Abstract
As cells age and are exposed to genotoxic stress, preservation of the genomic code requires multiple DNA repair pathways to remove single or double- strand breaks. Loss of function somatic genomic aberrations or germline deficiency in genes involved in DNA repair can result in acute cell death or following a latency period cellular transformation. Therapeutic exploitation of DNA repair by inhibition of poly (adenosine diphosphate [ADP]) ribose polymerases (PARP), a family of enzymes involved in the repair of single-strand and in some cases double-strand breaks, has become a novel cancer treatment. While the application of PARP inhibitors (PARPis) was initially focused on tumors with BRCA1 or BRCA2 deficiency, our current knowledge has extended synthetic susceptibilities of PARPi to deficiencies in proteins and pathways involved in DNA damage repair (DDR) in particular those that repair double-strand breaks using homologous recombination (HR). There is an increasing appreciation that genitourinary (GU) malignancies, including bladder, and especially prostate cancers, contain subsets of patients with germline and somatic alterations in HR genes that may reflect increased response to PARPis. In this review, we describe the mechanisms and rationale of PARPi use in GU cancers, summarize previously reported pre-clinical and clinical trials, and identify ongoing trials to determine how PARPis and strategies targeted at HR repair can be applied for widespread application in GU cancers.
Keywords: PARP1 inhibition, prostate adenocarcinoma, urothelial cell carcinoma, bladder cancer, prostate cancer, renal cell carcinoma, DNA repair defects, targeted therapy
Introduction
PARP Function Inhibition and Synthetic Lethality in Homologous Recombination Deficient Cells
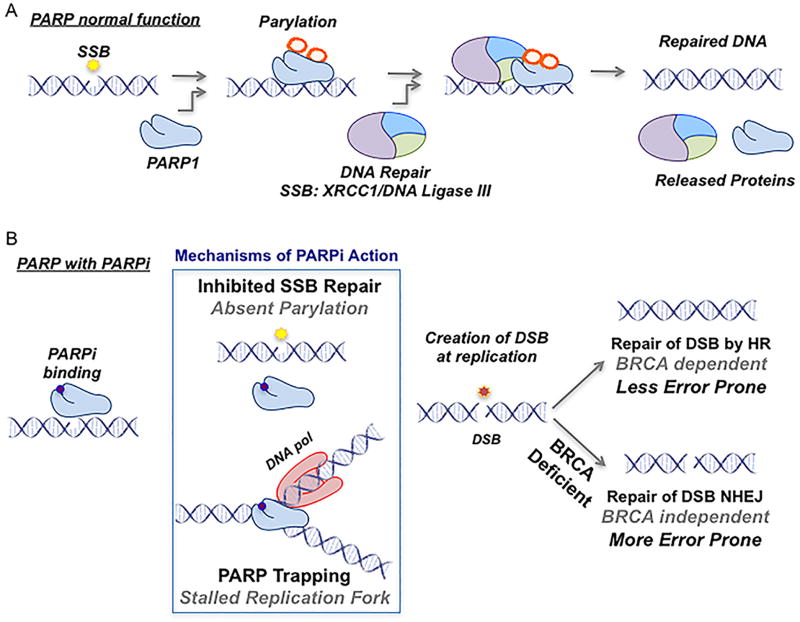
DNA is constantly under repair due to the damaging byproducts of cellular metabolism, de novo disruption of chemical bonds, and exposure to environmental agents1. The most common DNA aberration is discontinuity in one strand, also known as a single-strand break (SSB), which is predominantly repaired via base-excision repair (BER) mechanisms, but also by nucleotide excision repair (NER) and DNA mismatch repair (MMR) to a much lesser extent. SSB repair requires the orchestration of several proteins, including polyadenosine diphosphate [ADP] ribose polymerase-1 (PARP1) and -2 (PARP2). PARP1 and PARP2 catalyze the addition of poly (ADP)-ribose (also known as PARylation) from nicotinamide adenine dinucleotide (NAD+) molecules to client proteins in malignant and non-malignant cells and to PARP1/2 themselves (Fig 1)2. There are 17 members of the PARP family, of which PARP1 was the first to be described3. PARP1 is responsible for the majority of PARylation in malignant and non-malignant cells which is essential for the efficiency of cellular functions including DNA repair and chromatin regulation4, 5.
Figure 1.
PARP Function and Mechanism
PARP1 acts as a DNA damage sensor and binds to the site of SSBs, increasing its enzymatic activity as much as 500-fold (Fig 1A)6. PARP1 binding results in the recruitment and PARylation of a number of different DNA repair proteins including DNA polymerase beta and x-ray repair cross complimenting protein 1 (XRCC1) to repair the SSB (Fig 1)7. Recruitment of these proteins causes destabilization and disassociation of the PARP-DNA interaction, which is necessary for DNA repair to proceed8. If the repair of SSBs is deficient or disabled, SSBs can be converted to double-strand breaks (DSB) due to collapse or blockage of DNA replication forks during the S phase of the cell cycle9. In this situation, the mechanisms of DSB repair will be activated and attempt to repair the lesion10. Cells have evolved sophisticated mechanisms to repair DSBs to maintain genetic fidelity as unrepaired or misrepaired DSBs can result in senescence, genetic instability, apoptosis, and transformation10. PARP1 knockout mice, for example, are deficient in the repair of SSBs but accomplish DNA repair through redundantly utilizing, preferentially, one arm of the DSB repair pathway known as homologous recombination (HR). Due to this failsafe, PARP1 knockout mice are viable with no visible phenotypes of infertility, or increased risk of malignancy11. Most of DSB repair is accomplished by two mechanisms: homologous recombination (HR) and non-homologous end joining (NHEJ) (Fig 1B). HR is slower and more accurate because it utilizes the sister chromatid as a template during the S/G2 phases of the cell cycle12. NHEJ is faster but more error-prone than HR as damaged ends are directly ligated12.
BRCA Loss, DNA Repair, and Synthetic Lethality
BRCA1 and BRCA2 are large nuclear proteins that play an integral role in the HR pathway13. Loss of function of BRCA1 and/or BRCA2 disrupts HR and DSB repair resulting in utilization of more error-prone pathways (NHEJ or the single-strand annealing [SSA] sub-pathway of HR) (Fig 1B)14. Women who inherit a BRCA1/2 mutation have an estimated lifetime risk of 40%–80% of developing breast cancer and an 11%–40% risk of developing ovarian cancer15. In patients with germline BRCA1/2 carrier mutations, their tumors typically have a somatic loss of heterozygosity with de novo DNA structural aberration of the remaining functional BRCA allele16. DSB repair in BRCA-deficient tumors is largely dependent on NHEJ for the repair of DSBs (Fig 1B)14. The polymerase POLQ has also been recently shown to be redundant to HR and facilitates an essential bypass mechanism for DSBs in HR-deficient tumors17, 18. Although cells deficient in either PARP or BRCA alone are viable, the loss of both results in a lethal phenotype. This concept, known as ‘synthetic lethality’, was first described by Dobzhansky in 1946, and in 1997 by Hartwell et. al., who first suggested its application to cancer therapeutics19, 20. In 2005, two groups demonstrated that treatment of BRCA1 or BRAC2 deficient normal and tumor cells with PARP inhibitors (PARPis) resulted in synergistic cell death associated with unrepaired DSBs21, 22. Creation of synthetic lethality, by combining PARPis with BRCA loss intrinsic to the tumor, proved that a genetic vulnerability could be exploited for selective treatment to minimize off-target toxicity. The relatively low frequency of germline BRCA1/2 mutations associated with prostate adenocarcinoma and bladder urothelial cell carcinoma initially appeared to limit the clinical applicability of PARPi to GU tumors23, 24. However, it was hypothesized that this synthetic lethal approach with PARPis was not solely dependent on inherited BRCA1 or BRAC2 loss alone but could possibly extend to other germline or somatic deficiencies in the HR pathway.
