Abstract
Background
The objective of this study was to identify molecular alterations in the human blood serum related to bipolar disorder, using nuclear magnetic resonance (NMR) spectroscopy and chemometrics.
Methods
Metabolomic profiling, employing 1H-NMR, 1H-NMR T2-edited, and 2D-NMR spectroscopy and chemometrics of human blood serum samples from patients with bipolar disorder (n = 26) compared with healthy volunteers (n = 50) was performed.
Results
The investigated groups presented distinct metabolic profiles, in which the main differential metabolites found in the serum sample of bipolar disorder patients compared with those from controls were lipids, lipid metabolism-related molecules (choline, myo-inositol), and some amino acids (N-acetyl-l-phenyl alanine, N-acetyl-l-aspartyl-l-glutamic acid, l-glutamine). In addition, amygdalin, α-ketoglutaric acid, and lipoamide, among other compounds, were also present or were significantly altered in the serum of bipolar disorder patients. The data presented herein suggest that some of these metabolites differentially distributed between the groups studied may be directly related to the bipolar disorder pathophysiology.
Conclusions
The strategy employed here showed significant potential for exploring pathophysiological features and molecular pathways involved in bipolar disorder. Thus, our findings may contribute to pave the way for future studies aiming at identifying important potential biomarkers for bipolar disorder diagnosis or progression follow-up.
Electronic supplementary material
The online version of this article (doi:10.1186/s40345-017-0088-2) contains supplementary material, which is available to authorized users.
Keywords: 1H-NMR, Biomarkers, Bipolar disorder, Metabolic profiling, Chemometrics, 2D NMR
Background
Bipolar disorder (BD) is a potentially debilitating mental disorder affecting 1–3% of the population worldwide (Goodwin and Jamison 2007; Marohn 2011; Nierenberg et al. 2013; Phillips and Kupfer 2013). Previous studies have suggested a number of theories to explain the etiology of BD, including the influence of genetic factors, deficits in neurodevelopmental processes, implication of neurodegenerative pathologies, changes in monoamine neurotransmission, and abnormalities in neuroplasticity (van Enkhuizen et al. 2015; Goes 2016; Passos et al. 2016). A number of medications have been used for several decades to treat this disorder and/or to ameliorate the symptoms, though the mechanisms of action of most of mood stabilizers still remain to be fully elucidated (Malhi et al. 2013; Yatham et al. 2013). Increased accuracy in early diagnosis of BD is the key to improve the mental health outcomes, to ameliorate the clinical course, and to make better the treatment response of patients with this disorder. However, the diagnosis of BD, especially in early stages and in predominantly depressive presentations, remains challenging (Brietzke et al. 2016). These challenges could possibly be overcome by the identification of differentially expressed biomarkers, which could reflect or convey the pathophysiological processes (Oswald et al. 2007; Marmol 2008).
Hydrogen-1 nuclear magnetic resonance (1H-NMR) spectroscopy-based metabolomics can be used to monitor a wide range of metabolites in biological samples, allowing for a sensitive, high-throughput molecular screening (Beckonert et al. 2007). 1H-NMR has an exceptional reproducibility and is quantitative to the extent that a given peak area is directly proportional to the concentration of the corresponding metabolite, turning this technique a well-established approach for metabolomics (Maher et al. 2008). Although many platforms for metabolic profiling (for instance, the LC–MS, GC–MS, NMR, capillary electrophoresis-MS) are available, none technology alone provides the complete coverage of the metabolome. Previous employment of these techniques to study the brain of patients with psychiatric disorders was reported (Lan et al. 2009; Strzelecki et al. 2015), but it was scarcely applied for serum metabolome studies of individuals with mental illnesses.
In this study, a NMR metabolomics profiling approach was used to identify molecular alterations in the human blood serum of BD patients. Our intention was to conduct analysis to distinguish the individuals of type 1 BD patients and healthy control groups, based on their metabolic profiles. Also, as previously reported by us (Sussulini et al. 2009), with time distance and new sampling, NMR-based metabolomics has been applied as to verify if it is possible to be used for precise classification of aforementioned illness.
Methods
Studied population
Twenty-six euthymic outpatients with bipolar disorder (BD) type 1 (BD group) and 50 healthy control (HC) individuals (control group) were included in the research. All individuals in the BD group, with age between 18 and 65 years old, fulfilled the DSM-IV (Diagnostic and Statistical Manual of Mental Disorders) criteria according to SCID-I (Structured Clinical Interview for DSM Disorder). Euthymia was defined as not fulfill criteria for any mood episode and present, at the same time, with scores below 8 in Young Mania Rating Scale (YMRS) and Hamilton Depression Rating Scale (HDRS)-17 items. Exclusion criteria were as follows: be acutely suicidal, comorbidity with substance use disorders, except nicotine, presence of acute or unstable or chronic general medical comorbidity, inability to read and understand study procedures, pregnancy, and postpartum period. All subjects were inquired on medical history, including lifetime use of any medication. Body mass index (BMI) was also measured using the formula BMI = Weight (kg)/Height (m)2.
