induced Ligand Exchange. Nat Protoc. 2006;1:324–336. doi: 10.1038/nprot.2006.51. [DOI] [PubMed] [Google Scholar]
- 102.Stoeva SI, Lee JS, Smith JE, Rosen ST, Mirkin CA. Multiplexed Detection of Protein Cancer Markers with Biobarcoded Nanoparticle Probes. J Am Chem Soc. 2006;128:8378–8379. doi: 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- 103.Tang S, Zhao J, Storhoff JJ, Norris PJ, Little RF, Yarchoan R, Stramer SL, Patno T, Domanus M, Dhar A, et al. Early Detection of Human Immunodeficiency Type 1 Capsid (P24) Antigen. JAIDS J Acquired Immune Defic Syndr. 2007;46:231–237. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- 104.Kim EY, Stanton J, Korber BT, Krebs K, Bogdan D, Kunstman K, Wu S, Phair JP, Mirkin CA, Wolinsky SM. Detection of Hiv-1 P24 Gag in Plasma by a Nanoparticle-Based Bio-Barcode-Amplification Method. Nanomedicine (London, U K) 2008;3:293–303. doi: 10.2217/17435889.3.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee JS, Ulmann PA, Han MS, Mirkin CA. A DNA-Gold Nanoparticle-Based Colorimetric Competition Assay for the Detection of Cysteine. Nano Lett. 2008;8:529–533. doi: 10.1021/nl0727563. [DOI] [PubMed] [Google Scholar]
- 106.Xu X, Zhao Z, Qin L, Wei W, Levine JE, Mirkin CA. Fluorescence Recovery Assay for the Detection of Protein-DNA Binding. Anal Chem. 2008;80:5616–5621. doi: 10.1021/ac8007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zheng G, Daniel WL, Mirkin CA. A New Approach to Amplified Telomerase Detection with Polyvalent Oligonucleotide Nanoparticle Conjugates. J Am Chem Soc. 2008;130:9644–9645. doi: 10.1021/ja803035p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goluch ED, Stoeva SI, Lee JS, Shaikh KA, Mirkin CA, Liu C. A Microfluidic Detection System Based Upon a Surface Immobilized Biobarcode Assay. Biosens Bioelectron. 2009;24:2397–2403. doi: 10.1016/j.bios.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim YP, Daniel WL, Xia Z, Xie H, Mirkin CA, Rao J. Bioluminescent Nanosensors for Protease Detection Based Upon Gold Nanoparticle-Luciferase Conjugates. Chem Commun (Cambridge, U K) 2010;46:76–78. doi: 10.1039/b915612g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Surinova S, Schiess R, Huttenhain R, Cerciello F, Wollscheid B, Aebersold R. On the Development of Plasma Protein Biomarkers. J Proteome Res. 2011;10:5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 111.Findeisen P, Neumaier M. Functional Protease Profiling for Diagnosis of Malignant Disease. Proteomics: Clin Appl. 2012;6:60–78. doi: 10.1002/prca.201100058. [DOI] [PubMed] [Google Scholar]
- 112.Hakomori S. Tumor-Associated Carbohydrate Antigens Defining Tumor Malignancy: Basis for Development of Anti-Cancer Vaccines. Adv Exp Med Biol. 2001;491:369–402. doi: 10.1007/978-1-4615-1267-7_24. [DOI] [PubMed] [Google Scholar]
- 113.Fuzery A, Levin J, Chan M, Chan D. Translation of Proteomic Biomarkers into FDA Approved Cancer Diagnostics: Issues and Challenges. Clin Proteomics. 2013;10:13. doi: 10.1186/1559-0275-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ueda K. Glycoproteomic Strategies: From Discovery to Clinical Application of Cancer Carbohydrate Biomarkers. Proteomics: Clin Appl. 2013;7:607–617. doi: 10.1002/prca.201200123. [DOI] [PubMed] [Google Scholar]
- 115.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 116.Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA Array Detection with Nanoparticle Probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 117.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman Spectroscopic Fingerprints for DNA and RNA Detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 118.Park SJ, Taton TA, Mirkin CA. Array-Based Electrical Detection of DNA with Nanoparticle Probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 119.Bailey RC, Nam JM, Mirkin CA, Hupp JT. Real-Time Multicolor DNA Detection with Chemoresponsive Diffraction Gratings and Nanoparticle Probes. J Am Chem Soc. 2003;125:13541–13547. doi: 10.1021/ja035479k. [DOI] [PubMed] [Google Scholar]
- 120.Nam JM, Stoeva SI, Mirkin CA. Bio-Bar-Code-Based DNA Detection with Pcr-Like Sensitivity. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 121.Cao YC, Jin R, Thaxton CS, Mirkin CA. A Two-Color-Change, Nanoparticle-Based Method for DNA Detection. Talanta. 2005;67:449–455. doi: 10.1016/j.talanta.2005.06.063. [DOI] [PubMed] [Google Scholar]
- 122.Gibbs JM, Park SJ, Anderson DR, Watson KJ, Mirkin CA, Nguyen ST. Polymer-DNA Hybrids as Electrochemical Probes for the Detection of DNA. J Am Chem Soc. 2005;127:1170–1178. doi: 10.1021/ja046931i. [DOI] [PubMed] [Google Scholar]
- 123.Pokorski JK, Nam JM, Vega RA, Mirkin CA, Appella DH. Cyclopentane-Modified Pna Improves the Sensitivity of Nanoparticle-Based Scanometric DNA Detection. Chem Commun (Cambridge, U K) 2005:2101–2103. doi: 10.1039/b418383e. [DOI] [PubMed] [Google Scholar]
- 124.Jin R, Cao YC, Thaxton CS, Mirkin CA. Glass-Bead-Based Parallel Detection of DNA Using Composite Raman Labels. Small. 2006;2:375–380. doi: 10.1002/smll.200500322. [DOI] [PubMed] [Google Scholar]
- 125.Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Multiplexed DNA Detection with Biobarcoded Nanoparticle Probes. Angew Chem, Int Ed. 2006;45:3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 126.Thaxton CS, Georganopoulou DG, Mirkin CA. Gold Nanoparticle Probes for the Detection of Nucleic Acid Targets. Clin Chim Acta. 2006;363:120–126. doi: 10.1016/j.cccn.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 127.Hill HD, Vega RA, Mirkin CA. Nonenzymatic Detection of Bacterial Genomic DNA Using the Bio Bar Code Assay. Anal Chem. 2007;79:9218–9223. doi: 10.1021/ac701626y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lytton-Jean AK, Han MS, Mirkin CA. Microarray Detection of Duplex and Triplex DNA Binders with DNA-Modified Gold Nanoparticles. Anal Chem. 2007;79:6037–6041. doi: 10.1021/ac070635h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xu X, Georganopoulou DG, Hill HD, Mirkin CA. Homogeneous Detection of Nucleic Acids Based Upon the Light Scattering Properties of Silver-Coated Nanoparticle Probes. Anal Chem. 2007;79:6650–6654. doi: 10.1021/ac070867g. [DOI] [PubMed] [Google Scholar]
- 130.Schwarzenbach H, Hoon DS, Pantel K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 131.Alhasan AH, Kim DY, Daniel WL, Watson E, Meeks JJ, Thaxton CS, Mirkin CA. Scanometric Microrna Array Profiling of Prostate Cancer Markers Using Spherical Nucleic Acid-Gold Nanoparticle Conjugates. Anal Chem. 2012;84:4153–4160. doi: 10.1021/ac3004055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lutz AM, Willmann JK, Cochran FV, Ray P, Gambhir SS. Cancer Screening: A Mathematical Model Relating Secreted Blood Biomarker Levels to Tumor Sizes. PLoS Med. 2008;5:e170. doi: 10.1371/journal.pmed.0050170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hori SS, Gambhir SS. Mathematical Model Identifies Blood Biomarker-Based Early Cancer Detection Strategies and Limitations. Sci Transl Med. 2011;3:109ra116. doi: 10.1126/scitranslmed.3003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hull LC, Farrell D, Grodzinski P. Highlights of Recent Developments and Trends in Cancer Nanotechnology Research–View from Nci Alliance for Nanotechnology in Cancer. Biotechnol Adv. 2014;32:666–678. doi: 10.1016/j.biotechadv.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 135.Li D, Chan DW. Proteomic Cancer Biomarkers from Discovery to Approval: It’s Worth the Effort. Expert Rev Proteomics. 2014;11:135–136. doi: 10.1586/14789450.2014.897614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wisdom GB. Enzyme-Immunoassay. Clin Chem. 1976;22:1243–1255. [PubMed] [Google Scholar]
- 137.Gold P, Freedman SO. Specific Carcinoembryonic Antigens of the Human Digestive System. J Exp Med. 1965;122:467–481. doi: 10.1084/jem.122.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gold P, Freedman SO. Demonstration of Tumor-Specific Antigens in Human Colonic Carcinomata by Immunological Tolerance and Absorption Techniques. J Exp Med. 1965;121:439–462. doi: 10.1084/jem.121.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shitrit D, Zingerman B, Shitrit AB, Shlomi D, Kramer MR. Diagnostic Value of Cyfra 21–1, Cea, Ca 19–9, Ca 15–3, and Ca 125 Assays in Pleural Effusions: Analysis of 116 Cases and Review of the Literature. Oncologist. 2005;10:501–507. doi: 10.1634/theoncologist.10-7-501. [DOI] [PubMed] [Google Scholar]
- 140.Tapia FJ, Polak JM, Barbosa AJ, Bloom SR, Marangos PJ, Dermody C, Pearse AG. Neuron-Specific Enolase Is Produced by Neuroendocrine Tumours. Lancet. 1981;317:808–811. doi: 10.1016/s0140-6736(81)92682-9. [DOI] [PubMed] [Google Scholar]
- 141.Harding M, McAllister J, Hulks G, Vernon D, Monie R, Paul J, Kaye SB. Neurone Specific Enolase (Nse) in Small Cell Lung Cancer: A Tumour Marker of Prognostic Significance? Br J Cancer. 1990;61:605–607. doi: 10.1038/bjc.1990.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lamberts SW, Hofland LJ, Nobels FR. Neuroendocrine Tumor Markers. Front Neuroendocrinol. 2001;22:309–339. doi: 10.1006/frne.2001.0218. [DOI] [PubMed] [Google Scholar]
- 143.Portela-Gomes GM, Hacker GW, Weitgasser R. Neuroendocrine Cell Markers for Pancreatic Islets and Tumors. Appl Immunohistochem Mol Morphol. 2004;12:183–192. doi: 10.1097/00129039-200409000-00001. [DOI] [PubMed] [Google Scholar]
- 144.Li H, Cao Z, Zhang Y, Lau C, Lu J. Simultaneous Detection of Two Lung Cancer Biomarkers Using Dual-Color Fluorescence Quantum Dots. Analyst. 2011;136:1399–1405. doi: 10.1039/c0an00704h. [DOI] [PubMed] [Google Scholar]
- 145.Cao Z, Li H, Lau C, Zhang Y. Cross-Talk-Free Simultaneous Fluoroimmunoassay of Two Biomarkers Based on Dual-Color Quantum Dots. Anal Chim Acta. 2011;698:44–50. doi: 10.1016/j.aca.2011.04.045. [DOI] [PubMed] [Google Scholar]
- 146.Geho D, Lahar N, Gurnani P, Huebschman M, Herrmann P, Espina V, Shi A, Wulfkuhle J, Garner H, Petricoin E, 3rd, et al. Pegylated, Steptavidin-Conjugated Quantum Dots Are Effective Detection Elements for Reverse-Phase Protein Microarrays. Bioconjugate Chem. 2005;16:559–566. doi: 10.1021/bc0497113. [DOI] [PubMed] [Google Scholar]
- 147.Gokarna A, Jin LH, Hwang JS, Cho YH, Lim YT, Chung BH, Youn SH, Choi DS, Lim JH. Quantum Dot-Based Protein Micro- and Nanoarrays for Detection of Prostate Cancer Biomarkers. Proteomics. 2008;8:1809–1818. doi: 10.1002/pmic.200701072. [DOI] [PubMed] [Google Scholar]
- 148.Gu B, Xu C, Yang C, Liu S, Wang M. Zno Quantum Dot Labeled Immunosensor for Carbohydrate Antigen 19–9. Biosens Bioelectron. 2011;26:2720–2723. doi: 10.1016/j.bios.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 149.Jin LH, Li SM, Cho YH. Enhanced Detection Sensitivity of Pegylated Cdse/Zns Quantum Dots-Based Prostate Cancer Biomarkers by Surface Plasmon-Coupled Emission. Biosens Bioelectron. 2012;33:284–287. doi: 10.1016/j.bios.2011.12.043. [DOI] [PubMed] [Google Scholar]
- 150.Kerman K, Endo T, Tsukamoto M, Chikae M, Takamura Y, Tamiya E. Quantum Dot-Based Immunosensor for the Detection of Prostate-Specific Antigen Using Fluorescence Microscopy. Talanta. 2007;71:1494–1499. doi: 10.1016/j.talanta.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 151.Mukundan H, Kubicek JZ, Holt A, Shively JE, Martinez JS, Grace K, Grace WK, Swanson BI. Planar Optical Waveguide-Based Biosensor for the Quantitative Detection of Tumor Markers. Sens Actuators, B. 2009;138:453–460. [Google Scholar]
- 152.Mukundan H, Xie H, Anderson AS, Grace WK, Shively JE, Swanson BI. Optimizing a Waveguide-Based Sandwich Immunoassay for Tumor Biomarkers: Evaluating Fluorescent Labels and Functional Surfaces. Bioconjugate Chem. 2009;20:222–230. doi: 10.1021/bc800283e. [DOI] [PubMed] [Google Scholar]
- 153.Kuriyama M, Wang MC, Lee CI, Papsidero LD, Killian CS, Inaji H, Slack NH, Nishiura T, Murphy GP, Chu TM. Use of Human Prostate-Specific Antigen in Monitoring Prostate Cancer. Cancer Res. 1981;41:3874–3876. [PubMed] [Google Scholar]
- 154.Andriole GL, Crawford ED, Grubb RL, 3rd, Buys SS, Chia D, Church TR, Fouad MN, Isaacs C, Kvale PA, Reding DJ, et al. Prostate Cancer Screening in the Randomized Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial: Mortality Results after 13 Years of Follow-Up. J Natl Cancer Inst. 2012;104:125–132. doi: 10.1093/jnci/djr500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Barry MJ. Clinical Practice. Prostate-Specific-Antigen Testing for Early Diagnosis of Prostate Cancer. N Engl J Med. 2001;344:1373–1377. doi: 10.1056/NEJM200105033441806. [DOI] [PubMed] [Google Scholar]
- 156.Heijnsdijk EA, Wever EM, Auvinen A, Hugosson J, Ciatto S, Nelen V, Kwiatkowski M, Villers A, Paez A, Moss SM, et al. Quality-of-Life Effects of Prostate-Specific Antigen Screening. N Engl J Med. 2012;367:595–605. doi: 10.1056/NEJMoa1201637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al. Prostate-Cancer Mortality at 11 Years of Follow-Up. N Engl J Med. 2012;366:981–990. doi: 10.1056/NEJMoa1113135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Thaxton CS, Elghanian R, Thomas AD, Stoeva SI, Lee JS, Smith ND, Schaeffer AJ, Klocker H, Horninger W, Bartsch G, et al. Nanoparticle-Based Bio-Barcode Assay Redefines “Undetectable” Psa and Biochemical Recurrence after Radical Prostatectomy. Proc Natl Acad Sci U S A. 2009;106:18437–18442. doi: 10.1073/pnas.0904719106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Thompson IM, Pauler DK, Goodman PJ, Tangen CM, Lucia MS, Parnes HL, Minasian LM, Ford LG, Lippman SM, Crawford ED, et al. Prevalence of Prostate Cancer among Men with a Prostate-Specific Antigen Level < or = 4.0 Ng Per Milliliter. N Engl J Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 160.D’Amico AV, Chen MH, Roehl KA, Catalona WJ. Preoperative Psa Velocity and the Risk of Death from Prostate Cancer after Radical Prostatectomy. N Engl J Med. 2004;351:125–135. doi: 10.1056/NEJMoa032975. [DOI] [PubMed] [Google Scholar]
- 161.Ulmert D, Serio AM, O’Brien MF, Becker C, Eastham JA, Scardino PT, Bjork T, Berglund G, Vickers AJ, Lilja H. Long-Term Prediction of Prostate Cancer: Prostate-Specific Antigen (Psa) Velocity Is Predictive but Does Not Improve the Predictive Accuracy of a Single Psa Measurement 15 Years or More before Cancer Diagnosis in a Large, Representative, Unscreened Population. J Clin Oncol. 2008;26:835–841. doi: 10.1200/JCO.2007.13.1490. [DOI] [PubMed] [Google Scholar]
- 162.Arrabito G, Reisewitz S, Dehmelt L, Bastiaens PI, Pignataro B, Schroeder H, Niemeyer CM. Biochips for Cell Biology by Combined Dip-Pen Nanolithography and DNA-Directed Protein Immobilization. Small. 2013;9:4243–4249. doi: 10.1002/smll.201300941. [DOI] [PubMed] [Google Scholar]
