Abstract
Matrix metalloproteinase 2 (MMP2) is a potent protumorigenic, proangiogenic, and prometastatic enzyme that is overexpressed in metastatic cancer. Although there have been various studies on the MMP2 gene, further studies of regulatory factors are required to achieve inhibition of MMP2 enzyme activities. MicroRNAs (miRNAs) play key roles in tumor metastasis. However, the specific functions of miRNAs in metastasis are unclear. In this study, we assessed the function of the microRNA-29 family (miR-29s) in HT1080 human fibrosarcoma cells and examined the regulatory mechanisms of these miRNAs on MMP2 activation. Using miRanda, TargetScan, and PicTar databases, miR-29s were identified as candidate miRNAs targeting MMP2. Gain-of-function studies showed that overexpression of miR-29s could inhibit the invasion of HT1080 cells, suggesting their tumor-suppressive roles in HT1080 cells. In addition, dual luciferase reporter assays indicated that miR-29s could inhibit the expression of the luciferase gene containing the 3'-untranslated region of MMP2 mRNA. Ectopic expression of miR-29s down-regulated the expression of MMP2. Moreover, ectopic expression of miR-29s reduced MMP2 enzyme activity. These results suggested that miR-29s could decrease the invasiveness of HT1080 cells by modulating MMP2 signaling. Taken together, our results demonstrated that miR-29s may serve as therapeutic targets to control tumor metastasis.
Keywords: Fibrosarcoma, Matrix Metalloproteinase 2, MicroRNAs, Neoplasm Invasiveness
INTRODUCTION
Tumor metastasis is the last stage of tumorigenesis in which malignant cancer cells metastasize to organs distant from the primary tumor.1 During tumor development, tumor cells are released from the primary tumor, enter the surrounding tissues, migrate to distant organs through the circulation system, distribute to the organs via vascular outflow, and form new tumors through cell proliferation in secondary organs.1,2
During the stages of tumor development, tumor cell infiltration into the surrounding tissue is regulated by various matrix metalloproteinases (MMPs).2,3 MMPs are zinc-dependent endopeptidases that can modulate extracellular matrix (ECM) proteins and a variety of molecules on the cell surface. MMPs contain four domains: the prodomain, the catalytic domain, the hinge region, and the hemotaxin domain. Structurally, the prodomain of MMPs has a thiol group (-SH), which binds to the zinc ion of the catalytic domain and maintains the MMP as an inactive zymogen.4,5 In order for zymogens to be activated, various proteolytic enzymes must cleave the binding between the thiol of the prodomain and the zinc ion of the catalytic domain. In particular, for activation of MMP2, membrane type 1 MMP (MT1-MMP) and tissue inhibitor of metalloproteinase 2 (TIMP2) must interact in the cell membrane.4,5 Conversely, MMPs are inhibited by a variety of endogenous proteolytic enzymes, including α2-macroglobulin, α1-proteinase inhibitor, α1-chymotrypsin, and TIMPs.6 TIMP2 was first identified as an inhibitor of MMP2.4 TIMP2 can also activate pro-MMP2.7 When TIMP2 is present at high concentrations in the cell, TIMP2 inhibits pro-MMP2 activation by binding to all MT1-MMPs on the cell surface. However, when TIMP2 is present at low concentrations, TIMP2 promotes the activation of pro-MMP2 by acting as an adjunct to the binding of pro-MMP2 to MT1-MMP on the cell surface.6,7
Recently, miRNA research has shown that miRNAs regulate MMP activity.8 In general, miRNAs are small noncoding RNAs composed of 18-21 nucleotides that bind to mRNAs and inhibit translation or mRNA degradation in cells.9,10 In addition, miRNAs play an important role in various regulatory processes in vivo, such as cell proliferation, neuronal differentiation, apoptosis, and regulation of cell development. miRNAs are also known to be key elements of tumorigenesis. In particular, miR-29b, a member of the miR-29 family, has been shown to inhibit metastasis. The miR-29 family which contains three isotypes: miR-29a, miR-29b, and miR-29c. miR-29a and miR-29b1 are located on chromosome 7q32.3, whereas miR-29b2 and miR-29c are located on chromosome 1q32.2.11,12 The sequences of miR-29b1 and miR-29b2 are identical. However, they can be distinguished based on their differences in loci. According to a recent report, miR-29s have various functions by negatively regulating multiple genes. For example, these miRNAs can regulate the methylation of lung cancer by targeting of DNA methyltransferases 3A and 3B13 and can also suppress tumor angiogenesis, invasion, and metastasis.14 In addition, miR-29s regulate the processing of β-amyloid precursor protein by decreasing β-secretase expression and activate p53 via suppressing p85 alpha and CDC42, both of which negatively regulate p53.15 HT1080, a highly metastatic human fibrosarcoma cell line, secretes many different MMPs. Thus, this cell line is suitable for studying invasion inhibitors.