For comparison, a control group of 50 healthy volunteers was recruited from the community. To be eligible for the HC group, individuals were required to have age between 18 and 65 years old, negative history of any psychiatric condition current or along lifetime, and never made use of psychiatric medication. In addition, only volunteers with no family history of major mental disorders (schizophrenia-spectrum disorders, mood disorders, and suicide) were included. The same exclusion criteria of BD group were applied to HC group.
Clinical assessment included SCID-I for confirmation of diagnosis and assessment of psychiatric comorbidities. YMRS and HDRS-17 items were used to determine severity of manic and depressive symptoms, respectively. Functioning was estimated by General Assessment of Functioning (GAF).
Serum collection and storage
All blood samples were taken in the morning (between 8 and 10 a.m.) after 12 h of fasting. Blood was drawn into Vacutainer tubes, and immediately allowed to clot for at least 30 min at room temperature, before it was centrifuged at 1500×g, for 5 min. The serum was then aliquoted, transferred into clean polypropylene tubes, and stored at −80 °C until use. The maximum period of storage was of two weeks.
NMR spectroscopy analyses: 1H-NMR, 1H-NMR T2-edited, and 2D NMR
For NMR spectroscopy analyses, serum samples were thawed and centrifuged at 12,300×g, for 10 min, at 4 °C to separate any precipitate. Aliquots of 250 μL of the supernatants were diluted with 250 μL of deuterated water (D2O) or PBS buffer (250 μL containing 10% D2O) and transferred into 5.0 mm diameter NMR tubes. NMR tubes were placed into a Bruker 600 NMR spectrometer (Bruker Advance III, Bruker GmBH, Rheinestennen, Germany) using TBI (Triple resonance Broadband Inverse) probe at 25 °C. 1H-NMR spectra were recorded as three independent measurements for each sample using the CH3-lactate signal (δ1.33, 3H, d, 3 J = 7.0 Hz) as a reference, and applying the pulse sequence WATERGATE (p3919gp) with 128 ns (Piotto et al. 1992). T2-edited NMR spectra were recorded using the CPMG (Carr-Purcell-Meiboom-Gill) sequence (Meiboom and Gill 1958), where a fixed total spin–spin relaxation delay 2nτ of 100 ms was used to attenuate the broad NMR signals from slowly tumbling molecules such as proteins and lipids retaining those from low molecular weight compounds and some lipid components. For each spectrum, 128 transients were acquired into 32,000 data points, with a spectral width of 12 kHz. To confirm the assignments made from 1H-NMR and T2-edited spectra, some blood serum samples were also examined using 2D [1H–13C] HSQC. For each 2D spectrum, 256 increments with 64 transients per increment were collected and extended to 4 K data points. The signal assignments were based on the literature and/or databases such as Biological Magnetic Data Bank (BMRB) (Ulrich et al. 2008) and Human Metabolome Database (HMDB) (Wishart et al. 2007) and are indicated on the T2-edited spectrum and confirmed by the 2D fully assigned [1H–13C] HSQC data.
Chemometrics of 1H NMR spectral data
1H-NMR data were transported to a data matrix, and chemometrics analysis, based on principal component analysis (PCA) and partial least-squares discriminant analysis (PLS-DA), were performed using MATLAB (The MathWorks, Natick, MA) or Pirouette (Infometrix, USA) software. T2-edited spectra were not used in chemometric analysis. The spectral region of chemical shifts, from 1.00 to 4.40 ppm, was used in chemometrics (Nørgaard 2009). PLS-DA was used as a supervision method for classification (Sena et al. 2004). All variables were auto scaled.
Results
The summary of the collected sample characteristics is presented in Table 1. A predominance of females over males was observed in our BD sample, and the percentages were statistically different between the BD and HC groups. In addition, we observed, as expected, a high prevalence of tobacco smoking individuals in our BD group compared with HC group. Functioning was also impaired in the BD group compared with HC group, which reflects a well-known incomplete association between mood symptoms and functionality in individuals with BD. In addition, there was a clear predominance of chronic patients, with a mean duration of illness of about 15 years.
Table 1.