- 163.Ginger DS, Zhang H, Mirkin CA. The Evolution of Dip-Pen Nanolithography. Angew Chem, Int Ed. 2004;43:30–45. doi: 10.1002/anie.200300608. [DOI] [PubMed] [Google Scholar]
- 164.Kim JD, Ahn DG, Oh JW, Park WJ, Jung HI. Ribosome Display and Dip-Pen Nanolithography for the Fabrication of Protein Nanoarrays. Adv Mater. 2008;20:3349. [Google Scholar]
- 165.Kim KH, Kim JD, Kim YJ, Kang SH, Jung SY, Jung H. Protein Immobilization without Purification Via Dip-Pen Nanolithography. Small. 2008;4:1089–1094. doi: 10.1002/smll.200700519. [DOI] [PubMed] [Google Scholar]
- 166.Lee KB, Lim JH, Mirkin CA. Protein Nanostructures Formed Via Direct-Write Dip-Pen Nanolithography. J Am Chem Soc. 2003;125:5588–5589. doi: 10.1021/ja034236p. [DOI] [PubMed] [Google Scholar]
- 167.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein Nanoarrays Generated by Dip-Pen Nanolithography. Science. 2002;295:1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 168.Lee SW, Oh BK, Sanedrin RG, Salaita K, Fujigaya T, Mirkin CA. Biologically Active Protein Nanoarrays Generated Using Parallel Dip-Pen Nanolithography. Adv Mater. 2006;18:1133. [Google Scholar]
- 169.Lim JH, Ginger DS, Lee KB, Heo J, Nam JM, Mirkin CA. Direct-Write Dip-Pen Nanolithography of Proteins on Modified Silicon Oxide Surfaces. Angew Chem, Int Ed. 2003;42:2309–2312. doi: 10.1002/anie.200351256. [DOI] [PubMed] [Google Scholar]
- 170.Mirkin CA. The Power of the Pen: Development of Massively Parallel Dip-Pen Nanolithography. ACS Nano. 2007;1:79–83. doi: 10.1021/nn700228m. [DOI] [PubMed] [Google Scholar]
- 171.Nam JM, Han SW, Lee KB, Liu XG, Ratner MA, Mirkin CA. Bioactive Protein Nanoarrays on Nickel Oxide Surfaces Formed by Dip-Pen Nanolithography. Angew Chem, Int Ed. 2004;43:1246–1249. doi: 10.1002/anie.200353203. [DOI] [PubMed] [Google Scholar]
- 172.Piner RD, Zhu J, Xu F, Hong SH, Mirkin CA. “Dip-Pen” Nanolithography. Science. 1999;283:661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 173.Salaita K, Wang YH, Mirkin CA. Applications of Dip-Pen Nanolithography. Nat Nanotechnol. 2007;2:145–155. doi: 10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- 174.Senesi AJ, Rozkiewicz DI, Reinhoudt DN, Mirkin CA. Agarose-Assisted Dip-Pen Nanolithography of Oligonucleotides and Proteins. ACS Nano. 2009;3:2394–2402. doi: 10.1021/nn9005945. [DOI] [PubMed] [Google Scholar]
- 175.Wu CC, Reinhoudt DN, Otto C, Velders AH, Subramaniam V. Protein Immobilization on Ni(Ii) Ion Patterns Prepared by Microcontact Printing and Dip-Pen Nanolithography. ACS Nano. 2010;4:1083–1091. doi: 10.1021/nn901270c. [DOI] [PubMed] [Google Scholar]
- 176.Zheng ZJ, Daniel WL, Giam LR, Huo FW, Senesi AJ, Zheng GF, Mirkin CA. Multiplexed Protein Arrays Enabled by Polymer Pen Lithography: Addressing the Inking Challenge. Angew Chem, Int Ed. 2009;48:7626–7629. doi: 10.1002/anie.200902649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Jakeway SC, de Mello AJ, Russell EL. Miniaturized Total Analysis Systems for Biological Analysis. Fresenius’ J Anal Chem. 2000;366:525–539. doi: 10.1007/s002160051548. [DOI] [PubMed] [Google Scholar]
- 178.Ying L, Wang Q. Microfluidic Chip-Based Technologies: Emerging Platforms for Cancer Diagnosis. BMC Biotechnol. 2013;13:76. doi: 10.1186/1472-6750-13-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Saylor RA, Lunte SM. A Review of Microdialysis Coupled to Microchip Electrophoresis for Monitoring Biological Events. J Chromatogr A. 2015;1382:48. doi: 10.1016/j.chroma.2014.12.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yan J, Hu M, Li D, He Y, Zhao R, Jiang X, Song S, Wang L, Fan C. A Nano- and Micro-Integrated Protein Chip Based on Quantum Dot Probes and a Microfluidic Network. Nano Res. 2008;1:490–496. [Google Scholar]
- 181.Jokerst JV, Raamanathan A, Christodoulides N, Floriano PN, Pollard AA, Simmons GW, Wong J, Gage C, Furmaga WB, Redding SW, et al. Nano-Bio-Chips for High Performance Multiplexed Protein Detection: Determinations of Cancer Biomarkers in Serum and Saliva Using Quantum Dot Bioconjugate Labels. Biosens Bioelectron. 2009;24:3622–3629. doi: 10.1016/j.bios.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hu M, He Y, Song S, Yan J, Lu HT, Weng LX, Wang LH, Fan C. DNA-Bridged Bioconjugation of Fluorescent Quantum Dots for Highly Sensitive Microfluidic Protein Chips. Chem Commun (Cambridge, U K) 2010;46:6126–6128. doi: 10.1039/c0cc01608j. [DOI] [PubMed] [Google Scholar]
- 183.Hu M, Yan J, He Y, Lu H, Weng L, Song S, Fan C, Wang L. Ultrasensitive, Multiplexed Detection of Cancer Biomarkers Directly in Serum by Using a Quantum Dot-Based Microfluidic Protein Chip. ACS Nano. 2010;4:488–494. doi: 10.1021/nn901404h. [DOI] [PubMed] [Google Scholar]
- 184.Zhang H, Liu L, Fu X, Zhu Z. Microfluidic Beads-Based Immunosensor for Sensitive Detection of Cancer Biomarker Proteins Using Multienzyme-Nanoparticle Amplification and Quantum Dots Labels. Biosens Bioelectron. 2013;42:23–30. doi: 10.1016/j.bios.2012.10.076. [DOI] [PubMed] [Google Scholar]
- 185.McIntire KR, Vogel CL, Princler GL, Patel IR. Serum Alpha-Fetoprotein as a Biochemical Marker for Hepatocellular Carcinoma. Cancer Res. 1972;32:1941–1946. [PubMed] [Google Scholar]
- 186.Rich N, Singal AG. Hepatocellular Carcinoma Tumour Markers: Current Role and Expectations. Best Pract Res Clin Gastroenterol. 2014;28:843–853. doi: 10.1016/j.bpg.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 187.Canney PA, Moore M, Wilkinson PM, James RD. Ovarian Cancer Antigen Ca125: A Prospective Clinical Assessment of Its Role as a Tumour Marker. Br J Cancer. 1984;50:765–769. doi: 10.1038/bjc.1984.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Jacobs I, Bast RC., Jr The Ca 125 Tumour-Associated Antigen: A Review of the Literature. Hum Reprod. 1989;4:1–12. doi: 10.1093/oxfordjournals.humrep.a136832. [DOI] [PubMed] [Google Scholar]
- 189.Kenemans P, Yedema CA, Bon GG, von Mensdorff-Pouilly S. Ca 125 in Gynecological Pathology–a Review. Eur J Obstet Gynecol Reprod Biol. 1993;49:115–124. doi: 10.1016/0028-2243(93)90135-y. [DOI] [PubMed] [Google Scholar]
- 190.Li Z, Wang Y, Wang J, Tang Z, Pounds JG, Lin Y. Rapid and Sensitive Detection of Protein Biomarker Using a Portable Fluorescence Biosensor Based on Quantum Dots and a Lateral Flow Test Strip. Anal Chem. 2010;82:7008–7014. doi: 10.1021/ac101405a. [DOI] [PubMed] [Google Scholar]
- 191.Yang Q, Gong X, Song T, Yang J, Zhu S, Li Y, Cui Y, Li Y, Zhang B, Chang J. Quantum Dot-Based Immunochromatography Test Strip for Rapid, Quantitative and Sensitive Detection of Alpha Fetoprotein. Biosens Bioelectron. 2011;30:145–150. doi: 10.1016/j.bios.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 192.Cheng X, Pu X, Jun P, Zhu X, Zhu D, Chen M. Rapid and Quantitative Detection of C-Reactive Protein Using Quantum Dots and Immunochromatographic Test Strips. Int J Nanomed. 2014;9:5619–5626. doi: 10.2147/IJN.S74751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Estes GB, Munoz M, Burdash NM, Virella G. A Quantitative Immunofluorescence Test for the Detection of Anti-Candida Antibodies. J Immunol Methods. 1980;35:105–113. doi: 10.1016/0022-1759(80)90155-6. [DOI] [PubMed] [Google Scholar]
- 194.Ngom B, Guo Y, Wang X, Bi D. Development and Application of Lateral Flow Test Strip Technology for Detection of Infectious Agents and Chemical Contaminants: A Review. Anal Bioanal Chem. 2010;397:1113–1135. doi: 10.1007/s00216-010-3661-4. [DOI] [PubMed] [Google Scholar]
- 195.Shahjamali MM, Bosman M, Cao SW, Huang X, Saadat S, Martinsson E, Aili D, Tay YY, Liedberg B, Loo SCJ, et al. Gold Coating of Silver Nanoprisms. Adv Funct Mater. 2012;22:849–854. [Google Scholar]
- 196.Allin KH, Nordestgaard BG. Elevated C-Reactive Protein in the Diagnosis, Prognosis, and Cause of Cancer. Crit Rev Clin Lab Sci. 2011;48:155–170. doi: 10.3109/10408363.2011.599831. [DOI] [PubMed] [Google Scholar]
- 197.Wei Q, Lee M, Yu X, Lee EK, Seong GH, Choo J, Cho YW. Development of an Open Sandwich Fluoroimmunoassay Based on Fluorescence Resonance Energy Transfer. Anal Biochem. 2006;358:31–37. doi: 10.1016/j.ab.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 198.Chen MJ, Wu YS, Lin GF, Hou JY, Li M, Liu TC. Quantum-Dot-Based Homogeneous Time-Resolved Fluoroimmunoassay of Alpha-Fetoprotein. Anal Chim Acta. 2012;741:100–105. doi: 10.1016/j.aca.2012.06.042. [DOI] [PubMed] [Google Scholar]
- 199.Ge S, Ge L, Yan M, Song X, Yu J, Liu S. A Disposable Immunosensor Device for Point-of-Care Test of Tumor Marker Based on Copper-Mediated Amplification. Biosens Bioelectron. 2013;43:425–431. doi: 10.1016/j.bios.2012.12.047. [DOI] [PubMed] [Google Scholar]
- 200.Wegner KD, Jin Z, Linden S, Jennings TL, Hildebrandt N. Quantum-Dot-Basedforster Resonance Energy Transfer Immunoassay for Sensitive Clinical Diagnostics of Low-Volume Serum Samples. ACS Nano. 2013;7:7411–7419. doi: 10.1021/nn403253y. [DOI] [PubMed] [Google Scholar]
- 201.Kim J, Kwon S, Park JK, Park I. Quantum Dot-Based Immunoassay Enhanced by High-Density Vertical Zno Nanowire Array. Biosens Bioelectron. 2014;55:209–215. doi: 10.1016/j.bios.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 202.Jou AF, Lu CH, Ou YC, Wang SS, Hsu SL, Willner I, Ho JA. Diagnosing the Mir-141 Prostate Cancer Biomarker Using Nucleic Acid-Functionalized Cdse/Zns Qds and Telomerase. Chem Sci. 2015;6:659–665. doi: 10.1039/c4sc02104e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Qian J, Wang C, Pan X, Liu S. A High-Throughput Homogeneous Immunoassay Based on Forster Resonance Energy Transfer between Quantum Dots and Gold Nanoparticles. Anal Chim Acta. 2013;763:43–49. doi: 10.1016/j.aca.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 204.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 205.Choi Y, Pinto M. Estrogen Receptor Beta in Breast Cancer: Associations between Erbeta, Hormonal Receptors, and Other Prognostic Biomarkers. Appl Immunohistochem Mol Morphol. 2005;13:19–24. doi: 10.1097/00129039-200503000-00004. [DOI] [PubMed] [Google Scholar]
- 206.Omoto Y, Iwase H. Clinical Significance of Estrogen Receptor Beta in Breast and Prostate Cancer from Biological Aspects. Cancer Sci. 2015;106:337. doi: 10.1111/cas.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gao YC, Wu J. Microrna-200c and Microrna-141 as Potential Diagnostic and Prognostic Biomarkers for Ovarian Cancer. Tumor Biol. 2015;36:4843. doi: 10.1007/s13277-015-3138-3. [DOI] [PubMed] [Google Scholar]
- 208.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. Microrna Signatures in Human Ovarian Cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 209.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, et al. Circulating Micrornas as Stable Blood-Based Markers for Cancer Detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zuo QF, Zhang R, Li BS, Zhao YL, Zhuang Y, Yu T, Gong L, Li S, Xiao B, Zou QM. Microrna-141 Inhibits Tumor Growth and Metastasis in Gastric Cancer by Directly Targeting Transcriptional Co-Activator with Pdz-Binding Motif, Taz. Cell Death Dis. 2015;6:e1623. doi: 10.1038/cddis.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Duffy MJ, Shering S, Sherry F, McDermott E, O’Higgins N. Ca 15-3: A Prognostic Marker in Breast Cancer. Int J Biol Markers. 2000;15:330–333. doi: 10.1177/172460080001500410. [DOI] [PubMed] [Google Scholar]
- 212.Keshaviah A, Dellapasqua S, Rotmensz N, Lindtner J, Crivellari D, Collins J, Colleoni M, Thurlimann B, Mendiola C, Aebi S, et al. Ca15-3 and Alkaline Phosphatase as Predictors for Breast Cancer Recurrence: A Combined Analysis of Seven International Breast Cancer Study Group Trials. Ann Oncol. 2007;18:701–708. doi: 10.1093/annonc/mdl492. [DOI] [PubMed] [Google Scholar]
- 213.Nath S, Mukherjee P. Muc1: A Multifaceted Oncoprotein with a Key Role in Cancer Progression. Trends Mol Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zhu YD, Peng J, Jiang LP, Zhu JJ. Fluorescent Immunosensor Based on Cus Nanoparticles for Sensitive Detection of Cancer Biomarker. Analyst. 2014;139:649–655. doi: 10.1039/c3an01987j. [DOI] [PubMed] [Google Scholar]
- 215.Ward AM, Catto JWF, Hamdy FC. Prostate Specific Antigen: Biology, Biochemistry and Available Commercial Assays. Ann Clin Biochem. 2001;38:633–651. doi: 10.1258/0004563011901055. [DOI] [PubMed] [Google Scholar]
- 216.Akter R, Rahman MA, Rhee CK. Amplified Electrochemical Detection of a Cancer Biomarker by Enhanced Precipitation Using Horseradish Peroxidase Attached on Carbon Nanotubes. Anal Chem. 2012;84:6407–6415. doi: 10.1021/ac300110n. [DOI] [PubMed] [Google Scholar]
- 217.Liu F, Zhang Y, Ge S, Lu J, Yu J, Song X, Liu S. Magnetic Graphene Nanosheets Based Electrochemiluminescence Immunoassay of Cancer Biomarker Using Cdte Quantum Dots Coated Silica Nanospheres as Labels. Talanta. 2012;99:512–519. doi: 10.1016/j.talanta.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 218.Gao Z, Xu M, Hou L, Chen G, Tang D. Magnetic Bead-Based Reverse Colorimetric Immunoassay Strategy for Sensing Biomolecules. Anal Chem. 2013;85:6945–6952. doi: 10.1021/ac401433p. [DOI] [PubMed] [Google Scholar]
- 219.Romslo I. Prostatic Acid Phosphatase. Br Med J. 1971;1:406. doi: 10.1136/bmj.1.5745.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Markovic O, Markovic N. Cervical Acid Phosphatase: A Biomarker of Cervical Dysplasia and a Potential Surrogate Endpoint for Colposcopy. Dis Markers. 2004;19:279–286. doi: 10.1155/2004/845798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Taira A, Merrick G, Wallner K, Dattoli M. Reviving the Acid Phosphatase Test for Prostate Cancer. Oncology. 2007;21:1003–1010. [PubMed] [Google Scholar]
- 222.You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UH, Rotello VM. Detection and Identification of Proteins Using Nanoparticle-Fluorescent Polymer ‘Chemical Nose’ Sensors. Nat Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 223.Peterson JE, Zurakowski D, Italiano JE, Jr, Michel LV, Connors S, Oenick M, D’Amato RJ, Klement GL, Folkman J. Vegf, Pf4 and Pdgf Are Elevated in Platelets of Colorectal Cancer Patients. Angiogenesis. 2012;15:265–273. doi: 10.1007/s10456-012-9259-z. [DOI] [PubMed] [Google Scholar]
- 224.Liu J, Liu C, Qiu L, Li J, Zhang P, Sun Y. Overexpression of Both Platelet-Derived Growth Factor-Bb and Vascular Endothelial Growth Factor-C and Its Association with Lymphangiogenesis in Primary Human Non-Small Cell Lung Cancer. Diagn Pathol. 2014;9:128. doi: 10.1186/1746-1596-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Talaat RM, Salem TA, El-Masry S, Imbarek A, Mokhles M, Abdel-Aziz A. Circulating Pro- and Anti-Angiogenic Mediators in Patients Infected with Hepatitis C at Different Stages of Hepatocellular Carcinoma. J Med Virol. 2014;86:1120–1129. doi: 10.1002/jmv.23932. [DOI] [PubMed] [Google Scholar]
