Abstract
Purpose
To compare the accuracy and precision of four different T1 estimation algorithms for MOLLI.
Methods
Four T1 estimation algorithms, including the original fit, Inversion group(IG) fit, Instantaneous signal loss simulation(InSiL) and Bloch equation simulation with slice profile correction(BLESSPC) were studied. T1 estimation accuracy, precision, reproducibility and sensitivity to heart rate(HR), flip angle(FA) and acquisition scheme(AcS) variations were compared in simulation, phantom, and volunteer studies.
Results
T1 estimation accuracy of IG (−2.4%±3.9%) and original fit (−3.2%±1.4%) were worse than BLESSPC (0.2%±1.5%) and InSiL (−0.7%±2.1%). The original fit had the best precision for T1 from 409ms-1884ms for the same FA (0.67%±0.16% vs. 0.90%±0.23% using IG, 0.78%±0.11% using InSiL, 0.77%±0.12% using BLESSPC). BLESSPC generated the most consistent in vivo T1 values over different FAs and AcS, and the T1 estimation reproducibility was similar (p>0.3) among the four methods when FA=35°. When using FA=50°, the reproducibility was significantly improved only when using BLESSPC (1.6%±0.9 vs. 2.6%±1.9%, p<0.05).
Conclusion
BLESSPC has superior accuracy and is the least sensitive to FA, HR, and AcS variations. T1 estimation using BLESSPC and FA=50° is superior to conventional MOLLI with FA=35° in accuracy and precision. Further clinical studies in varying pathological conditions are warranted to confirm our findings.
Keywords: T1 mapping, MOLLI, myocardial fibrosis, cardiac MRI
INTRODUCTION
Myocardial T1 mapping is a rapidly evolving technique that allows for quantitative tissue characterization in a range of cardiac diseases (1–5). Various pulse sequences for myocardial T1 mapping have been proposed (6–9). Among these techniques, the Modified Look-Locker Inversion Recovery (MOLLI) sequence is widely used due to its high precision and reproducibility (10–12). The MOLLI sequence samples a group of balanced steady-state free precession (bSSFP) images at end-diastole using a “L(m)M(n)N” or a “L(m)M” acquisition scheme, where 3 or 2 inversion sets are acquired and each set is composed of L, M and N (or L and M) ECG-triggered single-shot images, respectively. Between each inversion image set, there are several resting heartbeats to allow for sufficient recovery of the longitudinal magnetization before the next inversion, which is indicated by the numbers (m and n) in parentheses. All acquired images with different inversion times (TI) are then used in a pixel-wise 3-parameter exponential fitting algorithm to generate the T1 map. A potential drawback of the MOLLI approach is its sensitivity to intrinsic biologic parameters such as tissue T1, T2, and heart rate as well as technical factors including flip angle and imperfect inversion (13,14).
After the original MOLLI 3(3)3(3)5 sequence was proposed (6), some studies have been performed to optimize the MOLLI sequence and improve its accuracy. For example, the nominal radiofrequency (RF) excitation flip angle (FA) has been reduced from 50° to 35° (15), which becomes a standard FA for the MOLLI sequence. Kellman et al. (16) proposed to use the 5(3)3 acquisition scheme for native myocardial T1 mapping and the 4(1)3(1)2 acquisition scheme for post-contrast T1 mapping, which reduces the breath-hold time from 17 heart beats to 11 heart beats. Furthermore, an improved adiabatic inversion pulse has been used to improve the inversion efficiency from 92% to 96% for the MOLLI sequence (14). Assuming the inversion factor δ is known, a T1 correction algorithm (T1corrected = T1/δ) can be applied to improve the MOLLI T1 estimation accuracy further (14). Despite the above modifications, MOLLI T1 estimation accuracy is limited by its T1 estimation algorithm, which is sensitive to FA variations and incomplete signal recovery prior to the inversion pulses, etc.
Recently, several novel T1 calculation algorithms were proposed for the MOLLI sequence, including instantaneous signal loss simulation (InSiL) T1 estimation (17), inversion group (IG) fit (18), and Bloch equation simulation with slice profile correction (BLESSPC) T1 estimation (19). Compared to the original fit for the MOLLI sequence, InSiL generates better T1 mapping accuracy and is less sensitive to heart rate (HR) variations in tissues with longer T1 values (17). However, the sensitivity of InSiL to acquisition scheme and FA variations has not been evaluated in vivo. In prior work, IG fit generated consistent T1 estimations for arbitrary acquisition schemes, including schemes without rest periods that are required in the original MOLLI fit (18). However, the sensitivity of IG fit to HR and FA variations has not been evaluated. Although BLESSPC was originally proposed as a T1 estimation algorithm for FLASH-MOLLI (a MOLLI sequence with spoiled gradient readout) (19), it is also possible to apply the BLESSPC T1 estimation to the conventional MOLLI sequence with a bSSFP readout.
Currently, there is no standard T1 mapping approach for clinical cardiac MR protocols. The systematic evaluation of each approach regarding accuracy, precision, and reproducibility is crucial to reach a consensus and has high impact with regards to radiogenomics’ contribution to precision medicine. Although the MOLLI sequence with original fit has been evaluated against three other T1 mapping techniques in term of accuracy, precision, and reproducibility (11), no comparison has been performed across these recently developed T1 estimation algorithms for the MOLLI sequence. The goal of this work was to evaluate and compare the accuracy, precision, and reproducibility of the four T1 estimation algorithms (BLESSPC, InSiL, IG fit, original fit) for the MOLLI sequence and to evaluate the impact of HR, FA and acquisition scheme variations on T1 estimation.
METHODS
Pulse sequence
The MOLLI pulse sequence was implemented in the Siemens Works-in-progress package (WIP780B) and installed on a 1.5T MRI scanner (Avanto Fit Siemens Healthcare; Erlangen, Germany). The T1 estimation accuracy, precision, reproducibility and sensitivity to variations in HR, FA, and T2 using four different T1 estimation algorithms (BLESSPC, InSiL, IG fit, original fit) were evaluated using the MOLLI 5(3)3 scheme, which is one of the MOLLI acquisition schemes proposed by Kellman et al. (16). To evaluate the sensitivity to variations in MOLLI acquisition schemes, two additional acquisition schemes were studied, including 5(0)3, which represents an acquisition scheme without using dummy heart beats, and 4(1)3(1)2 (16).