PARP-inhibitors: Mechanism of Action
Small molecule PARPis have existed for nearly 30 years and have progressed through several generations of chemical modification. Structural modifications improved selectivity and potency, but all PARPis have a nicotinamide pharmacore that results in competitive inhibition by binding/blocking the catalytic domain of PARP1 and PARP225. PARP2 is far less abundant and is responsible for 5–10% of total PARylation in response to DNA damage26. All currently studied PARPis bind to both PARP1 and PARP2 (Table 1). The original primary mechanism of action for cytotoxicity of PARPis was attributed to the generation of SSBs and a resultant overwhelming number of DSBs via catalytic inhibition of PARP122. A second and possibly equally important mechanism of PARPi action was shown by Murai et. al. in which PARPis trap PARP1/2 at the sites of DNA damage, known as “PARP-trapping.” The interaction between PARP1/2 and DNA is normally reversible, however, PARPis may induce stabilization of the interaction27. The irreversibly bound PARP-DNA complex prevents DNA repair, transcription, and replication, and is ultimately cytotoxic28. Evidence for the importance of PARP trapping is demonstrated by the greater cytotoxic effect of PARPis compared to PARP depletion (PARP1/2−/−) in BRCA-deficient cells as well as the development of resistance to PARPi treatment by decreased PARP1 expression29. A third hypothesis combines both mechanisms suggesting that catalytic inhibition of PARP1 via PARPis prevents automodification of the PARP1 enzyme, resulting in a stabilization of the PARP-DNA interaction30. Research regarding the precise mechanisms of PARPi action and their effect on tolerability will be an important part of their application to specific tumor types.
Table 1.
Current PARP inhibitors undergoing clinical evaluation
| Inhibitor | Other name(s) | Company |
|---|---|---|
| Olaparib | AZD2281, KU0059436 | AstraZeneca |
| Veliparib | ABT-888 | Abbvie |
| Niraparib | MK-4827 | Tesaro |
| Talazoparib | BMN-673 | BioMarin Pharmaceuticals |
| Rucaparib | AG-14699 | Clovis Oncology |
| CEP-9722 | Teva Pharmaceutical Industries | |
| SC10914 | Jiangxi Qingfeng Pharmaceutical Co. Ltd. |
PARPis: Early Clinical Results
The hypothesis-driven development of PARPis led to rapid initiation of clinical trials utilizing single-agent PARPis to treat BRCA-deficient tumors, namely breast and ovarian cancers. Single-agent use of AZD-2281 (Olaparib) was recently approved by the FDA for the treatment of women with recurrent ovarian cancer carrying a germline BRCA mutation treated with three or more prior lines of chemotherapy. This approval was partly based on the results of a phase II, international, multicenter, single-arm trial of 193 heavily pre-treated (averaging 4.3 lines of prior therapy) women with confirmed germline BRCA-mutated ovarian cancer. Patients were given 400 mg Olaparib twice daily until toxicity or progression. They reported an overall response rate of 31%, stable disease (at 8 weeks) in 40% and a median OS of 16.6 months31. Ledermann et al. also conducted a phase II, randomized, placebo-controlled study of maintenance treatment with Olaparib in patients with platinum-sensitive, relapsed, high-grade serous ovarian cancer who had previously received two or more platinum-based regimens and had a partial or complete response to their most recent regimen. BRCA1/2 carrier mutation status was not required. Patients were randomly assigned to receive 400 mg Olaparib twice daily or placebo within 8 weeks of completion of their last dose of platinum-based chemotherapy until disease progression. Progression free survival was significantly longer in the Olaparib arm (median, 8.4 months vs. 4.8 months, p<0.001), though there was no difference in overall survival at the time of interim analysis (38% maturity) (HR 0.94; 95% CI, 0.63–1.39; p=0.75). Demographics and germline BRCA mutation status were well balanced between the two groups. The lower risk of disease progression was present in all sub-groups analyzed (though there were too few BRCA1/2wt patients for analysis). The toxicity profile was similar to previous reports and there were no significant differences between the groups regarding health-related quality of life32.
While single-agent PARPis have been effective in select patients, their tumoricidal effect may be enhanced when administered with DNA damaging chemotherapy. Multiple pre-clinical studies confirm the ability of PARPis to potentiate DNA damaging agents such as cisplatin and carboplatin in BRCA1−/− or BRCA2−/− cell culture and xenograft tumor models33–35. Platinum based agents, such as cisplatin or carboplatin induce intra (repaired via NER) and inter-strand DNA crosslinks (repaired partly via HR)37. It is hypothesized that combinatorial therapy with PARPi and platinum based agents, particularly in a system deficient in HR, results in an increased accumulation of DNA damage and a subsequent dependence on PARP1 mediated DNA repair38. Patients with BRCA1/2 deficient breast and ovarian tumors appear to have an inherent sensitivity to platinum-containing chemotherapeutic regimens. This was demonstrated by improved progression-free survival, overall survival, and higher partial and complete response rates compared to patients lacking these mutations39–45. PARPi combined with platinum based chemotherapy may exploit this inherent sensitivity and has been tested in a randomized, open-label, phase II study in patients with recurrent high-grade serous ovarian cancer who had received up to three previous courses of platinum-based chemotherapy with >6 months to disease progression following their last treatment. Germline BRCA mutation status for these patients was known and patients were randomized to receive Olaparib plus paclitaxel and carboplatin or paclitaxel and carboplatin without additional therapy. Though this cohort of patients was heavily pre-treated, progression free survival was significantly longer in the Olaparib plus chemotherapy group (median 12.2 months [95% CI 9.7 – 15] vs. 9.6 months [95% CI 9.1–9.7], HR 0.51 [95% CI 0.34–0.77]; p=0.0012), especially when patients were sub-stratified by their germline BRCA mutation status (HR 0.21 [0.08–0.55]; p=0.0015)46. A clinical trial (NCT02470585) to evaluate combination therapy (veliparib + carboplatin + paclitaxel vs. carboplatin + paclitaxel) in previously untreated patients with stage III or IV high grade serous epithelial ovarian cancer is currently recruiting.