- 226.Shaw VE, Lane B, Jenkinson C, Cox T, Greenhalf W, Halloran CM, Tang J, Sutton R, Neoptolemos JP, Costello E. Serum Cytokine Biomarker Panels for Discriminating Pancreatic Cancer from Benign Pancreatic Disease. Mol Cancer. 2014;13:114. doi: 10.1186/1476-4598-13-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Huang CC, Chiu SH, Huang YF, Chang HT. Aptamer-Functionalized Gold Nanoparticles for Turn-on Light Switch Detection of Platelet-Derived Growth Factor. Anal Chem. 2007;79:4798–4804. doi: 10.1021/ac0707075. [DOI] [PubMed] [Google Scholar]
- 228.Lokeshwar VB, Cerwinka WH, Isoyama T, Lokeshwar BL. Hyal1 Hyaluronidase in Prostate Cancer: A Tumor Promoter and Suppressor. Cancer Res. 2005;65:7782–7789. doi: 10.1158/0008-5472.CAN-05-1022. [DOI] [PubMed] [Google Scholar]
- 229.Lokeshwar VB, Cerwinka WH, Lokeshwar BL. Hyal1 Hyaluronidase: A Molecular Determinant of Bladder Tumor Growth and Invasion. Cancer Res. 2005;65:2243–2250. doi: 10.1158/0008-5472.CAN-04-2805. [DOI] [PubMed] [Google Scholar]
- 230.Pham HT, Block NL, Lokeshwar VB. Tumor-Derived Hyaluronidase: A Diagnostic Urine Marker for High-Grade Bladder Cancer. Cancer Res. 1997;57:778–783. [PubMed] [Google Scholar]
- 231.Cheng D, Han W, Yang K, Song Y, Jiang M, Song E. One-Step Facile Synthesis of Hyaluronic Acid Functionalized Fluorescent Gold Nanoprobes Sensitive to Hyaluronidase in Urine Specimen from Bladder Cancer Patients. Talanta. 2014;130:408–414. doi: 10.1016/j.talanta.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 232.Takahashi Y, Kitadai Y, Bucana CD, Cleary KR, Ellis LM. Expression of Vascular Endothelial Growth Factor and Its Receptor, Kdr, Correlates with Vascularity, Metastasis, and Proliferation of Human Colon Cancer. Cancer Res. 1995;55:3964–3968. [PubMed] [Google Scholar]
- 233.Hyodo I, Doi T, Endo H, Hosokawa Y, Nishikawa Y, Tanimizu M, Jinno K, Kotani Y. Clinical Significance of Plasma Vascular Endothelial Growth Factor in Gastrointestinal Cancer. Eur J Cancer. 1998;34:2041–2045. doi: 10.1016/s0959-8049(98)00282-2. [DOI] [PubMed] [Google Scholar]
- 234.Locopo N, Fanelli M, Gasparini G. Clinical Significance of Angiogenic Factors in Breast Cancer. Breast Cancer Res Treat. 1998;52:159–173. doi: 10.1023/a:1006175504673. [DOI] [PubMed] [Google Scholar]
- 235.Singhal S, Vachani A, Antin-Ozerkis D, Kaiser LR, Albelda SM. Prognostic Implications of Cell Cycle, Apoptosis, and Angiogenesis Biomarkers in Non-Small Cell Lung Cancer: A Review. Clin Cancer Res. 2005;11:3974–3986. doi: 10.1158/1078-0432.CCR-04-2661. [DOI] [PubMed] [Google Scholar]
- 236.Cho H, Yeh EC, Sinha R, Laurence TA, Bearinger JP, Lee LP. Single-Step Nanoplasmonic Vegf165 Aptasensor for Early Cancer Diagnosis. ACS Nano. 2012;6:7607–7614. doi: 10.1021/nn203833d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hsu PI, Chen CH, Hsieh CS, Chang WC, Lai KH, Lo GH, Hsu PN, Tsay FW, Chen YS, Hsiao M, et al. Alpha1-Antitrypsin Precursor in Gastric Juice Is a Novel Biomarker for Gastric Cancer and Ulcer. Clin Cancer Res. 2007;13:876–883. doi: 10.1158/1078-0432.CCR-06-1404. [DOI] [PubMed] [Google Scholar]
- 238.Khazanov E, Yavin E, Pascal A, Nissan A, Kohl Y, Reimann-Zawadzki M, Rubinstein A. Detecting a Secreted Gastric Cancer Biomarker Molecule by Targeted Nanoparticles for Real-Time Diagnostics. Pharm Res. 2012;29:983–993. doi: 10.1007/s11095-011-0638-8. [DOI] [PubMed] [Google Scholar]
- 239.Budd GT, Cristofanilli M, Ellis MJ, Stopeck A, Borden E, Miller MC, Matera J, Repollet M, Doyle GV, Terstappen LWMM, et al. Circulating Tumor Cells Versus Imaging - Predicting Overall Survival in Metastatic Breast Cancer. Clin Cancer Res. 2006;12:6403–6409. doi: 10.1158/1078-0432.CCR-05-1769. [DOI] [PubMed] [Google Scholar]
- 240.Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, Matera J, Allard WJ, Doyle GV, Terstappen LWWM. Circulating Tumor Cells at Each Follow-up Time Point During Therapy of Metastatic Breast Cancer Patients Predict Progression-Free and Overall Survival. Clin Cancer Res. 2006;12:4218–4224. doi: 10.1158/1078-0432.CCR-05-2821. [DOI] [PubMed] [Google Scholar]
- 241.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, et al. Circulating Tumor Cells: A Novel Prognostic Factor for Newly Diagnosed Metastatic Breast Cancer. J Clin Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 242.Pantel K, Brakenhoff RH, Brandt B. Detection, Clinical Relevance and Specific Biological Properties of Disseminating Tumour Cells. Nat Rev Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 243.Lian W, Litherland SA, Badrane H, Tan WH, Wu DH, Baker HV, Gulig PA, Lim DV, Jin SG. Ultrasensitive Detection of Biomolecules with Fluorescent Dye-Doped Nanoparticles. Anal Biochem. 2004;334:135–144. doi: 10.1016/j.ab.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 244.Chiang AC, Massague J. Molecular Origins of Cancer Molecular Basis of Metastasis. N Engl J Med. 2008;359:2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Meacham CE, Morrison SJ. Tumour Heterogeneity and Cancer Cell Plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Burrell RA, McGranahan N, Bartek J, Swanton C. The Causes and Consequences of Genetic Heterogeneity in Cancer Evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 247.Almendro V, Marusyk A, Polyak K. Cellular Heterogeneity and Molecular Evolution in Cancer. Annu Rev Pathol: Mech Dis. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 248.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Janicke F, et al. Detection of Circulating Tumor Cells in Peripheral Blood of Patients with Metastatic Breast Cancer: A Validation Study of the Cellsearch System. Clin Cancer Res. 2007;13:920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 249.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating Tumor Cells: Approaches to Isolation and Characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sherr CJ. Cancer Cell Cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 251.Evan GI, Vousden KH. Proliferation, Cell Cycle and Apoptosis in Cancer. Nature. 2001;411:342–348. doi: 10.1038/35077213. [DOI] [PubMed] [Google Scholar]
- 252.Bishop JM. Molecular Themes in Oncogenesis. Cell. 1991;64:235–248. doi: 10.1016/0092-8674(91)90636-d. [DOI] [PubMed] [Google Scholar]
- 253.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-Mesenchymal Transition in Breast Cancer Relates to the Basal-Like Phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 254.Kalluri R, Weinberg RA. The Basics of Epithelial-Mesenchymal Transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Zajchowski DA, Bartholdi MF, Gong Y, Webster L, Liu HL, Munishkin A, Beauheim C, Harvey S, Ethier SP, Johnson PH. Identification of Gene Expression Profiles That Predict the Aggressive Behavior of Breast Cancer Cells. Cancer Res. 2001;61:5168–5178. [PubMed] [Google Scholar]
- 256.May CD, Sphyris N, Evans KW, Werden SJ, Guo WJ, Mani SA. Epithelial-Mesenchymal Transition and Cancer Stem Cells: A Dangerously Dynamic Duo in Breast Cancer Progression. Breast Cancer Res. 2011;13:202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Thiery JP. Epithelial-Mesenchymal Transitions in Tumour Progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 258.Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-Mesenchymal Transitions in Development and Disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 259.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem Cells, Cancer, and Cancer Stem Cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 260.Visvader JE, Lindeman GJ. Cancer Stem Cells in Solid Tumours: Accumulating Evidence and Unresolved Questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 261.Wicha MS, Liu SL, Dontu G. Cancer Stem Cells: An Old Idea - a Paradigm Shift. Cancer Res. 2006;66:1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 262.Haun JB, Devaraj NK, Hilderbrand SA, Lee H, Weissleder R. Bioorthogonal Chemistry Amplifies Nanoparticle Binding and Enhances the Sensitivity of Cell Detection. Nat Nanotechnol. 2010;5:660–665. doi: 10.1038/nnano.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.He H, Xie C, Ren J. Nonbleaching Fluorescence of Gold Nanoparticles and Its Applications in Cancer Cell Imaging. Anal Chem. 2008;80:5951–5957. doi: 10.1021/ac8005796. [DOI] [PubMed] [Google Scholar]
- 264.Huang SS, Li RN, Qu YX, Shen J, Liu J. Fluorescent Biological Label for Recognizing Human Ovarian Tumor Cells Based on Fluorescent Nanoparticles. J Fluoresc. 2009;19:1095–1101. doi: 10.1007/s10895-009-0509-4. [DOI] [PubMed] [Google Scholar]
- 265.Li K, Liu YT, Pu KY, Feng SS, Zhan RY, Liu B. Polyhedral Oligomeric Silsesquioxanes-Containing Conjugated Polymer Loaded Plga Nanoparticles with Trastuzumab (Herceptin) Functionalization for Her2-Positive Cancer Cell Detection. Adv Funct Mater. 2011;21:287–294. [Google Scholar]
- 266.Mi Y, Li K, Liu Y, Pu KY, Liu B, Feng SS. Herceptin Functionalized Polyhedral Oligomeric Silsesquioxane - Conjugated Oligomers - Silica/Iron Oxide Nanoparticles for Tumor Cell Sorting and Detection. Biomaterials. 2011;32:8226–8233. doi: 10.1016/j.biomaterials.2011.07.040. [DOI] [PubMed] [Google Scholar]
- 267.Rizvi SB, Rouhi S, Taniguchi S, Yang SY, Green M, Keshtgar M, Seifalian AM. Near-Infrared Quantum Dots for Her2 Localization and Imaging of Cancer Cells. Int J Nanomed. 2014;9:1323–1337. doi: 10.2147/IJN.S51535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Song EQ, Hu J, Wen CY, Tian ZQ, Yu X, Zhang ZL, Shi YB, Pang DW. Fluorescent-Magnetic-Biotargeting Multifunctional Nanobioprobes for Detecting and Isolating Multiple Types of Tumor Cells. ACS Nano. 2011;5:761–770. doi: 10.1021/nn1011336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sun P, Zhang HY, Liu C, Fang J, Wang M, Chen J, Zhang JP, Mao CB, Xu SK. Preparation and Characterization of Fe3o4/Cdte Magnetic/Fluorescent Nanocomposites and Their Applications in Immuno-Labeling and Fluorescent Imaging of Cancer Cells. Langmuir. 2010;26:1278–1284. doi: 10.1021/la9024553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Tao L, Song CJ, Sun YJ, Li XH, Li YY, Jin BQ, Zhang ZJ, Yang K. A Fluorescent and Chemiluminescent Difunctional Mesoporous Silica Nanoparticle as a Label for the Ultrasensitive Detection of Cancer Cells. Anal Chim Acta. 2013;761:194–200. doi: 10.1016/j.aca.2012.11.046. [DOI] [PubMed] [Google Scholar]
- 271.Wang M, Mi CC, Wang WX, Liu CH, Wu YF, Xu ZR, Mao CB, Xu SK. Immunolabeling and Nir-Excited Fluorescent Imaging of Hela Cells by Using Nayf4:Yb,Er Upconversion Nanoparticles. ACS Nano. 2009;3:1580–1586. doi: 10.1021/nn900491j. [DOI] [PubMed] [Google Scholar]
- 272.Wang M, Mi CC, Zhang YX, Liu JL, Li F, Mao CB, Xu SK. Nir-Responsive Silica-Coated Naybf4:Er/Tm/Ho Upconversion Fluorescent Nanoparticles with Tunable Emission Colors and Their Applications in Immunolabeling and Fluorescent Imaging of Cancer Cells. J Phys Chem C. 2009;113:19021–19027. doi: 10.1021/jp906394z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Welsher K, Liu Z, Daranciang D, Dai H. Selective Probing and Imaging of Cells with Single Walled Carbon Nanotubes as near-Infrared Fluorescent Molecules. Nano Lett. 2008;8:586–590. doi: 10.1021/nl072949q. [DOI] [PubMed] [Google Scholar]
- 274.Wu XY, Liu HJ, Liu JQ, Haley KN, Treadway JA, Larson JP, Ge NF, Peale F, Bruchez MP. Immunofluorescent Labeling of Cancer Marker Her2 and Other Cellular Targets with Semiconductor Quantum Dots. Nat Biotechnol. 2002;21:41–46. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 275.Yang J, Lim EK, Lee HJ, Park J, Lee SC, Lee K, Yoon HG, Suh JS, Huh YM, Haam S. Fluorescent Magnetic Nanohybrids as Multimodal Imaging Agents for Human Epithelial Cancer Detection. Biomaterials. 2008;29:2548–2555. doi: 10.1016/j.biomaterials.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 276.Sukhanova A, Devy M, Venteo L, Kaplan H, Artemyev M, Oleinikov V, Klinov D, Pluot M, Cohen JHM, Nabiev I. Biocompatible Fluorescent Nanocrystals for Immunolabeling of Membrane Proteins and Cells. Anal Biochem. 2004;324:60–67. doi: 10.1016/j.ab.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 277.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Quantum Dot - Aptamer Conjugates for Synchronous Cancer Imaging, Therapy, and Sensing of Drug Delivery Based on Bi-Fluorescence Resonance Energy Transfer. Nano Lett. 2007;7:3065–3070. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 278.Chu TC, Shieh F, Lavery LA, Levy M, Richards-Kortum R, Korgel BA, Ellington AD. Labeling Tumor Cells with Fluorescent Nanocrystal-Aptamer Bioconjugates. Biosens Bioelectron. 2006;21:1859–1866. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 279.Deng T, Li J, Zhang LL, Jiang JH, Chen JN, Shen GL, Yu RQ. A Sensitive Fluorescence Anisotropy Method for the Direct Detection of Cancer Cells in Whole Blood Based on Aptamer-Conjugated near-Infrared Fluorescent Nanoparticles. Biosens Bioelectron. 2010;25:1587–1591. doi: 10.1016/j.bios.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 280.Hwang DW, Ko HY, Lee JH, Kang H, Ryu SH, Song IC, Lee DS, Kim S. A Nucleolin-Targeted Multimodal Nanoparticle Imaging Probe for Tracking Cancer Cells Using an Aptamer. J Nucl Med. 2010;51:98–105. doi: 10.2967/jnumed.109.069880. [DOI] [PubMed] [Google Scholar]
- 281.Liu HY, Xu SM, He ZM, Deng AP, Zhu JJ. Supersandwich Cytosensor for Selective and Ultrasensitive Detection of Cancer Cells Using Aptamer-DNA Concatamer-Quantum Dots Probes. Anal Chem. 2013;85:3385–3392. doi: 10.1021/ac303789x. [DOI] [PubMed] [Google Scholar]
- 282.Daniel MC, Astruc D. Gold Nanoparticles: Assembly, Supramolecular Chemistry, Quantum-Size-Related Properties, and Applications toward Biology, Catalysis, and Nanotechnology. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 283.Link S, El-Sayed MA. Shape and Size Dependence of Radiative, Non-Radiative and Photothermal Properties of Gold Nanocrystals. Int Rev Phys Chem. 2000;19:409–453. [Google Scholar]
- 284.Maier SA, Brongersma ML, Kik PG, Meltzer S, Requicha AAG, Atwater HA. Plasmonics - a Route to Nanoscale Optical Devices. Adv Mater. 2001;13:1501. [Google Scholar]
- 285.Bajaj A, Miranda OR, Kim IB, Phillips RL, Jerry DJ, Bunz UHF, Rotello VM. Detection and Differentiation of Normal, Cancerous, and Metastatic Cells Using Nanoparticle-Polymer Sensor Arrays. Proc Natl Acad Sci U S A. 2009;106:10912–10916. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Di Corato R, Bigall NC, Ragusa A, Dorfs D, Genovese A, Marotta R, Manna L, Pellegrino T. Multifunctional Nanobeads Based on Quantum Dots and Magnetic Nanoparticles: Synthesis and Cancer Cell Targeting and Sorting. ACS Nano. 2011;5:1109–1121. doi: 10.1021/nn102761t. [DOI] [PubMed] [Google Scholar]