Accordingly, in this study, we used HT1080 cells to screen and identify miRNAs that could regulate MMP2 activity. We also investigated the functions of miRNAs that could regulate MMP2 using HT1080 cells.
MATERIALS AND METHODS
1. Cell culture and transfection
HT1080 human fibrosarcoma cells were purchased from American Type Culture Collection (ATCC, VA, USA) and grown in Earl's minimum essential media (EMEM) supplemented with 10% fetal bovine serum (Gibco, NY, USA). Cells were cultured at 37℃ in a humidified incubator with an atmosphere containing 5% CO2. miRNAs were purchased from Bioneer (Bioneer; Daejeon, Korea). Fluorescein isothiocyanate (FITC)-labeled control miRNA (Bioneer, Daejeon, Korea) was used as a control. HT1080 cells were seeded into 24-well cell culture plates at a density of 1×105 cells/well in 300 µL growth medium. After reaching 70% confluence, cells were then transfected with FITC-labeled control miRNA or miRNA mimics (50 nM) using Lipofectamine 2000 Transfection Reagent (Invitrogen, NY, USA) according to the manufacturer's protocol and then incubated for 48-72 h.
2. Gelatin zymography
To measure MMP activity, we used gelatin zymography. Forty-eight hours after transfection of miRNAs, cells were washed with phosphate-buffered saline (PBS) and then cultured in 300 µL serum-free medium for 24 h. Subsequently, cell-conditioned media was collected and centrifuged at 12,000× g for 10 min at 4℃. The resulting supernatant was mixed with 3 µL zymogram sample buffer (Bio-Rad, CA, USA), and 6 µL of the mixture was loaded onto 8% acrylamide:bisacrylamide (29:1) (Bio-Rad, CA, USA) separating gels containing 0.625 mg/mL gelatin. After electrophoresis at 95 V for 2.5 h, the gels were incubated in renaturation buffer (2.5% Triton X-100) for 2 h at room temperature to remove sodium dodecyl sulfate (SDS). Subsequently, the zymo-gels were washed three times with water, incubated at 37℃ overnight in development buffer (50 mM Tris HCl, pH 7.5, 0.2 M NaCl, 5 mM CaCl, 0.02% Brij 35). After incubation, zymogram gels were stained with 0.25% (w/v) Coomassie brilliant blue R-250 (Bio-Rad, CA, USA) and then destained with destaining buffer (10% acetic acid and 20% methanol).
3. Western blot analysis
Medium without serum was collected from HT1080 cells at 48 h after transfection with miRNAs. Cells were lysed in M-PER mammalian protein extraction reagent (Thermo Scientific, IL, USA) containing protease inhibitor cocktail (Roche, Mannheim, Germany). The same amount of protein from conditioned media or cell lysates was mixed with an equal volume of sample loading buffer, boiled for 5 min, and then subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 10% gels. Subsequently, separated proteins were transferred to polyvinylidene fluoride membranes (PALL Life Sciences, FL, USA). The membranes were then soaked with 5% skim milk in TBST (0.01 M Tris [pH 7.6], 0.1 M NaCl, and 0.1% Tween-20) for 1-2 h at room temperature, washed with TBST, and incubated with primary antibodies targeting MMP2, MMP9, TIMP2 (R&D Systems, MN, USA), TIMP1 (Neomarkers Inc., CA, USA), MT1-MMP (Millipore, MA, USA), reversion-inducing-cysteine-rich protein with kazal motifs (RECK), E-cadherin, zonula occludens-1 (ZO-1), β-catenin, N-cadherin, vimentin, Snail, Twist (Cell Signaling Technology, MA, USA), and Actin (Santa Cruz Biotechnology, TX, USA). The membranes were then washed three times each for 10 min with TBST, followed by incubation with horseradish peroxidase-conjugated secondary antibodies. Immunoreactivity was detected by incubation of the membranes to enhanced chemiluminescent (ECL) reagents (BIONOTE, Gyeonggi-do, Korea) for 5 min before exposure to X-ray film (Fuji, Tokyo, Japan).