Though these results were promising, the relative low frequency of germline BRCA mutations in other solid tumors seemed to initially limit the clinical application of PARPi. Fortunately, advances in next-generation sequencing technology have enabled the analysis of the genetic complement of thousands of tumors. This led to the identification of novel somatic mutations in genes encoding for components involved in HR that can functionally mimic BRCA loss and synergize with PARPi treatment. This concept was termed “BRCAness”, defined as a tumor with HR deficiency without deficiency in BRCA1/247. McCabe et. al. demonstrated this with the discovery that deficiency in one of a number of proteins involved in HR (RAD51, RAD54, DSS1, RPA1, NBS1, ATR, ATM, CHK1, CHK2, FANCD2, FANCA, and FANCC) induced sensitivity to PARPi48. The list of genes that, when mutated, may be involved in HR deficiency continues to expand (Table 2). Mutations in genes associated with transcriptional regulation and cell cycle control that affect DNA repair have also been shown to confer sensitivity to PARPi (Table 2)49. Clinical data to support this notion that PARPis have more broad anti-tumor activity has recently been reported with Niraparib as maintenance treatment for women with platinum-sensitive, recurrent ovarian cancer. Although women with germline BRAC1/2 mutations and those with HR deficiency benefited the most from Niraparib, women with neither of these predictive features still benefited compared to placebo (progression-free survival of 9.3 months versus 3.9 months, p<0.001)50 These discoveries may potentially lead to an expanded application of PARPi in the treatment of tumors genetically wild-type for BRCA1/2 but deficient in HR due to somatic or germline mutations in other genes associated with HR-mediated DNA repair.
Table 2.
Common BRCAness Genes and Evidence for PARPi sensitivity
DNA repair genes associated with homologous recombination and PARPi sensitivity
| Gene | Function in DSD repair | Evidence for PARPi sensitivity | Reference |
|---|---|---|---|
| ATM | protein kinase, DNA damage sensor, involved in repair of DSBs in heterochromatin, V(D)J class switching, meiotic recombination | +++ | Mateo J. et al., N. Engl. J. Med. 2015. |
| ATR | protein kinase, DNA damage sensor, phosporylates CHK1, BRCA1, BLM, FANCD2/FANCI, absence results in deficient HR | ++ | DeFranco C. et al., Cancer Res. 2016. |
| BAP1 | deubiquitinase required for efficient assembly of BRCA1/RAD51 at site of DSB | ++ | Goga A. et al., J Clin Oncol. 2016/Affar E. et al., Proc. Natl. Acad. Sci. USA. 2013. |
| BRCA1 | phosphoprotein that assists in 5′ to 3′ resection of DSBs, loading of RAD51 | +++ | Ledermann J. et al., N Engl J Med. 2012/Mateo J. et al., N. Engl. J. Med. 2015. |
| BRCA2 | phosphoprotein that assists with RAD51 loading | +++ | Ledermann J. et al., N Engl J Med. 2012/Mateo J. et al., N. Engl. J. Med. 2015. |
| CDK12 | regulates expression of BRCA1, FANCI, FANCD2, ATR | ++ | Ashwoth A. et al., Cancer Res. 2014./Shapiro G. et al, Cell Rep. 2016. |
| CHK2 | recruits ATM/ATR to cell cycle effectors and DNA repair machinery, phosphorylates BRCA1 | + | Fulda S. et al., Oncotarget. 2016 |
| FANCA | member of FA core complex which recruits and activates FANCD2/FANCI that co-localizes with BRCA1 | + | McCabe N. et al., Cancer Res. 2006 |
| FANCC | member of FA core complex which recruits and activates FANCD2/FANCI that co-localizes with BRCA1 | ++ | Wells S. et al., Clin Cancer Res. 2015 |
| FANCD2 | co-localizes with BRCA1 during process of homologous recombination | + | McCabe N. et al., Cancer Res. 2006/Villalona-Calero, M., Front Oncol. 2014 |
| FANCF | member of FA core complex which recruits and activates FANCD2/FANCI that co-localizes with BRCA1 | + | McCabe N. et al., Cancer Res. 2006 |
| PALB2 | promotes BRCA2 function and formation of BRCA complex | +++ | Mateo J. et al., N. Engl. J. Med. 2015. |
| NBS1 | localizes to sites of DSBs, complexes with MRE11a/Rad50, activates ATM | +++ | McNeish I. et al., Lancet Oncol. 2016 |
| WRN | 3′ to 5′ exonuclease promotes repair of DSBs | + | Helleday, T. et al., Caner Res. 2010 |
| RAD51c | assists with recruitment, stablization, and loading of RAD51 | +++ | McNeish I. et al., Lancet Oncol. 2016/Bang Y. et al., Mol Cancer Ther. 2013. |
| RAD51d | assists with recruitment, stablization of RAD51 | +++ | McNeish I. et al., Lancet Oncol. 2016 |
| MRE11A | localizes to sites of DSBs, complexes with NBS1/Rad50, activates ATM | ++ | Li D. et al., Clin Cancer Res. 2014 |
| CHK1 | protein kinase, phosporylates BLM, FANCE, FANCD2, RAD51 promoting DSB repair | ++ | Dent P. et al. Mol Pharmaol. 2012./Dent P. et al. Cancer Biol Ther. 2013 |
| BLM | RecQ helicase involved in DSB resection | + | Helleday, T. et al., Caner Res. 2010 |
cell culture
patient derived xenografts/animal models
clinical trial
List of genes whose deficiency is associated with homologous repair deficiency. Evidence for PARPi sensitivity based on response to PARPi in presence of deleterious mutation(s) or inhibition of listed gene without associated mutations in BRCA1/2 or other BRCAness genes
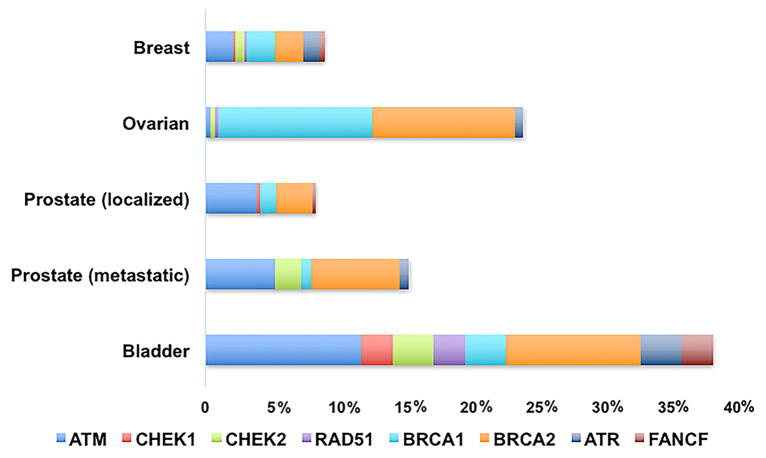
PARPi holds a great deal of promise in the management of genitourinary (GU) cancers such as prostate and urothelial cancer. The genomes of prostate adenocarcinoma (PCa) and urothelial carcinoma (UC) have been shown to contain reasonable subsets of patient tumors with somatic mutations in genes implicated in the HR pathway in both early- and late-stage tumors (Fig 2)51–53. In this article, we summarize the scientific rational for targeting PARP1 in GU cancers, previously reported pre-clinical and clinical trials, and ongoing trials attempting to determine how PARPis can best be used in the management of GU cancers, particularly those with mutations in genes associated with HR.
Figure 2.