- 287.Lee K, Lee H, Bae KH, Park TG. Heparin Immobilized Gold Nanoparticles for Targeted Detection and Apoptotic Death of Metastatic Cancer Cells. Biomaterials. 2010;31:6530–6536. doi: 10.1016/j.biomaterials.2010.04.046. [DOI] [PubMed] [Google Scholar]
- 288.Rosenholm JM, Meinander A, Peuhu E, Niemi R, Eriksson JE, Sahlgren C, Linden M. Targeting of Porous Hybrid Silica Nanoparticles to Cancer Cells. ACS Nano. 2009;3:197–206. doi: 10.1021/nn800781r. [DOI] [PubMed] [Google Scholar]
- 289.Shi XG, Wang SH, Meshinchi S, Van Antwerp ME, Bi XD, Lee IH, Baker JR. Dendrimer-Entrapped Gold Nanoparticles as a Platform for Cancer-Cell Targeting and Imaging. Small. 2007;3:1245–1252. doi: 10.1002/smll.200700054. [DOI] [PubMed] [Google Scholar]
- 290.Shi XY, Wang SH, Shen MW, Antwerp ME, Chen XS, Li C, Petersen EJ, Huang QG, Weber WJ, Baker JR. Multifunctional Dendrimer-Modified Multiwalled Carbon Nanotubes: Synthesis, Characterization, and in Vitro Cancer Cell Targeting and Imaging. Biomacromolecules. 2009;10:1744–1750. doi: 10.1021/bm9001624. [DOI] [PubMed] [Google Scholar]
- 291.Shi XY, Wang SH, Van Antwerp ME, Chen XS, Baker JR., Jr Targeting and Detecting Cancer Cells Using Spontaneously Formed Multifunctional Dendrimer-Stabilized Gold Nanoparticles. Analyst. 2009;134:1373–1379. doi: 10.1039/b902199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-Specific Targeting of Nanoparticles by Multivalent Attachment of Small Molecules. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 293.Xie HY, Zuo C, Liu Y, Zhang ZL, Pang DW, Li XL, Gong JP, Dickinson C, Zhou WZ. Cell-Targeting Multifunctional Nanospheres with Both Fluorescence and Magnetism. Small. 2005;1:506–509. doi: 10.1002/smll.200400136. [DOI] [PubMed] [Google Scholar]
- 294.Yang SJ, Lin FH, Tsai KC, Wei MF, Tsai HM, Wong JM, Shieh MJ. Folic Acid-Conjugated Chitosan Nanoparticles Enhanced Protoporphyrin Ix Accumulation in Colorectal Cancer Cells. Bioconjugate Chem. 2010;21:679–689. doi: 10.1021/bc9004798. [DOI] [PubMed] [Google Scholar]
- 295.Wang SH, Shi XY, Van Antwerp M, Cao ZY, Swanson SD, Bi XD, Baker JR. Dendrimer-Functionalized Iron Oxide Nanoparticles for Specific Targeting and Imaging of Cancer Cells. Adv Funct Mater. 2007;17:3043–3050. [Google Scholar]
- 296.Mi CC, Zhang JP, Gao HY, Wu XL, Wang M, Wu YF, Di YQ, Xu ZR, Mao CB, Xu SK. Multifunctional Nanocomposites of Superparamagnetic (Fe3o4) and Nir-Responsive Rare Earth-Doped up-Conversion Fluorescent (Nayf4: Yb, Er) Nanoparticles and Their Applications in Biolabeling and Fluorescent Imaging of Cancer Cells. Nanoscale. 2010;2:1141–1148. doi: 10.1039/c0nr00102c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wang BZ, Zhu C, Liu L, Lv F, Yang Q, Wang S. Synthesis of a New Conjugated Polymer for Cell Membrane Imaging by Using an Intracellular Targeting Strategy. Polym Chem. 2013;4:5212–5215. [Google Scholar]
- 298.Pu KY, Li K, Liu B. A Molecular Brush Approach to Enhance Quantum Yield and Suppress Nonspecific Interactions of Conjugated Polyelectrolyte for Targeted Far-Red/near-Infrared Fluorescence Cell Imaging. Adv Funct Mater. 2010;20:2770–2777. [Google Scholar]
- 299.Wang X, Li YD. Monodisperse Nanocrystals: General Synthesis, Assembly, and Their Applications. Chem Commun (Cambridge, U K) 2007:2901–2910. doi: 10.1039/b700183e. [DOI] [PubMed] [Google Scholar]
- 300.West JL, Halas NJ. Engineered Nanomaterials for Biophotonics Applications: Improving Sensing, Imaging, and Therapeutics. Annu Rev Biomed Eng. 2003;5:285–292. doi: 10.1146/annurev.bioeng.5.011303.120723. [DOI] [PubMed] [Google Scholar]
- 301.Crochet J, Clemens M, Hertel T. Quantum Yield Heterogeneities of Aqueous Single-Wall Carbon Nanotube Suspensions. J Am Chem Soc. 2007;129:8058–8059. doi: 10.1021/ja071553d. [DOI] [PubMed] [Google Scholar]
- 302.Carlson LJ, Maccagnano SE, Zheng M, Silcox J, Krauss TD. Fluorescence Efficiency of Individual Carbon Nanotubes. Nano Lett. 2007;7:3698–3703. doi: 10.1021/nl072014+. [DOI] [PubMed] [Google Scholar]
- 303.Tsyboulski DA, Rocha JDR, Bachilo SM, Cognet L, Weisman RB. Structure-Dependent Fluorescence Efficiencies of Individual Single-Walled Cardon Nanotubes. Nano Lett. 2007;7:3080–3085. doi: 10.1021/nl071561s. [DOI] [PubMed] [Google Scholar]
- 304.Herr JK, Smith JE, Medley CD, Shangguan DH, Tan WH. Aptamer-Conjugated Nanoparticles for Selective Collection and Detection of Cancer Cells. Anal Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 305.Medley CD, Bamrungsap S, Tan WH, Smith JE. Aptamer-Conjugated Nanoparticles for Cancer Cell Detection. Anal Chem. 2011;83:727–734. doi: 10.1021/ac102263v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Smith JE, Medley CD, Tang ZW, Shangguan D, Lofton C, Tan WH. Aptamer-Conjugated Nanoparticles for the Collection and Detection of Multiple Cancer Cells. Anal Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 307.Lee JH, Lee K, Moon SH, Lee Y, Park TG, Cheon J. All-in-One Target-Cell-Specific Magnetic Nanoparticles for Simultaneous Molecular Imaging and Sirna Delivery. Angew Chem, Int Ed. 2009;48:4174–4179. doi: 10.1002/anie.200805998. [DOI] [PubMed] [Google Scholar]
- 308.Chen HW, Medley CD, Sefah K, Shangguan D, Tang ZW, Meng L, Smith JE, Tan WH. Molecular Recognition of Small-Cell Lung Cancer Cells Using Aptamers. Chem Med Chem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, Rotello VM. Cell Surface-Based Differentiation of Cell Types and Cancer States Using a Gold Nanoparticle-Gfp Based Sensing Array. Chem Sci. 2010;1:134–138. [Google Scholar]
- 310.Latorre A, Somoza A. DNA-Mediated Silver Nanoclusters: Synthesis, Properties and Applications. Chem Bio Chem. 2012;13:951–958. doi: 10.1002/cbic.201200053. [DOI] [PubMed] [Google Scholar]
- 311.Yuan Z, Chen YC, Li HW, Chang HT. Fluorescent Silver Nanoclusters Stabilized by DNA Scaffolds. Chem Commun (Cambridge, U K) 2014;50:9800–9815. doi: 10.1039/c4cc02981j. [DOI] [PubMed] [Google Scholar]
- 312.Yeh HC, Sharma J, Han JJ, Martinez JS, Werner JH. A DNA-Silver Nanocluster Probe That Fluoresces Upon Hybridization. Nano Lett. 2010;10:3106–3110. doi: 10.1021/nl101773c. [DOI] [PubMed] [Google Scholar]
- 313.Yeh HC, Sharma J, Shih IM, Vu DM, Martinez JS, Werner JH. A Fluorescence Light-up Ag Nanocluster Probe That Discriminates Single-Nucleotide Variants by Emission Color. J Am Chem Soc. 2012;134:11550–11558. doi: 10.1021/ja3024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Li J, Zhong X, Zhang H, Le XC, Zhu JJ. Binding-Induced Fluorescence Turn-on Assay Using Aptamer-Functionalized Silver Nanocluster DNA Probes. Anal Chem. 2012;84:5170–5174. doi: 10.1021/ac3006268. [DOI] [PubMed] [Google Scholar]
- 315.Liu JJ, Song XR, Wang YW, Zheng AX, Chen GN, Yang HH. Label-Free and Fluorescence Turn-on Aptasensor for Protein Detection Via Target-Induced Silver Nanoclusters Formation. Anal Chim Acta. 2012;749:70–74. doi: 10.1016/j.aca.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 316.Sharma J, Yeh HC, Yoo H, Werner JH, Martinez JS. Silver Nanocluster Aptamers: In Situ Generation of Intrinsically Fluorescent Recognition Ligands for Protein Detection. Chem Commun (Cambridge, U K) 2011;47:2294–2296. doi: 10.1039/c0cc03711g. [DOI] [PubMed] [Google Scholar]
- 317.Seidel CAM, Schulz A, Sauer MHM. Nucleobase-Specific Quenching of Fluorescent Dyes 0.1. Nucleobase One-Electron Redox Potentials and Their Correlation with Static and Dynamic Quenching Efficiencies. J Phys Chem. 1996;100:5541–5553. [Google Scholar]
- 318.Heinlein T, Knemeyer JP, Piestert O, Sauer M. Photoinduced Electron Transfer between Fluorescent Dyes and Guanosine Residues in DNA-Hairpins. J Phys Chem B. 2003;107:7957–7964. [Google Scholar]
- 319.Walczak S, Morishita K, Ahmed M, Liu JW. Towards Understanding of Poly-Guanine Activated Fluorescent Silver Nanoclusters. Nanotechnology. 2014;25:155501. doi: 10.1088/0957-4484/25/15/155501. [DOI] [PubMed] [Google Scholar]
- 320.Yin J, He X, Wang K, Xu F, Shangguan J, He D, Shi H. Label-Free and Turn-on Aptamer Strategy for Cancer Cells Detection Based on a DNA-Silver Nanocluster Fluorescence Upon Recognition-Induced Hybridization. Anal Chem. 2013;85:12011–12019. doi: 10.1021/ac402989u. [DOI] [PubMed] [Google Scholar]
- 321.Bergers G, Benjamin LE. Tumorigenesis and the Angiogenic Switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 322.Hood JD, Cheresh DA. Role of Integrins in Cell Invasion and Migration. Nat Rev Cancer. 2002;2:91. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 323.Hong H, Shi J, Yang YA, Zhang Y, Engle JW, Nickles RJ, Wang XD, Cai WB. Cancer-Targeted Optical Imaging with Fluorescent Zinc Oxide Nanowires. Nano Lett. 2011;11:3744–3750. doi: 10.1021/nl201782m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Yezhelyev MV, Al-Hajj A, Morris C, Marcus AI, Liu T, Lewis M, Cohen C, Zrazhevskiy P, Simons JW, Rogatko A, et al. In Situ Molecular Profiling of Breast Cancer Biomarkers with Multicolor Quantum Dots. Adv Mater. 2007;19:3146. [Google Scholar]
- 325.Chen XL, Estevez MC, Zhu Z, Huang YF, Chen Y, Wang L, Tan WH. Using Aptamer-Conjugated Fluorescence Resonance Energy Transfer Nanoparticles for Multiplexed Cancer Cell Monitoring. Anal Chem. 2009;81:7009–7014. doi: 10.1021/ac9011073. [DOI] [PubMed] [Google Scholar]
- 326.Huang YF, Chang HT, Tan WH. Cancer Cell Targeting Using Multiple Aptamers Conjugated on Nanorods. Anal Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 327.Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-Flares: Probes for Transfection and Mrna Detection in Living Cells. J Am Chem Soc. 2007;129:15477. doi: 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Briley W, Halo TL, Randeria PS, Alhasan AH, Auyeung E, Hurst SJ, Mirkin CA. Biochemistry and Biomedical Applications of Spherical Nucleic Acids (Snas) Acs Sym Ser. 2012;1119:1–20. [Google Scholar]
- 329.Choi CHJ, Hao LL, Narayan SP, Auyeung E, Mirkin CA. Mechanism for the Endocytosis of Spherical Nucleic Acid Nanoparticle Conjugates. Proc Natl Acad Sci U S A. 2013;110:7625–7630. doi: 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Cutler JI, Auyeung E, Mirkin CA. Spherical Nucleic Acids. J Am Chem Soc. 2012;134:1376–1391. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 331.Giljohann DA, Seferos DS, Patel PC, Millstone JE, Rosi NL, Mirkin CA. Oligonucleotide Loading Determines Cellular Uptake of DNA-Modified Gold Nanoparticles. Nano Lett. 2007;7:3818–3821. doi: 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 333.Patel PC, Giljohann DA, Daniel WL, Zheng D, Prigodich AE, Mirkin CA. Scavenger Receptors Mediate Cellular Uptake of Polyvalent Oligonucleotide-Functionalized Gold Nanoparticles. Bioconjugate Chem. 2010;21:2250–2256. doi: 10.1021/bc1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett. 2009;9:308–311. doi: 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Levsky JM, Singer RH. Fluorescence in Situ Hybridization: Past, Present and Future. J Cell Sci. 2003;116:2833–2838. doi: 10.1242/jcs.00633. [DOI] [PubMed] [Google Scholar]
- 336.Massich MD, Giljohann DA, Schmucker AL, Patel PC, Mirkin CA. Cellular Response of Polyvalent Oligonucleotide-Gold Nanoparticle Conjugates. ACS Nano. 2010;4:5641–5646. doi: 10.1021/nn102228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Massich MD, Giljohann DA, Seferos DS, Ludlow LE, Horvath CM, Mirkin CA. Regulating Immune Response Using Polyvalent Nucleic Acid-Gold Nanoparticle Conjugates. Mol Pharmaceutics. 2009;6:1934–1940. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Prigodich AE, Randeria PS, Briley WE, Kim NJ, Daniel WL, Giljohann DA, Mirkin CA. Multiplexed Nanoflares: Mrna Detection in Live Cells. Anal Chem. 2012;84:2062–2066. doi: 10.1021/ac202648w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Prigodich AE, Seferos DS, Massich MD, Giljohann DA, Lane BC, Mirkin CA. Nano-Flares for Mrna Regulation and Detection. ACS Nano. 2009;3:2147–2152. doi: 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Zheng D, Seferos DS, Giljohann DA, Patel PC, Mirkin CA. Aptamer Nano-Flares for Molecular Detection in Living Cells. Nano Lett. 2009;9:3258–3261. doi: 10.1021/nl901517b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Li N, Chang C, Pan W, Tang B. A Multicolor Nanoprobe for Detection and Imaging of Tumor-Related Mrnas in Living Cells. Angew Chem, Int Ed. 2012;51:7426–7430. doi: 10.1002/anie.201203767. [DOI] [PubMed] [Google Scholar]
- 342.Yang B, Zhang XB, Kang LP, Huang ZM, Shen GL, Yu RQ, Tan WH. Intelligent Layered Nanoflare: “Lab-on-a-Nanoparticle” for Multiple DNA Logic Gate Operations and Efficient Intracellular Delivery. Nanoscale. 2014;6:8990–8996. doi: 10.1039/c4nr01676a. [DOI] [PubMed] [Google Scholar]
- 343.Halo TL, McMahon KM, Angeloni NL, Xu Y, Wang W, Chinen AB, Malin D, Strekalova E, Cryns VL, Cheng C, et al. Nanoflares for the Detection, Isolation, and Culture of Live Tumor Cells from Human Blood. Proc Natl Acad Sci U S A. 2014;111:17104–17109. doi: 10.1073/pnas.1418637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Briley WE, Bondy MH, Randeria PS, Dupper TJ, Mirkin CA. Sticky-flares: A Platform for the Quantification and Real-Time Tracking of RNA in Live Cells. Proc Natl Acad Sci U S A. 2015;112:9591–9595. doi: 10.1073/pnas.1510581112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Lahm H, Doppler S, Dressen M, Werner A, Adamczyk K, Schrambke D, Brade T, Laugwitz KL, Deutsch MA, Schiemann M, et al. Live Fluorescent Rna-Based Detection of Pluripotency Gene Expression in Embryonic and Induced Pluripotent Cells of Different Species. Stem Cells. 2015;33:392. doi: 10.1002/stem.1872. [DOI] [PubMed] [Google Scholar]
- 346.Mehta A, Sequiera GL, Ramachandra CJA, Sudibyo Y, Chung YY, Sheng JW, Wong KY, Tan TH, Wong P, Liew R, et al. Re-Trafficking of Herg Reverses Long Qt Syndrome 2 Phenotype in Human Ips-Derived Cardiomyocytes. Cardiovasc Res. 2014;102:497–506. doi: 10.1093/cvr/cvu060. [DOI] [PubMed] [Google Scholar]
