Abstract
Introduction
The continual Middle East respiratory syndrome (MERS) threat highlights the importance of developing effective antiviral therapeutics to prevent and treat MERS coronavirus (MERS-CoV) infection. A surface spike (S) protein guides MERS-CoV entry into host cells by binding to cellular receptor dipeptidyl peptidase-4 (DPP4), followed by fusion between virus and host cell membranes. MERS-CoV S protein represents a key target for developing therapeutics to block viral entry and inhibit membrane fusion.
Areas covered
This review illustrates MERS-CoV S protein’s structure and function, particularly S1 receptor-binding domain (RBD) and S2 heptad repeat 1 (HR1) as therapeutic targets, and summarizes current advancement on developing anti-MERS-CoV therapeutics, focusing on neutralizing monoclonal antibodies (mAbs) and antiviral peptides.
Expert opinion
No anti-MERS-CoV therapeutic is approved for human use. Several S-targeting neutralizing mAbs and peptides have demonstrated efficacy against MERS-CoV infection, providing feasibility for development. Generally, human neutralizing mAbs targeting RBD are more potent than those targeting other regions of S protein. However, emergence of escape mutant viruses and mAb’s limitations make it necessary for combining neutralizing mAbs recognizing different neutralizing epitopes and engineering them with improved efficacy and reduced cost. Optimization of the peptide sequences is expected to produce next-generation anti-MERS-CoV peptides with improved potency.
Keywords: MERS, MERS-CoV, spike protein, receptor-binding domain, membrane fusion, monoclonal antibodies, peptides, therapeutics
1. Introduction
First identified in Saudi Arabia in June 2012, Middle East respiratory syndrome (MERS) is caused by MERS coronavirus (MERS-CoV) [1]. MERS is an acute respiratory disease and often leads to pneumonia and renal failure, very similar to severe acute respiratory syndrome (SARS), a worldwide epidemic in 2003 caused by another coronavirus, SARS-CoV [1–5]. Most MERS cases have been found in countries of the Middle East, including Saudi Arabia, Qatar, and the United Arab Emirates [6–10]. However, a most recent MERS outbreak occurred in South Korea, where 186 cases could all be traced back to a 68-year-old South Korean man travelling from the Middle East [11–15]. The MERS outbreak in South Korea demonstrated that close contact with MERS-CoV-infected patients led to efficient human-to-human transmission, mainly resulting from high population density and insufficient healthcare system [15–17]. Globally, MERS-CoV has caused at least 1,813 human infections, including 645 deaths, as of 11 November 2016 (mortality rate ~36%), in 27 countries worldwide (http://www.who.int/emergencies/mers-cov/en/). Development of effective intervention strategies to curb the spread of MERS-CoV is, therefore, urgently needed.
Like SARS-CoV, MERS-CoV is a zoonotic virus transmitted from animals to humans [18–21]. Bats are the likely natural reservoir of MERS-CoV, and two mutations appeared to play critical roles in the eventual bat-to-human transmission of MERS-CoV [22–28]. Dromedary camels are believed to be an important reservoir host of MERS-CoV and they appear to be the only animal host responsible for human infections [29]. It is demonstrated that camels in the Middle East, as well as East and North Africa, have high seropositive rates for MERS-CoV [29,30]. In addition, MERS-CoV isolates from dromedaries and humans show almost identical genetic and clinical characteristics [19,20,31–34]. Furthermore, dromedary camels developed primarily upper respiratory tract infection upon MERS-CoV inoculation and people become infected with MERS-CoV after close contact with sick camels, providing evidence for camel-to-camel and camel-to-human transmission of MERS-CoV [19–21,33]. However, another report suggested that camel-to-human transmission is rare [35].
MERS-CoV is a novel beta-coronavirus phylogenetically related to bat coronaviruses HKU4 and HKU5, the two prototype species in lineage C of the beta-coronavirus genus [2,36–38]. Unlike HKU4 and HKU5, MERS-CoV is the first human coronavirus in the group C species of the genus beta-coronavirus, and the sixth coronavirus to cause human infections [36,39]. Similar to the genomes of other coronaviruses, the MERS-CoV genome is a single, positive-stranded RNA encoding at least 10 open reading frames (ORFs), nine of which are expressed from seven subgenomic mRNAs (sg mRNAs), which are then translated into four major viral structural proteins, including spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as several accessory proteins, such as 3, 4a, 4b, 5, and 8b with unknown origins and functions. The ORF1a and ORF1 b genomic RNAs at the 5′-end are translated into virus replication-related proteins and cleaved to produce 16 functional nonstructural proteins (nsps) that are related to viral RNA synthesis and recombination [39–41]. The life cycle of MERS-CoV replication is described in Figure 1 [8,39,42–44]. Different from some other beta-coronaviruses, the MERS-CoV genome does not encode a hemagglutinin-esterase (HE) protein [1]. Genomic analysis of MERS-CoV implies a potential for occurring genetic recombination during a MERS-CoV outbreak [45].
Figure 1.
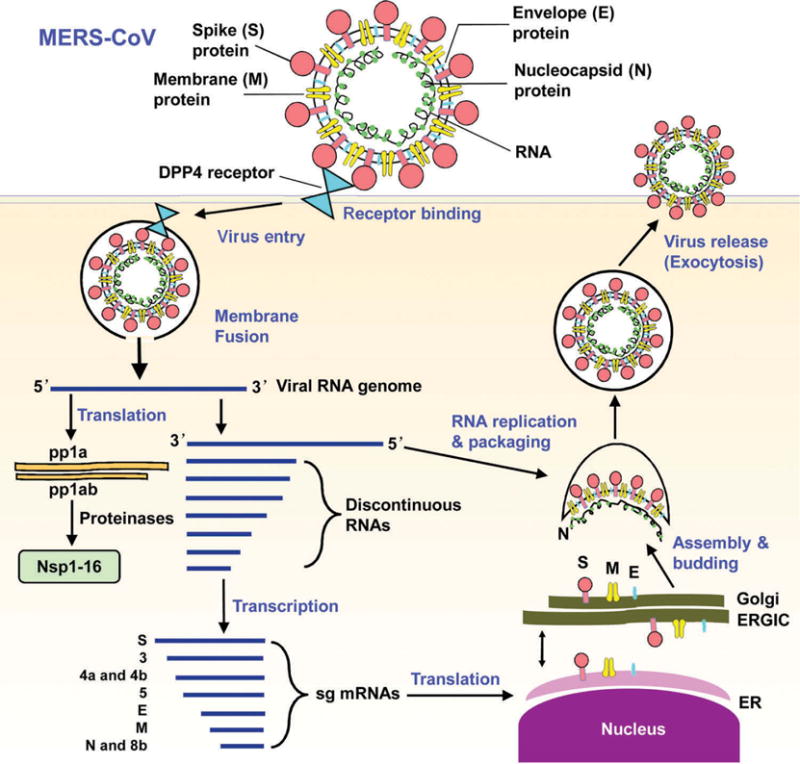
Schematic diagram of MERS-CoV life cycle [8,39,42,43]. MERS-CoV binds to its cellular receptor DPP4 via the S protein and then enters target cells, followed by fusion of the cell and virus membranes and release of the viral RNA genome into the cytoplasm. The open reading frame (ORF), 1a and 1b, in the viral genomic RNA is translated into replicase polyproteins pp1a and pp1ab, respectively, and then potentially cleaved by papain-like protease (PLpro), 3 C-like cysteine protease (3CLpro, main protease), and other viral proteinases into 16 nonstructural proteins (nsp1–16). A negative-strand genomic-length RNA is synthesized as the template for replicating viral genomic RNA. Negative-strand subgenome-length mRNAs (sg mRNAs) are formed from the viral genome as discontinuous RNAs and used as the template to transcribe sg mRNAs. Viral N protein is assembled with the genomic RNA in the cytoplasm. The synthesized S, M and E proteins are gathered in the endoplasmic reticulum (ER) and transported to the ER-Golgi intermediate compartment (ERGIC) where they interact with the RNA-N complex and assemble into viral particles. The viral particles are maturated in the Golgi body and then released from the cells.
Among encoded coronavirus proteins, the S protein is responsible for receptor binding and subsequent viral entry into host cells, and it is, therefore, a major therapeutic target [44,46–48]. This review introduces the structure and function of MERS-CoV S protein, illustrates different regions of this protein as important therapeutic targets, and summarizes the advancement of developing antiviral therapeutics targeting MERS-CoV S protein. It is anticipated for the readers to gain a broad understanding of the crucial roles of MERS-CoV S protein in antiviral development.