Mutation frequency DNA repair/Homologous Recombination genes. The mutation frequency of somatic mutations is listed by occurrence in the TCGA of each provisional histology.
PARPi and Prostate Adenocarcinoma
PCa is the most common cancer in men with 180,000 new diagnosis and 26,000 deaths in the US in 201654. Metastatic PCa is lethal with no curative therapies. Androgen deprivation is first-line therapy for hormone sensitive metastatic PCa, though several large phase III clinical trials (CHAARTED and STAMPEDE) have recently demonstrated improved survival combining docetaxel and androgen deprivation in hormone sensitive patients55, 56. Unfortunately, most patients with metastasis will progress to castration resistant PCa (CRPC) despite androgen blockade within 2–3 years57. Although additional treatments exist for metastatic CRPC, including docetaxel, enzalutamide, abiraterone, radium-223, sipuleucel-T and cabizitaxel, these therapies are not effective in all men. Responses are not durable, and provide only a modest overall survival benefit58. The comprehensive genetic characterization of PCa identified a significant number of germline and somatic mutations in DNA repair genes, specifically those involved in HR59, 60. Given the frequency of these HR gene mutations (up to 17–19% in metastatic CRPC)61, the paradigm of PARPi synthetic lethality will likely be an important addition to the management of prostate cancer, particularly in the metastatic setting.
Genetic Characterization of Prostate Cancer and DNA Damage Repair Deficiency
Prostate cancer is now firmly recognized as a cancer that is predisposed in men carrying germline BRCA mutations. The relative risk of developing prostate cancer for men with germline BRCA1 mutations <age 65 is 1.8 (95% CI 1.01–3.29) and in men with germline BRCA2 mutations the relative risk is 8.6 (95% CI 5.1–12.6)62. BRCA mutation carriers are significantly more likely to present with a Gleason score of 8 or higher, T3/T4 disease, nodal involvement, and metastasis then non-carriers63. Pritchard et al utilized whole-exome sequencing or targeted next-generation sequencing assays in a large population of men with biopsy-proven metastatic prostate cancer and found 11.8% had at least one presumed pathogenic germline mutation in a gene involved in DNA repair of which the majority of these genes were implicated in HR52. BRCA1/2 germline mutations were found in 6% of metastatic patients which contrasts with <1% of men with localized prostate cancer. This provocative data suggests enrichment of germline BRCA1/2 mutation carriers and other germline DNA repair mutation carriers in metastatic versus localized disease.
Somatic HR gene mutations including BRAC1/2 and ATM also appear to be present in a significant subset of PCa cases. The Cancer Genome Atlas (TCGA) performed a comprehensive molecular characterization of localized prostate cancer specimens following radical prostatectomy and found that 9% of tumors had somatic mutations (truncating or missense) in genes associated with HR and the repair of DSBs (BRCA1/2, FANCD2, CKD12, ATM)64. The mutation frequency of these genes is even greater in patients with more aggressive or later stage disease. Robinson et al. identified alterations in DNA repair genes (BRCA1/2, ATM, CDK12, FANCA, RAD51B/C) in 15% of 150 metastatic CRPC biopsy samples with many of these gene products associated with HR61. The existing germline and somatic genetic data suggests that DNA repair, specifically HR, may have metastasis suppressor properties indirectly and/or perhaps directly in prostate cancer. This data makes a compelling argument that men with metastatic prostate cancer should consider undergoing both germline and somatic mutation HR gene testing.
Unique Features of PARP1 in Prostate Cancer
Molecular studies suggest that PARP1 may have a functional role in prostate cancer progression. PARP1 directly regulates the androgen receptor (AR), and PARPi decreases recruitment of the AR to target promoters65. PARP1 may also be involved in regulation of AR activity at later stages of androgen-independent action. PARP1 activity is significantly increased in CRPC cells compared to hormone sensitive prostate cancer cells, and PARPi causes depletion of both the AR and PARP1 on chromatin in CRPC cells65. Conversely, the AR itself may also be involved in the regulation of DSB repair66, 67. These results indicate that PARPi may have a unique multimodal mechanism of action in the treatment of prostate cancer.
The most common gene fusions in PCa involve TMPRSS2-ERG found in over 50% of cases of metastatic PCa61, 64, 53. ERG is an oncogene of the ETS family of transcription factors and TMPRSS2 is an androgen-regulated gene preferentially expressed in the prostate. The TMPRSS2-ERG fusion can result in androgen regulated overexpression of ERG. The clinical significance of the TMPRSS2-ERG fusion is unknown, but the presence of the TMPRSS2-ERG gene fusion is associated with PARPi sensitivity in vitro and in vivo68. The mechanistic rationale was described by Brenner et. al. who demonstrated that ERG interacts with PARP1 via DNA-dependent protein kinase, catalytic subunit (DNA-PKcs) and that PARP1 is required for ERG-mediated transcription. Overexpression of ERG increases the accumulation of DSBs and treatment with ERG siRNA led to a reduction in detectable DSBs. Treatment of ETS positive tumors with Olaparib led to significantly diminished growth in xenograft models compared to ETS negative tumors68.
Somatic loss of function mutations in the tumor suppressor PTEN (found in15% and 40% of localized and metastatic prostate tumors), results in impaired HR capacity and confers PARPi sensitivity similar to BRCA1/2 deficiency in prostate cancer cells61, 64, 69, 70. Though clinical evidence to date has not revealed an association between these features and response to PARPi, these unique interactions may provide the basis for other possible therapeutic implications for PARPi in prostate cancer in combination with androgen deprivation, chemotherapy, and/or radiotherapy71, 72.
PARP1 Inhibition Clinical Trials in Prostate Cancer
One of the first studies to demonstrate a clinical benefit of PARPi in PCa was a phase I trial of Olaparib in 60 solid tumor patients who were germline BRCA mutation carriers73. Of the three patients with CRPC, one was a BRCA2 mutation carrier and had resolution of their bony metastasis and a >50% decrease in their PSA73. These preliminary results enabled further trials in patients with advanced PCa. Hussain et al. performed a single-arm pilot study to determine the safety and efficacy of Temozolomide (TMZ) combined with Veliparib in patients with metastatic CRPC who had progressed following treatment with docetaxel74. TMZ is an alkylating agent that generates DNA adducts, which can produce SSBs during the process of their repair. PARP1 can bind to these SSBs, and in the presence of PARPis, facilitate PARP1 trapping75. For these reasons, TMZ has shown success in combination with PARPi in other tumor types76. Veliparib increased the sensitivity of prostate cancer cells to TMZ in vitro and in vivo34, 77. Veliparib also reversed resistance to TMZ in a mouse model of prostate cancer resulting in improved overall survival77. Of the 25 patients evaluated in this study, two had a partial response (8%), 13 (52%) had a stable PSA, and 10 (40%) had progression. Patients were not pre-screened for somatic or germline HR-mediated DNA repair gene mutations, though 8 of the 25 patients were assessed for TMPRSS2:ERG fusion (present in only 1 patient). The combination was tolerable, but demonstrated limited clinical activity72. The lack of pre-screening for HR deficiency status, the low dose of Veliparib, the weak ability of Veliparib to trap PARP1 and the unproven benefit of TMZ in metastatic CRPC likely contributed to these modest results.