Clinical and demographic characteristics of the sample
| Healthy controls (n = 50) | BD (n = 26) | Test | ||
|---|---|---|---|---|
| U or X 2 | p value | |||
| Age in years; mean (SD) | 35.69 (13.03) | 40.38 (12.66) | 187.000 | 0.261 |
| Sex female; N(%) | 17 (34) | 17 (65) | 5.114 | 0.023 |
| Ethnicitycaucasian; N(%) | 18 (36) | 14 (53) | 0.391 | 0.531 |
| Years of education; mean (SD) | 12.1 (3.8) | 12.4 (3.6) | 181.500 | 0.865 |
| BMI in kg/m2; mean (SD) | 26.2 (4.43) | 28.06 (7.58) | 170.500 | 0.507 |
| Hours of physical exercise per week; mean (SD) | 1.88 (2.44) | 2 (2.93) | 176.500 | 0.949 |
| Current smoking; N(%) | 3 (11.5) | 4 (23.5) | 1.084 | 0.297 |
| Young mania rating scale; mean (SD) | 0.5 (1.06) | 0.7 (1.35) | 208.500 | 0.661 |
| Hamilton depression rating scale; mean (SD) | 1.53 (1.96) | 3.23 (3.84) | 167.500 | 0.167 |
| General functioning assessment (symptoms); mean (SD) | 88.84 (6.97) | 80 (10.15) | 109.500 | 0.004 |
| General functioning assessment (functioning); mean (SD) | 87.69 (4.52) | 75.88 (16.12) | 127.000 | 0.011 |
| Age at onset in years; mean (SD) | – | 24.83 (9.83) | – | – |
| Duration of illness in years; mean (SD) | – | 15.44 (12.69) | – | – |
| Number of total mood episodes; mean (SD) | – | 9.05 (9.52) | – | – |
| Number of past manic episodes; mean (SD) | – | 3.38 (3.82) | – | – |
| Number of past hypomanic episodes; mean (SD) | – | 1.61 (4.11) | – | – |
| Number of past depressive episodes; mean (SD) | – | 4.05 (5.47) | – | – |
| Number of past mixed episodes; mean (SD) | – | 0 (0) | – | – |
| Age of diagnosis; mean (SD) | – | 31.16 (11.11) | – | – |
| Age of first use of psychiatric medication; mean (SD) | – | 26.55 (10.03) | – | – |
| Age of first use of mood stabilizer; mean (SD) | – | 31.58 (11.78) | – | – |
1H NMR spectroscopy data analysis
To explore all the potential differences in the metabolic profiles between the BD and HC groups, the 1H-NMR spectra (Fig. 1a) were subjected to PCA, i.e., the spectral data obtained for 76 blood serum samples that were acquired as 228 spectra were used in PCA. T2-edited spectra were not used in chemometrics analysis because these spectra show altered peak areas in relation to the peaks of 1H-NMR spectra. Based on the PCA results, it was possible to observe a distinction of the samples into two groups: BD and HC using the spectral region δ 1.00 to 4.40 (Fig. 2).
Fig. 1.
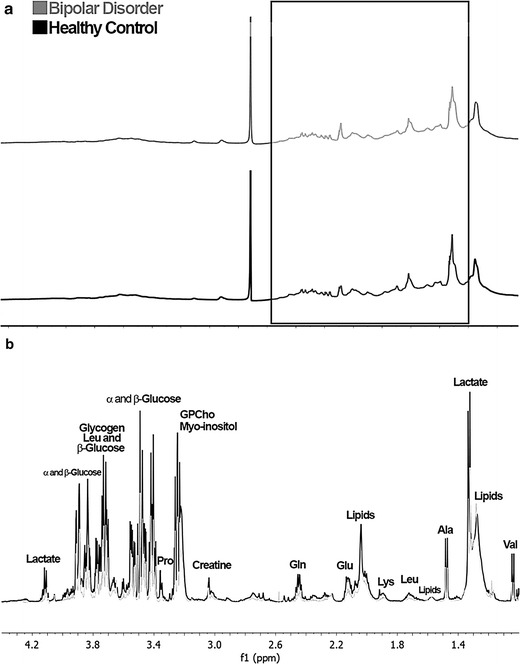
a 1H NMR Spectra with water suppression (Pulse sequence: p3919gp, ns = 128) blood serum sample from HC (black), and BD group (gray). Spectral region highlighted δ 4.40 to 1.00 with higher loadings. b 1H NMR Spectra recorded with T2-filter (Pulse sequence: cpmgpr1d, ns = 128) blood serum sample from HC (black), and BD group (gray). Assignments: Lactate (δ 1.33 and 4.11), glucose (δ 3.41, 3.46, 3.77 and 3.82), leucine (Leu; δ 1.71 and 3.73), proline (Pro; δ 3.34), glycerylphosphocholine (GPCho; δ 3.24), myo-inositol (δ 3.25), creatine (δ 3.03), glutamine (Gln; δ 2.44), glutamate (Glu; δ 2.14), lipids (δ 1.29 and 2.04), lysine (Lys; δ 1.68), alanine (Ala; δ 1.47), valine (Val; δ 1.04)
Fig. 2.