- 347.Khare S, Ratsimandresy RA, de Almeida L, Cuda CM, Rellick SL, Misharin AV, Wallin MC, Gangopadhyay A, Forte E, Gottwein E, et al. The Pyrin Domain-Only Protein Pop3 Inhibits Alr Inflammasomes and Regulates Responses to Infection with DNA Viruses. Nat Immunol. 2014;15:343. doi: 10.1038/ni.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Kratz A, Beguin P, Kaneko M, Chimura T, Suzuki AM, Matsunaga A, Kato S, Bertin N, Lassmann T, Vigot R, et al. Digital Expression Profiling of the Compartmentalized Translatome of Purkinje Neurons. Genome Res. 2014;24:1396–1410. doi: 10.1101/gr.164095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Seftor EA, Seftor REB, Weldon DS, Kirsammer GT, Margaryan NV, Gilgur A, Hendrix MJC. Melanoma Tumor Cell Heterogeneity: A Molecular Approach to Study Subpopulations Expressing the Embryonic Morphogen Nodal. Semin Oncol. 2014;41:259–266. doi: 10.1053/j.seminoncol.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Postovit LM, Margaryan NV, Seftor EA, Hendrix MJ. Role of Nodal Signaling and the Microenvironment Underlying Melanoma Plasticity. Pigm Cell Melanoma Res. 2008;21:348–357. doi: 10.1111/j.1755-148X.2008.00463.x. [DOI] [PubMed] [Google Scholar]
- 351.Strizzi L, Hardy KM, Kirsammer GT, Gerami P, Hendrix MJ. Embryonic Signaling in Melanoma: Potential for Diagnosis and Therapy. Lab Invest. 2011;91:819–824. doi: 10.1038/labinvest.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Strizzi L, Postovit LM, Margaryan NV, Lipavsky A, Gadiot J, Blank C, Seftor RE, Seftor EA, Hendrix MJ. Nodal as a Biomarker for Melanoma Progression and a New Therapeutic Target for Clinical Intervention. Expert Rev Dermatol. 2009;4:67–78. doi: 10.1586/17469872.4.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Topczewska JM, Postovit LM, Margaryan NV, Sam A, Hess AR, Wheaton WW, Nickoloff BJ, Topczewski J, Hendrix MJ. Embryonic and Tumorigenic Pathways Converge Via Nodal Signaling: Role in Melanoma Aggressiveness. Nat Med. 2006;12:925–932. doi: 10.1038/nm1448. [DOI] [PubMed] [Google Scholar]
- 354.Choi Y, Kim HS, Woo J, Hwang EH, Cho KW, Kim S, Moon WK. Real-Time Imaging of the Epithelial-Mesenchymal Transition Using Microrna-200a Sequence-Based Molecular Beacon-Conjugated Magnetic Nanoparticles. PLoS One. 2014;9:e102164. doi: 10.1371/journal.pone.0102164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Pan W, Yang HJ, Zhang TT, Li YH, Li N, Tang B. Dual-Targeted Nanocarrier Based on Cell Surface Receptor and Intracellular Mrna: An Effective Strategy for Cancer Cell Imaging and Therapy. Anal Chem. 2013;85:6930–6935. doi: 10.1021/ac401405n. [DOI] [PubMed] [Google Scholar]
- 356.Pan W, Zhang TT, Yang HJ, Diao W, Li N, Tang B. Multiplexed Detection and Imaging of Intracellular Mrnas Using a Four-Color Nanoprobe. Anal Chem. 2013;85:10581–10588. doi: 10.1021/ac402700s. [DOI] [PubMed] [Google Scholar]
- 357.Piao Y, Liu F, Seo TS. A Novel Molecular Beacon Bearing a Graphite Nanoparticle as a Nanoquencher for in Situ Mrna Detection in Cancer Cells. ACS Appl Mater Interfaces. 2012;4:6785–6789. doi: 10.1021/am301976r. [DOI] [PubMed] [Google Scholar]
- 358.Qiao GM, Gao Y, Li N, Yu ZZ, Zhuo LH, Tang B. Simultaneous Detection of Intracellular Tumor Mrna with Bi-Color Imaging Based on a Gold Nanoparticle/Molecular Beacon. Chem - Eur J. 2011;17:11210–11215. doi: 10.1002/chem.201100658. [DOI] [PubMed] [Google Scholar]
- 359.Qiao GM, Zhuo LH, Gao Y, Yu LJ, Li N, Tang B. A Tumor Mrna-Dependent Gold Nanoparticle-Molecular Beacon Carrier for Controlled Drug Release and Intracellular Imaging. Chem Commun (Cambridge, U K) 2011;47:7458–7460. doi: 10.1039/c1cc11490e. [DOI] [PubMed] [Google Scholar]
- 360.Xue JP, Shan LL, Chen HY, Li Y, Zhu HY, Deng DW, Qian ZY, Achilefu S, Gu YQ. Visual Detection of Stat5b Gene Expression in Living Cell Using the Hairpin DNA Modified Gold Nanoparticle Beacon. Biosens Bioelectron. 2013;41:71–77. doi: 10.1016/j.bios.2012.06.062. [DOI] [PubMed] [Google Scholar]
- 361.Agrawal P, Strijkers GJ, Nicolay K. Chitosan-Based Systems for Molecular Imaging. Adv Drug Delivery Rev. 2010;62:42–58. doi: 10.1016/j.addr.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 362.Alitalo A, Detmar M. Interaction of Tumor Cells and Lymphatic Vessels in Cancer Progression. Oncogene. 2012;31:4499–4508. doi: 10.1038/onc.2011.602. [DOI] [PubMed] [Google Scholar]
- 363.Altinoglu EI, Russin TJ, Kaiser JM, Barth BM, Eklund PC, Kester M, Adair JH. Near-Infrared Emitting Fluorophore-Doped Calcium Phosphate Nanoparticles for in Vivo Imaging of Human Breast Cancer. ACS Nano. 2008;2:2075–2084. doi: 10.1021/nn800448r. [DOI] [PubMed] [Google Scholar]
- 364.Ballou B, Ernst LA, Andreko S, Harper T, Fitzpatrick JAJ, Waggoner AS, Bruchez MP. Sentinel Lymph Node Imaging Using Quantum Dots in Mouse Tumor Models. Bioconjugate Chem. 2007;18:389–396. doi: 10.1021/bc060261j. [DOI] [PubMed] [Google Scholar]
- 365.Cheng LA, Yang K, Shao MW, Lee ST, Liu ZA. Multicolor in Vivo Imaging of Upconversion Nanopaticles with Emissions Tuned by Luminescence Resonance Energy Transfer. J Phys Chem C. 2011;115:2686–2692. [Google Scholar]
- 366.Choi KY, Min KH, Yoon HY, Kim K, Park JH, Kwon IC, Choi K, Jeong SY. Pegylation of Hyaluronic Acid Nanoparticles Improves Tumor Targetability in Vivo. Biomaterials. 2011;32:1880–1889. doi: 10.1016/j.biomaterials.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 367.Chou LY, Chan WC. Fluorescence-Tagged Gold Nanoparticles for Rapidly Characterizing the Size-Dependent Biodistribution in Tumor Models. Adv Healthcare Mater. 2012;1:714–721. doi: 10.1002/adhm.201200084. [DOI] [PubMed] [Google Scholar]
- 368.Cohen S, Margel S. Engineering of near IR Fluorescent Albumin Nanoparticles for in Vivo Detection of Colon Cancer. J Nanobiotechnol. 2012;10:36. doi: 10.1186/1477-3155-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Cohen S, Pellach M, Kam Y, Grinberg I, Corem-Salkmon E, Rubinstein A, Margel S. Synthesis and Characterization of near Ir Fluorescent Albumin Nanoparticles for Optical Detection of Colon Cancer. Mater Sci Eng, C. 2013;33:923–931. doi: 10.1016/j.msec.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 370.Deng D, Zhang D, Li Y, Achilefu S, Gu Y. Gold Nanoparticles Based Molecular Beacons for in Vitro and in Vivo Detection of the Matriptase Expression on Tumor. Biosens Bioelectron. 2013;49:216–221. doi: 10.1016/j.bios.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 371.Fortin PY, Genevois C, Koenig A, Heinrich E, Texier I, Couillaud F. Detection of Brain Tumors Using Fluorescence Diffuse Optical Tomography and Nanoparticles as Contrast Agents. J Biomed Opt. 2012;17:126004. doi: 10.1117/1.JBO.17.12.126004. [DOI] [PubMed] [Google Scholar]
- 372.Hong GS, Robinson JT, Zhang YJ, Diao S, Antaris AL, Wang QB, Dai HJ. In Vivo Fluorescence Imaging with Ag2s Quantum Dots in the Second near-Infrared Region. Angew Chem, Int Ed. 2012;51:9818–9821. doi: 10.1002/anie.201206059. [DOI] [PubMed] [Google Scholar]
- 373.Janes KA, Calvo P, Alonso MJ. Polysaccharide Colloidal Particles as Delivery Systems for Macromolecules. Adv Drug Delivery Rev. 2001;47:83–97. doi: 10.1016/s0169-409x(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 374.Key J, Cooper C, Kim AY, Dhawan D, Knapp DW, Kim K, Park JH, Choi K, Kwon IC, Park K, et al. In Vivo Nirf and Mr Dual-Modality Imaging Using Glycol Chitosan Nanoparticles. J Controlled Release. 2012;163:249–255. doi: 10.1016/j.jconrel.2012.07.038. [DOI] [PubMed] [Google Scholar]
- 375.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, et al. Near-Infrared Fluorescent Type Ii Quantum Dots for Sentinel Lymph Node Mapping. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Lee CM, Jang D, Kim J, Cheong SJ, Kim EM, Jeong MH, Kim SH, Kim DW, Lim ST, Sohn MH, et al. Oleyl-Chitosan Nanoparticles Based on a Dual Probe for Optical/Mr Imaging in Vivo. Bioconjugate Chem. 2011;22:186–192. doi: 10.1021/bc100241a. [DOI] [PubMed] [Google Scholar]
- 377.Lee H, Lee K, Kim IK, Park TG. Synthesis, Characterization, and in Vivo Diagnostic Applications of Hyaluronic Acid Immobilized Gold Nanoprobes. Biomaterials. 2008;29:4709–4718. doi: 10.1016/j.biomaterials.2008.08.038. [DOI] [PubMed] [Google Scholar]
- 378.Liu JN, Liu Y, Bu WB, Bu JW, Sun Y, Du JL, Shi JL. Ultrasensitive Nanosensors Based on Upconversion Nanoparticles for Selective Hypoxia Imaging in Vivo Upon near-Infrared Excitation. J Am Chem Soc. 2014;136:9701–9709. doi: 10.1021/ja5042989. [DOI] [PubMed] [Google Scholar]
- 379.Nam T, Park S, Lee SY, Park K, Choi K, Song IC, Han MH, Leary JJ, Yuk SA, Kwon IC, et al. Tumor Targeting Chitosan Nanoparticles for Dual-Modality Optical/Mr Cancer Imaging. Bioconjugate Chem. 2010;21:578–582. doi: 10.1021/bc900408z. [DOI] [PubMed] [Google Scholar]
- 380.Papkovsky DB, Dmitriev RI. Biological Detection by Optical Oxygen Sensing. Chem Soc Rev. 2013;42:8700–8732. doi: 10.1039/c3cs60131e. [DOI] [PubMed] [Google Scholar]
- 381.Perrault SD, Chan WCW. In Vivo Assembly of Nanoparticle Components to Improve Targeted Cancer Imaging. Proc Natl Acad Sci U S A. 2010;107:11194–11199. doi: 10.1073/pnas.1001367107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Popovic Z, Liu WH, Chauhan VP, Lee J, Wong C, Greytak AB, Insin N, Nocera DG, Fukumura D, Jain RK, et al. A Nanoparticle Size Series for in Vivo Fluorescence Imaging. Angew Chem, Int Ed. 2010;49:8649–8652. doi: 10.1002/anie.201003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Schadlich A, Caysa H, Mueller T, Tenambergen F, Rose C, Gopferich A, Kuntsche J, Mader K. Tumor Accumulation of Nir Fluorescent Peg Pla Nanoparticles: Impact of Particle Size and Human Xenograft Tumor Model. ACS Nano. 2011;5:8710–8720. doi: 10.1021/nn2026353. [DOI] [PubMed] [Google Scholar]
- 384.Schadlich A, Rose C, Kuntsche J, Caysa H, Mueller T, Gopferich A, Mader K. How Stealthy Are Peg-Pla Nanoparticles? An Nir in Vivo Study Combined with Detailed Size Measurements. Pharm Res. 2011;28:1995–2007. doi: 10.1007/s11095-011-0426-5. [DOI] [PubMed] [Google Scholar]
- 385.Sharker SM, Jeong CJ, Kim SM, Lee JE, Jeong JH, In I, Lee H, Park SY. Photo- and Ph-Tunable Multicolor Fluorescent Nanoparticle-Based Spiropyran-and Bodipy-Conjugated Polymer with Graphene Oxide. Chem - Asian J. 2014;9:2921–2927. doi: 10.1002/asia.201402399. [DOI] [PubMed] [Google Scholar]
- 386.Shen JW, Wang JN, Kong DL, Yan XP. Sub-20 Nm Sandwich-Structured Nagdf4:Yb/Tm@Naluf4:Yb/Tm@Nayf4 Nanocrystals for in Vivo Upconversion Luminescence/Computed Tomography Imaging. RSC Adv. 2014;4:5088–5091. [Google Scholar]
- 387.von Maltzahn G, Park JH, Lin KY, Singh N, Schwoppe C, Mesters R, Berdel WE, Ruoslahti E, Sailor MJ, Bhatia SN. Nanoparticles That Communicate in Vivo to Amplify Tumour Targeting. Nat Mater. 2011;10:545–552. doi: 10.1038/nmat3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Wilson WR, Hay MP. Targeting Hypoxia in Cancer Therapy. Nat Rev Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 389.Wissmann C, Detmar M. Pathways Targeting Tumor Lymphangiogenesis. Clin Cancer Res. 2006;12:6865–6868. doi: 10.1158/1078-0432.CCR-06-1800. [DOI] [PubMed] [Google Scholar]
- 390.Wu X, He X, Wang K, Xie C, Zhou B, Qing Z. Ultrasmall near-Infrared Gold Nanoclusters for Tumor Fluorescence Imaging in Vivo. Nanoscale. 2010;2:2244–2249. doi: 10.1039/c0nr00359j. [DOI] [PubMed] [Google Scholar]
- 391.Yi ZG, Zeng SJ, Lu W, Wang HB, Rao L, Liu HR, Hao JH. Synergistic Dual-Modality in Vivo Upconversion Luminescence/X-Ray Imaging and Tracking of Amine-Functionalized Naybf4:Er Nanoprobes. ACS Appl Mater Interfaces. 2014;6:3839–3846. doi: 10.1021/am500383m. [DOI] [PubMed] [Google Scholar]
- 392.Zhang J, Li C, Zhang X, Huo S, Jin S, An FF, Wang X, Xue X, Okeke CI, Duan G, et al. In Vivo Tumor-Targeted Dual-Modal Fluorescence/Ct Imaging Using a Nanoprobe Co-Loaded with an Aggregation-Induced Emission Dye and Gold Nanoparticles. Biomaterials. 2015;42:103–111. doi: 10.1016/j.biomaterials.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Chen YY, Ma PA, Yang DM, Wu Y, Dai YL, Li CX, Lin J. Multifunctional Core-Shell Structured Nanocarriers for Synchronous Tumor Diagnosis and Treatment in Vivo. Chem - Asian J. 2014;9:506–513. doi: 10.1002/asia.201301262. [DOI] [PubMed] [Google Scholar]
- 394.Choi KY, Jeon EJ, Yoon HY, Lee BS, Na JH, Min KH, Kim SY, Myung SJ, Lee S, Chen X, et al. Theranostic Nanoparticles Based on Pegylated Hyaluronic Acid for the Diagnosis, Therapy and Monitoring of Colon Cancer. Biomaterials. 2012;33:6186–6193. doi: 10.1016/j.biomaterials.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Corsi F, Fiandra L, De Palma C, Colombo M, Mazzucchelli S, Verderio P, Allevi R, Tosoni A, Nebuloni M, Clementi E, et al. Her2 Expression in Breast Cancer Cells Is Downregulated Upon Active Targeting by Antibody-Engineered Multifunctional Nanoparticles in Mice. ACS Nano. 2011;5:6383–6393. doi: 10.1021/nn201570n. [DOI] [PubMed] [Google Scholar]
- 396.Fu A, Wilson RJ, Smith BR, Mullenix J, Earhart C, Akin D, Guccione S, Wang SX, Gambhir SS. Fluorescent Magnetic Nanoparticles for Magnetically Enhanced Cancer Imaging and Targeting in Living Subjects. ACS Nano. 2012;6:6862–6869. doi: 10.1021/nn301670a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Hollis CP, Weiss HL, Evers BM, Gemeinhart RA, Li T. In Vivo Investigation of Hybrid Paclitaxel Nanocrystals with Dual Fluorescent Probes for Cancer Theranostics. Pharm Res. 2014;31:1450–1459. doi: 10.1007/s11095-013-1048-x. [DOI] [PubMed] [Google Scholar]
- 398.Kim K, Kim JH, Park H, Kim YS, Park K, Nam H, Lee S, Park JH, Park RW, Kim IS, et al. Tumor-Homing Multifunctional Nanoparticles for Cancer Theragnosis: Simultaneous Diagnosis, Drug Delivery, and Therapeutic Monitoring. J Controlled Release. 2010;146:219–227. doi: 10.1016/j.jconrel.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 399.Lu RM, Chang YL, Chen MS, Wu HC. Single Chain Anti-C-Met Antibody Conjugated Nanoparticles for in Vivo Tumor-Targeted Imaging and Drug Delivery. Biomaterials. 2011;32:3265–3274. doi: 10.1016/j.biomaterials.2010.12.061. [DOI] [PubMed] [Google Scholar]
- 400.Santra S, Kaittanis C, Perez JM. Cytochrome C Encapsulating Theranostic Nanoparticles: A Novel Bifunctional System for Targeted Delivery of Therapeutic Membrane-Impermeable Proteins to Tumors and Imaging of Cancer Therapy. Mol Pharmaceutics. 2010;7:1209–1222. doi: 10.1021/mp100043h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Wang S, Kim G, Lee YE, Hah HJ, Ethirajan M, Pandey RK, Kopelman R. Multifunctional Biodegradable Polyacrylamide Nanocarriers for Cancer Theranostics–a “See and Treat” Strategy. ACS Nano. 2012;6:6843–6851. doi: 10.1021/nn301633m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Wu X, Sun X, Guo Z, Tang J, Shen Y, James TD, Tian H, Zhu W. In Vivo and in Situ Tracking Cancer Chemotherapy by Highly Photostable Nir Fluorescent Theranostic Prodrug. J Am Chem Soc. 2014;136:3579–3588. doi: 10.1021/ja412380j. [DOI] [PubMed] [Google Scholar]
- 403.Baker M. The Whole Picture. Nature. 2010;463:977–980. doi: 10.1038/463977a. [DOI] [PubMed] [Google Scholar]
