Capsule summary
A retrospective, multi-center study of adults with and without chronic rhinosinusitis (CRS) identifies a significant association between rs6967330 in the viral receptor CDHR3 – known to be associated with wheezing and asthma in children – and the development of CRS.
Keywords: Chronic rhinosinusitis, asthma, genetics, rhinovirus, CDHR3
To the Editor
Rhinoviruses (RV) are the most frequent virus isolated in chronic rhinosinusitis (CRS)1 and asthma exacerbations.2 CRS and asthma are both heterogeneous inflammatory disorders of the airway; however, their frequent co-occurrence suggests a unified airway endotype with a common pathophysiology. A recent genome-wide association study (GWAS) identified a missense single nucleotide polymorphism (SNP) in the Cadherin related family member 3 gene (CDHR3) that results in a cysteine to tyrosine substitution at position 529 (rs6967330). This rs6967330 risk allele (A) was found to be a significant additive risk factor for acute hospitalizations secondary to asthma in the first 6 years of life.3 Further mechanistic studies identified CDHR3 as the receptor for RV-C, an especially virulent subtype of RV highly correlated with severe airway disease.4 Given that sinonasal epithelia is the primary site for RV infection,5 we hypothesized that the combination of the rs6967330 genetic SNP with RV-C infections could be responsible for the development of CRS in adults.
We performed a retrospective case-control study of non-Hispanic white adults recruited from two different tertiary academic rhinology centers: the University of Arizona (UofA) and the University of Pennsylvania (UPenn) with diagnosed CRS (UofA n=76, UPenn n=267) and healthy controls (UofA n=52, UPenn n=337). Further details are available in the online methods and the participant’s characteristics are shown in Table E1. In the UPenn group, those with CRS were significantly older and more likely to be males compared to the controls. At both UofA and UPenn the prevalence of asthma was approximately 2-fold greater in the CRS compared to the control group.
DNA was extracted from buccal swabs or saliva samples collected from all study participants and rs6967330 was genotyped. In the control group for both populations, the allele frequency of the minor A allele was similar to that seen in the general non-Hispanic white population.3 There were no significant differences in subjective or objective measures of CRS including nasal polyp status in individuals with CRS related to CDHR3 genotype. We calculated the risk for CRS associated with rs6967330, stratified by location and adjusted for both sex and age in additive and dominant models. Additive models included genotype as an ordinal variable with three categories (0, 1, 2 for GG, AG, and AA genotypes) and dominant models included genotype as a binary variable (0, 1 for GG vs. AA+AG genotypes). Using an additive model, rs6967330 significantly increased the odds for CRS by 2.08 at the UofA (95% CI, 1.02–2.41, p=0.043) for each additional ‘A’ allele and remained significant when adjusting for sex and age. Using a dominant model, rs6967330 significantly increased the odds for CRS by 1.5 at UPenn (95% CI, 1.0–2.1, p=0.022) if an individual carried one or two copies of the minor A allele of rs6967330. Meta-analysis estimates of the UofA and UPenn populations demonstrated significantly increased risk for CRS in both additive and dominant models, which remained essentially unchanged after adjustment for age and sex. (Table 1) Excluding adults with co-occurring asthma did not significantly change this risk (mOR=1.55, 95% C, 1.11–2.17; p=0.011), suggesting that this association was not confounded by the presence of asthma (Figure 1).
Table 1.
Association between rs6967330 and CRS at UofA and UPenn
| Unadjusted | Adjusted for sex and age | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| All Participants | n | OR | 95%CI | P | PHet* | n | adjOR | 95%CI | P | PHet* |
| Additive Model | ||||||||||
| UofA | 128 | 2.08 | 1.02, 4.21 | 0.043 | 126 | 2.14 | 1.04, 4.42 | 0.039 | ||
| UPenn | 604 | 1.31 | 0.97, 1.78 | 0.077 | 595 | 1.28 | 0.91, 1.80 | 0.162 | ||
| Meta | 732 | 1.41 | 1.07, 1.86 | 0.015 | NS | 721 | 1.40 | 1.03, 1.92 | 0.032 | NS |
| Dominant Model | ||||||||||
| UofA | 128 | 2.00 | 0.94, 4.23 | 0.071 | 126 | 2.06 | 0.95, 4.46 | 0.068 | ||
| UPenn | 604 | 1.50 | 1.06, 2.11 | 0.022 | 595 | 1.46 | 0.99, 2.17 | 0.057 | ||
| Meta | 732 | 1.57 | 1.15, 2.15 | 0.005 | NS | 721 | 1.57 | 1.11, 2.23 | 0.012 | NS |
| Non-Asthmatic Participants | ||||||||||
| Additive Model | ||||||||||
| UofA | 95 | 2.09 | 0.93, 4.72 | 0.076 | 93 | 2.05 | 0.86, 4.78 | 0.095 | ||
| UPenn | 442 | 1.48 | 1.02, 2.15 | 0.038 | 435 | 1.43 | 0.95, 2.15 | 0.087 | ||
| Meta | 537 | 1.57 | 1.12, 2.20 | 0.009 | NS | 528 | 1.53 | 1.06, 2.21 | 0.023 | NS |
| Dominant Model | ||||||||||
| UofA | 95 | 2.04 | 0.88, 4.74 | 0.095 | 93 | 2.01 | 0.84, 4.81 | 0.115 | ||
| UPenn | 442 | 1.85 | 1.22, 2.80 | 0.004 | 435 | 1.77 | 1.12, 2.81 | 0.014 | ||
| Meta | 537 | 1.88 | 1.30, 2.73 | 0.001 | NS | 528 | 1.82 | 1.22, 2.74 | 0.004 | NS |
Heterogeneity p-value based on chi-squared (NS indicates no heterogeneity between studies)
Figure 1.
