Abstract
In this review article, emphasis is placed on the critical survey of available data concerning modified nucleobase and 2-deoxyribose products that have been identified in cellular DNA following exposure to a wide variety of oxidizing species and agents including, hydroxyl radical, one-electron oxidants, singlet oxygen, hypochlorous acid and ten-eleven translocation enzymes. In addition, information is provided about the generation of secondary oxidation products of 8-oxo-7,8-dihydroguanine and nucleobase addition products with reactive aldehydes arising from the decomposition of lipid peroxides. It is worth noting that the different classes of oxidatively generated DNA damage that consist of single lesions, intra- and interstrand cross-links were unambiguously assigned and quantitatively detected on the basis of accurate measurements involving in most cases high performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. The reported data clearly show that the frequency of DNA lesions generated upon severe oxidizing conditions, including exposure to ionizing radiation is low, at best a few modifications per 106 normal bases. Application of accurate analytical measurement methods has also allowed the determination of repair kinetics of several well-defined lesions in cellular DNA that however concerns so far only a restricted number of cases.
Keywords: Single base oxidation modifications, DNA oxidative degradation pathways, measurement of oxidatively generated DNA damage, repair of oxidized bases
Graphical Abstract
Introduction
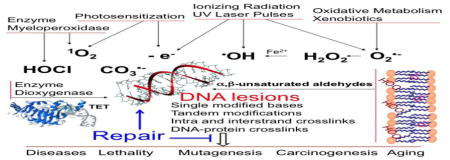
Oxidatively generated damage to cellular DNA is ubiquitous, being associated with oxidative, metabolism through the initial generation of the low reactive superoxide anion radical (O2•−), the product of one-electron reduction of oxygen during aerobic respiration [1]. Subsequently, O2•− can be converted through enzymatic dismutation to hydrogen peroxide (H2O2) that, like its precursor, exhibits a very low reactivity toward biomolecules while it is involved in redox signaling. However, transition metals such as ferrous ions are able to reduce H2O2 according to the Fenton reaction giving rise to highly reactive hydroxyl radicals (•OH) that are implicated in most oxidative degradation pathways of cellular DNA under physiological may be exacerbated during inflammation and under various conditions. The formation of O2•− pathological situations including arteriosclerosis, type 2 diabetes, cancer and neurodegenerative diseases [2–5]. This is often accompanied by the activation of NO-synthases that triggers the release of nitric oxide (•NO) another poorly reactive radical although it can induce the generation of a powerful oxidant peroxynitrite (ONOO−), a reactive nitrogen species (RNS), through the efficient combination of •NO and O2•− [7]. The UVA component of solar radiation, a major environmental factor, is also able to damage DNA mostly through the formation of singlet oxygen (1O2), another reactive oxygen species (ROS) [8,9]. Ionizing radiation acting as both a generator of •OH and a strong one-electron oxidant has been considered as the most biologically relevant oxidizing agent of DNA for more than 20 years since mid-50’s [10–12]. Bystander effects that are mediated by the delayed generation of •OH and •NO have been shown to damage DNA in neighboring cells despite the fact that they are not directly hit with UV and ionizing radiations [13–16].
Earlier studies on the formation of oxidatively-induced damage to DNA have focused on pyrimidine bases mostly for analytical reasons. The discovery and tentative characterization of two isomers of 5(6)-hydroperoxy-6(5)-hydroxy-5,6-dihydrothymine in 1959 [17] provided a strong impetus toward mechanistic studies on oxidative degradation pathways of thymine, uracil and cytosine in the subsequent 20 years. Efforts were also made on the basis of gas chromatography-mass spectrometry measurements to gain information on the main •OH-mediated reactions of 2-deoxyribose moiety involved in the formation of DNA strand breaks [18,19]. The advent of high performance liquid chromatography (HPLC) a powerful separation tool largely replacing paper and thin-layer chromatography in the mid 70’s allowed for the complete separation of complex mixtures of oxidized nucleosides including thymidine hydroperoxides, polar degradation products of 2’-deoxyguanosine and modified short oligonucleotides. The development of other spectrometric methods included both 1-D and 2-D high field 1H, 13C and 15N NMR and soft ionization mass spectrometry methods such as electrospray ionization. These advances permitted the structural assignment of complex oxidation products arising from polar purine components and more relevant DNA substrates such as short oligonucleotides. In addition, efforts were made to delineate the reactivity of defined base or sugar radicals upon their mild generation usually by UV activation of a photolabile precursor at a specific site within a defined sequence DNA fragment [20,21]. Theoretical calculation approaches gradually emerged through the development of highly performant ab initio DFT based computational and simulation methods, all of which have provided relevant information about the reactivity and energy requirements of key intermediates, which is not accessible experimentally [22–25]. Based on detailed structural and mechanistic studies, one may propose comprehensive oxidative degradation pathways of the main nucleic acid components upon exposure to 1O2, •OH, one-electron oxidants, halogenating agents and cellular dioxygenases; this information has been recently critically reviewed in exhaustive surveys. In parallel, there have been numerous attempts to search for the presence of some of the oxidation products in cells and tissues that were previously characterized in model studies. As will be briefly reviewed in a subsequent section, most of the early assays that were designed and applied for the analysis of certain products for decades suffer from major drawbacks, often leading to an overestimation of the levels of base oxidatively induced modifications of up to two or three orders of magnitude. Several of these inaccurate measurements are still being used to support putative correlations involving possible biomarkers of oxidatively induced DNA lesions and biological end-points related to aging, neurological disease and cancer risk. The main aim of the present review article is to provide an updated overview of the current situation concerning the accurate characterization and quantitative formation of mostly oxidatively induced base modifications in nuclear DNA of cells and tissues. Emphasis is placed on HPLC coupled to electrospray ionization-tandem mass spectrometry (ESI-MS/MS) as the gold standard of analysis because of its accuracy, versatility and reproducibility, taking into consideration that DNA extraction and subsequent work-up remain critical sources of spurious oxidation of inherent and overwhelmingly abundant DNA components present in the analysis..
Main reactive endogenous and exogenous cellular oxidizing species and agents
Oxidatively generated damage to DNA may occur directly or indirectly through the action of various oxidizing species that result from metabolic processes and/or exposure to diverse chemical and physical agents. A brief description of the main biologically relevant oxidants that are able to trigger DNA degradation pathways is provided below together with information on their mode of action and preferred targets.
Reactive oxygen species
Hydroxyl radicals (•OH) are endogenously produced in cells by Fenton type-driven reactions and they react efficiently with most molecules at the site of generation. This explains why •OH cannot be scavenged effectively, in contrast to numerous incorrect claims that examine the addition of antioxidants to the cellular medium [26]. A convenient source of •OH used to investigate radical-mediated oxidation reactions of cellular DNA involves the indirect effect of ionizing radiation through the radiolysis of bound water molecules. It is well documented from extensive model studies that •OH reacts predominantly by adding to the 5,6-pyrimidine and 7,8-purine double bonds of nucleobases giving rise to radical intermediates to which O2 may subsequently add or act as an one-electron oxidizing agent [27]. A second prominent oxidation reaction for thymine and 5-methylcytosine consists of •OH-mediated hydrogen abstraction from the methyl group leading to the formation of 5-(uracilyl) and 5-(cytosyl) methyl radicals, respectively [28,29]. Another efficient competitive •OH reaction when considering the nucleotide monomeric units is the abstraction of a hydrogen atom from the three methine carbons and the 5-hydroxymethyl group of 2-deoxyribose, all being reactive sites due to the presence of a vicinal O atom [27]. The formation of carbon centered radicals at C3 and C5 followed in aerated aqueous solutions by the generation of peroxyl radical intermediates lead to the quantitative formation of strand breaks. This is not the case for the abstraction reaction initiated at C1 that gives rise to 2-deoxyribonolactone, also called an oxidized abasic site [19,30–32]. It may be noted that chemical reactions initiated by the formation of C4 radicals are more complex. One of the degradation products has been identified as 2-deoxypentos-4-ulose, another oxidized abasic site. In addition, a competitive oxidative pathway leads to the formation of DNA strand break bearing a 3’-phosphoglycolate residues with the concomitant release of malondialdehyde and non- modified nucleobase [31, 33,34]. However, large disparities have been observed in the quantitative formation of the various sugar oxidation products measured in isolated DNA using either an isotope-dilution gas chromatography-mass spectrometry assay [32] or a more recently designed HPLC-based method [34].
As already pointed out, H2O2 and O2•− together with its conjugate acid, the related minor hydroperoxide radical (HO2•), do not exhibit any significant reactivity toward nucleobase and 2-deoxyribose DNA components in aqueous solutions DNA. However, it has been shown that O2•− but not O2 [35,36] is able to react with the highly oxidizing radicals G(-H)• that arise from deprotonation of the guanine radical cation by two competitive reactions [37]. Predominant addition of O2•− to G(-H)• gives rise through a complex sequence of reactions leading to 2,2,4-triamino-5(2H)-oxazolone (Oz) [38] while relatively minor one-electron reduction leads to restoration of the guanine base in a chemical repair process. Similar O2•− addition reactions have also been observed for other highly oxidizing radicals that are derived from the one-electron oxidation of tyrosine and tryptophan, respectively [39,40].
Singlet oxygen (1O2) in the 1Δ state is another biologically relevant ROS that is mostly generated in cells by type II photosensitization mechanism through energy transfer from triplet excited molecules to molecular oxygen. One typical reaction involves endogenous photosensitizers, which upon excitation by the UVA component of solar radiation, lead to the predominant generation of 1O2 as the main source of cellular DNA oxidation. Evidence has been provided from extensive model studies that the recombination of peroxyl radicals of various biomolecules can lead to the release of 1O2 as the result of decomposition of transient tetroxides according to the concerted Russell mechanism [41]. However, no evidence for the occurrence of such a mechanism has been found so far in cells. As a dienophile, 1O2 reacts preferentially with unsaturated compounds that possess electron rich double bonds. This is the case with guanine as being the exclusive nucleic acid component susceptible to react with 1O2 in aqueous solutions. Interestingly, synthetic naphthalene endoperoxides that are able to release pure 1O2 upon gentle warming, and that can be labeled with stable isotopes (i.e., 18O2), are available for mechanistic studies in both aqueous solution and cellular DNA [42,43].
Pyrimidine peroxyl radicals
In general, peroxyl radicals are weak oxidants and they tend to react very slowly with DNA. However, the situation is different for pyrimidine peroxyl radicals attached to oligonucleotides which, upon an initial radical reaction triggered by either •OH or one-electron oxidants in aqueous solution, can induce additional damage as shown initially by the pioneering work of Box et al [43]. Evidence has been provided through detailed experimental and theoretical mechanistic studies that 5-(6)-hydroperoxy-5,6-dihydropyrimidyl radicals are able to add to vicinal guanine bases in a sequence dependent manner with a strong preference toward purines located on the 5’-end [45–47]. The use of [18O]-labeled molecular oxygen showed that an 18O atom is transferred from the peroxyl group of thymine to the C8 of vicinal guanine giving rise to a tandem nucleobase lesion consisting of formylamine and 8-oxo-7,8-dihydroguanine (8-oxoGua) [46]. It was estimated from an extensive mechanistic study that about 50% of 8-oxo-7,8-dihydropurine bases, including predominantly 8-oxoGua and to a lesser extent 8-oxo-7,8-dihydroadenine (8-oxoAde), are generated as part of tandem base lesions upon exposure of DNA to •OH in aerated aqueous solutions [48]. As an initial study addressing the repair of oxidatively generated tandem base lesions, the two sequence isomers of vicinal 8-oxoGua and formylamine clustered damage were partly refractory to enzymatic removal by common base excision repair (BER) enzymes: endonuclease III and formamidopyrimidine DNA N-glycosylase [49]. Evidence was provided that 5-hydroperoxy-6-hydroxy-5,6-dihydrothyminyl, one of the two main peroxyl radicals induced by one-electron oxidation of a pyrimidine base in a thymine doublet, is able to abstract a hydrogen atom from the methyl group of the vicinal nucleobase. This was found to give rise to tandem base lesions consisting of 5,6-dihydroxy-5,6-dihydrothymine (ThyGly) on one side and either 5-formyluracil (5-FoUra) or 5-hydroxymethyluracil (5-HmUra) on the adjacent nucleobase [50]. Other examples of formation of oxidatively generated tandem lesions through the initial formation of defined peroxyl pyrimidine radicals in synthetic oligonucleotides using suitable photo-labile radical precursors are available. For example, evidence was provided that pyrimidine peroxyl radicals are also able to add to 3’ or 5’-contiguous thymine nucleobases, thus generating tandem base modifications [51,52]. As a competitive minor reaction, peroxyl pyrimidine radicals were shown to abstract a hydrogen atom from the anomeric proton of the adjacent nucleoside leading to the formation of 2-deoxyribonolactone as part of a tandem sugar-nucleobase lesion. It was also demonstrated, in more recent model studies, that the peroxyl radical specifically generated in double stranded DNA fragments from appropriate photochemical precursor at C4 of a 2-deoxyribose moiety is able to induce the formation of DSBs [53,54]. However, no reports have so far been reported on the formation of the above described oxidatively-induced tandem lesions in cellular DNA. This may be at least partly explained by the lack of sufficient sensitivity using standard HPLC-ESI-MS/MS instruments.
One-electron oxidants
Ionizing radiation, through its direct interaction with DNA components in the solid state, was the first physical agent used to abstract an electron from purine nucleobases and generate the corresponding base radical cations [55,56]. However, a different strategy was applied in aqueous solutions to gain mechanistic insights into one-electron oxidation degradation pathways of nucleobases whose susceptibility to undergo ionization decreases in the following order guanine > adenine > thymine > cytosine as an inverse correlation with their reduction potential [57]. Several type I photosensitizers including 2-methyl-1,4-naphthoquinone, riboflavin and benzophenone were selected for performing one-electron oxidation experiments [8,28]. This approach has allowed detailed mechanistic studies of the radical cations of purine and pyrimidine nucleobases in aqueous solution either as free components or when inserted into DNA. Another suitable photochemical technique to generate base radical cations consists of exposing DNA or its constituents to high intensity 266 nm nanosecond laser pulses. Increase in the pulse intensity favors bi-photonic ionization of the bases at the expense of [2+2] photocycloaddition of pyrimidine nucleobases. Increasing interest is devoted to the carbonate anion radical (CO3•−) as a biologically relevant one-electron oxidant that is likely efficiently produced during inflammation. Favorable coupling of O2•− and •NO, emanating from the activation of NO-synthase gives rise to peroxynitrite that upon reaction with CO2 generates nitrosoperoxycarbonate, the precursor of CO3•− and •NO2 [58,59]. Purine and pyrimidine radical cations, thereby generated by these different agents, are able to undergo two main competitive reactions including deprotonation and hydration. Interestingly, the pathways leading to stable products from one-electron oxidants of nucleobases in aerated aqueous solutions converge with those of •OH-induced decomposition. As a consequence, many of the products of the two pathways are identical although there are marked differences in the yield of unstable intermediates and the final distribution of products. However, a peculiar property of the guanine radical cations, in addition to their ability to undergo hydration [60], is their tendency to react with other nucleophiles. A likely nucleophile in chromatin involves histone abundant lysine residues bearing a primary amino group, that can give rise to DNA-protein crosslinks [61,62]. Alternatively, the N3 atom of vicinal intrastrand thymine and opposite cytosine can yield intra and interstrand crosslinks [63–65]. Here, we will only discuss GT intra-strand cross-links that have been characterized so far in cellular DNA upon exposure to high intensity UVC laser irradiation [66].
One electron oxidation of adenine generates the adenine radical cation, which rapidly undergoes deprotonation to the neutral N6-adeninyl radical [67]. One- and two-photon photolysis of DNA-A tracts with energies inferior to the ionization potential were recently shown to generate N6-centered radicals nearly quantitatively in low temperate glasses [68]. The reaction of hypochlorite with adenine and cytosine in DNA, RNA and polynucleotides also lead to N-centered radicals through the decomposition of intermediate chloramines [69]. The chemistry of N-centered radicals of 2’-deoxyadenosine (dA) was studied by exposure of the corresponding photolabile phenylhydrazone derivatives to near-UV light [70]. The major reactions of the nucleoside involved H-atom abstraction to regenerate dA, deamination to give 2’-deoxyinosine, and the formation of dA-dA dimers as determined by mass spectrometry analyses. Mason and co-workers [71] have developed an immune spin-trapping technique to detect DNA radicals as DMPO-DNA nitrone adducts by electron spin resonance (ESR). Using this method, they identified the same N6-adduct of adenine as above by ESR and MS analyses upon exposure of purified DNA and cellular DNA to CuCl2-H2O2 [72]. The characterization in cellular DNA of one-electron oxidation products of adenine with the exception however of 8-oxoAde and 4,6-diamino-5-formamidopyrimidine (FapyAde) awaits further experiments.
Peroxidases
Several mammalian heme peroxidases including myeloperoxidase (MPO), lactoperoxidase (LPO) and eosinophil peroxidase (EPO) are able to trigger the formation of hypohalous acid in inflammatory cells through the H2O2-mediated oxidation of related halides and pseudo halides including thiocyanate[73–76]. MPO-mediated oxidation reactions released from activated leukocytes catalyze the formation of hypochlorous acid (pKa = 7.46) in equilibrium with hypochlorite (OCl−), andare very efficient to kill invading pathogens [77]. Evidence has been provided that both HOCl and hypobromous acid (HOBr) are efficient halogenating agents of cytosine, adenine and guanine in model compounds and isolated DNA and RNA [78–80]. N-Chloramines that may be generated by the reaction of HOCl with the amino groups of amino are also able to chlorinate nucleobases albeit with a lower efficiency than HOCl [81].
Measurement of oxidatively generated damage in cellular DNA
Much effort has been made during the last five decades to develop assays aimed at monitoring the formation of oxidatively in cellular DNA following. Early attempts at the beginning of the 70’s focused on the measurement of radiation-induced 5,6-dihydroxy-5,6-dihydrothymine (ThyGly) also called “thymine glycol”. For this purpose cellular DNA was pre-labeled with [3H]- or [14C]-thymidine before being subjected to gamma or X-irradiation. The formation of ThyGly was assessed indirectly as either 2-methylglycerol or acetol that arises from borohydride reduction [82] and alkali degradation treatment, respectively [83]. Thus, various attempts were made to monitor the formation and repair of ThyGly in bacterial and mammalian cells exposed to ionizing radiation [82,84,85]. These authors reported that ThyGly was generated in cellular DNA upon exposure to either UV radiation [83,85] or hematoporphyrin photosensitization [86]; however, these conditions do not trigger thymine oxidation. It was then confirmed that both assays suffered from major flaws. In particular, the occurrence of self-radiolysis most likely led to an overestimation of ThyGly by three to four orders of magnitude, thus explaining the previously mentioned inconsistencies [87]. Since then, numerous chromatographic based assays together with more global methods including immunological approaches and enzymatic assays were developed. More recently with the discovery of 5-hydroxymethylcytosine (5-HmCyt) as a relevant epigenetic mark, numerous efforts have been devoted to the design of next-generation sequencing and array-based hybridization methods capable of mapping this modified base and its oxidation products at the nucleoside level in the genome [88–90]. Detailed information on these different methods is available in several recently published comprehensive review articles [91–94]. In the present contribution, a brief summary of the respective strengths and limitations of the main methods that have been recently or are currently utilized is provided.
Immunoassays
Many attempts have been made over the years to develop immunological based methods including among others, the enzyme linked immunosorbent assay (ELISA) and the radioimmunoassay (RIA) for measuring oxidatively generated base damage in DNA. Initial efforts focused on the preparation of monoclonal and polyclonal antibodies directed against ThyGly [95–96]. Subsequently, interest was devoted to design immunoassays for detecting 8-oxoGua [97–99] after discovery of this guanine oxidation product [100]. As a common observation, the main limitation of the designed ELISA and radioimmunoassays (RIA) is the lack of specificity due to the weak antigenicity of single oxidized bases in DNA that show a notable cross-reactivity with the overwhelming natural base precursors within the 10−4 to 10−5 range. This is insufficient to detect low physiological levels of ThyGly, 8-oxoGua as well as most other oxidized bases in cellular DNA on the order of a few modifications per 106 normal nucleosides. This has also been demonstrated for a monoclonal antibody directed against 5’,8-cyclo-2’-deoxyadenosine (cdA), which is a bulkier tandem base-sugar modification [101]. In contrast, immunological detection of 5-HmCyt enzymatically generated in embryonic and neuronal cells is routinely achieved with 0.1 % of this modification with respect to cytosine [102–103]. Similarly, immunological detection of 5-formylcytosine (5-FoCyt) and 5-carboxycytosine (5-CaCyt) generated through iterative oxidation is not accurate because the modifications are present at least 100-fold less than 5-HmCyt. Antibodies against 8-oxoGua are still widely used for sensitive histochemical immunofluorescence detection in human and animal tissues with various clinical applications and basic research involving DNA repair [104–109]. However the specificity of these antibodies remains to be established under in vivo conditions. It may also be pointed out that the measurement of 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-oxodG) in human fluids including urine, plasma and saliva has been achieved by competitive ELISA using N45.1 monoclonal antibody [110,111]. It was recently confirmed that the immunological detection of 8-oxodG in urine is less quantitative than that achieved by HPLC-ESI-MS/MS due to the presence of interfering contaminants such as D-glucose and D-galactose [112,113].
Enzymatic assays
Enzymatic based methods have become available with the isolation, characterization and cloning of DNA repair enzymes with emphasis on DNA N-glycosylases that remove oxidatively generated nucleobase lesions. The aim of enzymatic approaches, which include the alkaline single cell gel electrophoresis assay, also called “comet assay” [114–117], the alkaline elution technique [118,119] and the alkaline unwinding assay [120] is to convert oxidized nucleobases in a more or less specific way into strand breaks through the transient formation of alkali-sensitive abasic sites. The quantitation of the strand breaks generated during the analytical step is achieved in a very sensitive manner under alkaline conditions using fluorescence emission measurement after appropriate dye staining. The assays in their initial version allows for the quantitative detection of direct DNA nicks and so-called alkali-labile lesions including normal and oxidized abasic sites, ThyGly, 5-foUra, Oz), FapyAde and to a lesser extent 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) [121]. Additional strand breaks can be induced upon incubation of released oxidized DNA with the BER enzymes. Thus, bacterial formamidopyrimidine DNA N-glycosylase (Fpg) is used to recognize and indirectly measure 8-oxoGua, FapyGua and FapyAde [114,122]. The use of human 8-oxoguanine glycosylase 1 (hOGG1) made the detection of oxidized purine bases more specific since only 8-oxoGua and FapyGua are substrates for this enzyme [123]. In a complementary way, endonuclease III (endo III) is able to cleave the N-glycosidic bond of several oxidized pyrimidine nucleobases including ThyGly and 5-methyl-5-hydroxyhydantoin (HydThy) and related common oxidation products of cytosine such as 5,6-dihydroxy-5,6-dihydrouracil (UraGly) and related cytosine oxidation products. Usually, external calibration of the assays whose application requires only a small number of cells is carried out in parallel using ionizing radiation as a standard source of oxidatively generated damage to DNA assuming that the exposure of cellular DNA to 1 Gy gives rise to 3 single strand breaks and alkali-labile sites per 106 nucleosides [124]. One of the main advantages of these enzymatic assays with respect to HPLC-ESI-MS/MS assays is their high sensitivity, which benefits from the almost lack of adventitious oxidation of DNA samples during DNA release and subsequent analysis. This largely explains why the steady-state levels of 8-oxoGua in human cells obtained by DNA repair glycosylases are about 7 to 10 lower than those measured by either HPLC-ECD or HPLC-ESI-MS/MS [125–127]. It was also shown that Fpg-sensitive sites accumulate with age in the DNA of primary embryo fibroblast cultures [126] and hepatocytes from ogg1−/− nul mice [128]. Another relevant application dealt with the measurement of radiation-induced formation of Fpg- and endo III-sensitive sites in the DNA of human monocytes showing a linear increase within the dose range 0.1 – 0.5 Gy [129]. Enzymatic detection assays have several shortcomings that may slightly affect their general suitability. One problem concerns the lack of specificity of repair enzymes since quantitative information is provided on classes of damage and not on individual lesions. There are also uncertainties about the efficiency of the release of enzyme-sensitive sites since the ability to repair oxidized bases as part of tandem lesions is not as efficient as that of isolated oxidized bases [48,49]. In addition to applications already mentioned, the measurement of Fpg and endo III-sensitive sites has enlightened detailed mechanistic studies on oxidatively induced damage to cellular by UVA and visible light in which endogenous photosensitizers are implicated [8,9,115,130,131]. Increasing interest has also been devoted to human biomonitoring of genotoxic effects of environmental agents in particular nanomaterials [132] using the modified comet assay.