2. Structure and function of MERS-CoV S protein
The S protein mediates viral attachment to host cells and virus-cell membrane fusion, thereby playing pivotal roles in MERS-CoV infection. During the infection process, the S protein of MERS-CoV is cleaved into a receptor-binding subunit S1 and a membrane-fusion subunit S2 [48–51]. The functional domains in MERS-CoV S protein and amino acid residues covering respective regions are listed in Figure 2(a).
Figure 2.
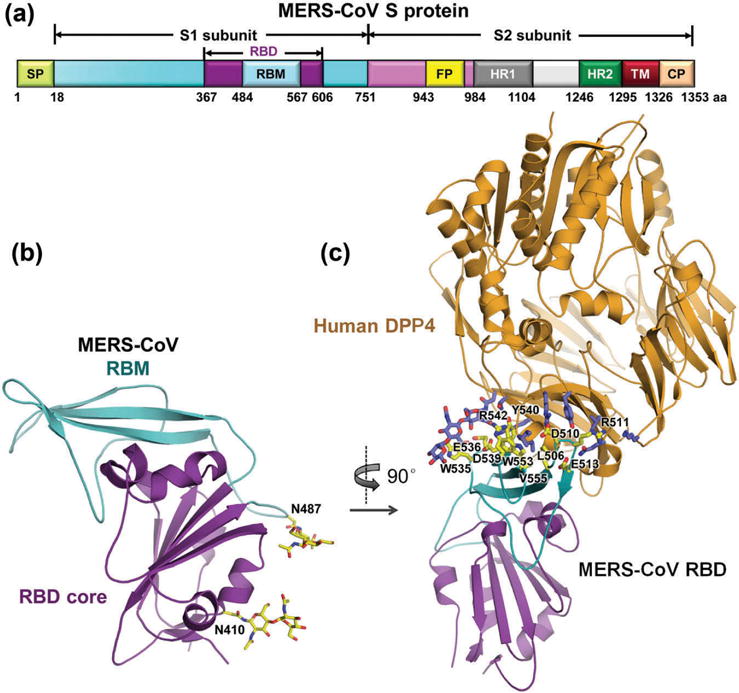
Functional domains of MERS-CoV S protein and structural basis of MERS-CoV receptor binding [49,51,52]. (a) Schematic diagram of MERS-CoV S protein. S contains S1 and S2 subunits. SP, signal peptide; RBD, receptor-binding domain; RBM, receptor-binding motif; FP, fusion peptide; HR1 and HR2, heptad repeat region 1 and 2; TM, transmembrane; CP, cytoplasmic tail. (b) Crystal structure of MERS-CoV RBD. Core structure is in purple, and RBM is in cyan (PDB ID: 4KQZ). The two N-linked glycans are labeled in black. (c) Crystal structure of MERS-CoV RBD in complex with its receptor human hDPP4 (orange) (PDB ID: 4KR0). Contacting residues in RBM are shown as yellow sticks and labeled in black. Full color available online.
2.1. Structure and function of MERS-CoV S1 subunit
2.1.1. MERS-CoV S1-RBD-mediated receptor binding
Unlike SARS-CoV, which requires angiotensin-converting enzyme 2 (ACE2) as its receptor for binding target cells, MERS-CoV utilizes dipeptidyl peptidase 4 (DPP4, also known as CD26) as its cellular receptor [53,54]. The MERS-CoV S1 subunit contains a receptor-binding domain (RBD) that binds DPP4, mediating viral attachment to target cells [48,49,55,56]. MERS-CoV RBD may bind DPP4 from different hosts, including humans, camels, ferrets, and bats, and the binding affinity is different in the hosts carrying different DPP4s, determining host species restriction and susceptibility of MERS-CoV [28,57,58].
2.1.2. MERS-CoV RBD and RBD/DPP4 complex structures
Similar to SARS-CoV RBD, MERS-CoV RBD (residues 367–588) is composed of a core subdomain and a receptor-binding motif (RBM). Although the RBDs of MERS-CoV and SARS-CoV share a high degree of structural similarity in their core subdomains, their RBMs are quite different, which explains the different receptors noted above [51,52]. The core subdomain contains a five-stranded antiparallel β-sheet and several connecting helices, which are stabilized by three disulfide bonds [49,51,52]. The RBM consists of a four-stranded antiparallel β-sheet being connected to the core via intervening loops [49,52]. Two N-linked glycans (N410 and N487) are located in the core and RBM, respectively (Figure 2(b)) [52]. In particular, the RBM (residues 484–567) is responsible for interacting with the extracellular β-propeller domain of DPP4 (Figure 2(c)) [49,51].
2.2. Structure and function of MERS-CoV S protein S2 subunit
2.2.1. MERS-CoV S2-mediated membrane fusion mechanism
Similar to the S2 of other coronaviruses, such as SARS-CoV, MERS-CoV S2 subunit is responsible for membrane fusion. In this process, heptad repeat 1 (HR1) and 2 (HR2) regions of S2 play indispensable and complementary roles [50,59]. S2 mediates membrane fusion by undergoing dramatic conformational changes [44,50,59,60]. Prior to membrane fusion, the S protein presents as a native trimeric structure on the viral surface. During the membrane fusion process, S2 dissociates from S1, and the two heptad repeat regions in S2, designated HR1 and HR2, form a 6-helix bundle (6-HB) fusion core, exposing a hydrophobic fusion peptide inserted into the host membrane and bringing the viral and host membranes into proximity for fusion (Figure 3(a)). Understanding of this fusion core structure will guide rational design of MERS-CoV fusion inhibitors and anti-MERS-CoV therapeutics specifically targeting S2.
Figure 3.
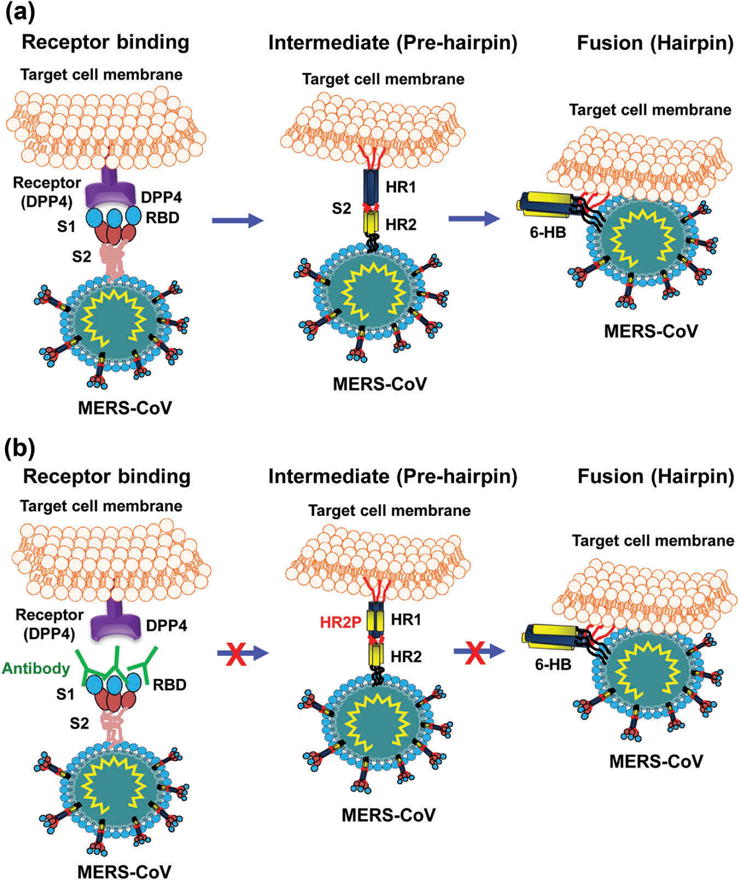
Schematic diagrams of MERS-CoV S protein S2-mediated membrane fusion and MERS-CoV S-targeting mAbs and peptides [59,61]. (a) Schematic diagram of MERS-CoV S2-mediated membrane fusion. The following major processes are involved in MERS-CoV membrane fusion. In receptor binding stage, S protein, which exists as a trimer, binds to the cellular receptor DPP4 via S1-RBD. This binding triggers conformational changes of S protein, leading to dissociation of S1 from S2 with exposed HR1-trimer and HR2-trimer, thus entering intermediate (pre-hairpin) stage. In fusion (hairpin) stage, HR1 and HR2 helices associate with each other to form a 6-helix bundle (6-HB) fusion core, and bring the membranes of virus and cell into close proximity for fusion. (b) Schematic diagram of mechanism of action of MERS-CoV S1-RBD-targeting neutralizing mAbs and S2-HR1-targeting peptides. The RBD-specific antibody (IgG or Fab) binds to viral S1-RBD and interrupts the binding between RBD and DPP4, thus blocking virus infection. HR1-targeting HR2 peptide (e.g. HR2P) binds to the HR1-trimer to form a heterologous 6-HB, thus interferes with subsequent 6-HB fusion core formation and virus-cell membrane fusion, resulting in the inhibition of MERS-CoV infection.