Both phantom and healthy volunteer studies were performed. The typical parameters for the MOLLI sequence in phantom and in vivo studies are as follows unless specified otherwise. For 5(3)3 and 5(0)3 acquisition schemes, the shortest TIs were 120 ms and 200 ms for the two inversion groups, respectively. For the 4(1)3(1)2 acquisition scheme, the shortest TIs in each inversion were 100 ms, 180 ms, and 260 ms, respectively. In the MOLLI sequence, single-shot bSSFP images were acquired using nominal FA = 35°, ten dummy start-up RF pulses with linear FA increments, and linear phase-encoding order, matrix = 192 × 124, interpolated to 192 × 154, Field of view (FOV) = 340 × 273 mm2, interpolated pixel size 1.8 × 1.8 mm2, TR/TE=2.5ms/1.1ms, 6/8 partial Fourier acquisition, and 2X GRAPPA with 24 k-space auto-calibration lines. Protocol printout of the standard MOLLI 5(3)3 acquisition is provided online (Supporting material S1).
T1 estimation algorithms
Four different T1 estimation algorithms were applied to generate T1 maps based on acquired MOLLI data. The original T1 estimation algorithm merges all of the images acquired in different inversion groups into a single set and a 3-parameter exponential curve fitting (i.e. y = A - B exp(−TI/T1*)) is performed for each pixel. T1 is estimated using the conventional Look-Locker correction T1 = T1* (B/A - 1). Assuming the inversion factor δ is known, the inversion factor correction algorithm T1corrected = T1/δ (14) is applied to improve the T1 estimation accuracy. In this manuscript, the original T1 estimation algorithm with Look-Locker correction and inversion factor correction is referred to as the “original fit” method.
IG fit is a MOLLI fitting algorithm recently proposed by Marshall et al (18), which allows arbitrary acquisition schemes and rest periods (including no rest period). In IG fit, the 3-parameter exponential equation (y = A - Bi exp(−TI/T1*)) is used for each inversion group with the same parameters A and T1, but different Bi (i=1, 2, …, the number of inversion groups) (18). The final T1 is calculated using T1 = T1* (B1/A - 1)/δ.
The InSiL technique is a T1 estimation algorithm described by Shao et al (17) to improve the T1 evaluation accuracy for the MOLLI sequence. InSiL simulates the signal evolution of the MOLLI sequence using the Bloch equation. The effect of longitudinal signal perturbation due to single-shot bSSFP readout is parameterized as an instantaneous signal loss in longitudinal magnetization by an unknown factor of C (0⩽C⩽1) at the time point when the k-space center line is acquired during the single-shot acquisition. Hence, a detailed Bloch simulation of the single-shot balanced SSFP readout was not performed in InSiL T1 estimation. Assuming the inversion factor δ is known, the three unknowns (M0, T1, and C) in InSiL can be solved for each pixel by matching the simulated signal to the acquired signal using a least squares fitting algorithm. Detailed descriptions of the InSiL algorithm has been previously described (17).
The BLESSPC T1 estimation algorithm was proposed for the FLASH-MOLLI sequence, which is a variation of the MOLLI sequence that was modified by incorporating a spoiled gradient echo readout to avoid the off-resonance banding artifacts due to bSSFP readout at 3.0T (19). BLESSPC simulates the signal evolution of a sequence using Bloch equations by considering the effect of each individual RF excitation and the non-ideal slice profile, and solves the three free parameters, T1, M0 and apparent flip angle α, by minimizing the mean square error between the simulated signal and the measured signal for each pixel (19). In this work, we will extend the BLESSPC algorithm to calculate T1 values based on the standard bSSFP-based MOLLI sequence wherein the signal evolution of bSSFP readouts are simulated using the Bloch equation. To simulate the signal evolution of bSSFP, tissue T2 information is needed. In this work, a fixed T2 = 45 ms was used in BLESSPC for the MOLLI sequence.
To improve T1 estimation accuracy, the inversion factor δ needs to be appropriately estimated (17,20,21). Previous studies have shown that the actual in vivo δ can be significantly different from the theoretical value or phantom results (17,19). Therefore, we used a previous proposed “MOLLI+M0” sequence (17) to measure the average inversion factor for phantoms and in vivo. The “MOLLI+M0” sequence acquires an additional proton density weighted image without an inversion pulse 3 seconds following the MOLLI 3(3)3(3)5 acquisition in a single breath-hold (17). The imaging parameters, including matrix size, pixel size etc., were the same as MOLLI 4(1)3(1)2 acquisitions described in the pulse sequence section. Based on data from the "MOLLI+M0" sequence, the BLESSPC 4-parameter fitting (T1, M0, α, and δ) was used to estimate the average δ for phantoms and the average δ in vivo. The two measured average δ for phantoms and for in vivo were subsequently used to calculate T1 maps for each T1 estimation algorithm.
Simulations
To compare the T1 estimation accuracy of the four T1 estimation algorithm - BLESSPC, InSiL, IG fit and original fit - for the MOLLI sequence, and their sensitivity to T1, T2, FA, and acquisition scheme, Bloch equation simulations of the MOLLI sequence were performed in MATLAB (the Mathworks, Natick, MA) for various T1s, T2s, FAs, and acquisition schemes using actual parameters of the MOLLI sequence listed in the pulse sequence section. Specifically, the simulations were performed with MOLLI 5(3)3 acquisition, T1 = 1200 ms, FA = 35°, T2 = 50 ms, HR = 60 bpm, and one of the parameters was varied in each of the following five scenarios : (1) T1 values varied from 220 ms, 300 ms to 1800 ms (100 ms increments); (2) FA values varied from 20° to 50° (5° increments); (3) T2 values varied from 20 ms to 100 ms (10 ms increments); (4) heart rates varied from 40 bpm to 100 bpm (10 bpm increments); (5) various acquisition schemes including MOLLI 5(0)3 and 4(1)3(1)2. In scenarios (1) and (2), the T1 estimation precision was estimated using Monte-Carlo simulation with 65,536 trials by adding Gaussian noise with the same standard deviation, which corresponded to a 2% noise relative to the M0 weighted bSSFP signal at FA= 35°. Gaussian noise was used because a phase sensitive (PSIR) reconstruction (22) was used to restore the sign and the real component of the background phase corrected image was used, which results in normally distributed noise (16). In Bloch simulations, the effects of imperfect (non-rectangular) slice profiles of the 2D excitation pulse [duration = 480 μs, time bandwidth product = 1.6] were included, and the inversion pulse was assumed to be instantaneous with an inversion factor of 0.96.
Phantom Study
Nine 50 ml agar and CuSO4 gel phantoms with a range of T1 = 217 ms - 1884 ms, T2 = 35 ms - 72 ms were used. Reference T1 and T2 values for each gel phantom were determined by a standard inversion recovery spin echo technique with 12 TIs (TI = 50 ms - 5000 ms), TR/TE = 10s/4.6ms using a three-parameter fitting, i.e., M(t) = A - B exp(−TI/T1) with matrix size 128 × 128 (printed protocol provided in supporting material S2). Reference T2 values were calculated using a standard spin-echo technique with 11 TEs (TE = 5 ms - 250 ms), with TR = 10s using a three-parameter fitting, i.e., M(t) = M0exp(−TE/T2) and the same matrix size (printed protocol provided in supporting material S3). A region of interest (ROI) was manually drawn for each tube and the average T1, T2 values were used as reference T1 and T2 values.
