Summary
Replication stress is a crucial driver of genomic instability. Understanding the mechanisms of replication stress response is instrumental to improve diagnosis and treatment of human disease. Electron microscopy (EM) is currently the only technique that allows to directly visualize a high number of replication intermediates and to monitor their remodeling upon stress. At the same time, DNA fiber analysis is useful to gain mechanistic insight on how genotoxic agents perturb replication fork dynamics genome-wide at single-molecule resolution. Combining these techniques has proven invaluable to achieve a comprehensive view of the mechanisms that ensure error-free processing of damaged replication forks. Here, we review how EM and single-molecule DNA fiber approaches can be used together to shed light into the mechanisms of replication stress response and discuss important cautions to be taken into account when comparing results obtained by EM and DNA fiber.
Graphical Abstract
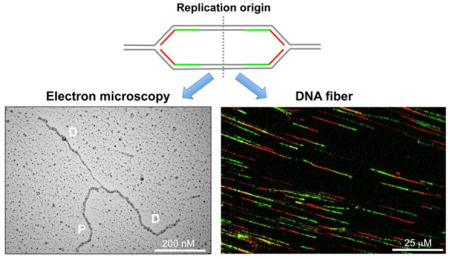
Aberrant DNA replication is one of the leading causes of mutations and chromosome rearrangements associated with several cancer related pathologies [1]. An accurate response to replication insults is mandatory for the faithful transmission of genetic information to daughter cells [2, 3]. Replication forks are constantly challenged and arrested by DNA lesions induced by endogenous and exogenous agents. In addition to DNA lesions, intrinsic replication fork obstacles such as transcribing RNA polymerases, unusual DNA structures, tightly-bound protein-DNA complexes, and oncogene activation challenge DNA replication fork progression. At the same time, agents that stall or damage DNA replication forks are widely used for chemotherapy, in the attempt to selectively target highly proliferating cancer cells [4]. Thereby, understanding the mechanisms of replication stress response following genotoxic stress induction is rapidly emerging as a central theme in cell survival and human disease.
Replication stress can be defined as the transient slowing or stalling of replication forks due to genotoxic insults. These insults might perturb replication fork structure, for example by promoting accumulation of single-strand DNA (ssDNA) regions where the template is not promptly replicated (Figure 1). ssDNA at replication fork junctions might originate from physical uncoupling of the replicative helicase that continues to unwind the DNA duplex after the polymerase stalls [5–8]. Furthermore, ssDNA gaps can be transferred behind a damaged replication fork, if replication restarts before the lesion is repaired or as a consequence of a faulty replication stress response mechanism [6, 9–11]. ssDNA accumulation upon replication stress is also contributed by nucleases, which play key roles in processing stalled replication intermediates [12–16]. They promote the limited degradation of nascent DNA strands required for efficient fork restart [12, 15, 17, 18]. However, they can also promote an extensive and uncontrolled degradation of stalled replication intermediates under pathological conditions. For example, the MRE11 nuclease is involved in the extensive resection of stalled replication forks in the absence of selected Fanconi Anemia (FA) and Homologous Recombination (HR) factors, including the Fanconi Anemia Complementation Group D2 (FANCD2) factor and the Breast Cancer Susceptibility factors BRCA2 and BRCA1 [13, 14, 16]. This extended fork degradation leads to long ssDNA stretches and is one of the leading causes of chemosensitivity of BRCA1- or BRCA2-deficient tumors [19].
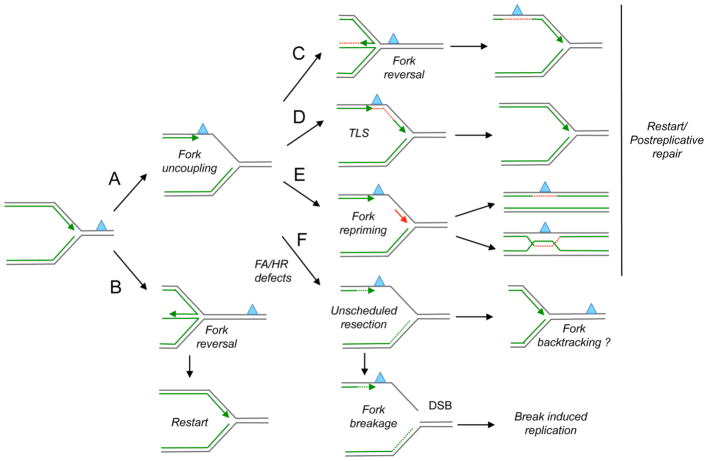
Figure 1. Mechanisms of replication fork processing and restart.