4. Quantitative real-time polymerase chain reaction (PCR)
RNA extraction was carried out using an RNAeasy mini kit (Qiagen, CA, USA). Three micrograms of total RNA was then reverse transcribed using an M-MLV cDNA synthesis system (Invitrogen, CA, USA). The reverse-transcribed cDNA was used for real-time PCR. First-strand cDNA was amplified using SYBR Premix Ex Taq II (Takara, Kusatsu, Japan) with indicated primers. Each reaction (20 µL) contained 10 ng of cDNA and 5 pmol of each primer (MMP2, MMP9, TIMP2, MT1-MMP, p53, RECK, 18S RNA; Qiagen, CA, USA). Real-time qPCR was run on a Roter-Gene instrument (Qiagen, CA, USA). Cycling conditions were as follows: one cycle of denaturation at 95℃ for 30 s followed by 40 two-segment cycles of amplification (95℃ for 5 s and 60℃ for 30 s).
5. Luciferase assay
Plasmid psiCHECK-2 (Promega, WI, USA) containing the 3'-UTR of MMP2 was used in luciferase reporter assays (Fig. 1B). These plasmids contained Renilla luciferase whose 3' end was fused to the 3'-UTR of human MMP2. Firefly luciferase was included for normalization. One day before transfection, HT1080 cells were seeded into 24-well plates with 300 µL growth medium. For miRNA transfection, 500 ng of plasmid DNA and 50 nM miRNA were transiently cotransfected into HT1080 cells. At 48-72 h after transfection, luciferase activity was measured using a Dual-luciferase Reporter Assay System (Promega, WI, USA) according to the manufacturer's protocol. Unlike other luciferase reporter vectors, the psiCHECK-2 Vector also contained the firefly luciferase gene. Since firefly luciferase was used to standardize the transfection, there was no need to transfect a vector control.
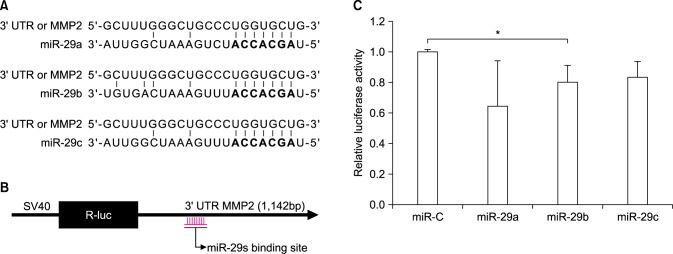
FIG. 1. miR-29s directly targeted MMP2 by interaction with the 3'-UTR of MMP2. (A) Predicted consequential pairing of the target region. The sequences of miR-29s binding sites within the 3'-UTR of MMP2 and their seed regions (bold). (B), (C) HT1080 cells were cotransfected with miR-29s mimics or control miRNA (50 nM) and the plasmid psiCHECK-2 (500 ng) containing the wild-type 3'-UTR of MMP2. After 48 h, luciferase assays were performed. Firefly luciferase was used to normalize transfections and eliminated the need to transfect a second vector control (*p<0.05).
6. Matrigel assay
Cell invasion was assessed using Matrigel Invasion Chambers with 8.0-µm polyethylene terephthalate (PET) membranes in two 24-well plates (BD Biosciences, MA, USA). Cells (1.0×104) transfected with miR-29s were seeded with 500 µL serum-free medium on a transwell chamber precoated with Matrigel. Medium (750 µL) containing 1% fetal bovine serum in the lower chamber served as a chemoattractant. After incubation at 37℃ for 24 h, the noninvading cells were removed using a cotton swab. Invasive cells attached to the lower surface of the membrane were fixed with methanol at room temperature for 2 min and then stained with toluidine blue. The membranes were washed three times with water to remove background staining. Finally, the number of stained invasive cells on the lower surface of the membrane was counted under a microscope and statistically analyzed.
7. Statistical analysis
Data were analyzed using one-tailed Student's t-test. Differences with p-values of less than 0.05 were considered statistically significant.