The Trial of PARP Inhibition in Prostate Cancer (TOPARP-A) sought to determine whether patients with metastatic CRPC responded to treatment with full dose Olaparib71. TOPARP-A was a phase II study in which 49 patients with metastatic CRPC were treated with Olaparib, 400 mg twice daily. Prior therapies included docetaxel (49 men, 98%), abiraterone or enzalutamide (49 men, 98%), and cabazitaxel (29 men, 58%). Before treatment, men were biopsied, and their tumors sequenced with exome and transcriptome analysis. In 16 of the 49 patients (33%), homozygous deleterious mutations were found in HR repair genes (BRCA1/2, ATM, CHEK2, FANCA). Of these patients, 88% had a response to Olaparib. Only two of the 33 (6%) HRD-negative patients had a clinical response. Median overall survival was 13.8 months in the HR deficiency group vs. 7.5 months in the HR wild-type group (p=0.05). Dose reductions to 300 mg twice daily were required in 13 patients, and anemia was the most common cause of dose reduction (7 patients). These results highlight the potential application of a biomarker (i.e. HR deficiency) for pre-selection of men that may benefit from PARPis. This study resulted in the FDA granting Olaparib breakthrough therapy designation for patients with BRCA1/2 or ATM-mutated metastatic CRPC who had received a prior taxane-based chemotherapy and at least one next generation androgen receptor directed therapy (enzalutamide or abiraterone).
NCI9012 was a phase II study of metastatic CRPC patients in which metastatic tumors were biopsied and assessed for ETS fusions (ie TMPRSS2:ERG), stratified by their ETS fusion status, and then randomized to receive Abiraterone or Abiraterone plus Veliparib. The primary endpoint was PSA response. Secondary endpoints included safety, objective response rate, and progression-free survival. In the study, 185 eligible patients underwent biopsy (89 soft tissue, 96 bone); 159 (86 %) had adequate tissue, 35% were ETS+. Exploratory analysis from sequencing biopsy tissue (N=75) was performed in which 19 patients (25%) had DNA repair gene deficiency (DRD; homozygous deletions or deleterious mutations: BRCA 1, BRCA 2, ATM, FANCA, PALB2, RAD51B, and RAD51C). Preliminary results reveal a trend in favor of Abiraterone + Veliparib for PSA response (71% vs. 64%, p=0.33), objective response rate (53% vs 43%, p=0.32), and median PFS(11 vs 8.8 mo, p=.87)72. Regardless of treatment, patients with somatic deleterious DRD mutations had longer median PFS (13.5 [95% CI: 8.2-NR] vs. 5.8 [95% CI: 4.2–8.2] months, p=0.01) and a higher PSA response rate (89% vs. 57%, p=0.02) than those without somatic DRD mutations. This data suggests that patients with DRD mutations may have an improved response to either PARPi or ADT, through unexplained mechanisms. The increase in frequency in frequency of DRD mutations found in CRPC tumors compared to early confined cancers, suggest that these tumors may undergo selection during progression. Future studies that attempt identify the mechanism of interaction between DRD mutations, AR signaling and PARP inhibition will be important to optimize treatment. A summary of ongoing clinical trials with PARPis for the treatment of PCa is listed in Table 3.
Table 3.
Current Clinical Trials of PARP1 Inhibitors in Prostate Cancer
| Study # | Phase/Design | Disease State | Agent | Prior Treatment | Primary Outcome |
|---|---|---|---|---|---|
| NCT01972217 | II, double blind, placebo controlled, RCT | Metastatic CRPC | Abiraterone + Olaparib vs. Abiraterone + placebo | Docetaxel | Radiologic PFS |
| NCT02324998 | I, open label RCT | Localized intermediate/high risk prostate cancer | Neoadjuvant Olaparib vs. neoadjuvant Olaparib + Degarelix | none | Degree of PARP inhibition |
| NCT01078662 | II, open label non-randomized, non-controlled | Advanced ovarian, breast, prostate, pancreatic, advanced tumors w/BRCA1 and/or BRCA2 mutations | Olaparib | All ok but previous PARPi | Tumor response rate |
| NCT02500901 | I, open label, non-randomized, non-controlled | Metastatic prostate cancer | Enzalutamide + Niraparib | All ok but prior PARPi, ezalutamide or other next generation AR targeted therapy | Maximum tolerated dose |
| NCT01576172 | II, open label RCT | Metastatic CRPC | Abiraterone + prednisone + Veliparib vs. Abiraterone + prednisone | All ok | PSA response rate |
| NCT02952534 | II, open label | Metastatic CRPC w/HR deficiency (BRCA1/2, ATM, or molecular evidence of HR deficiency) | Rucaparib | Abiraterone and/or Enzalutamide and 1 prior taxane based chemotherapy. No prior PARPi, mitoxantrone, cyclophosphomide, platinum-based chemotherapy. | ORR and PSA response |
| NCT02975934 | III, open label, RCT | Metastatic CRPC w/HR deficiency (BRCA1/2, ATM) | Rucaparib vs. standard therapy (enzalutamide, abiraterone + prednisone, or docetaxel) | 1 prior next generation androgen receptor targeted therapy. No prior PARPi | rPFS |
PARPi and Bladder Urothelial Carcinoma
Over 77,000 new cases of urothelial cancer (UC) will be diagnosed in 2016 in the United States, representing 4.6% of all new cancer cases54. Approximately 30% of newly diagnosed patients present with muscle invasive bladder cancer (MIBC), of whom 50% will progress to distant metastasis, and 5% will initially present with metastatic disease78. Unfortunately, neither the management nor the mortality of UC has changed significantly in over 30 years. UC is responsive to systemic chemotherapy of multi-drug platinum combinations. MVAC (methotrexate, vinblastine, doxorubicin, and cisplatin) was first applied to UC in 198579, and is still utilized today with a slight modification of dose and schedule (dose dense MVAC, ddMVAC). Prior to 2016, the only change in systemic therapy for UC was the combination of gemcitabine and cisplatin that became an alternative to MVAC in the 2000’s after it demonstrated similar outcomes with less toxicity80. For over 25 years no new agents were FDA approved. PD-L1 inhibitors, Atezolizumab and Nivolumab, were approved by the FDA in 2016 and granted breakthrough status as second-line immunotherapy for patients with locally advanced or metastatic disease that have progressed during or following platinum-containing chemotherapy. These decisions were based on studies revealing similar, sufficient safety and efficacy (median PFS: Atezolizumab, 2.1 months [95% CI, 2.1–2.1], Nivolumab, 2.8 months [95% CI, 1.5–5.9], median OS: Atezolizumab, 7.9 months [95% CI, 6.6–9.3], Nivolumab 9.7 months [95% CI, 7.3–16.2])81, 82. Unfortunately, only 20–30% of patients with metastatic UC will achieve a partial or complete response to checkpoint immunotherapy and there are currently no reliable methods to predict response81. Thus, further research to identify new strategies for treatment of UC remain a critical focus.