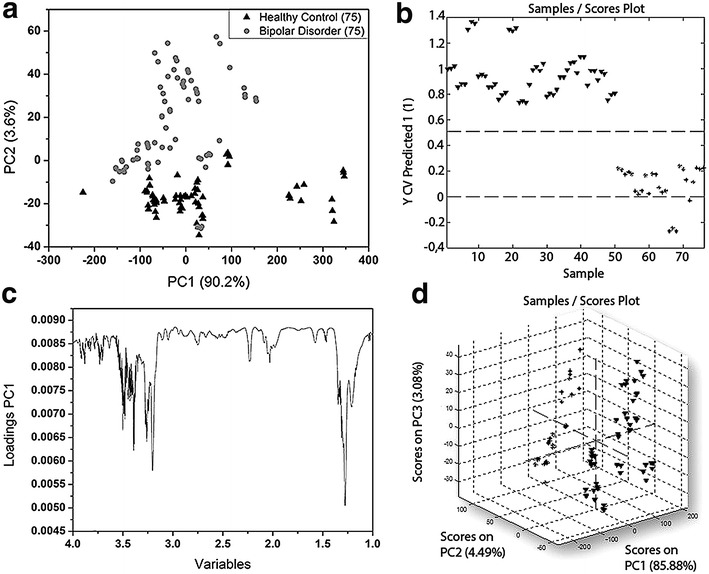
Metabolomics discriminate bipolar disorder patients from healthy individuals. a Plots of cross-validated PCA scores, 50 samples (25 BD + 25 HC) is equivalent to 150 spectra (75 BD + 75 HC). b 2D samples scores plot of PLS-DA, 76 samples (26 BD + 50 HC) is equivalent to 228 (78 BD + 150 HC). c Loadings Graph of spectral region δ 1.00 to 4.40 used in chemometrics. d 3D samples scores plot of PLS-DA
Considering the PCA results and the previous observations about the spectral regions not relevant for analyses, PLS-DA was performed using the same spectral range employed in the PCA. PLS-DA of 1H NMR spectroscopy data revealed an excellent separation between BD and HC groups.
Through the assignments from 1H-NMR edited with T2 filter (Fig. 1b) and the correlations from the HSQC contour maps (Fig. 3), it was possible to compare the obtained data with databases. Thereby, based mainly on spectral data (chemical shift, coupling constants, and multiplicity) and in accordance with biochemical knowledge, seven key-metabolites (Fig. 4) were found to be crucial for the two-group separation. Furthermore, it was observed that there are significant differences in some metabolite concentrations when the two investigated groups were compared, in which important significant variations in some metabolites were observed, and compounds were then identified through T2-edited spectra. The blood serum samples from the BD group had shown to be richer in lipids, whose amounts were up to 1/3 higher compared with those from the HC group. Also, metabolites such as d-glucose, l-alanine, and lactate were more pronounced in BD group compared with HC group. All these metabolites were found to be more abundant in BD group than in HC group, as their peaks areas were at most presenting a ratio of about 3:2 (Additional file 1: Table S3).
Fig. 3.
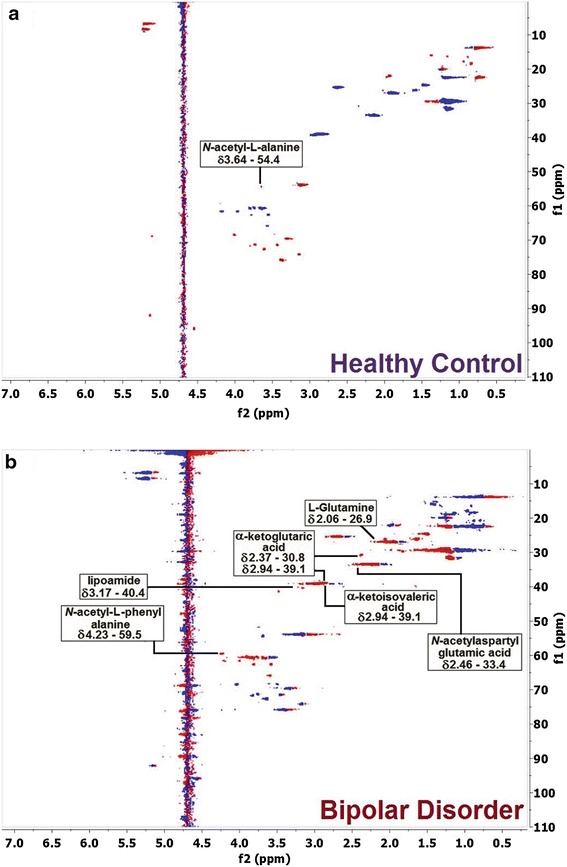
Two-dimensional NMR Spectra: Contour maps of HSQC (pulse sequence: hsqcedetgpsp.3) blood serum sample from (a) HC (ns = 64, SD = 16), and b BD group (ns = 64, SD = 16)
Fig. 4.