2.2.2. MERS-CoV S2-based fusion core structure
The fusion core structure of MERS-CoV is similar to that of SARS-CoV, but it is distinct from that of the other coronaviruses, such as mouse hepatitis virus (MHV) and HCoV-NL63 [59,62–64]. X-ray crystallography has identified a stable 6-HB fusion core structure [59], which contains a parallel trimeric coiled coil of three longer HR1 helices and three shorter HR2 chains surrounding it in an oblique antiparallel manner [50,59]. The 6-HB helices of MERS-CoV are formed by residues 987–1,062 in the HR1 region and residues 1,263–1,279 in the HR2 region, respectively. In addition, residues 1,283–1,285 in the S2 form a one-turn 310 helix at the C-terminus of HR1-L6-HR2 fusion protein. The interaction between HR1 and HR2 helices is predominantly hydrophobic, consisting of a number of hydrogen bonds formed through key residues and mainly located around the N- and C-terminal regions of the HR2 helices [59].
Based on the crystal structure of the 6-HB of MERS-CoV, two peptides, designated HR1P and HR2P, which span residues 998–1,039 in HR1 and residues 1,251–1,286 in HR2 domains, respectively, were designed to investigate the interaction between HR1 and HR2 in the 6-HB and determine its stability. Results showed that HR1P interacted with HR2P to form a 6-HB and that HR1P/HR2P mixture at equimolar concentration constituted a helical complex with strong thermal stability [59].
2.3. Structure and function of other regions of MERS-CoV S protein
2.3.1. Protease-dependent activation of MERS-CoV S protein
MERS-CoV S protein needs to be activated for entry into target cells. Such activation can be directed by one or more of the following proteases: TMPRSS2, endosomal cathepsins (B/L), and proprotein convertases, depending on the host cell type and tissues [65–67]. For example, both TMPRSS2 and cathepsin L may activate MERS-CoV S protein for viral entry in naturally susceptible cells, such as Caco-2 [65]. In addition, using bioinformatics and peptide cleavage assays, two cleavage sites were identified for furin protease at the S1/S2 boundary and within S2, respectively [68]. It is indicated that MERS-CoV S protein was proteolyzed by furin during protein biosynthesis for the S1/S2 cleavage site, and virus entry for the S2 cleavage site, respectively. This two-step furin-mediated protease cleavage of S protein suggests the importance of furin activation in MERS-CoV fusion and infection.
3. Targets in MERS-CoV S protein for development of therapeutics
The structure and function of MERS-CoV S protein determine the key role of this protein as an important target to develop therapeutics against MERS. Specific targets in the S protein include N-terminal domain (NTD), RBD and other regions in S1 subunit, and HR1 and HR2 in S2 subunit, as well as other targets related to the function of MERS-CoV S protein. The therapeutic agents currently developed based on these targets are characterized as anti-MERS-CoV neutralizing mAbs, anti-DPP4 mAbs, DPP4 antagonists, peptidic fusion inhibitors, protease inhibitors, siRNA, and other molecules. The anti-MERS-CoV agents mainly block receptor binding or membrane fusion, thus leading to the inhibition of MERS-CoV infection. None of the anti-MERS-CoV therapeutic agents are approved for human use. The S proteintargeting anti-MERS-CoV therapeutic agents, including neutralizing mAbs and peptides, are summarized in Tables 1–2.
Table 1.
Summary of MERS-CoV S protein-targeting neutralizing mAbsa.
| MAb name | Target region in S protein | Antiviral mechanism | In vivo protection | Crystal structures available | Ref. |
|---|---|---|---|---|---|
| Mouse mAbs | |||||
| G2 | S1 outside RBD |
Has weaker binding affinity to S1; recognizes neutralizing epitopes in S1 outside the RBD; potently neutralizes infection of pseudotyped MERS-CoV (England1 and Bisha1 strains) | Not reported | N/A | [69] |
| Mersmab1 | RBD | Recognizes neutralizing epitopes at RBD residues D510, R511, and W553; disrupts RBD-hDPP4 receptor binding; inhibits S-mediated pseudotypted MERS-CoV entry; potently neutralizes infection of pseudotyped and live MERS-CoV (EMC2012 strain) | Not reported | No | [70] |
| 2E6 4C2 |
RBD | Recognizes conformational epitopes at RBD residues Y397-N398, K400, L495-K496, P525, V527-S532, W535-E536, and D539-Q544, which overlap with hDPP4-binding sites at RBD residues W535, E536, D539, Y540, and R542 (for 4C2); blocks RBD-hDPP4 receptor binding and virus entry; neutralizes infection of pseudotyped and live MERS-CoV (EMC2012) | Not reported | No Yes, RBD/mAb-Fab complex |
[71] |
| F11 D12 |
RBD | Recognizes neutralizing epitopes at RBD residues D509 (for F11), W535 and E536 (for D12); disrupts RBD-hDPP4 receptor binding; potently neutralizes infection of pseudotyped MERS-CoV (England1 and/or Bisha1) | Not reported | No Yes, RBD/mAb-Fab complex |
[69] |
| G4 | S2 | Recognizes neutralizing epitopes in S2; low neutralizing potency against infection of pseudotyped MERS-CoV (England1 and Bisha1) | Not reported | N/A | [69] |
| Human mAbs | |||||
| LCA60 | S1-NTD & RBD | Recognizes neutralizing epitopes at S1-NTD (residue V33) and RBD (residues T489, K493, E536, and E565); interferes with RBD-hDPP4 receptor binding; potently neutralizes infection of 3 live MERS-CoV strains (EMC2012, London1, and Jordan-N3) | Prophylactically and therapeutically protects Ad5/hDPP4-transduced wild-type or IFNAR-KO mice from challenge of MERS-CoV (EMC2012 and London1) | No | [72] |
| MERS-4 MERS-27 |
RBD | Recognizes neutralizing epitopes at RBD residues D455, E513, R542 (for MERS-4), W535 and D539 (for MERS-27); blocks RBD-hDPP4 receptor interactions; inhibits S-mediated pseudotyped MERS-CoV entry; neutralizes pseudotyped and live MERS-CoV infection | Not reported | No Yes, RBD/mAb-Fab complex |
[73,74] |
| REGN3051 REGN3048 |
RBD | Blocks S-mediated pseudotyped MERS-CoV entry; neutralizes infection of divergent strains of pseudotyped and live MERS-CoV (EMC2012) | Protects HuDPP4 mice from MERS-CoV challenge | No | [75] |
| 1E9 1F8 3A1 3B12 3B11 3B11-N 3C12 M14D3 |
RBD | Recognizes at least 3 distinct neutralizing epitope groups (including residues L506, T512, Y540, R542, P547); blocks RBD-hDPP4 receptor binding; inhibits S-expressing pseudotyped MERS-CoV attaching to target cells; neutralizes pseudotyped and live MERS-CoV infection with different potency | 3B11-N therapeutically protects rhesus monkeys from MERS-CoV (Jordan-N3) infection, reducing lung pathology | No | [76,77] |
| m336 m337 m338 |
RBD | Recognizes neutralizing epitopes of RBD overlapping with the DPP4-binding site, including residues F506, D510, W535, D539, Y540, R542, W553 (for m336); strongly blocks RBD-hDPP4 receptor binding; potently inhibits pseudotyped and live MERS-CoV (EMC2012) infection | m336 protects hDPP4-Tg mice and rabbits from MERS-CoV infection | Yes, RBD/mAb-Fab complex No No |
[61,78–80] |
| hMS-1 | RBD | Recognizes neutralizing epitopes at RBD residues 510, 511 and 553; blocks RBD-hDPP4 receptor binding; neutralizes infection of pseudotyped MERS-CoV expressing S protein of at least 8 strains and live MERS-CoV (EMC2012) | Protects hDPP4-Tg mice from lethal MERS-CoV (EMC2012) infection | No | [81] |
| 4C2 h | RBD | Blocks RBD-hDPP4 receptor binding; neutralizes pseudotyped and live MERS-CoV infection | Protects Ad5/hDPP4-transduced mice from MERS-CoV (EMC2012) infection | No | [71] |
N/A: not applicable. Fab: antigen-binding fragment of mAb; hDPP4: human dipeptidyl peptidase 4; hDPP4-Tg mice: human DPP4-transgenic mice; HuDPP4 mice: humanized DPP4 mice; mAb: monoclonal antibody; MERS-CoV: Middle East respiratory syndrome coronavirus; RBD: receptor-binding domain; S1-NTD: S1 subunit N-terminal domain.