Genetic Characterization of Bladder Cancer and DNA Damage Repair Deficiency
The characterization of the genomic landscape of UC has identified targetable genomic alterations, including a high number of loss of function mutations in DNA damage repair genes39, 67, 68. In 2004, Bentley et al. hypothesized that a deficiency in the repair of DSBs may be responsible for the high frequency of chromosomal instability observed in bladder cancer. Prior to the identification and isolation of specific proteins involved in DNA damage repair, crude nuclear extracts from high grade bladder carcinoma were found to be more error-prone compared to extracts from normal urothelial cells for in vitro DNA repair83. The TCGA analysis of bladder UC provided a possible explanation for these results as truncating and missense mutations in genes associated with the BRCAness phenotype and genes known to confer PARPi sensitivity (CHEK1/2, RAD51, BRCA1/2, ATM, ATR, MDC1, FANCF) were identified in 34% of tumors51, 84–87(TCGA, Provisional 2015). Similar to BRCA-deficient breast and ovarian cancer, sensitivity to neoadjuvant platinum-based chemotherapy is significantly associated with mutations in genes involved in HR repair. Plimack et al. found response to neoadjuvant cisplatin chemotherapy was associated with mutations in general DNA repair genes (p < 0.001), some of which are strongly associated with HR (ATM, and FANCC). Of the patients who responded, 13/15 (87%) contained deleterious mutations in genes associated with DNA repair, whereas none of the non-responders contained these mutations. The presence of these mutations was also associated with significantly improved progression-free (p = 0.0085) and overall survival (p = 0.007). The rate of pT0 or complete response was 41% in DRD-positive patients88. The prevalence of these somatic mutations in HR genes as well as their association with platinum sensitivity suggests PARP as a rational target for treatment of UC in select patients.
PARP1 Inhibition in Bladder Cancer
The only published data regarding PARPi in UC is in pre-clinical cell culture and xenograft models. Jian et al. evaluated the effect of the PARPi CEP-9722 and its active metabolite, CEP-8983 in human UC cell lines and xenograft models of nude mice. Cells and xenografts were treated with cisplatin, CEP-8983 alone, or a combination of cisplatin and CEP-8983. Western blot and immunofluorescence were used to assess DNA-repair proteins (ATM, BRCA1, MRE11, RAD51, RAD50, CHK1, CHK2) and to quantify HR-mediated DNA DSB repair. Radiation induced RAD51 nuclear localization was utilized as functional marker of HR repair activity. As the authors hypothesized, reduced capacity for HR repair (decreased RAD51 nuclear localization) was associated with increased sensitivity to PARPi. The combination of PARPi and cisplatin caused a significant increase in DNA damage vs cisplatin alone89. Though these pre-clinical results are encouraging, there are no clinical trials of PARPi in bladder UC.
Future directions
Continuing larger trials with PARPis in patients with PCa will be essential to validate their clinical benefit. Expanding preclinical in vitro and in vivo models in UC to validate the efficacy of PARPi monotherapy as well as combination therapy is the first step toward clinical trials in selected patients with metastatic UC and HR deficiency. Given the early preliminary data, the development of accurate, timely, and cost efficient methods of predicting response to PARPi is desperately needed. Though a number of genes involved in DNA repair, and more specifically HR have been identified to confer sensitivity to PARPi, limiting PARPi use to this small cohort of HR genes could significantly limit the patient population that may benefit from these medications. Multiple approaches are underway to develop biomarkers for identification of patients who will respond to PARPi. Larsen et al. for example, analyzed 55 familial BRCA1 or BRCA2 breast tumors and 128 sporadic breast tumors to identify a transcriptional signature of DNA repair deficiency90. Others have been working to develop functional HR assays via immunostaining using surrogates such like RAD51 or MRE11 localization91, 92. One assay that combines tumor sequencing with DNA cytogenetics is the Myriad Homologous Recombination Deficiency (HRD) test. The Myriad HRD test is performed on formalin-fixed, paraffin-embedded (FFPE) tumor samples evaluating for 54,091 single nucleotide polymorphisms as well as 43 genes involved in HR, including BRCA1 and BRCA2. A HR deficiency score is calculated based on loss of heterozygosity (LOH), large scale transitions (LST), and telomeric-allelic imbalance (TAI). The Myriad HRD test has been validated in breast cancer93, and recently in ovarian cancer. Using the Myriad HRD test, Timms et al. analyzed samples from the NOVA study, a phase III trial of Niraparib in platinum-sensitive ovarian cancer patients who either had a germline BRCA1/2 mutation or responded to their most recent platinum-based chemotherapeutic regimen. All patients in the germline BRCA1/2 mutation cohort met the cutoff score for HR deficiency, as did 55% of patients in the non-germline BRCA1/2 mutated cohort94. Patients who met the cut off score in the non-germline BRCA1/2 mutated cohort also had a significant improvement in progression-free survival compared to placebo (median 12.9 months vs. 3.8 months, p < 0.0001)94. A positive HRD score, or meeting the cut off for HR deficiency, has also been shown to predict pathologic response to neoadjuvant chemotherapy in triple-negative and BRCA1/2 mutated breast cancer95. Pre-clinical studies are currently underway to validate the accuracy of these tools to predict HR deficiency and PARPi sensitivity in PCa and bladder UC. For example, we are currently obtaining whole genome sequencing and HRD scores of bladder UC patient derived xenografts (PDX). These PDX models are then exposed to placebo, PARPi alone, cisplatin alone, or PARPi + cisplatin, to determine if the HRD score can predict response (change in size of tumor).
After establishment of PARPi efficacy in GU cancers and identification of predictive biomarkers for selecting PARPi candidates, our next challenge will be to determine how to apply PARPis in GU cancers to achieve the maximum benefit. Ultimately, patients may not only benefit in the metastatic or adjuvant setting but also from the neoadjuvant application of PARPi. For example, patients that are cisplatin ineligible may benefit from neoadjuvant use of PARPi combined with carboplatin chemotherapy96. Adjuvant use of PARPi in patients with high risk or micro-metastatic PCa could potentially render them disease free or be combined with androgen deprivation in node-positive patients. Developing trials to answer these questions could progress rapidly with the application of new biomarkers and next-generation sequencing.
Acknowledgments
Financial Support: No financial support was provided for this study.
Funding: PTT is funded by the Nesbitt Family, a Kimmel Translational Science Award (SKF-13-021), an ACS Scholar award (122688-RSG-12-196-01-TBG), American Lung Association (LCD-339465), Movember-PCF and the NIH (R01CA166348). JJM is supported by a grant from the VHA BX003692-01.
Footnotes
Conflict of Interest: No authors have a relevant conflict of interest, Dr. Hussain has research support with AstraZeneca for clinical trials outside of the described work. Dr. Meeks has research funding from the VHA outside of the described work.
