Abstract
Spinach is an important leafy vegetable enriched with multiple necessary nutrients. Here we report the draft genome sequence of spinach (Spinacia oleracea, 2n=12), which contains 25,495 protein-coding genes. The spinach genome is highly repetitive with 74.4% of its content in the form of transposable elements. No recent whole genome duplication events are observed in spinach. Genome syntenic analysis between spinach and sugar beet suggests substantial inter- and intra-chromosome rearrangements during the Caryophyllales genome evolution. Transcriptome sequencing of 120 cultivated and wild spinach accessions reveals more than 420 K variants. Our data suggests that S. turkestanica is likely the direct progenitor of cultivated spinach and spinach domestication has a weak bottleneck. We identify 93 domestication sweeps in the spinach genome, some of which are associated with important agronomic traits including bolting, flowering and leaf numbers. This study offers insights into spinach evolution and domestication and provides resources for spinach research and improvement.
Spinach is an economically important vegetable crop but previous genomic resources were of limited use for comparative and functional analyses. Here, Xu et al. present a high quality draft spinach genome and transcriptome data for multiple Spinacia accessions providing insight into Caryophyllales genome evolution.
Spinach (Spinacia oleracea L., 2n=2 × =12) is an important and nutritious green leafy vegetable and a rich source of carotenoids, folate, vitamin C, calcium and iron. It is commonly used as a salad, a cooked vegetable or as an ingredient in fresh or cooked meat and vegetable dishes1. Spinach is increasing in popularity and is cultivated in more than 60 countries with production increased nearly tenfold in the past 40 years (annual production of ∼2.5 million tonnes in 1974 and ∼23 million in 2013; http://faostat3.fao.org/).
Spinach is an annual or biennial plant, and is commonly considered as dioecious or occasionally monoecious2. It is native to central Asia and thought to have originated in Persia (Iran)1. The genus Spinacia contains two wild species, S. turkestanica Ilj. and S. tetrandra Stev., which are considered the probable ancestors of cultivated spinach3. Spinach has a relatively recent domestication history4. In addition to common horticultural traits (for example, high yields, fast growth, slow bolting and leaf type), disease resistance breeding has been the major focus of spinach improvement programmes, especially resistance to the downy mildew pathogen, Peronospora farinosa f.sp. spinaciae (Pfs)1,4. Several studies have also demonstrated that wide variations in important traits including carotenoid and folate concentrations1,5 and oxalate and nitrate contents6,7 exist in spinach germplasm, providing new breeding targets for spinach improvement. Nevertheless, genetic diversity among spinach germplasm remains largely unexplored at the molecular level with only limited studies spanning useful but relatively small collections of spinach accessions8,9,10,11. Furthermore, due to its minor crop status, few genomic resources have been developed, hindering the understanding and utilization of the extant germplasm resources.
Spinach is a member of the Amaranthaceae family, which is composed of about 180 genera and 2,500 species including important crops such as beets and quinoa, and represents the most species-rich lineage within Caryophyllales, the basal order of core eudicots12. Previously a draft spinach genome assembly was generated, which was mainly used to provide evidence to support the separation of Caryophyllales from rosids and asterids12. However, the assembly (498 Mb) represents only about half of the genome with an estimated size of 989 Mb (ref. 13) and contains many short assembled fragments (N50=19 kb), thus limiting its utility as a reference for comparative, evolutionary and functional genomic studies.
Here we report a high-quality genome assembly of a Chinese spinach cultivar, Sp75, in addition to transcriptome sequences of 120 cultivated and wild spinach accessions. These sequences provide insights into the structure and evolution of the spinach genome, the phylogeny and genetic diversity of spinach populations, genomic signatures underlying spinach domestication, and the underlying molecular basis of specific agronomically important traits. The spinach genome sequence represents a solid foundation for comparative genomic studies in Caryophyllales and eudicots, and together with the transcriptome variation data, provide valuable resources for facilitating spinach research and improvement.
Results
Spinach genome sequencing and assembly
An inbred spinach line, Sp75, was selected for reference genome sequencing using the whole genome shotgun approach. Illumina paired-end libraries with insert sizes of 150 bp, 200 bp, 300 bp, 500 bp and 1 kb, and mate-pair libraries with insert sizes of 3 kb, 10 kb and 15 kb were constructed and sequenced, which generated a total of 169 Gb high-quality cleaned sequences (Supplementary Table 1), representing approximately 168-fold coverage of the spinach genome that has an estimated size of 1,009 Mb based on k-mer analysis of the Illumina sequences (Supplementary Fig. 1) and 989 Mb based on flow cytometry13. De novo assembly of the Illumina sequences resulted in a draft genome of ∼870 Mb with an N50 scaffold length of ∼319.5 kb and the longest scaffold of 3.3 Mb (Table 1). To further improve the assembly, we constructed spinach genome maps using the BioNano Irys system14. A total of 808,135 molecules (>150 kb) with a total length of 214.9 Gb were collected, representing ∼213 × coverage of the spinach genome. De novo assembly of these molecules resulted in a total of 898 genome consensus maps with a total length of 1,087.6 Mb and individual map lengths ranging from 49.8 kb to 8.9 Mb. Reconstruction of the genome assembly using BioNano consensus maps resulted in super-scaffolds with a total length of ∼996 Mb, N50 of 919,290 bp, and the longest of 9.3 Mb (Supplementary Fig. 2). Furthermore, using a genetic map we constructed, we were able to anchor a total of 439 scaffolds covering 463.4 Mb (47%) of the assembled genome to the six linkage groups (Fig. 1, Supplementary Note 1 and Supplementary Figs 3–5).