- 404.Hahn MA, Singh AK, Sharma P, Brown SC, Moudgil BM. Nanoparticles as Contrast Agents for in-Vivo Bioimaging: Current Status and Future Perspectives. Anal Bioanal Chem. 2011;399:3–27. doi: 10.1007/s00216-010-4207-5. [DOI] [PubMed] [Google Scholar]
- 405.Rudin M, Weissleder R. Molecular Imaging in Drug Discovery and Development. Nat Rev Drug Discovery. 2003;2:123–131. doi: 10.1038/nrd1007. [DOI] [PubMed] [Google Scholar]
- 406.Weissleder R. A Clearer Vision for in Vivo Imaging. Nat Biotechnol. 2001;19:316–317. doi: 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- 407.Cho K, Wang X, Nie S, Chen ZG, Shin DM. Therapeutic Nanoparticles for Drug Delivery in Cancer. Clin Cancer Res. 2008;14:1310–1316. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 408.Danhier F, Feron O, Preat V. To Exploit the Tumor Microenvironment: Passive and Active Tumor Targeting of Nanocarriers for Anti-Cancer Drug Delivery. J Controlled Release. 2010;148:135–146. doi: 10.1016/j.jconrel.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 409.Bertrand N, Wu J, Xu X, Kamaly N, Farokhzad OC. Cancer Nanotechnology: The Impact of Passive and Active Targeting in the Era of Modern Cancer Biology. Adv Drug Delivery Rev. 2014;66:2–25. doi: 10.1016/j.addr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor Vascular Permeability and the Epr Effect in Macromolecular Therapeutics: A Review. J Controlled Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- 411.Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. Challenges and Key Considerations of the Enhanced Permeability and Retention Effect for Nanomedicine Drug Delivery in Oncology. Cancer Res. 2013;73:2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Frank D, Tyagi C, Tomar L, Choonara YE, du Toit LC, Kumar P, Penny C, Pillay V. Overview of the Role of Nanotechnological Innovations in the Detection and Treatment of Solid Tumors. Int J Nanomed. 2014;9:589–613. doi: 10.2147/IJN.S50941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Maeda H. Toward a Full Understanding of the Epr Effect in Primary and Metastatic Tumors as Well as Issues Related to Its Heterogeneity. Adv Drug Delivery Rev. 2015 doi: 10.1016/j.addr.2015.01.002.. [DOI] [PubMed] [Google Scholar]
- 414.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an Emerging Platform for Cancer Therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 415.Matsumura Y, Maeda H. A New Concept for Macromolecular Therapeutics in Cancer Chemotherapy: Mechanism of Tumoritropic Accumulation of Proteins and the Antitumor Agent Smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 416.Jain RK. Transport of Molecules across Tumor Vasculature. Cancer Metastasis Rev. 1987;6:559–593. doi: 10.1007/BF00047468. [DOI] [PubMed] [Google Scholar]
- 417.Prabhakar U, Maeda H, Jain RK, Sevick-Muraca EM, Zamboni W, Farokhzad OC, Barry ST, Gabizon A, Grodzinski P, Blakey DC. Challenges and Key Considerations of the Enhanced Permeability and Retention Effect for Nanomedicine Drug Delivery in Oncology. Cancer Res. 2013;73:2412–2417. doi: 10.1158/0008-5472.CAN-12-4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Fang J, Nakamura H, Maeda H. The Epr Effect: Unique Features of Tumor Blood Vessels for Drug Delivery, Factors Involved, and Limitations and Augmentation of the Effect. Adv Drug Delivery Rev. 2011;63:136–151. doi: 10.1016/j.addr.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 419.Corbin IR, Chen J, Li H, Cao W, Zheng G. Functionalizing Low-Density Lipoprotein Nanoparticles for in Vivo near-Infrared Optical Imaging of Cancer. Proc SPIE. 2007;6626:66260B. [Google Scholar]
- 420.Cedervall T, Lynch I, Foy M, Berggard T, Donnelly SC, Cagney G, Linse S, Dawson KA. Detailed Identification of Plasma Proteins Adsorbed on Copolymer Nanoparticles. Angew Chem, Int Ed. 2007;46:5754–5756. doi: 10.1002/anie.200700465. [DOI] [PubMed] [Google Scholar]
- 421.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Understanding the Nanoparticle-Protein Corona Using Methods to Quantify Exchange Rates and Affinities of Proteins for Nanoparticles. Proc Natl Acad Sci U S A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S, Dawson KA. The Nanoparticle-Protein Complex as a Biological Entity; a Complex Fluids and Surface Science Challenge for the 21st Century. Adv Colloid Interface Sci. 2007;134–135:167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 423.Lynch I, Salvati A, Dawson KA. Protein-Nanoparticle Interactions: What Does the Cell See? Nat Nanotechnol. 2009;4:546–547. doi: 10.1038/nnano.2009.248. [DOI] [PubMed] [Google Scholar]
- 424.Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, Elia G, Dawson K. The Evolution of the Protein Corona around Nanoparticles: A Test Study. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 425.Mahmoudi M, Lynch I, Ejtehadi MR, Monopoli MP, Bombelli FB, Laurent S. Protein-Nanoparticle Interactions: Opportunities and Challenges. Chem Rev. 2011;111:5610–5637. doi: 10.1021/cr100440g. [DOI] [PubMed] [Google Scholar]
- 426.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle Size and Surface Properties Determine the Protein Corona with Possible Implications for Biological Impacts. Proc Natl Acad Sci U S A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Ehrenberg MS, Friedman AE, Finkelstein JN, Oberdorster G, McGrath JL. The Influence of Protein Adsorption on Nanoparticle Association with Cultured Endothelial Cells. Biomaterials. 2009;30:603–610. doi: 10.1016/j.biomaterials.2008.09.050. [DOI] [PubMed] [Google Scholar]
- 428.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Mediating Tumor Targeting Efficiency of Nanoparticles through Design. Nano Lett. 2009;9:1909–1915. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- 429.Walkey CD, Chan WC. Understanding and Controlling the Interaction of Nanomaterials with Proteins in a Physiological Environment. Chem Soc Rev. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 430.Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. Nanoparticle Size and Surface Chemistry Determine Serum Protein Adsorption and Macrophage Uptake. J Am Chem Soc. 2012;134:2139–2147. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 431.Walkey CD, Olsen JB, Song F, Liu R, Guo H, Olsen DW, Cohen Y, Emili A, Chan WC. Protein Corona Fingerprinting Predicts the Cellular Interaction of Gold and Silver Nanoparticles. ACS Nano. 2014;8:2439–2455. doi: 10.1021/nn406018q. [DOI] [PubMed] [Google Scholar]
- 432.Chinen AB, Guan CM, Mirkin CA. Spherical Nucleic Acid Nanoparticle Conjugates Enhance G-Quadruplex Formation and Increase Serum Protein Interactions. Angew Chem, Int Ed. 2015;54:527–531. doi: 10.1002/anie.201409211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Lane LA, Qian X, Smith AM, Nie S. Physical Chemistry of Nanomedicine: Understanding the Complex Behaviors of Nanoparticles in Vivo. Annu Rev Phys Chem. 2015;66:521. doi: 10.1146/annurev-physchem-040513-103718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Mahmoudi M, Sheibani S, Milani AS, Rezaee F, Gauberti M, Dinarvand R, Vali H. Crucial Role of the Protein Corona for the Specific Targeting of Nanoparticles. Nanomedicine (London, U K) 2015;10:215–226. doi: 10.2217/nnm.14.69. [DOI] [PubMed] [Google Scholar]
- 435.Otsuka H, Nagasaki Y, Kataoka K. Pegylated Nanoparticles for Biological and Pharmaceutical Applications. Adv Drug Delivery Rev. 2003;55:403–419. doi: 10.1016/s0169-409x(02)00226-0. [DOI] [PubMed] [Google Scholar]
- 436.Karakoti AS, Das S, Thevuthasan S, Seal S. Pegylated Inorganic Nanoparticles. Angew Chem, Int Ed. 2011;50:1980–1994. doi: 10.1002/anie.201002969. [DOI] [PubMed] [Google Scholar]
- 437.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle Pegylation for Imaging and Therapy. Nanomedicine. 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Yoo JW, Chambers E, Mitragotri S. Factors That Control the Circulation Time of Nanoparticles in Blood: Challenges, Solutions and Future Prospects. Curr Pharm Des. 2010;16:2298–2307. doi: 10.2174/138161210791920496. [DOI] [PubMed] [Google Scholar]
- 439.Howard MD, Jay M, Dziubla TD, Lu XL. Pegylation of Nanocarrier Drug Delivery Systems: State of the Art. J Biomed Nanotechnol. 2008;4:133–148. [Google Scholar]
- 440.Amoozgar Z, Yeo Y. Recent Advances in Stealth Coating of Nanoparticle Drug Delivery Systems. Wires Nanomed Nanobi. 2012;4:219–233. doi: 10.1002/wnan.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Abuchowski A, McCoy JR, Palczuk NC, van Es T, Davis FF. Effect of Covalent Attachment of Polyethylene Glycol on Immunogenicity and Circulating Life of Bovine Liver Catalase. J Biol Chem. 1977;252:3582–3586. [PubMed] [Google Scholar]
- 442.Bhadra D, Bhadra S, Jain S, Jain NK. A Pegylated Dendritic Nanoparticulate Carrier of Fluorouracil. Int J Pharm. 2003;257:111–124. doi: 10.1016/s0378-5173(03)00132-7. [DOI] [PubMed] [Google Scholar]
- 443.Cheng TL, Chen BM, Chan LY, Wu PY, Chern JW, Roffler SR. Poly(Ethylene Glycol) Modification of Beta-Glucuronidase-Antibody Conjugates for Solid-Tumor Therapy by Targeted Activation of Glucuronide Prodrugs. Cancer Immunol Immunother. 1997;44:305–315. doi: 10.1007/s002620050387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Francis GE, Fisher D, Delgado C, Malik F, Gardiner A, Neale D. Pegylation of Cytokines and Other Therapeutic Proteins and Peptides: The Importance of Biological Optimisation of Coupling Techniques. Int J Hematol. 1998;68:1–18. doi: 10.1016/s0925-5710(98)00039-5. [DOI] [PubMed] [Google Scholar]
- 445.Jevprasesphant R, Penny J, Jalal R, Attwood D, McKeown NB, D’Emanuele A. The Influence of Surface Modification on the Cytotoxicity of Pamam Dendrimers. Int J Pharm. 2003;252:263–266. doi: 10.1016/s0378-5173(02)00623-3. [DOI] [PubMed] [Google Scholar]
- 446.Moffatt S, Cristiano RJ. Uptake Characteristics of Ngr-Coupled Stealth Pei/Pdna Nanoparticles Loaded with Plga-Peg-Plga Tri-Block Copolymer for Targeted Delivery to Human Monocyte-Derived Dendritic Cells. Int J Pharm. 2006;321:143–154. doi: 10.1016/j.ijpharm.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 447.Niidome T, Yamagata M, Okamoto Y, Akiyama Y, Takahashi H, Kawano T, Katayama Y, Niidome Y. Peg-Modified Gold Nanorods with a Stealth Character for in Vivo Applications. J Controlled Release. 2006;114:343–347. doi: 10.1016/j.jconrel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 448.Peracchia MT, Fattal E, Desmaele D, Besnard M, Noel JP, Gomis JM, Appel M, d’Angelo J, Couvreur P. Stealth Pegylated Polycyanoacrylate Nanoparticles for Intravenous Administration and Splenic Targeting. J Controlled Release. 1999;60:121–128. doi: 10.1016/s0168-3659(99)00063-2. [DOI] [PubMed] [Google Scholar]
- 449.Kale AA, Torchilin VP. Enhanced Transfection of Tumor Cells in Vivo Using “Smart” Ph-Sensitive Tat-Modified Pegylated Liposomes. J Drug Target. 2007;15:538–545. doi: 10.1080/10611860701498203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Xiao RZ, Zeng ZW, Zhou GL, Wang JJ, Li FZ, Wang AM. Recent Advances in Peg-Pla Block Copolymer Nanoparticles. Int J Nanomed. 2010;5:1057–1065. doi: 10.2147/IJN.S14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Cho WS, Cho M, Jeong J, Choi M, Cho HY, Han BS, Kim SH, Kim HO, Lim YT, Chung BH, et al. Acute Toxicity and Pharmacokinetics of 13 Nm-Sized Peg-Coated Gold Nanoparticles. Toxicol Appl Pharmacol. 2009;236:16–24. doi: 10.1016/j.taap.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 452.Schipper ML, Iyer G, Koh AL, Cheng Z, Ebenstein Y, Aharoni A, Keren S, Bentolila LA, Li J, Rao J, et al. Particle Size, Surface Coating, and Pegylation Influence the Biodistribution of Quantum Dots in Living Mice. Small. 2009;5:126–134. doi: 10.1002/smll.200800003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Lipka J, Semmler-Behnke M, Sperling RA, Wenk A, Takenaka S, Schleh C, Kissel T, Parak WJ, Kreyling WG. Biodistribution of Peg-Modified Gold Nanoparticles Following Intratracheal Instillation and Intravenous Injection. Biomaterials. 2010;31:6574–6581. doi: 10.1016/j.biomaterials.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 454.Khlebtsov N, Dykman L. Biodistribution and Toxicity of Engineered Gold Nanoparticles: A Review of in Vitro and in Vivo Studies. Chem Soc Rev. 2011;40:1647–1671. doi: 10.1039/c0cs00018c. [DOI] [PubMed] [Google Scholar]
- 455.Simpson CA, Agrawal AC, Balinski A, Harkness KM, Cliffel DE. Short-Chain Peg Mixed Monolayer Protected Gold Clusters Increase Clearance and Red Blood Cell Counts. ACS Nano. 2011;5:3577–3584. doi: 10.1021/nn103148x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Abu Lila AS, Kiwada H, Ishida T. The Accelerated Blood Clearance (Abc) Phenomenon: Clinical Challenge and Approaches to Manage. J Controlled Release. 2013;172:38–47. doi: 10.1016/j.jconrel.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 457.Park K. To Pegylate or Not to Pegylate, That Is Not the Question. J Controlled Release. 2010;142:147–148. doi: 10.1016/j.jconrel.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 458.Ishida T, Kiwada H. Accelerated Blood Clearance (Abc) Phenomenon Upon Repeated Injection of Pegylated Liposomes. Int J Pharm. 2008;354:56–62. doi: 10.1016/j.ijpharm.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 459.Ishida T, Kashima S, Kiwada H. The Contribution of Phagocytic Activity of Liver Macrophages to the Accelerated Blood Clearance (Abc) Phenomenon of Pegylated Liposomes in Rats. J Controlled Release. 2008;126:162–165. doi: 10.1016/j.jconrel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 460.Ishida T, Kiwada H. Accelerated Blood Clearance (Abc) Phenomenon Induced by Administration of Pegylated Liposome. Yakugaku Zasshi. 2008;128:233–243. doi: 10.1248/yakushi.128.233. [DOI] [PubMed] [Google Scholar]
- 461.Tagami T, Nakamura K, Shimizu T, Ishida T, Kiwada H. The Relationship between Pegylated Sirna-Lipoplex and Anti-Peg Igm on the Induction of Accelerated Blood Clearance (Abc) Phenomenon. Yakugaku Zasshi. 2008;128:109–110. [Google Scholar]
- 462.Chen SF, Zheng J, Li LY, Jiang SY. Strong Resistance of Phosphorylcholine Self-Assembled Monolayers to Protein Adsorption: Insights into Nonfouling Properties of Zwitterionic Materials. J Am Chem Soc. 2005;127:14473–14478. doi: 10.1021/ja054169u. [DOI] [PubMed] [Google Scholar]
- 463.Bernards MT, Cheng G, Zhang Z, Chen SF, Jiang SY. Nonfouling Polymer Brushes Via Surface-Initiated, Two-Component Atom Transfer Radical Polymerization. Macromolecules. 2008;41:4216–4219. [Google Scholar]
- 464.Chen SF, Jiang SY. A New Avenue to Nonfouling Materials. Adv Mater. 2008;20:335–338. [Google Scholar]
- 465.Cheng G, Xue H, Zhang Z, Chen SF, Jiang SY. A Switchable Biocompatible Polymer Surface with Self-Sterilizing and Nonfouling Capabilities. Angew Chem, Int Ed. 2008;47:8831–8834. doi: 10.1002/anie.200803570. [DOI] [PubMed] [Google Scholar]
