Abstract
The multi-domain protein XRCC1 is without catalytic activity, but can interact with a number of known repair proteins. The interaction between the N-terminal domain (NTD) of XRCC1 and DNA polymerase β (pol β) is critical for recruitment of pol β to sites of DNA damage and repair. Crystallographic and NMR approaches have identified oxidized and reduced forms of the XRCC1 NTD, and the corresponding forms of XRCC1 have been identified in cultured mouse fibroblast cells. Both forms of NTD interact with pol β, but the interaction is much stronger with the oxidized form. The potential for formation of the C12-C20 oxidized conformation can be removed by alanine substitution at C12 (C12A) cells expressing C12A XRCC1 (XRE8) with those expressing wild-type XRCC1 (XC5). Reduced C12A XRCC1 is detected at sites of micro-irradiation DNA damage, but provides slower recruitment of pol β. Expression of reduced XRCC1 does not affect sensitivity to MMS or H2O2. In contrast, further oxidative stress imposed by glutathione depletion results in increased sensitization of reduced XRCC1-expressing cells to H2O2 compared with wild-type XRCC1-expressing cells. There is no indication of enhanced H2O2-generated free radicals or DNA strand breaks in XRE8 cells. However, elevated cellular PAR is found following H2O2 exposure, suggesting BER deficiency of H2O2-induced damage in the C12A expressing cells.
Keywords: XRCC1, DNA polymerase β, methyl methanesulfonate, hydrogen peroxide, glutathione depletion, PAR
Introduction
Repair of base lesions in cellular DNA occurs primarily by base excision repair (BER). PARP-1 protein recognizes and binds BER intermediates [1] stimulating synthesis of PAR polymers onto itself and other repair proteins [2], and mediating recruitment of XRCC1. XRCC1 is a multi-domain protein without catalytic activity, but can interact with a number of known repair proteins [3]. The interaction between the N-terminal domain (NTD) of XRCC1 and DNA polymerase β (pol β) is important for recruitment of pol β to sites of BER intermediates [4]. Methyl methanesulfonate (MMS)-induced methylated DNA bases are removed by a damage specific monofunctional glycosylase producing abasic (AP) sites that are cleaved by AP endonuclease 1. Pol β dRP lyase activity is then critical for removing the 5′-deoxyribose (dRP) flap, and the polymerase domain performs gap-filling DNA synthesis leaving a DNA nicked substrate suitable for DNA ligation. Pol β- and XRCC1-deficient mouse fibroblasts are hypersensitive to MMS and this is linked to a repair deficiency as measured by strand breaks [5] and accumulation of PAR [4, 6].
XRCC1 recruitment also occurs following cellular treatment with H2O2 [7]. H2O2 has been widely utilized as a representative oxidizing agent in studies of oxidative stress in mammalian cells leading to an oxidized environment, cellular DNA damage and finally cell death [8]. As an intracellular oxidant, H2O2 can generate highly reactive hydroxyl radicals by the Fe2+-dependent Fenton reaction [9]. H2O2 treatment results in production of free radicals in the iron rich area in close proximity to DNA [10, 11]. It was demonstrated in cultured rat astrocytes that iron chelators significantly protected cells against H2O2 toxicity confirming the importance of endogenous iron and hydroxyl radical formation in H2O2-mediated cytotoxicity [12, 13]. Additionally, the identified modified DNA bases (at least 10 base products) and single-strand breaks are consistent with hydroxyl radical-mediated damage [14]. For example, the interaction of the hydroxyl radical with guanine in DNA generates 8-oxo-7,8-dihydroguanine (8-oxoG) and 2,6-diamino-5-formamido-4-hydroxypyrimidine (FAPy-G) [15]. XRCC1-dependent BER of oxidatively damaged DNA, e.g., 8-oxoG, is frequently mediated by a bifunctional glycosylase/lyase. In the case of the incised AP site generated by OGG1 β-elimination [16], the resulting 3′-dRP group can be removed by APE or Tdp1/PNKP prior to gap filling [17]. Alternatively, NEIL1 glycosylase catalyzes βδ-elimination at AP sites leaving a 3′-phosphate to be removed by PNKP [18].
The reduced glutathione tripeptide (L-γ-glutamyl-L-cysteinylglycine) is an essential cellular small thiol anti-oxidant important for control and regulation of redox-mediated signaling and damage [19–21]. Glutathione can act as a scavenger utilizing its free cysteine sulfhydryl group in reduction and conjugation reactions to inactivate free radicals and reactive species including the hydroxyl radical [22]. Alternate glutathione-dependent cellular mechanisms for direct detoxification of H2O2 may counteract formation of iron-catalyzed free radicals. For example, reduced glutathione is a required cofactor and electron donor for the enzymatic reduction of H2O2 to water catalyzed by the anti-oxidant enzyme glutathione peroxidase, with formation of the oxidized glutathione dimer (GSSG) [13]. Reduced thiol-GSH (98% of glutathione in the cell) is readily regenerated from GSSG by the activity of glutathione reductase [23]. The enzyme catalase catalyzes the decomposition of H2O2 to water by a glutathione independent mechanism. Peroxiredoxins, a small group of thioredoxin peroxidases involved in oxidant defense and redox regulation, can also detoxify H2O2 [20].
Studies of cellular H2O2 exposure have frequently utilized L-buthionine sulfoximine (BSO) and the anti-oxidant N-acetyl cysteine [24]. BSO is known to be a specific inhibitor of γ-glutamylcysteine synthetase, the rate-limiting enzyme critical for synthesis and maintenance of intracellular glutathione [25]. Treatment of cells with BSO was reported to increase ROS levels in H2O2-treated cells [24]. Additionally, BSO has been demonstrated to enhance cytotoxicity of the bifunctional alkylating agent melphalan and the cross-linking agent cisplatin in cellular, animal and patient studies [26, 27]. It is proposed that decreased glutathione-mediated detoxification results in increased cytotoxic DNA damage.
Crystallographic and NMR approaches have identified oxidized and reduced forms of the XRCC1 NTD [28]. Both forms of NTD interact with pol β, but the interaction is much stronger (50-fold) with the oxidized form. The potential for formation of the C12-C20 oxidized conformation can be removed by alanine substitution at C12 (C12A) leading to stabilized reduced XRCC1 with a lower pol β binding affinity. Stable transfection of C12A XRCC1 into Xrcc1−/− cells has been described [4]. In our previous studies, expression of this XRCC1 mutant was fully able to complement the MMS hypersensitivity of Xrcc1−/− cells, but there was lesser complementation of H2O2 sensitivity [4], conditions under which production of the oxidized form of wild-type XRCC1 would be anticipated [4, 28]. In the current study, we further characterize the capability of reduced and oxidized XRCC1 to protect cells from MMS, H2O2 and extreme oxidative stress occurring as a result of BSO-mediated cellular glutathione depletion.
2. Materials and methods
2.1. Isolation of cells complemented with wild-type and the reduced form of XRCC1
Xrcc1+/+ p53−/− and Xrcc1−/− p53−/− (Table 1) mouse embryonic fibroblasts (MEFs) were obtained from Dr. Robert Tebbs [29]. It has not been possible to establish Xrcc1−/− MEFs in a p53-positive background. The cells were maintained at 37 °C in low glucose DMEM (Invitrogen) supplemented with 10% FBS. Mycoplasma testing was performed routinely using a MycoAlert® Mycoplasma detection kit (Lonza, Rockland, ME). All XRCC1 variant cell lines utilized in these studies are p53-negative (Table 1).
Table 1.