Table 1. Summary of spinach genome assembly.
|
Contig |
Scaffold |
Super scaffold |
||||
|---|---|---|---|---|---|---|
| Size (bp) | Number | Size (bp) | Number | Size (bp) | Number | |
| N90 | 1,554 | 71,235 | 5,121 | 6,093 | 11,883 | 3,878 |
| N80 | 4,762 | 40,488 | 81,129 | 2,246 | 103,609 | 1,409 |
| N70 | 8,537 | 27,590 | 155,870 | 1,489 | 205,174 | 730 |
| N60 | 12,418 | 19,540 | 229,174 | 1,033 | 395,765 | 370 |
| N50 | 16,570 | 13,759 | 319,471 | 711 | 919,290 | 201 |
| N25 | 31,281 | 4,483 | 626,780 | 218 | 3,106,702 | 51 |
| Longest | 185,618 | 1 | 3,292,865 | 1 | 9,343,782 | 1 |
| Total | 830,856,911 | 215,350 | 869,796,885 | 78,264 | 996,306,834 | 77,702 |
Only contigs and scaffolds ≥500 bp were included in the genome assembly.
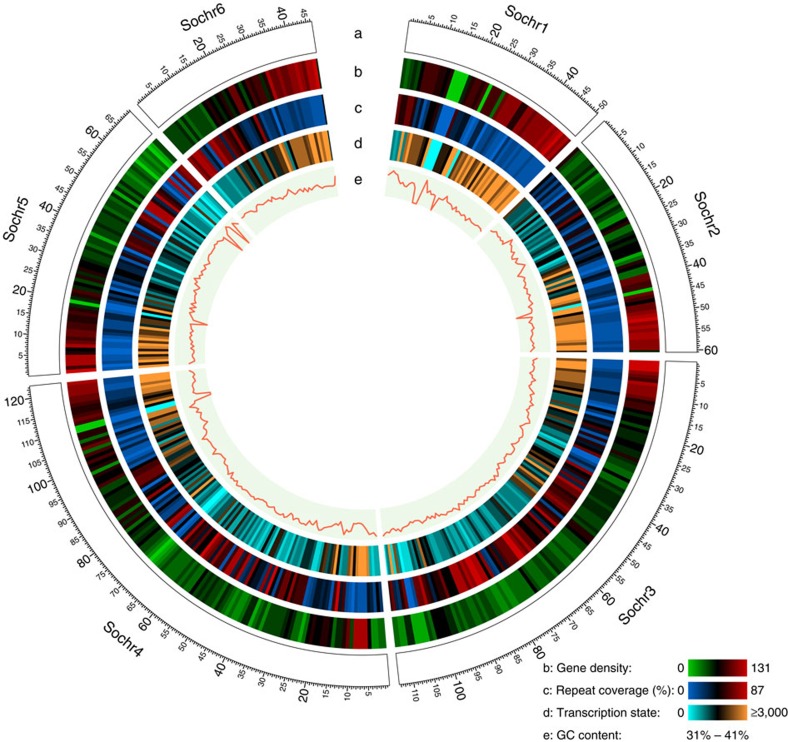
Figure 1. Spinach genome landscape.
(a) Ideogram of the six spinach pseudochromosomes (in Mb scale). (b) Gene density represented as number of genes per Mb. (c) Percentage of coverage of repeat sequences per Mb. (d) Transcription state. The transcription level was estimated by read counts per million mapped reads in 1-Mb windows. (e) GC content in 1-Mb windows. The six spinach pseudo-chromosomes represented 47% of the genome assembly. This figure was generated using Circos (http://circos.ca/).
The quality of the spinach genome assembly was first assessed using BUSCO15. The analysis revealed that 97.1% of the core eukaryotic genes were detected in the spinach genome, and 95.7% of them were completely covered. Furthermore, RNA-Seq data sets generated from the 107 S. oleracea samples were used to evaluate the quality of the assembled spinach genome. Overall, ∼95% of the RNA-Seq reads could be mapped to the assembled spinach genome (Supplementary Data 1). In summary, the high coverage of the core eukaryotic genes and the high mapping rate of RNA-Seq reads indicated the high quality of the assembled genome.
Transposable elements and gene models
The assembled spinach genome contains a total of ∼618 Mb (74.4%) of repeat sequences, higher than that in most eudicot genomes reported to date. Class I transposons are the major source of repeat sequences, covering 59.5% of the genome. The predominant subgroup in the class I transposon family is the long terminal repeat (LTR) retrotransposons, which accounts for 56.6% of the genome assembly (Supplementary Table 2). In contrast, class II or DNA transposons are much less prevalent (‘cut-and-paste' transposons, 6.0%; miniature inverted repeat transposable elements (MITEs), 2.9%; and helitron, 0.1%). Although LTR retrotransposons can be eliminated efficiently from the genome, upon proliferation or transposition burst, they can substantially increase the genome size in a short evolutionary time16. Sugar beet, another Caryophyllales species closely related to spinach, has a smaller genome size (760 Mb) and a lower content of repetitive sequence (42%) in its assembled genome12. We identified 976 and 498 full-length LTR retrotransposons in our spinach assembly and the sugar beet genome, respectively. We found that there was a more recent burst of LTR retrotransposons in spinach, which was estimated to occur at ∼1.5 million years ago (Mya), compared to that in sugar beet occurring at ∼2.7 Mya (Supplementary Fig. 6). This may partially explain the larger genome size and higher repeat content in spinach as compared to sugar beet. Transposon amplification has also been reported to contribute to the differences in genome sizes of other closely related species, such as melon and cucumber17.