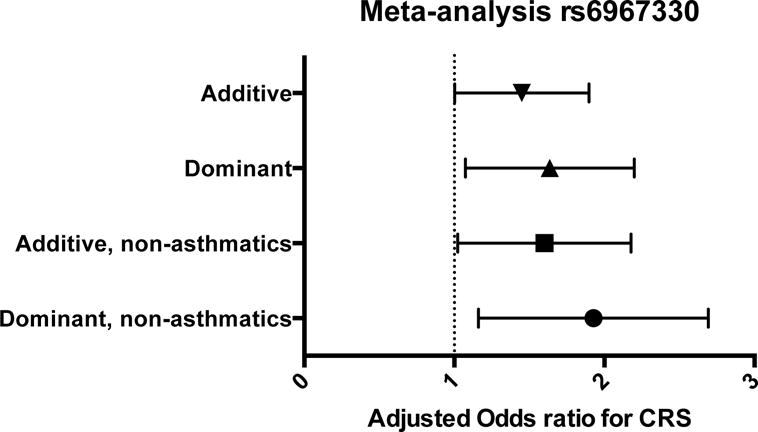
Meta-analysis of rs6967330 from UofA and UPenn. The rs6967330 SNP is significantly associated with CRS in both additive and dominant models. This association remains significant in those without self-reported asthma (non-asthmatics). mOR was adjusted for sex and age.
RV infections are the most frequent cause of upper respiratory infections (URIs), otherwise known as the common cold. In most individuals, URIs resolve within 5–7 days. However, it is unknown why in adults, symptoms persist and can progress into CRS or trigger asthma exacerbations. The primary site of RV infection is the sinonasal epithelia. Until recently, the only known RV subtypes (A and B) bound to the low-density lipoprotein receptor (LDLR)5 and the intercellular adhesion molecule-1 (ICAM-1) receptors expressed on the apical surface of ciliated airway epithelia.6 In 2006, a third subtype, RV-C, was discovered using molecular detection techniques4, and found to be associated with more severe airway disease. RV-C isolates were identified with significantly higher frequency in the nasal washes of individuals with CRS,1 and determined to be a trigger for acute wheezing lower respiratory illnesses (WLRI) and the subsequent development of childhood asthma.7
CDHR3 was first identified as a candidate gene for early childhood asthma with severe exacerbations in a large GWAS3 The CDHR3 protein is highly expressed in both upper and lower airway epithelia, and when plasmid DNA containing the mutation domain 5 (C529Y) was transfected into 293T cells it resulted in a marked increase of CDHR3 trafficking to the cell membrane compared to the wild-type (G) major allele. Moreover, CDHR3 was identified as the putative receptor for RV-C, with the homozygous presence of the rs6967330 risk allele increasing RV-C binding by ten-fold in a transduced HeLA cell line stably expressing CDHR3-Y529 compared to control HeLa cells.8
In summary, we have determined that carriers of rs6967330 are at significantly increased risk for CRS. The association between rs6967330 and airway disease was first reported in a study of acute wheezing lower respiratory illnesses (WLRI) in young children3. It is now well established that WLRI caused by rhinovirus in this age group are strongly associated with the subsequent development of asthma.7 Similarly, CRS usually starts with acute URI episodes, most often caused by rhinoviruses, and progresses into a disease characterized by quiescent periods followed by acute flare-ups. CDHR3 rs6967330 may be a common genetic risk factor between viral-associated childhood asthma and CRS.
Acknowledgments
Sources of support: NIH –K08 DE021413 (EHC), P30 DC011735 (DRR), R01DC013588 (NAC), R01 HL132523 (EHC, SG, FDM)
Abbreviations
- CRS
chronic rhinosinusitis
- CDHR3
Cadherin related family member 3 gene
- RV
Rhinovirus
- URIs
upper respiratory infections
- LDLR
low-density lipoprotein receptor
- ICAM-1
intercellular adhesion molecule-1
- WLRI
wheezing lower respiratory illnesses
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cho GS, Moon B-J, Lee B-J, Gong C-H, Kim NH, Kim Y-S, et al. High rates of detection of respiratory viruses in the nasal washes and mucosae of patients with chronic rhinosinusitis. J Clin Microbiol. 2013;51:979–84. doi: 10.1128/JCM.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol. 2010;84:7418–26. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bønnelykke K, Sleiman P, Nielsen K, Kreiner-Møller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–5. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 4.Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, et al. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardin PG, Johnston SL, Sanderson G, Robinson BS, Pickett MA, Fraenkel DJ, et al. Detection of rhinovirus infection of the nasal mucosa by oligonucleotide in situ hybridization. Am J Respir Cell Mol Biol. 1994;10:207–13. doi: 10.1165/ajrcmb.10.2.8110476. [DOI] [PubMed] [Google Scholar]
- 6.Greve JM, Davis G, Meyer AM, Forte CP, Yost SC, Marlor CW, et al. The major human rhinovirus receptor is ICAM-1. Cell. 1989;56:839–47. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- 7.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–72. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, et al. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112:5485–90. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, et al. European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol Suppl. 2012 3pprecedingtableofcontents–1–298. [PubMed] [Google Scholar]
- 10.Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, Brook I, Ashok Kumar K, Kramper M, et al. Clinical practice guideline (update): adult sinusitis. Otolaryngol Head Neck Surg. 2015;152:S1–S39. doi: 10.1177/0194599815572097. [DOI] [PubMed] [Google Scholar]