Different mechanisms may resume DNA synthesis when replication forks are stalled by a leading strand lesion (blue triangle). (A) Fork uncoupling: Replication fork uncoupling leads to ssDNA accumulation at the fork junction through functional dissociation of the MCM helicase and the stalled polymerase. Alternatively, fork uncoupling may result from nuclease-mediated resection of stalled forks. (B) Fork reversal: Replication forks might reverse before encountering the lesion giving time for DNA repair in the duplex template before forks are restarted. (C) Alternatively, fork reversal may promote lesion bypass via template switching. (D) Translesion Synthesis (TLS): Low fidelity TLS polymerases may function at stalled replication forks to ensure continued DNA synthesis through damaged templates. (E) Fork repriming: DNA synthesis can be reprimed (red arrow) and reinitiated ahead of a lesion or block. The resulting gaps are repaired post-replicatively by a recombination-based mechanism or by specific Translesion Synthesis (TLS) polymerases. (F) Unscheduled resection: FA/HR proteins, including BRCA1, BRCA2 and FANCD2, regulate the stability of stalled replication forks, and prevent extended nucleolytic degradation of nascent strands. Uncontrolled nuclease activity may lead to extended nascent strand degradation, and the resulting nuclease-dependent ssDNA gaps that form behind the forks could promote reannealing of the parental strands and “fork backtracking”. Alternatively, prolonged fork stalling may promote “fork breakage” by structure-specific endonucleases. Broken forks are able to resume DNA synthesis by the error-prone Break-Induced Replication mechanism.
Genotoxic insults can also lead to remodeling of the canonical three-way junctions present at replication forks into four-way junctions, called reversed replication forks [20]. Fork reversal is a remarkably frequent mechanism of replication stress response, that allows replication forks to reverse their course in response to genotoxic insults—including a variety of chemotherapeutic treatments—thereby preventing fork collision with the drug-induced DNA damage [8, 21, 22]. In this respect, detecting changes in fork architecture and the local accumulation of ssDNA is pivotal to define the mechanisms by which replication forks deal with genotoxic insults. Interestingly, the same FA/HR factors that control MRE11 nuclease activity and ssDNA accumulation are also emerging as key players involved in different aspects of fork remodeling [8, 23, 24], making this structural analysis instrumental to understand clinically relevant phenomena and thereby to potentially improve cancer chemotherapeutic treatments.
The development of new single-molecule DNA fiber and EM approaches and their use to answer key mechanistic questions on replication stress has provided a unique set of tools to detect ssDNA discontinuities, reversed replication forks and other types of replication perturbations [22, 25–30]. Most approaches routinely used to study DNA replication dynamics and perturbations provide parameters obtained from an ensemble of cells. Conversely, DNA fiber analysis and EM provide data on the status of replication forks at the genome-wide level by cumulative analysis of individual DNA molecules [27, 31, 32]. Here, we discuss the possible applications of these two techniques to the field of replication stress response. We also discuss how these two techniques might lead to apparently controversial results if data are not properly interpreted and the new directions that could be taken to increase their applications in cancer research.
Single molecule DNA fiber
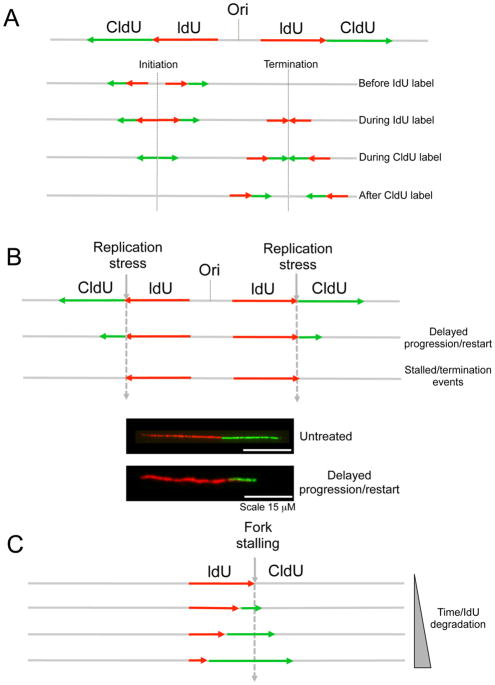
This single-molecule analysis of replication exploits the ability of many organisms to incorporate halogenated pyrimidine nucleoside analogs into replicating DNA and provides a powerful tool to monitor genome-wide replication perturbations at single-molecule resolution [25, 26, 28, 31, 33–35]. Briefly, ongoing replication events are sequentially labeled with two thymidine analogs—e.g., iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU)—and individual two-color labeled DNA tracts are visualized on stretched DNA fibers by immunofluorescence. Following the pulse-labeling, the individual DNA molecules are stretched into fibers either using the combing [25, 26] or spreading [28] techniques (for a detailed description of the protocols utilized for the single-molecule analysis of DNA replication intermediates see [32]). This simple labeling scheme allows monitoring several key replication parameters. First, measurements of the length of consecutive IdU-CldU tracts provide the speed of ongoing replication forks, whereas analysis of IdU-only tracts provides information on the frequency of stalled/terminated replication events (Figure 2A). In addition, an estimation of the number of newly initiated forks can be obtained by measuring frequency of CldU-only or CldU-IdU-CldU tracts. New initiation events can be quantified more accurately by measuring the inter-origin distance, which is the distance between two adjacent two-color labeled replication events [30]. To measure the inter-origin distance, it is however crucial to counterstain the entire DNA filaments to confirm that two replication events belong to the same molecule and that DNA fibers are not broken. Alternatively, global replication fork density can be estimated even more accurately by dividing the total number of two-color labeled forks by the total length of counterstained DNA fiber analyzed [36]. This density is then divided by the fraction of cells in S phase to determine the number of forks that are active at any time during the S phase. Counterstaining the DNA filaments is also useful to simultaneously monitor the synchronized progression of two sister forks emanating from a give origin. In this context, fork asymmetry can be measured from the ratio between the two sister forks and used as a parameter to estimate the frequency of fork stalling/collapse [37, 38]. For these types of analyses the combing approach is certainly favorable [33, 35, 37, 38], as it aligns all molecules along a single axis, preventing excessive crossing of DNA fibers.