Cell lines used in these studies.
| Cell line name | Genotype |
|---|---|
| Xrcc1+/+ | Xrcc1+/+ p53−/− [29] |
| Xrcc1−/− | Xrcc1−/− p53−/− [29] |
| XC5 | Xrcc1−/− p53−/− complemented with wild-type XRCC1* |
| XRE8 | Xrcc1−/− p53−/− complemented with C12A XRCC1 [4] |
this study.
Complementing wild-type XRCC1 was expressed in Xrcc1−/− cells using lentivirus. Lentiviral plasmid constructs for stable expression of mouse XRCC1 were created by PCR amplification of the full length XRCC1 gene (Invitrogen) from the pDONR221 vector [30] with primers that introduced NheI and NotI sites. The amplified XRCC1 was cloned into the pCDH-EF1-MCS-IRES-Puro (System Biosciences, Mountain View, CA) to produce pCDH532-XRCC1, which was sequenced verified. Lentivirus particles were produced by the NIEHS Viral Vector Core and packaged in HEK293T/17 cells (ATCC # CRL-11268) according to published Current Protocols in Neuroscience by P. Salmon and D. Trono. Briefly, 293T cells were transiently transfected with pMD2G, psPAX2 and transfer vector containing pCDH532-XRCC1 using Lipofectamine 2000. Supernatant was collected 48 h after transfection and concentrated by centrifugation at 50,000 × g for 2 h. Pellets were resuspended in phosphate-buffered saline (PBS) and used for infection. All titers were determined by quantitative PCR to measure the number of lentiviral particles that integrated into the host genome. In addition, biological titration of viruses co-expressing fluorescent moieties was determined by flow cytometry. Xrcc1−/− cells were transduced with lentiviral particles at multiplicity of infection (MOI) of 20 per 50,000 cells, and stable cell lines were recovered after puromycin selection (6 μg/ml, Life Technologies). After characterization by western blot analysis, single cell wild-type XRCC1 clones demonstrating an XRCC1 expression level similar to Xrcc1+/+ cells were isolated. One clone (XC5, Table 1) was chosen for further study.
Preparation of C12A mutant XRCC1-expressing cells (XRE8, Table 1) has been described previously [4]. The C12A mutation was introduced by site-directed mutagenesis of XRCC1 cDNA in the pDONR221 vector and subcloned into pEF-DEST51 vector using Gateway technology (Invitrogen). The mammalian cell expression vectors were sequence verified, transfected into Xrcc1−/− cells as described [4], and single cell clones were selected in blasticidin (10 μg/ml), then screened by western blotting. By prevention of the C12-C20 oxidized conformation, this mutant allows expression of stabilized reduced XRCC1 with a lower pol β binding affinity. The only other free cysteine residue in the NTD of XRCC1 is C132. Since C132 is buried and approximately 15Å distant from C20, it is unlikely that a disulfide bond would form between these residues. The possibility that C20 could participate in an aberrant disulfide bond with another protein has not been detected in previous analysis [4], but cannot be ruled out.
2.2. Micro-irradiation and immunofluorescence
Cells were seeded on 35 mm glass bottomed petri dishes containing an etched grid (MatTek, Ashland, MA) at 2 × 105 cells per dish and incubated in growth medium supplemented with 10 μM BrdU for 24 h. After 24 h, DNA single strand breaks (SSBs) and oxidized DNA base lesions were introduced by UV laser micro-irradiation at 355 nm with intensity equivalent to 0.176 μJ as evaluated in previous publications [6, 31]. Samples were imaged using a 40x C-Apochromat NA 1.2 Korr FCS M27 water immersion objective coupled to a Zeiss LSM780 confocal microscope (Carl Zeiss MicroImaging).
Previous studies have established that peak XRCC1 and pol β recruitment occurs 1 min after micro-irradiation in MEFs [6, 31]. In general, cells were fixed in 4% paraformaldehyde 1 min after irradiation. For time course experiments, cells were fixed at the repair time indicated. After fixation, cells were permeabilized with 0.25% Triton X-100 in PBS for 10 min, washed three times in PBS, then further permeabilized in 1% SDS for 5 min at 37 °C, washed six time in PBS, then blocked with 3% BSA in PBS for 60 min. Cells were then incubated with anti-XRCC1 antibody (1:50; Abcam ab1838), anti-PADPR antibody (1:100; Abcam ab14460) and anti-pol β antibody (1:200, Abcam ab26343) for 1 h. Cells were washed three times with PBS, then incubated in Alexa 488 conjugated anti-mouse, Alexa 647 conjugated anti-chicken or Alexa 546 conjugated anti-rabbit (1:2,000; Life Technologies) secondary antibodies for 1 h as appropriate. Cells were again washed three times with PBS. Fluorescence images were acquired with 512 × 512 pixels, bidirectional mode averaging of 2 lines, zoom 1.0 and pixel dwell time 3.5 μs with the same 40x water immersion objective on the Zeiss LSM780 microscope controlled by Zen 2012 SP2 software. Recruitment of XRCC1 and pol β, and synthesis of PAR at the site of DNA damage was analyzed using IMAGEJ as described previously [6]. Each experiment was repeated on at least four cells, and images are representative.
For pol β recruitment studies, XC5 and XRE8 cells were transiently transfected 24 h after plating with human C-terminal TurboGFP-tagged human pol β (RG210765) purchased from Origene (Rockville, MD) using Lipofectamine™ 2000. Transfection medium was supplemented with 10 μM BrdU (Sigma-Aldrich). 24 h after transfection, medium was changed to fresh growth medium without BrdU and DNA SSBs and DNA base lesions were introduced by micro-irradiation with a 355 nm laser at maximum power (60 mW, 100% output) using a one pixel width strip (0.45 μM) per nucleus and an irradiation time of 200 μs. For image acquisition in live-cell experiments with GFP-tagged reporters, a 488 nm laser was used at 1% intensity to minimalize photo bleaching. Fluorescence emission was detected in the range of 491–580 nm using a master gain lower than 800 V for all image acquisition with pinhole of 40 μm. Only cells with similar signal intensities were analyzed and fresh room temperature medium was added prior to irradiation. Three pre-bleach images were acquired to establish a base line prior to damage induction. Images were then acquired at room temperature every 3 s after the bleaching event for 3–10 min. Relative recruitment of GFP-pol β was determined by measuring the signal intensity of GFP at the induced damage site using a manually drawn fixed 2.25 μm width region of interest (ROI). Each experiment was repeated on nineteen cells and analyzed using SigmaPlot (Systat Software Inc.). Time-lapse recruitment curves were corrected by subtracting the post bleach signal intensity of the entire nucleus excluding the ROI. After intensity normalization setting the amplitudes of both curves in the range 0–100%, data were plotted as mean relative recruitment ± SEM and fitted to a single exponential. Statistical significance between the two cell lines was calculated using a Mann-Whitney Rank Sum test.
2.3. Cytotoxicity studies by growth inhibition assay
Cells were seeded (10,000 – 30,000 cells per well in six-well dishes) in medium without selection antibiotic. The next day, they were treated for 1 h with a range of concentrations of MMS (Sigma) or H2O2 (Fisher Scientific). In some studies, cells were pre-treated for 16 h with 15 μM of BSO (Cayman Chemical Company) prior to MMS or H2O2 dosing. After washing as appropriate, growth medium was added. Triplicate wells for each drug concentration were counted by a cell lysis procedure [30, 32] when untreated cells were 80% confluent, and results expressed as % control growth. Fold sensitization was determined at IC90 concentrations, the dose required for 90% decrease in cell growth. For experiments with MEFs, we find this growth inhibition assay to be more reliable and consistent than clongenic or short-term cytotoxicity assays. Results obtained are in agreement with alternate assay methods [30].