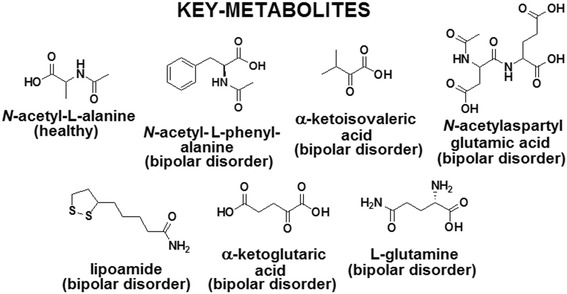
Chemical structures of seven key-metabolites (1 from HC group + 6 from BD group) identified by 2D NMR spectroscopy
Discussion
1H-NMR-based metabolomics were used to chemometric analysis. PCA and PLS-DA analyses were employed to examine the metabolic profiles of blood serum of individuals with BD compared with HC volunteers. The loading plot (Fig. 2) highlighted the most significant variables with the highest loading values, which corresponded to the spectral region from δ 1.00 to 4.40. The 1H-NMR spectra of BD blood serum samples are characterized by overlaps that resulted in broad and intense peaks. So, we acquired T2-edited 1H-NMR spectra (Fig. 1b) to achieve a better spectral resolution and assign peaks referred to the metabolites with the low molecular weight (<1500 Da), such as: l-valine, l-alanine, and creatine. The peaks assignments were performed in comparison with chemical shifts values that were published previously (Misra and Bajpai 2009). It is important to mention that the peaks in T2-edited spectra are not appropriate for integration. Through comparative analysis of the 2D NMR spectra, we observed important differences in metabolic profiles among serum samples from the two, BD and HC, groups. These differences are indicated in Fig. 3, in which it is possible to observe that some metabolites such as lipoamide, l-glutamine, among others, were detected only in BD group, while N-acetyl-l-alanine was observed only in HC group (Fig. 4).
The metabolites that had their peaks assigned based on the 2D NMR and that were classified as “healthy” means that these metabolites have a higher concentration in blood serum samples from the HC (healthy control) group compared with the BD group individuals (Additional file 1: Table S1). The same reasoning can be applied to the BD group. However, it is important to remark that we have not quantified the identified biomarkers up to the present moment.
Despite the large overlapping between the peaks in the 1H NMR spectra, it was possible to estimate the concentration ratios for some metabolites, such as d-glucose, l-alanine, lactate, and lipids, through comparison of the mean value rates of peaks integration of 1H-NMR spectra (Additional file 1: Table S3). For comparison of HC and BD profiles, the greatest variation in concentration of metabolites was observed for the lipids.
Partially, our results are in accordance with numerous previous studies that have demonstrated changes in brain metabolites in BD by in vivo 1H NMR. Davanzo et al. (2001) have shown an increase in myo-inositol in anterior cingulate cortex in children with BD. Acute lithium(I) treatment was associated to a significant reduction in the myo-inositol/creatine ratio, especially among responders to this medication (Davanzo et al. 2001). This result was also reproduced in adults (Machado-Vieira et al. 2015). Several other studies produced mixed results with choline, GABA, and glutamate, probably influenced by methodological issues, chosen brain areas or differences in subpopulations of individuals with the disorder (Bustillo 2013).
The increase of risk of hyperlipidemia in BD patients was reported by Hsu et al. (2015), which corroborates with our results, in which increases in the lipids levels in BD were also noticed (Fig. 1b). Furthermore, currently Yoshimi et al. (2016) studied the metabolism of BD patients through analysis of blood serum samples using capillary electrophoresis-time-of-flight mass spectrometry (CE-TOF/MS). They reported changes in amino acids metabolism (valine, alanine, glutamine) and metabolites belonging to the citric acid and urea cycles, such as the α-ketoglutarate and N-acetylglutamic acid whose levels were increased in BD patients. In our analyses, the presence of α-ketoglutaric acid, α-ketoisovaleric acid and N-acetyl-l-aspartyl-l-glutamic acid (NAAG) was observed, as pointed in Fig. 4.