A total of 25,495 protein-coding genes were predicted from the spinach genome, which is comparable to the number of genes predicted in sugar beet (26,923; RefBeet-1.2) (ref. 18). Among the predicted spinach protein-coding genes, 22,860 (89.7%) were supported by our RNA-Seq data. The coding sequences in spinach have an average length of 1,157 bp and the predicted genes have an average of 5.3 exons; both are similar to sugar beet (Supplementary Table 3). A total of 22,103 (86.7%) spinach proteins have homologues in at least one protein database including nr, TrEMBL, Swiss-Prot and TAIR10, 19,620 (77.0%) and 18,725 (73.5%) contain InterPro and Pfam domains, respectively, and 17,744 (69.6%) can be assigned with Gene Ontology (GO) terms (Supplementary Table 4). Furthermore, a total of 1,202 transcription factors were identified in the spinach genome, slightly more than sugar beet but less than non-Caryophyllales plant species (Supplementary Table 5). Moreover, the spinach genome encodes 892 protein kinases and 68% of them belong to the receptor-like kinase family. One receptor-like kinase group, the WAK family, was found to be expanded in basal eudicots spinach and sugar beet and monocot rice as compared to other analysed eudicot species (Supplementary Table 6).
Comparative genomics
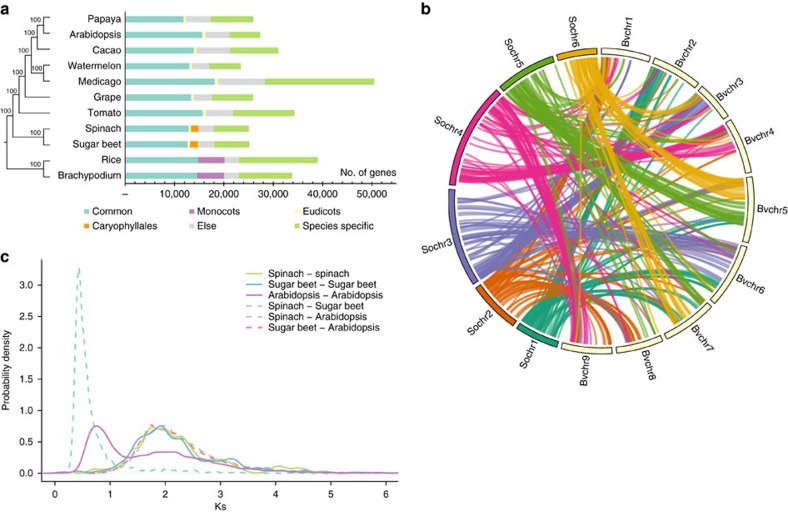
Comparison of gene content between spinach and other ten representative plant species, including sugar beet, tomato, grape, Medicago, watermelon, cacao, Arabidopsis, papaya, Brachypodium and rice (Supplementary Data 2), indicated that 17,968 (71.7%) spinach genes were present with homologues in at least one of the other ten species. A total of 1,520 (6.1%) spinach genes were Caryophyllales specific (unique to spinach and sugar beet), and 7,097 (28.3%) were spinach specific (having no homologues in the other ten species). A phylogenetic tree constructed using single-copy genes from all 11 species confirmed that spinach is a sister taxon of sugar beet, and supported that these two species constituted the clade which is the most basal in core eudicots12 (Fig. 2a). Despite that the spinach pseudo-chromosomes only represented 47% of the genome assembly, our analysis strongly indicated that the genomes of spinach and sugar beet shared many syntenic regions, whose pattern clearly suggested that substantial inter- and intra-chromosome rearrangements have occurred during the Caryophyllales genome evolution (Fig. 2b).
Figure 2. Comparative genomic analysis among spinach and other plant species.
(a) Phylogenetic relationship and gene clusters of 11 plant species. A maximum parsimony (MP) species tree (left) was constructed using protein sequences of the 2,047 single-copy genes. Bars (right) represent the number of genes in different categories for each species. Common: genes that are found in at least 10 of the 11 species. Monocots: genes that are only found in the two monocots, rice and Brachypodium; Eudicots: genes that are found in at least eight of nine eudicots but not in the two monocots; Caryophyllales: genes that are only found in spinach and sugar beet; Species-specific: genes with no homologues in other species. (b) Syntenic relationships between spinach and sugar beet genomes. (c) Ks distribution of homologous gene pairs in spinach, sugar beet and Arabidopsis. The probability density of Ks was estimated using the ‘density' function in R.
Whole genome duplication is prevalent in flowering plants and is an important driving force for genome evolution and gene neofunctionalization19. We identified homologous pairs in the syntenic genomic regions within and between spinach, sugar beet and Arabidopsis. The distribution of synonymous substitution rate (Ks) of the homologous pairs indicated that as expected an ancient whole genome triplication (the γ event), which is shared by core eudicots, had occurred in the evolutionary history of the spinach genome. However, no recent whole genome duplication event was found in spinach, nor in sugar beet12 (Fig. 2c). In addition, the divergence of spinach and sugar beet occurred approximately 38.4 Mya (95% confidence interval: 38.1–38.7 Mya). The common ancestor of spinach and sugar beet diverged from Arabidopsis soon after the ancient triplication event (γ) (Fig. 2c).