2.4. Measurement of total cellular glutathione
Cellular glutathione was assayed using a GSH-Glo kit (Promega) as outlined by the manufacturer. Cells (20,000) were seeded in triplicate in a white clear-bottom 96-well plate. The following day, they were treated with MMS (0.15 or 1 mM) or H2O2 (350 or 500 μM) for 1 h. In some studies, cellular glutathione was significantly depleted by treatment of cells for 16 h with 15 μM BSO. Cells were then washed, and incubated with control medium for 3 h. A 1x solution of GSH-Glo Reagent was prepared by adding 100 μl of Luciferin-NT substrate, 100 μl of glutathione-S-transferase, and 100 μl of 100 mM TCEP (final concentration of 1 mM) to 10 ml of GSH-Glo Reaction Buffer. Medium was removed from the cells and GSH-Glo reagent (100 μl) was added to each well and incubated at room temp for 30 min. Reconstituted Luciferin Detection Reagent (100 μl) was then added to each well and incubated at room temperature for 15 min. Luminescence was measured using a Tecan Infinite M100 Pro microplate reader. To determine background levels, 10 μl of H2O was added to an empty well. A standard curve was used to calculate total glutathione, subtracting out the background reading from all sample readings.
2.5. Detection of cellular oxidative stress
A CellROX® Green Flow Cytometry kit (Molecular Probes) was used for oxidative stress detection according to the manufacturer’s directions. Once oxidized, the CellROX® Green reagent binds to DNA, thus signal is localized primarily in the nucleus and mitochondria. XC5 and XRE8 cells were seeded in 100 mm dishes at a density of 1 × 106 and were pretreated with 15 μM BSO later the same day. After 16 h, cells were treated with 500 μM H2O2 for 1 h. Cells were washed then incubated with control medium until harvesting 3 h later. After trypsinization, cells were centrifuged, washed with PBS, and the pellet obtained was prepared for flow cytometry. Cells were stained with 500 nM of CellROX Green Detection Reagent for 15 min at 37°C protected from light followed by 5 nM SYTOX Red Dead Cell Stain for 15 min at 37°C. Cytometric analysis utilized 488 nm excitation for CellROX Green and 639 nm excitation for SYTOX red stain. Viable cells were gated to exclude dead cells using an LSRII flow cytometer (BD Biosciences).
2.6. Flow cytometric detection of γ-H2A.X
Appearance of phosphorylated histone H2A.X (γ-H2A.X) is used as a biomarker of response to DNA damage that may include formation of double strand breaks [33]. Flow cytometric analysis of cellular γ-H2A.X has been fully described previously [34]. XC5 and XRE8 cells (1×106) were seeded in 100 mm dishes, allowed to attach, then treated with 15 μM BSO for 16 h. The following day, cells were dosed with 500 μM H2O2 for 1 h, washed once, and then incubated a further 3 h. Cells were isolated by trypsinization, centrifuged, and washed with PBS. The cell pellet was prepared for flow cytometry using the H2A.X Phosphorylation Assay kit (EMD Millipore) and samples were read on an LSR II (BD Biosciences) and analyzed using FACSDiva software (BD Biosciences).
2.7. Determination of total cellular PAR
Cellular PAR levels were quantified using the PARP in vivo Pharmacodynamic Assay 2nd Generation (PDA II) kit (4520-096- K, Trevigen) following the kit protocol as described previously [4]. Briefly, XC5 and XRE8 cells were seeded in 60 mm dishes at 106 cells/dish and treated the next day with 0.5 to 2 mM H2O2 at 4 °C in growth medium supplemented with 25 mM HEPES. After 20 min exposure, they were washed in ice-cold HBSS and fresh, warm growth medium was added. Dishes were incubated for 10 min at 37 °C in a 10% CO2 incubator, then immediately placed on ice and lysed according to the kit protocol. Control cells were treated similarly. After cell lysis and DNA digestion, total protein amounts were determined for each sample using a Pierce BCA Protein Assay kit and then 3 μg of XC5 and XRE8 cell extract were added to pre-coated capture antibody plates and incubated overnight at 4 °C. The following morning, wells were washed four times in PBS with Tween-20 (PBST), and then a 1:250 dilution of PAR polyclonal detecting antibody was added and incubated at room temperature for 2 h. Wells were again washed four times in PBST, then a 1:250 dilution of goat anti-rabbit IgG-HRP was added and incubated for 1 h. Cells were washed again four times in PBST, then a 1:1 mixture of PARP PeroxyGlow™ A and B was added and luminescence was measured with an Infinite M100 Pro microplate reader (Tecan).
3. Results
3.1. Immunofluorescent imaging of repair proteins in XC5 and XRE8 cells
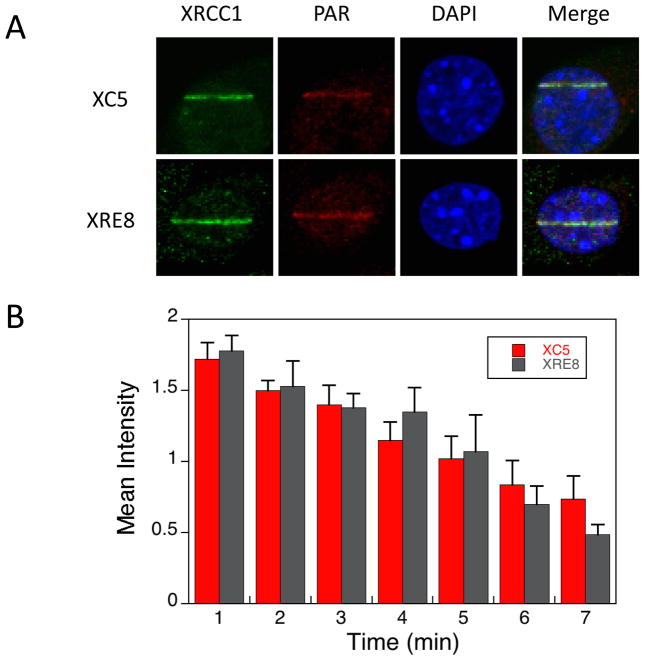
Recruitment of XRCC1 and synthesis of PAR at the site of micro-irradiation DNA damage in mouse fibroblasts were analyzed by immunofluorescence. Representative results with XC5 (expressing both reduced and oxidized forms of wild-type XRCC1), and XRE8 cells (expressing the reduced form of XRCC1), are shown in Figure 1A. These images reveal that at 1 min after damage, there was little difference in PAR synthesis and XRCC1 recruitment (also observed in Fig. 1B) between the two cell lines. The results suggest that both wild-type and C12A XRCC1 are rapidly recruited to the sites of DNA damage. A time course experiment using immunofluorescent XRCC1 staining every minute confirmed similar recruitment and disassociation of XRCC1 in XC5 and XRE8 cells after micro-irradiation damage (Fig. 1B).
Fig. 1.
Immunofluorescent imaging of XRCC1 and PAR in XC5 and XRE8 cells. A) Cells were micro-irradiated in stripes to initiate XRCC1 recruitment and PAR synthesis and were fixed after 1 min repair time. Immunofluorescent staining methods (anti-XRCC1 antibody with Alexa 488 conjugated anti-mouse secondary antibody and anti-PAR antibody with Alexa 647 conjugated anti-chicken secondary antibody) are outlined in Section 2.2. Representative cells are shown. B) Time course of recruitment to damaged sites of endogenous XRCC1 in wild-type complemented XC5 cells and C12A reduced XRCC1 in XRE8 cells after fixing cells at the repair times specified and staining as outlined above. At least 5 cells were analyzed at each time point, error bars represent SEM.