Gas chromatography-mass spectrometry (GC-MS)
Capillary gas chromatography coupled to mass spectrometry in the selective ion monitoring mode was introduced in the mid 80’s for measuring the formation of radiation-induced modifications of nucleobases including 12 oxidation products in DNA and model compounds. This method involves the strong acidic hydrolysis of DNA with concentrated formic acid (88%) at 150°C for 30 to 40 min in order to liberate DNA modifications as a free base [133].
Subsequently, the overwhelming normal nucleobases and related minor oxidation products are converted to trimethylsilylated derivatives by heating at 140°C for 30 min before GC-MS analysis. A different protocol at least in the initial step [134] was developed to monitor the formation of 5’,8-cyclo-2’-deoxyguanosine (cdG) in isolated [135] and cellular DNA [136]. This involved the enzymatic release of the purine 5’,8-cyclonucleosides using a cocktail of exonucleases and exonucleases. The acid based protocol was applied to the measurement of oxidatively generated base damage in cellular DNA a few years later [137]. Rapidly, major discrepancies up to 2 orders of magnitude became apparent between the yield of 8-oxoGua measured by GC-MS and HPLC coupled to electrochemical detection (ECD) [138], a method that is discussed in the next section. Several groups showed that the derivatization step appeared to be a major source of artefactual oxidation giving rise to the usual •OH-mediated oxidation products with a frequency of close to 10−4 (for a review article, see Cadet et al 1997 [139]. Another major drawback of acid hydrolysis of DNA is that it leads to the degradation of many oxidatively generated bases including FapyGua and FapyAde as it was unambiguously demonstrated [140]. Therefore, a mild acidic treatment involving hydrogen fluoride stabilized in pyridine has designed and when combined with a HPLC pre-purification step was shown to render quantitative GC-MS determination of both FapyAde and FapyGua in cellular DNA. An alternative version of the GC-MS assay allows one to overcome these two major shortcomings [141]. This version involves an alternative step to acid hydrolysis that is the enzymatic release of oxidized nucleobases by DNA repair N-glycosylases. Although this assay is very promising, there have only been a few recent applications [142,143]. Unfortunately, the analyses of DNA oxidatively modified lesions until mid-2000s’ were dominated by the method using acid hydrolysis and GC-MS analyses giving way to a flood of unreliably high values and misleading correlations between the level of oxidized nucleobases and biological end-points, such cancer risk, aging and repair [144–147].
It is surprising that GC-MS analysis despite its well-documented shortcomings has received recent notice with questionable frequencies of 48 8-oxoG and 35 FapyG per 106 bases in the DNA of wild type strains of Caenorhabditis elegans [148].
High performance liquid chromatography-electrochemical detection (HPLC-ECD)
An assay for the sensitive detection of 8-oxodG, an oxidation product of 2’-deoxyguanosine [100] was developed using HPLC coupled to electrochemical detection (HPLC-ECD) operating in the oxidative mode [149]. This analytical method provided a strong impetus to the analysis of oxidatively induced lesions in cellular DNA. Very rapidly, this method became suitable for the detection of 8-oxodG in cellular DNA upon exposure to oxidizing agents [150,151] and in kidney DNA of rats treated with potassium bromate [152] a well-documented one-electron oxidant agent once metabolized. These studies together with numerous HPLC-ECD measurements during the following 15 years have fully established 8-oxodG as a relevant biomarker of DNA. This received further support from the comparative evaluation of the available methods for measuring 8-oxodG by the European Standard Committee on Oxidative DNA Damage (ESCODD), a consortium of 25 laboratories [127,153]. ESCODD concluded that both HPLC-ECD and HPLC-MS/MS unlike GC-MS in its basic version constitute robust methods. However, and this will be discussed in the next section, DNA extraction and the subsequent work-up before HPLC measurements represent critical steps in which adventitious oxidative processes can occur with natural and relatively highly abundant nucleobases. In this respect, attempts were made to optimize the extraction step using the chaotropic method (i.e., with NaI as a salting out agent) and to minimize artefactual contribution [154]. It may be added that other electrochemically active modified nucleobases and nucleosides including 5-hydroxyuracil (5-OHUra), 5-hydroxycytosine (5-OHCyt), 8-oxoAde and FapyGua can also be detected by HPLC-ECD using higher oxidation potentials [155–157].
High performance liquid chromatography - mass spectrometry (HPLC-MS)
Early methods using one quadrupole (HPLC-MS) for the measurement of several oxidized 2’-deoxyribonucleosides usually show a limit of detection in the picomol range. This is clearly insufficient to detect oxidatively generated damage to cellular DNA under conditions where 30 to 50 μg of DNA are available and where the frequency of individual modifications reaches a few lesions per 106 nucleosides, a level that may only concern the most important modifications such as 8-oxoGua and ThyGly. Accordingly, it is rather surprising that attempts were made to monitor the formation of several radiation-induced oxidized nucleosides in cellular DNA through HPLC-MS measurements. The levels of reported 8-oxoGua and 8-oxoAde obtained by single quadrupole HPLC-MS, however, were between 50- and 100-fold greater [158,159] than those determined by HPLC-MS/MS [160,161]. Similarly, relatively high values for the 5R and 5S diastereomers of minor 5’,8-cyclo-2’-deoxyribonucleosides were reported on the basis of HPLC-MS measurements[162]. Other questionable results still obtained by HPLC-MS analysis suggested that Sp, a secondary oxidation of 8-oxoGua, was generated preferentially at the expense of its precursor in Escherichia (E.) coli strains exposed to one-electron oxidants [163]. The apparent formation of elevated values of 8-oxodG, 8-oxodA and 5’,8-cyclodA are explained by the erroneous detection provided by the use of only one quadrupole. Under these conditions, the possibility of detecting independent fragments arising from electrospray ionization of trace levels of impurities may give misleading and false identification of the targeted lesions in the selective ion monitoring (SIM) mode. So far, the only oxidized 2’-deoxyribonucleoside that has been accurately detected by HPLC-MS is 5-(hydroxymethyl)-2’-deoxycytidine (5-HmdC), the ten-eleven translocation (TET) mediated oxidation product of 5-methyl-2’-deoxycytidine (5-mdC) that accumulates in cellular DNA to the level of a few lesions per 103 nucleosides in embryonic stem cells (ESC) and brain tissues as detected by HPLC-MS [164,165]. However, attempts to detect 5-formyl-2’-deoxycytidine (5-FodC), the iterative enzymatic oxidation product that is present at the level of about one tenth to one hundred lower yields are problematic. This can be successfully achieved through by HPLC-ESI-MS/MS measurement [166] as is also the case for 5-carboxy-2’-deoxycytidine (5-CadC) [167] the more extensive oxidation product of 5-FodU.
High performance liquid chromatography – tandem mass spectrometry (HPLC-MS/MS)
HPLC-ESI-MS/MS and the sensitive HPLC-MS3 version are recognized as gold standard methods for monitoring the formation of modified nucleosides in biological samples, including nuclear DNA and biological fluids such as urine, plasma and saliva [91–93,168–172].The success of this method lies in the versatility and sensitivity of electrospray ionization that allows the efficient detection of a large number of oxidized DNA components with an optimum sensitivity that, on the average, extends into the femtomole range for modified 2’-deoxyribonucleosides using accurate multiple reaction monitoring mode [91,92]. Furthermore, the availability of synthetic [13C]- and [15N]-labeled internal standards allows for the use of isotope dilution as a means to correct for any losses during DNA digestion and changes in ionization efficiency during analysis [173–175]. Numerous successful HPLC-ESI-MS/MS measurements have been made to monitor the formation of several single and tandem oxidatively generated damage in cellular DNA following the initial detection of 8-oxodG in nuclear DNA and urine [176,177]. Although the method is highly accurate, there remains one major difficulty that restricts the use of HPLC-ESI-MS/MS. As already mentioned for other chromatographic methods, this is due to the unavoidable spurious oxidation of normal DNA bases (likely through the involvement of Fenton type reactions during DNA extraction and subsequent work-up) in particular during enzymatic digestion and release of individual DNA components. It should be noted that DNA is an excellent metal chelator, and thus, will bind potentially redox active metal ions during DNA extraction and release them during DNA digestion. Therefore, special attention was given during the ESCODD trial and subsequent studies [178–180] to develop methods to minimize the occurrence of artefactual oxidation. So far, this was partly achieved by the addition of transition metal chelators and anti-oxidants to the DNA sample extracts and digests prior to HPLC-MS/MS analysis. One major critical parameter that requires special attention is the amount of DNA used for the measurements in view of results indicating an inverse correlation between the level of artefactual oxidation and the quantity of extracted DNA [181]. Therefore, a minimum of 30 μg of DNA is required to minimize the contribution of spurious oxidation to levels that are lower than physiologically relevant values without at this stage being able to totally prevent artifactual autooxidation. This explains the difference of about 7 to 10 fold in the levels of 8-oxodGuo measured by chromatographic methods and the frequency of Fpg-sensitive site determined by either the comet assay or the alkaline elution technique, even if in the latter cases, there is a risk of underestimation. Such artefactual oxidation is the main reason why HPLC-MS/MS measurements are only useful for DNA samples in which the level of damage has been significantly increased by several fold, with respect to the steady-state levels, by exposure to either endogenous or exogenous oxidants as further illustrated in the following sections. It is also clear that HPLC-MS/MS is not an appropriate method to monitor the formation of oxidatively generated damage to DNA in mitochondrial DNA since there is a high risk of artifactual oxidation due to the relatively low amount of DNA available, usually in the low μg range with reasonable quantities of cell and tissue samples. The measurement of 8-oxoG and 8-oxodGuo, which is free of methodological artefactual oxidation, in human and animal fluids by HPLC-MS/MS will not be discussed in detail in this review (for recent reviews, see [111,171,172]. The origin of released 8-oxodG in urine still remains unclear particularly with purines since their metabolism involves enzymatic oxidation [182]. As a final remark, one should note the lack of accurate data on the formation of oxidatively induced damage to 2-deoxyribose in cellular DNA. So far only one attempt has been made to measure 2-deoxyribonolactone and 2-deoxyribonucleoside 5’-aldehyde in the DNA of TK6 human lymphoblastoid cells upon exposure to gamma rays using a GC-MS method with isotopic dilution [32]. However, the high yield of the latter oxidized 2’-deoxyribonucleoside, i. e. 0.22 lesions per 106 2’-deoxyribonucleosides and per Gy, or about 10-fold higher than that of 8-oxodG, suggests that the yield of this damage may be overestimated.
Single base modifications
Singlet oxygen
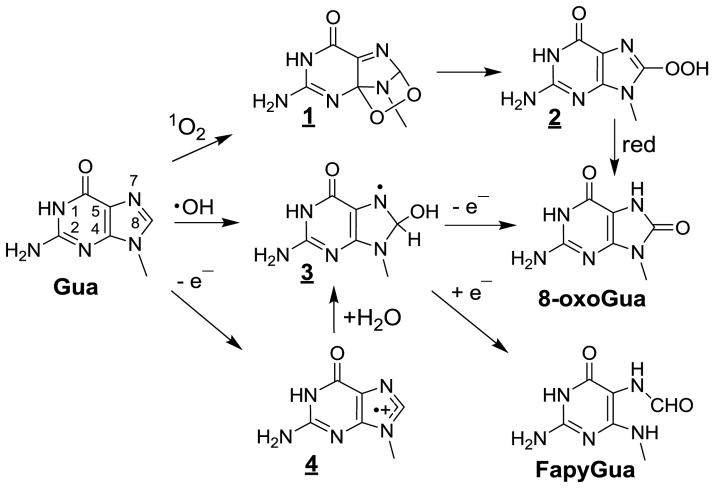
The search of 1O2 oxidation products in cellular DNA has been greatly facilitated by extensive and detailed mechanistic studies [8,183,184] using a clean source of 18[1O2] through the thermolysis of DHPN18O2 [42,185]. Abundant information is available on the 1O2-mediated oxidation reactions of the guanine moiety of isolated 2’-deoxyribonucleoside and short oligonucleotides. This reaction is rationalized in terms of initial Diels-Alder [4+2] cycloaddition across the 4- and 8-carbons of the imidazole ring [186] (Fig 1). A relatively minor pathway involves rearrangement of guanine endoperoxides 1 that transform into 8-hydroperoxyguanine 2, both which are relatively unstable [187,188]. Reduction of the 8-hydroperoxide group is likely to give rise to 8-hydroxyguanine [189–193] that is in dynamic equilibrium with the predominant 8-oxoGua tautomer in solution [121,186]. The reactivity of 1O2 towards guanine bases in double stranded DNA has been shown to be much lower than that for either single stranded DNA or isolated 2’-deoxyguanosine [193,194]. In addition, a major change was observed in the product distribution of 1O2 guanine oxidation products in isolated calf thymus DNA upon either exposure to type II photosensitizer [195–197] or incubation with DHPNO2 [183,198]. The reaction with DNA appears to be far simpler than that observed for the isolated nucleoside. Thus, 8-oxoGua is predominantly product generated in DNA, whereas in sharp contrast, spiroiminodihydantoin (Sp) is predominantly formed at the expense of 8-oxoGua upon oxidation of the nucleoside [199]. This can be rationalized in terms of steric factors that prevent formation of the quinonoid intermediate and its subsequent hydration. It may be added that no evidence for the formation of FapyGua, a typical one-electron oxidation degradation product of guanine was observed [200]; thus, this excludes any significant contribution of charge transfer reaction as proposed earlier [201]. Incubation of human monocytes cells with [18O]-labeled DHPNO2 that has been shown to accumulate intracellularly led to the generation of [18O]-labeled 8-oxodGuo in genomic DNA [200]. Levels as high as 2 lesions of 8-oxodG per 106 2’-deoxyribonucleosides, which represents about 12,000 lesions per nuclear DNA, were accurately quantified by HPLC-ESI-MS/MS using the multiple reaction monitoring (MRM) mode with [M+5] labeled 8-oxodG as the internal standard [91,173]. This clearly established the ability of 1O2 to react with the guanine base in cellular DNA producing 8-oxodG according to the previously characterized mechanism involving initial [4+2] cycloaddition of 1O2 across the 4 and 8 carbons of the purine ring. The possible formation of 8-oxoGua through radical reactions once DHPNO2 penetrates into cells was ruled out. Further support for the specific guanine oxidation by 1O2 in cells incubated with DHPNO2 was provided by the detection of nicks induced in extracted DNA by bacterial Fpg [202], a DNA repair enzyme that is able to cleave the N-glycosidic bond of 8-oxoGua residues. On the other hand, there was no significant increase in the levels of strand breaks induced upon incubation of DNA with endonuclease III that essentially recognizes oxidized pyrimidine bases. It was also shown that the steady-state level of strand breaks and alkali-labile sites did not increase upon incubation of the cells with DHPNO2. It should be remembered that 1O2 is mostly at the origin of nuclear 8-oxoGua measured in numerous types of cells [8,203] and human skin explants [204–206] exposed to UVA radiation as the result of a predominant type II photosensitization mechanism [205]. With respect to monocytes exposed to UVA, the relative contribution of 1O2 compared to •OH in the formation of 8-oxoGua was about 4:1 [115,207]. This ratio is likely lower in the DNA of UVA-irradiated melanocytes [208] due to the ability for melanin to photosensitize the generation of O2•−, a potential precursor of •OH.
Figure 1.
Oxidation of guanine by OH, one-electron oxidants and singlet oxygen (1O2).
A relevant topic has recently emerged with assessment of the photosensitizing ability of 6-thioguanine (6-TGua) and related analogues including azathioprine and 6-mercaptopurine, which are well documented anticancer, immunosuppressant and anti-inflammatory agents that are efficiently incorporated into DNA [209–211]. It was reported that UVA irradiation of GM5399 primary human fibroblasts pre-incubated with azathioprine led to the induction of 8-oxodG in nuclear DNA as the result of photosensitized generation of 1O2 [212]. It was recently estimated from a detailed photophysical study that the quantum yield of 1O2 generation through energy transfer from triplet excited 6-TGua to 3O2 was 24% [213], an approximate two-fold lower value than previously estimated [214]. UVA irradiation of 6-TGua in double-stranded DNA predominantly gives rise to guanine-6-sulfinate [215] as the result of efficient 1O2 oxidation through the intermediacy of a peroxy precursor, as inferred from a theoretical study [216]². It would be of interest to search for the formation of both 8-oxoGua and guanine-6-sufinated in the DNA of cells pre-incubated with 6-TGua prior to UVA irradiation.
Hydroxyl radical
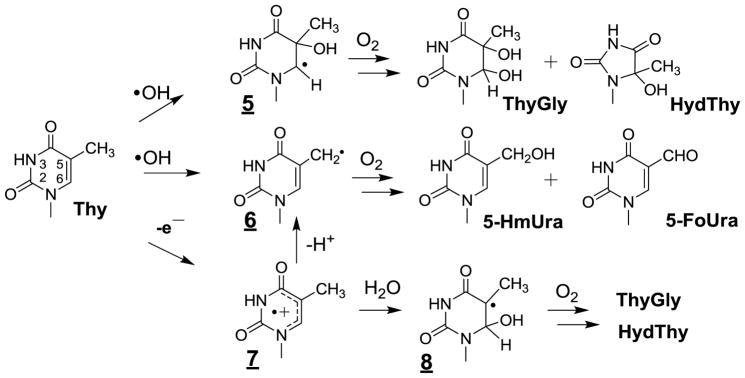
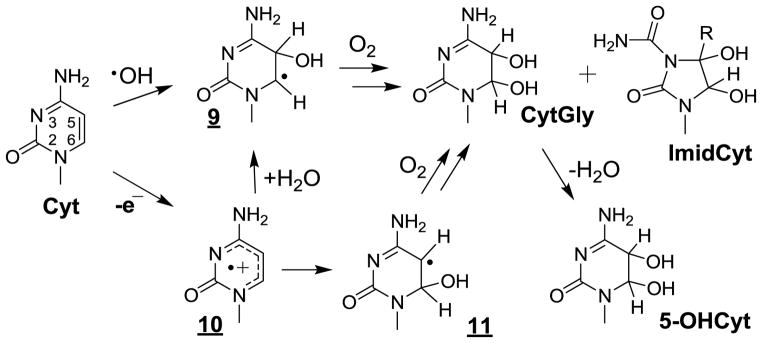
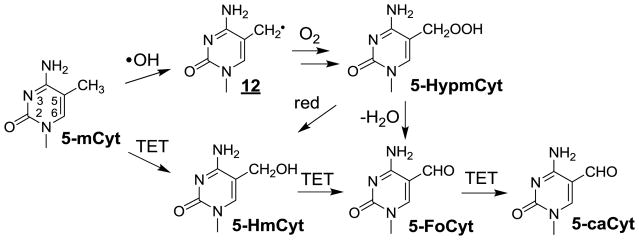
Relevant information has been gained concerning the formation of the major •OH-mediated oxidation products of the pyrimidine bases including thymine, cytosine and 5-methylcytosine in cellular DNA. This was achieved on the basis of HPLC-ESI-MS/MS measurements of modified 2’-deoxyribonucleosides and/or nucleobases that were enzymatically or chemically released from nuclear DNA from either human monocytes [160,161,173] or Fischer F98 glioma cells [217] subsequent to exposure to gamma rays. As depicted in Figure 2 the main •OH-mediated oxidation products of thymine (Thy) involved the cis and trans isomers of ThyGly, 5-hydroxymethylhydantoin (HydThy), 5-hydroxymethyluracil (5-HmUra) and 5-formyluracil (5-FoUra) as previously characterized in model studies including isolated DNA [28,65,218]. The formation of ThyGly is rationalized in terms of the initial addition of •OH at C5 and to a lesser extent at C6 giving rise to transient 5 and 9 [27,219]. In a subsequent step, fast O2 addition leads to 5,6-hydroxyhydroperoxides through related peroxyl intermediates. The formation of ThyGly is explained by reduction of the peroxide function whereas competitive rearrangement of the pyrimidine ring, through opening of the 5,6-bond and subsequent recyclization, leads to HydThy. The methyl group of Thy is also a major target for •OH reaction such that it is subject to hydrogen abstraction thereby generating the 5-(uracilyl)methyl radical (6). Reduction and dehydration of transient 5-hydroperoxymethyluracil formed after O2 addition to 6 gives rise to 5-HmUra and 5-FoUra, respectively. The oxidation reactions of cytosine (Cyt) triggered by •OH present similarities and differences with respect to those identified for Thy (Fig 3) [29]. The 5,6-double bond of Cyt is the exclusive site of reaction with •OH leading to the formation of about 90% of 5-hydroxy-5,6-dihydrocytosyl (9) and 10% of 6-hydroxy-5,6-dihydrocytos-6-yl radical (11). Relatively unstable 5,6-hydroxyhydroperoxides are expected to be generated after addition of O2 to 9 and 11. As observed for Thy, reduction of the Cyt hydroperoxides leads to the formation of 5,6-dihydroxy-5,6-dihydrocytosine products that in contrast to ThyGly are unstable [220,221]. Dehydration gives rise to 5-hydroxycytosine (5-OHCyt) whereas competitive deamination generates 5,6-dihydroxy-5,6-dihydrouracil (UraGly). In addition, opening of the 5,6-bond of Cyt 5,6-hydroxyhydroperoxides followed by hydrolysis and recyclization of the transient ureid likely comprise the rearrangement pathways involved in the formation of 5-hydroxyhydantoin (HydUra). Another peculiar feature of •OH-mediated oxidation reactions of Cyt deals with the intramolecular cyclization of 6-hydroperoxy-5-hydroxyl-5,6-dihydrocytosine [222] that, after opening of the endoperoxide and subsequent rearrangement, gives rise to 1-carbamoyl-4,5-dihydroxy-2-oxoimidazolidine (ImidCyt) (Fig 3) [29,223]. Related 5-mCyt oxidation products including 5,6-dihydroxy-5,6-dihydro-5-methylcytosine and 1-carbamoyl-4,5-dihydroxy-5-methyl-2-oxoimidazolidine have been identified in isolated DNA exposed to OH [217]. However, the latter modifications have not been accurately detected so far by HPLC-ESI-MS/MS in cellular DNA exposed to ionizing radiation likely due to the relatively low yields of formation. In contrast, 5-FoCyt and 5-HmCyt, which arise from the formation of 5-hydroperoxymethylcytosine (5-HypmCyt) following O2 addition to initially generated 5-(cytosyl)methyl radicals (12)(Fig 4), were detected in a relative ratio of 10 to 1 respectively.