Table 2.
Summary of MERS-CoV S protein-targeting peptides and other therapeutic agentsa.
| Therapeutic name | Target region | Mechanism of inhibition of virus infection | In vivo protection | Crystal structures available | Ref. |
|---|---|---|---|---|---|
| Anti-DPP4 agents targeting RBD | |||||
| Anti-DPP4 mAbs 2F9, 1F7, YS110 |
RBD-binding region in DPP4 | Blocks RBD binding to hDPP4, thus preventing virus entry into target cells and inhibiting MERS-CoV infection | Not reported | N/A | [82] |
| DPP4 antagonist | RBD-binding region in DPP4 | Competes with RBD binding to hDPP4, inhibiting MERS-CoV infection | Not reported | N/A | [83] |
| Peptides targeting S2-HR1 | |||||
| HR2L HR2P HR2P-M1 HR2P-M2 |
S2-HR1 | Blocks 6-HB formation; inhibits MERS-CoV S-mediated cell-cell fusion at lower micromolar; inhibits pseudotyped and live MERS-CoV infection | HR2P-M2 protects Ad5/hDPP4-transduced mice and hDPP4-Tg mice from challenge of MERS-CoV (EMC2012) with or without mutations | Yes, HR1/HR2-6-HB complex | [59,84,85] |
| P1 | S2-HR1 | Inhibits pseudotyped MERS-CoV infection | Not reported | Yes, HR1/HR2-6-HB complex | [50] |
| Others affecting the function of MERS-CoV S protein | |||||
| Cathepsin inhibitors | Cathepsin | Blocks MERS-CoV S-mediated cell entry and virus-cell membrane fusion | Not reported | N/A | [66,86] |
| TMPRSS2 inhibitors | TMPRSS2 | Blocks MERS-CoV S-mediated cell entry and virus-cell membrane fusion | Not reported | N/A | [65] |
| Furin inhibitors | Furin | Blocks MERS-CoV S-mediated cell entry and virus infection | Not reported | N/A | [68] |
| siRNA | Furin | Silences furin activity, thus decreasing MERS-CoV S-mediated cell entry | Not reported | N/A | [68] |
| IFITM proteins | S? | Blocks MERS-CoV S-mediated cell entry potentially through mechanisms other than endosomal cholesterol accumulation | Not reported | N/A | [87] |
N/A: not applicable. DPP4: dipeptidyl peptidase 4; hDPP4-Tg mice: human DPP4-transgenic mice; 6-HB: 6-helix bundle; HR1 and HR2: heptad repeat region 1 and 2; MERS-CoV: Middle East respiratory syndrome coronavirus; RBD: receptor-binding domain; S: spike protein.
3.1. MERS-CoV S1-NTD or S1 outside RBD as therapeutic target
MERS-CoV S1-NTD and S1 outside the RBD may serve as potential targets to develop therapeutic countermeasures against MERS-CoV infection. A few neutralizing mAbs are identified to target these regions [69,72]. For example, a mouse mAb G2 that recognizes neutralizing epitopes in S1 outside the RBD may neutralize pseudotyped MERS-CoV infection [69]. In addition, human mAb LCA60, which recognizes a neutralizing epitope at residue V33 of S1-NTD, can also neutralize infection of pseudotyped MERS-CoV [72]. The neutralizing mAbs specific for S1-NTD or S1 outside the RBD could be used to supplement RBD-specific mAbs to increase their anti-MERS-CoV activity.
3.2. MERS-CoV RBD as therapeutic target
The RBD is a major target for anti-MERS-CoV therapeutics. Most MERS-CoV neutralizing antibodies target the RBD, and RBD-specific mAbs have more potent neutralizing activity than those targeting the S1 region outside RBD or the S2 region (Table 1), suggesting that MERS-CoV RBD could be served as a main neutralizing target for developing antibody-based therapeutics.
3.2.1. MERS-CoV RBD-targeting mouse neutralizing mAbs
Several mouse-derived, RBD-targeting MERS-CoV neutralizing mAbs have been generated from hybridomas of mice immunized with MERS-CoV S-encoding DNA and S1 or RBD proteins, and most of them maintain MERS-CoV neutralizing ability by blocking RBD-DPP4 receptor binding [69–71]. These mouse neutralizing antibodies may inhibit DPP4 binding at, or near, the key RBD residues D510, R511, W535, E536, D539, Y540, R542, and W553 (Figure 4(a), Table 1), and potently neutralize infection of divergent pseudotyped or live MERS-CoV [69,71].
Figure 4.
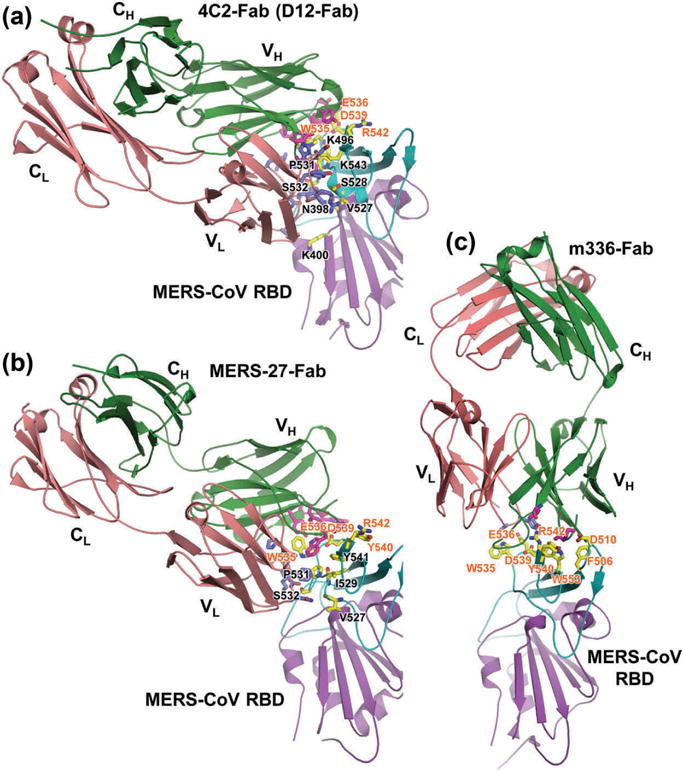
Structural basis of MERS-CoV infection inhibited by RBD-specific neutralizing antibodies [61,69,71,74]. (a) Crystal structures of MERS-CoV RBD in complex with mouse neutralizing mAb 4C2-Fab (PDB ID: 5DO2) or D12-Fab (PDB ID: 4ZPT). Crystal structures of RBD in complex with human neutralizing mAbs MERS-27-Fab (PDB ID: 4ZS6) (b) and m336-Fab (PDB ID: 4XAK) (c). MERS-CoV RBD core structure is colored purple, and RBM is in cyan. The mAb-Fab light (L) and heavy (H) chains are in red and green, respectively. VH, CH, VL, and CL indicate variable heavy, constant heavy, variable light, and constant light chains, respectively. Contacting residues at the RBD-binding interface in Fab-VL and VH chains are shown as blue and magenta sticks, respectively, and those in RBM are shown as yellow sticks. Contacting residues in the RBM involved in both human hDPP4-binding and Fab-binding are labeled in red, and the selected RBD residues at the Fab-binding interface are in black. Full color available online.
It is shown that Mersmab1, a conformation-dependent neutralizing mAb, efficiently blocked MERS-CoV RBD binding to its receptor DPP4 in soluble and cell-associated forms, and that the binding epitopes were critical at residues D510, R511, and W553 of RBD. Mersmab1 also inhibited S-mediated pseudotyped MERS-CoV entry into hDPP4-expressing cells and potently neutralized pseudotyped and live MERS-CoV infection [70]. It is also revealed that mouse mAbs 4C2 and 2E6, which neutralized MERS-CoV infection with high efficiency, interfered with RBD/DPP4 interactions by competing with each other for RBD binding [71]. In addition to their competitive effects, mouse mAbs capable of recognizing various RBD neutralizing epitopes may have synergistic effects on neutralizing MERS-CoV infection. For example, F11 and D12 mAbs, which recognize neutralizing epitopes at, or near, residues 509, or W535 and E536, respectively, the opposite sides of RBD, have different profiles in neutralizing a panel of eight pseudotyped MERS-CoVs- bearing S protein. While F11 was unable to neutralize pseudotyped MERS-CoV expressing S protein of Bisha1 or England1 strain containing D509G mutation, D12 could neutralize both pseudotyped MERS-CoVs irrespective of the mutation [69]. Nevertheless, the protective and therapeutic abilities of such mouse neutralizing mAbs, if not humanized, have not been evaluated in appropriate animal models.