3.2. Recruitment of pol β after laser irradiation
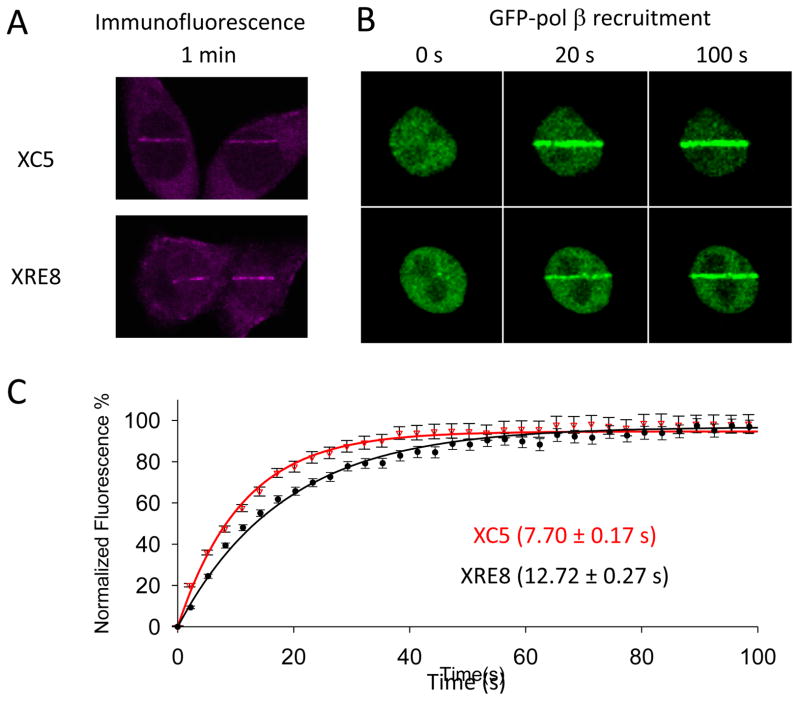
Immunofluorescent imaging of endogenous pol β in XC5 and XRE8 cells revealed similar levels of recruitment at 1 min after micro-irradiation (Fig. 2A). Pol β recruitment was further examined using transiently transfected GFP-pol β. Representative cells after 20 and 100 s recruitment of GFP-pol β are shown in Figure 2B. Next, relative recruitment of GFP-pol β was determined by measuring the signal intensity of GFP at the induced damage site every 3 s using a defined region of interest (ROI). Subtracting background and the signal intensity of the entire nucleus (excluding the damage line ROI) corrects the intensity measurements for acquiring a time-lapse curve. Curves were normalized using intensity taken at the beginning of recruitment and maximal intensity values. The results are shown in Figure 2C where recruitment data are fitted to a single exponential. The maximal signal amplitude was not significantly different between the two cell lines, Imax of 4.8 ± 0.4 and 4.29 ± 0.3 in XC5 and XRE8, respectively. The half-time of pol β recruitment to DNA damage in XC5 cells, 7.70 ± 0.17 s (n=19) was statistically different from the half-time in XRE8 cells of 12.72 ± 0.27 s (n=19) (p<0.001, Mann-Whitney Rank Sum Test). Since pol β recruitment is in part dependent on binding to XRCC1, these results indicate faster initial pol β recruitment to wild-type XRCC1, present as both oxidized and reduced forms [4], than to the reduced form of XRCC1. It is likely that the recruitment curve represents the combined binding and disassociation of pol β and XRCC1. The oxidized form of XRCC1 has a significantly higher affinity for pol β [28, 35], resulting in slower pol β disassociation and the more rapid overall recruitment observed.
Fig. 2.
Pol β recruitment to UV micro-irradiation damage sites. (A) XC5 and XRE8 cells were irradiated in stripes and endogenous pol β recruitment was analyzed by immunofluorescence (anti-pol β antibody with Alexa 546 conjugated ant-rabbit secondary antibody) after 1 min. (B) Recruitment of transiently expressed GFP-pol β (see section 2.2.) was followed for at least 3 min after DNA damage. Images of representative cells after recruitment for 20 and 100 s are shown. (C) Plot of time course (100 s) of recruitment of GFP-pol β in XC5 and XRE8 cells. Experiments and data analysis are described in Section 2.2. Fluorescence data were normalized using intensity at the beginning of recruitment and maximal intensity values and fitted to a single exponential. Nineteen cells were analyzed in each case, and error bars represent SEM. The half-time of pol β recruitment in XC5 cells (7.70 ± 0.17 s) was statistically different (p<0.001) to the that in XRE8 cells (12.72 ± 0.27 s).
3.3. Effect of BSO pre-treatment on cellular MMS sensitivity
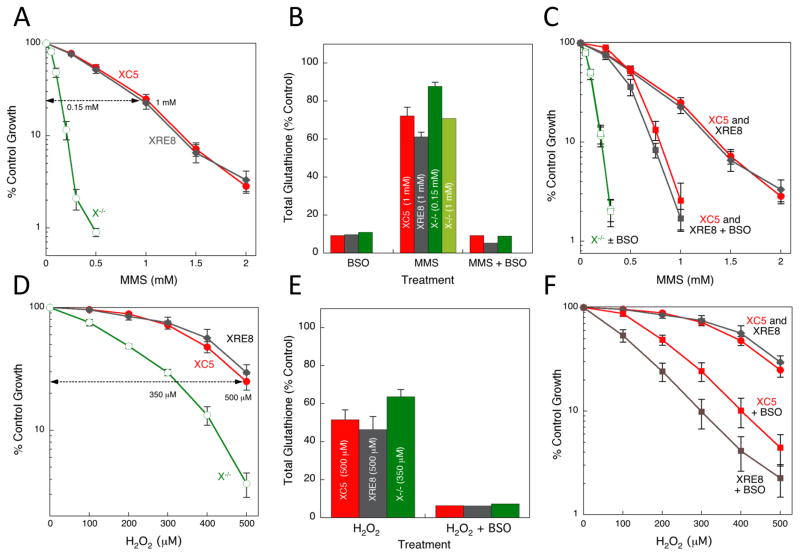
The sizeable hypersensitivity of Xrcc1−/− cells after a 1 h exposure to the DNA methylating agent MMS (Fig. 3A) has been demonstrated previously [4, 5]. Additionally, similar MMS resistance (Fig. 3A) was documented in wild-type XRCC1-expressing cells (XC5) and cells expressing C12A disulfide-blocked reduced XRCC1 (XRE8) [4]. Here, we examined glutathione levels in these cells and found that a 16 h treatment of XC5, XRE8 and Xrcc1−/− cells with 15 μM BSO was relatively non-toxic, yet resulted in approximately 90% depletion of total cellular glutathione (Fig. 3B). A 1 h treatment with 1 mM MMS, and analysis after 4 h revealed 30–40% depletion of cellular glutathione in all 3 cell types (Fig. 3B). For comparison, Xrcc1−/− cells were also treated with a lower (0.15 mM) equitoxic dose of MMS (see Fig. 3A) and a lesser 10% glutathione depletion was observed (Fig. 3B).
Fig. 3.
