Key Clinical Message
We reported a case of a t(2;14) balanced reciprocal translocation carrier mother that conceived by IVF accompanied by PGD/PGS using array‐CGH; however, de novo t(2;3) was detected in the prenatal diagnosis. A healthy baby was delivered, and careful observation is needed for PGD/PGS cases.
Keywords: Balanced translocation carrier, de novo balanced reciprocal translocation, infertility, preimplantation genetic diagnosis, recurrent implantation failure
Reciprocal translocations are common and detected in approximately 1:500 to 1:625 newborns 1, 2. Most cases can be expected to come from translocation families. In contrast, de novo balanced reciprocal translocations are rare, and investigations of these translocations may improve the understanding of chromosomal rearrangements. Until now, only a few studies of de novo balanced reciprocal translocations have been reported; however, the parents of these cases were cytogenetically normal.
Here, we report a very rare case of de novo apparently balanced reciprocal translocations in a fetus conceived from a balanced reciprocal translocation carrier mother.
Case Report
A 32‐year‐old nulli‐gravida and her husband, who was 34 years old, visited our medical center to seek treatment for primary infertility over the course of 2 years. The cause of infertility was unexplained. The patient underwent intrauterine insemination three times, in vitro fertilization‐embryo transfer (IVF‐ET) twice, and thawing ET twice; however, she did not conceive. Therefore, conventional cytogenetic analyses of the couples were performed on lymphocytes from phytohemagglutinin (PHA)‐stimulated peripheral blood cultures for the evaluation of the recurrent implantation failure. GTG‐banded chromosomes were obtained according to a standard protocol, and 20 metaphases were examined and karyotyped using the Cytovision system version 3.6 (Applied Imaging, UK). The chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature (ISCN) 2009. Karyotype analysis of the patient revealed 46,XX,t(2;14)(q35;q32.1).ish t(2;14)(tel14q+;tel14q‐) (Fig. 1), and the result for her husband was 46,XY. During 1 year, three cycles of IVF accompanied by preimplantation genetic diagnosis (PGD) and aneuploid screening (PGS) were performed using BAC‐array comparative genomic hybridization (BAC‐array CGH). For the first two cycles, a total of 39 embryos (blastomere biopsies from 29 cleavages and trophectoderm biopsies from 10 blastocysts) were analyzed using the P scanning human BAC‐array (Macrogen Inc., Korea). Only five euploid embryos were transferred;, however, the patient did not conceive. During December 2015, 20 oocytes were retrieved, and six blastocysts were cryopreserved using vitrification. In the subsequent cycle, some trophectoderm cells (approximately five to ten cells) were biopsied from six warmed blastocysts and the samples were analyzed with an Illumina BlueGnome 24Sure DNA microarray (BlueGnome LTD, Cambridge, UK) following the BlueGnome 24sure V3 protocol (Fig. 2) 3. Two euploid embryos were transferred, and a vital fetus was confirmed in the fifth gestational week. The integrated test for Down syndrome was abnormal (1:61), and amniocentesis was performed at 17 weeks of gestation. Cytogenetic analysis revealed a karyotype of 46,OO,t(2;3)(q31;q27). No genomic imbalance and no loss or increase in the dosage of genetic probes specific for chromosomes 2 and 3 were confirmed by a single‐nucleotide polymorphism (SNP)‐array (CytoScan® HD Arrays, Affymetrix, Santa Clara, CA) using cultured amniocytes (Fig 3). A level II ultrasound revealed no structural abnormalities. After genetic counseling, the parents decided to continue the pregnancy, and the pregnancy was well maintained. At 40 weeks of gestation, a healthy male baby was delivered with a body weight of 3280 g. Pediatric examination of the infant showed no phenotypic abnormalities.
Figure 1.
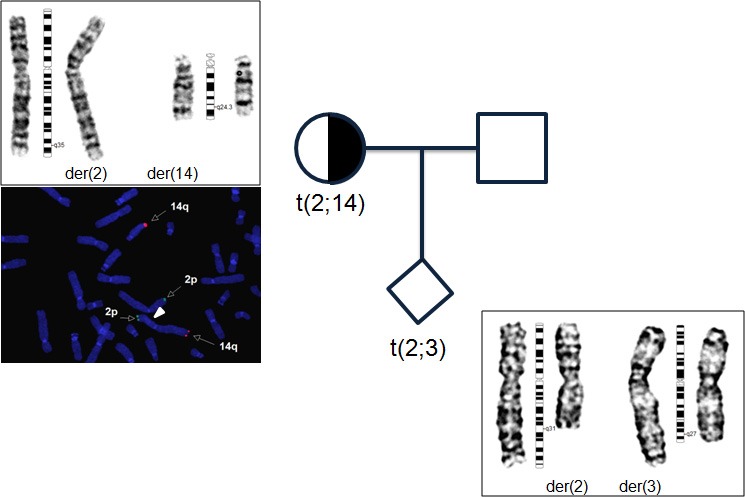
Pedigree chart and cytogenetic analysis. The left figure indicates the karyotype that shows 46,XX,t(2;14)(q35;q32.1) in the peripheral blood of the patient, and FISH analysis using a subtelomere 2p probe (Tel 2p: Vysis) labeled in green and a subtelomere 14q probe (Tel 14q: Vysis) labeled in orange. The right lower figure demonstrates the karyotype that shows 46,OO,t(2;3)(q31;q27) in the amniocytes.