Notably, crystal structures of two mouse neutralizing mAbs, 4C2 and D12, and MERS-CoV RBD-binding complexes are available, in which neutralizing epitopes are identified in the RBD that are critical for mAb binding [69,71] (Figure 4(a), Table 1). The characterized RBD-mAb crystal structures and the identified neutralizing epitopes will provide useful guidance for humanizing MERS-CoV mAbs, based on which to develop effective mAb- based anti-MERS-CoV therapeutic agents.
3.2.2. MERS-CoV RBD-targeting human neutralizing mAbs
Human neutralizing mAbs targeting MERS-CoV RBD have been extensively studied. These mAbs can be generated using screening of B cells derived from convalescent patients, nonimmune human antibody phage display libraries, or humanized mouse mAbs [71–73,75–78]. By competing with MERS-CoV RBD residues for hDPP4 binding, the RBD-targeting human mAbs may efficiently block RBD binding to the DPP4 receptor and subsequent virus entry into target cells, thus inhibiting MERS-CoV infection (Figure 3(b)). Several key residues critical for mAb binding, including F/L506, D510, T512, W535, E536, D539, Y540, R542, W553, and E565, have been identified in the RBD of MERS-CoV (Figure 4(b,c), Table 1).
LCA60 is the first human neutralizing mAb isolated from a MERS-CoV-infected individual. In addition to binding MERS-CoV S1-NTD, this mAb can also strongly bind RBD at residues T489, K493, E565, and E536, and thus interfered with the binding of RBD to viral cellular receptor DPP4, leading to potent neutralization of infection of three MERS-CoV strains isolated in 2012, including EMC2012, London1/2012, and Jordan-N3/2012 [72]. Likewise, MERS-4 and MERS-27 mAbs inhibited infection of pseudotyped and live MERS-CoV by blocking RBD-DPP4 interaction at the cell surface and preventing S-DPP4-mediated syncytia formation [73]. By inhibiting MERS-CoV entry into susceptible cells, REGN3048 and/or REGN3051 mAbs efficiently neutralized infectivity of live MERS-CoV EMC2102 strain and pseudoviruses expressing S protein of different strains with mutations at A431P, S457G, S460 F, A482 V, L506 F, D509G, and V534A, respectively, of the RBD [75]. Similar to mouse neutralizing mAbs, some human neutralizing mAbs, such as MERS-4 and MERS-27, also demonstrate synergistic effects in preventing MERS-CoV infection [73]. In such cases, mAbs can still neutralize naturally occurring or escape mutant MERS-CoVs generated from different antibody epitope groups although they might not neutralize those from the same epitope group [76].
Crystal structures of mAb-Fab/MERS-CoV RBD complexes are available for two human neutralizing mAbs [61,74]. Analyses of MERS-27-Fab/RBD complex revealed two critical residues at W535 and D539 positions of RBD that are important for RBD recognition by this mAb and binding with hDPP4 receptor (Figure 4(b)). By disrupting protein–protein and protein–carbohydrate interactions between RBD and hDPP4, MERS-27 effectively inhibited S-mediated pseudotyped MERS-CoV entry and infection [74]. Interestingly, as noted in the crystal structure of m336-Fab/RBD complex, the epitopes of mAb m336 are mapped to the RBD residues that overlap with the hDPP4-binding site (Figure 4(c)) [61], indicating that hDPP4 and m336 recognize identical RBD epitopes, thus explaining the potent neutralizing activity conferred by this human neutralizing mAb.
Currently, several RBD-specific human neutralizing mAbs have been evaluated for therapeutic effects in MERS-CoV- infected animal models, including Ad5/hDPP4-transduced mice, humanized DPP4 (HuDPP4) mice, and hDPP4-transgenic (hDPP4-Tg) mice, as well as rabbits and rhesus monkeys, demonstrating their protective ability against MERS-CoV infection [71,72,75,77,79–81]. For example, LCA60 mAb can prophylactically and therapeutically protect Ad5/hDPP4-transduced mice from infection of two MERS-CoV strains, EMC2012 and London1. It also protected INF-α/β receptor-deficient (IFNARKO) mice from challenge by these viruses [72]. In addition, REGN3048 and REGN3051 mAbs blocked MERS-CoV infection and disease in HuDPP4 mice, protecting them against virus challenge [75]. Furthermore, it is recently shown that m336 mAb reduced viral RNAs in rabbits infected with MERS-CoV post-treatment and protected hDPP4-Tg mice before and after MERS-CoV infection [79,80], and that 3B11-N mAb reduced lung pathology in rhesus monkeys infected with MERS-CoV before treatment [77].
3.3. Receptor DPP4-based anti-MERS-CoV therapeutics
DPP4 is an identified receptor for MERS-CoV. DPP4-targeting therapeutic agents, including antibodies specific to DPP4 and DPP4 antagonist, can block the binding or interaction between MERS-CoV RBD and DPP4, and thus inhibit MERS-CoV infection (Table 2). Utilizing the aforementioned mechanisms, anti-DPP4 (CD26) antibodies 2F9,1F7, and YS110 prevent MERS-CoV entry into susceptible cells, significantly blocking virus infection. The epitopes of these anti-DPP4 mAbs appear to be mapped to residue 358 or covering a region at residues 248–358 of S1 [82]. In addition, DPP4-binding protein adenosine deaminase (ADA) competes with MERS-CoV RBD binding to DPP4, especially at crucial residues Q286 and L294, determining its role as a naturally occurring antagonist of MERS-CoV infection [83]. These identified anti-MERS-CoV agents can be utilized as alternatives to neutralizing mAbs in preventing MERS-CoV infection.
3.4. MERS-CoV HR1/HR2 in S2 as therapeutic target
Like SARS-CoV, the HR1 domain in S2 subunit of MERS-CoV S protein is an important target for developing fusion inhibitors against MERS-CoV (Table 2) [50,59,60,88]. As noted, peptides derived from the S2 subunit HR2 domain exhibited good anti- MERS-CoV activity, while those from the HR1 had low, to no, inhibitory activity against MERS-CoV infection, possibly because that the HR1 peptides cannot form a soluble and stable trimer, but have tendency to aggregate in the physiological solution or PBS [50,59,60,88]. Indeed, it is indicated that HR2-derived peptide HR2P potently inhibited MERS-CoV replication and S-mediated cell-cell fusion at 50% inhibitory concentration (IC50) of ~0.6 and 0.8 μM, respectively, while HR1-derived peptides, including HR1P, HR1L, and HR1M, had no inhibitory activity at all, even at high concentrations [59].
Further studies have demonstrated that antiviral activity of HR2-derived peptides correlates with the peptide length, in which longer sequences have better antiviral activity than those with shorter sequences. This may be due to the fact that longer HR2 peptides tend to form more stable 6-HB coiled-coil structure with HR1 peptides [59]. For example, the 36-mer HR2P and 45-mer HR2L peptides maintained strong inhibitory activity against MERS-CoV S-mediated cell-cell fusion at IC50 of 0.8 and 0.5 μM, respectively, while a 19-mer HR2S short peptide exhibited no anti-MERS-CoV activity [59].
Previous studies on HIV-1 have indicated that the stability, solubility, and antiviral activity of anti-HIV peptides can be improved by introducing charged residues to peptide C34 of the gp41 HR2 [89]. Similarly, site-mutating residues, including those forming intramolecular salt-bridges, were introduced into the HR2P peptides without blocking interactions between the HR1 and HR2 of MERS-CoV S2. It is shown that introduction of: (1) T1263E and L1267R mutations to HR2P-M1 peptide, and (2) T1263E, L1267K, S1268K, Q1270E, Q1271E, A1275K, and N1277E mutations to HR2P-M2 peptide significantly increased the peptides’ stability and water solubility (69-fold for HR2P-M1 and 1,786-fold for HR2P-M2) and enhanced their inhibitory activity on MERS-CoV S-mediated cell-cell fusion (9% for HR2P-M1 and 41% for HR2P-M2) [59]. In particular, the HR2P-M2 peptide interacted with the HR1 peptide to form a stable α-helical complex, blocking 6-HB formation between the HR1 and HR2 of MERS-CoV S2 and subsequent membrane fusion (Figure 3(b)), thus potently inhibiting infection of S protein-expressing pseudotyped MERS-CoV with or without mutation at residue Q1020 of HR1 [59]. These studies confirm the possibility to further improve the stability, solubility, and antiviral potency of the HR2 peptide HR2P-M2.