Disease resistance genes
Currently, resistance against downy mildew is the major focus of spinach breeding1. Recognition of pathogen effector is mainly mediated by plant disease resistance (R) genes, which encode nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins20. We identified 139 NBS-LRR genes in the spinach genome and classified them into six categories based on their Toll/Interleukin-1 receptor domains, coiled coil (CC) motifs and LRRs (Supplementary Data 3). The number of NBS-LRR genes in spinach is lower than that in most other plant species (Supplementary Fig. 7a). The two most abundant groups of NBS-LRR genes in spinach are CNL (coiled coil, NBS and LRR) and NL (NBS and LRR), representing 40 and 36% of the NBS-LRR genes, respectively. Comparison of NBS-LRR genes between spinach and other five representative plants (sugar beet, tomato, Arabidopsis, rice and moss) in different orders confirmed that NBS-LRR genes are generally expanded in higher plants, but to different extents and in different subclasses (Supplementary Fig. 7a). Interestingly, same as reported in Dohm et al.12, we also found that the two Caryophyllales, spinach and sugar beet, harboured only one TNL gene, while the TNL family is highly expanded in other eudicots, for example, 19 members in tomato and 78 in Arabidopsis (Supplementary Fig. 7a–b).
It was reported that a sequence characterized amplified region (SCAR) marker, DM-1, was tightly linked to the Pfs-1 locus and could distinguish spinach genotypes that were homozygous resistant, heterozygous resistant or homozygous susceptible to race 6 of Pfs21. We mapped the DM-1 marker to the spinach genome and identified five NBS-LRR genes close to the marker, of which two pairs were tandemly duplicated (Supplementary Fig. 7c). As such, these are potentially candidate genes for spinach resistance to race 6 of Pfs.
Transcriptome diversity and population analysis
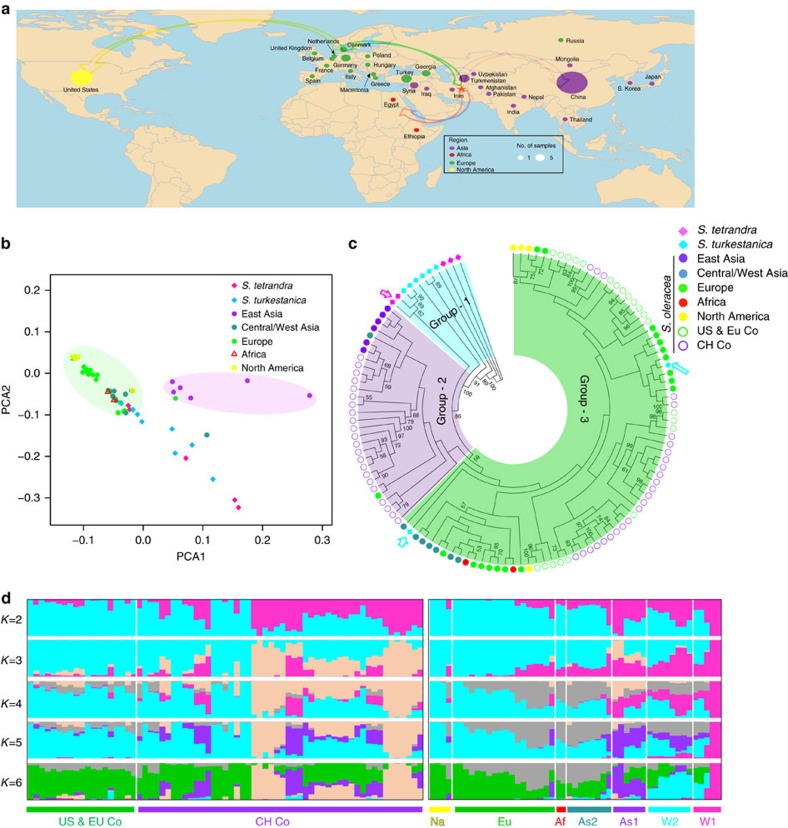
Unlike other major crops, spinach has a much smaller collection of genetic stocks1, especially for the wild species, with only a small number of accessions available in public gene banks3. To assess genetic diversity and infer population structure of spinach germplasm, we performed transcriptome sequencing of 120 spinach accessions including 107 cultivated S. oleracea and 13 wild accessions (5 S. tetrandra and 8 S. turkestanica). The cultivated S. oleracea accessions originated from diverse sources including Asia (14), Europe (18), Africa (2), North America (4), and 49 more provided by seed companies in China and 20 by companies in Europe or America (Fig. 3a and Supplementary Data 1). We generated a total of approximately one billion RNA-Seq reads (∼100 Gb in total) and more than 90% of the accessions had more than 5 million reads (Supplementary Data 1). Mapping of this large-scale RNA-Seq data set to the spinach reference genome identified a total of 420,545 single nucleotide polymorphisms (SNPs) and 12,618 small insertions and deletions (indels) (Supplementary Table 7). A previous study by Fujito et al.22 indicated that two accessions, Sp39 (Ames 23664) and Sp40 (PI 608712), were wrongly classified as S. tetrandra. Our phylogenetic analysis (see below), combined with findings from Fujito et al.22, suggested that Sp39 and Sp40 might belong to S. turkestanica. Nonetheless, we excluded these two accessions in counting population SNPs and genetic diversities. S. oleracea, S. tetrandra and S. turkestanica possessed ∼192.5 k, ∼117.3 k and ∼52.0 k SNPs, respectively (Supplementary Table 7). As expected, most SNPs (65%) were located in the coding regions, among which 43% were nonsynonymous substitutions (Supplementary Table 8), and the remaining SNPs (35%) were located in untranslated regions, introns of alternatively spliced transcripts and intergenic regions corresponding to non-coding RNAs. Moreover, 6,080 SNPs were located at either start or stop codons, or splice site acceptors or donors, impacting 4,415 genes. In addition, we also identified 1,358 genes that harbour small indels causing frame shifts or indels of amino acid sequences.
Figure 3. Geographic distribution and population structure of the 120 spinach accessions.