Figure 2. Studying DNA replication fork dynamics by DNA fiber analysis.
(A) Schematic of the labeling patterns obtained for a bidirectionally progressing fork with equal pulse labeling with ldU (red) and CIdU (green). Upon detection of the ldU and CIdU tracts, different types of labeling patterns might be obtained corresponding to ongoing or terminated/stalled forks. (B). Schematic of the labeling patterns obtained when a replication inhibitor is added after IdU incorporation. Forks that are able to restart following replication inhibition are characterized by a contiguous red-green signal, whereas forks that are stall upon replication inhibition will display only a red signal. The progression rates after replication inhibition can be derived by comparing the length of the CldU (green) tracts in the presence and absence of the replication inhibitor. Below, representative image of an ongoing replication fork in the presence and absence of a replication inhibitor. (C) Schematic of the labeling pattern used to study replication fork degradation following replication inhibition. Nucleolytic resection of stalled forks will result in shortening of the IdU (red) tract where the thymidine analog was incorporated before replication stress induction.
DNA fiber analysis can be also used to study how specific genotoxic agents perturb replication fork progression [8, 21, 22, 39, 40]. In this case, cells are generally pulse-labeled with the first thymidine analog IdU (red label), followed by treatment with a selected replication inhibitor or DNA damaging agent, and concomitant (or subsequent) labeling with the second thymidine analog, CldU (green label) (Figure 2B). The labeling times for the first and second label can vary depending on the particular cell type or genotoxic agent used for these experiments (generally between 15 and 60 min). If treatment with the selected genotoxic agent decreases the rate of fork progression, the mean tract length of the second label (green) will be smaller compared to the untreated control (Figure 2B). In addition, the ability of replication forks to restart following damage induction can be measured by quantifying the frequency of IdU-CldU (red-green) tracts (restarting forks) [19, 40, 41]. Conversely, forks containing only the first label correspond to termination events and stalled forks that were unable to restart following drug treatment.
Nucleolytic resection following replication fork stalling can be monitored using the same labeling scheme described above [13, 14, 16, 19, 42, 43]. In this case, nucleolytic resection of both strands of a stalled replication forks results in shortening of the first tract (the one where the thymidine analog was incorporated before drug treatment) (Figure 2C). This approach has been recently used to demonstrate that the MRE11 nuclease promotes the extended degradation of unprotected replication forks in the absence of BRCA1 or BRCA2 [13, 14, 16, 19]. An important caution to use in this analysis is that shortening of the first tract should be measured only on forks characterized by contiguous IdU-CldU signals (and not on forks that have only the IdU label) to ensure that the shortening phenotype is indeed due to nucleolytic resection of stalled replication forks that can resume DNA synthesis and not to premature termination events (which would carry only the first label). In addition, the results might be significantly affected by the kind of drug used to stall or slow down replication forks. For example, hydroxyurea, which transiently inhibits DNA synthesis by causing an imbalance in the deoxyribonucleotide pool, is a widely used for this kind of experiments because it causes a global perturbation on all ongoing replication forks [13, 14, 19]. Conversely, drugs that induce specific DNA lesions, such as interstrand cross-linking agents or UV light, can be used for these experiments with the limitation that forks in close proximity to the lesion might be differentially perturbed compared to replication forks that are distant. In this regard, a recently developed quantum dot technology allowed visualization of the cross-linked sites on genomic DNA [44]. Using the quantum dot technique, the authors elegantly demonstrated that replication forks that meet the cross-linked site are able to traverse the damaged site and continue replicating, although with some delay [44]. Whether forks that are in close vicinity to DNA lesions are differentially perturbed compared to distant forks remains an open question in the field. To properly address this question, future studies should extend the quantum dot technology to other drugs or replication inhibitors by using custom-made antibodies or novel strategies that specifically detect DNA lesions on genomic DNA.