Four groups of MOLLI scans were performed to evaluate the accuracy, precision, and the impact of varying HR, FA, and acquisition scheme. In group I, the MOLLI 5(3)3 with FA=35° at each simulated heart rate (40 bpm - 100 bpm, 20 bpm increments) were acquired. In group II, the MOLLI 5(3)3 scans were acquired using FA = 20° and 50° at simulated HR = 60 bpm. In group III, MOLLI 5(0)3 and 4(1)3(1)2 were acquired with FA=35°. The scans were repeated five times in each group.
A “MOLLI+M0” scan (17) was performed once to measure the average inversion factor for each phantom. After the average inversion factor had been measured for all the phantoms, four T1 maps were generated for each MOLLI scan using BLESSPC, InSiL, IG fit and original fit using the measured average inversion factor. After T1 maps were generated using any one of the four T1 estimation algorithm, average T1 maps and standard deviation (SD) T1 maps were calculated pixel-by-pixel over the repeated measurements. The region of interests (ROIs) on average T1 maps and SD T1 maps were selected to determine the mean average T1 values (T1mean) and mean SD T1 values (T1SD). The T1 estimation errors were calculated using T1err = T1mean - T1reference and T1err(%) = (T1mean - T1reference)/ T1reference×100%, where T1reference is the reference T1 value for each phantom. To evaluate T1 estimation precision, the coefficient of variation (CoV) for each phantom was calculated using CoV = (T1SD/ T1mean) ×100%.
In Vivo Study
The study was approved by the Institutional Review Board and was compliant with the Health Insurance Portability and Accountability Act. All subjects provided written informed consent. Eight healthy volunteers (5 males, age 28.9±4.3 years) underwent non-contrast MRI. Standard cardiac shimming was applied to reduce off-resonance variations in the heart region. The “MOLLI+M0” pulse sequence was performed on four of the eight volunteers (3 males, age 28.3 ±3.0 years, weight 74.8±21.7 kg, ranges from 50 kg to 102 kg) as a preliminary group to establish the average in vivo inversion factor for the MOLLI sequence. Images of the mid-left ventricular (mid-LV) short-axis were acquired at end-expiration using the MOLLI sequence with the follow acquisition schemes and parameters: (1) 5(3)3, FA=20°, 35° and 50°; (2) 5(0)3 and 4(1)3(1)2, FA=35° (Experiment 1). The MOLLI 5(3)3 with FA = 35° and 50° were repeated once (Experiment 2) to evaluate intra-scan reproducibility. The subject was then brought outside of the scanner room and taken back into the scanner after 5 minutes. The MOLLI sequence with 5(3)3 acquisition scheme with FA = 35° and 50° were repeated once again (Experiment 3) to evaluate the inter-scan reproducibility. The matrix size at in vivo study was the same as the matrix size in our phantom studies. The FOVs were adjusted according to each subject’s body size and were in range 300 × 241 mm2 - 440 × 353 mm2.
The average inversion factor at in vivo was calculated by drawing an ROI covering the entire left ventricular (LV) myocardial region for each volunteer in the preliminary group. For each in vivo MOLLI scan, four T1 maps were generated using BLESSPC, InSiL, IG fit and the original fit after motion correction of the image data (22). For a fair comparison, the same inversion factor was used for all the four T1 estimation algorithms and was set to the average inversion factor measured in the preliminary group. An ROI was drawn at the inter-ventricular septal region on one of the four T1 maps (randomly selected) and copied to the other three T1 maps. Care was taken to avoid blood pool and the epicardial surface. The average T1 values using BLESSPC, InSiL, IG fit, and original fit were then calculated using values from the selected ROIs. For each T1 estimation algorithm, the measured myocardial T1 values by MOLLI with different acquisition schemes and flip angles were compared to those measured by the standard MOLLI 5(3)3 with FA= 35°. To evaluate the sensitivity to flip angle variations, CoVs of myocardial T1 values across different flip angles using CoV(FAs) = SD(T1mean values of 20°, 35° and 50°) / Mean(T1mean values of 20°, 35° and 50°). Similarly, the CoVs across different acquisition schemes were calculated. Intra and inter-scan reproducibility were calculated as the absolute difference in the ROI-averaged T1 (dT1) divided by the average of experiments 1 and 2, and of experiments 1 and 3, respectively.
Statistical Analysis
One-way ANOVA was used for comparisons of the precision and reproducibility of the four methods in MATLAB (the Mathworks, Natick, MA). Two-tailed Student’s t tests were used for pair-wise comparisons. A p-value < 0.05 was considered statistically significant. Bonferroni correction was used for pair-wise comparison of the four T1 estimation algorithms, which resulted in a statistical significance threshold of p < 0.008.
RESULTS
Simulations
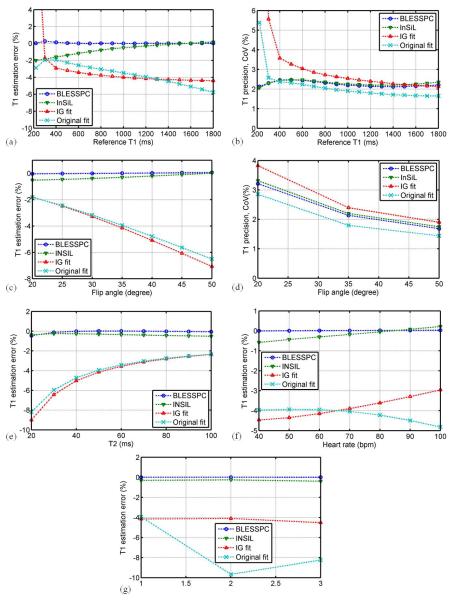
Figure 1 shows the simulation results of T1 estimation accuracy at different simulation scenarios (various T1s, FAs, T2s, HRs and acquisition schemes) and T1 estimation precision at different T1 values and flip angles using BLESSPC, InSiL, IG fit and original fit for the MOLLI sequence. Overall, BLESSPC and InSiL were more accurate and were less sensitive to flip angle, tissue T2, heart rate, and acquisition scheme variations when compared to IG fit and original fit, with BLESSPC being slightly better than InSiL. Regarding T1 estimation precision, the original fit is superior to the other three methods for T1 > 400 ms, while for T1<400 ms, BLESSPC and InSiL were more precise (Figure 1b). IG fit resulted in the lowest precision for T1 < 1000 ms compared to the other three methods (Figure 1b). Higher flip angles (20° - 50°) resulted in better T1 estimation precision (lower CoV) for all four methods (Figure 1d).