Figure 2.
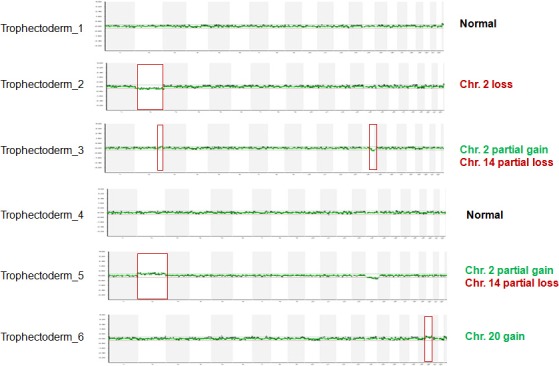
The PGD results. Embryos 1 and 4 were transferred.
Figure 3.
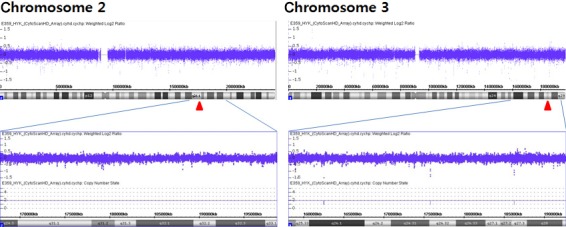
Partial results of SNP‐arrays of the cultured amniocytes. No significant genomic changes were identified in the regions of the chromosome break points.
Written informed consent was obtained from the couple, and this report was approved by the Institutional Review Board of CHA Gangnam Medical Center.
Discussion
Reciprocal translocation is the most frequent structural anomalies in humans and usually harmless; however, they are associated with an increased risk of unbalanced gametes and abnormal progeny 4. A few translocations are associated with a high risk of having an abnormal child, many translocations are associated with an intermediate level of risk, and some carriers have a low risk. In general, the carriers have high potential miscarriage rate. Stern et al. reported that chromosomal translocations were found in 3.2% of couples with recurrent implantation failure 5. In this case, chromosomal analysis was performed due to recurrent implantation failure, and the wife was confirmed to be a 2, 14 balanced reciprocal translocation carrier.
Preimplantation genetic diagnosis using array‐CGH was chosen. The genomic coverage of the 24sure+arrays has an approximate screening resolution of 0.5 Mb across the genome and 0.25 Mb in peri‐centromeric and subtelomeric regions, and it enables better detection and characterization of segmental chromosome imbalances in a single cell. Of the embryos diagnosed, 15.6% (7/45) were euploidy. Although this percentage was remarkably lower than the euploidy rate of other PGD cases with chromosomal abnormal carriers in our center (27.9%, 2 years of data from our center, data not published), it was consistent with literature 6.
Most of the balanced reciprocal translocations are inherited. De novo balanced reciprocal translocations are detected in approximately one in 800–1000 cases 7, 8. The mechanisms of the formation of de novo reciprocal translocations in humans have not yet been identified. The first step for determining the mechanisms underlying these translocations is to identify the break points. A study in prenatal diagnosis reported that chromosomes 3, 2, 7, 1, and 9 were involved more frequently in apparently balanced de novo rearrangement 8. In the present case, we confirmed the break points at 2q31 and 3q27. This finding coincides with the location of common fragile sites. Next, determining the parental origin of translocation is important for understanding the mechanisms underlying this translocation. A few studies regarding the parental origin of de novo balanced reciprocal translocations have been reported, and most of these cases were predominantly of paternal origin 9, 10, 11. In this case, the break points detected in the amniocytes were completely different from maternal regions, and we suggest that this de novo chromosomal rearrangement was of paternal origin.
A total of 40~100% of the reported cases had a genomic imbalance detectable by array comparative genomic hybridization (aCGH) or SNP‐array and is associated with serious congenital anomalies 6, 12. Even if there is no detectable imbalance, a de novo apparently balanced translocation has the possibility of an abnormal phenotype because of other mechanisms such as position effect 6. According to the literature, the reported risk of congenital anomalies ranged from 6.1 to 12.5% 7, 13, 14. A recent study with a long‐term follow‐up of children with prenatally diagnosed de novo apparently balanced translocation exhibited similar health, developmental, and behavioral outcomes to those of the normal population 14. In this current case, a high‐resolution array‐CGH of the trophectoderm and a SNP‐array of the amniocytes were performed, and a resolution of at least 1 Mb and 1 Kb was achieved with the array‐CGH and SNP‐array, respectively. Genomic imbalance or, loss or gain in the dosage of the genetic probes at the break points was not detected; moreover, no phenotypic abnormalities were manifested at birth. However, assessment of long‐term health, developmental, or behavioral outcomes is needed. Although this case has limitations, to the best of our knowledge, this is the first case of a de novo balanced reciprocal translocation conceived using PGS from a balanced reciprocal translocation carrier mother.