References
- 1.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 2.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochemical Journal. 1999;342( Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 3.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 4.Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes and Development. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 5.Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Molecular Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sukhanova MV, Abrakhi S, Joshi V, et al. Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging. Nucleic Acids Research. 2016;44:e60. doi: 10.1093/nar/gkv1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Underhill C, Toulmonde M, Bonnefoi H. A review of PARP inhibitors: from bench to bedside. Ann Oncol. 2011;22:268–279. doi: 10.1093/annonc/mdq322. [DOI] [PubMed] [Google Scholar]
- 8.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 9.Caldecott KW. Single-strand break repair and genetic disease. Nature Reviews: Genetics. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 10.Iarovaia OV, Rubtsov M, Ioudinkova E, Tsfasman T, Razin SV, Vassetzky YS. Dynamics of double strand breaks and chromosomal translocations. Molecular Cancer. 2014;13:249. doi: 10.1186/1476-4598-13-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Murcia JM, Niedergang C, Trucco C, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao Z, Bozzella M, Seluanov A, Gorbunova V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst) 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 14.Tutt A, Bertwistle D, Valentine J, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrucelli N, Daly MB, Feldman GL. Hereditary breast and ovarian cancer due to mutations in BRCA1 and BRCA2. Genet Med. 2010;12:245–259. doi: 10.1097/GIM.0b013e3181d38f2f. [DOI] [PubMed] [Google Scholar]
- 16.Welcsh PL, King MC. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Human Molecular Genetics. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 17.Ceccaldi R, Liu JC, Amunugama R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 20.Dobzhansky T. Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics. 1946;31:269–290. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 22.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 23.Cavanagh H, Rogers KM. The role of BRCA1 and BRCA2 mutations in prostate, pancreatic and stomach cancers. Hereditary Cancer in Clinical Practice. 2015;13:16. doi: 10.1186/s13053-015-0038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mersch J, Jackson MA, Park M, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. 2015;121:269–275. doi: 10.1002/cncr.29041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steffen JD, Brody JR, Armen RS, Pascal JM. Structural Implications for Selective Targeting of PARPs. Front Oncol. 2013;3:301. doi: 10.3389/fonc.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menissier de Murcia J, Ricoul M, Tartier L, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO Journal. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murai J, Huang SY, Renaud A, et al. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 2014;13:433–443. doi: 10.1158/1535-7163.MCT-13-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang YQ, Wang PY, Wang YT, Yang GF, Zhang A, Miao ZH. An Update on Poly(ADP-ribose)polymerase-1 (PARP-1) Inhibitors: Opportunities and Challenges in Cancer Therapy. Journal of Medicinal Chemistry. 2016 doi: 10.1021/acs.jmedchem.6b00055. [DOI] [PubMed] [Google Scholar]
- 29.Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkins TA, Shi Y, Rodriguez LE, et al. Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors. Molecular Cancer Research. 2015;13:1465–1477. doi: 10.1158/1541-7786.MCR-15-0191-T. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman B, Shapira-Frommer R, Schmutzler RK, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ledermann J, Harter P, Gourley C, et al. Olaparib maintenance therapy in patients with platinum-sensitive relapsed serous ovarian cancer: a preplanned retrospective analysis of outcomes by BRCA status in a randomised phase 2 trial. Lancet Oncol. 2014;15:852–861. doi: 10.1016/S1470-2045(14)70228-1. [DOI] [PubMed] [Google Scholar]
- 33.Rottenberg S, Jaspers JE, Kersbergen A, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donawho CK, Luo Y, Luo Y, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 35.Xu K, Chen Z, Cui Y, Qin C, He Y, Song X. Combined olaparib and oxaliplatin inhibits tumor proliferation and induces G2/M arrest and gamma-H2AX foci formation in colorectal cancer. Onco Targets Ther. 2015;8:3047–3054. doi: 10.2147/OTT.S89154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JM, Hays JL, Annunziata CM, et al. Phase I/Ib study of olaparib and carboplatin in BRCA1 or BRCA2 mutation-associated breast or ovarian cancer with biomarker analyses. J Natl Cancer Inst. 2014;106:dju089. doi: 10.1093/jnci/dju089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, Li L. DNA crosslinking damage and cancer - a tale of friend and foe. Transl Cancer Res. 2013;2:144–154. doi: 10.3978/j.issn.2218-676X.2013.03.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drean A, Lord CJ, Ashworth A. PARP inhibitor combination therapy. Crit Rev Oncol Hematol. 2016;108:73–85. doi: 10.1016/j.critrevonc.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 39.Tan DS, Rothermundt C, Thomas K, et al. “BRCAness” syndrome in ovarian cancer: a case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J Clin Oncol. 2008;26:5530–5536. doi: 10.1200/JCO.2008.16.1703. [DOI] [PubMed] [Google Scholar]
- 40.Alsop K, Fereday S, Meldrum C, et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol. 2012;30:2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Byrski T. Germline mutation of BRCA1 gene in Polish families with strong aggregation of breast and/or ovarian cancer based on coding sequence analysis using the SSCP method. Ann Acad Med Stetin. 2003;49:27–43. [PubMed] [Google Scholar]
- 42.Byrski T, Dent R, Blecharz P, et al. Results of a phase II open-label, non-randomized trial of cisplatin chemotherapy in patients with BRCA1-positive metastatic breast cancer. Breast Cancer Res. 2012;14:R110. doi: 10.1186/bcr3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pal T, Permuth-Wey J, Kapoor R, Cantor A, Sutphen R. Improved survival in BRCA2 carriers with ovarian cancer. Familial Cancer. 2007;6:113–119. doi: 10.1007/s10689-006-9112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vencken PM, Kriege M, Hoogwerf D, et al. Chemosensitivity and outcome of BRCA1- and BRCA2-associated ovarian cancer patients after first-line chemotherapy compared with sporadic ovarian cancer patients. Ann Oncol. 2011;22:1346–1352. doi: 10.1093/annonc/mdq628. [DOI] [PubMed] [Google Scholar]
- 45.Sun C, Li N, Ding D, et al. The role of BRCA status on the prognosis of patients with epithelial ovarian cancer: a systematic review of the literature with a meta-analysis. PLoS One. 2014;9:e95285. doi: 10.1371/journal.pone.0095285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oza AM, Cibula D, Benzaquen AO, et al. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 2015;16:87–97. doi: 10.1016/S1470-2045(14)71135-0. [DOI] [PubMed] [Google Scholar]
- 47.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nature Reviews: Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 48.An update on PARP inhibitors for the treatment of cancer McCabe N, Turner NC, Lord CJ, et al. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140.