Electron microscopy
EM has proven extremely useful to gain mechanistic insight on how different kinds of genotoxic stress perturb DNA replication and is the only technique that allows direct visualization and quantification of DNA replication intermediates to date [8, 15, 20]. The fine architecture of replication intermediates is inspected using a combination of in vivo psoralen cross-linking and EM, as already described [27]. The crosslinking step is required to preserve the DNA structures from living cells during the extraction and enrichment procedures that precede EM visualization (for a detailed description of the experimental procedure see [27]). EM analysis allows distinguishing duplex DNA—which is expected to appear as a 10 nm thick fiber, after the platimun/carbon coating step necessary for EM visualization—from ssDNA, which has a reduced thickness of 5–7 nm. EM detection of ssDNA stretches could be assisted by adding specific ssDNA binding proteins to specifically “label” the ssDNA regions [6, 45]. The unequivocal identification of replication fork structures requires high magnification images to differentiate a true junction from occasional overlaps of two DNA molecules. In addition, replication forks should be normally characterized by two arms of equal length resulting from enzymatic digestion of replicating genomic DNA.
EM can be used to identify the presence of ssDNA gaps either at fork junctions or along replicated duplexes (Figure 3). This analysis has been instrumental to confirm that the formation of ssDNA stretches at replication fork junctions is indeed a common structural determinant linked with replication stalling following treatment with a wide variety of genotoxic agents [8]. Conversely, accumulation of “internal” unreplicated ssDNA gaps could reflect repriming mechanisms that can reinitiate DNA synthesis beyond a polymerase-blocking lesion, possibly coupled to processing and extension of the resulting ssDNA gap [24].
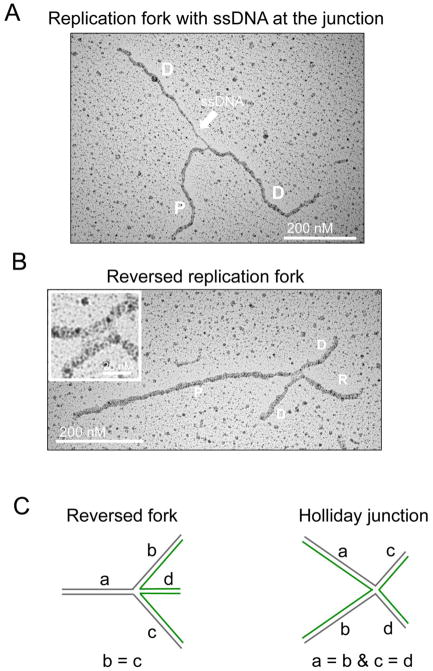
Figure 3. EM analysis of replication intermediates.
(A) Electron micrograph of a replication fork with a ssDNA gap at the junction. P, Parental strand; D, Daughter strand (B) Electron micrograph of a reversed replication fork. R, Reversed arm. The inset shows a magnified image of the four-way junction with the rhomboid structure at the junction itself. (C) Schematic drawing of a reversed fork and a Holliday Junction and their expected features in terms of the lengths of the different strands.
Another structure typically detected by EM—which is not unambiguously identified by biochemical or molecular biology approaches—is a reversed replication fork (Figure 3B). The criteria used for the unequivocal assignment of reversed forks are similar to those previously discussed for the identification of three-way junctions forks [27]. In particular, the presence of a rhomboid structure at the junction itself provides a clear indication that the junction is opened up and that the four-way junction structure is not simply the result of the occasional crossing of two DNA molecules. In addition, the length of the two arms corresponding to the newly replicated duplex should be equal (b=c), whereas the length of the parental arm and the regressed arm can vary (a ≠ b = c ≠ d). Conversely, canonical Holliday junction structures will be characterized by arms of equal length two by two (a = b, c = d). For a more detailed description of the parameters that need to be taken into account for a reliable assignment of reversed fork structures we refer the interested reader to [27]. Despite some skepticism on the existence and the physiological role of these structures, the use of EM to detect reversed forks has recently allowed to establish fork reversal as a remarkably frequent mechanism that enables DNA replication forks to assist DNA damage repair and tolerance in metazoan cells [8]. In addition, genetic knockdown studies have started to reveal key cellular factors required both for the formation and the resolution of reversed replication forks [8, 15, 21, 22, 46].