(a) Geographic information of the 120 accessions. The number of samples is represented by the dot size on the world map. The red star indicates the suggested origin location of spinach, and the arrows suggest the domestication history of spinach. Commercial cultivars provided by Chinese seed companies are plotted in China and commercial cultivars provided by American/European companies are plotted in United States. (b) PCA plot of non-commercial spinach accessions. (c) Phylogenetic tree of all spinach accessions inferred from transcriptome SNPs, with S. tetrandra Sp42 and Sp43 as the outgroup. The pink arrow indicates the two S. tetrandra (Sp39 and Sp40) that were grouped to S. turkestanica and the blue arrows indicate the two S. turkestanica (Sp47 and Sp48) that were clustered with cultivars. (d) Model-based clustering analysis of the 51 non-commercial (right) and 69 commercial (left) spinach accessions, given different number of groups (K=2 to 6). The y axis quantifies subgroup membership, and the x axis shows different accessions. W1: S. tetrandra; W2: S. turkestanica; As1: East Asia; As2: Central/West Asia; Eu: Europe; Af: Africa; Na: North America; CH Co: companies in China; US & EU Co: companies in United States and Europe.
We then estimated the nucleotide diversity (π) in transcriptomes of spinach populations. The average values of π for S. oleracea, S. turkestanica and S. tetrandra were 0.67 × 10−3, 0.83 × 10−3 and 6.40 × 10−3, respectively (Supplementary Table 9). The nucleotide diversity in the group of wild S. tetrandra was almost ten times of that in the group of S. oleracea or S. turkestanica, indicating a highly diverse gene pool in S. tetrandra that contains valuable genetic diversity for spinach improvement.
Principal component analysis of spinach accessions (excluding those from commercial companies due to their ambiguous geographic background) showed that those from East Asia and those from other regions were roughly divided into two groups, and S. turkestanica accessions were generally more closely related to the cultivated ones than S. tetrandra (Fig. 3b). The observation was further supported by the phylogenetic analysis, where spinach accessions were clustered into three major groups (Fig. 3c). The first group consisted of S. turkestanica and S. tetrandra accessions, the second group contained cultivars from East Asia, Chinese commercial varieties and two cultivars from Pakistan and Russia, and the third group included cultivars from Central/West Asia, Europe, North America and Africa, as well as the remaining commercial cultivars (Fig. 3c). These results suggest that cultivated spinach was likely domesticated directly from the wild species S. turkestanica. The East Asian cultivars and cultivars from Central/West Asia had a common ancestor but evolved separately. Moreover, the phylogenetic tree indicates that the European spinaches were derived from the varieties from Central/West Asia while the North American spinaches were descendants of European ones (Fig. 3c). We found two S. turkestanica samples (Sp47 and Sp48) that were clustered with varieties from Europe, America, Africa and West Asia (Fig. 3c). Sp47 was re-classified as a S. hybr recently in the National Plant Germplasm System (http://www.ars-grin.gov/npgs), and Sp48 showed mixed traits of wild and cultivated spinaches, indicating it could also be a hybrid between a wild and a cultivated accession.
We further used the model-based program STRUCTURE23 to estimate individual ancestry and admixture proportions using the SNP data. Our analysis indicated that K=2 ancestral types best explained the current population structure (Supplementary Fig. 8). Compared to other annual crops (for example, tomato, watermelon and cucumber), wild and cultivated spinach populations have more complex subpopulation structures, which might be partially due to the outcrossing nature of spinach. However, the cultivated spinaches, especially those from Europe and North America, were genetically more homogeneous than the wild groups (Fig. 3d). The ancestor–descendant relationships among the Central/West Asian, European/African and North American groups, as well as the reduced structure complexity during the distribution of spinach from its origin to the other parts of the world, are in agreement with the domestication history of spinach, which was native to Central Asia in Iran (Persia), introduced to North Africa and Europe around 1,100 A.D. by Arabs, and then brought to North America by the early colonists4 (Fig. 3a).
We investigated the linkage disequilibrium (LD) decay patterns of spinach, and found that the decay of LD with the physical distance between SNPs occurred at 5 kb in cultivated S. oleracea and 4 kb in its progenitor (S. turkestanica) accessions (r2=0.2) (Supplementary Fig. 9). The relatively rapid LD decay in cultivated spinach indicates a weak bottleneck during its domestication.