Glutamate and glutamine are well-known markers (Manji et al. 2003). Also, Pålsson et al. (2015) reported the increase in the levels of these metabolites in blood serum and cerebrospinal fluid (CSF) of BD patients using HPLC with fluorescence detection. This increase can be due to mitochondrial dysfunction or alterations in the metabolism of the cell (Hsu et al. 2015).
However, to our knowledge, the 1H-NMR spectroscopy-based metabolomics analysis of serum was less studied. Sussulini et al. (2009) investigated differential metabolites in human serum sample of patients with BD (n = 25) under different drug treatments: lithium(I) (n = 15) versus other medications (n = 10). This strategy showed significant potential for exploring pathophysiological and toxicological features involved in BD. The investigated groups (HC and patients with BD under different treatments) could be distinguished according to their metabolic profiles, and the main differential metabolites found were glycoprotein lipids, mono- and polyunsaturated lipids, acetate, choline, glutamate, and myo-inositol.
The results of this research should be interpreted at light of some limitations. First, the small sample size precludes subgroup analysis including potential medication effects in metabolic profile. In addition, the inclusion of only euthymic individuals makes impossible to know which metabolic changes are related to traits and which are related to mood states in BD. The different proportion of men and women between the BD and HC groups is a result of a female predominance in the mental health service where the study was conducted. On the other hand, no significant differences in metabolic profile between female and male were noticed here. Strengths of the current study included the very careful selection of cases and controls, including the inclusion of patients without any significant comorbidities and HCs without any evidence of personal or family mental illness history.
Conclusion
Considering the main results of this study, we can conclude that 1H NMR spectroscopy-based metabolomics analysis of serum could be a useful strategy to investigate the pathophysiology of BD, as well as to identify and validate potential biomarkers for diagnosis and/or for treatment follow-up.
In this study, it was possible to the identify seven key-metabolites, which showed to be in accordance with some other works described in the literature that used different analytical platforms for monitoring BD. Although the greatest differences between the two studied groups were observed in lipid ratios (more abundant in the BD group), due to the overlapping, some integration of the peak areas can not be taken as sufficiently precise for quantification of some metabolites. Nevertheless, it is important to mention that the spectral region used was similar to the work of Sussulini et al. (2009) indicating a greater accuracy in this spectral range when looking for the biomarkers and also showing the high reproducibility of the NMR spectroscopy technique in metabolomic profiling of BD individuals.
Authors’ contributions
SS participated in data collection, data analysis, and writing of manuscript. MP, ACC, EA, QC, and MNN participated in recruitment and assessment of patients and controls, data analysis, and writing. LR, MZG, and CDM participated in data collection, analysis, and writing. GMP and AJMB participated in NMR data collection, analysis, and writing. AL participated in conception, fund raising, and writing. MAFH, RP, LT, and EB participated in all the steps of conception, execution, and diffusion of results. All authors read and approved the final manuscript.
Acknowledgements
None.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The Local Ethics Committee approved this study (Numbers 35376414.8.0000.5505), and all subjects provided written informed consents before their inclusion in the study. The subjects consented in publication of data preserving anonymity.
Funding
We thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brasília, Brazil) for financial support and fellowships. SS received a Young Talent Scholarship from the CNPq. We also acknowledge the scientific grant from FAPESP (Process Number 2014/18938-8).
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1. HSQC and Description of 1H-NMR spectra for individuals with bipolar disorder and for healthy controls, HSQC contour map correlations and identification of possible key metabolites.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s40345-017-0088-2) contains supplementary material, which is available to authorized users.
Contributor Information
Sumit Sethi, Phone: +55 11 5576-4990, Email: sumitsethi92@gmail.com.
Mariana Pedrini, Email: mariana.pedrini@uebel.com.br.
Lucas B. Rizzo, Phone: +55 11 5576-4990, Email: lucas-rizzo@hotmail.com
Maiara Zeni-Graiff, Phone: +55 11 5576-4990, Email: ma_graiff@hotmail.com.
Caroline Dal Mas, Phone: +55 11 5576-4447, Email: carolinedalmas88@gmail.com.
Ana Cláudia Cassinelli, Phone: +55 11 3466-2100, Email: acmcassinelli@yahoo.com.br.
Mariane N. Noto, Phone: +55 11 5576-4990, Email: marianenoto@hotmail.com
Elson Asevedo, Phone: +55 11 5576-4990, Email: elsonmed@gmail.com.