More in general, EM can be used to monitor any kind of alterations in the genomic DNA structure including the formation of ssDNA bubbles, loops or alternative DNA structures [29, 47]. In addition, EM can be used to derive information on nucleosome positioning and dynamics at replication forks based on the principle that nucleosomal DNA is inaccessible to the cross-liking agents used to stabilize replication intermediates prior extraction of the genomic DNA [27]. In particular, upon psoralen crosslinking in vivo and denaturation of deproteinized DNA prior to EM visualization, the DNA strands would separate in-between crosslinks, appearing as a string of “ssDNA bubbles” separated by cross-linked sites. Each of those ssDNA bubbles represents the position of a nucleosome in vivo, providing precious information on chromatin structure on replicating genomic DNA [27, 48, 49].
Combining DNA fiber and electron microscopy results
One of the major advantages of the DNA fiber technique is that allows genome-wide monitoring of replication fork dynamics by cumulative analysis of individual replication molecules over a prolonged period of time. However, stretched DNA fibers have a stretching range from 2 to 4kb/μm depending on the stretching technique [26, 30, 50], thereby limiting the resolution of the DNA fiber technique to few kilobases of DNA. The other relevant limitation of the DNA fiber technology is that it does not allow distinction of the two newly replicated tracts because they are both equally labeled with the thymidine analogs and they are “collapsed” in a single fiber upon DNA stretching. The EM technique has a much higher resolution—i.e., 30–50 base pairs [8, 27]. In addition, EM images show the actual Y-shaped structure of a replication fork with the two daughter strands branching from the three-way junction, even though they do not allow distinction of the leading and lagging strands. When comparing DNA fiber and EM results, another important aspect to the taken into consideration is that EM is a “static” method, which only takes snapshots of a reaction by “freezing” the replication intermediates with the cross-linking step. Conversely, the DNA fiber technology allows monitoring the dynamics of replication perturbation for a prolonged period of time, corresponding to the timing of the thymidine-analog incorporation on replicating DNA.
These differences between the two techniques become particularly relevant when studying the nucleolytic degradation of stalled replication intermediates. The nuclease-mediated degradation of stalled forks must occur on both strands in order to be detected as a shortening phenotype by DNA fiber because both tracts are labeled by the same thymidine analogs, as already discussed. In addition, this degradation must lead to the loss of several kilobases of DNA in order to be detected by DNA fiber because of the lower resolution of this technique. Conversely, the higher resolution power of the EM technique allows detecting short ssDNA discontinuities present in either of the newly replicated tracts. For example, the EM technique was instrumental to show that the RAD51 recombinase is required to prevent formation of short ssDNA gaps on newly replicated DNA in Xenopus egg extracts [24]. In particular, RAD51 limits the MRE11-dependent resection of internal ssDNA gaps that might form when persistent DNA lesions move behind the fork to be repaired postreplicatively. These internal ssDNA gaps are present on both nascent strands and are in most cases shorter than 300 nucleotides. This limited nascent DNA degradation can only be detected by EM, but could also give rise to a more extensive degradation on both nascent strands, a condition necessary to detect complete loss of signal by fiber analysis.
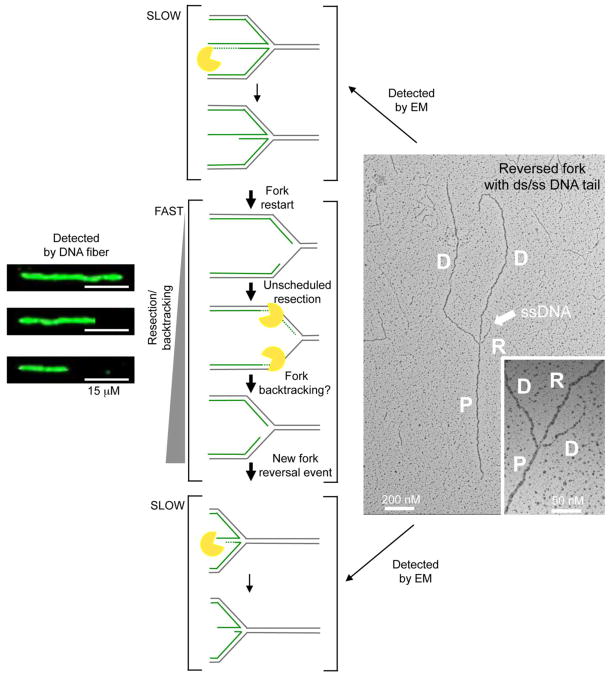
Along the same line, recent studies showed that the human DNA2 helicase promotes the limited degradation of reversed replication forks to mediate their restart after prolonged replication arrest [15]. This conclusion was again mainly derived from EM experiments showing that reversed forks accumulate in the absence of DNA2 and that DNA2-mediated resection is directed to completely or partially digest one strand of the reversed arm (Figure 4). However, DNA fiber analysis performed under similar conditions of prolonged replication arrest suggested that DNA2 can degrade stalled replication intermediates for several kilobases beyond the maximum length of the reversed arms measured by EM. A possible interpretation of these results is that after the initial DNA2-mediated regressed arm degradation is complete, other nucleolytic activities or DNA2 itself may co-degrade both sides of the replication fork under conditions of prolonged replication arrest, thus leading to extensive degradation events detectable by DNA fibers. In this scenario, the EM images likely represent snapshots of the “slow step” of this reaction—i.e., the DNA2-mediated degradation of the regressed arms—resulting in the drastic increase in reversed fork frequency observed in the absence of DNA2. Once the regressed arm has been resolved, the nucleolytic degradation might quickly proceed to degrade nascent strands behind the junction finally leading to reannealing of the parental strands and “backtracking” of the fork (Figure 4). A new reversal event may occur when this extensive degradation leads to asymmetric ssDNA accumulation at the fork, resetting the backtracked fork in the “slow step” of the process. This sequence of events would be effectively detected by fibers as fork backtracking, while EM would simply enrich for snapshots of the “slow steps” of a more extensive reaction.