Figure 2.
Oxidation of thymine by OH and one-electron oxidation . Initial attack of OH occurs at both C5 and C6 positions to give the corresponding OH adducts (e.g. 5). Analogous reactions occur for 5-methylcytosine (N3-C4=O → N3=C4-NH2).
Figure 3.
Oxidation of cytosine by OH and one-electron oxidation.
Figure 4.
Oxidation of 5-methycytosine by •OH and TET enzymes.
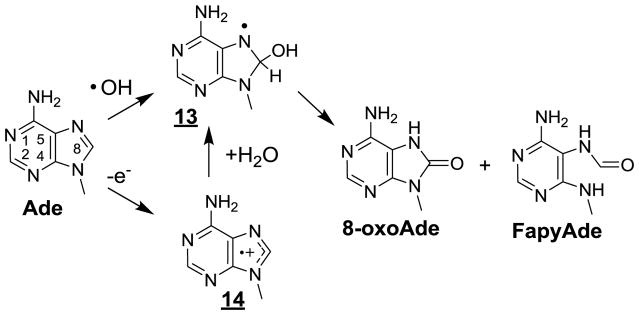
As already mentioned a significant proportion of •OH-mediated degradation products of guanine in cellular DNA are expected to result from reactions induced by pyrimidine peroxyl radicals either by addition to the vicinal bases or one-electron oxidation. However, •OH reacts efficiently with guanine as it does with other nucleobases in cellular DNA. A relatively minor fraction involves the addition of •OH to the C8 position of the purine moiety (close to 17% for free 2’-deoxyguanosine) giving rise to the reducing 8-hydroxy-7,8-dihydroguanyl radical 2 (Fig. 1) [219,224]. In contrast to what is observed with pyrimidine radicals, molecular oxygen is not able to add to intermediate 2 which rather is subject to one-electron oxidation by O2 leading to the formation of 8-oxoGua. Interestingly, 2 tends to undergo competitive one-electron reduction in cellular DNA, likely by thiol compounds, thereby generating FapyGua [224] with a yield that is about two-fold higher than that of 8-oxoGua [160]. Similar degradation pathways are involved in the •OH-induced formation of 8-oxoAde and FapyAde (Fig 5) that are generated with about a 10-fold lower efficiency than that seen for related guanine degradation products. The lower efficiency of adenine degradation may largely be rationalized by the lack of a significant contribution of oxidation reactions triggered by pyrimidine peroxyl radicals that predominantly target guanine in double stranded DNA.
Figure 5.
Oxidation of adenine by OH and one-electron oxidants.
Oz, another •OH-mediated or one-electron oxidation productof guanine [38] that is formed by the addition of superoxide anion radical to highly oxidizing G(-H)• has been detected in liver DNA of diabetic rats [225]. The yield of Oz was found to be 10-fold lower than that of 8-oxoGua. It has been recently reported that 5-carboxamide-5-formamido-2-iminohydantoin is a major •OH-mediated oxidation product of Gua in free nucleoside and oligonucleotides [226].
This should stimulate further studies aimed at searching for the formation of this rearrangement product of the purine ring of Gua in cellular DNA.
Attempts to search for the formation of 2-hydroxy-2’-deoxyadenosine, one of the main radiation-induced degradation products of the adenine moiety of cellular DNA [227] tentatively measured by GC-MS, however failed. Therefore, the relevance of 2-hydroxy-2’-deoxyadenosine as an adenine oxidation product is questionable since it was not detected using accurate HPLC-MS/MS in the DNA of THP1 cells exposed to γ –rays at doses up to 200 Gy [228].
One-electron oxidation
Overwhelming 8-oxoGua and several oxidized bases including ThyGly, 5-HmUra, 5-FoUra and 5-OHCyt have been detected by HPLC-ESI-MS/MS as the corresponding 2’-deoxyribonucleosides in cellular DNA upon ionization by UVC nanosecond high intensity laser pulses [66,161]. The formation of these base oxidation products may be rationalized in terms of the initial generation of purine and pyrimidine radical cations [28,186] followed by subsequent hydration and/or deprotonation reactions. Hydration reactions are highly specific giving rise to the 8-hydroxy-7,8-dihydroguanyl radical (3) from 4 (Fig 1) and the 6-hydroxy-5,6-dihydropyrimidin-5-yl radical (8) from 7 (Fig 2). In addition, competitive deprotonation of the thymine radical cation 7 leads to 6 [229,230] a radical which similar to 1, 7 and 13 are generated by •OH. In addition, 8-oxoAde and FapyAde have been also measured in about 10-fold lower yield compared to related guanine degradation products. Their formation is rationalized in terms of initial formation of the radical cation 14 that is converted into the 8-hydroxy-7,8-dihydroadenyl radical 13 followed by either one-electron oxidation into 8-oxoAde or one-electron reduction into FapyAde (Fig 5). Therefore, it is not surprising that the five oxidized bases resulting from the fate of radicals 3, 5, 6, 8 and 13 under aerobic conditions have also been identified as •OH-induced degradation products. However, there is a strong bias in the distribution of the oxidation products induced by ionization with respect to •OH. It was found that •OH-induced 8-oxoGua was formed with efficiency similar or even lower to that of 5-HmUra, 5-FoUra or 5-OHCyt. In contrast, 8-oxoGua generated by photoionization exhibits at least a 10-fold higher yield than any of the other four base oxidation products. The predominant formation of 8-oxoGua cannot be accounted for by the preferential ionization of guanine since photophysical studies showed that the four main base radical cations were induced with a similar efficiency upon laser irradiation. Therefore, it is reasonable to propose that subsequent to ionization of the bases, a redistribution of the radical cations thus produced takes place along the oligonucleotide chains, through long- and short-charge transfer mechanisms, as proposed for isolated DNA [231–233]. Guanine nucleobase, which exhibits the lowest ionization potential among DNA components, is the preferential sink for positive holes before they are converted into final degradation products.
Secondary oxidation reactions of 8-oxo-7,8-dihydroguanine
Several oxidized nucleobases including 8-oxoGua, 5-OHCyt and 5-hydroxyuracil, that show a much lower oxidation potential than normal bases, are susceptible to further degradation by one-electron oxidants including type I photosensitizers, CO3•− and organic radicals as demonstrated in model studies [234–243]. Major attention has been given to 8-oxoGua because it is about 100-fold more prone to one-electron oxidation and possesses a lower oxidation potential by about 0.5 eV compared to the parent molecule [234,235]. In neutral solution, spiroiminodihydantoin (Sp) was the main final degradation product of 8-oxoGua [236–238,241,242] which arises from an 1,2-acyl shift rearrangement of transient 5-hydroxy-7,8-dihydro-8-oxoguanine [244] initially proposed as a stable compound [245].The formation of guanidinohydantoin from the same intermediate was favored at acidic pH [246]. Interestingly, these studies have provided a strong stimulus to delineate the biochemical features of Sp, including the mutagenic potential [247,248] and associated repair by glycosylases [249–253]. The absolute configuration of 4R and 4S diastereomers of Sp 2’-deoxyribonucleosides was recently assigned [254,255]. However, only a few attempts have been made to search for the occurrence of secondary oxidation of 8-oxoGua in cellular DNA. In an initial study, the steady state level of Sp of 200 lesions per 106 Gua was reported to be at least one order of magnitude higher than 8-oxoGua in the DNA of wild type E.coli [163]. The same authors reported increases of Sp in Nei deficient E. coli compared to the wild type WT strain and unusually high increases when the cells were treated with 500 μM Cr(VI); the levels of Sp actually reached as much as 6,000 lesions per 106 Gua. It should be noted that the measurement of Sp was performed by HPLC-MS operating in the SIM mode, a method with shortcomings and well-documented lack of compound selectivity as already discussed (vide supra). The above extremely high value of Sp has to be compared with the relatively low level of secondary oxidation products of 8-oxoGua that were measured in the DNA of liver and colon tissues of Rag2−/− mice infected with Helicobacter hepaticus using the relevant and accurate HPLC-ESI-MS/MS method [256,257]. The frequency of Sp and Gh was found to be within the range of 1 to 7 lesions per 108 nucleosides, which is close to the limit of detection. These data show that the secondary one-electron oxidation of 8-Gua is modest even under conditions of infection in which inflammation triggers a massive production of ONOO−, CO3•−, and •OH. This is in agreement with chemical considerations that question the possibility that 8-oxoGua, present at a steady-state level of a only few residues per 106, can be a target for further oxidation among overwhelming normal bases when hole transfer process within DNA helix is restricted to 20 bp. This also explains why the proposed sacrificial role for 8-oxoGua [258,259] by acting as a preferential target of one-oxidation agents may protect canonical bases has been ruled out [260].
Halogenation of nucleobases
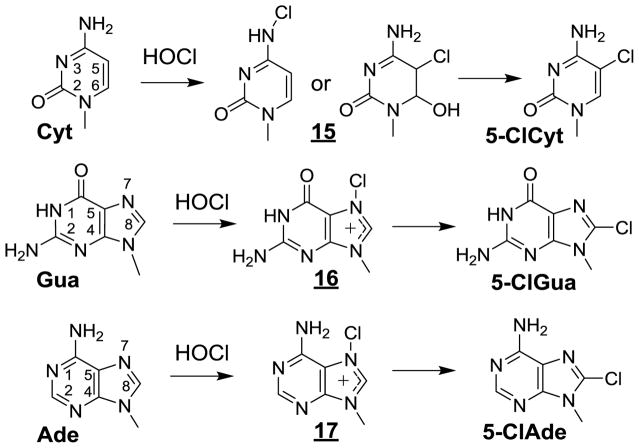
Hypochlorous acid (HOCl), a one-electron oxidizing and chlorinating agent is enzymatically generated by myeloperoxidase released from activated neutrophils during inflammation [261–264]. Exposure of SKM-1 cells to HOCl has been shown to induce the formation in DNA and RNA of the nucleosides of 5-chlorocytosine (5-ClCyt), 8-chloroguanine (5-ClGua) and 8-chloroadenine (5-ClAde) that were accurately measured by HPLC-ESI-MS/MS [265]. This may be rationalized by the transient formation of chloramines through the reaction of HOCl with the exocyclic amino group and subsequent rearrangement into the corresponding chlorinated bases (Fig 6). 5-ClCyt is formed predominantly over 5-ClGua and 5-ClAde in the DNA and RNA in SKM-1 cells, with the latter biopolymer showing a higher susceptibility to HOCl reactions [265]. In a subsequent study, the halogenated pyrimidine base 5-ClCyt was higher in the DNA of diabetic patients than in healthy volunteers, suggesting that this product may be used as a relevant biomarker of inflammation [266]. This idea is supported by the [265] observation of a significant increase in the steady-state level of highly mutagenic 5-ClCyt [267] in the colon and liver of Rag2−/− mice that were subjected to chronic inflammation as the result of infection with Helicobacter hepaticus [256]. Similarly high values of this biomarker of inflammation, half of which were close or more than 10 5-ClCyt lesions per 108 normal bases, were assessed by HPLC-ESI-MS/MS in the human colon of patients suffering from inflammatory bowel disease [268].
Figure 6.
Halogenation of DNA bases (Cyt, Gua and Ade)
Aldehyde adducts to amino-substituted bases
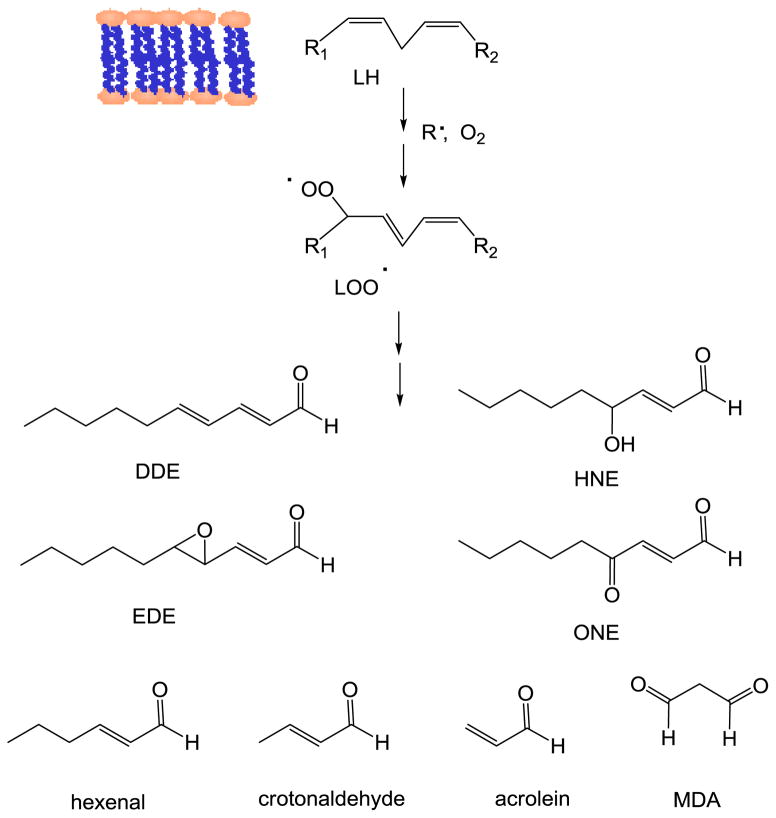
Lipid peroxidation is a crucial redox stress event, which has been associated with the development of a number of pathologies such as cancer, neurodegenerative and inflammatory diseases. The lipid peroxidation process generates a complex mixture of phospholipid products including hydroperoxides that can decompose leading to electrophilic derivatives such as aldehydes and epoxyaldehydes [269]. Lesions resulting from the reaction of DNA with breakdown products of lipid peroxides including malonyldialdehyde (MDA), 4-hydroxy-2-nonenal (HNE), 4-oxo-(2E)-nonenal (ONE), 2,4-decadienal (DDE), 4,5-epoxy-(2E)-decenal (EDE), hexenal, acrolein, and crotonaldehyde, have been detected at basal levels in human tissues and in clinical situations associated with redox stress disorders [270–272] (Fig 7).
Figure 7.
Main α,β-unsaturated aldehydes arising from the decomposition of lipid peroxides.
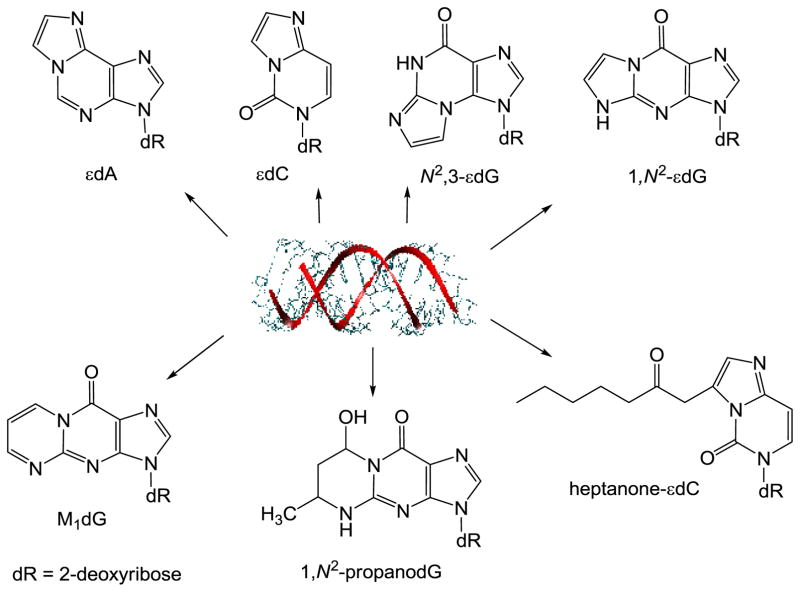
MDA is one of the best studied lipid peroxidation products. This dialdehyde reacts with 2′-deoxyguanosine (dG), 2′-deoxyadenosine (dA), and 2′-deoxycytidine (dC), yielding the cyclic pyrimidopurinone 3-(2-deoxy-β-D-erythro-pentofuranosyl) pyrimido[1,2-a]purin-10(3H)-one (M1dG), the acyclic N6-(3-oxo-1-propenyl)-2′-deoxyadenosine (M1dA) and N4-(3-oxo-1-propenyl)-2′-deoxycytidine (M1dC) adducts [273–275] (Fig 8). Basal levels of the promutagenic pyrimidopurinone adduct M1dG have been detected in different human tissues [276–279]. Interestingly, it has been shown that the intake of dietary polyunsaturated fatty acids correlates with the formation M1dG in female leukocytes [280]. Levels of M1dGo from 0.004 to 9.15 adducts per 108 nucleotides were reported by Ma et al. [281] (2015) in human leukocyte DNA using a methodology based on LC/nano electrospray ionization high-resolution tandem mass spectrometry (HRMS/MS). Interestingly, M1dG is oxidized to 6-oxo-M1dG in genomic DNA of intact cells indicating a possible role of 6-oxo-M1dG in the cellular consequences attributed to M1dG [282].
Figure 8.
Main adducts of α,β-unsaturated aldehydes to amino-substituted bases of 2’-deoxyribonucleosides
The reaction of DNA bases with α,β-unsaturated aldehydes, yields cyclic-substituted propano adducts as the 1,N2-propano-2′-deoxyguanosine (1,N2–propanodG) formed by Michael addition at the exocyclic amino group followed by ring closure [272,283]. Background levels of 1,N2–propanodG adducts resulting from acrolein, crotonaldehyde, and HNE reactions have been detected in DNA from different rodent and human tissues [284–286]. Using a method involving online HPLC/ESI/MS-MS, accurate determinations of 1,N2–propanodG levels in DNA extracts of human cultured cells (3.43 ± 0.33 /108 dG) and rat tissue (liver, 4.61 ± 0.69 /108 dG; brain, 5.66 ± 3.70 /108 dG; and lung, and 2.25 ± 1.72 /108 dG) have been performed [287]. Recently, 1,N2–propanodG adducts with levels of 2.4–3.5 adducts per 108 nucleotides were detected in untreated human MRC5 cells. Cell treatment with crotonaldehyde increases the levels of propano adducts in a concentration-dependent manner [288]. Levels of 1,N2–propanodG were shown to be higher in the liver DNA of glutathione-depleted rats [289] indicating that they are persistently formed by endogenous pathways.
High levels of 1,N2-propanodG (20.8 fmol of 1,N2-propanodG/mg creatinine) were present in urine samples from individuals exposed to urban air pollution [290]. Knowing that 1,N2-propanodG promotes DNA miscoding in human cells, largely through G → T transversions, and can inhibit DNA replication [291], the monitoring of 1,N2–propanodG levels may help protecting the health of urban populations.
Epoxidized α,β-unsaturated aldehydes, formed during the lipid peroxidation processes, can generate ethano or etheno derivatives upon reaction with DNA [270,271] (Fig 8). Several etheno adducts, 1,N6-etheno-2′-deoxyadenosine (εdA), 3,N4-etheno-2′-deoxycytidine (εdC),N2,3-etheno-2′-deoxyguanosine (N2,3-εdG), and 1,N2-etheno-2′-deoxyguanosine (1,N2-εdG), have been detected in cells as well as in rodent and human tissues [270,292–294]. Higher levels of εdA and εdC were found in chronic infections and inflammation [270,295]. Etheno adducts have been used as biomarkers for DNA damage resulting from the reactions of endogenous lipid peroxidation end products [296].
Etheno adducts are genotoxic and mutagenic as shown in vitro systems by primer extension assays and in vivo by site-specific mutagenesis in cells [297]. Heptanone-substituted 3,N4-etheno-2′-deoxycytidine (heptanone-εdCyd) blocked DNA synthesis and increased miscoding in both bacteria and human cells [298]. Recently, Patra et al have analyzed incorporation events using steady-state kinetics and LC-MS analysis of primers extended opposite 1,N6-ethenodA by recombinant human DNA polymerase [299]. The results showed a preference for purine pairing opposite 1,N6-ethenodA and for -1 frameshifts.
Ten-eleven translocation (TET) oxidation
The methylation of Cyt to 5-mCyt plays a pivotal role in epigenetics as a chemical modification or a mark that regulates gene expression. For example, genes are silenced by the methylation of cytosine predominantly occurring in CpG dinucleotides and gene promoter regions [300]. The discovery of unusually high levels of 5-hydroxymethylcytosine (5-HmCyt) in Purkinje neurons [301], and the initial characterization of ten eleven translocation (TET) family enzymes that oxidize 5-mCyt to 5-HmCyt [302], led to a surge of research inaugurating a novel pathway that operates to remove 5mC marks in the genome during cell differentiation and tissue homeostasis [303]. TET enzymes (I-III) are α–ketoglutarate and Fe(II)-dependent dioxygenases that oxidize 5-mCyt by an iterative process to give 5-HmCyt, 5-FoCyt and 5-CaCyt in double stranded DNA [304] (Fig 9). Interestingly, vitamin C acts as a catalyst and enhances TET activity in cells and animals [305]. In the past 8 years, numerous approaches have been applied to quantify 5-HmCyt and related modifications in genomic DNA [306]. These approaches utilize various methods to separate 5-HmC from complex biological mixtures, including TLC, GC, CE, LC, with anion exchange, reverse phase and HILIC solid phase chromatography, in combination with mainly mass spectrometry methods of detection [306,307]. Isotopic labels of 5-mCyt nucleoside and its oxidation products for MS analyses are either commercially available or fairly easy to synthesize starting with labeled dThd [217,308]. Thereby, the levels of 5-HmCyt vary appreciably from one tissue to another in mice, with the highest levels usually observed in the brain [165]. In humans, he levels of 5-hmCyt in blood DNA varies from 0.01 to 0.03% Cyt [309] while that in normal brain DNA varies from 0.54–1.34% Cyt [310,311]. It should be noted that the levels of 5-HmCyt in healthy cells and tissues are at least 100-fold higher than those of other oxidatively induced lesions modifications such as 8-oxoGua making quantification relatively easy without the problem of artifactual oxidation during sample preparation. Although it is possible to increase the formation of 5-HmCyt upon exposure of cells to ionizing radiation, the increase obtained at lethal doses of ionizing radiation is minor compared to the level of endogenous TET-mediated 5-HmC in cellular DNA [217]. The quantification of 5-HmCyt by LC-MS/MS may be achieved on a routine basis generally with about 1–2 μg of purified DNA; this corresponds to the extracted DNA from about 100,000 cells. The analyses of individual nucleobases or nucleosides of 5-HmCyt together with certain related modifications have been extended to the DNA of other organisms, mitochondrial DNA [311] mammalian RNA [312], and human urine [313]. Depending on the method and the instrument of analysis, the limit of detection of 5-HmCyt in cellular DNA is in the low- or sub-fmol range [314]. Indeed, a recent method with derivatization and MS detection reports the possibility of measuring 5-HmCyt from 20 cells [315]. The levels of 5-HmCyt in transformed cells in culture and cancerous tissue attain relatively low levels that may lead to difficulties in quantification using current methodologies. For example, the level of 5-HmCyt in brain tumors varies from 0.03 to 0.75 5-HmCyt/Cyt% [310,311]. Likewise the levels of related modifications, including 5-FoCyt, 5-HmUra, 5-HmUra, are usually 100-fold lower than those of 5-HmCyt under physiological conditions. As mentioned in previous sections, the analysis of 5-FoCyt and 5-CaCyt nucleosides by HPLC-ESI-MS/MS may be problematic because of the low levels present in certain cases. A recent study suggests that 5-HmUra may be produced by random enzymatic oxidation of thymine in DNA by TET [316]; however, the levels of 5-HmUra remain low compared to 5-HmCyt likely because of active and efficient DNA repair by single-strand selective monofunctional uracil DNA glycosylase [321].