Domestication and population differentiation of spinach
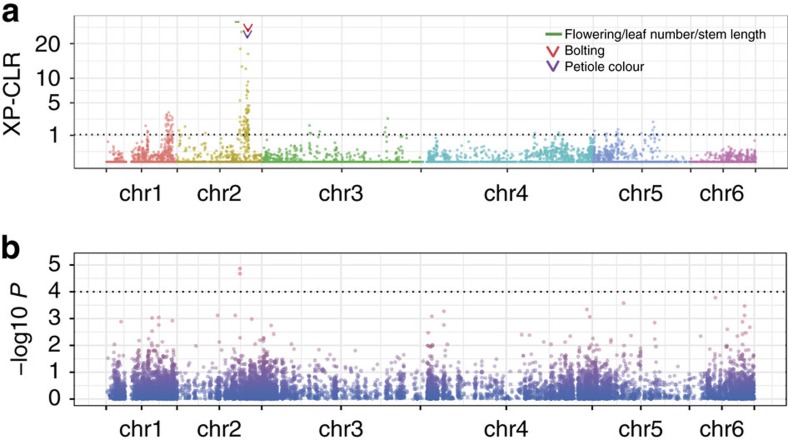
According to multiple lines of evidence, domestication of spinach started at ∼2,000 years ago24. Unfortunately, no direct historical record or research of early domesticated traits is currently available. Spinach is thought to have originated in Persia and then introduced and cultivated worldwide. During the spread of spinach to East Asia, Europe and North America, local gardeners and breeders started to improve traits such as yield, leaf quality and bolting resistance25. These improvements may have arisen independently in various spinach-growing areas4. Domestication often imposes a significant reduction of genetic diversity and enriches genetic properties favoured by humans26. However, in spinach we found a very small difference of genome-wide genetic diversity between S. oleracea (π=0.67 × 10−3) and its progenitor S. turkestanica (π=0.83 × 10−3), further supporting a weak bottleneck during its domestication. Nonetheless using the transcriptome variant data, we were able to identify potential selective signals in the spinach genome by scanning extreme allele frequency differentiation over extended linked regions using an across-population likelihood method implemented in XP-CLR27. A total of 93 regions (∼2.3 Mb in total), ranging from 10 kb to 150 kb in length (24 kb on average), were identified as candidate domestication sweeps that involved 261 (1.0%) protein-coding genes (Fig. 4a and Supplementary Data 4). Genes in the selected regions were significantly enriched with those having the fucosidase activity (GO:0015928, adjusted P=0.00142, hypergeometric distribution test), receptor signalling protein serine/threonine kinase activity (GO:0004702, adjusted P=0.00919), or signal transducer activity (GO:0004871, adjusted P=0.01593), implying their potentially important roles in spinach domestication.
Figure 4. Genome-wide scan of selective sweeps and GWAS of bolting.
(a) Distribution of XP-CLR scores across the spinach genomes. The black horizontal dashed line refers to the top 1% threshold. Arrows and the short interval indicate positions of known SNP markers and QTL, respectively, for different traits. (b) Manhattan plot of the GWAS for spinach bolting trait. The significance threshold (1 × 10−4) is indicated by the black horizontal dashed line.
A large number of sweeps with strong selection signals were located at a region from 44.7 to 50.5 Mb of chromosome 2. Interestingly, this region contains a number of known markers and quantitative trait loci (QTLs) associated with several potential domestication traits in spinach such as bolting (Fig. 4a and Supplementary Data 5). We further performed a genome-wide association study for the bolting trait in 59 spinach accessions. The strongest association signals also resided in this region and overlapped with the selective sweep with the second highest XP-CLR score (Fig. 4b). In addition, one flowering QTL and one gene (Spo00403) showing high homology to the Arabidopsis AGAMOUS-like 20, which has been reported to control flowering time28, were also covered by the sweeps in this region (Fig. 4a and Supplementary Data 4 and 5). Furthermore, QTLs for leaf number and stem length, and an SNP marker associated with petiole colour, were also found in the sweeps (Fig. 4a and Supplementary Data 5). Taken together, our data provide insights into the genomic basis of the potential domestication traits in spinach.
We next investigated the population differentiation using the FST statistic for each sliding-window between S. oleracea and S. turkestanica. The windows with top 1% FST values (>0.3907) were merged into 103 genomic regions, covering 222 genes (Supplementary Data 6). QTLs for spinach flowering, and SNP markers associated with bolting, petiole colour and erectness traits were covered by these highly divergent regions (Supplementary Fig. 10).
Discussion
We report a high-quality draft genome sequence of spinach, which was assembled using high coverage of Illumina sequences and BioNano genome maps. The spinach genome sequence provides an important resource for future comparative genomic and evolutionary studies, especially in the order Caryophyllales that constitutes the basal clade in core eudicots12. The spinach genome is among the most highly repetitive plant genomes reported to date, and much larger than the genome of its closely related species sugar beet, likely due to a recent burst of LTR retrotransposons after speciation. However, the two genomes show high collinearity and no recent whole genome duplications have occurred during the evolution of both genomes. Furthermore, our analysis indicates substantial chromosome fissions, fusions and rearrangements during the evolution of the Caryophyllales genomes.
Currently little information is available on spinach evolution and domestication. Through the analysis of transcriptome variants from a large collection of cultivated and wild spinach accessions, we infer that wild S. turkestanica could be the direct progenitor of cultivated S. oleracea and spinach domestication has a very weak bottleneck. A genome-wide scan for domestication signatures revealed 93 selective sweeps in the spinach genome that contain a number of QTLs and markers that are known to be associated with potential domestication traits in spinach such as bolting, flowering, leaf number and stem length. Furthermore, a set of highly divergent genomic regions were identified between wild and cultivated spinach species, which also contain QTLs and markers associated with important spinach traits, providing important information for facilitating spinach breeding.
Our comprehensive genome and transcriptome analyses provide novel insights into spinach genome architecture and evolution, disease-resistance and horticultural traits, and spinach genetic diversity, evolution and domestication. Furthermore, the genome sequence and transcriptome variant data provide valuable resources for facilitating spinach research and improvement.
Methods
Plant materials and library preparation and sequencing
A sibling inbred line, Sp75, was grown under standard greenhouse conditions with a 16-h light (27 °C) and 8-h dark (19 °C) cycle. Young leaves from 20-days-old plants were collected, immediately frozen in liquid nitrogen and stored at −80 °C. DNA was extracted using the QIAGEN DNeasy Plant Mini Kit following the manufacturer's instructions (QIAGEN, Valencia, CA, USA). DNA quality was evaluate via agarose gel electrophoresis and its quantity was determined on a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA). Paired-end libraries with insert sizes of 150 bp, 200 bp, 300 bp, 500 bp and 1 kb and mate-pair libraries with insert sizes of 3, 10 and 15 kb were constructed using the Genomic DNA Sample Prep kit and the Nextera Mate Pair Sample Preparation kit (Illumina, San Diego, CA, USA), respectively, following the manufacturer's instructions, and sequenced on an Illumina HiSeq 2500 platform with paired-end mode.