Figure 1.
Simulation - comparison of four T1 estimation algorithms for the MOLLI sequence in terms of (a, b) T1 estimation accuracy and precision for T1 range from 220 ms - 1800 ms, (c, d) T1 estimation accuracy and precision at different radio-frequency excitation flip angles for T1 = 1200 ms, and their sensitivity to tissue T2 variation (e), heart rate variation (f) and acquisition schemes variation (g). The standard acquisition scheme 5(3)3 is used for results from a - f. Overall, BLESSPC and InSiL generated more accurate native myocardial T1 values and were less sensitive to flip angle, tissue T2, and heart rate, and acquisition scheme variations when compared to IG fit and original fit. BLESSPC was slightly better than InSiL. The original fit had superior precision over the other three methods for T1 > 400 ms, while for T1<400 ms, BLESSPC and InSiL were more precise. Using IG fit resulted in much lower precision for T1 < 1000 ms compared to the other three methods. The CoV of IG fit for T1 = 220 ms > 10%, which is out of the display range in (b).
Phantom study
The average inversion factor measured using the “MOLLI+M0” pulse sequence was 0.96±0.01 (range 0.94 - 0.97), which is close to theoretical inversion factor value of the improved adiabatic inversion pulse used in the MOLLI sequence (14). Therefore, an inversion factor of 0.96 was used to generate all T1 maps in our phantom studies.
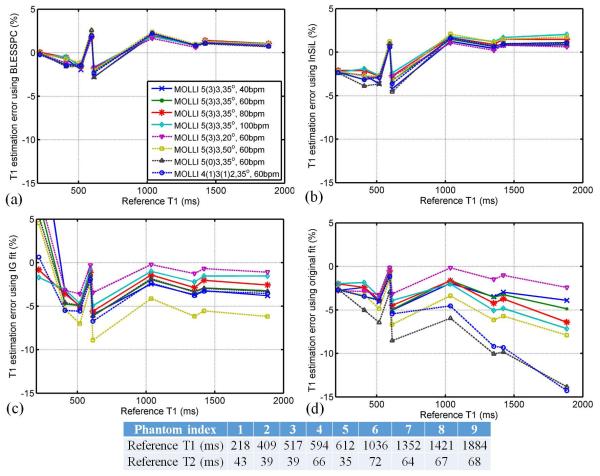
Figure 2 shows the T1 estimation errors relative to reference T1 values (i.e. accuracy) using BLESSPC, InSiL, IG fit and original fit for MOLLI acquisitions in phantom studies at different heart rates (group I), different flip angles (group II) and different acquisition schemes (group III). Consistent with simulation results, BLESSPC and InSiL provided more accurate T1 values than IG fit and original fit. For the standard MOLLI 5(3)3 sequence with FA =35°, the average T1 estimation error were 5.7±12.1 ms (percentile error 0.2%±1.5%) using BLESSPC, 1.2±14.6 ms (percentile error −0.7%±2.1%) using InSiL, −27.0±22.4 ms (percentile error −2.4%±3.9%) using IG fit and −30.8±27.7 ms (percentile error −3.2%±1.4%) using the original fit for all phantoms. Figure 2 showed that, compared with the other three methods, BLESSPC generated the most consistent T1 values for the MOLLI sequence at different heart rates, flip angles and acquisition schemes (maximum absolute T1 changes < 5 ms relative to 5(3)3 with FA=35° and HR=60 bpm).
Figure 2.
Phantom study to assess the accuracy of T1 estimation by four fitting algorithms BLESSPC (a), InSiL (b), IG fit (c), and Original fit (d) in groups I-III. An inversion factor of 0.96 was used for all approaches. Phantom reference T1 values were obtained using standard inversion recovery spin echo and shown in bottom panel. The BLESSPC fitting, InSiL fitting, IG fit and the original fit were applied to the MOLLI sequence. Overall, BLESSPC and InSiL generated more accurate T1 values when compared to IG fit and original fit and were less sensitive to flip angle, heart rate, and acquisition scheme variations.
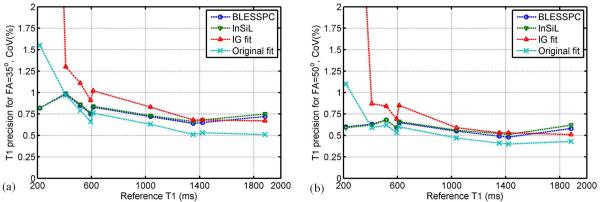
Figure 3 shows the corresponding T1 estimation precisions using BLESSPC, InSiL, IG fit and original fit for MOLLI acquisitions in phantom studies for MOLLI 5(3)3 acquisitions at FA=35° and 50°. For FA =35°, the original fit generated the best precision for T1 > 400 ms (Average CoV = 0.67%±0.16%). In comparison, the average CoV was 0.77%±0.12% using BLESSPC, was 0.78%±0.11% using InSiL, and was 0.90%±0.23% using IG fit. For the very short T1 phantom (T1= 218 ms), BLESSPC (CoV = 0.8%) and InSiL (CoV = 0.8%) were more precise than IG fit (CoV = 10.9%) and original fit (CoV = 1.6%). For the MOLLI 5(3)3 sequence with FA =50°, T1 estimation precision was significantly improved (~ 30% improvement on average) compared to that with FA =35° using any of the four T1 estimation algorithms (p < 0.001). However, using FA = 50° in the MOLLI sequence resulted in larger T1 errors using IG fit (−4.6%±3.8%) and original fit (−4.7%±2.1%). In contrast, BLESSPC (0.5%±1.6%) and InSiL (−0.4%±2.3%) maintained accurate T1 estimation. IG fit resulted in lowest T1 estimation precision for T1 < 1000 ms. These results were consistent with simulation results. Compared to the original fit with FA =35°, using FA = 50° and BLESSPC T1 estimation provided better accuracy (0.5%±1.6% vs. −3.2%±1.4%) and precision (0.58%±0.07% vs. 0.77%±0.33%) for T1 values ranges from 218 ms - 1884 ms.
Figure 3.
Phantom study to assess the precision of T1 estimation by four fitting algorithms (BLESSPC, InSiL, IG fit, and Original fit) for MOLLI 5(3)3 acquisitions at FA=35° and FA=50°. Precision is represented using coefficients of variation (CoV) based on 5 repeated scans. For T1 > 400ms, the original fit was the most precise, while for very short T1 (T1 = 217.5 ms), BLESSPC and InSiL were more precise than IG fit and the original fit. IG fit was the least precise method for phantoms with T1 < 1000 ms. The CoVs of IG fit for T1 = 218 ms > 5%, which are out of the display range in (a) and (b).