Figure 9.
Additional reactions of guanine radical cation 3.
Complex modifications
Intrastrand base lesions (G*-T*)
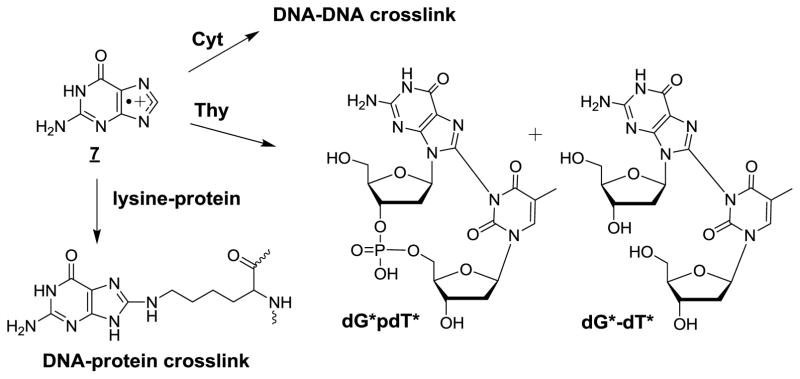
The formation of intrastrand cross-links (ICLs) involving one-electron oxidation of the guanine radical cation 3 was recently reported in cellular DNA [66]. This reaction constitutes a novel relevant example of nucleophilic addition reactions to 3 upon exposing HeLa cells to high intensity nanosecond UVC laser pulses followed by analyses of the damage by suitable enzymatic hydrolysis and subsequent HPLC-ESI-MS/MS measurement. The crosslink involved the formation of a covalent bond between C8 of guanine and N3 of thymine separated by an undamaged nucleotide residing between the modified bases (Fig 9). Importantly, the level of dG*-dT* crosslinks was found to increase with the intensity of laser pulses. The formation of dG*-dT* was a minor process with respect to the generation of other base oxidation products that arise from either hydration or deprotonation of the corresponding radical cations. For example, the yield of dG*-dT* was about 800-fold lower than the predominant yield of 8-oxoGua. In addition, either 5-FoUra or 5-HmUra, the two methyl oxidation products of thymine were formed with a yield of 60-fold higher than dG*-dT*. Interestingly, the ability of 4 to form dG*-dT* and to a lesser extent dG*pdT*in which both modified bases are vicinal was previously observed in model studies involving single-stranded oligonucleotides [68] and DNA duplexes [318]. It was recently shown that DNA repair glycosylases were able to cleave the N-glycosidic bond of dG*-dT* and dG*dC-dT* [319] which were site-specifically inserted into oligodeoxynucleotides with a defined sequence [63] (Crean et al 2008).
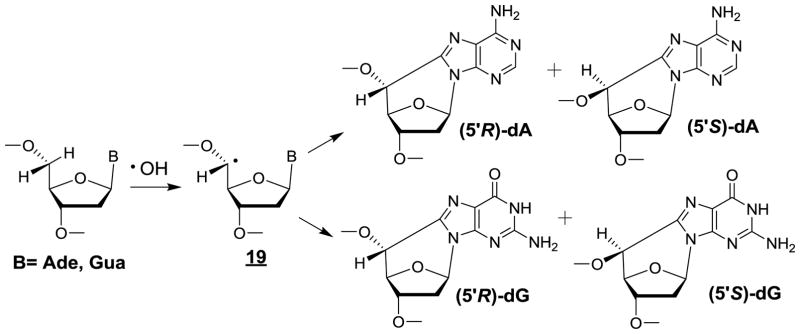
Purine 5’,8-cyclo-2’-deoxyribonucleosides
Support for •OH-mediated intramolecular cyclization of the C5’-yl sugar radical 15 of adenosine 5’-monophosphate and 2’-deoxyadenosine to the C8 position of the purine base moiety was gained from isolation and characterization of the corresponding cyclic ribonucleotide [320] and 2’-deoxyribonucleoside [321]. These modifications exist as either 5’R or 5’S diastereomers [322]. Similar reactions that give rise to (5’R) and (5’S)-5’,8-cyclo-2’-deoxyguanosine (cdG) (Fig 10) were subsequently found to occur in free 2’-deoxyguanosine and isolated DNA [137]. A resurgence of interest for these intranucleotide crosslinks was noted after the 5’R and 5’S diastereomers of 5’,8-cyclo-2’-deoxyadenosine (cdA) were found to be relevant substrates for the nucleotide excision repair pathway [323], an observation that was confirmed by another research group a few months later [324]. This discovery stimulated further mechanistic studies on the formation of both cdA and cdG [325,326] while major efforts were devoted toward better understanding the underlying repair mechanisms [327] and to delineate the effects of these tandem lesions on DNA replication and transcription [328,329]. In addition, attempts were made to search for the formation of purine 5’,8-cyclo-2’-deoxyribonucleosides in cellular DNA. For this purpose, the two diastereomers of cdG and cdA were measured by GC-MS [330] and HPLC-MS [331,332] respectively: two methods that suffer from major drawbacks as already discussed (vide supra). Thus, (5’S)-cdA was detected in the DNA of primary keratinocytes of two normal human subjects upon exposure to a low dose of 5 Gy of X-rays with a frequency of 14 lesions/109 normal 2’-deoxyribonucleosides per Gy [162,333]. This yield appears to be highly overestimated because it is comparable to the yield of a much more common oxidation product, 8-oxodG, which was generated in gamma-irradiated human monocytes with a similar efficiency of 20 lesions/109 2’-deoxyribonucleosides per Gy [160]. It should be noted that the formation of (5’S)-cdA in isolated aqueous solutions of DNA under aerated conditions is a minor pathway and that the amount of (5’S)-cdA induced by •OH represents only 1% of 8-oxoG [325]. Furthermore, a dose of 2kGy of gamma rays was necessary, using an accurate HPLC-ESI-MS/MS method, to barely detect traces of (5’S)-cdA whose yield was estimated to be at most 0.2 lesion/109 2’-deoxyribonucleosides per Gy [325]. The very low efficiency of •OH induced formation of purine 5’,8-cyclonucleosides in cellular DNA thus raises a question about their biological relevance; this idea actually received further confirmation from a recent study [334]. None of the four diastereomers of cdA and cdG was detected by HPLC-ESI-MS/MS measurements in the DNA of radiation-resistant hyperthermophilic Thermococcus gammatolerans archeon strains exposed to 5 kGy of gamma radiation [334]! The questionable formation of purine 5’,8-cyclo-2’-deoxyribonucleosides in cells is consistent with the low efficiency for •OH to generate C5’-yl radicals at either adenine or guanine nucleotides (less than 3% for each), which for the most part are efficiency scavenged by O2 [325].
Figure 10.
Formation of purine 5’,8-cyclo-2’-deoxyribonucleosides by •OH-mediated hydrogen atom abstraction at C5’.
Other confusing aspects concern the steady-levels in nuclear DNA of both cdA and cdG that, in the past, have been measured by either capillary HPLC associated with MS3 detection or, more recently, by nanoLC-NSI-MS/MS in the brain, kidney and liver tissues of several animals including healthy Long-Evans Agouti (LEA) rats and Long-Evans Cinnamon rats (LEC) bearing a genetic dysfunction as in human Wilson’s disease [335,336]. Other studies have involved Ercc−/Δ mice in which mutations led to a reduced expression of XPF-ERCC1 that affects NER efficiency and presumably leads to accumulation of both cdA and cdG with age [337,338]. Surprisingly, the steady-state levels of purine 5’,8-cyclo-2’-deoxyribonucleosides were elevated reaching 1 cdA/106 2’-deoxyribonucleosides in the liver and brain tissues of LEA rats [336]. Again, the levels were comparable to the average levels of 8-oxodG, a more frequently oxidatively generated damage to DNA. It seems that the levels of cdA and CdG are overestimated by at least two orders of magnitude as also suggested by the elevated values of 5-FoUra and 5-HmUra that were measured in the DNA of LEA rats [335]. Thus, the level of 20 FoUra/106 2’-deoxyribonucleosides assessed in hepatic tissue corresponds to the amount observed in cellular DNA exposed to 1 kGy of gamma rays! Clearly, further studies are necessary to resolve these apparent discrepancies.
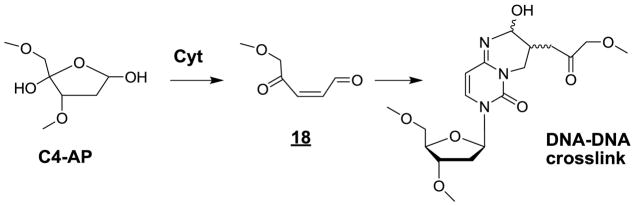
Interstrand cross-links
Insight into the formation of intrastrand crosslinks was gained from model studies that led to the isolation and characterization of four diastereomers of 6-(2-deoxy-β-D-erythro-pentofuranosyl)-2-hydroxy-3(3-hydroxy-2-oxopropyl)-2,6-dihydroimidazo[1,2-c]-pyrimidin-5(3H)-one in isolated DNA upon exposure to •OH in aerated aqueous solutions [339,340]. Interestingly, the four Cyt adducts were detected in the DNA of human THP1 monocytes exposed to gamma rays. Further information concerning the mechanism of generation of Cyt adducts was inferred from incubation of isolated and cellular DNA with bleomycin, a radiometric enediyne that is known to predominantly abstract the C4’ hydrogen atom in double-stranded DNA [341]. The resulting carbon centred C4’ radical similar to that generated by •OH is converted among other degradation pathways into the corresponding C4’ oxidized abasic site (C4-AP) [33] after O2 addition and elimination of the nucleobase (Fig 11). The abasic site or more likely its acyclic form [342] is prone to subsequent β-elimination, which is enhanced when either a cytosine or adenine base is located on the opposite strand [343]. This results in cleavage of the 3’-phosphodiester bond with the concomitant generation of 18, a highly reactive conjugated keto-aldehyde, which can add to 4-amino group of 2’-deoxycytidine on the opposite strand giving rise to interstrand DNA cross-links (Fig 11). The proposed mechanism of ICL formation has received further support from subsequent model studies in which a photolabile precursor of the C4’ radical was site-specifically inserted into DNA duplexes [343,344]. The formation of Cyt adducts that are present in untreated cells at a level of about 1–3 lesions per 108 nucleosides was found to increase upon exposure to gamma rays with a relative occurrence of 1% with respect to 8-oxoG. In contrast, incubation of cells with bleomycin leads to a significant 4-fold increase in the frequency of the ICLs while the level of 8-oxoG is not affected. The search for the formation of similar cycloadducts to adenine in cellular DNA expected to be generated from other model studies [344] awaits further investigation.
Figure 11.
Formation of a DNA interstrand crosslink by initial •OH-mediated hydrogen atom abstraction at C4’.
Repair of oxidatively damaged cellular DNA
There is an abundance of literature concerning the enzymatic activity, substrate specificity and mechanistic aspects of diverse prokaryotic and eukaryotic DNA N-glycosylases that act on oxidized bases through the predominant base excision repair pathway as reviewed in several contributions to this issue. In contrast, there is a paucity of accurate information on the repair of individual oxidatively generated base damage in cellular DNA. This is mostly explained by difficulties that have been encountered in measuring well-defined DNA oxidatively induced modifications in isolated cells or animal tissues, particularly under mild oxidative stress conditions. Most relevant data concern the repair of 8-oxoG on the basis of HPLC-ECD measurements. In addition, an example of repair that does not implicate BER pathway is available [340].
Base excision repair of 8-oxoGua
In pioneering work, Kasai and co-workers were able to accurately monitor the formation of 8-oxoG by HPLC-CED in the hepatic DNA of mice after whole-animal exposure to γ-rays [151]. The average radiation-induced yield of 8-oxoG (20 lesions/109 bases per Gy) is similar to that measured by HPLC-MS/MS in γ-irradiated human monocytes, being three orders of magnitude lower than that observed in isolated DNA. Interestingly, evidence for the efficient repair of 8-oxoG was provided by a decrease of this oxidized base within a few hours during post-irradiation incubation. A similar observation was reported in mouse FM3A cells in which 8-oxoG was specifically induced in nuclear DNA by riboflavin sensitization to UVA radiation [345]. It was found that about 2/3 of the initial amount of 8-oxoG was repaired by about 2h post-irradiation. Other studies have confirmed that 8-oxoG was efficiently repaired in the DNA of animal tissues exposed to various oxidizing agents, including UVA radiation [346], ferric nitrilotriacetate [347,348], diesel exhaust particles [349] and asbestos [350]. The repair kinetics of 1O2-specifically generated 8-oxoG in the DNA of liver and immortalized embryonic fibroblast cells lines from mice was determined from the analysis of Fpg-sensitive sites [128]. The removal of 50% of 8-oxoG lesions in both cell lines obtained from healthy mice required 4 h. This time-course compares with a 12 h period that was required in order to achieve a similar extent of repair in cells generated from ogg1−/− null mice that are partly deficient in BER excision ability.
Repair of Cyt adducts as part of interstrand DNA crosslink
The kinetics of removal of interstrand clustered DNA lesions generated by initial hydrogen atom abstraction at C4’ of the 2-deoxyribose moiety was assessed using HPLC-ESI-MS/MS by monitoring the gradual decrease of the levels of Cyt adducts generated in human monocytes upon treatment with bleomycin [340]. The repair rate of the Cyt adducts showed a typical biphasic phase and appears to be two-fold slower than that of 8-oxoG. Evidence has recently been gained, from model studies involving bacterial UvrABC complex, that nucleotide excision repair is also involved in the removal of the above interstrand lesions giving rise to double strand breaks (DSBs) [351,352]. This constitutes a novel example of DSB generation arising from one radical hit as the result of DNA repair processing of a Cyt adduct containing interstrand crosslink. This involves incision of the damaged strand opposite to the nick that was initially generated by chemical reactions giving rise to the clustered lesion (Fig 11)
Conclusions and perspectives
Major progress has been made during the last two decades in the identification of oxidatively-induced damage to DNA and the mechanisms of formation of damage products, which include complex lesions such as tandem modifications and interstrand cross-links, DNA protein cross-links, in addition to predominant oxidized single bases. The development of HPLC coupled to tandem mass spectrometry that became available during the 90’s has made it possible to accurately detect modifications in cellular DNA. In agreement with earlier measurements using either HPLC-ECD or enzymatic-sensitive sites, the steady-state level of 8-oxoG, an ubiquitous DNA oxidation modification, lies within the range of a few lesions per 107 2’-deoxyribonucleosides. This applies as well to other forms of oxidatively generated base modifications that have been measured in cellular DNA by HPLC-MS/MS methods. Another interesting conclusion is that the mechanism of formation of DNA products established from model studies in aerated aqueous solutions may be extrapolated in part to cellular DNA. The availability of capillary liquid chromatography associated with nanoelectrospray ionization tandem mass spectrometry (LC-NSI-MS/MS) displays a significant increase in the sensitivity of detection and represents a powerful tool for further investigations. A recent application of this method to mitochondrial DNA [353] has however shown the limitation of the approach when low amounts of nucleic acids are analyzed. The high values of 8-oxodG that varied within the range of 130–400 lesions/106 are likely due at least partly to the occurrence of artifactual oxidation which explains so far the lack of accurate data on the steady-state levels of 8-oxoGua in mitochondrial DNA. Other interesting potential applications of LC-NSI-MS/MS should focus on the search in cellular DNA of tandem base lesions that arise from the reactions of peroxyl pyrimidine radical with vicinal nucleobases. As discussed above, it remains to be established whether the occurrence of secondary oxidation products, most likely arising from one-electron oxidant-mediated reactions, is of biological significance. Another current conflicting issue that needs to be adequately addressed concerns the reported steady-state levels of purine 2’-deoxyribonucleosides that range widely between 0.2 and 10 S-cdA per 107 nucleosides, depending on the method employed [336,355]. It would be surprising that such large differences in the yield of S-cdA of up to a factor of 50 may only be explained by the diverse sources of DNA subjected to analysis.
Table 1.
Formation of oxidatively generated damage in cellular DNA.
| DNA lesions | •OH | One-e oxidant | 1O2 | TET |
|---|---|---|---|---|
| 5,6-Dihydroxy-5,6-dihydrothymine (ThyGly) | ++ | + | ||
| 5-Hydroxy-5-methylhydantoin (ThyHyd) | + | |||
| 5-Hydroxymethyluracil (5-HmUra) | ++ | + | ||
| 5-Formyluracil (5-FoUra) | ++ | + | ||
| 5-Hydroxycytosine (5-OHCyt) | + | + | ||
| 5,6-Dihydroxy-5,6-dihydrouracil (UraGly) | + | |||
| 1-Carbamoyl-4,5-dihydroxy-2-oxoimidazolidine (ImidCyt) | + | |||
| 5-Hydroxyhydantoin (UraHyd) | + | |||
| 5-Hydroxymethylcytosine (HmCyt) | + | + | +++ | |
| 5-Formylcytosine (5-FoCyt) | + | + | ++ | |
| 5-Carboxylcytosine (5-CaCyt) | + | |||
| 8-oxo-7,8-dihydroguanine (8-oxoGua) | ++ | ++ | ++ | |
| 2,6-Diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) | ++ | |||
| 8-Oxo-7,8-dihydroadenine (8-oxoAde) | + | + | ||
| 2,2,4-Triamino-5(2H)-oxazolone (Oz) | + | |||
| Guanine-cytosine adduct Interstrand-crosslink (sugar-cytosine) | + | + | ||
| (5’S)-5’,8-cyclo-2’-deoxyadenosine (cdA) | ? |
Major damage: may reach a few lesions per 104 nucleobases.
Fair damage: between 10 and 100 lesions/108 nucleobases per Gy of ionizing radiation.
Minor damage: lower than 10/108 nucleobases per Gy of ionizing radiation.
Highlights.
HPLC coupled to either ESI-tandem mass spectrometers or an electrochemical detector is appropriate for accurately monitoring oxidatively damage to cellular DNA.
Seventeen single base lesions and several intra- and interstrand cross-links have been identified in cellular DNA as chemical or enzymatic oxidation products.
Other base modifications arising from oxidative reactions included halogenated nucleobases and base adducts with reactive aldehydes from lipid peroxides.
There is paucity of reliable data on the base excision repair kinetics of oxidized bases in cells at the exception of 8-oxo-7,8-dihydroguanine.