In conclusion, we have presented the prenatal diagnosis of a de novo apparently balanced reciprocal translocation. Our case represents the first case of a de novo balanced reciprocal translocation conceived using PGD from a balanced translocation carrier mother. We suggest close observation and long‐term follow‐up in all PGD/PGS cases.
Authorship
JWK: performed in data collection and wrote the manuscript. SHS: participated in analysis and interpretation the samples and revised the manuscript. WSL: collected the samples, interpreted the data, revised, and finally approved the article.
Conflict of Interest
The authors declare no conflict of interests.
Acknowledgments
This work was supported by Priority research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF‐2009‐0093821).
Clinical Case Reports 2017; 00(00): 000–000
The authors consider that the first two authors should be regarded as joint first authors.
References
- 1. Mackie Ogilvie, C. , and Scriven P. N.. 2002. Meiotic outcomes in reciprocal translocation carriers ascertained in 3‐day human embryos. Eur. J. Hum. Genet. 10:801–806. [DOI] [PubMed] [Google Scholar]
- 2. Oliver‐Bonet, M. , Navarro J., Carrera M., Egozcue J., and Benet J.. 2002. Aneuploid and unbalanced sperm in two translocation carriers: evaluation of the genetic risk. Mol. Hum. Reprod. 8:958–963. [DOI] [PubMed] [Google Scholar]
- 3. Liu, J. , Wang W., Wun X., Liu L., Jin H., Li M., et al. 2012. DNA microarray reveals that high proportions of human blastocysts from women of advanced maternal age are aneuploid and mosaic. Biol. Reprod. 87:148. [DOI] [PubMed] [Google Scholar]
- 4. Nussbaum, R. L. , McIness R. R., and Willard H. F.. 2007. Genetics in medicine, 7th ed. Thompson & Thompson, Philadelphia. [Google Scholar]
- 5. Stern, C. , Pertile M., Norris H., Hale L., and Baker H. W. 1999. Chromosome translocations in couples with in‐vitro fertilization implantation failure. Hum. Reprod. 14:2097–2101. [DOI] [PubMed] [Google Scholar]
- 6. Gardner, R. J. M. , Sutherland G. R., and Shaffer L. G.. 2003. Chromosome abnormalities and genetic counseling, 3rd ed Oxford University Press, New York. [Google Scholar]
- 7. Peng, H. H. , Chao A. S., Wang T. H., Chang Y. L., and Chang S. D.. 2006. Prenatally diagnosed balanced chromosome rearrangements: eight years' experience. J. Reprod. Med. 51:699–703. [PubMed] [Google Scholar]
- 8. Giardino, D. , Corti C., Ballarati L., Colombo D., Sala E., Villa N., et al. 2009. De novo balanced chromosome rearrangements in prenatal diagnosis. Prenat. Diagn. 29:257–265. [DOI] [PubMed] [Google Scholar]
- 9. Thomas, N. S. , Morris J. K., Baptista J., Ng B. L., Crolla J. A., and Jacobs P. A.. 2010. De novo apparently balanced translocations in man are predominantly paternal in origin and associated with a significant increase in paternal age. J. Med. Genet. 47:112–115. [DOI] [PubMed] [Google Scholar]
- 10. Grossmann, V. , Hockner M., Karmous‐Benailly H., Liang D., Puttinger R., Quadrelli R., et al. 2010. Parental origin of apparently balanced de novo complex chromosomal rearrangements investigated by microdissection, whole genome amplification, and microsatellite‐mediated haplotype analysis. Clin. Genet. 78:548–553. [DOI] [PubMed] [Google Scholar]
- 11. Hockner, M. , Spreiz A., Fruhmesser A., Tzschach A., Dufke A., Rittinger O., et al. 2012. Parental origin of de novo cytogenetically balanced reciprocal non‐Robertsonian translocations. Cytogenet. Genome. Res. 136:242–245. [DOI] [PubMed] [Google Scholar]
- 12. Baptista, J. , Mercer C., Prigmore E., Gribble S. M., Carter N. P., Maloney V., et al. 2008. Breakpoint mapping and array CGH in translocations: comparison of a phenotypically normal and an abnormal cohort. Am. J. Hum. Genet. 82:927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. De Warburton, D. 1991. novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am. J. Hum. Genet. 49:995–1013. [PMC free article] [PubMed] [Google Scholar]
- 14. Sinnerbrink, I. B. , Sherwan A., Meiser B., Halliday J., Amor D. J., Waters E., et al. 2013. Long‐term health and development of children diagnosed prenatally with a de novo apparently balanced chromosomal rearrangement. Prenat. Diagn. 33:831–838. [DOI] [PubMed] [Google Scholar]