MMS and H2O2 sensitivity, and glutathione depletion in XRCC1 cells. (A) XC5, XRE8 and Xrcc1−/− cells were treated with a range of concentrations of MMS for 1 h as described in Section 2.3. Sensitivity was measured by a growth inhibition assay. Plotted are mean ± SEM values obtained from at least 3 independent experiments. (B) Total cellular glutathione was measured using a GSH-Glo kit as outlined by the manufacturer (see Section 2.4.). Cell lines as in (A) were pre-treated with BSO (15 μM for 16 h), with MMS at the equitoxic concentrations indicated for 1 h, or a combination of BSO pre-treatment (for 16 h) followed by MMS. Glutathione was analyzed 3 h after the MMS exposure. (C) Cells were pre-treated with BSO (15 μM for 16 h) followed by a 1 h exposure to MMS as described in Section 2.3. Plotted are mean ± SEM values obtained from at least 3 independent experiments. (D) XC5, XRE8 and Xrcc1−/− cells were treated with a range of concentrations of H2O2 for 1 h. Plotted are mean ± SEM values obtained from at least 3 independent experiments. (E) Total cellular glutathione was measured in the same cell lines after treatment with H2O2 at the equitoxic concentrations indicated for 1 h, or a combination of BSO pre-treatment (15 μM for 16 h), followed by H2O2. Glutathione was analyzed 3 h after the H2O2 exposure. (F) XC5 and XRE8 cells were pre-treated with BSO (15 μM for 16 h) followed by a 1 h exposure to H2O2. Plotted are mean ± SEM values obtained from at least 3 independent experiments.
The substantial MMS-induced depletion of glutathione observed at survival study relevant concentrations in XC5 and XRE8 cells may be regarded as a marker of oxidative stress and suggests the possibility of an oxidative mechanism of MMS-induced damage in these cell lines. A highly interconnected MMS survival network has been described in mouse cells where MMS caused a dose-dependent increase in the oxidatively generated DNA lesion 8-oxoG implying that MMS results in formation of both methylated and oxidized base lesions [36]. It should be noted that a direct MMS detoxification mechanism by thiols could also result in the observed cellular glutathione depletion. MMS rapidly methylates glutathione in vitro yielding S-methyl glutathione and providing an alternate methylation target [37]. MMS-mediated glutathione depletion has been documented previously in cultured hepatocytes, and, in these studies MMS toxicity was proposed to be a result of increased oxidative damage (lipid peroxidation) in addition to methylation of DNA [38, 39].
Cellular treatment with BSO followed by MMS produced a similar glutathione depletion as treatment with BSO alone (Fig. 3B) and resulted in approximately 1.8-fold sensitization to MMS in both XC5 and XRE8 cells at the IC90 dose (Fig. 3C). Similar sensitization to MMS following BSO-induced glutathione depletion serves as an indicator of similar levels of cytotoxic oxidative intermediates in both the XRCC1-expressing cell lines. The results suggest formation of MMS-induced oxidatively damaged DNA, in addition to the expected DNA methylation. Alternatively, glutathione depletion by BSO may preclude direct detoxification mechanisms also resulting in increased MMS sensitivity. There was no evidence for BSO-mediated sensitization in Xrcc1−/− cells (Fig. 3C). The XRCC1-dependence of the BSO-mediated sensitization to MMS in cells where there was known glutathione depletion (Fig. 3B) points to the absence of repair in Xrcc1−/− cells.
3.4. Cellular H2O2 sensitivity and effect of BSO pre-treatment
XRE8 cells have similar H2O2 sensitivity to the wild-type expressing XC5 cells, and Xrcc1−/− cells demonstrate hypersensitivity to H2O2 treatment (Fig. 3D). There was lesser hypersensitivity to H2O2 than to MMS in Xrcc1−/− cells (compare Fig. 3A and 3D), suggesting substantial protection in the absence of XRCC1-dependent repair. Treatment of cells for 16 h with equitoxic doses of H2O2 (500 μM for XC5 and XRE8, and 350 μM for Xrcc1−/−) resulted in 40–50% depletion of total cellular glutathione (Fig. 3E). This significant glutathione depletion is consistent with glutathione-dependent cellular mechanisms for both free radical inactivation and direct detoxification of H2O2. Pre-treatment with BSO followed by H2O2 treatment resulted in strong and similar glutathione depletion in all three cell types (Fig. 3E).
It has been suggested that extremely high cellular glutathione levels would be required to protect DNA against the damaging effects of H2O2-induced hydroxyl radicals that react at the site where they are formed [40]. However, depletion of glutathione by BSO did result in sensitization of both XC5 and XRE8 cells to H2O2 (Fig. 3F). One possibility is that H2O2 sensitivity would be increased under depleted conditions where glutathione is no longer sufficient for direct oxidant inactivation. However, XRE8 cells expressing the reduced form of XRCC1 were more highly sensitized to H2O2 by BSO than XC5 cells expressing the wild-type protein (Fig. 3F) despite similar control glutathione levels and glutathione depletion by H2O2 and BSO in both cell types (Fig. 3E). These data suggest that production of the oxidized form of XRCC1, known to occur as a result of H2O2 exposure [28], allows for enhanced repair of oxidatively damaged DNA mediated by H2O2 exposure under conditions of glutathione depletion.
3.5. Analysis of cellular oxidative stress induced by H2O2
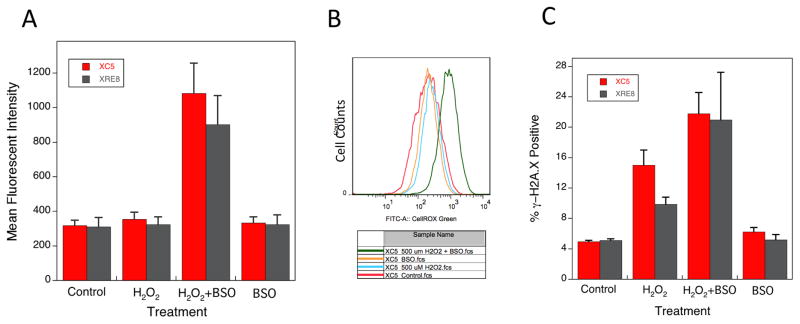
Oxidative stress in cells was detected using flow cytometry and the CellROX® Green reagent that in its oxidized state binds to DNA. Thus, the fluorescent signal is localized primarily in the nucleus and mitochondria and is expressed as mean fluorescent intensity (Fig. 4A). Cells were treated for 1 h with H2O2, with or without pre-treatment with BSO, and the signal was measured after 4 h. The fluorescent signals in control, H2O2- and BSO-treated XC5 and XRE8 cells were similar. Thus, hydroxyl radicals localized in close proximity to DNA as a result of 500 μM H2O2 treatment alone were not detectable beyond control values, most likely because of their rapid interaction with DNA. Yet, a large increase in signal was observed after co-treatment with H2O2 and BSO, possibly the result of the lesser availability of glutathione for oxidant inactivation, but there was no observable difference between XC5 and XRE8 cells (Fig. 4A). An alternate method of data presentation also revealed a strong increase in staining when XC5 cells were co-treated with H2O2 and BSO (Fig. 4B), and similar data were generated with XRE8 cells (not shown). Therefore, the hypersensitivity of XRE8 cells to the H2O2 and BSO combination (Fig. 3F) could not be attributed to a detectable difference in H2O2-derived cellular hydroxyl radicals. Reduced XRCC1 is unlikely to enhance formation of hydroxyl radicals, but could adversely affect the repair of cytotoxic oxidatively generated DNA damage.
Fig. 4.