Acknowledgments
Paolo Di Mascio thanks PFAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo, No. 2012/12663-1, CEPID Redoxoma No. 2013/07937-8), CNPq (Conselho Nacional para o Desenvolvimento Científico e Tecnológico, No. 301307/2013-0) and PRPUSP (Pro-Reitoria de Pesquisa da Universidade de São Paulo, NAP Redoxoma No. 2011.1.9352.1.8) for financial support. Kelvin J. A. Davies was supported by grant # ES003598 from the National Institute of Environmental Health Sciences of the US National Institutes of Health. J. Richard Wagner thanks the Natural Sciences and Engineering Research Council of Canada for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kalyanaraman B. Teaching the basics of redox biology to medical and graduate students: Oxidants, antioxidants and disease mechanisms. Redox Biol. 2013;1:244–257. doi: 10.1016/j.redox.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pillay CS, Eagling BD, Driscoll SR, Rohwer JM. Quantitative measures for redox signaling. Free Radic Biol Med. 2016;96:290–303. doi: 10.1016/j.freeradbiomed.2016.04.199. [DOI] [PubMed] [Google Scholar]
- 3.Jha JC, Banal C, Chow BS, Cooper ME, Jandeleit-Dahm K. Diabetes and kidney disease: role of oxidative stress. Antioxid Redox Signal. 2016 Apr 1; doi: 10.1089/ars.2016.6664. [Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perrotta I, Aquila S. The role of oxidative stress and autophagy in atherosclerosis. Oxid Med Cell Longev. 2015;2015:130315. doi: 10.1155/2015/130315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leandro GS, Sykora P, Bohr VA. The impact of base excision DNA repair in age-related neurodegenerative diseases. Mutat Res. 2015;776:31–39. doi: 10.1016/j.mrfmmm.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tafani M, Sansone L, Limana F, Arcangeli T, De Santis E, Polese M, Fini M, Russo MA. The interplay of reactive oxygen species, hypoxia, inflammation, and sirtuins in cancer initiation and progression. Oxid Med Cell Longev. 2016;2016:3907147. doi: 10.1155/2016/3907147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radi R. Peroxynitrite, a stealthy biological oxidant. J Biol Chem. 2013;288:26464–26472. doi: 10.1074/jbc.R113.472936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadet J, Sage E, Douki T. Ultraviolet radiation-mediated damage to cellular DNA. Mutat Res. 2005;571:3–17. doi: 10.1016/j.mrfmmm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Epe B. DNA damage spectra induced by photosensitization. Photochem Photobiol Sci. 2012;11:98–106. doi: 10.1039/c1pp05190c. [DOI] [PubMed] [Google Scholar]
- 10.Scholes G, Weiss J, Wheeler CM. Formation of hydroperoxides from nucleic acids by irradiation with x-rays in aqueous systems. Nature. 1956;178:157. doi: 10.1038/178157a0. [DOI] [PubMed] [Google Scholar]
- 11.Scholes G, Ward JF, Weiss J. Mechanism of the radiation-induced degradation of nucleic acids. J Mol Biol. 1960;2:379–391. doi: 10.1016/s0022-2836(60)80049-6. [DOI] [PubMed] [Google Scholar]
- 12.Scholes G. The radiation chemistry of aqueous solutions of nucleic acids and nucleoproteins. Prog Biophys Mol Biol. 1963;13:59–104. doi: 10.1016/s0079-6107(63)80014-0. [DOI] [PubMed] [Google Scholar]
- 13.Little JB, Azzam EI, de Toled SM, Nagasawa H. Bystander effects: intercellular transmission of radiation damage signals. Radiat Prot Dosimetry. 2002;99:159–162. doi: 10.1093/oxfordjournals.rpd.a006751. [DOI] [PubMed] [Google Scholar]
- 14.Widel M, Krzywon A, Gajda K, Skonieczna M, Rzeszowska-Wolny J. Induction of bystander effects by UVA, UVB, and UVC radiation in human fibroblasts and the implication of reactive oxygen species. Free Radic Biol Med. 2014;68:278–287. doi: 10.1016/j.freeradbiomed.2013.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Yakovlev VA. Role of nitric oxide in the radiation-induced bystander effect. Redox Biol. 2015;6:396–400. doi: 10.1016/j.redox.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiteside JR, McMillan TJ. A bystander effect is induced in human cells treated with UVA radiation but not UVB radiation. Radiat Res. 2009;171:204–211. doi: 10.1667/RR1508.1. [DOI] [PubMed] [Google Scholar]
- 17.Ekert B, Monier R. Structure of thymine hydroperoxide produced by x-irradiation. Nature. 1959;184:58–59. [PubMed] [Google Scholar]
- 18.Dizdaroglu M, Schulte-Frohlinde D, von Sonntag C. Radiation chemistry of DNA. II. Strand breaks and sugar release by gamma-irradiation of DNA in aqueous solution. The effect of oxygen. Z NaturforschC. 1975;30:826–828. doi: 10.1515/znc-1975-11-1225. [DOI] [PubMed] [Google Scholar]
- 19.Dizdaroglu M, Schulte-Frohlinde D, von Sonntag C. Isolation of 2-deoxy-D-erythro-pentonic acid from an alkali-labile site in gamma-irradiated DNA. Int J Radiat Biol. 1977;32:481–483. doi: 10.1080/09553007714551241. [DOI] [PubMed] [Google Scholar]
- 20.Romieu A, Bellon S, Gasparutto D, Cade J. Synthesis and UV photolysis of oligodeoxynucleotides that contain 5-(phenylthiomethyl)-2'-deoxyuridine: a specific photolabile precursor of 5-(2'-deoxyuridilyl)methyl radical. Org Lett. 2000;2:1085–1088. doi: 10.1021/ol005643y. [DOI] [PubMed] [Google Scholar]
- 21.San Pedro JM, Greenberg MM. 5,6-Dihydropyrimidine peroxyl radical reactivity in DNA. J Am Chem Soc. 2014;136:3928–393. doi: 10.1021/ja412562p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labet V, Morell C, Grand A, Cadet J, Cimino P, Barone V. Formation of cross-linked adducts between guanine and thymine mediated by hydroxyl radical and one-electron oxidation: a theoretical study. Org Biomol Chem. 2008;6:3300–3305. doi: 10.1039/b805589k. [DOI] [PubMed] [Google Scholar]
- 23.Munk BH, Burrows CJ, Schlegel HB. An exploration of mechanisms for the transformation of 8-oxoguanine to guanidinohydantoin and spiroiminodihydantoin by density functional theory. J Am Chem Soc. 2008;130:5245–5256. doi: 10.1021/ja7104448. [DOI] [PubMed] [Google Scholar]
- 24.Labet V, Morell C, Tognetti V, Syzgantseva OA, Joubert L, Jorge N, Grand A, Cadet J. Characterization of the chemical reactivity and selectivity of DNA bases through the use of DFT-based descriptors. In: De Proft F, Geerlings P, editors. Structure, Bonding and Reactivity of Heterocyclic Compounds. Berlin, Heidelberg: Springer; 2014. pp. 35–70. Vol 38 of Topics in Heterocyclic Chemistry. [Google Scholar]
- 25.Dumont E, Monari A. Understanding DNA under oxidative stress and sensitization: the role of molecular modeling. Front Chem. 2015;3:43. doi: 10.3389/fchem.2015.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forman HJ, Augusto O, Brigelius-Flohe R, Dennery PA, Kalyanaraman B, Ischiropoulos H, Mann GE, Radi R, Roberts LJ, 2nd, Vina J, Davies KJA. Even free radicals should follow some rules: a guide to free radical research terminology and methodology. Free Radic Biol Med. 2015;78:233–235. doi: 10.1016/j.freeradbiomed.2014.10.504. [DOI] [PubMed] [Google Scholar]
- 27.von Sonntag C. The Chemical Basis of Radiation Biology. Taylor Francis; London: 1987. [Google Scholar]
- 28.Cadet J, Douki T, Ravanat J-L. Oxidatively generated base damage to cellular DNA. Free Radic Biol Med. 2010;49:9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 29.Wagner JR, Cadet J. Oxidation reactions of cytosine DNA components by hydroxyl radical and one-electron oxidants in aerated aqueous solutions. Acc Chem Res. 2010;43:564–571. doi: 10.1021/ar9002637. [DOI] [PubMed] [Google Scholar]
- 30.Roginskaya M, Razskazovskiy Y, Bernhard WA. 2-Deoxyribonolactone lesions in X-ray-irradiated DNA: quantitative determination by catalytic 5-methylene-2-furanone release. Angew Chem Int Ed Engl. 2005;44:6210–6213. doi: 10.1002/anie.200501956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dedon PC. The chemical toxicology of 2-deoxyribose oxidation in DNA. Chem Res Toxicol. 2008;21:206–219. doi: 10.1021/tx700283c. [DOI] [PubMed] [Google Scholar]
- 32.Chan W, Chen B, Wang L, Taghizadeh K, Demott MS, Dedon PC. Quantification of the 2-deoxyribonolactone and nucleoside 5'-aldehyde products of 2-deoxyribose oxidation in DNA and cells by isotope-dilution gas chromatography mass spectrometry: differential effects of gamma-radiation and Fe2+-EDTA. J Am Chem Soc. 2010;132:6145–6153. doi: 10.1021/ja910928n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B, Zhou X, Taghizadeh K, Chen J, Stubbe J, Dedon PC. GC/MS methods to quantify the 2-deoxypentos-4-ulose and 3'-phosphoglycolate pathways of 4' oxidation of 2-deoxyribose in DNA: application to DNA damage produced by gamma radiation and bleomycin. Chem Res Toxicol. 2007;20:1701–1708. doi: 10.1021/tx700164y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roginskaya M, Mohseni R, Moore TJ, Bernhard WA, Razskazovskiy Y. Identification of the C4'-oxidized abasic site as the most abundant 2-deoxyribose lesion in radiation-damaged DNA using a novel HPLC-based approach. Radiat Res. 2014;181:131–137. doi: 10.1667/RR12993.1. [DOI] [PubMed] [Google Scholar]
- 35.Willson R, Wardman P, Asmus KD. Interaction of dGMP radical with cysteamine and promethazine as possible model of DNA repair. Nature. 1974;252:323–324. doi: 10.1038/252323a0. [DOI] [PubMed] [Google Scholar]
- 36.Candeias LP, Steenken S. Reaction of HO* with guanine derivatives in aqueous solution: formation of two different redox-active OH-adduct radicals and their unimolecular transformation reactions. Properties of G(-H)*. Chemistry. 2000;6:475–484. doi: 10.1002/(sici)1521-3765(20000204)6:3<475::aid-chem475>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 37.Misiaszek R, Crean C, Joffe A, Geacintov NE, Shafirovich V. Oxidative DNA damage associated with combination of guanine and superoxide radicals and repair mechanisms via radical trapping. J Biol Chem. 2004;279:32106–32115. doi: 10.1074/jbc.M313904200. [DOI] [PubMed] [Google Scholar]
- 38.Cadet J, Berger M, Buchko GW, Joshi P, Raoul S, Ravanat J-L. 2,2-Diamino-4-[(3,5-di-O-acetyl-2'-deoxy-β-D-erythro-pentofuranosyl)amino]-5-(2H)-oxazolone. J Am Chem Soc. 1994;116:7403–7404. [Google Scholar]
- 39.Das AB, Nauser T, Koppenol WH, Kettle AJ, Winterbourn CC, Nagy P. Rapid reaction of superoxide with insulin-tyrosyl radicals to generate a hydroperoxide with subsequent glutathione addition. Free Radic Biol Med. 2014;70:86–95. doi: 10.1016/j.freeradbiomed.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Michalski R, Zielonka J, Gapys E, Marcinek A, Joseph J, Kalyanaraman B. Real-time measurements of amino acid and protein hydroperoxides using coumarin boronic acid. J Biol Chem. 2014;289:22536–22553. doi: 10.1074/jbc.M114.553727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Mascio P, Martinez GR, Miyamoto S, Ronsein GE, Medeiros MHG, Cadet J. Singlet molecular oxygen: Düsseldorf - São Paulo, the Brazilian connection. Arch Biochem Biophys. 2016;595:161–175. doi: 10.1016/j.abb.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 42.Martinez GR, Ravanat J-L, Medeiros MHG, Cadet J, Di Mascio P. Synthesis of a naphthalene endoperoxide as a source of 18 O-labeled singlet oxygen for mechanistic studies. J Am Chem Soc. 2000;122:10212–10213. [Google Scholar]
- 43.Pierlot C, Aubry J-M, Briviba K, Sies H, Di Mascio P. Naphthalene endoperoxides as generators of singlet oxygen in biological media. Methods Enzymol. 2000;319:3–20. doi: 10.1016/s0076-6879(00)19003-2. [DOI] [PubMed] [Google Scholar]
- 44.Box HC, Freund HG, Budzinski EE, Wallace JC, Maccubbin AE. Free radical-induced double base lesions. Radiat Res. 1995;141:91–94. [PubMed] [Google Scholar]
- 45.Bourdat AG, Douki T, Frelon S, Gasparutto D, Cadet J. Tandem base lesions are generated by hydroxyl radical within isolated DNA in aerated aqueous solution. J Am Chem Soc. 2000;122:4549–4556. [Google Scholar]
- 46.Douki T, Rivière J, Cadet J. DNA tandem lesions containing 8-oxo-7,8-dihydroguanine and formamido residues arise from intramolecular addition of thymine peroxyl radical to guanine. Chem Res Toxicol. 2002;15:445–454. doi: 10.1021/tx0155909. [DOI] [PubMed] [Google Scholar]
- 47.Dupont C, Patel C, Ravanat J-L, Dumont E. Addressing the competitive formation of tandem DNA lesions by a nucleobase peroxyl radical: a DFT-D screening. Org Biomol Chem. 2013;11:3038–3045. doi: 10.1039/c3ob40280k. [DOI] [PubMed] [Google Scholar]
- 48.Bergeron F, Auvré F, Radicella JP, Ravanat J-L. HO* radicals induce an unexpected high proportion of tandem base lesions refractory to repair by DNA glycosylases. Proc Natl Acad Sci USA. 2010;107:5528–5533. doi: 10.1073/pnas.1000193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bourdat AG, Gasparutto D, Cadet J. Synthesis and enzymatic processing of oligodeoxynucleotides containing tandem base damage. Nucleic Acids Res. 1999;27:1015–1024. doi: 10.1093/nar/27.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh A, Joy A, Schuster GB, Douki T, Cadet; J. Selective one-electron oxidation of duplex DNA oligomers: reaction at thymines. Org Biomol Chem. 2008;6:916–928. doi: 10.1039/b717437c. [DOI] [PubMed] [Google Scholar]
- 51.Carter KN, Greenberg MM. Tandem lesions are the major products resulting from a pyrimidine nucleobase radical. J Am Chem Soc. 2003;125:13376–13378. doi: 10.1021/ja036629u. [DOI] [PubMed] [Google Scholar]
- 52.Hong IS, Carter KN, Sato K, Greenberg MM. Characterization and mechanism of formation of tandem lesions in DNA by a nucleobase peroxyl radical. J Am Chem Soc. 2007;129:4089–4098. doi: 10.1021/ja0692276. [DOI] [PubMed] [Google Scholar]
- 53.Taverna Porro ML, Greenberg MM. DNA double strand cleavage via interstrand hydrogen atom abstraction. J Am Chem Soc. 2013;135:16368–16371. doi: 10.1021/ja409513q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taverna Porro ML, Greenberg MM. Double-strand breaks from a radical commonly produced by DNA-damaging agents. Chem Res Toxicol. 2015;28:810–816. doi: 10.1021/acs.chemrestox.5b00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gregoli S, Olast M, Bertinchamps A. Free radical formation in deoxyguanosine-5'-monophosphate gamma-irradiated in frozen solution. A computer-assisted analysis of temperature-dependent ESR spectra. Radiat Res. 1977;72:201–217. [PubMed] [Google Scholar]
- 56.Hüttermann J. Solid-state radiation chemistry of DNA and its constituents. Ultramicroscopy. 1982;10:25–40. doi: 10.1016/0304-3991(82)90184-x. [DOI] [PubMed] [Google Scholar]
- 57.Crespo-Hernández CE, Close D, Gorb LM, Leszczynski J. Determination of redox potentials for the Watson-Crick base pairs, DNA nucleosides, and relevant nucleosides analogues. J Phys Chem B. 2007;111:5386–5395. doi: 10.1021/jp0684224. [DOI] [PubMed] [Google Scholar]
- 58.Denicola A, Freeman BA, Trujillo M, Radi R. Peroxynitrite reaction with carbon dioxide/bicarbonate: kinetics and influence on peroxynitrite-mediated oxidations. Arch Biochem Biophys. 1996;333:49–58. doi: 10.1006/abbi.1996.0363. [DOI] [PubMed] [Google Scholar]
- 59.Medinas DB, Cerchiaro G, Trindade DF, Augusto O. The carbonate radical and related oxidants derived from bicarbonate buffer. IUBMB Life. 2007;59:255–262. doi: 10.1080/15216540701230511. [DOI] [PubMed] [Google Scholar]
- 60.Kasai H, Yamaizumi Z, Berger M, Cadet J. Photosensitized formation of 7,8-dihydro-8-oxo-2'-deoxyguanosine (8-hydroxy-2'-deoxyguanosine) in DNA by riboflavin: a non singlet oxygen mediated reaction. J Am Chem Soc. 1992;114:9692–9694. [Google Scholar]
- 61.Perrier S, Hau J, Gasparutto D, Cadet J, Favier A, Ravanat J-L. Characterization of lysine-guanine cross-links upon one-electron oxidation of a guanine-containing oligonucleotide in the presence of a trilysine peptide. J Am Chem Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 62.Silerme S, Bobyk L, Taverna-Porro M, Cuier C, Saint-Pierre C, Ravanat J-L. DNA-polyamine cross-links generated upon one electron oxidation of DNA. Chem Res Toxicol. 2014;27:1011–1018. doi: 10.1021/tx500063d. [DOI] [PubMed] [Google Scholar]
- 63.Crean C, Uvaydov Y, Geacintov NE, Shafirovich V. Oxidation of single-stranded oligonucleotides by carbonate radical anions: generating intrastrand cross-links between guanine and thymine bases separated by cytosines. Nucleic Acids Res. 2008;36:742–755. doi: 10.1093/nar/gkm1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cadet J, Ravanat J-L, TavernaPorro M, Menoni H, Angelov D. Oxidatively generated complex DNA damage: tandem and clustered lesions. Cancer Lett. 2012;327:5–15. doi: 10.1016/j.canlet.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 65.Cadet J, Wagner JR. Oxidatively generated base damage to cellular DNA by hydroxyl radical and one-electron oxidants: similarities and differences. Arch Biochem Biophys. 2014;557:47–54. doi: 10.1016/j.abb.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 66.Madugundu GS, Wagner JR, Cadet J, Kropachev K, Yun BH, Geacintov NE, Shafirovich V. Generation of guanine-thymine cross-links in human cells by one-electron oxidation mechanisms. Chem Res Toxicol. 2013;26:1031–1033. doi: 10.1021/tx400158g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Candeias LP, Steenken S. Ionization of purine nucleosides and nucleotides and their components by 193-nm laser photolysis in aqueous solution: model studies for oxidative damage of DNA. J Am Chem Soc. 1992;114:699–704. [Google Scholar]
- 68.Banyasz A, Ketola TM, Muñoz-Losa A, Rishi S, Adhikary A, Sevilla MD, Martinez-Fernandez L, Improta R, Markovitsi D. UV-induced adenine radicals induced in DNA A-tracts: spectral and dynamical characterization. J Phys Chem Lett. 2016;22:3949–3953. doi: 10.1021/acs.jpclett.6b01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hawkins CL, Davies MJ. Hypochlorite-induced damage to DNA, RNA, and polynucleotides: formation of chloramines and nitrogen-centered radicals. Chem Res Toxicol. 2002;15:83–92. doi: 10.1021/tx015548d. [DOI] [PubMed] [Google Scholar]
- 70.Kuttappan-Nair V, Samson-Thibault F, Wagner JR. Generation of 2'-deoxyadenosine N6-aminyl radicals from the photolysis of phenylhydrazone derivatives. Chem Res Toxicol. 2010;23:48–54. doi: 10.1021/tx900268r. [DOI] [PubMed] [Google Scholar]
- 71.Summers FA, Mason RP, Ehrenshaft M. Development of immunoblotting techniques for DNA radical detection. Free Radic Biol Med. 2013;56:64–71. doi: 10.1016/j.freeradbiomed.2012.10.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhattacharjee S, Chatterjee S, Jiang J, Sinh BK, Mason RP. Detection and imaging of the free radical DNA in cells--site-specific radical formation induced by Fenton chemistry and its repair in cellular DNA as seen by electron spin resonance, immuno-spin trapping and confocal microscopy. Nucleic Acids Res. 2012;40:5477–5486. doi: 10.1093/nar/gks180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiss SJ, Test ST, Eckmann CM, Roos D, Regiani S. Brominating oxidants generated by human eosinophils. Science. 1986;234:200–203. doi: 10.1126/science.3018933. [DOI] [PubMed] [Google Scholar]
- 74.Mayeno AN, Curran AJ, Roberts RL, Foote CS. Eosinophils preferentially use bromide to generate halogenating agents. J Biol Chem. 1889;264:5660–5668. [PubMed] [Google Scholar]
- 75.Furtmüller PG, Obinger C, Hsuanyu Y, Dunford HB. Mechanism of reaction of myeloperoxidase with hydrogen peroxide and chloride ion. Eur J Biochem. 267:5858–5864. doi: 10.1046/j.1432-1327.2000.01491.x. [DOI] [PubMed] [Google Scholar]
- 76.Davies MJ, Hawkins CL, Pattison DI, Rees MD. Mammalian heme peroxidases: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:1199–1234. doi: 10.1089/ars.2007.1927. [DOI] [PubMed] [Google Scholar]
- 77.Hurst JK, Barrette WC., Jr Leukocytic oxygen activation and microbicidal oxidative toxins. Crit Rev Biochem Mol Biol. 1989;24:271–328. doi: 10.3109/10409238909082555. [DOI] [PubMed] [Google Scholar]
- 78.Henderson JP, Byun J, Heinecke JW. Molecular chlorine generated by the myeloperoxidase-hydrogen peroxide-chloride system of phagocytes produces 5-chlorocytosine in bacterial RNA. J Biol Chem. 1999;274:33440–33448. doi: 10.1074/jbc.274.47.33440. [DOI] [PubMed] [Google Scholar]
- 79.Henderson JP, Byun J, Williams MV, McCormick ML, Parks WC, Ridnour LA, Heinecke JW. Bromination of deoxycytidine by eosinophil peroxidase: a mechanism for mutagenesis by oxidative damage of nucleotide precursors. Proc Natl Acad Sci USA. 2001;98:1631–1636. doi: 10.1073/pnas.041146998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Masuda M, Suzuki T, Friesen MD, Ravanat J-L, Cadet J, Pignatelli B, Nishino H, Ohshima H. Chlorination of guanosine and other nucleosides by hypochlorous acid and myeloperoxidase of activated human neutrophils. Catalysis by nicotine and trimethylamine. J Biol Chem. 2001;276:40486–40496. doi: 10.1074/jbc.M102700200. [DOI] [PubMed] [Google Scholar]
- 81.Stanley NR, Pattison DL, Hawkins CL. Ability for hypochlorous acid and N-chloramines to chlorinate DNA and its constituents. Chem Res Toxicol. 2010;23:1293–1302. doi: 10.1021/tx100188b. [DOI] [PubMed] [Google Scholar]
- 82.Hariharan PV, Cerutti PA. Formation and repair of gamma ray induced thymine damage in Micrococcus radiodurans. J Mol Biol. 1972;66:65–81. doi: 10.1016/s0022-2836(72)80006-8. [DOI] [PubMed] [Google Scholar]
- 83.Hariharan PV, Cerutti PA. Formation of products of the 5,6-dihydroxydihydrothymine type by ultraviolet light in HeLa cells. Biochemistry. 1977;16:2791–2795. doi: 10.1021/bi00631a032. [DOI] [PubMed] [Google Scholar]
- 84.Hariharan PV, Remsen JF, Cerutti PA. Excision-repair of gamma-ray-damaged thymine in bacterial and mammalian systems. Basic Life Sci. 1975;5A:51–59. doi: 10.1007/978-1-4684-2895-7_8. [DOI] [PubMed] [Google Scholar]
- 85.Targovnik HS, Hariharan PV. Excision repair of 5,6-dihydroxydihydrothymine from the DNA of micrococcus radiodurans. Radiat Res. 1980;83:360–363. [PubMed] [Google Scholar]
- 86.Hariharan PV, Courtney J, Eleczko S. Production of hydroxyl radicals in cell systems exposed to haematoporphyrin and red light. Int J Radiat Biol. 1980;37:691–694. [PubMed] [Google Scholar]