Figure 4. Different structures detected by EM and DNA fibers.
Schematic of the different structures detected by EM and DNA fibers under conditions of prolonged replication arrest. EM is a “static” method, which enriches for snapshots of the “SLOW steps” of a reaction (i.e., partially resected reversed forks). After fork restart, the nucleolytic degradation quickly proceeds to degrade nascent strands behind the junction under conditions of prolonged replication arrest (FAST step). Reannealing of the parental strands leads to “backtracking” of the fork. A new reversal event arises as a consequence of asymmetric degradation and thus ssDNA accumulation in proximity to the fork. Backtracking is easily detected by DNA fiber, but not by EM because a reversed fork formed after degradation and backtracking is indistinguishable from the original reversed fork present before initial degradation. (Left) Representative DNA fiber experiment showing nascent strand degradation/backtracking. Scale bar 15 μM. (Right) Electron micrograph of a partially single stranded reversed replication fork. The white arrow points to ssDNA region on the reversed arm. Inset, magnified four-way junction at the reversed replication fork. D = Daughter strand, P = Parental strand; R, Reversed arm.
Future directions and concluding remarks
The single-molecule DNA fiber and electron microscopy techniques provide a unique set of tools to investigate the molecular mechanisms of replication stress response and how these mechanisms impact on cancer and aging. The recent development of novel technologies to specifically detect DNA lesions open new avenues to improve even further the potential applications of DNA fiber and EM in the replication field. In particular, a novel generation of antibodies is now available to probe specific kinds of DNA lesions—e.g, interstrand crosslinks [44], R-loops [51–53], or topoisomerase I-DNA covalent complexes [54]. These antibodies can be gold-labeled or detected using highly fluorescent quantum dots, thus allowing a direct visualization of the DNA lesion on genomic DNA at single molecule resolution. The combination of these novel lesion-detection techniques with DNA fiber and EM will answer several important biological questions. For example, this approach has recently been used to study forks that face site of DNA crosslinks [44]. These studies highlighted a novel mechanism by which forks can “traverse” inter-strand DNA crosslinks instead of being stalled as originally assumed. More in general, combining these lesion-detection approaches with the DNA fiber technology will be invaluable to establish whether forks in the proximity of DNA lesions are differentially perturbed compared to distant forks. Moreover, combining the same techniques with EM will be useful to determine which is the minimal distance between a three-way junction fork and a lesion necessary to activate the fork reversal mechanism.
Another unexplored area in the field relates to nucleosome deposition on reversed replication forks, or more in general at stressed forks. Histones are evicted ahead of the fork during normal DNA replication and this process is accompanied with their recycling and new histone deposition on the recently replicated daughter strands to restore chromatin on newly synthesized DNA [55]. Interestingly, replication stress interferes with histone recycling with potential impact on epigenetic stability [56, 57]. However, how replication stress interferes with nucleosome deposition is still poorly understood. Moreover, whether the regressed arm of a reversed replication fork is coated by histones and how this eventually affects fork remodeling and restart is still unknown. An important avenue for future EM studies will be to derive information on nucleosome positioning and dynamics at replication forks from denaturing EM spreadings, by looking at string of “ssDNA bubbles”—representing the position of a nucleosome—separated by cross-linked sites.
We envision that DNA fiber and EM techniques will be increasingly used together to shed light onto the constantly growing number of DNA replication stress response mechanisms. Moreover, the combination of these techniques with the recently developed approaches to study the loading of factors on newly replicated DNA—i.e., isolation of proteins on nascent DNA (iPOND) [58, 59] and chromatin immunoprecipitation (ChIP) sequencing [60, 61]—will likely lead to major breakthrough discoveries in the near future.
Highlights.