RESULTS
1. MMP2 was a putative target of miR-29s
To identify miRNAs that could regulate MMP2 activity, miRNAs that could bind to MMP2 mRNA were predicted using TargetScan, PicTar, and miRanda. The following miRNAs were found to have a conserved binding site in the 3'-UTR of MMP2: miR-29s, miR-124, miR-506, miR-519, miR-17-5p, miR-20-5p, miR-93-5p, and miR-106-5p. Among these miRNAs, miR-29b have been shown to be down-regulated in cancer and is significantly associated with poor recurrence-free survival in patients with cancer.14 Therefore, subsequent experiments were conducted to investigate whether miR-29s could regulate the invasion of cancer cells through inhibiting MMP2. Consequential pairing showed that miRNA target complementarity in the seed may might influence the efficacy of miRNA targeting (Fig. 1A).
2. miR-29s directly targeted the 3'-UTR of MMP2 and inhibited MMP2 expression
The interaction between miR-29s and the 3'-UTR of MMP2 was then examined by luciferase reporter assays. First, we cloned a construct by inserting the 3'-UTR of MMP2 into the psiCHECK-2 vector. This plasmid contains renilla luciferase gene with 3'-UTR MMP2, so that renilla luciferase activity can be used to determine whether miR-29s targets 3-'UTR MMP2. The psiCHECK-2 vector also contains the firefly luciferase gene to normalize transfections. Normalization of the transfection was possible without co-transfection of the control vector. Ectopic expression of miR-29s significantly decreased luciferase activity, demonstrating that miR-29s targeted the 3'-UTR of MMP2 directly based on the decreased luciferase activity (Fig. 1C).
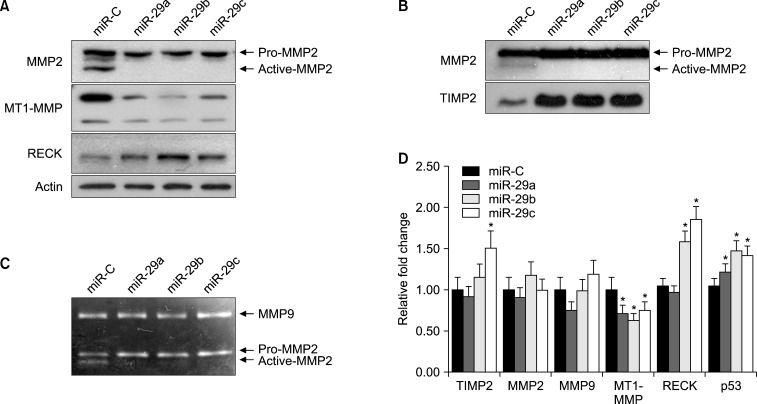
Next, we assessed the capability of miR-29s to regulate endogenous MMP2 expression by western blotting in both conditioned medium and cell lysates. As shown in Fig. 2A, ectopic expression of miR-29s significantly decreased endogenous expression of pro-MMP2 and active MMP2 in whole-cell lysates. However, miR-29s did not alter MMP2 mRNA expression, as shown in Fig. 2D, indicating that miR-29s targeted MMP2 in the translation step, not in the transcription step. In conditioned medium, the expression of active MMP2 was decreased. Intriguingly, the expression of TIMP2, an MMP2 inhibitor, was enhanced by miR-29s (Fig. 2B). These data indicated that MMP2 was directly and negatively regulated by miR-29s.
FIG. 2. miR-29s downregulated MMP2 and reduced MMP2 activity. (A) HT1080 cells were transfected with control miRNA or miR-29s mimics (50 nM) for 72 h. For western blotting, cells lysates were prepared and analyzed using the indicated antibodies. Actin was used as a loading control. (B) After 72 h, conditioned medium from the same experiment was harvested and used for western blotting. (C) Conditioned medium from the same experiment was processed for gelatin zymography. The band at 72 kDa represents the active form of MMP2, and the band at 92 kDa represents pro-MMP2. (D) Quantitative real-time PCR. HT1080 cells were transfected with 50 nM of miR-29s mimics. After incubation for 48 h, total RNA was isolated and analyzed by SYBR quantitative real-time PCR. The results were normalized to 18s RNA expression and are presented as the relative expression level (*p<0.05).