- 87.Cadet J, Berger M. Radiation-induced decomposition of the purine bases within DNA and related model compounds. Int J Radiat Biol. 1985;47:127–143. doi: 10.1080/09553008514550201. [DOI] [PubMed] [Google Scholar]
- 88.Plongthongkum N, Diep DH, Zhang K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genet. 2014;15:647–661. doi: 10.1038/nrg3772. [DOI] [PubMed] [Google Scholar]
- 89.Shull AY, Noonepalle SK, Lee EJ, Choi JH, Shi H. Sequencing the cancer methylome. Methods Mol Biol. 2015;1238:627–651. doi: 10.1007/978-1-4939-1804-1_33. [DOI] [PubMed] [Google Scholar]
- 90.Kizaki S, Chandran A, Sugiyama H. Identification of sequence specificity of 5-methylcytosine oxidation by tet1 protein with high-throughput sequencing. Chembiochem. 2016;17:403–406. doi: 10.1002/cbic.201500646. [DOI] [PubMed] [Google Scholar]
- 91.Cadet J, Douki T, Ravanat J-L. Measurement of oxidatively generated base damage in cellular DNA. Mutat Res. 2011;711:3–12. doi: 10.1016/j.mrfmmm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 92.Cadet J, Douki T, Ravanat J-L, Wagner JR. Measurement of oxidatively generated base damage to nucleic acids in cells: facts and artefacts. Bioanal Rev. 2012;4:55–74. [Google Scholar]
- 93.Ravanat J-L, Cadet J, Douki T. Oxidatively generated DNA lesions as potential biomarkers of in vivo oxidative stress. Curr Mol Med. 2012;12:655–671. doi: 10.2174/156652412800792651. [DOI] [PubMed] [Google Scholar]
- 94.Cadet J, Wagner JR. TET enzymatic oxidation of 5-methylcytosine, 5-hydroxymethylcytosine and 5-formylcytosine. Mutat Res. 2014;764–765:18–35. doi: 10.1016/j.mrgentox.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 95.Leadon SA, Hanawalt PC. Monoclonal antibody to DNA containing thymine glycol. Mutat Res. 1983;112:191–200. doi: 10.1016/0167-8817(83)90006-8. [DOI] [PubMed] [Google Scholar]
- 96.Rajagopalan R, Melamede RJ, Laspia MF, Erlanger BF, Wallace SS. Properties of antibodies to thymine glycol, a product of the radiolysis of DNA. Radiat Res. 1984;97:499–510. [PubMed] [Google Scholar]
- 97.Ide H, Kow YW, Chen BX, Erlanger BF, Wallace SS. Antibodies to oxidative DNA damage: characterization of antibodies to 8-oxopurines. Cell Biol Toxicol. 1997;13:405–417. doi: 10.1023/a:1007467726635. [DOI] [PubMed] [Google Scholar]
- 98.Toyokuni S, Tanaka T, Hattori Y, Nishiyama Y, Yoshida A, Uchida K, Hiai H, Ochi H, Osawa T. Quantitative immunohistochemical determination of 8-hydroxy-2'-deoxyguanosine by a monoclonal antibody N45.1: its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab Invest. 1997;76:365–374. [PubMed] [Google Scholar]
- 99.Mitchell DL, Meador J, Paniker L, Gasparutto D, Jeffrey WH, Cadet J. Development and application of a novel immunoassay for measuring oxidative DNA damage in the environment. Photochem Photobiol. 2002;75:257–265. doi: 10.1562/0031-8655(2002)075<0257:daaoan>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 100.Kasai H, Nishimura S. Hydroxylation of deoxyguanosine at the C-8 position by ascorbic acid and other reducing agents. Nucleic Acids Res. 1984;12:2137–2145. doi: 10.1093/nar/12.4.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Iwamoto T, Brooks PJ, Nishiwaki T, Nishimura K, Kobayashi N, Sugiura S, Mori T. Quantitative and in situ detection of oxidatively generated DNA damage 8,5'-cyclo-2'-deoxyadenosine using an immunoassay with a novel monoclonal antibody. Photochem Photobiol. 2014;90:829–836. doi: 10.1111/php.12239. [DOI] [PubMed] [Google Scholar]
- 102.Yang Z, Jiang W, Liu F, Zhou Y, Yin H, Ai S. A novel electrochemical immunosensor for the quantitative detection of 5-hydroxymethylcytosine in genomic DNA of breast cancer tissue. Chem Commun. 2015;51:14671–14673. doi: 10.1039/c5cc05921f. [DOI] [PubMed] [Google Scholar]
- 103.Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, Koseki H, Sasaki H, Hajkova P. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol. 2016;18:225–233. doi: 10.1038/ncb3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee SF, Pervaiz S. Assessment of oxidative stress-induced DNA damage by immunoflourescent analysis of 8-oxodG. Methods Cell Biol. 2011;103:99–113. doi: 10.1016/B978-0-12-385493-3.00005-X. [DOI] [PubMed] [Google Scholar]
- 105.Sattarova EA, Sinitsyna OI, Vasyunina EA, Duzhak AB, Kolosova NG, Zharkov DO, Nevinsky GA. Age-dependent guanine oxidation in DNA of different brain regions of Wistar rats and prematurely aging OXYS rats. Biochim Biophys Acta. 2013;1830:3542–3552. doi: 10.1016/j.bbagen.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 106.Habib SL, Liang S. Hyperactivation of Akt/mTOR and deficiency in tuberin increased the oxidative DNA damage in kidney cancer patients with diabetes. Oncotarget. 2014;5:2542–2550. doi: 10.18632/oncotarget.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Y, Shi X, Zhang J, Zhang X, Martin RC. Hepatic protection and anticancer activity of curcuma: a potential chemopreventive strategy against hepatocellular carcinoma. Int J Oncol. 2014;44:505–513. doi: 10.3892/ijo.2013.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu J, Li X, Zhang M, Chen Z, Wang Y, Zeng C, Liu Z, Chen H. Regulation of MUTYH, a DNA repair enzyme, in renal proximal tubular epithelial cells. Oxid Med Cell Longev. 2015;2015:682861. doi: 10.1155/2015/682861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Suzuki M, Bandoski C, Bartlett JD. Fluoride induces oxidative damage and SIRT1/autophagy through ROS-mediated JNK signaling. Free Radic Biol Med. 2015;89:369–378. doi: 10.1016/j.freeradbiomed.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cooke MS, Olinski R, Loft S European Standards Committee on Urinary (DNA) Lesion Analysis. Measurement and meaning of oxidatively modified DNA lesions in urine. Cancer Epidemiol Biomarkers Prev. 2008;17:3–14. doi: 10.1158/1055-9965.EPI-07-0751. [DOI] [PubMed] [Google Scholar]
- 111.Barregard L, Møller P, Henriksen T, Mistry V, Koppen G, Rossner P, Jr, Sram RJ, Weimann A, Poulsen HE, Nataf R, Andreoli R, Manini P, Marczylo T, Lam P, Evans MD, Kasai H, Kawai K, Li YS, Sakai K, Singh R, Teichert F, Farmer PB, Rozalski R, Gackowski D, Siomek A, Saez GT, Cerda C, Broberg K, Lindh C, Hossain MB, Haghdoost S, Hu CW, Chao MR, Wu KY, Orhan H, Senduran N, Smith RJ, Santella RM, Su Y, Cortez C, Yeh S, Olinski R, Loft S, Cooke MS. Human and methodological sources of variability in the measurement of urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine. Antioxid Redox Signal. 2013;18:2377–2391. doi: 10.1089/ars.2012.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Møller P, Cooke MS, Collins A, Olinski R, Rozalski R, Loft S. Harmonising measurements of 8-oxo-7,8-dihydro-2'-deoxyguanosine in cellular DNA and urine. Free Radic Res. 2012;46:541–553. doi: 10.3109/10715762.2011.644241. [DOI] [PubMed] [Google Scholar]
- 113.Rossner P, Jr, Orhan H, Koppen G, Sakai K, Santella RM, Ambroz A, Rossnerova A, Sram RJ, Ciganek M, Neca J, Arzuk E, Mutlu N, Cooke MS. Urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine analysis by an improved ELISA: An inter-laboratory comparison study. Free Radic Biol Med. 2016;95:169–179. doi: 10.1016/j.freeradbiomed.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 114.Collins AR, Duthie SJ, Dobson VL. Direct enzymic detection of endogenous oxidative base damage in human lymphocyte DNA. Carcinogenesis. 1993;14:1733–1735. doi: 10.1093/carcin/14.9.1733. [DOI] [PubMed] [Google Scholar]
- 115.Pouget JP, Douki T, Richard MJ, Cadet J. DNA damage induced in cells by gamma and UVA radiation as measured by HPLC/GC-MS and HPLC-EC and Comet assay. Chem Res Toxicol. 2000;13:541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
- 116.Collins AR. The use of bacterial repair endonucleases in the comet assay. Methods Mol Biol. 2011;691:137–147. doi: 10.1007/978-1-60761-849-2_8. [DOI] [PubMed] [Google Scholar]
- 117.Azqueta A, Collins AR. The essential comet assay: a comprehensive guide to measuring DNA damage and repair. Arch. Toxicol. 2013;87:949–968. doi: 10.1007/s00204-013-1070-0. [DOI] [PubMed] [Google Scholar]
- 118.Pflaum M, Will O, Epe B. Determination of steady-state levels of oxidative DNA base modifications in mammalian cells by means of repair endonucleases. Carcinogenesis. 1997;18:2225–2231. doi: 10.1093/carcin/18.11.2225. [DOI] [PubMed] [Google Scholar]
- 119.Pflaum M, Will O, Mahler HC, Epe B. DNA oxidation products determined with repair endonucleases in mammalian cells: types, basal levels and influence of cell proliferation. Free Radic Res. 1998;29:585–594. doi: 10.1080/10715769800300631. [DOI] [PubMed] [Google Scholar]
- 120.Hartwig A, Dally H, Schlepegrell R. Sensitive analysis of oxidative DNA damage in mammalian cells: use of the bacterial Fpg protein in combination with alkaline unwinding. Toxicol Lett. 1996;88:85–90. doi: 10.1016/0378-4274(96)03722-8. [DOI] [PubMed] [Google Scholar]
- 121.Cadet J, Loft S, Olinski R, Evans MD, Bialkowski K, Wagner JR, Dedon PC, Møller P, Greenberg MM, Cooke MS. Biologically relevant oxidants and terminology, classification and nomenclature of oxidatively generated damage to nucleobases and 2-deoxyribose in nucleic acids. Free Radic Res. 2012;4:367–381. doi: 10.3109/10715762.2012.659248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Collins AR. Measuring oxidative damage to DNA and its repair with the comet assay. Biochim Biophys Acta. 2014;1840:794–80. doi: 10.1016/j.bbagen.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 123.Smith CC, O'Donovan MR, Martin EA. hOGG1 recognizes oxidative damage using the comet assay with greater specificity than FPG or ENDOIII. Mutagenesis. 2006;21:185–190. doi: 10.1093/mutage/gel019. [DOI] [PubMed] [Google Scholar]
- 124.Kohn KW, Erickson LC, Ewig RA, Friedman CA. Fractionation of DNA from mammalian cells by alkaline elution. Biochemistry. 1976;15:4629–4637. doi: 10.1021/bi00666a013. [DOI] [PubMed] [Google Scholar]
- 125.Collins A, Cadet J, Epe B, Gedik C. Problems in the measurement of 8-oxoguanine in human DNA. Report of a workshop, DNA oxidation, held in Aberdeen, UK, 19–21 January. 1997. Carcinogenesis. 18:1833–1836. doi: 10.1093/carcin/18.9.1833. [DOI] [PubMed] [Google Scholar]
- 126.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Collins AR, Cadet J, Möller L, Poulsen HE, Viña J. Are we sure we know how to measure 8-oxo-7,8-dihydroguanine in DNA from human cells? Arch Biochem Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 128.Osterod M, Hollenbach S, Hengstler JG, Barnes DE, Lindahl T, Epe B. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis. 2001;22:1459–1463. doi: 10.1093/carcin/22.9.1459. [DOI] [PubMed] [Google Scholar]
- 129.Sauvaigo S, Petec-Calin C, Caillat S, Odin F, Cadet J. Comet assay coupled to repair enzymes for the detection of oxidative damage to DNA induced by low doses of gamma-radiation: use of YOYO-1, low-background slides, and optimized electrophoresis conditions. Anal Biochem. 2002;303:107–109. doi: 10.1006/abio.2001.5559. [DOI] [PubMed] [Google Scholar]
- 130.Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- 131.Pflaum M, Kielbassa C, Garmyn M, Epe B. Oxidative DNA damage induced by visible light in mammalian cells: extent, inhibition by antioxidants and genotoxic effects. Mutat Res. 1998;408:137–146. doi: 10.1016/s0921-8777(98)00029-9. [DOI] [PubMed] [Google Scholar]
- 132.Collins A, El Yamani N, Dusinska M. Sensitive detection of oxidative damage in DNA induced by nanomaterials. Free Radic Biol Med. 2017 doi: 10.1016/j.freeradbiomed.2017.02.001. this issue. [DOI] [PubMed] [Google Scholar]
- 133.Dizdaroglu M. Application of capillary gas chromatography-mass spectrometry to chemical characterization of radiation-induced base damage of DNA: implications for assessing DNA repair processes. Anal Biochem. 1985;144:593–603. doi: 10.1016/0003-2697(85)90158-7. [DOI] [PubMed] [Google Scholar]
- 134.Dizdaroglu M. Characterization of free-radical induced damage to DNA by the combined use of enzymatic hydrolysis and gas-chromatography-mass spectrometry. J Chromatogr. 1986;367:357–366. doi: 10.1016/s0021-9673(00)94856-8. [DOI] [PubMed] [Google Scholar]
- 135.Dizdaroglu M. Free-radical-induced formation of an 8,5'-cyclo-2'-deoxyguanosine moiety in deoxyribonucleic acid. Biochem J. 1986;238:247–254. doi: 10.1042/bj2380247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dizdaroglu M, Dirksen ML, Jiang HX, Robbins JH. Ionizing-radiation-induced damage in the DNA of cultured human cells: identification of 8,5-cyclo-2-deoxyguanosine. Biochem J. 1987;241:929–932. doi: 10.1042/bj2410929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dizdaroglu M, Nackerdien Z, Chao BC, Gajewski E, Ra G. Chemical nature of in vivo DNA damage in hydrogen peroxide-treated mammalian cells. Arch Biochem Biophys. 1991;285:388–390. doi: 10.1016/0003-9861(91)90378-v. [DOI] [PubMed] [Google Scholar]
- 138.Halliwell B, Dizdaroglu M. The measurement of oxidative damage to DNA by HPLC and GC-MS techniques. Free Radic Res Commun. 1992;16:75–87. doi: 10.3109/10715769209049161. [DOI] [PubMed] [Google Scholar]
- 139.Cadet J, Douki T, Ravanat J-L. Artifacts associated with the measurement of oxidized DNA bases. Environ Health Perspect. 1997;105:1034–1039. doi: 10.1289/ehp.105-1470384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Douki T, Martini R, Ravanat J-L, Turesky RJ, Cadet J. Measurement of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 8-oxo-7,8-dihydroguanine in isolated DNA exposed to gamma radiation in aqueous solution. Carcinogenesis. 1997;18:2385–2391. doi: 10.1093/carcin/18.12.2385. [DOI] [PubMed] [Google Scholar]
- 141.Jaruga P, Speina E, Gackowski D, Tudek B, Olinski R. Endogenous oxidative DNA base modifications analysed with repair enzymes and GC/MS technique. Nucleic Acids Res. 2000;28:E16. doi: 10.1093/nar/28.6.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gassman NR, Coskun E, Stefanick DF, Horton JK, Jaruga P, Dizdaroglu M, Wilson SH. Bisphenol a promotes cell survival following oxidative DNA damage in mouse fibroblasts. PLoS One. 2015;10:e0118819. doi: 10.1371/journal.pone.0118819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gassman NR, Coskun E, Jaruga P, Dizdaroglu M, Wilson SH. Combined effects of high-dose bisphenol a and oxidizing agent (KBrO3) on cellular microenvironment, gene expression, and chromatin structure of Ku70-deficient mouse embryonic fibroblasts. Environ Health Perspect. 2016;124:1241–1252. doi: 10.1289/EHP237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Malins DC, Hellstrom KE, Anderson KM, Johnson PM, Vinson MA. Antioxidant-induced changes in oxidized DNA. Proc Natl Acad Sci USA. 2002;99:5937–5941. doi: 10.1073/pnas.082111199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Malins DC, Johnson PM, Barker EA, Polissar NL, Wheeler TM, Anderson KM. Cancer-related changes in prostate DNA as men age and early identification of metastasis in primary prostate tumors. Proc Natl Acad Sc USA. 2003;100:5401–5406. doi: 10.1073/pnas.0931396100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jaruga P, Dizdaroglu M. Repair of products of oxidative DNA base damage in human cells. Nucleic Acids Res. 1996;24:1389–1394. doi: 10.1093/nar/24.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lipinski LJ, Hoehr N, Mazur SJ, Dianov GL, Sentürker S, Dizdaroglu M, Bohr VA. Repair of oxidative DNA base lesions induced by fluorescent light is defective in xeroderma pigmentosum group A cells. Nucleic Acids Res. 1999;27:3153–3158. doi: 10.1093/nar/27.15.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Arczewska KD, Tomazella GG, Lindvall JM, Kassahun H, Maglioni S, Torgovnick A, Henriksson J, Matilainen O, Marquis BJ, Nelson BC, Jaruga P, Babaie E, Holmberg CI, Bürglin TR, Ventura N, Thiede B, Nilsen H. Active transcriptomic and proteomic reprogramming in the C. elegans nucleotide excision repair mutant xpa-1. Nucleic Acids Res. 2013;41:5368–5381. doi: 10.1093/nar/gkt225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Floyd RA, Watson JJ, Wong PK, Altmiller DH, Rickard RC. Hydroxyl free radical adduct of deoxyguanosine: sensitive detection and mechanism of formation. Free Radic Res Commun. 1986;1:163–172. doi: 10.3109/10715768609083148. [DOI] [PubMed] [Google Scholar]
- 150.Floyd RA, Watson JJ, Harris J, West M, Won PK. Formation of 8-hydroxydeoxyguanosine, hydroxyl free radical adduct of DNA in granulocytes exposed to the tumor promoter, tetradecanoylphorbol acetate. Biochem Biophys Res Commun. 1986;137:841–846. doi: 10.1016/0006-291x(86)91156-3. [DOI] [PubMed] [Google Scholar]
- 151.Kasai H, Crain PF, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- 152.Kasai H, Nishimura S, Kurokawa Y, Hayashi Y. Oral administration of the renal carcinogen, potassium bromate, specifically produces 8-hydroxydeoxyguanosine in rat target organ DNA. Carcinogenesis. 1987;8:1959–1961. doi: 10.1093/carcin/8.12.1959. [DOI] [PubMed] [Google Scholar]
- 153.Gedik CM, Collins AR ESCODD. Establishing the background level of base oxidation in human lymphocyte DNA: results of an inter-laboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 154.Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc Natl Acad Sci USA. 1998;95:288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wagner JR, Hu CC, Ames BN. Endogenous oxidative damage of deoxycytidine in DNA. Proc Natl Acad Sci USA. 1992;89:3380–3384. doi: 10.1073/pnas.89.8.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kaur H, Halliwell B. Measurement of oxidized and methylated DNA bases by HPLC with electrochemical detection. Biochem J. 1996;318(Pt 1):21–23. doi: 10.1042/bj3180021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kasai H. Analysis of a form of oxidative DNA damage, 8-hydroxy-2'-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat Res. 1997;387:147–163. doi: 10.1016/s1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]
- 158.Tuo J, Jaruga P, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J Biol Chem. 2002;277:30832–30837. doi: 10.1074/jbc.M204814200. [DOI] [PubMed] [Google Scholar]
- 159.Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- 160.Pouget J-P, Frelon S, Ravanat J-L, Testard I, Odin F, Cadet J. Formation of modified DNA bases in cells exposed either to gamma radiation or to high-LET particles. Radiat Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 161.Douki T, Ravanat J-L, Pouget J-P, Testard I, Cadet J. Minor contribution of direct ionization to DNA base damage induced by heavy ions. Int J Radiat Biol. 2006;82:119–127. doi: 10.1080/09553000600573788. [DOI] [PubMed] [Google Scholar]
- 162.D’Errico M, Parlanti E, Teson M, Bernades de Jesus BM, Degan P, Calcagnile A, Jaruga P, Bjørås M, Crescenzi M, Pedrini AM, Egly J-M, Zambruno G, Stefanini M, Dizdaroglu M, Dogliotti E. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 2006;25:4305–4315. doi: 10.1038/sj.emboj.7601277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem Res Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Münzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, Müller M, Biel M, Carell T. Quantification of the sixth DNA base hydroxymethylcytosine in the brain. Angew Chem Int Ed Engl. 2010;49:5375–5377. doi: 10.1002/anie.201002033. [DOI] [PubMed] [Google Scholar]
- 165.Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koc S, Brückl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pfaffeneder T, Hackner B, Truss M, Münzel M, Müller M, Deiml CA, Hagemeier C, Carell T. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 167.Gackowski D, Zarakowska E, Starczak M, Modrzejewska M, Olinski R. Tissue-specific differences in DNA modifications (5-hydroxymethylcytosine, 5-formylcytosine, 5-carboxylcytosine and 5-hydroxymethyluracil) and their interrelationships. PLoS One. 2015;10:e0144859. doi: 10.1371/journal.pone.0144859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cadet J, Poulsen H. Measurement of oxidatively generated base damage in cellular DNA and urine. Free Radic Biol Med. 2010;48:1457–1459. doi: 10.1016/j.freeradbiomed.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 169.Tretyakova N, Villalta PW, Kotapati S. Mass spectrometry of structurally modified DNA. Chem Rev. 2013;113:2395–2436. doi: 10.1021/cr300391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liu S, Wang Y. Mass spectrometry for the assessment of the occurrence and biological consequences of DNA adducts. Chem Soc Rev. 2015;44:7829–7854. doi: 10.1039/c5cs00316d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Poulsen HE, Nada LL, Broedbaek K, Nielse PE, Weimann A. Detection and interpretation of 8-oxodG and 8-oxoGua in urine, plasma and cerebrospinal fluid. Biochim Biophys Acta. 2014;1840:801–808. doi: 10.1016/j.bbagen.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 172.Hu CW, Cooke MS, Tsai YH, Chao MR. 8-Oxo-7,8-dihydroguanine and 8-oxo-7,8-dihydro-2'-deoxyguanosine concentrations in various human body fluids: implications for their measurement and interpretation. Arch Toxicol. 2015;89:201–210. doi: 10.1007/s00204-014-1255-1. [DOI] [PubMed] [Google Scholar]
- 173.Frelon S, Douki T, Ravanat J-L, Pouget J-P, Tornabene C, Cadet J. High-performance liquid chromatography-tandem mass spectrometry measurement of radiation-induced base damage to isolated and cellular DNA. Chem Res Toxicol. 2000;13:1002–1010. doi: 10.1021/tx000085h. [DOI] [PubMed] [Google Scholar]
- 174.Glintborg B, Weimann A, Kensler TW, Poulsen HE. Otipraz chemoprevention trial in Qidong, People's Republic of China: unaltered oxidative biomarkers. Free Radic Biol Med. 2006;41:1010–1014. doi: 10.1016/j.freeradbiomed.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 175.Singh R, Teichert F, Verschoyle RD, Kaur B, Vives M, Sharma RA, Steward WP, Gescher AJ, Farmer PB. Simultaneous determination of 8-oxo-2'-deoxyguanosine and 8-oxo-2'-deoxyadenosine in DNA using online column-switching liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:151–160. doi: 10.1002/rcm.3866. [DOI] [PubMed] [Google Scholar]