Cellular oxidative stress and DNA damage as a result of H2O2 treatment. Oxidative stress was analyzed using flow cytometry and CellROX® Green reagent that binds to DNA upon oxidation as described in Section 2.5. (A) Mean fluorescent intensity in XC5 and XRE8 cells treated with H2O2 (500 μM for 1 h), BSO (15 μM pre-treatment for 16 h) or a combination of the two agents. Cells were analyzed 3 h after H2O2 dosing. Plotted are values from at least 3 experiments ± SEM. (B) Histogram overlay showing CellRox® Green fluorescence intensity (FITC-A) of XC5 cells after the same treatments as described in (A). (C) Phosphorylation of histone H2A.X, an indicator of DNA strand breaks, was analyzed using flow cytometry and the EMD Millipore H2A.X Phosphorylation Assay kit as described in Section 2.6. Cells treated with H2O2 (500 μM for 1 h), BSO (15 μM pre-treatment for 16 h) or a combination of the two agents. Cells were analyzed 3 h after H2O2 dosing. Plotted are % γ-H2A.X positive cells after cell treatments as indicated.
3.6. Analysis of histone H2A.X phosphorylation following treatment with H2O2
Phosphorylation of histone H2A.X (γ-H2A.X) is an indicator of response to DNA damage with formation of DNA strand breaks. The level of γ-H2A.X in control, untreated cells was set to a value of 5% (Fig. 4C). Treatment of cells for 1 h with H2O2 resulted in increased formation of phosphorylated H2A.X after the 3 h repair incubation used above. Co-treatment with H2O2 and BSO further increased formation of γ-H2A.X in both cell lines, but formation was no higher in the XRE8 cells. Thus, the hypersensitivity of XRE8 cells when treated with H2O2 and BSO (Fig. 3F) could not be ascribed to an increase in the level of DNA strand breaks derived from repair of damaged DNA.
3.7. Total cellular PAR following treatment with H2O2
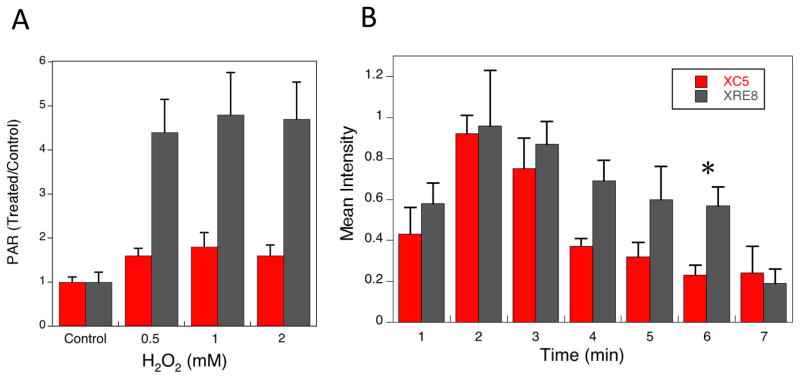
Elevation of cellular PAR has been proposed as an indicator of BER deficiency [6]. Total cellular PAR was measured in cells treated for 20 min with 0.5 – 2 mM H2O2 in cold medium. Warm medium was added after washing and cells were allowed to repair for 10 min at 37 °C. A significant increase in PAR relative to untreated controls was identified in XRE8 cells at all H2O2 doses analyzed (Fig. 5A), but a lesser increase in PAR was found in XC5 cells. A BER deficiency in XRE8 cells is consistent with the observed cellular hypersensitivity to H2O2 following glutathione depletion (Fig. 3F).
Fig. 5.
Analysis of cellular PAR after H2O2 exposure and following micro-irradiation. (A) Total cellular PAR levels were analyzed using the Trevigen PARP in vivo Pharmacodynamic Assay kit as described in Section 2.7. Cells were treated for 20 min at 4 °C with 0.5 – 2 mM H2O2 and allowed to repair for 10 min at 37 °C prior to analysis. Plotted are PAR values in H2O2-treated cells relative to untreated control cells of the same cell type, mean values from 3 experiments ± SEM. The control PAR values are 677 ± 56 and 337 ± 54 pg/ml in XC5 and XRE8, respectively. In XRE8, PAR values at all H2O2 concentrations were significantly higher than in control cells, whereas in XC5 there was no significant difference between control and treated (P<0.02). (B) Time course of PAR synthesis in XC5 cells (wild-type XRCC1) and in XRE8 cells (C12A XRCC1) at UV micro-irradiation damaged sites at the repair times specified (1–7 min). Representative images of PAR immunostaining at 1 min can be found in Figure 1A. Anti-PAR antibody with Alexa 647 conjugated anti-chicken secondary antibody were used for immunofluorescence analysis as described in section 2.2. At least 5 cells were analyzed at each time point, error bars represent SEM. The asterisk denotes statistical significance (P<0.05) between the two cell types at 6 min.
3.8. Time course of PAR synthesis
In view of the higher levels of PAR observed in H2O2-treated XRE8 cells (Fig. 5A), the time course of PAR synthesis after UV micro-irradiation damage (DNA single strand breaks and oxidized DNA base lesions) was analyzed by immunofluorescence and compared between the cell lines. It should be noted that the observed amount of PAR will be the net result of PAR synthesis by PARP and degradation of PAR chains by poly(ADP-ribose) glycohydrolase (PARG) [41]. The level of PAR in XC5 and XRE8 cells at 1 min after irradiation and at the peak level of 2 min were similar (Fig. 5B). After 2 min, PAR levels declined at a faster rate in XC5 than XRE8 cells. The difference between the two cell lines was statistically significant (P<0.05) at the 6 min time point (Fig. 5B). Representative images of PAR immunostaining throughout the time course can be found in Supplementary Figure 1A. These results are consistent with the elevated PAR data obtained with H2O2-treated XRE8 cells (Fig. 5A).
4. Discussion
Here, we describe further characterization of the phenotypes of Xrcc1−/− cells that express either wild-type XRCC1 (XC5 cells) or C12A XRCC1 locked in its reduced conformation (XRE8 cells) [4]. Both oxidized and reduced forms of the XRCC1 NTD interact with pol β, but the interaction is much stronger with the oxidized form [28, 35]. In wild-type mouse fibroblasts in culture, the proportion of reduced and oxidized forms of XRCC1 is similar [4]. Reduced XRCC1 protein is successfully recruited to sites of UV micro-irradiation DNA damage (Fig. 1B), but allows for a slower accumulation of pol β (Fig. 2C). MMS is a directly-acting DNA methylator and the MMS hypersensitivity observed in the absence of either pol β or XRCC1 has been ascribed to a deficiency in BER [5]. Here we found similar MMS sensitivities for XRE8 and XC5 cells despite the difference in XRCC1 and pol β binding affinity (Fig. 3A). In a previous study, the XRE8 cells were less sensitized to MMS by a PARP inhibitor than Xrcc1+/+ cells. Thus, these earlier results pointed to some type of alteration in the cellular repair response to DNA damage.
Expression of either wild-type or reduced XRCC1 results in similar sensitivity to H2O2 (Fig 3D). However, glutathione depletion by BSO results in a greater sensitization of XRE8 cells than XC5 cells to H2O2 (Fig. 3F). Significantly elevated cellular PAR detected in XRE8 cells following H2O2 exposure, suggests a BER deficiency of H2O2-induced damage in this cell line (Fig. 5A). Consistent with the elevated H2O2-induced PAR level in XRE8 cells, we find a more rapid decline in PAR levels following micro-irradiation damage in XC5 cells than in XRE8 cells (Fig. 5B and S1). Overall, these results indicate a role for the oxidized form of XRCC1 for maximal protection of cells against H2O2-induced cytotoxicity.