3. miR-29s inhibited MMP2 enzyme activity
In order to determine whether miR-29s affected the gelatinolytic activities of MMP2, HT1080 cells were transfected with miRNA mimics. Subsequently, conditioned medium was analyzed by gelatin zymography. MMP2 activity in HT1080 cells transfected with miR-29s showed lower enzyme activity than that in cells transfected with control miRNA (miR-C). Gelatin zymography showed that the band at 72 kDa representing MMP2 activity was reduced (Fig. 2C). However, the band at 92 kDa representing MMP9 activity was not changed. These results indicated that miR-29s could directly target MMP2 and negatively regulate MMP2 enzyme activity.
4. miR-29s regulated genes associated with MMP2 activation
miR-29s may inhibit tumor angiogenesis, invasion, and metastasis by modulating MMP2 signaling. To gain insights into these mechanisms, we investigated the effects of gain-of-function of miR-29s in HT1080 cells. Cells were transiently transfected with miR-29s or miR-C, as previously described. Real-time PCR was performed to measure the expression levels of genes associated with MMP2 activation. As shown in Fig. 2D, miR-29s tightly controlled genes associated with MMP2 activation. The expression levels of MMP9 and MT1-MMP, which activate MMP2, were decreased, whereas that of the MMP2 inhibitors TIMP2, RECK, and p53 were decreased (Fig. 2D). Furthermore, the expression of secreted TIMP2 was increased (Fig. 2B).
5. miR-29s inhibited cell invasion in vitro
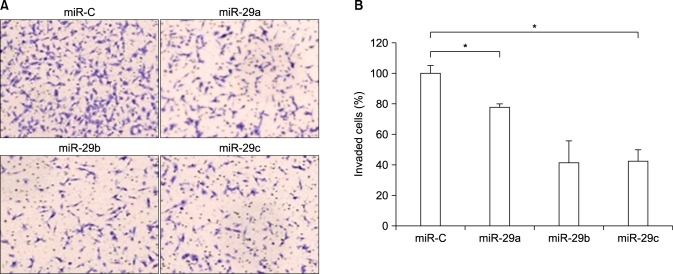
To test whether miR-29s may function as tumor suppressors, we examined whether miR-29s affected cell invasion. HT1080, a highly metastatic cells line, were used for the invasion analysis because of their well-characterized penetration into the matrigel membrane. HT1080 cells were transfected with miR-29s mimics and control miRNA. Subsequently, cells were plated in media without serum. Media supplemented with serum was used as a chemoattractant for evaluation of cell invasion by use of transwell assay. Significant differences in cell invasion were found between cells transfected with miR-29s and those transfected with control miRNA (Fig. 3A, B). This finding indicated that upregulation of miR-29s inhibited cell invasion and migration in vitro.
FIG. 3. miR-29s inhibited cell invasion in vitro. (A) Cellular invasion was evaluated in HT1080 cells using Matrigel Invasion Chambers with 8.0 mm PET membrane. Cells (1.0-104) transfected with miR-29s mimics were seeded with serum-free medium on transwell chambers precoated with Matrigel. Medium containing 1% fetal bovine serum was used as a chemoattractant. After incubation for 24 h, invaded cells were stained with toluidine blue. (B) The number of invaded cells was counted under a microscope. The data are presented as relative cell invasion (*p<0.05).