- 176.Serrano J, Palmeira CM, Wallace KB, Kuehl DW. Determination of 8-hydroxydeoxyguanosine in biological tissue by liquid chromatography/electrospray ionization-mass spectrometry/mass spectrometry. Rapid Commun Mass Spectrom. 1996;10:1789–1791. doi: 10.1002/(SICI)1097-0231(199611)10:14<1789::AID-RCM752>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 177.Ravanat J-L, Duretz B, Guiller A, Douki T, Cadet J. Isotope dilution high-performance liquid chromatography-electrospray tandem mass spectrometry assay for the measurement of 8-oxo-7,8-dihydro-2'-deoxyguanosine in biological samples. J Chromatogr B. 1998;715:349–356. doi: 10.1016/s0378-4347(98)00259-x. [DOI] [PubMed] [Google Scholar]
- 178.Ravanat J-L, Douki T, Duez P, Gremaud E, Herbert K, Hofer T, Lasserre L, Saint-Pierre C, Favier A, Cadet J. Cellular background level of 8-oxo-7,8-dihydro-2'-deoxyguanosine: an isotope based method to evaluate artefactual oxidation of DNA during its extraction and subsequent work-up. Carcinogenesis. 2012;23:1911–1918. doi: 10.1093/carcin/23.11.1911. [DOI] [PubMed] [Google Scholar]
- 179.Chao MR, Yen CC, Hu CW. Prevention of artifactual oxidation in determination of cellular 8-oxo-7,8-dihydro-2’-deoxyguanosine by isotope-dilution LC-MS/MS with automated solid-phase extraction. Free Radic Biol Med. 2008;44:464–473. doi: 10.1016/j.freeradbiomed.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 180.Mangal D, Vudathala D, Park J-H, Lee SH, Penning TM, Blair LA. Analysis of 7,8-dihydro-8-oxo-2’-deoxyguanosine in cellular DNA during oxidative stress. Chem Res Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Badouard C, Ménézo Y, Panteix G, Ravanat J-L, Douki T, Cadet J, Favier A. Determination of new types of DNA lesions in human sperm. Zygote. 2008;16:9–13. doi: 10.1017/S0967199407004340. [DOI] [PubMed] [Google Scholar]
- 182.Evans MD, Mistry V, Singh R, Gackowski D, Różalski R, Siomek-Gorecka A, Phillips DH, Zuo J, Mullender L, Pines A, Nakabeppu Y, Sakumi K, Sekiguchi M, Tsuzuki T, Bignami M, Oliński R, Cooke MS. Nucleotide excision repair of oxidised genomic DNA is not a source of urinary 8-oxo-7,8-dihydro-2'-deoxyguanosine. Free Radic Biol Med. 2016;99:385–391. doi: 10.1016/j.freeradbiomed.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 183.Ravanat J-L, Saint-Pierre C, Di Mascio P, Martinez GR, Medeiros MHG, Cadet J. Damage to isolated DNA mediated by singlet oxygen. Helv Chim Acta. 2001;84:3702–3709. [Google Scholar]
- 184.Cadet J, Di Mascio P. In: Peroxides in biological systems. Functional Groups in Organic Chemistry – The Chemistry of Peroxides. Rappoport Z, editor. Vol. 2. Wiley and Sons; New York: 2006. pp. 915–1000. [Google Scholar]
- 185.Martinez GR, Medeiros MHG, Ravanat J-L, Cadet J, Di Mascio P. Naphthalene endoperoxide as a source of [18O]-labeled singlet oxygen for oxidative DNA damage studies. Trends Photochem Photobiol. 2002;9: 25–39. [Google Scholar]
- 186.Cadet J, Douki T, Ravanat J-L. Oxidatively generated damage to the guanine moiety of DNA: Mechanistic aspects and formation in cells. Acc Chem Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 187.Sheu C, Foote CS. Endoperoxide formation in a guanosine derivative. J Am Chem Soc. 1993;115:10446–10447. [Google Scholar]
- 188.Sheu C, Kang P, Khan S, Foote CS. Low-temperature photosensitized oxidation of a guanosine derivative and formation of an imidazole ring-opened product. J Am Chem Soc. 2002;124:3905–3913. doi: 10.1021/ja011696e. [DOI] [PubMed] [Google Scholar]
- 189.Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin is the major product of 8-oxo 7, 8-dihydroguanosine with peroxynitrite in the presence of thiols and guanosine oxidation by methylene blue. Org Lett. 2001;3:963–936. [PubMed] [Google Scholar]
- 190.Martinez GR, Medeiros MHG, Ravanat J-L, Cadet J, Di Mascio P. [18O]-Labeled singlet oxygen as a tool for mechanistic studies of 8-oxo-7,8-dihydroguanine oxidative damage: detection of spiroiminodihydantoin, imidazolone and oxazolone derivatives. Biol Chem. 2002;383: 607–617. doi: 10.1515/BC.2002.063. [DOI] [PubMed] [Google Scholar]
- 191.Ravanat J-L, Martinez GR, Medeiros MHG, Di Mascio P, Cadet J. Singlet oxygen oxidation of 2'-deoxyguanosine. Formation and mechanistic insights. Tetrahedron. 2006;62: 10709–10715. [Google Scholar]
- 192.Fleming AM, Burrows CJ. G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA. Chem Res Toxicol. 2013;26:593–607. doi: 10.1021/tx400028y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ye Y, Muller JG, Luo W, Mayne CL, Shallop AJ, Jones RA, Burrows CJ. Formation of 13C-, 15N-, and 18O-labeled guanidinohydantoin from guanosine oxidation with singlet oxygen. Implications for structure and mechanism. J Am Chem Soc. 2003;125:13926–13927. doi: 10.1021/ja0378660. [DOI] [PubMed] [Google Scholar]
- 194.Sies H, Menck CF. Singlet oxygen induced DNA damage. Mutat Res. 1992;275: 367–375. doi: 10.1016/0921-8734(92)90039-r. [DOI] [PubMed] [Google Scholar]
- 195.Sies H. Damage to plasmid DNA by singlet oxygen and its protection. Mutat Res. 1993;299: 183–191. doi: 10.1016/0165-1218(93)90095-u. [DOI] [PubMed] [Google Scholar]
- 196.Raoul S, Cadet J. Photosensitized reaction of 8-oxo-7,8-dihydro-2‘-deoxyguanosine: identification of 1-(2-Deoxy-β-d-erythro-pentofuranosyl)cyanuric acid as the major singlet oxygen oxidation product. J Am Chem Soc. 1996;118: 1892–1898. [Google Scholar]
- 197.Ravanat J-L, Martinez GR, Medeiros MHG, Di Mascio P, Cadet J. Mechanistic aspects of the oxidation of DNA constituents mediated by singlet molecular oxygen. Arch Biochem Biophys. 2004;423: 23–30. doi: 10.1016/j.abb.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 198.Martinez GR, Ravanat J-L, Cadet J, de Medeiro MH, Di Mascio P. Spiroiminodihydantoin nucleoside formation from 2'-deoxyguanosine oxidation by [(18)O-labeled] singlet molecular oxygen in aqueous solution. J Mass Spectrom. 2007;42:1326–1332. doi: 10.1002/jms.1213. [DOI] [PubMed] [Google Scholar]
- 199.Ravanat JL, Di Mascio P, Martinez GR, Medeiros MHG, Cadet J. Singlet oxygen induces oxidation of cellular DNA. J Biol Chem. 2000;275:40601–40604. doi: 10.1074/jbc.M006681200. [DOI] [PubMed] [Google Scholar]
- 200.Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31:106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- 201.Ravanat J-L, Sauvaigo S, Caillat S, Martinez GR, Medeiros MHG, Di Mascio P, Favier A, Cadet J. Singlet-oxygen-mediated damage to cellular DNA determined by the comet assay associated with DNA repair enzymes. Biol Chem. 2004;385:7–20. doi: 10.1515/BC.2004.003. [DOI] [PubMed] [Google Scholar]
- 202.Cadet J, Douki T, Ravanat J-L. Oxidatively generated damage to cellular DNA by UVB and UVA radiation. Photochem Photobiol. 2015;91:140–155. doi: 10.1111/php.12368. [DOI] [PubMed] [Google Scholar]
- 203.Mouret S, Baudouin C, Charveron M, Favier A, Cadet J, Douki T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc Natl Acad Sci USA. 2006;103:13765–13770. doi: 10.1073/pnas.0604213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cadet J, Douki T, Ravanat J-L, Di Mascio P. Sensitized formation of oxidatively generated damage to cellular DNA by UV radiation. Photochem Photobiol Sci. 2009;8:903–911. doi: 10.1039/b905343n. [DOI] [PubMed] [Google Scholar]
- 205.Cadet J, Mouret S, Ravanat J-L, Douki T. Photoinduced damage to cellular DNA: direct and photosensitized reaction. Photochem Photobiol. 2012;88: 1048–1065. doi: 10.1111/j.1751-1097.2012.01200.x. [DOI] [PubMed] [Google Scholar]
- 206.Cadet J, Douki T, Pouget J-P, Ravanat J-L. Singlet oxygen DNA damage products: formation and measurement. Methods Enzymol. 2000;319: 143–153. doi: 10.1016/s0076-6879(00)19016-0. [DOI] [PubMed] [Google Scholar]
- 207.Mouret S, Forestier A, Douki T. The specificity of UVA-induced DNA damage in human melanocytes. Photochem Photobiol Sci. 2012;11:155–162. doi: 10.1039/c1pp05185g. [DOI] [PubMed] [Google Scholar]
- 208.O'Donovan P, Perrett CM, Zhang X, Montaner B, Xu YZ, Harwood CA, McGregor JM, Walker SL, Hanaoka F, Karran P. Azathioprine and UVA light generate mutagenic oxidative DNA damage. Science. 2005;309:1871–1874. doi: 10.1126/science.1114233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhang X, Jeffs G, Ren X, O'Donovan P, Montaner B, Perrett CM, Karran P, Xu YZ. Novel DNA lesions generated by the interaction between therapeutic thiopurines and UVA light. DNA Repair (Amst) 2007;6:344–354. doi: 10.1016/j.dnarep.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 210.Brem R, Karran P. Multiple forms of DNA damage caused by UVA photoactivation of DNA 6-thioguanine. Photochem Photobiol. 2012;88:5–13. doi: 10.1111/j.1751-1097.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- 211.Cooke MS, Duarte TL, Cooper D, Chen J, Nandagopal S, Evans MD. Combination of azathioprine and UVA irradiation is a major source of cellular 8-oxo-7,8-dihydro-2'-deoxyguanosine. DNA Repair (Amst) 2008;7:1982–1989. doi: 10.1016/j.dnarep.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 212.Pollum M, Ortiz-Rodríguez LA, Jockusch S, Crespo-Hernández CE. The triplet state of 6-thio-2'-deoxyguanosine: Intrinsic properties and reactivity toward molecular oxygen. Photochem Photobiol. 2016;92:286–292. doi: 10.1111/php.12563. [DOI] [PubMed] [Google Scholar]
- 213.Zhang Y, Zhu X, Smith J, Haygood MT, Gao R. Direct observation and quantitative characterization of singlet oxygen in aqueous solution upon UVA excitation of 6-thioguanines. J Phys Chem B. 2011;115:1889–1894. doi: 10.1021/jp109590t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Ren X, Li F, Jeffs G, Zhang X, Xu YZ, Karran P. Guanine sulphinate is a major stable product of photochemical oxidation of DNA 6-thioguanine by UVA irradiation. Nucleic Acids Res. 2010;38:1832–1840. doi: 10.1093/nar/gkp1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zou X, Zhao H, Yu Y, Su H. Formation of guanine-6-sulfonate from 6-thioguanine and singlet oxygen: a combined theoretical and experimental study. J Am Chem Soc. 2013;135:4509–4515. doi: 10.1021/ja400483j. [DOI] [PubMed] [Google Scholar]
- 216.Madugundu GS, Cadet J, Wagner JR. Hydroxyl-radical-induced oxidation of 5-methylcytosine in isolated and cellular DNA. Nucleic Acids Res. 2014;42:7450–7460. doi: 10.1093/nar/gku334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol. 2013;5:a012559. doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.von Sonntag C. Free-Radical-Induced DNA Damage and Its Repair. Springer-Verlag; Berlin, Heidelberg: 2006. [Google Scholar]
- 219.Tremblay S, Douki T, Cadet J, Wagner JR. 2'-Deoxycytidine glycols, a missing link in the free radical-mediated oxidation of DNA. J Biol Chem. 1999;274:20833–20838. doi: 10.1074/jbc.274.30.20833. [DOI] [PubMed] [Google Scholar]
- 220.Tremblay S, Wagner JR. Dehydration, deamination and enzymatic repair of cytosine glycols from oxidized poly(dG-dC) and poly(dI-dC) Nucleic Acids Res. 2008;36:284–293. doi: 10.1093/nar/gkm1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tremblay S, Gantchev T, Tremblay L, Lavigne P, Cadet J, Wagner JR. Oxidation of 2'-deoxycytidine to four interconverting diastereomers of N1-carbamoyl-4,5-dihydroxy-2-oxoimidazolidine nucleosides. J Org Chem. 2007;72:3672–3678. doi: 10.1021/jo062386n. [DOI] [PubMed] [Google Scholar]
- 222.Samson-Thibault F, Madugundu GS, Gao S, Cadet J, Wagner JR. Profiling cytosine oxidation in DNA by LC-MS/MS. Chem Res Toxicol. 2012;25:1902–1911. doi: 10.1021/tx300195f. [DOI] [PubMed] [Google Scholar]
- 223.Steenken S. Purine bases, nucleosides, and nucleotides: aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts. Chem Rev. 1989;89:503–520. [Google Scholar]
- 224.Matter B, Malejka-Giganti D, Csallany AS, Tretyakova N. Quantitative analysis of the oxidative DNA lesion, 2,2-diamino-4-[(-2-deoxy-β-D-erythro-pentofuranosyl)amino]-5(2H)-oxazolone (oxazolone) in vitro and in vivo by isotope dilution-capillary HPLC-ESI-MS/MS. Nucleic Acids Res. 2006;34:5449–5460. doi: 10.1093/nar/gkl596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Alshykhly OR, Fleming AM, Burrows CJ. 5-Carboxamido-5-formamido-2-iminohydantoin, in addition to 8-oxo-7,8-dihydroguanine, is the major product of the iron-Fenton or X-ray radiation-induced oxidation of guanine under aerobic reducing conditions in nucleoside and DNA contexts. J Org Chem. 2015;80:6996–7007. doi: 10.1021/acs.joc.5b00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mori T, Hori Y, Dizdaroglu M. DNA base damage generated in vivo in hepatic chromatin of mice upon whole body γ–irradiation. Int J Radiat Biol. 1993;64:645–650. doi: 10.1080/09553009314551881. [DOI] [PubMed] [Google Scholar]
- 227.Frelon S, Douki T, Cadet J. Radical oxidation of the adenine moiety of nucleoside and DNA: 2-hydroxy-2'-deoxyadenosine is a minor decomposition product. Free Radic Res. 2002;36:499–508. doi: 10.1080/10715760290025889. [DOI] [PubMed] [Google Scholar]
- 228.Wagner JR, van Lier JE, Berger M, Cadet J. Synthesis, NMR and decomposition of thymine hydroperoxides: Primary products of radical oxidation of thymidine and DNA. J Am Chem Soc. 1994;116:2235–2242. [Google Scholar]
- 229.Cadet J, Berger M, Douki T, Ravanat J-L. Oxidative damage to DNA: Formation, measurement and biological significance. Reviews Physiol Biochem Pharmacol. 1997;131:1–8. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 230.Genereux JC, Barton JK. Mechanisms for DNA charge transport. Chem Rev. 2010;110:1642–1662. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kanvah S, Joseph J, Schuster GB, Barnett RN, Cleveland CL, Landman U. Oxidation of DNA: damage to nucleobases. Acc Chem Res. 2010;43:280–287. doi: 10.1021/ar900175a. [DOI] [PubMed] [Google Scholar]
- 232.Kawai K, Majima T. Hole transfer kinetics of DNA. Acc Chem Res. 2013;46:2616–2625. doi: 10.1021/ar400079s. [DOI] [PubMed] [Google Scholar]
- 233.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solutions. J Am Chem Soc. 1997;119: 617–618. [Google Scholar]
- 234.Bernstein R, Prat F, Foote CS. On the mechanism of DNA cleavage by fullerenes investigated in model systems. Electron transfer from guanosine and 8-oxoguanosine to C60. J Am Chem Soc. 1999;121:464–465. [Google Scholar]
- 235.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 236.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem Res Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 237.Luo W, Mulle JG, Burrow CJ. The pH-dependent role of superoxide in riboflavin-catalyzed photooxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2001;3:2801–2804. doi: 10.1021/ol0161763. [DOI] [PubMed] [Google Scholar]
- 238.Ravanat J-L, Saint-Pierre C, Cadet J. One-electron oxidation of the guanine moiety of 2'-deoxyguanosine: influence of 8-oxo-7,8-dihydro-2'-deoxyguanosine. J Am Chem Soc. 2003;125:2030–2031. doi: 10.1021/ja028608q. [DOI] [PubMed] [Google Scholar]
- 239.Rivière J, Bergeron F, Tremblay S, Gasparutto D, Cadet J, Wagner JR. Oxidation of 5-hydroxy-2'-deoxyuridine into isodialuric acid, dialuric acid, and hydantoin products. J Am Chem Soc. 2004;126:6548–6549. doi: 10.1021/ja049438f. [DOI] [PubMed] [Google Scholar]
- 240.Rivière J, Klarskov K, Wagner JR. Oxidation of 5-hydroxypyrimidine nucleosides to 5-hydroxyhydantoin and its alpha-hydroxy-ketone isomer. Chem Res Toxicol. 2005;18:1332–1338. doi: 10.1021/tx050121i. [DOI] [PubMed] [Google Scholar]
- 241.Crean C, Geacintov NE, Shafirovich V. Oxidation of 8-oxo-7,8-dihydroguanine by carbonate radical anions; insight from oxygen-18 labeling experiments. Angew Chem Int Ed Engl. 2005;44:5057–5060. doi: 10.1002/anie.200500991. [DOI] [PubMed] [Google Scholar]
- 242.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 243.McCallum JEB, Kuniyoshi CY, Foote CS. Characterization of 5-hydroxy-8-oxo-7,8-dihydroguanosine in the photosensitized oxidation of 8-oxo-7,8-dihydroguanosine and its rearrangement to spiroiminodihydantoin. J Am Chem Soc. 2004;126:16777–16782. doi: 10.1021/ja030678p. [DOI] [PubMed] [Google Scholar]
- 244.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ye Y, Munk BH, Muller JG, Cogbill A, Burrows CJ, Schlegel HB. Mechanistic aspects of the formation of guanidinohydantoin from spiroiminodihydantoin under acidic conditions. Chem Res Toxicol. 2009;22:526–535. doi: 10.1021/tx800402y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 247.Delaney S, Delaney JC, Essigmann JM. Chemical-biological fingerprinting: probing the properties of DNA lesions formed by peroxynitrite. Chem Res Toxicol. 2007;20:1718–1729. doi: 10.1021/tx700273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Leipold MD, Workman H, Muller JG, Burrows CJ, David SS. Biochemistry 2003 Sep Recognition and removal of oxidized guanines in duplex DNA by the base excision repair enzymes hOGG1, yOGG1, and yOGG2. Biochemistry. 2003;42:11373–11381. doi: 10.1021/bi034951b. [DOI] [PubMed] [Google Scholar]
- 249.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 250.Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43:4039–4954. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Shafirovich V, Kropachev K, Anderson T, Liu Z, Kolbanovskiy M, Martin BD, Sugden K, Shim Y, Chen X, Min JH, Geacintov NE. Base and nucleotide excision repair of oxidatively generated guanine lesions in DNA. J Biol Chem. 2016;291:5309–5319. doi: 10.1074/jbc.M115.693218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Fleming AM, Burrows CJ. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic Biol Med. 2017 doi: 10.1016/j.freeradbiomed.2016.11.030. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Karwowski B, Dupeyrat F, Bardet M, Ravanat J-L, Krajewski P, Cadet J. Nuclear magnetic resonance studies of the 4R and 4S diastereomers of spiroiminodihydantoin 2'-deoxyribonucleosides: absolute configuration and conformational features. Chem Res Toxicol. 2006;19:1357–1365. doi: 10.1021/tx060088f. [DOI] [PubMed] [Google Scholar]
- 254.Fleming AM, Oren AM, He Y, Zhu, Dukor RK, Burrows CJ. Reconciliation of chemical, enzymatic, spectroscopic and computational data to assign the absolute configuration of the DNA base lesion spiroiminodihydantoin. J Am Chem Soc. 2013;135:18191–18204. doi: 10.1021/ja409254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Mangerich A, Dedon PC, Fox JG, Tannenbaum SR, Wogan GN. Chemistry meets biology in colitis-associated carcinogenesis. Free Radic Res. 2013;47:958–986. doi: 10.3109/10715762.2013.832239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Friedman KA, Keller A. On the non-uniform distribution of guanine in introns of human genes: possible protection of exons against oxidation by proximal introns poly-G sequences. J Phys Chem B. 2001;105:11859–11865. [Google Scholar]
- 258.Friedman KA, Keller A. Guanosine distribution and oxidation resistance in eight eukaryotic genomes. J Am Chem Soc. 2004;126:2368–237. doi: 10.1021/ja038217r. [DOI] [PubMed] [Google Scholar]
- 259.Kanvah S, Schuster GB. The sacrificial role of easily oxidizable sites in the protection of DNA from damage. Nucleic Acids Res. 2005;33:5133–5138. doi: 10.1093/nar/gki801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Winterbourn CC, Kettle AJ. Biomarkers of myeloperoxidase-derived hypochlorous acid. Free Radic Biol Med. 2000;29:403–409. doi: 10.1016/s0891-5849(00)00204-5. [DOI] [PubMed] [Google Scholar]
- 261.Hawkins CL, Davies MJ. Hypochlorite-induced damage to DNA, RNA, and polynucleotides: formation of chloramines and nitrogen-centered radicals. Chem Res Toxicol. 2002;15:83–92. doi: 10.1021/tx015548d. [DOI] [PubMed] [Google Scholar]
- 262.Gomez-Mejiba SE, Zhai Z, Gimenez MS, Ashby MT, Chilakapati J, Kitchin K, Mason RP, Ramirez DC. Myeloperoxidase-induced genomic DNA-centered radicals. J Biol Chem. 2010;285:20062–20071. doi: 10.1074/jbc.M109.086579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Winterbourn CC, Kettle AJ, Hampton MB. Reactive oxygen species and neutrophil function. Annu Rev Biochem. 2016;85:765–792. doi: 10.1146/annurev-biochem-060815-014442. [DOI] [PubMed] [Google Scholar]
- 264.Badouard C, Masuda M, Nishino H, Cadet J, Favier A, Ravanat J-L. Detection of chlorinated DNA and RNA nucleosides by HPLC coupled to tandem mass spectrometry as potential biomarkers of inflammation. J Chromatogr B. 2005;827:26–31. doi: 10.1016/j.jchromb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 265.Badouard C, Douki T, Faure P, Halimi S, Cadet J, Favier A, Ravanat J-L. DNA lesions as biomarkers of inflammation and oxidative stress: a preliminary evaluation. In: Grune T, editor. Free Radicals and Diseases, Gene Expression, Cellular Metabolism and Pathophysiology. IOS Press; 2005. pp. 1–9. [Google Scholar]
- 266.Fedeles BI, Freudenthal BD, Yau E, Singh V, Chang SC, Li D, Delaney JC, Wilson SH, Essigmann JM. Intrinsic mutagenic properties of 5-chlorocytosine: A mechanistic connection between chronic inflammation and cancer. Proc Natl Acad Sci USA. 2015;112:E4571–4580. doi: 10.1073/pnas.1507709112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Knutson CG, Mangerich A, Zeng Y, Raczynski AR, Liberman RG, Kang P, Ye W, Prestwich EG, Lu K, Wishnok JS, Korzenik JR, Wogan GN, Fox JG, Dedon PC, Tannenbaum SR. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proc Natl Acad Sci USA. 2013;110:E2332–E2341. doi: 10.1073/pnas.1222669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.West JD, Marnett LJ. Reactive intermediates as modulators of cell signaling and cell death. Chem Res Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 269.Nair N, Bartsch H, Nair H. Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: A review of published adduct types and levels in humans. Free Radic Biol Med. 2007;43:1109–1120. doi: 10.1016/j.freeradbiomed.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 270.Blair IA. DNA adducts with lipid peroxidation products. J Biol Chem. 2008;283:15545–15549. doi: 10.1074/jbc.R700051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Medeiros MHG. Exocyclic DNA adducts as biomarkers of lipid oxidation and predictors of disease. Challenges in developing sensitive and specific methods for clinical studies. Chem Res Toxicol. 2009;22:419–425. doi: 10.1021/tx800367d. [DOI] [PubMed] [Google Scholar]