MMS-induced depletion of glutathione observed in XC5 and XRE8 cells may be regarded as a marker of oxidative stress and suggests the possibility of an oxidative mechanism of MMS-induced damage in these cell lines. However, direct MMS detoxification by thiols could also result in cellular glutathione depletion. Sensitization to MMS following BSO-induced glutathione depletion similarly suggests formation of MMS-induced oxidatively damaged DNA, in addition to the expected DNA methylation. Alternatively, glutathione depletion by BSO may preclude direct detoxification mechanisms and result in increased MMS sensitivity. The XRCC1-dependence of the BSO-mediated sensitization to MMS (Fig. 3C) in cells where there was known glutathione depletion (Fig. 3B) indicates the absence of repair in Xrcc1−/− cells. Taken together, the data obtained following MMS exposure supports the hypothesis of increased formation of cytotoxic oxidatively generated DNA damage under low glutathione conditions with DNA repair by an XRCC1-dependent mechanism.
Glutathione is believed to have a low ability to efficiently scavenge the highly reactive hydroxyl radical. It has been proposed that extremely high cellular glutathione levels would be required to protect DNA against the damaging effects of H2O2-induced hydroxyl radicals that react at the site where they are formed [40]. Yet, depletion of glutathione by BSO did result in H2O2 sensitization of both XC5 and XRE8 cells. Enhanced H2O2 cytotoxicity could also result under conditions where glutathione is no longer sufficient for direct oxidant inactivation. XRE8 cells were more highly sensitized to H2O2 by BSO than XC5 cells (Fig. 3F) despite similar control glutathione levels and glutathione depletion by H2O2 and BSO in both cell types (Fig. 3E). These results support the idea that formation of the oxidized conformation of XRCC1, known to occur as a result of H2O2 exposure [28], allows for enhanced XRCC1-dependent BER of oxidatively damaged DNA mediated by H2O2 exposure under conditions of glutathione depletion. The elevated PAR in H2O2-treated XRE8 cells (Fig. 5A) is consistent with a BER deficiency in XRE8 cells and with the observed cellular hypersensitivity to H2O2 following glutathione depletion (Fig. 3F).
Concluding Statement
In summary, in previously published experiments, the level of total cellular PAR after MMS treatment was higher in reduced XRCC1-expressing XRE8 cells. We proposed a disruption of PARP-1, XRCC1 and pol β interactions during repair [4]. The results of the present study are consistent with this proposal. Taken together, these results illustrate that the redox conformational transition in the N-terminal domain of XRCC1 can influence the repair activity of XRCC1. Explanation of the differential survival phenotypes after MMS versus H2O2 treatment may be related to the distinct BER requirements for alkylated base repair and oxidized base repair, respectively. In the latter case, gap trimming of the BER intermediate requires Tdp1 and PNKP, and decreased stability of the C12A XRCC1-pol β complex may alter activities of these enzymes.
Supplementary Material
Immunofluorescent imaging of PAR in XC5 and XRE8 cells. Cells were micro-irradiated in stripes to initiate PAR synthesis and were fixed after 1–7 min repair time. Immunofluorescent staining methods (anti-PAR antibody with Alexa 647 conjugated anti-chicken secondary antibody) are outlined in Section 2.2. Representative cells are shown for each time point. The mean data in this experiment are presented as a bar graph in Figure 5B.
Highlights.
Reduced XRCC1 recruits to DNA damage, initial recruitment of pol β is slower.
Enhanced H2O2 sensitivity of C12A-expressing XRE8 cells with glutathione depletion.
No indication of enhanced H2O2-generated free radicals or DNA breaks in XRE8 cells.
PAR elevated following H2O2 exposure, suggests BER deficiency in XRE8 cells.
Acknowledgments
The authors thank the NIEHS Protein Expression Core Facility for assistance with expression vector construction, C. Jeff Tucker and Erica Scappini at the NIEHS Fluorescence Microscopy and Imaging Center for their expert assistance with micro-irradiation studies, Negin Martin in the Viral Vector Core for assistance with the lentivirus expression vector and the NIEHS Flow Cytometry Center for help with flow cytometric analysis. We appreciate guidance on figure preparation and statistical analysis by Dr. William Beard.
Grant Support
This work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (project number Z01 ES050158 and ES050159).
Footnotes
Conflict of Interest Statement:
The authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lavrik OI, Prasad R, Sobol RW, Horton JK, Ackerman EJ, Wilson SH. Photoaffinity labeling of mouse fibroblast enzymes by a base excision repair intermediate. Evidence for the role of poly(ADP-ribose) polymerase-1 in DNA repair. J Biol Chem. 2001;276:25541–25548. doi: 10.1074/jbc.M102125200. [DOI] [PubMed] [Google Scholar]
- 2.Amé JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 3.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst) 2003:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 4.Horton JK, Stefanick DF, Gassman NR, Williams JG, Gabel SA, Cuneo MJ, Prasad R, Kedar PS, DeRose EF, Hou EW, London RE, Wilson SH. Preventing oxidation of cellular XRCC1 affects PARP-mediated DNA damage responses. DNA Repair (Amst) 2013;12:774–785. doi: 10.1016/j.dnarep.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase β in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gassman NR, Stefanick DF, Kedar PS, Horton JK, Wilson SH. Hyperactivation of PARP triggers nonhomologous end-joining in repair-deficient mouse fibroblasts. PLoS ONE. 2012;7:e49301. doi: 10.1371/journal.pone.0049301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng CF, Schafer FQ, Buettner GR, Rodgers VG. The rate of cellular hydrogen peroxide removal shows dependency on GSH: mathematical insight into in vivo H2O2 and GPx concentrations. Free Radic Res. 2007;41:1201–1211. doi: 10.1080/10715760701625075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Bahr SM. Biochemistry of free radicals and oxidative stress. Science Internat. 2013;1:111–117. [Google Scholar]
- 10.Halliwell B, Gutteridge JMC. Methods Enzymol. Academic Press; 1990. Role of free radicals and catalytic metal ions in human disease: An overview; pp. 1–85. [DOI] [PubMed] [Google Scholar]
- 11.Jaruga P, Dizdaroglu M. Repair of products of oxidative DNA base damage in human cells. Nucleic Acids Res. 1996;24:1389–1394. doi: 10.1093/nar/24.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liddell JR, Robinson SR, Dringen R. Endogenous glutathione and catalase protect cultured rat astrocytes from the iron-mediated toxicity of hydrogen peroxide. Neurosci Lett. 2004;364:164–167. doi: 10.1016/j.neulet.2004.04.042. [DOI] [PubMed] [Google Scholar]
- 13.Liddell JR, Hoepken HH, Crack PJ, Robinson SR, Dringen R. Glutathione peroxidase 1 and glutathione are required to protect mouse astrocytes from iron-mediated hydrogen peroxide toxicity. J Neurosci Res. 2006;84:578–586. doi: 10.1002/jnr.20957. [DOI] [PubMed] [Google Scholar]