DISCUSSION
In this study, we identified miR-29s targeting MMP2 using bioinformatics tools and determined whether MMP2 was a direct target of miR-29s. Moreover, we found that miR-29s decreased MMP2 activity by modulating genes regulating MMP2 activation and repressing the expression of prometastatic genes, thereby inhibiting the invasiveness of fibrosarcoma cells. Fibrosarcoma is a malignant mesenchymal tumor derived from fibrous connective tissue. There is little research on miRNA function associated with fibrosarcoma.16 Notably, miR-520c and miR-373 can activate the Ras/mitogen-activated protein kinase pathway by activating MMP9 in fibrosarcoma cells.17 Other studies have shown that miR-409-3p, a potential tumor-suppressive miRNA, targets angiogenin to suppress fibrosarcoma cell proliferation, invasion, and metastasis.18
In 2009, Welch and colleagues proposed the “metastamir” concept, referring to a group of miRNAs associated with metastasis. Recently, several metastamirs have been shown to play roles in cancer metastasis. For example, a metastasis-promoting metastamir containing miR-10b, miR-373, miR-520c, miR-21, miR-143, miR-182 was identified.19 Metastasis-supressing metastamirs containing miR-335, miR-206, miR-146a/b, and miR-31 have also been identified.20
The metastamir containing miR-29s is involved in invasion, metastasis, the epithelial-to-mesenchymal transition (EMT), and differentiation of skeletal muscle cells and osteoblasts. In particular, miR-29b can inhibit metastasis by inhibiting MMP2 expression in prostate cancer cells.21 Activation of MMP2 causes degradation of the ECM, which promotes vascularization and metastasis of cancer cells. In addition, MMP2 promotes cancer cell migration and proliferation by stimulating remodeling of the ECM and release of ECM-binding growth factors.14 In addition, MMP2 overexpression has been observed in several malignant tumors.22
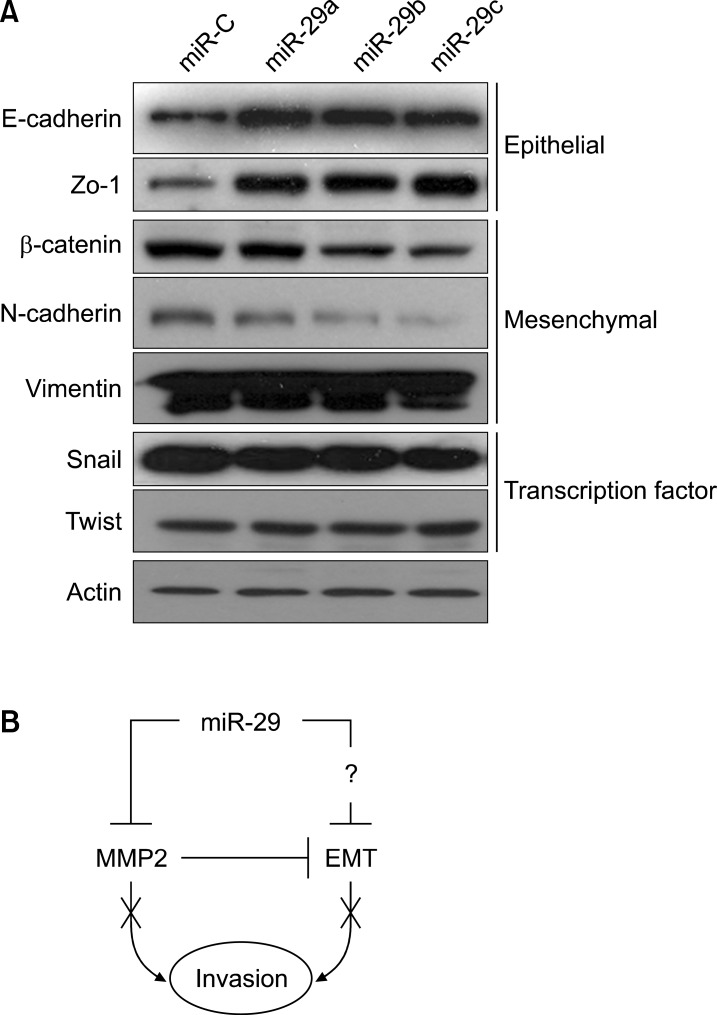
Metastasis is the spread of malignant tumors in which tumor cells migrate, settle, and proliferate in unrelated microenvironments.23 In early stages of metastasis, adhesion to the epithelial cells is weakened, which suggests changes in EMT signaling, such as changes in the expression of E-cadherin and N-cadherin.24 When a cell acquires invasion or migration capability, EMT epithelial markers decrease, whereas mesenchymal markers increase.25 Ectopic expression of miR-29s in fibrosarcoma cells induced the expression of E-cadherin and ZO-1 (Fig. 4A) but decreased the expression of the mesenchymal markers N-cadherin and β-catenin, suggesting that the properties of less-invasive epithelial cells had been acquired and that the EMT was reversed (Fig. 4A). However, Twist and Snail, the key transcription factors modulating the EMT, were not altered. Because of the complexity of the EMT pathway, we could not rule out the possibility that miR-29s may affect other regulatory elements. Therefore, further studies are needed to assess whether miR-29s can target a variety of other genes associated with the EMT.
FIG. 4. miR-29s suppressed invasion by regulating MMP2 activity and the EMT. (A) HT1080 cells were transfected with control miRNA or miR-29s mimics (50 nM) for 72 h. Total cell lysates were analyzed by western blotting with antibodies targeting EMT markers. Actin was used as a loading control. (B) Schematic diagram of the effects of miR-29s on invasion.