The in vivo protective efficacy of MERS-CoV HR1-targeting peptide was evaluated in Ad5/hDPP4-transduced mice [90] and hDPP4-Tg mice [91]. Intranasal (i.n.) administration of HR2P-M2 peptide before viral challenge effectively protected the challenged mice from infection of MERS-CoV with or without HR1-Q1020 mutation, as evidenced by the reduced viral loads in lung tissues [84], and the decreased mortality of mice challenged with lethal dose of MERS-CoV [85,91]. Protection could be enhanced by combining this peptide with interferon β, a cytokine that potently inhibits MERS-CoV infection and virus clearance, both before and after virus challenge [84]. These studies suggest that the HR2P-M2 peptide in a nasal spray formulation could be applied to protect high-risk populations, such as family members of MERS patients, healthcare workers, and others having close contact with MERS patients, or in combination with other antiviral agents for treatment of MERS patients [92,93].
3.5. Other targets related to function of MERS-CoV S protein
As discussed above, the S protein of MERS-CoV must be cleaved by some cellular proteases (e.g. TMPRSS2) into S1 and S2 subunits with the functions to bind the receptor and mediate membrane fusion, respectively [65,66]. Therefore, the related cellular proteases can serve as targets for developing inhibitors of S protein-mediated viral fusion and entry into the target cells (Table 2). Since endosomal cathepsins (B/L) and transmembrane serine protease TMPRSS2 can activate MERS-CoV S-mediated virus-cell entry and uptake, treatment of cells with cathepsin (B/L) inhibitors, such as MDL28170 and teicoplanin, or TMPRSS2 inhibitor camostat mesylate can block MERS-CoV entry into target cells [65,66,86]. It is demonstrated that furin, a ubiquitously expressed protease, plays a key role in protease-activated MERS-CoV S-based fusion. Thus, treatment of MERS-CoV-permissive or DPP4-expressing cells with furin inhibitor (dec-RVKR-CMK) inhibits MERS-CoV S-mediated entry and virus infection in a dose-dependent manner [68]. Moreover, siRNA silencing of furin activity decreases MERS-CoV S-mediated entry. Accordingly, blockage of furin cleavage at the S cleavage sites significantly reduces virus infection. Different from the proteases processing S protein at the stage of virus uptake, proprotein convertases utilize S protein as a substrate and their processing is dispensable for the activation of S protein. Therefore, blockade of this protease does not affect S protein-driven cell-cell and virus-cell fusion and MERS-CoV infectivity, although it can reduce the processing of S protein in infected cells. It is therefore advisable that host cell protease-targeting anti-MERS-CoV agents should be focused on enzymes processing S protein in the virus uptake stage [94].
Other regions of MERS-CoV S protein, if identified, can serve as supplemental targets of anti-MERS-CoV therapeutics. For instance, mouse mAb G4 is demonstrated to bind MERS-CoV S2 subunit, neutralizing infection of pseudotyped MERS-CoV bearing S protein with low neutralizing activity (Table 1) [69], but its specific epitopes have not been clearly defined. Studies have also found that interferon-induced transmembrane proteins (IFITMs) may inhibit entry of S-mediated MERS-CoV into IFITM-transduced 293T cells (Table 2). However, its inhibition activity to MERS-CoV is less efficient than that observed in other human coronaviruses, such as 229E-CoV and NL63-CoV [87].
4. Conclusion
The continual increase of MERS cases, coupled with the continuous MERS outbreak resulting from zoonotic sources and possible human-to-human transmission of MERS-CoV, highlights the importance of developing effective antiviral agents to control MERS. As noted above, the structure and function of MERS-CoV S protein in virus entry and virus-cell membrane fusion effectively determine the crucial role of S protein as an important therapeutic target. Indeed, a variety of neutralizing mAbs and therapeutic agents has already been developed based on the viral S protein, and most of them have demonstrated protective efficacy against MERS-CoV infection, although none is currently approved for application in humans. The summary of available preclinical therapeutics will guide further development of effective anti-MERS-CoV therapeutic agents for human use.
5. Expert opinion
MERS-CoV S protein is a key target for developing therapeutics against MERS-CoV infection. Since the emergence of MERS, a number of S-targeting anti-MERS-CoV therapeutic agents, including mouse and human neutralizing mAbs and anti- MERS-CoV peptides, have been identified with in vitro efficacy in cell culture and/or in vivo efficacy in animal models, providing feasibility for further development. In addition, the available crystal structures of mAb/RBD complexes and HR1/HR2 6-HB complex will be applicable to elucidate the mechanisms of action of S-targeting anti-MERS-CoV neutralizing mAbs and peptides, thus being helpful for engineering new neutralizing antibodies and designing novel peptides with improved protective efficacy against MERS-CoV infection.
Notwithstanding the high potency of mouse neutralizing mAbs in in vitro testing, they, if not humanized, will not be appropriate for direct use in humans due to the potential risks of human-anti mouse antibody responses and other side effects [95]. Nevertheless, the neutralizing epitopes identified from mouse neutralizing mAbs will guide the design of humanized MERS-CoV mAbs and development of effective antiviral therapeutics and vaccines against MERS-CoV. Remarkably, human neutralizing mAbs have shown efficacy to protect against MERS-CoV infection in several animal models, demonstrating high potential for further development as effective prophylactic and therapeutic agents for human use. Since intravenous (i.v.) or intraperitoneal (i.p.) injection of MERS-CoV S-RBD-targeting humanized and human neutralizing mAbs conferred complete protection of animals from MERS-CoV infection without causing side effects and pathology [80,81], it is thus suggested that these routes can be selected to deliver anti-MERS-CoV-S mAbs to MERS-CoV-infected patients. Compared with human neutralizing mAbs targeting other regions of MERS-CoV S protein, those targeting the RBD are more potent, and could recognize key residues that are responsible for DPP4 binding. It should be noted that change of one or several of these critical residues in RBD may lead to the generation of escape mutant virus strains, in which neutralizing mAbs targeting the original residues in RBD will reduce or lose neutralizing activity against the new strains. This phenomenon has further elucidated the need for combinational application of two or more potent human neutralizing mAbs that target different neutralizing epitopes in RBD or other functional regions in MERS-CoV S protein, or have different mechanisms of action, in order to improve their antiviral activity against divergent virus strains, including those escape mutant strains. Attention should also be paid to other challenges of developing therapeutic mAbs, such as poor tissue penetration (resulting from large molecular size), high production cost, and inadequate pharmacokinetics [96–98]. These challenges will make it necessary to use antibody engineering technologies to generate MERS-CoV S-targeting antibody fragments with smaller molecule weight, reduced production cost, and good tissue penetration, but maintaining sufficient protective efficacy.
Similar to anti-HIV peptides [89], the stability, solubility, and antiviral activity of MERS-CoV S-specific peptides can be improved by introducing intramolecular salt-bridges or increasing peptide length. Therefore, further optimization of the HR2-derived peptides is expected to produce next-generation anti-MERS-CoV peptides with improved inhibitory efficacy. As an alternative and promising approach, combining the HRI-targeting peptide inhibitors with the RBD-specific neutralizing mAbs may result in synergistic antiviral effect against divergent MERS-CoV strains, including those resistant to S-targeting neutralizing mAbs and peptides.
In view of the key role of MERS-CoV S protein in virus infection and the ability of this protein to serve as an important target, it is expected that more effective and safer MERS-CoV S-based antiviral therapeutics can be developed and evaluated in appropriate animal models, moving them forward to human clinical trials. Importantly, the strategies applied to the development of anti-MERS-CoV therapeutics, as detailed in this review, can be used for the development of antiviral agents against future emerging pathogenic coronaviruses and other life-threatening enveloped viruses with class I membrane fusion proteins, such as Ebola and influenza viruses.
Article highlights.
MERS-CoV binds dipeptidyl peptidase 4 (DPP4) via receptor-binding domain (RBD) in spike (S) protein S1 subunit and then mediates virus entry into target cells via S2 subunit. Therefore, S protein plays pivotal roles in MERS-CoV infection and serves as an important therapeutic target.
Specific therapeutic targets in S protein include N-terminal domain and RBD in S1, and heptad repeat 1 (HR1) and 2 (HR2) in S2. S1-RBD and S2-HR1 are major targets for developing anti-MERS-CoV therapeutic monoclonal antibodies (mAbs) and peptidic fusion inhibitors, respectively.