In Vivo Study
The average inversion factor of native myocardial tissues for the inversion pulse used in the in vivo study was 0.88±0.01 (range 0.87 - 0.89) in the preliminary group of 4 subjects. Subsequently, an inversion factor of 0.88 was used to generate all in vivo T1 maps of the MOLLI sequence. The offline processing time for the MOLLI T1 map reconstruction (matrix = 192 × 154) using a general-purpose desktop computer (Inter Core i5-3450 CPU, 3.10GHz) was ~2.5 sec when using the original fit, ~5 sec when using the IG fit, ~3 sec when using InSiL, and ~2 min when using BLESSPC.
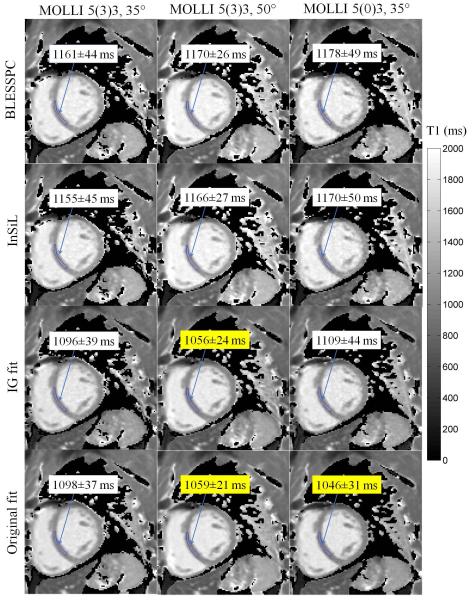
Table 1 lists the native myocardial T1 values in 8 healthy volunteers calculated with BLESSPC, InSiL, IG fit and original fit for the MOLLI sequence using different acquisition schemes [5(3)3, 5(0)3, and 4(1)3(1)2] and flip angles (FA = 20°, 35°, 50°). Myocardial T1 maps of a healthy subject generated using BLESSPC, InSiL, IG fit and Original fit with varying acquisition schemes and FAs are shown in Figure 4. Using the conventional MOLLI acquisition scheme 5(3)3 and FA=35°, the average native myocardial T1 values were 1151.3±22.7 ms using BLESSPC and 1148.4±22.6 ms using InSiL. In comparison, the average native myocardial T1 values were 1092.7±24.2 ms using the IG fit and 1095.9±22.4 ms using the original fit. The difference in native T1 values was not significant between the original fit and IG fit (p=0.14) whereas native T1 values derived using any other pair of methods were significantly different (p<0.002). Relative to the standard MOLLI 5(3)3, FA=35° acquisition scheme, BLESSPC, InSiL, and IG fit generated consistent myocardial T1 values with mean discrepancies of less than 1% even though different MOLLI acquisition schemes were used. Further, the original fit generated lower T1 values using acquisition schemes without sufficient rest heart beats (up to 8.7%±1.7% T1 decrease at 5(0)3 relative to 5(3)3 acquisition scheme). For MOLLI scans with different flip angles, the CoVs measured across three scans (FA=20°, 35°, 50°) were 1.2%±0.6% (BLESSPC), 1.6%±0.6% (InSiL), 3.3%±0.6% (IG fit), and 2.7%±0.3% (original fit), respectively (Table 1). Both original fit and IG fit resulted in up to 3.4%±0.9% and 3.7%±0.9% T1 decrease at FA = 50° relative to FA=35°, respectively. In contrast, the native myocardial T1 values were not significantly changed at FA = 50° relative to FA = 35° using BLESSPC (1174.5±27.7 ms for FA=50°, p=0.20) and InSiL (1177.3±29.2 for FA=50°, p=0.07).
Table 1.
Myocardial T1 values at selected ROI (LV inter-septal region) in 8 healthy volunteers measured by the MOLLI sequence using four different T1 estimations algorithms.
| MOLLI | Myocardium | BLESSPC | InSiL | IG fit4 | Original fit4 |
|---|---|---|---|---|---|
| 5(3)3, 35° | T1 (ms) | 1167.9±23.1 | 1165.5±22.9 | 1092.7±24.2 | 1095.9±22.4 |
| 5(0)3, 35° | T1 (ms) | 1166.2±34.8 | 1165.2±34.8 | 1089.9±31.1 | 1000.4±31.6* |
| Difference 1 | (−0.2%±1.8%) | (−0.0%±1.8%) | (−0.3%±1.2%) | (−8.7%±1.7%) | |
| 4(1)3(1)2, 35° | T1 (ms) | 1161.6±23.7 | 1157.9±23.3 | 1090.1±22.4 | 1039.3±26.5* |
| Difference 1 | (−0.5%±1.2%) | (−0.6%±1.2%) | (−0.2%±1.9%) | (−5.2%±2.0%) | |
| 5(3)3, 20° | T1 (ms) | 1150.8±26.9* | 1145.0±27.0* | 1123.0±31.3* | 1117.1±26.1* |
| Difference 1 | (−1.5%±1.3%) | (−1.8%±1.3%) | (2.8%±1.3%) | (1.9%±0.7%) | |
| 5(3)3, 50° | T1 (ms) | 1174.5±27.7 | 1177.3±29.2 | 1052.6±23.9* | 1059.2±25.6* |
| Difference 1 | (0.6%±1.1%) | (1.0%±1.3%) | (−3.7%±0.9%) | (−3.4%±0.9%) | |
| CoV of 3 schemes 2 | 0.9%±0.5%d | 1.0%±0.5%d | 1.0%±0.7% d | 4.7%±1.0%abc | |
| CoV of 3 FAs 3 | 1.2%±0.6%bcd | 1.6%±0.6%acd | 3.3%±0.6%ab | 2.7%±0.3%ab | |
Mean myocardial T1 fractional difference compared to the standard MOLLI 5(3)3, FA=35° acquisition.
the 3 schemes include 5(3)3, 5(0)3 and 4(1)3(1)2;
the 3 flip angles (FAs) include FA=20°, 35° and 50° using the 5(3)3 acquisition scheme;
For a fair comparison with BLESSPC and InSiL T1 values, the reported T1 values for IG fit and original fit were T1 values after inversion factor correction, which is T1corrected = T1/δ, and δ=0.88.
indicate significant T1 estimation difference compared to the standard MOLLI 5(3)3, FA=35° acquisition (p<0.05),
indicate significant difference compared to BLESSPC (p<0.008),
indicate significant difference compared to InSiL (p<0.008),
indicate significant difference compared to IG fit (p<0.008),
indicate significant difference compared to original fit (p<0.008).
Figure 4.