Genome assembly and quality evaluation
Raw Illumina reads were processed to collapse duplicated read pairs into unique read pairs. Duplicated read pairs were defined as those having identical bases at positions of 14 to 90 in both left and right reads. The resulting reads were further processed to remove adaptor and low quality sequences using Trimmomatic29 (v0.33; parameters ‘SLIDINGWINDOW:4:20 LEADING:3 TRAILING:3 MINLEN:40'). Furthermore, sequences of the junction adaptor used in the mate-pair library construction were identified and removed together with the trailing bases using the ShortRead package30. Reads shorter than 40 bp were discarded. Finally, sequencing errors in paired-end reads were corrected with QuorUM31 (parameter ‘--kmer-len 24').
The high-quality cleaned paired-end reads were first assembled into contigs using Platanus32 (v1.2.1; parameters ‘-k 32 -s 10 -c 2 -a 10.0 -u 0.1 -d 0.5'). The resulting contigs were first processed by a module ‘Prepare' released with SOAPdenovo2 (v2.04; ref. 33) with ‘-D' option, then connected to scaffolds with SOAPdenovo2 using all paired-end and mate-paired reads. To further improve the assembly, we generated three additional assemblies using different combinations of contig assembly and scaffolding programs: (1) contig assembly with Platanus and scaffolding with SSPACE34 (parameters ‘-x 0 -m 32 -o 20 -t 0 -k 5 -a 0.70 -n 15'); (2) both contig assembly and scaffolding with Platanus; and (3) both contig assembly and scaffolding with SOAPdenovo2 (parameters ‘-K 60 -m 127 -M 2 -d 1 -R'). Reads from the 15-kb insert mate-pair library were then used to break any potential scaffolding errors in these four assemblies with REAPR35 (v1.0.17; parameter ‘-score f=10'). Using the first assembly (Platanus+SOAPdenovo2) as the backbone, the other three assemblies were integrated. First, the three assemblies were aligned to the first assembly using LAST36 (v604; parameters ‘-r 1 -q 3 -a 7 -b 1 –e 80'), and based on the alignments local collinear blocks were determined by Mugsy37 (v1.2.3; parameters ‘-d 1000 -c 30 -duplications 0'). Scaffolds from the first assembly were then connected if they were aligned adjacently in the same scaffold in the other assemblies without any conflicts. Gaps in the final consensus scaffolds were filled using gapcloser (v1.12; parameter ‘-p 25') in the SOAPdenovo2 package33 with paired-end reads. The assembly was further polished to correct base errors using Pilon38 (v1.8) with paired-end reads.
BioNano genome map construction and integration
High molecular weight DNA was extracted from spinach Sp75 seedlings by Amplicon Express (Pullman, WA). Labeling of high molecular weight DNA molecules and BioNano map data collection were performed according to Pendleton et al.39 using the IrysPrep Reagent Kit (BioNano Genomics, San Diego, CA, USA). Single molecule maps longer than 150 kb were used for de novo assembly using IrysView (BioNano Genomics) with p value threshold set to 1 × 10−8. The resulting consensus maps (CMAPs) were then used to join the assembled spinach scaffolds to super-scaffolds using the ‘Sewing machine pipeline'40 with parameters of ‘--f_con 13 --f_algn 30 --s_con 8 --s_algn 90 -T 1e-8'. The gap lengths between assembled genome scaffolds were estimated by BioNano CMAPs.
Annotation of transposable elements
A de novo LTR retrotransposon library and a MITE library were constructed by screening the assembled spinach genome using LTRharvest41 (v1.5.7; parameters ‘-minlenltr 100 -maxlenltr 6000 -mindistltr 1500 -maxdistltr 25000 -mintsd 5 -maxtsd 5 -motif tgca -vic 10') and MITE-Hunter42 (v11-2011; parameters ‘-n 20 -c 50'), respectively. The spinach genome was then masked using RepeatMasker (v4.0.5; http://www.repeatmasker.org/) with the LTR retrotransposon and MITE libraries, and the unmasked sequences were further searched for repeat elements using RepeatModeler (v1.0.8; http://www.repeatmasker.org/RepeatModeler.html). All the repetitive sequences generated above were combined into a single repeat library, and then compared against the Swiss-Prot database (http://www.uniprot.org/) using BLAST with an e-value cutoff of 0.01. Sequences matching any non-TE proteins in the database were removed from the repeat library. TEs in the library were classified using REPCLASS43 (v1.0.1). The repeat library was then used to identify TEs in the assembled spinach genome with RepeatMasker (parameters ‘-s -x -nolow -norna -no_is -a').
LTR retrotransposon insertion time analysis
We used LTRharvest41 (v1.5.7) to de novo detect full length LTR retrotransposons in both sugar beet and spinach genomes with parameters of ‘-motif tgca -motifmis 1 -minlenltr 100 -maxlenltr 3000 -mintsd 4 -maxtsd 20'. Domains located in internal regions of LTR retrotransposons including reverse transcriptase, protease, ribonuclease H and integrase were identified using LTRdigest44. Only LTR retrotransposons containing all four domains were used for the insertion time analysis. The two ends of these LTR retrotransposons were aligned with MUSCLE45 (v3.8.31) and the distance was calculated using the distmat program in the EMBOSS package46 (v6.5.7) with parameter of ‘-nucmethod 2'. The insertion time (T) of an LTR retrotransposon was calculated using the formula T=K/2r, where K is the distance and r is the rate of nucleotide substitution, which was set to 7.0 × 10−9 substitutions per site per year according to Ossowski et al.47.