- 466.Vaisocherova H, Yang W, Zhang Z, Cao ZQ, Cheng G, Piliarik M, Homola J, Jiang SY. Ultralow Fouling and Functionalizable Surface Chemistry Based on a Zwitterionic Polymer Enabling Sensitive and Specific Protein Detection in Undiluted Blood Plasma. Anal Chem. 2008;80:7894–7901. doi: 10.1021/ac8015888. [DOI] [PubMed] [Google Scholar]
- 467.Depp V, Alikhani A, Grammer V, Lele BS. Native Protein-Initiated Atrp: A Viable and Potentially Superior Alternative to Pegylation for Stabilizing Biologics. Acta Biomater. 2009;5:560–569. doi: 10.1016/j.actbio.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 468.Schramm OG, Pavlov GM, van Erp HP, Meier MAR, Hoogenboom R, Schubert US. A Versatile Approach to Unimolecular Water-Soluble Carriers: Atrp of Pegma with Hydrophobic Star-Shaped Polymeric Core Molecules as an Alternative for Pegylation. Macromolecules. 2009;42:1808–1816. [Google Scholar]
- 469.Jiang SY, Cao ZQ. Ultralow-Fouling, Functionalizable, and Hydrolyzable Zwitterionic Materials and Their Derivatives for Biological Applications. Adv Mater. 2010;22:920–932. doi: 10.1002/adma.200901407. [DOI] [PubMed] [Google Scholar]
- 470.Barz M, Luxenhofer R, Zentel R, Vicent MJ. Overcoming the Peg-Addiction: Well-Defined Alternatives to Peg, from Structure-Property Relationships to Better Defined Therapeutics. Polym Chem. 2011;2:1900–1918. [Google Scholar]
- 471.Estephan ZG, Schlenoff PS, Schlenoff JB. Zwitteration as an Alternative to Pegylation. Langmuir. 2011;27:6794–6800. doi: 10.1021/la200227b. [DOI] [PubMed] [Google Scholar]
- 472.Keefe AJ, Jiang SY. Poly(Zwitterionic)Protein Conjugates Offer Increased Stability without Sacrificing Binding Affinity or Bioactivity. Nat Chem. 2012;4:59–63. doi: 10.1038/nchem.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Styslinger TJ, Zhang N, Bhatt VS, Pettit N, Palmer AF, Wang PG. Site-Selective Glycosylation of Hemoglobin with Variable Molecular Weight Oligosaccharides: Potential Alternative to Pegylation. J Am Chem Soc. 2012;134:7507–7515. doi: 10.1021/ja300893t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Besheer A, Liebner R, Meyer M, Winter G. Challenges for Pegylated Proteins and Alternative Half-Life Extension Technologies Based on Biodegradable Polymers. ACS Sym Ser. 2013;1135:215–233. [Google Scholar]
- 475.Coady DJ, Sardon H, Yang YY, Hedrick JL. Biodegradable Water Soluble Polymers as an Alternative to PEG. Abstracts of Papers, 245th National Meeting of the American Chemical Society; New Orleans, LA. April 7–11, 2013; Washington, DC: American Chemical Society; 2013. [Google Scholar]
- 476.DeAngelis PL. Heparosan Polysaccharide for Drug Delivery; a Biosuperior Alternative to Pegylation. Glycobiology. 2013;23:1347. [Google Scholar]
- 477.Schlapschy M, Binder U, Borger C, Theobald I, Wachinger K, Kisling S, Haller D, Skerra A. Pasylation: A Biological Alternative to Pegylation for Extending the Plasma Half-Life of Pharmaceutically Active Proteins. Protein Eng, Des Sel. 2013;26:489–501. doi: 10.1093/protein/gzt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Sun R, Du XJ, Sun CY, Shen S, Liu Y, Yang XZ, Bao Y, Zhu YH, Wang J. A Block Copolymer of Zwitterionic Polyphosphoester and Polylactic Acid for Drug Delivery. Biomater Sci. 2015;3:1105–1113. doi: 10.1039/c4bm00430b. [DOI] [PubMed] [Google Scholar]
- 479.Almeida JP, Chen AL, Foster A, Drezek R. In Vivo Biodistribution of Nanoparticles. Nanomedicine (London, U K) 2011;6:815–835. doi: 10.2217/nnm.11.79. [DOI] [PubMed] [Google Scholar]
- 480.Choi HS, Liu W, Liu F, Nasr K, Misra P, Bawendi MG, Frangioni JV. Design Considerations for Tumour-Targeted Nanoparticles. Nat Nanotechnol. 2010;5:42–47. doi: 10.1038/nnano.2009.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Liao WY, Li HJ, Chang MY, Tang AC, Hoffman AS, Hsieh PC. Comprehensive Characterizations of Nanoparticle Biodistribution Following Systemic Injection in Mice. Nanoscale. 2013;5:11079–11086. doi: 10.1039/c3nr03954d. [DOI] [PubMed] [Google Scholar]
- 482.Torosean S, Flynn B, Axelsson J, Gunn J, Samkoe KS, Hasan T, Doyley MM, Pogue BW. Nanoparticle Uptake in Tumors Is Mediated by the Interplay of Vascular and Collagen Density with Interstitial Pressure. Nanomedicine. 2013;9:151–158. doi: 10.1016/j.nano.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Minchinton AI, Tannock IF. Drug Penetration in Solid Tumours. Nat Rev Cancer. 2006;6:583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 484.Blouw B, Song H, Tihan T, Bosze J, Ferrara N, Gerber HP, Johnson RS, Bergers G. The Hypoxic Response of Tumors Is Dependent on Their Microenvironment. Cancer Cell. 2003;4:133–146. doi: 10.1016/s1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 485.Manzoor AA, Lindner LH, Landon CD, Park JY, Simnick AJ, Dreher MR, Das S, Hanna G, Park W, Chilkoti A, et al. Overcoming Limitations in Nanoparticle Drug Delivery: Triggered, Intravascular Release to Improve Drug Penetration into Tumors. Cancer Res. 2012;72:5566–5575. doi: 10.1158/0008-5472.CAN-12-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Huang X, Li L, Liu T, Hao N, Liu H, Chen D, Tang F. The Shape Effect of Mesoporous Silica Nanoparticles on Biodistribution, Clearance, and Biocompatibility in Vivo. ACS Nano. 2011;5:5390–5399. doi: 10.1021/nn200365a. [DOI] [PubMed] [Google Scholar]
- 487.Akiyama Y, Mori T, Katayama Y, Niidome T. Conversion of Rod-Shaped Gold Nanoparticles to Spherical Forms and Their Effect on Biodistribution in Tumor-Bearing Mice. Nanoscale Res Lett. 2012;7:565. doi: 10.1186/1556-276X-7-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Harris BJDP. Particle Shape Effects in Vitro and in Vivo. Front Biosci Scholar Ed. 2012;S4:1344–1353. doi: 10.2741/s336. [DOI] [PubMed] [Google Scholar]
- 489.Christian DA, Cai S, Garbuzenko OB, Harada T, Zajac AL, Minko T, Discher DE. Flexible Filaments for in Vivo Imaging and Delivery: Persistent Circulation of Filomicelles Opens the Dosage Window for Sustained Tumor Shrinkage. Mol Pharmaceutics. 2009;6:1343–1352. doi: 10.1021/mp900022m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Shape Effects of Filaments Versus Spherical Particles in Flow and Drug Delivery. Nat Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Lee S, Yun HS, Kim SH. The Comparative Effects of Mesoporous Silica Nanoparticles and Colloidal Silica on Inflammation and Apoptosis. Biomaterials. 2011;32:9434–9443. doi: 10.1016/j.biomaterials.2011.08.042. [DOI] [PubMed] [Google Scholar]
- 492.Tarn D, Ashley CE, Xue M, Carnes EC, Zink JI, Brinker CJ. Mesoporous Silica Nanoparticle Nanocarriers: Biofunctionality and Biocompatibility. Acc Chem Res. 2013;46:792–801. doi: 10.1021/ar3000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Shahbazi MA, Hamidi M, Makila EM, Zhang H, Almeida PV, Kaasalainen M, Salonen JJ, Hirvonen JT, Santos HA. The Mechanisms of Surface Chemistry Effects of Mesoporous Silicon Nanoparticles on Immunotoxicity and Biocompatibility. Biomaterials. 2013;34:7776–7789. doi: 10.1016/j.biomaterials.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 494.Barandeh F, Nguyen PL, Kumar R, Iacobucci GJ, Kuznicki ML, Kosterman A, Bergey EJ, Prasad PN, Gunawardena S. Organically Modified Silica Nanoparticles Are Biocompatible and Can Be Targeted to Neurons in Vivo. PLoS One. 2012;7:e29424. doi: 10.1371/journal.pone.0029424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.He Q, Zhang Z, Gao F, Li Y, Shi J. In Vivo Biodistribution and Urinary Excretion of Mesoporous Silica Nanoparticles: Effects of Particle Size and Pegylation. Small. 2011;7:271–280. doi: 10.1002/smll.201001459. [DOI] [PubMed] [Google Scholar]
- 496.Lee S, Kim MS, Lee D, Kwon TK, Khang D, Yun HS, Kim SH. The Comparative Immunotoxicity of Mesoporous Silica Nanoparticles and Colloidal Silica Nanoparticles in Mice. Int J Nanomed. 2013;8:147–158. doi: 10.2147/IJN.S39534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Hamad I, Al-Hanbali O, Hunter AC, Rutt KJ, Andresen TL, Moghimi SM. Distinct Polymer Architecture Mediates Switching of Complement Activation Pathways at the Nanosphere-Serum Interface: Implications for Stealth Nanoparticle Engineering. ACS Nano. 2010;4:6629–6638. doi: 10.1021/nn101990a. [DOI] [PubMed] [Google Scholar]
- 498.Huang J, Zhang H, Yu Y, Chen Y, Wang D, Zhang G, Zhou G, Liu J, Sun Z, Sun D, et al. Biodegradable Self-Assembled Nanoparticles of Poly (D,L-Lactide-Co-Glycolide)/Hyaluronic Acid Block Copolymers for Target Delivery of Docetaxel to Breast Cancer. Biomaterials. 2014;35:550–566. doi: 10.1016/j.biomaterials.2013.09.089. [DOI] [PubMed] [Google Scholar]
- 499.Marin E, Briceno MI, Caballero-George C. Critical Evaluation of Biodegradable Polymers Used in Nanodrugs. Int J Nanomed. 2013;8:3071–3090. doi: 10.2147/IJN.S47186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Yang Y, Pan D, Luo K, Li L, Gu Z. Biodegradable and Amphiphilic Block Copolymer-Doxorubicin Conjugate as Polymeric Nanoscale Drug Delivery Vehicle for Breast Cancer Therapy. Biomaterials. 2013;34:8430–8443. doi: 10.1016/j.biomaterials.2013.07.037. [DOI] [PubMed] [Google Scholar]
- 501.Liechty WB, Kryscio DR, Slaughter BV, Peppas NA. Polymers for Drug Delivery Systems. Annu Rev Chem Biomol Eng. 2010;1:149–173. doi: 10.1146/annurev-chembioeng-073009-100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Liechty WB, Peppas NA. Expert Opinion: Responsive Polymer Nanoparticles in Cancer Therapy. Eur J Pharm Biopharm. 2012;80:241–246. doi: 10.1016/j.ejpb.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Connor EE, Mwamuka J, Gole A, Murphy CJ, Wyatt MD. Gold Nanoparticles Are Taken up by Human Cells but Do Not Cause Acute Cytotoxicity. Small. 2005;1:325–327. doi: 10.1002/smll.200400093. [DOI] [PubMed] [Google Scholar]
- 504.Simpson CA, Huffman BJ, Gerdon AE, Cliffel DE. Unexpected Toxicity of Monolayer Protected Gold Clusters Eliminated by Peg-Thiol Place Exchange Reactions. Chem Res Toxicol. 2010;23:1608–1616. doi: 10.1021/tx100209t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Freese C, Uboldi C, Gibson MI, Unger RE, Weksler BB, Romero IA, Couraud PO, Kirkpatrick CJ. Uptake and Cytotoxicity of Citrate-Coated Gold Nanospheres: Comparative Studies on Human Endothelial and Epithelial Cells. Part Fibre Toxicol. 2012;9:23. doi: 10.1186/1743-8977-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Chen H, Dorrigan A, Saad S, Hare DJ, Cortie MB, Valenzuela SM. In Vivo Study of Spherical Gold Nanoparticles: Inflammatory Effects and Distribution in Mice. PLoS One. 2013;8:e58208. doi: 10.1371/journal.pone.0058208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Arnida, Malugin A, Ghandehari H. Cellular Uptake and Toxicity of Gold Nanoparticles in Prostate Cancer Cells: A Comparative Study of Rods and Spheres. J Appl Toxicol. 2010;30:212–217. doi: 10.1002/jat.1486. [DOI] [PubMed] [Google Scholar]
- 508.Favi PM, Gao M, Sepulveda Arango LJ, Ospina SP, Morales M, Pavon JJ, Webster TJ. Shape and Surface Effects on the Cytotoxicity of Nanoparticles: Gold Nanospheres Versus Gold Nanostars. J Biomed Mater Res, Part A. 2015 doi: 10.1002/jbm.a.35491.. [DOI] [PubMed] [Google Scholar]
- 509.Sultana S, Djaker N, Boca-Farcau S, Salerno M, Charnaux N, Astilean S, Hlawaty H, de la Chapelle ML. Comparative Toxicity Evaluation of Flower-Shaped and Spherical Gold Nanoparticles on Human Endothelial Cells. Nanotechnology. 2015;26:055101. doi: 10.1088/0957-4484/26/5/055101. [DOI] [PubMed] [Google Scholar]
- 510.Sun YN, Wang CD, Zhang XM, Ren L, Tian XH. Shape Dependence of Gold Nanoparticles on in Vivo Acute Toxicological Effects and Biodistribution. J Nanosci Nanotechnol. 2011;11:1210–1216. doi: 10.1166/jnn.2011.3094. [DOI] [PubMed] [Google Scholar]
- 511.Tarantola M, Pietuch A, Schneider D, Rother J, Sunnick E, Rosman C, Pierrat S, Sonnichsen C, Wegener J, Janshoff A. Toxicity of Gold-Nanoparticles: Synergistic Effects of Shape and Surface Functionalization on Micromotility of Epithelial Cells. Nanotoxicology. 2011;5:254–268. doi: 10.3109/17435390.2010.528847. [DOI] [PubMed] [Google Scholar]
- 512.Wang J, Xie YD, Wang LM, Tang JL, Li JY, Kocaefe D, Kocaefe Y, Zhang ZW, Li YP, Chen CY. In Vivo Pharmacokinetic Features and Biodistribution of Star and Rod Shaped Gold Nanoparticles by Multispectral Optoacoustic Tomography. RSC Adv. 2015;5:7529–7538. [Google Scholar]
- 513.Sengupta J, Datta P, Patra HK, Dasgupta AK, Gomes A. In Vivo Interaction of Gold Nanoparticles after Acute and Chronic Exposures in Experimental Animal Models. J Nanosci Nanotechnol. 2013;13:1660–1670. doi: 10.1166/jnn.2013.7113. [DOI] [PubMed] [Google Scholar]
- 514.Kwon KC, Ryu JH, Lee JH, Lee EJ, Kwon IC, Kim K, Lee J. Proteinticle/Gold Core/Shell Nanoparticles for Targeted Cancer Therapy without Nanotoxicity. Adv Mater. 2014;26:6436–6441. doi: 10.1002/adma.201401499. [DOI] [PubMed] [Google Scholar]
- 515.Zhang XD, Wu HY, Wu D, Wang YY, Chang JH, Zhai ZB, Meng AM, Liu PX, Zhang LA, Fan FY. Toxicologic Effects of Gold Nanoparticles in Vivo by Different Administration Routes. Int J Nanomed. 2010;5:771–781. doi: 10.2147/IJN.S8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Cheng L, Yang K, Shao M, Lu X, Liu Z. In Vivo Pharmacokinetics, Long-Term Biodistribution and Toxicology Study of Functionalized Upconversion Nanoparticles in Mice. Nanomedicine (London, U K) 2011;6:1327–1340. doi: 10.2217/nnm.11.56. [DOI] [PubMed] [Google Scholar]
- 517.Gu Z, Yan L, Tian G, Li S, Chai Z, Zhao Y. Recent Advances in Design and Fabrication of Upconversion Nanoparticles and Their Safe Theranostic Applications. Adv Mater. 2013;25:3758–3779. doi: 10.1002/adma.201301197. [DOI] [PubMed] [Google Scholar]
- 518.Zhou JC, Yang ZL, Dong W, Tang RJ, Sun LD, Yan CH. Bioimaging and Toxicity Assessments of near-Infrared Upconversion Luminescent Nayf4:Yb,Tm Nanocrystals. Biomaterials. 2011;32:9059–9067. doi: 10.1016/j.biomaterials.2011.08.038. [DOI] [PubMed] [Google Scholar]
- 519.Dong L, An D, Gong M, Lu Y, Gao HL, Xu YJ, Yu SH. Pegylated Upconverting Luminescent Hollow Nanospheres for Drug Delivery and in Vivo Imaging. Small. 2013;9:3235–3241. doi: 10.1002/smll.201300433. [DOI] [PubMed] [Google Scholar]
- 520.Peng J, Sun Y, Zhao L, Wu Y, Feng W, Gao Y, Li F. Polyphosphoric Acid Capping Radioactive/Upconverting Nalu-f4: Yb,Tm,153sm Nanoparticles for Blood Pool Imaging in Vivo. Biomaterials. 2013;34:9535–9544. doi: 10.1016/j.biomaterials.2013.07.098. [DOI] [PubMed] [Google Scholar]