Myocardial T1 maps of a healthy subject generated using BLESSPC, InSiL, IG fit and the original fit with the same three MOLLI scans. Acquisition schemes included 5(3)3 with FA = 35°, 5(3)3 with FA = 50° and 5(0)3 with FA=35°. ROIs, represented by blue regions at the interventricular septum, were selected for average T1 calculation. Average T1 values at the selected ROIs are shown in the white or yellow box. Both BLESSPC and InSiL generated relatively consistent T1 values for different MOLLI scans using varying parameters. Compared to BLESSPC and InSiL, IG fit generated lower T1 values at a higher flip angle while original fit generated lower T1 values at a higher flip angle or when using 5(0)3 acquisition scheme (indicated using the yellow box).
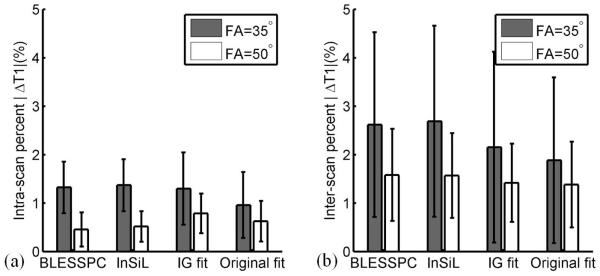
Figure 5 shows the intra- and inter-scan absolute percentile T1 difference at the mid-LV septal region in 8 healthy subjects using the MOLLI 5(3)3 acquisition with FA = 35° and 50° across four T1 estimation algorithms. There were no statistically significant differences among the four algorithms - BLESSPC, InSiL, IG and original fit - in terms of intra- and inter-scan reproducibility for MOLLI 5(3)3 acquisition with FA = 35°or with FA = 50° (p>0.3). The inter-scan reproducibility of BLESSPC was significantly improved (absolute percent difference = 2.6%±1.9% vs. 1.6%±0.9%, p<0.05) using an FA = 50° instead of FA= 35°. However, the same change in FA for InSiL only increased the inter-scan reproducibility modestly with a trend towards significance (absolute percent difference = 2.7%±2.0% vs. 1.6%±0.9%, p=0.051). The reproducibility for IG fit and the original fit was not significantly improved by changing the FA to 50° (p>0.1).
Figure 5.
Bar graphs show intra- and inter-scan reproducibility in healthy subjects by MOLLI 5(3)3 with nominal FA= 35° and 50° using BLESSPC, InSiL, IG and original T1 estimation algorithms. There were no statistically significant differences among the four algorithms in term of intra and inter-scan reproducibility (p>0.3) for the same FA. Using higher flip angle resulted in significant improved inter-scan reproducibility only when using BLESSPC (p<0.05).
DISCUSSION
In this study, we compared the accuracy, precision, reproducibility, and impact of varying flip angle, heart rate and acquisition schemes across four T1 estimation algorithms (BLESSPC, InSiL, IG fit and original fit) for the MOLLI sequence. BLESSPC and InSiL have superior accuracy compared to IG fit and original fit and are less sensitive to variations in flip angle, heart rate, and acquisition scheme. BLESSPC generates the most consistent T1 estimation in the presence of these variations, which were confirmed in vivo. For the commonly used 35° flip angle, the original fit has superior precision compared to BLESSPC, InSiL, and IG fit for T1 > 400 ms. We speculate this is because the BLESSPC model is slightly more sensitive to noise in the data than the original 3-parameter fitting model of MOLLI. In addition, the original MOLLI method tends to reduce the estimated T1 range because of larger T1 underestimation for longer T1 values, which could result in reduced apparent CoV (i.e. better precision) compared to BLESSPC. Nevertheless, BLESSPC and InSiL had a superior precision to IG fit for T1<1000 ms and superior precision to the original MOLLI fit for T1 < 400 ms. All four T1 estimation methods had similar reproducibility for native myocardial T1 mapping in healthy subjects. BLESSPC enables us to use the MOLLI sequence with a higher flip angle to improve T1 estimation precision and reproducibility without scarifying the T1 estimation accuracy. Based on our phantom and in vivo studies, the MOLLI technique, with a FA =50° and BLESSPC, yields better accuracy (phantom T1 error reduced from −3.2%±1.4% to 0.5%±1.6%), precision (~30% improvement on average) and in vivo reproducibility (>19% improvement) when compared to the original MOLLI fit with a FA =35°, which is currently widely used. Our original MOLLI fit T1 values were higher than previously reported (23) because we used a reduced inversion factor of 0.88, which resulted in elevated T1 values of 1095.9±22.4 ms. Without using inversion factor correction, the average myocardial T1 values using original fit were 964.4±19.7, which is similar to the previously reported values of 950 ± 21 ms (23).
For clinical applications, the dependence of the original MOLLI fit on T2 and MT effects constructively contribute to the measured T1 value difference between healthy myocardium and fibrosis (24). In the BLESSPC and InSiL methods, these constructive effects are not eliminated because they are accounted for in the inversion factor. The MT effect is a major reason for the lower measured in vivo inversion factor of 0.88 while the measured inversion factor in phantom was 0.96 using the same inversion pulse (Supporting material S4 and Figure S1). It is also the reason why the BLESSPC and InSiL in vivo T1 values were similar to previous methods such as the SASHA technique (7) that are known to be less sensitive to MT effects (24). A similar reduction of inversion factor in vivo has been found in previous studies (19,20). The greatly reduced inversion factor in vivo was not due to the body size of the volunteer, as the in vivo inversion factor had a very narrow range of 0.87-0.89 in spite of the wide range of body weight of the subjects (50kg – 102kg). In the BLESSPC and InSiL methods, a fixed inversion factor of 0.88 was used at in vivo and hence any changes in MT and T2 due to pathology would cause the actual inversion factor to deviate from 0.88 and would hence be reflected in the final T1 values. As the average in vivo inversion factor is relatively stable for different volunteers and close to the predicted inversion factor considering MT effect (Supporting material S4), we recommend to use the same in vivo inversion factor measured in this work for BLESSPC T1 estimation if the same adiabatic inversion pulse is used regardless of MRI systems used.
The B1 field and the actual local flip angle may vary considerably over the heart (from 0.75 to 1.0 in terms of scaling factors) at 1.5T. As all four T1 estimation algorithms do not use the flip angle as a known parameter, our experiment with different flip angles can be used to speculate the effects of B1 variation on T1 estimation. Our studies demonstrate that BLESSPC was least sensitive to flip angle and B1 field variations among the methods compared. For the MOLLI FA=50° case, even if the B1 field was reduced by 30% (FA reduced to 35°), our in vivo results showed that there are no significant T1 value changes in vivo using BLESSPC, but the T1 values increased by 3.5% using the original fit, and by 3.8% using the IG fit.