Quirino Cordeiro, Phone: +55 11 3466-2100, Email: qcordeiro@yahoo.com.
João G. M. Pontes, Phone: +55 19 3521-3042, Email: jgquimico@yahoo.com.br
Antonio J. M. Brasil, Phone: +55 19 3521-3042, Email: ajmbrasil1@gmail.com
Acioly Lacerda, Email: acioly@institutosinapse.org.
Mirian A. F. Hayashi, Phone: +55 11 5576-4447, Email: mhayashi.unifesp@gmail.com
Ronei Poppi, Phone: +55 19 3521-1106, Email: rjpoppi@gmail.com.
Ljubica Tasic, Phone: +55193521-1106, Email: ljubica@iqm.unicamp.br, Email: ljubica.tasic@gmail.com.
Elisa Brietzke, Email: elisabrietzke@hotmail.com.
References
- Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- Brietzke E, Rosa AR, Pedrini M, Noto MN, Kapczinski F, Scott J. Challenges and developments in research of the early stages of bipolar disorder. Rev Bras Psiquiatr. 2016;38:329–337. doi: 10.1590/1516-4446-2016-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustillo JR. Use of proton magnetic resonance spectroscopy in the treatment of psychiatric disorders: a critical update. Dialogues Clin Neurosci. 2013;15:329–337. doi: 10.31887/DCNS.2013.15.3/jbustillo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanzo P, Thomas MA, Yue K, Oshiro T, Belin T, Strober M, McCracken J. Decreased anterior cingulate myo-inositol/creatine spectroscopy resonance with lithium treatment in children with bipolar disorder. Neuropsychopharmacology. 2001;24:359–369. doi: 10.1016/S0893-133X(00)00207-4. [DOI] [PubMed] [Google Scholar]
- Goes FS. Genetics of bipolar disorder: recent update and future directions. Psychiatr Clin North Am. 2016;39:139–155. doi: 10.1016/j.psc.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin FK, Jamison KR. Manic-depressive illness: bipolar disorders and recurrent depression. 2. New York: Oxford University Press; 2007. [Google Scholar]
- Hsu JH, Chien IC, Lin CH. Increased risk of hyperlipidemia in patients with bipolar disorder: a population-based study. Gen Hosp Psychiatry. 2015;37:294–298. doi: 10.1016/j.genhosppsych.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Lan MJ, McLoughlin GA, Griffin JL, Tsang TM, Huang JT, Yuan P, Manji H, Holmes E, Bahn S. Metabolomic analysis identifies molecular changes associated with the pathophysiology and drug treatment of bipolar disorder. Mol Psychiatry. 2009;14:269–279. doi: 10.1038/sj.mp.4002130. [DOI] [PubMed] [Google Scholar]
- Machado-Vieira R, Gattaz WF, Zanetti MV, De Sousa RT, Carvalho AF, Soeiro-de-Souza MG, Leite CC, Otaduy MC. A longitudinal (6-week) 3T 1H-MRS study on the effects of lithium treatment on anterior cingulate cortex metabolites in bipolar depression. Eur Neuropsychopharmacol. 2015;25:2311–2317. doi: 10.1016/j.euroneuro.2015.08.023. [DOI] [PubMed] [Google Scholar]
- Maher AD, Crockford D, Toft H, Malmodin D, Faber JH, McCarthy MI, Barrett A, Allen M, Walker M, Holmes E, Lindon JC, Nicholson JK. Optimization of human plasma 1H NMR spectroscopic data processing for high-throughput metabolic phenotyping studies and detection of insulin resistance related to type 2 diabetes. Anal Chem. 2008;80:7354–7362. doi: 10.1021/ac801053g. [DOI] [PubMed] [Google Scholar]
- Malhi GS, Tanious M, Das P, Coulston CM, Berk M. Potential mechanisms of action of lithium in bipolar disorder. Current understanding. CNS Drugs. 2013;27:135–153. doi: 10.1007/s40263-013-0039-0. [DOI] [PubMed] [Google Scholar]
- Manji HK, Quiroz JA, Payne JL, Singh J, Lopes BP, Viegas JS, Zarate CA. The underlying neurobiology of bipolar disorder. World Psychiatry. 2003;2:136–146. [PMC free article] [PubMed] [Google Scholar]
- Marmol F. Lithium: bipolar disorder and neurodegenerative diseases Possible cellular mechanisms of the therapeutic effects of lithium. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:1761–1771. doi: 10.1016/j.pnpbp.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Marohn S. Natural medicine guide to bipolar disorder. Charlottesville: Hampton Roads Publishing Company; 2011. [Google Scholar]
- Meiboom S, Gill D. Modified spin-echo method for measuring nuclear relaxation times. Rev Sci Instrum. 1958;29:688–691. doi: 10.1063/1.1716296. [DOI] [Google Scholar]
- Misra D, Bajpai U. Metabolite characterization in serum samples from normal healthy human subjects by 1H and 13C NMR spectroscopy. Bull Chem Soc Ethiop. 2009;23:211–221. doi: 10.4314/bcse.v23i2.44964. [DOI] [Google Scholar]
- Nierenberg AA, Kansky C, Brennan BP, Shelton RC, Perlis R, Iosifescu DV. Mitochondrial modulators for bipolar disorder: a pathophysiologically informed paradigm for new drug development. Aust N Z J Psychiatry. 2013;47:26–42. doi: 10.1177/0004867412449303. [DOI] [PubMed] [Google Scholar]
- Nørgaard L. The iToolbox for MATLAB; KVL: Frederiksberg. 2009. http://www.models.life.ku.dk.