- 521.Derfus AM, Chan WCW, Bhatia SN. Probing the Cytotoxicity of Semiconductor Quantum Dots. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Tang Y, Han SL, Liu HM, Chen X, Huang L, Li XH, Zhang JX. The Role of Surface Chemistry in Determining in Vivo Biodistribution and Toxicity of Cdse/Zns Core-Shell Quantum Dots. Biomaterials. 2013;34:8741–8755. doi: 10.1016/j.biomaterials.2013.07.087. [DOI] [PubMed] [Google Scholar]
- 523.Galeone A, Vecchio G, Malvindi MA, Brunetti V, Cingolani R, Pompa PP. In Vivo Assessment of Cdse-Zns Quantum Dots: Coating Dependent Bioaccumulation and Genotoxicity. Nanoscale. 2012;4:6401–6407. doi: 10.1039/c2nr31826a. [DOI] [PubMed] [Google Scholar]
- 524.Hauck TS, Anderson RE, Fischer HC, Newbigging S, Chan WC. In Vivo Quantum-Dot Toxicity Assessment. Small. 2010;6:138–144. doi: 10.1002/smll.200900626. [DOI] [PubMed] [Google Scholar]
- 525.Ye L, Yong KT, Liu LW, Roy I, Hu R, Zhu J, Cai HX, Law WC, Liu JW, Wang K, et al. A Pilot Study in Non-Human Primates Shows No Adverse Response to Intravenous Injection of Quantum Dots. Nat Nanotechnol. 2012;7:453–458. doi: 10.1038/nnano.2012.74. [DOI] [PubMed] [Google Scholar]
- 526.Hong G, Robinson JT, Zhang Y, Diao S, Antaris AL, Wang Q, Dai H. In Vivo Fluorescence Imaging with Ag2s Quantum Dots in the Second near-Infrared Region. Angew Chem, Int Ed. 2012;51:9818–9821. doi: 10.1002/anie.201206059. [DOI] [PubMed] [Google Scholar]
- 527.Kim S, Bawendi MG. Oligomeric Ligands for Luminescent and Stable Nanocrystal Quantum Dots. J Am Chem Soc. 2003;125:14652–14653. doi: 10.1021/ja0368094. [DOI] [PubMed] [Google Scholar]
- 528.Ahn KY, Ko HK, Lee BR, Lee EJ, Lee JH, Byun Y, Kwon IC, Kim K, Lee J. Engineered Protein Nanoparticles for in Vivo Tumor Detection. Biomaterials. 2014;35:6422–6429. doi: 10.1016/j.biomaterials.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 529.Ding H, Yong KT, Roy I, Hu R, Wu F, Zhao LL, Law WC, Zhao WW, Ji W, Liu LW. Bioconjugated Plga-4-Arm-Peg Branched Polymeric Nanoparticles as Novel Tumor Targeting Carriers. Nanotechnology. 2011;22:165101. doi: 10.1088/0957-4484/22/16/165101. [DOI] [PubMed] [Google Scholar]
- 530.Miki K, Kimura A, Oride K, Kuramochi Y, Matsuoka H, Harada H, Hiraoka M, Ohe K. High-Contrast Fluorescence Imaging of Tumors in Vivo Using Nanoparticles of Amphiphilic Brush-Like Copolymers Produced by Romp. Angew Chem, Int Ed. 2011;50:6567–6570. doi: 10.1002/anie.201101005. [DOI] [PubMed] [Google Scholar]
- 531.Xiong LQ, Chen ZG, Tian QW, Cao TY, Xu CJ, Li FY. High Contrast Upconversion Luminescence Targeted Imaging in Vivo Using Peptide-Labeled Nanophosphors. Anal Chem. 2009;81:8687–8694. doi: 10.1021/ac901960d. [DOI] [PubMed] [Google Scholar]
- 532.Chen F, Nayak TR, Goel S, Valdovinos HF, Hong H, Theuer CP, Barnhart TE, Cai WB. In Vivo Tumor Vasculature Targeted Pet/Nirf Imaging with Trc105(Fab)-Conjugated, Dual-Labeled Mesoporous Silica Nanoparticles. Mol Pharmaceutics. 2014;11:4007–4014. doi: 10.1021/mp500306k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Cho YS, Yoon TJ, Jang ES, Hong KS, Lee SY, Kim OR, Park C, Kim YJ, Yi GC, Chang K. Cetuximab-Conjugated Magneto-Fluorescent Silica Nanoparticles for in Vivo Colon Cancer Targeting and Imaging. Cancer Lett. 2010;299:63–71. doi: 10.1016/j.canlet.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 534.Kolitz-Domb M, Grinberg I, Corem-Salkmon E, Margel S. Engineering of near Infrared Fluorescent Proteinoid-Poly(L-Lactic Acid) Particles for in Vivo Colon Cancer Detection. J Nanobiotechnol. 2014;12:30. doi: 10.1186/s12951-014-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Liu CY, Gao ZY, Zeng JF, Hou Y, Fang F, Li YL, Qiao RR, Shen L, Lei H, Yang WS, et al. Magnetic/Upconversion Fluorescent Nagdf4:Yb,Er Nanoparticle-Based Dual-Modal Molecular Probes for Imaging Tiny Tumors in Vivo. ACS Nano. 2013;7:7227–7240. doi: 10.1021/nn4030898. [DOI] [PubMed] [Google Scholar]
- 536.Takeda M, Tada H, Higuchi H, Kobayashi Y, Kobayashi M, Sakurai Y, Ishida T, Ohuchi N. In Vivo Single Molecular Imaging and Sentinel Node Navigation by Nanotechnology for Molecular Targeting Drug-Delivery Systems and Tailor-Made Medicine. Breast Cancer-Tokyo. 2008;15:145–152. doi: 10.1007/s12282-008-0037-0. [DOI] [PubMed] [Google Scholar]
- 537.Ding F, Guo S, Xie M, Luo W, Yuan C, Huang W, Zhou Y, Zhang XL, Zhou X. Diagnostic Applications of Gastric Carcinoma Cell Aptamers in Vitro and in Vivo. Talanta. 2015;134:30–36. doi: 10.1016/j.talanta.2014.09.036. [DOI] [PubMed] [Google Scholar]
- 538.Tong R, Coyle VJ, Tang L, Barger AM, Fan TM, Cheng JJ. Polylactide Nanoparticles Containing Stably Incorporated Cyanine Dyes for in Vitro and in Vivo Imaging Applications. Microsc Res Tech. 2010;73:901–909. doi: 10.1002/jemt.20824. [DOI] [PubMed] [Google Scholar]
- 539.Cao J, Xia JF, Deng DW, Gu YQ. Uniform Mesoporous Dye-Doped Silica Nanoparticles as a Multifunctional Drug Carrier for in Vivo Early Tumor Diagnosis. Proc SPIE. 2010;7845:78451R. [Google Scholar]
- 540.Liu JM, Chen JT, Yan XP. Near Infrared Fluorescent Trypsin Stabilized Gold Nanoclusters as Surface Plasmon Enhanced Energy Transfer Biosensor and in Vivo Cancer Imaging Bioprobe. Anal Chem. 2013;85:3238–3245. doi: 10.1021/ac303603f. [DOI] [PubMed] [Google Scholar]
- 541.Ma Y, Sadoqi M, Shao J. Biodistribution of Indocyanine Green-Loaded Nanoparticles with Surface Modifications of Peg and Folic Acid. Int J Pharm. 2012;436:25–31. doi: 10.1016/j.ijpharm.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 542.Rolfe BE, Blakey I, Squires O, Peng H, Boase NRB, Alexander C, Parsons PG, Boyle GM, Whittaker AK, Thurecht KJ. Multimodal Polymer Nanoparticles with Combined F-19 Magnetic Resonance and Optical Detection for Tunable, Targeted, Multimodal Imaging in Vivo. J Am Chem Soc. 2014;136:2413–2419. doi: 10.1021/ja410351h. [DOI] [PubMed] [Google Scholar]
- 543.Xiong LQ, Chen ZG, Yu MX, Li FY, Liu C, Huang CH. Synthesis, Characterization, and in Vivo Targeted Imaging of Amine-Functionalized Rare-Earth up-Converting Nanophosphors. Biomaterials. 2009;30:5592–5600. doi: 10.1016/j.biomaterials.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 544.Saravanakumar G, Min KH, Min DS, Kim AY, Lee CM, Cho YW, Lee SC, Kim K, Jeong SY, Park K, et al. Hydrotropic Oligomer-Conjugated Glycol Chitosan as a Carrier of Paclitaxel: Synthesis, Characterization, and in Vivo Biodistribution. J Controlled Release. 2009;140:210–217. doi: 10.1016/j.jconrel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 545.Chen J, Glaus C, Laforest R, Zhang Q, Yang M, Gidding M, Welch MJ, Xia Y. Gold Nanocages as Photothermal Transducers for Cancer Treatment. Small. 2010;6:811–817. doi: 10.1002/smll.200902216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Cheng L, Yang K, Li Y, Chen J, Wang C, Shao M, Lee ST, Liu Z. Facile Preparation of Multifunctional Upconversion Nanoprobes for Multimodal Imaging and Dual-Targeted Photothermal Therapy. Angew Chem, Int Ed. 2011;50:7385–7390. doi: 10.1002/anie.201101447. [DOI] [PubMed] [Google Scholar]
- 547.Choi WI, Kim JY, Kang C, Byeon CC, Kim YH, Tae G. Tumor Regression in Vivo by Photothermal Therapy Based on Gold-Nanorod-Loaded, Functional Nanocarriers. ACS Nano. 2011;5:1995–2003. doi: 10.1021/nn103047r. [DOI] [PubMed] [Google Scholar]
- 548.Dickerson EB, Dreaden EC, Huang X, El-Sayed IH, Chu H, Pushpanketh S, McDonald JF, El-Sayed MA. Gold Nanorod Assisted near-Infrared Plasmonic Photothermal Therapy (Pptt) of Squamous Cell Carcinoma in Mice. Cancer Lett. 2008;269:57–66. doi: 10.1016/j.canlet.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 549.El-Sayed IH, Huang X, El-Sayed MA. Selective Laser Photo-Thermal Therapy of Epithelial Carcinoma Using Anti-Egfr Antibody Conjugated Gold Nanoparticles. Cancer Lett. 2006;239:129–135. doi: 10.1016/j.canlet.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 550.Gobin AM, Lee MH, Halas NJ, James WD, Drezek RA, West JL. Near-Infrared Resonant Nanoshells for Combined Optical Imaging and Photothermal Cancer Therapy. Nano Lett. 2007;7:1929–1934. doi: 10.1021/nl070610y. [DOI] [PubMed] [Google Scholar]
- 551.Hessel CM, Pattani PV, Rasch M, Panthani MG, Koo B, Tunnell JW, Korgel BA. Copper Selenide Nanocrystals for Photothermal Therapy. Nano Lett. 2011;11:2560–2566. doi: 10.1021/nl201400z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Hirsch LR, Stafford RJ, Bankson JA, Sershen SR, Rivera B, Price RE, Hazle JD, Halas NJ, West JL. Nanoshell-Mediated near-Infrared Thermal Therapy of Tumors under Magnetic Resonance Guidance. Proc Natl Acad Sci U S A. 2003;100:13549–13554. doi: 10.1073/pnas.2232479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Huang X, El-Sayed IH, Qian W, El-Sayed MA. Cancer Cell Imaging and Photothermal Therapy in the near-Infrared Region by Using Gold Nanorods. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 554.Huang XH, El-Sayed IH, Qian W, El-Sayed MA. Cancer Cell Imaging and Photothermal Therapy in the near-Infrared Region by Using Gold Nanorods. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 555.Huang YF, Sefah K, Bamrungsap S, Chang HT, Tan W. Selective Photothermal Therapy for Mixed Cancer Cells Using Aptamer-Conjugated Nanorods. Langmuir. 2008;24:11860–11865. doi: 10.1021/la801969c. [DOI] [PubMed] [Google Scholar]
- 556.Jang B, Park JY, Tung CH, Kim IH, Choi Y. Gold Nanorod-Photosensitizer Complex for near-Infrared Fluorescence Imaging and Photodynamic/Photothermal Therapy in Vivo. ACS Nano. 2011;5:1086–1094. doi: 10.1021/nn102722z. [DOI] [PubMed] [Google Scholar]
- 557.Kim J, Park S, Lee JE, Jin SM, Lee JH, Lee IS, Yang I, Kim JS, Kim SK, Cho MH, et al. Designed Fabrication of Multifunctional Magnetic Gold Nanoshells and Their Application to Magnetic Resonance Imaging and Photothermal Therapy. Angew Chem, Int Ed. 2006;45:7754–7758. doi: 10.1002/anie.200602471. [DOI] [PubMed] [Google Scholar]
- 558.Lal S, Clare SE, Halas NJ. Nanoshell-Enabled Photothermal Cancer Therapy: Impending Clinical Impact. Acc Chem Res. 2008;41:1842–1851. doi: 10.1021/ar800150g. [DOI] [PubMed] [Google Scholar]
- 559.Loo C, Lin A, Hirsch L, Lee MH, Barton J, Halas N, West J, Drezek R. Nanoshell-Enabled Photonics-Based Imaging and Therapy of Cancer. Technol Cancer Res Treat. 2004;3:33–40. doi: 10.1177/153303460400300104. [DOI] [PubMed] [Google Scholar]
- 560.Loo C, Lowery A, Halas N, West J, Drezek R. Immunotargeted Nanoshells for Integrated Cancer Imaging and Therapy. Nano Lett. 2005;5:709–711. doi: 10.1021/nl050127s. [DOI] [PubMed] [Google Scholar]
- 561.Lu W, Singh AK, Khan SA, Senapati D, Yu H, Ray PC. Gold Nano-Popcorn-Based Targeted Diagnosis, Nanotherapy Treatment, and in Situ Monitoring of Photothermal Therapy Response of Prostate Cancer Cells Using Surface-Enhanced Raman Spectroscopy. J Am Chem Soc. 2010;132:18103–18114. doi: 10.1021/ja104924b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Moon HK, Lee SH, Choi HC. In Vivo near-Infrared Mediated Tumor Destruction by Photothermal Effect of Carbon Nanotubes. ACS Nano. 2009;3:3707–3713. doi: 10.1021/nn900904h. [DOI] [PubMed] [Google Scholar]
- 563.Robinson JT, Tabakman SM, Liang Y, Wang H, Sanchez Casalongue H, Vinh D, Dai H. Ultrasmall Reduced Graphene Oxide with High near-Infrared Absorbance for Photothermal Therapy. J Am Chem Soc. 2011;133:6825–6831. doi: 10.1021/ja2010175. [DOI] [PubMed] [Google Scholar]
- 564.Yang K, Hu L, Ma X, Ye S, Cheng L, Shi X, Li C, Li Y, Liu Z. Multimodal Imaging Guided Photothermal Therapy Using Functionalized Graphene Nanosheets Anchored with Magnetic Nanoparticles. Adv Mater. 2012;24:1868–1872. doi: 10.1002/adma.201104964. [DOI] [PubMed] [Google Scholar]
- 565.Yang K, Zhang S, Zhang G, Sun X, Lee ST, Liu Z. Graphene in Mice: Ultrahigh in Vivo Tumor Uptake and Efficient Photothermal Therapy. Nano Lett. 2010;10:3318–3323. doi: 10.1021/nl100996u. [DOI] [PubMed] [Google Scholar]
- 566.Qian J, Wang D, Cai FH, Zhan QQ, Wang YL, He SL. Photosensitizer Encapsulated Organically Modified Silica Nanoparticles for Direct Two-Photon Photodynamic Therapy and in Vivo Functional Imaging. Biomaterials. 2012;33:4851–4860. doi: 10.1016/j.biomaterials.2012.02.053. [DOI] [PubMed] [Google Scholar]
- 567.Singh AK, Hahn MA, Gutwein LG, Rule MC, Knapik JA, Moudgil BM, Grobmyer SR, Brown SC. Multi-Dye Theranostic Nanoparticle Platform for Bioimaging and Cancer Therapy. Int J Nanomed. 2012;7:2739–2750. doi: 10.2147/IJN.S28357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Cheng L, Yang K, Chen Q, Liu Z. Organic Stealth Nanoparticles for Highly Effective in Vivo near-Infrared Photothermal Therapy of Cancer. ACS Nano. 2012;6:5605–5613. doi: 10.1021/nn301539m. [DOI] [PubMed] [Google Scholar]
- 569.Bardhan R, Chen WX, Bartels M, Perez-Torres C, Botero MF, McAninch RW, Contreras A, Schiff R, Pautler RG, Halas NJ, et al. Tracking of Multimodal Therapeutic Nanocomplexes Targeting Breast Cancer in Vivo. Nano Lett. 2010;10:4920–4928. doi: 10.1021/nl102889y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Shi H, Ye XS, He XX, Wang KM, Cui WS, He DG, Li D, Jia XK. Au@Ag/Au Nanoparticles Assembled with Activatable Aptamer Probes as Smart “Nano-Doctors” for Image-Guided Cancer Thermotherapy. Nanoscale. 2014;6:8754–8761. doi: 10.1039/c4nr01927j. [DOI] [PubMed] [Google Scholar]
- 571.Yue CX, Liu P, Zheng MB, Zhao PF, Wang YQ, Ma YF, Cai LT. Ir-780 Dye Loaded Tumor Targeting Theranostic Nanoparticles for Nir Imaging and Photothermal Therapy. Biomaterials. 2013;34:6853–6861. doi: 10.1016/j.biomaterials.2013.05.071. [DOI] [PubMed] [Google Scholar]
- 572.Zheng MB, Zhao PF, Luo ZY, Gong P, Zheng CF, Zhang PF, Yue CX, Gao DY, Ma YF, Cai LT. Robust Icg Theranostic Nanoparticles for Folate Targeted Cancer Imaging and Highly Effective Photothermal Therapy. ACS Appl Mater Interfaces. 2014;6:6709–6716. doi: 10.1021/am5004393. [DOI] [PubMed] [Google Scholar]
- 573.Day ES, Bickford LR, Slater JH, Riggall NS, Drezek RA, West JL. Antibody-Conjugated Gold-Gold Sulfide Nanoparticles as Multifunctional Agents for Imaging and Therapy of Breast Cancer. Int J Nanomed. 2010;5:445–454. doi: 10.2147/ijn.s10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Loo C, Lin A, Hirsch L, Lee MH, Barton J, Halas NJ, West J, Drezek R. Nanoshell-Enabled Photonics-Based Imaging and Therapy of Cancer. Technol Cancer Res Treat. 2004;3:33–40. doi: 10.1177/153303460400300104. [DOI] [PubMed] [Google Scholar]
- 575.Lal S, Clare SE, Halas NJ. Nanoshell-Enabled Photothermal Cancer Therapy: Impending Clinical Impact. Acc Chem Res. 2008;41:1842–1851. doi: 10.1021/ar800150g. [DOI] [PubMed] [Google Scholar]
- 576.Bardhan R, Lal S, Joshi A, Halas NJ. Theranostic Nanoshells: From Probe Design to Imaging and Treatment of Cancer. Acc Chem Res. 2011;44:936–946. doi: 10.1021/ar200023x. [DOI] [PMC free article] [PubMed] [Google Scholar]