- 49.Michels J, Vitale I, Saparbaev M, Castedo M, Kroemer G. Predictive biomarkers for cancer therapy with PARP inhibitors. Oncogene. 2014;33:3894–3907. doi: 10.1038/onc.2013.352. [DOI] [PubMed] [Google Scholar]
- 50.Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 51.Cancer Genome Atlas Research N. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pritchard CC, Mateo J, Walsh MF, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med. 2016 doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Margulis V, Lin J, Yang H, Wang W, Wood CG, Wu X. Genetic susceptibility to renal cell carcinoma: the role of DNA double-strand break repair pathway. Cancer Epidemiol Biomarkers Prev. 2008;17:2366–2373. doi: 10.1158/1055-9965.EPI-08-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer Journal for Clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 55.Sweeney CJ, Chamberlain D. Insights into E3805: the CHAARTED trial. Future Oncol. 2015;11:897–899. doi: 10.2217/fon.14.310. [DOI] [PubMed] [Google Scholar]
- 56.James ND, Sydes MR, Clarke NW, et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet. 2016;387:1163–1177. doi: 10.1016/S0140-6736(15)01037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris WP, Mostaghel EA, Nelson PS, Montgomery B. Androgen deprivation therapy: progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat Clin Pract Urol. 2009;6:76–85. doi: 10.1038/ncpuro1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruch JM, Hussain MH. Evolving therapeutic paradigms for advanced prostate cancer. Oncology (Williston Park, NY) 2011;25:496–504. 508. [PubMed] [Google Scholar]
- 59.Leongamornlert D, Mahmud N, Tymrakiewicz M, et al. Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer. 2012;106:1697–1701. doi: 10.1038/bjc.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beltran H, Yelensky R, Frampton GM, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castro E, Eeles R. The role of BRCA1 and BRCA2 in prostate cancer. Asian J Androl. 2012;14:409–414. doi: 10.1038/aja.2011.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castro E, Goh C, Olmos D, et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J Clin Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas Research N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schiewer MJ, Goodwin JF, Han S, et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polkinghorn WR, Parker JS, Lee MX, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer Discov. 2013;3:1245–1253. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodwin JF, Schiewer MJ, Dean JL, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov. 2013;3:1254–1271. doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brenner JC, Ateeq B, Li Y, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dedes KJ, Wetterskog D, Mendes-Pereira AM, et al. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci Transl Med. 2010;2:53ra75. doi: 10.1126/scitranslmed.3001538. [DOI] [PubMed] [Google Scholar]
- 70.Mendes-Pereira AM, Martin SA, Brough R, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Molecular Medicine. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hussain MDS, et al. Co-targeting androgen receptor (AR) and DNA repair: A randomized ETS gene fusion-stratified trial of abiraterone + prednisone (Abi) +/− the PARP1 inhibitor veliparib for metastatic castration-resistant prostate cancer (mCRPC) patients (pts) (NCI9012)—A University of Chicago phase II consortium trial. J Clin Oncol. 2016;34(suppl) [Google Scholar]
- 73.Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 74.Hussain M, Carducci MA, Slovin S, et al. Targeting DNA repair with combination veliparib (ABT-888) and temozolomide in patients with metastatic castration-resistant prostate cancer. Invest New Drugs. 2014;32:904–912. doi: 10.1007/s10637-014-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murai J, Zhang Y, Morris J, et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther. 2014;349:408–416. doi: 10.1124/jpet.113.210146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Plummer R, Jones C, Middleton M, et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res. 2008;14:7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palma JP, Wang YC, Rodriguez LE, et al. ABT-888 confers broad in vivo activity in combination with temozolomide in diverse tumors. Clin Cancer Res. 2009;15:7277–7290. doi: 10.1158/1078-0432.CCR-09-1245. [DOI] [PubMed] [Google Scholar]
- 78.Milowsky MI, Rumble RB, Lee CT. Guideline on Muscle-Invasive and Metastatic Bladder Cancer (European Association of Urology Guideline): American Society of Clinical Oncology Clinical Practice Guideline Endorsement Summary. Journal of Oncology Practice. 2016;12:588–590. doi: 10.1200/JCO.2015.65.9797. [DOI] [PubMed] [Google Scholar]
- 79.Sternberg CN, Yagoda A, Scher HI, et al. Preliminary results of M-VAC (methotrexate, vinblastine, doxorubicin and cisplatin) for transitional cell carcinoma of the urothelium. J Urol. 1985;133:403–407. doi: 10.1016/s0022-5347(17)48996-8. [DOI] [PubMed] [Google Scholar]
- 80.von der Maase H, Hansen SW, Roberts JT, et al. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol. 2000;18:3068–3077. doi: 10.1200/JCO.2000.18.17.3068. [DOI] [PubMed] [Google Scholar]
- 81.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharma PBP, Kim JW, et al. Efficacy and safety of nivolumab monotherapy in metastatic urothelial cancer (mUC): results from the phase I/II CheckMate 032 study. J Clin Oncol. 2016;34 abstr 4501. [Google Scholar]
- 83.Bentley J, Diggle CP, Harnden P, Knowles MA, Kiltie AE. DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining. Nucleic Acids Res. 2004;32:5249–5259. doi: 10.1093/nar/gkh842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nickerson ML, Dancik GM, Im KM, et al. Concurrent alterations in TERT, KDM6A, and the BRCA pathway in bladder cancer. Clin Cancer Res. 2014;20:4935–4948. doi: 10.1158/1078-0432.CCR-14-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nickerson ML, Witte N, Im KM, et al. Molecular analysis of urothelial cancer cell lines for modeling tumor biology and drug response. Oncogene. 2016 doi: 10.1038/onc.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mullane SA, Werner L, Guancial EA, et al. Expression Levels of DNA Damage Repair Proteins Are Associated With Overall Survival in Platinum-Treated Advanced Urothelial Carcinoma. Clin Genitourin Cancer. 2016;14:352–359. doi: 10.1016/j.clgc.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Plimack ER, Dunbrack RL, Brennan TA, et al. Defects in DNA Repair Genes Predict Response to Neoadjuvant Cisplatin-based Chemotherapy in Muscle-invasive Bladder Cancer. Eur Urol. 2015;68:959–967. doi: 10.1016/j.eururo.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jian W, Xu HG, Chen J, et al. Activity of CEP-9722, a poly (ADP-ribose) polymerase inhibitor, in urothelial carcinoma correlates inversely with homologous recombination repair response to DNA damage. Anticancer Drugs. 2014;25:878–886. doi: 10.1097/CAD.0000000000000114. [DOI] [PubMed] [Google Scholar]
- 90.Larsen MJ, Kruse TA, Tan Q, et al. Classifications within molecular subtypes enables identification of BRCA1/BRCA2 mutation carriers by RNA tumor profiling. PLoS One. 2013;8:e64268. doi: 10.1371/journal.pone.0064268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mukhopadhyay A, Elattar A, Cerbinskaite A, et al. Development of a functional assay for homologous recombination status in primary cultures of epithelial ovarian tumor and correlation with sensitivity to poly(ADP-ribose) polymerase inhibitors. Clin Cancer Res. 2010;16:2344–2351. doi: 10.1158/1078-0432.CCR-09-2758. [DOI] [PubMed] [Google Scholar]
- 92.Lee JM, Gordon N, Trepel JB, Lee MJ, Yu M, Kohn EC. Development of a multiparameter flow cytometric assay as a potential biomarker for homologous recombination deficiency in women with high-grade serous ovarian cancer. Journal of Translational Medicine. 2015;13:239. doi: 10.1186/s12967-015-0604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Timms KM, Abkevich V, Hughes E, et al. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res. 2014;16:475. doi: 10.1186/s13058-014-0475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Timms K, Neff C, et al. Dna repair deficiencies in ovarian cancer: Genomic analysis of high grade serous ovarian tumors from the NOVA study. ESMO poster presentation; 2015. [Google Scholar]
- 95.Telli ML, Timms KM, Reid J, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tilki D, Svatek RS, Novara G, et al. Stage pT0 at radical cystectomy confers improved survival: an international study of 4,430 patients. J Urol. 2010;184:888–894. doi: 10.1016/j.juro.2010.04.081. [DOI] [PubMed] [Google Scholar]