In this work, we did not perform in vivo comparisons with the conditional reconstruction approach used in another widely used ShMOLLI technique (25) due to several reasons. First, despite the adaptive T1 reconstruction approach used in ShMOLLI, the fundamental T1 estimation algorithm used in ShMOLLI (i.e. 3-parameter exponential curve fitting) is the same as the original MOLLI. Since our main goal was to evaluate the accuracy and precision of several different T1 estimation algorithms rather than acquisition schemes, we chose to compare with the original MOLLI fit only and we speculate that our conclusion may be generalized to several other T1 mapping acquisition schemes, including ShMOLLI, which uses the original MOLLI fit. Second, the ShMOLLI sequence has already been previously compared with the original MOLLI sequence in terms of accuracy, precision, and reproducibility (11). Nevertheless, it is also possible to apply BLESSPC and InSiL T1 estimation algorithms to the ShMOLLI acquisition scheme. Based on our simulation results (Figure S2 in supporting material S5), compared to ShMOLLI, BLESSPC and InSiL is not only more accurate but also more precise for T1 < 1000 ms when they are applied to ShMOLLI acquisitions.
Our studies have limitations. First, this study was performed in a small cohort of healthy volunteers at mid-ventricular slice location only. Clinical patients with different pathologies will have a wider range of myocardial T1 values and much more confounding physiological parameters and conditions that could obscure any potential advantages associated with the techniques evaluated in this study. For example, the potential constructive contributory effects of MT (24) and T2 when differentiating healthy from pathological myocardial T1 values will likely be greater in clinical patients, and these are not investigated in our study. The simulation and phantom studies cannot reflect the complexity of the in vivo environment. The effects of partial volume effects of fat (26,27) and off resonance (28) have been shown to affect T1 values of the MOLLI sequence with the original fit. These effects together with other effects, such as motion and inflow, may also affect the accuracy of the new T1 reconstruction algorithms, all of which were not thoroughly evaluated in our study and all of which could be different in clinical patients. Hence the conclusion in our study cannot be easily generalized to clinical practice and substantial clinical studies work will be needed to confirm the benefits of the techniques in our study for patients with heart disease. Second, only three acquisition schemes of the MOLLI sequence were studied in this work, and the comparison of T1 estimation precision and reproducibility among the four T1 estimations algorithms were performed based on the MOLLI 5(3)3 acquisition. While the MOLLI 5(3)3 acquisition is one of the optimized acquisition schemes for native myocardial T1 mapping when using the original fit, the sequence itself may not be optimized for BLESSPC, InSiL or IG fit algorithms. Third, although not observed in our work, the MOLLI technique with a higher flip angle (FA=50°) may be subject to additional image artifacts compared to that with an FA=35°. This issue can potentially be solved by using a variable flip angle technique to smooth the signal in the transient state of bSSFP readout, which has been shown to reduce image artifacts for the SASHA (FA=70°) sequence (29).
CONCLUSIONS
Of the four T1 estimation algorithms for the MOLLI sequence tested, BLESSPC provided the most accurate and consistent T1 estimations at different heart rates and flip angles when compared to InSiL, IG fit, and original fit. BLESSPC, InSiL, and IG fit enabled the use of acquisition schemes with fewer rest heat beats, including no rest period. This flexibility may be used to shorten scan time and for acquisition scheme optimization to improve precision for T1 estimation. BLESSPC can be used with MOLLI at higher flip angles (FA=50°) to improve precision and reproducibility of T1 estimation without sacrificing T1 estimation accuracy. Further clinical studies in varying pathological conditions are warranted to confirm our findings.
Supplementary Material
Supporting material S1: Protocol printout of the standard MOLLI 5(3)3 sequence.
Supporting material S2: Protocol printout of the inversion recovery spin echo sequence with the inversion time = 50 ms as an example.
Supporting material S3: Protocol printout of the spin echo sequence with the echo time TE =5 ms as an example.
Supporting material S4: Bloch equation simulation to evaluate the effect of magnetization transfer (MT) and inversion factor on T1 estimations.
Supporting material S5: Simulation of the ShMOLLI sequence to compare BLESSPC and InSiL T1 estimation with ShMOLLI’s T1 estimation algorithm in terms of accuracy and precision.
Acknowledgments
Grant support: Research reported in this publication was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number R01HL127153.
REFERENCES
- 1.Moon JC, Messroghli DR, Kellman P, et al. Myocardial T1 mapping and extracellular volume quantification: a Society for Cardiovascular Magnetic Resonance (SCMR) and CMR Working Group of the European Society of Cardiology consensus statement. J. Cardiovasc. Magn. Reson. 2013;15:92. doi: 10.1186/1532-429X-15-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mewton N, Liu CY, Croisille P, Bluemke D, Lima JAC. Assessment of myocardial fibrosis with cardiovascular magnetic resonance. J. Am. Coll. Cardiol. 2011;57:891–903. doi: 10.1016/j.jacc.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferreira VM, Piechnik SK, Dall’Armellina E, et al. Native T1-mapping detects the location, extent and patterns of acute myocarditis without the need for gadolinium contrast agents. J. Cardiovasc. Magn. Reson. 2014;16:36. doi: 10.1186/1532-429X-16-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira VM, Piechnik SK, Dall’Armellina E, Karamitsos TD, Francis JM, Choudhury RP, Friedrich MG, Robson MD, Neubauer S. Non-contrast T1-mapping detects acute myocardial edema with high diagnostic accuracy: a comparison to T2-weighted cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 2012;14:42. doi: 10.1186/1532-429X-14-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burt JR, Zimmerman SL, Kamel IR, Halushka M, Bluemke DA. Myocardial T1 mapping: techniques and potential applications. Radiographics. 2014;34:377–95. doi: 10.1148/rg.342125121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn. Reson. Med. 2004;52:141–6. doi: 10.1002/mrm.20110. [DOI] [PubMed] [Google Scholar]
- 7.Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, Thompson RB. Saturation recovery single-shot acquisition (SASHA) for myocardial T(1) mapping. Magn. Reson. Med. 2014;71:2082–95. doi: 10.1002/mrm.24878. [DOI] [PubMed] [Google Scholar]