RBD-targeting neutralizing mAbs block receptor binding at key residues, leading to inhibition of MERS-CoV infection. Several RBD-specific human neutralizing mAbs have demonstrated therapeutic efficacy against MERS-CoV infection in Ad5/hDPP4 transduced, humanized DPP4 (HuDPP4), or hDPP4-transgenic (hDPP4-Tg) mice, rabbits, and rhesus monkeys.
HR2-derived, HR1-targeting peptides inhibit MERS-CoV infection by blocking HR1/HR2 to form 6-helix bundle structure, and prevent MERS-CoV challenge in Ad5/hDPP4-transduced or hDPP4-Tg mice.
There is a need to combine mAbs recognizing different neutralizing epitopes to improve their efficacy against escape mutant MERS-CoV strains, and engineer them with reduced cost and good tissue penetration. It is feasible to optimize HR1-targeting peptides with improved inhibitory efficacy, and combine them with neutralizing mAbs to enhance anti-MERS-CoV therapeutic efficacy.
This box summarizes key points contained in the article.
Acknowledgments
Funding
This paper was funded by NIH grants (R01AI089728, R01AI098775, R01AI110700, R21AI109094) and an intramural fund from the New York Blood Center (NYB000348).
Footnotes
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1•.Zaki AM, Van BS, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. This is a paper describing the first isolation of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. [DOI] [PubMed] [Google Scholar]
- 2.Bermingham A, Chand MA, Brown CS, et al. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 2012;17(40):20290. [PubMed] [Google Scholar]
- 3.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong NS, Zheng BJ, Li YM, et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362(9393):1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guery B, Poissy J, El ML, et al. Clinical features and viral diagnosis of two cases of infection with Middle East respiratory syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381(9885):2265–2272. doi: 10.1016/S0140-6736(13)60982-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majumder MS, Rivers C, Lofgren E, et al. Estimation of MERS-coronavirus reproductive number and case fatality rate for the Spring 2014 Saudi Arabia outbreak: insights from publicly available data. Plos Curr. 2014;6 doi: 10.1371/currents.outbreaks.98d2f8f3382d84f390736cd5f5fe133c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten C, Muth D, Corman VM, et al. An observational, laboratory-based study of outbreaks of Middle East respiratory syndrome coronavirus in Jeddah and Riyadh, Kingdom of Saudi Arabia, 2014. Clin Infect Dis. 2015;60(3):369–377. doi: 10.1093/cid/ciu812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386(9997):995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salamati P, Razavi SM. Be vigilant: New MERS-CoV outbreaks can occur in the Kingdom of Saudi Arabia. Travel Med Infect Dis. 2015;13(3):269–270. doi: 10.1016/j.tmaid.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kupferschmidt K. Emerging diseases. Soaring MERS cases in Saudi Arabia raise alarms. Science. 2014;344(6183):457–458. doi: 10.1126/science.344.6183.457. [DOI] [PubMed] [Google Scholar]
- 11.Choi JY. An outbreak of Middle East respiratory syndrome coronavirus infection in South Korea, 2015. Yonsei Med J. 2015;56(5):1174–1176. doi: 10.3349/ymj.2015.56.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SS, Wong NS. Probable transmission chains of Middle East respiratory syndrome coronavirus and the multiple generations of secondary infection in South Korea. Int J Infect Dis. 2015;38:65–67. doi: 10.1016/j.ijid.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ki M. MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol Health. 2015;2015(37):e2015033. doi: 10.4178/epih/e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizumoto K, Saitoh M, Chowell G, et al. Estimating the risk of Middle East respiratory syndrome (MERS) death during the course of the outbreak in the Republic of Korea, 2015. Int J Infect Dis. 2015;39:7–9. doi: 10.1016/j.ijid.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan A, Farooqui A, Guan Y, et al. Lessons to learn from MERS-CoV outbreak in South Korea. J Infect Dev Ctries. 2015;9(6):543–546. doi: 10.3855/jidc.7278. [DOI] [PubMed] [Google Scholar]
- 16.Jeon MH, Kim TH. Institutional preparedness to prevent future Middle East respiratory syndrome coronavirus-like outbreaks in Republic of Korea. Infect Chemother. 2016;48(2):75–80. doi: 10.3947/ic.2016.48.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho SY, Kang JM, Ha YE, et al. MERS-CoV outbreak following a single patient exposure in an emergency room in South Korea: an epidemiological outbreak study. Lancet. 2016;388(10048):994–1001. doi: 10.1016/S0140-6736(16)30623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Wong SK, Li F, et al. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol. 2006;80(9):4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemida MG, Elmoslemany A, Al-Hizab F, et al. Dromedary camels and the transmission of Middle East respiratory syndrome coronavirus (MERS-CoV) Transbound Emerg Dis. 2015 doi: 10.1111/tbed.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azhar EI, El-Kafrawy SA, Farraj SA, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370(26):2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 21.Memish ZA, Cotten M, Meyer B, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014;20(6):1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Liu C, Du L, et al. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J Virol. 2015;89(17):9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Qi J, Yuan Y, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16(3):328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Memish ZA, Mishra N, Olival KJ, et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19(11):1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ithete NL, Stoffberg S, Corman VM, et al. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis. 2013;19(10):1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23(8):468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munster VJ, Adney DR, Van DN, et al. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis) Sci Rep. 2016;6:21878. doi: 10.1038/srep21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Du L, Liu C, et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci USA. 2014;111(34):12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohd HA, Al-Tawfiq JA, Memish ZA. Middle East respiratory syndrome coronavirus (MERS-CoV) origin and animal reservoir. Virol J. 2016;13:87. doi: 10.1186/s12985-016-0544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reusken CB, Messadi L, Feyisa A, et al. Geographic distribution of MERS coronavirus among dromedary camels, Africa. Emerg Infect Dis. 2014;20(8):1370–1374. doi: 10.3201/eid2008.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hemida MG, Chu DK, Poon LL, et al. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis. 2014;20(7):1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu DK, Poon LL, Gomaa MM, et al. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis. 2014;20(6):1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adney DR, Van DN, Brown VR, et al. Replication and shedding of MERS-CoV in upper respiratory tract of inoculated dromedary camels. Emerg Infect Dis. 2014;20(12):1999–2005. doi: 10.3201/eid2012.141280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haagmans BL, Al Dhahiry SH, Reusken CB, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14(2):140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemida MG, Al-Naeem A, Perera RA, et al. Lack of Middle East respiratory syndrome coronavirus transmission from infected camels. Emerg Infect Dis. 2015;21(4):699–701. doi: 10.3201/eid2104.141949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan JF, Li KS, To KK, et al. Is the discovery of the novel human betacoronavirus 2c EMC/2012 (HCoV-EMC) the beginning of another SARS-like pandemic? J Infect. 2012;65(6):477–489. doi: 10.1016/j.jinf.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woo PC, Lau SK, Li KS, et al. Genetic relatedness of the novel human group C betacoronavirus to Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5. Emerg Microbes Infect. 2012;1(11):e35. doi: 10.1038/emi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woo PC, Wang M, Lau SK, et al. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81(4):1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van BS, De GM, Lauber C, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6):e00473–12. doi: 10.1128/mBio.00473-12. pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Almazan F, DeDiego ML, Sola I, et al. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. MBio. 2013;4(5):e00650–13. doi: 10.1128/mBio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scobey T, Yount BL, Sims AC, et al. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci USA. 2013;110(40):16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X, Chen X, Bian G, et al. Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J Gen Virol. 2014;95(Pt 3):614–626. doi: 10.1099/vir.0.059014-0. [DOI] [PubMed] [Google Scholar]