EM is currently the approach of choice to visualize replication intermediates
EM is a static approach, providing snapshots of the most abundant intermediates
DNA fiber monitors replication dynamics genome-wide at single-molecule resolution
Combining carefully EM and DNA fiber data is powerful to define replication stress
Acknowledgments
We would like to thank Ralph Zellweger for its contribution in the preparation of Figure 3. The work in the A.V. laboratory is supported by NIH grant R01GM108648 and by DOD BRCP Breakthrough Award BC151728. The work in the M.L. laboratory is supported by the SNF grants 31003A_169959 and 31003A_146924, and by the ERC Consolidator Grant 617102.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nature cell biology. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berti M, Vindigni A. Replication stress: getting back on track. Nature structural & molecular biology. 2016;23:103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harbor perspectives in biology. 2013;5:a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaillard H, Garcia-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 5.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & development. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Molecular cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Pages V, Fuchs RP. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science. 2003;300:1300–1303. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- 8.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, Herrador R, Vindigni A, Lopes M. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. The Journal of cell biology. 2015;208:563–579. doi: 10.1083/jcb.201406099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosal G, Chen J. DNA damage tolerance: a double-edged sword guarding the genome. Transl Cancer Res. 2013;2:107–129. doi: 10.3978/j.issn.2218-676X.2013.04.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 11.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhury I, Stroik DR, Sobeck A. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Molecular and cellular biology. 2014;34:3939–3954. doi: 10.1128/MCB.00457-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, Vujanovic M, Zellweger R, Moore H, Lee EH, Hendrickson EA, Cejka P, Stewart S, Lopes M, Vindigni A. DNA2 drives processing and restart of reversed replication forks in human cells. The Journal of cell biology. 2015;208:545–562. doi: 10.1083/jcb.201406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72:2814–2821. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- 17.Costanzo V. Brca2, Rad51 and Mre11: performing balancing acts on replication forks. DNA Repair (Amst) 2011;10:1060–1065. doi: 10.1016/j.dnarep.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Costanzo V, Robertson K, Bibikova M, Kim E, Grieco D, Gottesman M, Carroll D, Gautier J. Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Molecular cell. 2001;8:137–147. doi: 10.1016/s1097-2765(01)00294-5. [DOI] [PubMed] [Google Scholar]
- 19.Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, John S, Day A, Crespo AV, Shen B, Starnes LM, de Ruiter JR, Daniel JA, Konstantinopoulos PA, Cortez D, Cantor SB, Fernandez-Capetillo O, Ge K, Jonkers J, Rottenberg S, Sharan SK, Nussenzweig A. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- 21.Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, Marino F, Lucic B, Biasin V, Gstaiger M, Aebersold R, Sidorova JM, Monnat RJ, Jr, Lopes M, Vindigni A. RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition Human. Nature structural & molecular biology. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nature structural & molecular biology. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 23.Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic acids research. 2011;39:2153–2164. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nature structural & molecular biology. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 26.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, Bensimon A. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 27.Neelsen KJ, Chaudhuri AR, Follonier C, Herrador R, Lopes M. Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods in molecular biology. 2014;1094:177–208. doi: 10.1007/978-1-62703-706-8_15. [DOI] [PubMed] [Google Scholar]
- 28.Parra I, Windle B. High resolution visual mapping of stretched DNA by fluorescent hybridization. Nature genetics. 1993;5:17–21. doi: 10.1038/ng0993-17. [DOI] [PubMed] [Google Scholar]
- 29.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 30.Tuduri S, Tourriere H, Pasero P. Defining replication origin efficiency using DNA fiber assays. Chromosome research: an international journal on the molecular supramolecular and evolutionary aspects of chromosome biology. 2010;18:91–102. doi: 10.1007/s10577-009-9098-y. [DOI] [PubMed] [Google Scholar]
- 31.Herrick J, Bensimon A. Global regulation of genome duplication in eukaryotes: an overview from the epifluorescence microscope. Chromosoma. 2008;117:243–260. doi: 10.1007/s00412-007-0145-1. [DOI] [PubMed] [Google Scholar]
- 32.Techer H, Koundrioukoff S, Azar D, Wilhelm T, Carignon S, Brison O, Debatisse M, Le Tallec B. Replication dynamics: biases and robustness of DNA fiber analysis. Journal of molecular biology. 2013;425:4845–4855. doi: 10.1016/j.jmb.2013.03.040. [DOI] [PubMed] [Google Scholar]
- 33.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 34.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. The Journal of cell biology. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasero P, Bensimon A, Schwob E. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & development. 2002;16:2479–2484. doi: 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bialic M, Coulon V, Drac M, Gostan T, Schwob E. Analyzing the dynamics of DNA replication in Mammalian cells using DNA combing. Methods in molecular biology. 2015;1300:67–78. doi: 10.1007/978-1-4939-2596-4_4. [DOI] [PubMed] [Google Scholar]