- 14.Dizdaroglu M, Nackerdien Z, Chao BC, Gajewski E, Rao G. Chemical nature of in vivo DNA base damage in hydrogen peroxide-treated mammalian cells. Arch Biochem Biophys. 1991;285:388–390. doi: 10.1016/0003-9861(91)90378-v. [DOI] [PubMed] [Google Scholar]
- 15.Kawanishi S, Hiraku Y, Oikawa S. Mechanism of guanine-specific DNA damage by oxidative stress and its role in carcinogenesis and aging. Mutat Res. 2001;488:65–76. doi: 10.1016/s1383-5742(00)00059-4. [DOI] [PubMed] [Google Scholar]
- 16.Zharkov DO, Rosenquist TA, Gerchman SE, Grollman AP. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J Biol Chem. 2000;275:28607–28617. doi: 10.1074/jbc.M002441200. [DOI] [PubMed] [Google Scholar]
- 17.Izumi T, Hazra TK, Boldogh I, Tomkinson AE, Park MS, Ikeda S, Mitra S. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000;21:1329–1334. [PubMed] [Google Scholar]
- 18.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 20.Poole LB, Hall A, Nelson KJ. Overview of peroxiredoxins in oxidant defense and redox regulation. Curr Protoc Toxicol. 2011;49:7.9.1–7.9.15. doi: 10.1002/0471140856.tx0709s49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quintana-Cabrera R, Fernandez-Fernandez S, Bobo-Jimenez V, Escobar J, Sastre J, Almeida A, Bolanos JP. γ-Glutamylcysteine detoxifies reactive oxygen species by acting as glutathione peroxidase-1 cofactor. Nat Commun. 2012;3 doi: 10.1038/ncomms1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cacciatore I, Cornacchia C, Pinnen F, Mollica A, Di Stefano A. Prodrug approach for increasing cellular glutathione levels. Molecules. 2010;15:1242–1264. doi: 10.3390/molecules15031242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rushworth GF, Megson IL. Existing and potential therapeutic uses for N-acetylcysteine: the need for conversion to intracellular glutathione for antioxidant benefits. Pharmacol Ther. 2014;141:150–159. doi: 10.1016/j.pharmthera.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Park WH. The effect of MAPK inhibitors and ROS modulators on cell growth and death of H2O2-treated HeLa cells. Mol Med Rep. 2013;8:557–564. doi: 10.3892/mmr.2013.1551. [DOI] [PubMed] [Google Scholar]
- 25.Griffith OW. Mechanism of action, metabolism, and toxicity of buthionine sulfoximine and its higher homologs, potent inhibitors of glutathione synthesis. J Biol Chem. 1982;257:13704–13712. [PubMed] [Google Scholar]
- 26.Ozols RF, Louie KG, Plowman J, Behrens BC, Fine RL, Dykes D, Hamilton TC. Enhanced melphalan cytotoxicity in human ovarian cancer in vitro and in tumor-bearing nude mice by buthionine sulfoximine depletion of glutathione. Biochem Pharmacol. 1987;36:147–513. doi: 10.1016/0006-2952(87)90392-3. [DOI] [PubMed] [Google Scholar]
- 27.O’Dwyer PJ, Hamilton TC, LaCreta FP, Gallo JM, Kilpatrick D, Halbherr T, Brennan J, Bookman MA, Hoffman J, Young RC, Comis RL, Ozols R. Phase I trial of buthionine sulfoximine in combination with melphalan in patients with cancer. J Clin Oncol. 1996;14:249–256. doi: 10.1200/JCO.1996.14.1.249. [DOI] [PubMed] [Google Scholar]
- 28.Cuneo MJ, London RE. Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase β binding affinity. Proc Natl Acad Sci USA. 2010;107:6805–6810. doi: 10.1073/pnas.0914077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tebbs RS, Flannery ML, Meneses JJ, Hartmann A, Tucker JD, Thompson LH, Cleaver JE, Pedersen RA. Requirement for the Xrcc1 DNA base excision repair gene during early mouse development. Dev Biol. 1999;208:513–529. doi: 10.1006/dbio.1999.9232. [DOI] [PubMed] [Google Scholar]
- 30.Horton JK, Stefanick DF, Prasad R, Gassman NR, Kedar PS, Wilson SH. Base excision repair defects invoke hypersensitivity to PARP inhibition. Mol Cancer Res. 2014;12:1128–1139. doi: 10.1158/1541-7786.MCR-13-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masaoka A, Gassman NR, Kedar PS, Prasad R, Hou EW, Horton JK, Bustin M, Wilson SH. HMGN1 protein regulates poly(ADP-ribose) polymerase-1 (PARP-1) self-PARylation in mouse fibroblasts. J Biol Chem. 2012;287:27648–27658. doi: 10.1074/jbc.M112.370759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler WB. Preparing nuclei from cells in monolayer cultures suitable for counting and for following synchronized cells through the cell cycle. Anal Biochem. 1984;141:70–73. doi: 10.1016/0003-2697(84)90426-3. [DOI] [PubMed] [Google Scholar]
- 33.Revet I, Feeney L, Bruguera S, Wilson W, Dong TK, Oh DH, Dankort D, Cleaver JE. Functional relevance of the histone γH2Ax in the response to DNA damaging agents. Proc Natl Acad Sci USA. 2011;108:8663–8667. doi: 10.1073/pnas.1105866108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heacock ML, Stefanick DF, Horton JK, Wilson SH. Alkylation DNA damage in combination with PARP inhibition results in formation of S-phase-dependent double-strand breaks. DNA Repair (Amst) 2010;9:929–936. doi: 10.1016/j.dnarep.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabel SA, Smith CE, Cuneo MJ, Mueller GA, Kirby TW, DeRose EF, Krahn JM, London RE. Characterization of the redox transition of the XRCC1 N-terminal domain. Structure. 2014;22:1754–1763. doi: 10.1016/j.str.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravi D, Wiles AM, Bhavani S, Ruan J, Leder P, Bishop AJR. A network of conserved damage survival pathways revealed by a genomic RNAi screen. PLoS Genet. 2009;5:e1000527. doi: 10.1371/journal.pgen.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trezl L, Park KS, Kim S, Paik WK. Studies on in vitro S-methylation of naturally occurring thiol compounds with N-methyl-N-nitrosourea and methyl methanesulfonate. Environ Res. 1987;43:417–426. doi: 10.1016/s0013-9351(87)80042-7. [DOI] [PubMed] [Google Scholar]
- 38.Reitman FA, Shertzer HG, Berger ML. Toxicity of methylating agents in isolated hepatocytes. Biochem Pharmacol. 1988;37:3183–3188. doi: 10.1016/0006-2952(88)90318-8. [DOI] [PubMed] [Google Scholar]
- 39.Mizumoto K, Glascott PA, Jr, Farber JL. Roles for oxidative stress and poly(ADP-ribosyl)ation in the killing of cultured hepatocytes by methyl methanesulfonate. Biochem Pharmacol. 1993;46:1811–1818. doi: 10.1016/0006-2952(93)90587-m. [DOI] [PubMed] [Google Scholar]
- 40.Forman HJ, Augusto O, Brigelius-Flohe R, Dennery PA, Kalyanaraman B, Ischiropoulos H, Mann GE, Radi R, Roberts LJ, 2nd, Vina J, Davies KJ. Even free radicals should follow some rules: a guide to free radical research terminology and methodology. Free Radic Biol Med. 2015;78:233–235. doi: 10.1016/j.freeradbiomed.2014.10.504. [DOI] [PubMed] [Google Scholar]
- 41.Bonicalzi ME, Haince JF, Droit A, Poirier GG. Regulation of poly(ADP-ribose) metabolism by poly (ADP-ribose) glycohydolase: where and when? Cell Mol Life Sci. 2005;62:739–750. doi: 10.1007/s00018-004-4505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunofluorescent imaging of PAR in XC5 and XRE8 cells. Cells were micro-irradiated in stripes to initiate PAR synthesis and were fixed after 1–7 min repair time. Immunofluorescent staining methods (anti-PAR antibody with Alexa 647 conjugated anti-chicken secondary antibody) are outlined in Section 2.2. Representative cells are shown for each time point. The mean data in this experiment are presented as a bar graph in Figure 5B.