- 43.Durai P, Batool M, Shah M, et al. Middle East respiratory syndrome coronavirus: transmission, virology and therapeutic targeting to aid in outbreak control. Exp Mol Med. 2015;47:e181. doi: 10.1038/emm.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du L, He Y, Zhou Y, et al. The spike protein of SARS-CoV-a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Liu D, Shi W, et al. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. MBio. 2015;6:5. doi: 10.1128/mBio.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia S, Liu Q, Wang Q, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li F, Li W, Farzan M, et al. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 48.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu G, Hu Y, Wang Q, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500(7461):227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J, Lu G, Qi J, et al. Structure of the fusion core and inhibition of fusion by a heptad-repeat peptide derived from the S protein of MERS-CoV. J Virol. 2013;87:13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang N, Shi X, Jiang L, et al. Structure of MERS-CoV spike receptorbinding domain complexed with human receptor DPP4. Cell Res. 2013;23(8):986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y, Rajashankar KR, Yang Y, et al. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J Virol. 2013;87(19):10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54••.Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. This paper identifies dipeptidyl peptidase 4 (DPP4) as MERS-CoV’s receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang N, Jiang S, Du L. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev Vaccines. 2014;13(6):761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du L, Zhao G, Kou Z, et al. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol. 2013;87(17):9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barlan A, Zhao J, Sarkar MK, et al. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J Virol. 2014;88(9):4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van DN, Miazgowicz KL, Milne-Price S, et al. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J Virol. 2014;88(16):9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59••.Lu L, Liu Q, Zhu Y, et al. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun. 2014;5:3067. doi: 10.1038/ncomms4067. This paper describes formation of a 6-helix bundle core by HR1 and HR2 in MERS-CoV S2 and subsequent virus-cell membrane fusion, and discovery of a HR1-targeting anti-MERS-CoV HR2 peptidic fusion inhibitor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu S, Xiao G, Chen Y, et al. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363(9413):938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61••.Ying T, Prabakaran P, Du L, et al. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat Commun. 2015;6:8223. doi: 10.1038/ncomms9223. This paper describes the crystal structure of MERS-CoV RBD in complex with one of the RBD-targeting human neutralizing mAbs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu Y, Lou Z, Liu Y, et al. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem. 2004;279(47):49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu Y, Liu Y, Lou Z, et al. Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core. J Biol Chem. 2004;279(29):30514–30522. doi: 10.1074/jbc.M403760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng Q, Deng Y, Liu J, et al. Core structure of S2 from the human coronavirus NL63 spike glycoprotein. Biochemistry. 2006;45(51):15205–15215. doi: 10.1021/bi061686w. [DOI] [PubMed] [Google Scholar]
- 65.Gierer S, Bertram S, Kaup F, et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol. 2013;87(10):5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shirato K, Kawase M, Matsuyama S. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol. 2013;87(23):12552–12561. doi: 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qian Z, Dominguez SR, Holmes KV. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. Plos One. 2013;8(10):e76469. doi: 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci USA. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Shi L, Joyce MG, et al. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun. 2015;6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du L, Zhao G, Yang Y, et al. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol. 2014;88(12):7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y, Wan Y, Liu P, et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015;25(11):1237–1249. doi: 10.1038/cr.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72••.Corti D, Zhao J, Pedotti M, et al. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci USA. 2015;112(33):10473–10478. doi: 10.1073/pnas.1510199112. This paper describes the first human neutralizing mAb isolated from a MERS-CoV-infected individual, and demonstrates its in vivo protective efficacy against MERS-CoV infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Jiang L, Wang N, Zuo T, et al. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med. 2014;6(234):234ra59. doi: 10.1126/scitranslmed.3008140. This is one of the papers identifying MERS-CoV RBD-targeting human neutralizing mAbs against MERS-CoV infection. [DOI] [PubMed] [Google Scholar]
- 74.Yu X, Zhang S, Jiang L, et al. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci Rep. 2015;5:13133. doi: 10.1038/srep13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75•.Pascal KE, Coleman CM, Mujica AO, et al. Pre- and postexposure efficacy of fully human antibodies against spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci USA. 2015;112(28):8738–8743. doi: 10.1073/pnas.1510830112. This is one of the papers demonstrating in vivo therapeutic efficacy of MERS-CoV RBD-targeting human neutralizing mAbs against MERS-CoV infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76•.Tang XC, Agnihothram SS, Jiao Y, et al. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci USA. 2014;111(19):E2018–E2026. doi: 10.1073/pnas.1402074111. This is one of the papers identifying MERS-CoV RBD-targeting human neutralizing mAbs and elucidating their roles in virus adaptive evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson RF, Bagci U, Keith L, et al. 3B11-N, a monoclonal antibody against MERS-CoV, reduces lung pathology in rhesus monkeys following intratracheal inoculation of MERS-CoV Jordan-n3/2012. Virology. 2016;490:49–58. doi: 10.1016/j.virol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ying T, Du L, Ju TW, et al. Exceptionally potent neutralization of Middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol. 2014;88(14):7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Houser KV, Gretebeck L, Ying T, et al. Prophylaxis with a Middle East respiratory syndrome coronavirus (MERS-CoV)-specific human monoclonal antibody protects rabbits from MERS-CoV infection. J Infect Dis. 2016;213(10):1557–1561. doi: 10.1093/infdis/jiw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Agrawal AS, Ying T, Tao X, et al. Passive transfer of a germline-like neutralizing human monoclonal antibody protects transgenic mice against lethal Middle East respiratory syndrome coronavirus infection. Sci Rep. 2016;6:31629. doi: 10.1038/srep31629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu H, Sun S, Xiao H, et al. Single-dose treatment with a humanized neutralizing antibody affords full protection of a human transgenic mouse model from lethal Middle East respiratory syndrome (MERS)-coronavirus infection. Antiviral Res. 2016;132:141–148. doi: 10.1016/j.antiviral.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ohnuma K, Haagmans BL, Hatano R, et al. Inhibition of Middle East respiratory syndrome coronavirus Infection by anti-CD26 monoclonal antibody. J Virol. 2013;87(24):13892–13899. doi: 10.1128/JVI.02448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raj VS, Smits SL, Provacia LB, et al. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus. J Virol. 2014;88(3):1834–1838. doi: 10.1128/JVI.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84•.Channappanavar R, Lu L, Xia S, et al. Protective effect of intranasal regimens containing peptidic Middle East respiratory syndrome coronavirus fusion inhibitor against MERS-CoV infection. J Infect Dis. 2015;212(12):1894–1903. doi: 10.1093/infdis/jiv325. This paper provides evidence for in vivo therapeutic efficacy of MERS-CoV HR1-targeting peptidic fusion inhibitor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang S, Tao X, Xia S, et al. Intranasally administered peptidic viral fusion inhibitor protected hDPP4 transgenic mice from MERS-CoV infection. Lancet. 2015;386:S44. [Google Scholar]
- 86.Zhou N, Pan T, Zhang J, et al. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J Biol Chem. 2016;291(17):9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wrensch F, Winkler M, Pohlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014;6(9):3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bosch BJ, Martina BE, Van Der Zee R, et al. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc Natl Acad Sci USA. 2004;101(22):8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Otaka A, Nakamura M, Nameki D, et al. Remodeling of gp41-C34 peptide leads to highly effective inhibitors of the fusion of HIV-1 with target cells. Angew Chem Int Ed Engl. 2002;41(16):2937–2940. doi: 10.1002/1521-3773(20020816)41:16<2937::AID-ANIE2937>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 90.Zhao J, Li K, Wohlford-Lenane C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A. 2014;111(13):4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tao X, Garron T, Agrawal AS, et al. Characterization and demonstration of the value of a lethal mouse model of Middle East respiratory dyndrome coronavirus infection and disease. J Virol. 2015;90(1):57–67. doi: 10.1128/JVI.02009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu L, Xia S, Ying T, et al. Urgent development of effective therapeutic and prophylactic agents to control the emerging threat of Middle East respiratory syndrome (MERS) Emerg Microbes Infect. 2015;4(6):e37. doi: 10.1038/emi.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hafner S, Ojcius DM. MERS - A cautionary tale. Microbes Infect. 2015;17(8):542–544. doi: 10.1016/j.micinf.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gierer S, Muller MA, Heurich A, et al. Inhibition of proprotein convertases abrogates processing of the Middle Eastern respiratory syndrome coronavirus spike protein in infected cells but does not reduce viral infectivity. J Infect Dis. 2015;211(6):889–897. doi: 10.1093/infdis/jiu407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hansel TT, Kropshofer H, Singer T, et al. The safety and side effects of monoclonal antibodies. Nat Rev Drug Discov. 2010;9(4):325–338. doi: 10.1038/nrd3003. [DOI] [PubMed] [Google Scholar]
- 96.Chames P, Van RM, Weiss E, et al. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 2009;157(2):220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Foltz IN, Karow M, Wasserman SM. Evolution and emergence of therapeutic monoclonal antibodies: what cardiologists need to know. Circulation. 2013;127(22):2222–2230. doi: 10.1161/CIRCULATIONAHA.113.002033. [DOI] [PubMed] [Google Scholar]
- 98.Beck A, Wurch T, Bailly C, et al. Strategies and challenges for the next generation of therapeutic antibodies. Nat Rev Immunol. 2010;10(5):345–352. doi: 10.1038/nri2747. [DOI] [PubMed] [Google Scholar]