- 37.Conti C, Sacca B, Herrick J, Lalou C, Pommier Y, Bensimon A. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Molecular biology of the cell. 2007;18:3059–3067. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuduri S, Crabbe L, Conti C, Tourriere H, Holtgreve-Grez H, Jauch A, Pantesco V, De Vos J, Thomas A, Theillet C, Pommier Y, Tazi J, Coquelle A, Pasero P. Topoisomerase I suppresses genomic instability by preventing interference between replication and transcription. Nature cell biology. 2009;11:1315–1324. doi: 10.1038/ncb1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mouron S, Rodriguez-Acebes S, Martinez-Jimenez MI, Garcia-Gomez S, Chocron S, Blanco L, Mendez J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nature structural & molecular biology. 2013;20:1383–1389. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- 40.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Molecular cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vallerga MB, Mansilla SF, Federico MB, Bertolin AP, Gottifredi V. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E6624–6633. doi: 10.1073/pnas.1508543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Higgs MR, Reynolds JJ, Winczura A, Blackford AN, Borel V, Miller ES, Zlatanou A, Nieminuszczy J, Ryan EL, Davies NJ, Stankovic T, Boulton SJ, Niedzwiedz W, Stewart GS. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks. Molecular cell. 2015;59:462–477. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 43.Spies J, Waizenegger A, Barton O, Surder M, Wright WD, Heyer WD, Lobrich M. Nek1 Regulates Rad54 to Orchestrate Homologous Recombination and Replication Fork Stability. Molecular cell. 2016;62:903–917. doi: 10.1016/j.molcel.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang J, Liu S, Bellani MA, Thazhathveetil AK, Ling C, de Winter JP, Wang Y, Wang W, Seidman MM. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Molecular cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makhov AM, Griffith JD. Visualization of the annealing of complementary single-stranded DNA catalyzed by the herpes simplex virus type 1 ICP8 SSB/recombinase. Journal of molecular biology. 2006;355:911–922. doi: 10.1016/j.jmb.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 46.Ray Chaudhuri A, Ahuja AK, Herrador R, Lopes M. Poly(ADP-ribosyl) glycohydrolase prevents the accumulation of unusual replication structures during unperturbed S phase. Molecular and cellular biology. 2015;35:856–865. doi: 10.1128/MCB.01077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aze A, Sannino V, Soffientini P, Bachi A, Costanzo V. Centromeric DNA replication reconstitution reveals DNA loops and ATR checkpoint suppression. Nature cell biology. 2016;18:684–691. doi: 10.1038/ncb3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lucchini R, Sogo JM. Replication of transcriptionally active chromatin. Nature. 1995;374:276–280. doi: 10.1038/374276a0. [DOI] [PubMed] [Google Scholar]
- 49.Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, Lees M, Sandelin A, Pasero P, Lopes M, Groth A. New histone supply regulates replication fork speed and PCNA unloading. The Journal of cell biology. 2014;204:29–43. doi: 10.1083/jcb.201305017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sidorova JM, Li NZ, Schwartz DC, Folch A, Monnat RJ. Microfluidic-assisted analysis of replicating DNA molecules. Nature protocols. 2009;4:849–861. doi: 10.1038/nprot.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. doi: 10.1038/nature13374. [DOI] [PubMed] [Google Scholar]
- 52.Boguslawski SJ, Smith DE, Michalak MA, Mickelson KE, Yehle CO, Patterson WL, Carrico RJ. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. Journal of immunological methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 53.Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic acids research. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel AG, Flatten KS, Peterson KL, Beito TG, Schneider PA, Perkins AL, Harki DA, Kaufmann SH. Immunodetection of human topoisomerase I-DNA covalent complexes. Nucleic acids research. 2016;44:2816–2826. doi: 10.1093/nar/gkw109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gurard-Levin ZA, Quivy JP, Almouzni G. Histone chaperones: assisting histone traffic and nucleosome dynamics. Annual review of biochemistry. 2014;83:487–517. doi: 10.1146/annurev-biochem-060713-035536. [DOI] [PubMed] [Google Scholar]
- 56.Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Molecular cell. 2005;17:301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 57.Jasencakova Z, Scharf AN, Ask K, Corpet A, Imhof A, Almouzni G, Groth A. Replication stress interferes with histone recycling and predeposition marking of new histones. Molecular cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 58.Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nature protocols. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes & development. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lubelsky Y, MacAlpine HK, MacAlpine DM. Genome-wide localization of replication factors. Methods. 2012;57:187–195. doi: 10.1016/j.ymeth.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 61.Ostrow AZ, Viggiani CJ, Aparicio JG, Aparicio OM. ChIP-Seq to Analyze the Binding of Replication Proteins to Chromatin. Methods in molecular biology. 2015;1300:155–168. doi: 10.1007/978-1-4939-2596-4_11. [DOI] [PubMed] [Google Scholar]