- 8.Fitts M, Breton E, Kholmovski EG, Dosdall DJ, Vijayakumar S, Hong KP, Ranjan R, Marrouche NF, Axel L, Kim D. Arrhythmia insensitive rapid cardiac T1 mapping pulse sequence. Magn. Reson. Med. 2013;70:1274–82. doi: 10.1002/mrm.24586. [DOI] [PubMed] [Google Scholar]
- 9.Weingärtner S, Akçakaya M, Basha T, Kissinger KV, Goddu B, Berg S, Manning WJ, Nezafat R. Combined saturation/inversion recovery sequences for improved evaluation of scar and diffuse fibrosis in patients with arrhythmia or heart rate variability. Magn. Reson. Med. 2014;71:1024–34. doi: 10.1002/mrm.24761. [DOI] [PubMed] [Google Scholar]
- 10.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J. Cardiovasc. Magn. Reson. 2014;16:2. doi: 10.1186/1532-429X-16-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roujol S, Weingärtner S, Foppa M, Chow K, Kawaji K, Ngo LH, Kellman P, Manning WJ, Thompson RB, Nezafat R. Accuracy, precision, and reproducibility of four T1 mapping sequences: a head-to-head comparison of MOLLI, ShMOLLI, SASHA, and SAPPHIRE. Radiology. 2014;272:683–9. doi: 10.1148/radiol.14140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raman FS, Kawel-Boehm N, Gai N, Freed M, Han J, Liu C-Y, Lima JAC, Bluemke DA, Liu S. Modified look-locker inversion recovery T1 mapping indices: assessment of accuracy and reproducibility between magnetic resonance scanners. J. Cardiovasc. Magn. Reson. 2013;15:64. doi: 10.1186/1532-429X-15-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gai ND, Stehning C, Nacif M, Bluemke DA. Modified Look-Locker T1 evaluation using Bloch simulations: human and phantom validation. Magn. Reson. Med. 2013;69:329–36. doi: 10.1002/mrm.24251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kellman P, Herzka DA, Hansen MS. Adiabatic inversion pulses for myocardial T1 mapping. Magn. Reson. Med. 2014;71:1428–34. doi: 10.1002/mrm.24793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messroghli DR, Greiser A, Fröhlich M, Dietz R, Schulz-Menger J. Optimization and validation of a fully-integrated pulse sequence for modified look-locker inversion-recovery (MOLLI) T1 mapping of the heart. J. Magn. Reson. Imaging. 2007;26:1081–6. doi: 10.1002/jmri.21119. [DOI] [PubMed] [Google Scholar]
- 16.Kellman P, Arai AE, Xue H. T1 and extracellular volume mapping in the heart: estimation of error maps and the influence of noise on precision. J. Cardiovasc. Magn. Reson. 2013;15:56. doi: 10.1186/1532-429X-15-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao J, Nguyen K-L, Natsuaki Y, Spottiswoode B, Hu P. Instantaneous signal loss simulation (InSiL): an improved algorithm for myocardial T1 mapping using the MOLLI sequence. J. Magn. Reson. Imaging. 2015;41:721–9. doi: 10.1002/jmri.24599. [DOI] [PubMed] [Google Scholar]
- 18.Sussman MS, Yang IY, Fok K-H, Wintersperger BJ. Inversion group (IG) fitting: A new T1 mapping method for modified look-locker inversion recovery (MOLLI) that allows arbitrary inversion groupings and rest periods (including no rest period) Magn. Reson. Med. 2016;75(6):2232–40. doi: 10.1002/mrm.25829. [DOI] [PubMed] [Google Scholar]
- 19.Shao J, Rapacchi S, Nguyen K-L, Hu P. Myocardial T1 mapping at 3.0 tesla using an inversion recovery spoiled gradient echo readout and Bloch equation simulation with slice profile correction (BLESSPC) T1 estimation algorithm. J. Magn. Reson. Imaging. 2016;43:414–25. doi: 10.1002/jmri.24999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao J, Rashid S, Renella P, Nguyen K-L, Hu P. Myocardial T1 mapping for patients with implanted cardiac devices using wideband inversion recovery spoiled gradient echo readout. Magn. Reson. Med. 2016 doi: 10.1002/mrm.26223. [DOI] [PubMed] [Google Scholar]
- 21.Rodgers CT, Piechnik SK, Delabarre LJ, Van de Moortele P-F, Snyder CJ, Neubauer S, Robson MD, Vaughan JT. Inversion recovery at 7 T in the human myocardium: measurement of T(1), inversion efficiency and B(1)(+) Magn. Reson. Med. 2013;70:1038–46. doi: 10.1002/mrm.24548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xue H, Greiser A, Zuehlsdorff S, Jolly M-P, Guehring J, Arai AE, Kellman P. Phase-sensitive inversion recovery for myocardial T1 mapping with motion correction and parametric fitting. Magn. Reson. Med. 2013;69:1408–20. doi: 10.1002/mrm.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dabir D, Child N, Kalra A, et al. Reference values for healthy human myocardium using a T1 mapping methodology: results from the International T1 Multicenter cardiovascular magnetic resonance study. J. Cardiovasc. Magn. Reson. 2014;16:69. doi: 10.1186/s12968-014-0069-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robson MD, Piechnik SK, Tunnicliffe EM, Neubauer S. T1 measurements in the human myocardium: the effects of magnetization transfer on the SASHA and MOLLI sequences. Magn. Reson. Med. 2013;70:664–70. doi: 10.1002/mrm.24867. [DOI] [PubMed] [Google Scholar]
- 25.Piechnik SK, Ferreira VM, Dall’Armellina E, et al. Shortened Modified Look-Locker Inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J. Cardiovasc. Magn. Reson. 2010;12:69. doi: 10.1186/1532-429X-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mozes FE, Tunnicliffe EM, Pavlides M, Robson MD. Influence of fat on liver T1 measurements using modified Look-Locker inversion recovery (MOLLI) methods at 3T. J. Magn. Reson. Imaging. 2016;44:105–11. doi: 10.1002/jmri.25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellman P, Bandettini WP, Mancini C, et al. Characterization of myocardial T1-mapping bias caused by intramyocardial fat in inversion recovery and saturation recovery techniques. J. Cardiovasc. Magn. Reson. 2015;17:33. doi: 10.1186/s12968-015-0136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellman P, Herzka DA, Arai AE, Hansen MS. Influence of Off-resonance in myocardial T1-mapping using SSFP based MOLLI method. J. Cardiovasc. Magn. Reson. 2013;15:63. doi: 10.1186/1532-429X-15-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chow K, Spottiswoode BS, Pagano JJ, Thompson RB. Improved precision in SASHA T1 mapping with a variable flip angle readout. J. Cardiovasc. Magn. Reson. 2014;16:M9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting material S1: Protocol printout of the standard MOLLI 5(3)3 sequence.
Supporting material S2: Protocol printout of the inversion recovery spin echo sequence with the inversion time = 50 ms as an example.
Supporting material S3: Protocol printout of the spin echo sequence with the echo time TE =5 ms as an example.
Supporting material S4: Bloch equation simulation to evaluate the effect of magnetization transfer (MT) and inversion factor on T1 estimations.
Supporting material S5: Simulation of the ShMOLLI sequence to compare BLESSPC and InSiL T1 estimation with ShMOLLI’s T1 estimation algorithm in terms of accuracy and precision.