- Oswald P, Souery D, Kasper S, Lecrubier Y, Montgomery S, Wyckaert S, Zohar J, Mendlewicz J. Current issues in bipolar disorder: a critical review. Eur Neuropsychopharmacol. 2007;17:687–695. doi: 10.1016/j.euroneuro.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Pålsson E, Jakobsson J, Södersten K, Fujita Y, Sellgren C, Ekman CJ, Ågren H, Hashimoto K, Landén M. Markers of glutamate signaling in cerebrospinal fluid and serum from patients with bipolar disorder and healthy controls. Eur Neuropsychopharmacol. 2015;25:133–140. doi: 10.1016/j.euroneuro.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Passos IC, Mwangi B, Vieta E, Berk M, Kapczinski F. Areas of controversy in neuroprogression in bipolar disorder. Acta Psychiatr Scand. 2016;134:91–103. doi: 10.1111/acps.12581. [DOI] [PubMed] [Google Scholar]
- Phillips ML, Kupfer DJ. Bipolar disorder diagnosis: challenges and future directions. Lancet. 2013;381:1663–1671. doi: 10.1016/S0140-6736(13)60989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- Sena MM, Chaudhry ZF, Collins CH, Poppi RJ. Direct determination of diclofenac in pharmaceutical formulations containing B vitamins by using UV spectrophotometry and partial least squares regression. J Pharm Biomed Anal. 2004;36:743–749. doi: 10.1016/j.jpba.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Strzelecki D, Grzelak P, Podgórski M, Kałużyńska O, Stefańczyk L, Kotlicka-Antczak M, Gmitrowicz A. Comparison of metabolite concentrations in the left dorsolateral prefrontal cortex, the left frontal white matter, and the left hippocampus in patients in stable schizophrenia treated with antipsychotics with or without antidepressants. 1H-NMR spectroscopy study. Int J Mol Sci. 2015;16:24387–24402. doi: 10.3390/ijms161024387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussulini A, Prando A, Maretto DA, Poppi RJ, Tasic L, Banzato CE, Arruda MA. Metabolic profiling of human blood serum from treated patients with bipolar disorder employing 1H NMR spectroscopy and chemometrics. Anal Chem. 2009;81:9755–9763. doi: 10.1021/ac901502j. [DOI] [PubMed] [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Wenger RK, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Enkhuizen J, Janowsky DS, Olivier B, Minassian A, Perry W, Young JW, Geyer MA. The catecholaminergic-cholinergic balance hypothesis of bipolar disorder revisited. Eur J Pharmacol. 2015;753:114–126. doi: 10.1016/j.ejphar.2014.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, MacInnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatham LN, Kennedy SH, Parikh SV, Schaffer A, Beaulieu S, Alda M, O’Donovan C, Macqueen G, McIntyre RS, Sharma V, Ravindran A, Young LT, Milev R, Bond DJ, Frey BN, Goldstein BI. The evolution of CANMAT Bipolar Disorder Guidelines: past, present, and future. Bipolar Disord. 2013;15:58–60. doi: 10.1111/bdi.12038. [DOI] [PubMed] [Google Scholar]
- Yoshimi N, Futamura T, Kakumoto K, Salehi AM, Sellgren CM, Holmén-Larsson J, Jakobssong J, Pålssong E, Landén M, Hashimoto K. Blood metabolomics analysis identifies abnormalities in the citric acid cycle, urea cycle, and amino acid metabolism in bipolar disorder. BBA Clin. 2016;5:151–158. doi: 10.1016/j.bbacli.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]