- 272.Stone K, Uzieblo A, Marnett LJ. Studies of the reaction of malondialdehyde with cytosine nucleosides. Chem Res Toxicol. 1990;3:467–472. doi: 10.1021/tx00017a013. [DOI] [PubMed] [Google Scholar]
- 273.Chaudhary AK, Nokubo M, Reddy RG, Yeola SN, Morrow JD, Blair IA, Marnett LJ. Detection of endogenous malondialdehyde-deoxyguanosine adducts in human liver. Science. 1994;265:1580–1582. doi: 10.1126/science.8079172. [DOI] [PubMed] [Google Scholar]
- 274.Chaudhary AK, Reddy GR, Blair IA, Marnett LJ. Characterization of an N6-oxopropenyl-2′-deoxyadenosine adduct in malondialdehyde-modified DNA using liquid chromatography/electrospray ionization tandem mass spectrometry. Carcinogenesis. 1996;17:1167–1170. doi: 10.1093/carcin/17.5.1167. [DOI] [PubMed] [Google Scholar]
- 275.Vaca CE, Fang J-L, Mutanen M, Valsta L. 32P-Postlabeling determination of DNA adducts of malonaldehyde in humans: Total white blood cells and breast tissue. Carcinogenesis. 1995;16:1847–1851. doi: 10.1093/carcin/16.8.1847. [DOI] [PubMed] [Google Scholar]
- 276.Chaudhary AK, Nokubo M, Oglesby TD, Marnett LJ, Blair IA. Characterization of endogenous DNA adducts by liquid chromatography/electrospray ionization/tandem mass spectrometry. J Mass Spectrom. 1995;30:1157–116. [Google Scholar]
- 277.Wang M, Dhingra K, Hittleman WN, Liehr JG, de Andrade M, Li D. Lipid peroxidation-induced putative malondialdehyde-DNA adducts in human breast tissues. Cancer Epidemiol Biomarkers Prev. 1996;5:705–710. [PubMed] [Google Scholar]
- 278.Rouzer CA, Chaudhary AK, Nokubo M, Ferguson DM, Reddy GR, Blair IA, Marnett LJ. Analysis of themalondialdehyde-2′-deoxyguanosine adduct, pyrimidopurinone, in human leukocyte DNA by gas chromatography/electron capture negative chemical ionization/mass spectrometry. Chem Res Toxicol. 1997;10:181–188. doi: 10.1021/tx9601216. [DOI] [PubMed] [Google Scholar]
- 279.Fang JL, Vaca CE, Valsta LM, Mutanen M. Determination of DNA adducts of malonaldehyde in humans: Effects of dietary fatty acid composition. Carcinogenesis. 1996;17: 1035–1040. doi: 10.1093/carcin/17.5.1035. [DOI] [PubMed] [Google Scholar]
- 280.Ma M, Villalta PW, Balbo S, Stepanov I. Analysis of a malondialdehyde-deoxyguanosine adduct in human leukocyte DNA by liquid chromatography nanoelectrospray-high-resolution tandem mass spectrometry. Chem Res Toxicol. 2014;27:1829–1836. doi: 10.1021/tx5002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wauchope OR, Beavers WN, Galligan JJ, Mitchener MM, Kingsley PJ, Marnett LJ. Nuclear oxidation of a major peroxidation DNA adduct, M(1)dG, in the genome. Chem Res Toxicol. 2015;28:2334–2342. doi: 10.1021/acs.chemrestox.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Chung F-L, Young R, Hecht SS. Formation of cyclic 1,N2- propanodeoxyguanosine adducts in DNA upon reaction with acrolein or corotonaldehyde. Cancer Res. 1984;44:990–995. [PubMed] [Google Scholar]
- 283.Nath RG, Ocando JE, Chung F-L. Detection of 1,N2-propano-2′-deoxyguanosine adducts as potential endogenous DNA lesions in rodent and human tissues. Cancer Res. 1996;56: 452–456. [PubMed] [Google Scholar]
- 284.Chung F-L, Nath RG, Ocando JE, Nishikawa A, Zhang L. Deoxyguanosine adducts of trans-4-hydroxy-2-nonenal areendogenous DNA lesions in rodents and humans: Detection and potential sources. Cancer Res. 2000;60: 1507–1511. [PubMed] [Google Scholar]
- 285.Wacker M, Schuler D, Wanek P, Eder E. Development of a 32P-postlabeling method for the detection of 1,N2-propanodeoxyguanosine adducts of trans-4-hydroxy-2-nonenal in vivo. Chem Res Toxicol. 2000;13:1165–1173. doi: 10.1021/tx000058r. [DOI] [PubMed] [Google Scholar]
- 286.Garcia CCM, Freitas FP, Di Mascio P, Medeiros MHG. Ultrasensitive simultaneous quantification of 1,N2-etheno-2′-deoxyguanosine and 1,N2-propano-2′-deoxyguanosine in DNA by an online liquid chromatography-electrospray tandem mass spectrometry assay. Chem Res Toxicol. 2010;23:1245–1255. doi: 10.1021/tx1001018. [DOI] [PubMed] [Google Scholar]
- 287.Zang N, Song Y, Wu D, Xu T, Lu M, Zang W, Wang H. Detection of 1,N(2)-propano-2'-deoxyguanosine adducts in genomic DNA by ultrahigh performance liquid chromatography-electrospray ionization-tandem mass spectrometry in combination with stable isotope dilution. J Chromatogr A. 2016;1450: 36–44. doi: 10.1016/j.chroma.2016.04.067. [DOI] [PubMed] [Google Scholar]
- 288.Nath RG, Ocando JE, Richie JP, Jr, Chung FL. Effect of glutathione depletion on exocyclic adduct levels in the liver DNA of F344 rats. Chem Res Toxicol. 1997;10:1250–1253. doi: 10.1021/tx9701079. [DOI] [PubMed] [Google Scholar]
- 289.Garcia CCM, Freitas FP, Sanchez AB, Di Mascio P, Medeiros MHG. Levels in urinary samples from individuals exposed to urban air pollution. Chem Res Toxicol. 2013;26:1602–1604. doi: 10.1021/tx400273q. [DOI] [PubMed] [Google Scholar]
- 290.Stein S, Lao Y, Yang IY, Hecht SS, Moriya M. Genotoxicity of acetaldehyde- and crotonaldehyde- induced 1, N2-propanodeoxyguanosine DNA adducts in human cells. Mutat Res. 2006;608:1–7. doi: 10.1016/j.mrgentox.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 291.Doerge DR, Churchwell MI, Fang JL, Beland FA. Quantification of etheno DNA adducts using liquid chromatography, on-line sample processing, and electrospray tandem mass spectrometry. Chem Res Toxicol. 2000;13:1259–1264. doi: 10.1021/tx0001575. [DOI] [PubMed] [Google Scholar]
- 292.Morinello EJ, Ham A-J, Ranasinghe A, Sangaiah R, Swenberg JA. Simultaneous quantitation of N2,3-ethenoguanine and 1,N2-ethenoguanine with an immunoaffinity/gas chromatography/high-resolution mass spectrometry assay. Chem Res Toxicol. 2001;14:327–334. doi: 10.1021/tx0002076. [DOI] [PubMed] [Google Scholar]
- 293.Loureiro APM, Marques SA, Garcia CCM, Di Mascio P, Medeiros MHG. Development of an on-line liquid chromatography-electrospray tandem mass spectrometry assay to quantitatively determine 1,N2-etheno-2′-deoxyguanosine in DNA. Chem Res Toxicol. 2002;15:1302–1308. doi: 10.1021/tx025554p. [DOI] [PubMed] [Google Scholar]
- 294.Nair J, Gal A, Tamir S, Tannenbaum S, Wogan G, Bartsch H. Etheno adducts in spleen DNA of SJL mice stimulated to overproduce nitric oxide. Carcinogenesis. 1998;19:2081–2084. doi: 10.1093/carcin/19.12.2081. [DOI] [PubMed] [Google Scholar]
- 295.Arab K, Pedersen M, Nair J, Meerang M, Knudsen LE, Bartsch H. Typical signature of DNA damage in white blood cells: a pilot study on etheno adducts in Danish mother-newborn child pairs. Carcinogenesis. 2009;30:282–285. doi: 10.1093/carcin/bgn264. [DOI] [PubMed] [Google Scholar]
- 296.Gros L, Ishchenko AA, Saparbaev M. Enzymology of repair of etheno-adducts. Mutat Res. 2003;531:219–229. doi: 10.1016/j.mrfmmm.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 297.Pollack M, Yang IY, Kim HYH, Blair IA, Moriya M. Translesion DNA synthesis across the heptanone-etheno-2′-deoxycytidine adduct in cells. Chem Res Toxicol. 2006;19:1074–1079. doi: 10.1021/tx0600503. [DOI] [PubMed] [Google Scholar]
- 298.Patra A, Su Y, Zhang Q, Johnson KM, Guengerich FP, Egli M. Structural and kinetic analysis of miscoding opposite the DNA adduct 1,N6-ethenodeoxyadenosine by Human Translesion DNA Polymerase η. J Biol Chem. 2016;291:14134–14145. doi: 10.1074/jbc.M116.732487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Gen. 2005;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 300.Kriaucionis S, Heintz N. The Nuclear DNA Base 5-hydroxymethylcytosine Is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Tahiliani M, Koh KP, Shen YH, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Delatte B, Deplus R, Fuks F. Playing TETris with DNA modifications. EMBO J. 2014;33:1198–1211. doi: 10.15252/embj.201488290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Lu X, Zhao BS, He C. TET family proteins: Oxidation activity, interacting molecules, and functions in diseases. Chem Rev. 2015;115:2225–2239. doi: 10.1021/cr500470n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, Zhao C, Zhang D, Huang H, Gao J, Li Z, Jiao Y, Li C, Liu S, Wu D, Gu W, Yang YG, Xu GL, Wang H. Ascorbic acid enhances tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 305.Yuan BF, Feng YQ. Recent advances in the analysis of 5-methylcytosine and its oxidation products. TrAC Trends Anal Chem. 2014;54:24–35. [Google Scholar]
- 306.Liu MY, Denizio JE, Kohli RM. Quantification of oxidized 5-methylcytosine bases and TET enzyme activity. Methods Enzymol. 2016;573:365–385. doi: 10.1016/bs.mie.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Patel JP, Sowers ML, Herring JL, Theruvathu JA, Emmett MR, Hawkins BE, Zhang K, Dewitt DS, Prough DS, Sowers LC. Measurement of postreplicative DNA metabolism and damage in the rodent brain. Chem Res Toxicol. 2015;28:2352–2363. doi: 10.1021/acs.chemrestox.5b00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Xiong J, Jiang HP, Peng CY, Deng QY, Lan MD, Zeng H, Zheng F, Feng YQ, Yuan BF. DNA hydroxymethylation age of human blood determined by capillary hydrophilic-interaction liquid chromatography/mass spectrometry. Clin Epigenetics. 2015;7: 72. doi: 10.1186/s13148-015-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kraus TFJ, Globisch D, Wagner M, Eigenbrod S, Widmann D, Münzel M, Müller M, Pfaffeneder T, Hackner B, Feiden W, Schüller U, Carell T, Kretzschmar HA. Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int J Cancer. 2012;131:1577–1590. doi: 10.1002/ijc.27429. [DOI] [PubMed] [Google Scholar]
- 310.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 5-hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Tang Y, Chu JM, Huang W, Xiong J, Xing XW, Zhou X, Feng YQ, Yuan BF. Hydrophilic material for the selective enrichment of 5- hydroxymethylcytosine and its liquid chromatography-tandem mass spectrometry detection. Anal Chem. 2013;85:6129–6135. doi: 10.1021/ac4010869. [DOI] [PubMed] [Google Scholar]
- 312.Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, Pfeifer GP, Xu GL, Wang Y. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc. 2014;136:11582–11585. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Yin R, Mo J, Lu M, Wang H. Detection of human urinary 5-hydroxymethylcytosine by stable isotope dilution HPLC-MS/MS analysis. Anal Chem. 2015;87:1846–1852. doi: 10.1021/ac5038895. [DOI] [PubMed] [Google Scholar]
- 314.Zhang HY, Xiong J, Qi BL, Feng YQ, Yuan BF. The existence of 5-hydroxymethylcytosine and 5-formylcytosine in both DNA and RNA in mammals. Chem Commun. 2016;52:737–740. doi: 10.1039/c5cc07354e. [DOI] [PubMed] [Google Scholar]
- 315.Yuan F, Zhang XH, Nie J, Chen HX, Zhou YL, Zhang XX. Ultrasensitive determination of 5-methylcytosine and 5-hydroxymethylcytosine in genomic DNA by sheathless interfaced capillary electrophoresis-mass spectrometry. Chem Commun. 2016;52:698–2700. doi: 10.1039/c5cc10155g. [DOI] [PubMed] [Google Scholar]
- 316.Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, Schuermann D, Michalakis S, Kosmatchev O, Schiesser S, Steigenberger B, Raddaoui N, Kashiwazaki G, Müller U, Spruij CG, Vermeulen M, Leonhardt H, Schär P, Müller M, Carell T. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- 317.Boorstein RJ, Cummings A, Jr, Marenstein DR, Chan MK, Ma Y, Neubert TA, Brown SM, Teebor GW. Definitive identification of mammalian 5-hydroxymethyluracil DNA N-glycosylase activity as SMUG1. J Biol Chem. 2001;276:41991–41997. doi: 10.1074/jbc.M106953200. [DOI] [PubMed] [Google Scholar]
- 318.Yun B, Geacintov NE, Shafirovich V. Generation of guanine-thymidine cross-links in DNA by peroxynitrite/carbon dioxide. Chem Res Toxicol. 2011;24:1144–1152. doi: 10.1021/tx200139c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Talhaoui I, Shafirovich V, Liu Z, Saint-Pierre C, Akishev Z, Matkarimov BT, Gasparutto D, Geacintov NE, Saparbaev M. Oxidatively generated guanine(C8)-thymine(N3) intrastrand cross-links in double-stranded DNA are repaired by base excision repair pathways. J Biol Chem. 2015;290:14610–14617. doi: 10.1074/jbc.M115.647487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Keck K. Bildung von cyclonucleotiden bei bestrahlung wassiriger losungen von purinnucleotiden. Z Naturforsch. 1968;23b:1034–1043. [PubMed] [Google Scholar]
- 321.Mariaggi N, Cadet J, Téoule R. Cyclisation radicalaire de la désoxy-2’-adénosine en solution aqueuse, sous l’effet du rayonnement gamma. Tetrahedron. 1976;32:2385–2387. [Google Scholar]
- 322.Raleigh JA, Fuciarelli AF. Distribution of damage in irradiated 5′-AMP: 8,5′-cyclo-AMP, 8-hydroxy-AMP, and adenine release. Radiat Res. 1985;102:165–175. [Google Scholar]
- 323.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5',8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci USA. 2000;97:3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Brooks PJ, Wise DS, Berry DA, Kosmoski JV, Smerdon MJ, Somers RL, Mackie H, Spoonde AY, Ackerman EJ, Coleman K, Tarone RE, Robbins JH. The oxidative DNA lesion 8,5'-(S)-cyclo-2'-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275:22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 325.Belmadoui N, Boussicault F, Guerra M, Ravanat J-L, Chatgilialoglu C, Cadet J. Radiation-induced formation of purine 5',8-cyclonucleosides in isolated and cellular DNA: high stereospecificity and modulating effect of oxygen. Org Biomol Chem. 2010;8:3211–3219. doi: 10.1039/c004531d. [DOI] [PubMed] [Google Scholar]
- 326.Chatgilialoglu C, Ferreri C, Terzidis MA. Purine 5',8-cyclonucleoside lesions: chemistry and biology. Chem Soc Rev. 2011;40:1368–1382. doi: 10.1039/c0cs00061b. [DOI] [PubMed] [Google Scholar]
- 327.Kropachev K, Ding S, Terzidis MA, Masi A, Liu Z, Cai Y, Kolbanovskiy M, Chatgilialoglu C, Broyde S, Geacintov NE, Shafirovich V. Structural basis for the recognition of diastereomeric 5',8-cyclo-2'-deoxypurine lesions by the human nucleotide excision repair system. Nucleic Acids Res. 2014;42:5020–5032. doi: 10.1093/nar/gku162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Khan I, Suhasini AN, Banerjee T, Sommers JA, Kaplan DL, Kuper J, Kisker C, Brosh RM., Jr Impact of age-associated cyclopurine lesions on DNA repair helicases. PLoS One. 2014;9:e113293. doi: 10.1371/journal.pone.0113293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Brooks PJ. The cyclopurine deoxynucleotides: DNA repair, biological effects, mechanistic insights, and unanswered questions. Free Radic Biol Med. 2016 doi: 10.1016/j.freeradbiomed.2016.12.028. this issue. [DOI] [PubMed] [Google Scholar]
- 330.Jaruga P, Birincioglu M, Rodriguez H, Dizdaroglu M. Mass spectrometric assays for the tandem lesion 8,5’-cyclo-2’-deoxyguanosine in mammalian DNA. Biochemistry. 2002;41:3703–3711. doi: 10.1021/bi016004d. [DOI] [PubMed] [Google Scholar]
- 331.Dizdaroglu M, Jaruga P, Rodriguez H. Identification and quantification of 8,5’-cyclo-2’-deoxyadenosine in DNA by liquid chromatography mass spectrometry. Free Radic Biol Med. 2001;30:774–784. doi: 10.1016/s0891-5849(01)00464-6. [DOI] [PubMed] [Google Scholar]
- 332.Nyaga SG, Jaruga P, Lohani A, Dizdaroglu M, Evans MK. Accumulation of oxidatively induced DNA damage in human breast cancer cell lines following treatment with hydrogen peroxide. Cell Cycle. 2007;6:1472–1478. [PubMed] [Google Scholar]
- 333.D’Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM, Orioli D, Frosina G, Zambruno G, Dizdaroglu M, Stefanini M, Dogliotti E. Oncogene. 2007;26:4336–4343. doi: 10.1038/sj.onc.1210232. [DOI] [PubMed] [Google Scholar]
- 334.Barbier E, Lagorce A, Hachemi A, Dutertre M, Gorlas A, Morand L, Saint-Pierre C, Ravanat J-L, Douki T, Armengaud J, Gasparutto D, Confalonieri F, Breton J. Oxidative DNA damage and repair in the radioresistant archaeon Thermococcus gammatolerans. Chem Res Toxicol. 2016;29: 1796–1809. doi: 10.1021/acs.chemrestox.6b00128. [DOI] [PubMed] [Google Scholar]
- 335.Wang J, Yuan B, Guerrero C, Bahde R, Gupta S, Wang Y. Quantification of oxidative DNA lesions in tissues of Long-Evans Cinnamon rats by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope-dilution method. Anal Chem. 2011;83:2201–2209. doi: 10.1021/ac103099s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Yu Y, Guerrero CR, Liu S, Amato NJ, Sharma Y, Gupta S, Wang Y. Comprehensive assessment of oxidatively induced modifications of DNA in a rat model of human Wilson's disease. Mol Cell Proteomics. 2016;15:810–817. doi: 10.1074/mcp.M115.052696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Wang J, Clauson CL, Robbins PD, Niedernhofer LJ, Wang Y. The oxidative DNA lesions 8,5'-cyclopurines accumulate with aging in a tissue-specific manner. Aging Cell. 2012;11:714–716. doi: 10.1111/j.1474-9726.2012.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Tilstra JS, Robinson AR, Wang J, Gregg SQ, Clauson CL, Reay DP, Nasto LA, St Croix CM, Usas A, Vo N, Huard J, Clemens PR, Stolz DB, Guttridge DC, Watkins SC, Garinis GA, Wang Y, Niedernhofer LJ, Robbins PD. NF-κB inhibition delays DNA damage-induced senescence and aging in mice. J Clin Invest. 2012;122:2601–2612. doi: 10.1172/JCI45785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Regulus P, Spessotto S, Gateau M, Cadet J, Favier A, Ravanat J-L. Detection of new radiation-induced DNA degradation lesions by liquid chromatography coupled to tandem mass spectrometry. Rapid Commun Mass Spectrom. 2004;18:2223–2228. doi: 10.1002/rcm.1612. [DOI] [PubMed] [Google Scholar]
- 340.Regulus P, Duroux B, Bayle P-A, Favier A, Cadet J, Ravanat J-L. Oxidation of the sugar moiety of DNA by ionizing radiation or bleomycin could induce the formation of a cluster DNA lesion. Proc Natl Acad Sci USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Dedon PC, Goldberg IH. Free-radical mechanism involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem Res Toxicol. 1992;5:311–322. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- 342.Pogozelski WK, Tullius TD. Oxidative strand scission of nucleic acids: routes initiated by hydrogen abstraction from the sugar moiety. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 343.Sczepanski JT, Jacobs AC, Greenberg MM. Self promoted interstrand cross-link formation by an abasic site. J Am Chem Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- 344.Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. Scope and mechanism cross-link formation by the C4’-oxidized abasic site. J Am Chem Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Bessho T, Tano K, Nishmura S, Kasai H. Induction of mutations in mouse FM3A cells by treatment with riboflavin plus visible light and its possible relation with formation of 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) in DNA. Carcinogenesis. 1993;14:1069–1071. doi: 10.1093/carcin/14.5.1069. [DOI] [PubMed] [Google Scholar]
- 346.Hattori-Nakakuki Y, Nishigori C, Okamoto K, Imamura S, Hiai H, Toyokuni S. Formation of 8-hydroxy-2'-deoxyguanosine in epidermis of hairless mice exposed to near-UV. Biochem Biophys Res Commun. 1994;201:1132–113. doi: 10.1006/bbrc.1994.1823. [DOI] [PubMed] [Google Scholar]
- 347.Yamaguchi R, Hirano T, Asami S, Sugita A, Kasai H. Increase in the 8-hydroxyguanine repair activity in the rat kidney after the administration of a renal carcinogen, ferric nitrilotriacetate. Environ Health Perspect. 1996;104(Suppl 3):651–653. doi: 10.1289/ehp.96104s3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Yamaguchi R, Hirano T, Asami S, Chung MH, Sugita A, Kasai H. Increased 8-hydroxyguanine levels in DNA and its repair activity in rat kidney after administration of a renal carcinogen, ferric nitrilotriacetate. Carcinogenesis. 1996;17:2419–2422. doi: 10.1093/carcin/17.11.2419. [DOI] [PubMed] [Google Scholar]
- 349.Tsurudome Y, Hirano T, Yamato H, Tanaka I, Sagai M, Hirano H, Nagata N, Itoh H, Kasai H. Changes in levels of 8-hydroxyguanine in DNA, its repair and OGG1 mRNA in rat lungs after intratracheal administration of diesel exhaust particles. Carcinogenesis. 1999;20:1573–1576. doi: 10.1093/carcin/20.8.1573. [DOI] [PubMed] [Google Scholar]
- 350.Kim HN, Morimoto Y, Tsuda T, Ootsuyama Y, Hirohashi M, Hirano T, Tanaka I, Lim Y, Yun IG, Kasai H. Changes in DNA 8-hydroxyguanine levels, 8-hydroxyguanine repair activity, and hOGG1 and hMTH1 mRNA expression in human lung alveolar epithelial cells induced by crocidolite asbestos. Carcinogenesis. 2001;22:265–269. doi: 10.1093/carcin/22.2.265. [DOI] [PubMed] [Google Scholar]
- 351.Sczepanski JT, Jacobs AC, Van Houten B, Greenberg MM. Double-strand break formation during nucleotide excision repair of a DNA interstrand cross-link. Biochemistry. 2009;48:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Ghosh S, Greenberg MM. Nucleotide excision repair of chemically stabilized analogues of DNA interstrand cross-links produced from oxidized abasic sites. Biochemistry. 2014;53:5958–5965. doi: 10.1021/bi500914d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Ma B, Jin M, Villalta PW, Kapphahn RJ, Montezuma SR, Ferrington DA, Stepanov I. Simultaneous determination of 8-oxo-2'-deoxyguanosine and 8-oxo-2'-deoxyadenosine in human retinal DNA by liquid chromatography nanoelectrospray-tandem mass spectrometry. Sci Rep. 2016;6:22375. doi: 10.1038/srep22375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Maynard S, de Souza-Pinto NC, Scheibye-Knudsen M, Bohr VA. Mitochondrial base excision repair assays. Methods. 2010;51:416–425. doi: 10.1016/j.ymeth.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Kirkali G, de Souza-Pinto NC, Jaruga P, Bohr VA, Dizdaroglu M. Accumulation of (5'S)-8,5'-cyclo-2'-deoxyadenosine in organs of Cockayne syndrome complementation group B gene knockout mice. DNA Repair (Amst) 2009;8:274–278. doi: 10.1016/j.dnarep.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]