In summary, we demonstrated that miR-29s could regulate invasion by modulating MMP2 signaling (Fig. 4B). Our data suggested that miR-29s dysfunction accounted for one of the mechanisms of MMP2 overexpression in cancer cells. Our results also indicated that miR-29s deregulation may play an important role in the rapid growth and recurrence of cancer. Thus, restoration of miR-29s may be a promising strategy in anticancer chemotherapy.
ACKNOWLEDGEMENTS
This work was supported by the Health Fellowship Foundation.
Footnotes
CONFLICT OF INTEREST STATEMENT: None declared.
References
- 1.Baranwal S, Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. 2010;126:1283–1290. doi: 10.1002/ijc.25014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy G, Nagase H. Progress in matrix metalloproteinase research. Mol Aspects Med. 2008;29:290–308. doi: 10.1016/j.mam.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerkelä E, Saarialho-Kere U. Matrix metalloproteinases in tumor progression: focus on basal and squamous cell skin cancer. Exp Dermatol. 2003;12:109–125. doi: 10.1034/j.1600-0625.2003.120201.x. [DOI] [PubMed] [Google Scholar]
- 4.Amălinei C, Căruntu ID, Bălan RA. Biology of metalloproteinases. Rom J Morphol Embryol. 2007;48:323–334. [PubMed] [Google Scholar]
- 5.Strongin AY, Marmer BL, Grant GA, Goldberg GI. Plasma membrane-dependent activation of the 72-kDa type IV collagenase is prevented by complex formation with TIMP-2. J Biol Chem. 1993;268:14033–14039. [PubMed] [Google Scholar]
- 6.Roeb E, Matern S. Matrix metalloproteinases and colorectal cancer. Med Klin (Munich) 2003;98:763–770. doi: 10.1007/s00063-003-1322-5. [DOI] [PubMed] [Google Scholar]
- 7.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Li H. Role of microRNA-mediated MMP regulation in the treatment and diagnosis of malignant tumors. Cancer Biol Ther. 2013;14:796–805. doi: 10.4161/cbt.25936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 11.Garzon R, Heaphy CE, Havelange V, Fabbri M, Volinia S, Tsao T, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou J, Lin JH, Brenot A, Kim JW, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang JH, Zhou HC, Zeng C, Yang J, Liu Y, Huang X, et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 15.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 16.Varshney J, Subramanian S. MicroRNAs as potential target in human bone and soft tissue sarcoma therapeutics. Front Mol Biosci. 2015;2:31. doi: 10.3389/fmolb.2015.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu P, Wilson MJ. miR-520c and miR-373 upregulate MMP9 expression by targeting mTOR and SIRT1, and activate the Ras/Raf/MEK/Erk signaling pathway and NF-ĸB factor in human fibrosarcoma cells. J Cell Physiol. 2012;227:867–876. doi: 10.1002/jcp.22993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weng C, Dong H, Chen G, Zhai Y, Bai R, Hu H, et al. miR-409-3p inhibits HT1080 cell proliferation, vascularization and metastasis by targeting angiogenin. Cancer Lett. 2012;323:171–179. doi: 10.1016/j.canlet.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Xiao B, Tong H, Xie F, Zhang Z, Xiao GG. Regulation of breast cancer tumorigenesis and metastasis by miRNAs. Expert Rev Proteomics. 2012;9:615–625. doi: 10.1586/epr.12.64. [DOI] [PubMed] [Google Scholar]
- 21.Ru P, Steele R, Newhall P, Phillips NJ, Toth K, Ray RB. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol Cancer Ther. 2012;11:1166–1173. doi: 10.1158/1535-7163.MCT-12-0100. [DOI] [PubMed] [Google Scholar]
- 22.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 23.Nelson AR, Fingleton B, Rothenberg ML, Matrisian LM. Matrix metalloproteinases: biologic activity and clinical implications. J Clin Oncol. 2000;18:1135–1149. doi: 10.1200/JCO.2000.18.5.1135. [DOI] [PubMed] [Google Scholar]
- 24.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 25.Doerner AM, Zuraw BL. TGF-beta1 induced epithelial to mesenchymal transition (EMT) in human bronchial epithelial cells is enhanced by IL-1beta but not abrogated by corticosteroids. Respir Res. 2009;10:100. doi: 10.1186/1465-9921-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]