Protein-coding gene prediction and functional annotation
The repeat-masked spinach genome was used for gene prediction using MAKER48 (v2.31.6), which combines evidence from ab initio, transcript mapping and protein homology-based predictions to define the confident gene models. SNAP49 (v2009-02-03) and AUGUSTUS50 (v3.0.2) were used for ab initio gene predictions. For transcript mapping, RNA-Seq data were aligned to the spinach genome using Tophat51 (v2.0.13; parameters ‘--read-mismatches 1 --splice-mismatches 0 --min-intron-length 30') and then assembled using Cufflinks52 (v2.2.1; parameters ‘--no-effective-length-correction --min-intron-length 30 --min-frags-per-transfrag 5'). For homologous protein mapping, protein sequences from sugar beet, Arabidopsis, potato, tomato and Swiss-Prot were aligned to the genome using Spaln53 (v2.1.4; parameters ‘-Xk11 -H35'). For predicted genes without a transcript or homologous protein support, they were compared against the Pfam database (http://pfam.xfam.org) and those that did not contain any Pfam domains were discarded. Furthermore, genes highly homologous to transposable elements were also removed. Predicted genes with expression levels of at least one RPKM (reads per kilobase of exon model per million mapped reads) in at least one of the 120 spinach accessions were considered as supported by the RNA-Seq data.
To annotate the spinach predicted genes, their protein sequences were compared to GenBank nr, the Arabidopsis protein and UniProt (Swiss-Prot and TrEMBL; http://www.uniprot.org/) databases using BLAST (parameter ‘-evalue 1e-4'), as well as the InterPro database using InterProScan54 (v5.10–50.0). GO annotations were obtained using Blast2GO (ref. 55) (version 2.5.0) based on the BLAST results against the GenBank nr database and results from the InterProScan analysis. Functional descriptions were assigned to spinach genes using AHRD (v3.3; https://github.com/groupschoof/AHRD). Enzyme-encoding genes were extracted based on the AHRD result and Enzyme Commission (EC) information from the Blast2GO analysis. Transcription factors and protein kinases were identified and classified into different families using the iTAK pipeline56 (v1.6).
Comparative genomic analyses
Orthologous groups were constructed using OrthoMCL57 (v1.4; inflation parameter: 1.5) with an all -versus-all BLASTP comparison (parameters ‘-evalue 1e-5 -max_target_seqs 1000') of protein sequences from spinach and ten other plant species, including Brachypodium distachyon, rice, sugar beet, tomato, grape, Medicago truncatula, watermelon, cacao, Arabidopsis thaliana and papaya (Supplementary Data 2). Protein sequences of single-gene families were aligned by MUSCLE45 (v3.8.31). The alignments were trimmed by the trimAL program58 (v1.2rev59) with the parameter ‘gt' set to 0.9. The concatenated protein alignments were used to construct a phylogenetic tree using the maximum parsimony method implemented in MEGA6 (ref. 59) with 1,000 bootstrap replications. Syntenies within and between spinach, sugar beet and Arabidopsis genomes were detected using MCScanX60 (parameters ‘-s 5 -e 5'). Ks values of homologous gene pairs in the syntenic regions were calculated and the speciation time base on Ks values was derived using the equation T=Ks/2r with r=7.0 × 10−9 substitutions per site per year according to Ossowski et al.47.
Transcriptome sequencing of spinach germplasm collection
A total of 120 cultivated (S. oleracea) and wild (S. tetrandra and S. turkestanica) spinach accessions including nine that were recently reported by Xu et al.11 were collected, among which 51 were provided by the North Central Regional Plant Introduction Station (NCRPIS), Ames, Iowa, and 69 by seed companies (Supplementary Data 1). Sample collection, total RNA extraction and RNA-Seq library construction were same as described in Xu et al.11. Specifically, the entire 20-day-old seedlings were collected and stored at −80 °C till use. Total RNA was extracted using the QIAGEN RNeasy Plant Mini Kit following the manufacturer's instructions (QIAGEN, Valencia, CA, USA). The quality and quantity of RNA were assessed by electrophoresis on 1% agarose gels and by a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), respectively. The strand-specific RNA-Seq libraries were constructed for each accession following the protocol described in Zhong et al.61 and sequenced on an Illumina HiSeq 2500 platform (Illumina Inc., USA) using the single-end mode with the read length of 100 bp.
RNA-Seq data analysis and SNP calling
Raw RNA-Seq reads were processed using Trimmomatic29 (v0.33; parameters ‘SLIDINGWINDOW:4:20 LEADING:3 TRAILING:3 MINLEN:40') to remove adaptor and low quality sequences. Reads longer than 40 bp were kept and then aligned to the ribosomal RNA database (https://www.arb-silva.de/) using bowtie62 (v1.0.0; parameter ‘-v 3') and the aligned reads were discarded. For gene expression analysis, the resulting high-quality cleaned reads were aligned to the assembled spinach genome using TopHat51 allowing two mismatches. Following alignments, raw counts for each spinach gene were derived and normalized to RPKM.
To identify SNPs and small indels among the 120 spinach accessions, raw RNA-Seq reads were first processed to collapse duplicated reads into unique reads and remove adaptor and low quality sequences. The resulting reads were aligned to the spinach genome using BWA63 (v0.7.12-r1039; parameters ‘-n 0.04 -o 1 -e 2'). Only reads uniquely mapped (having a single best match) to the genome were kept. Following mapping, SNPs and small indels were identified based on the mpileup files generated by SAMtools64 (v1.1). The identified SNPs and small indels were supported by at least three distinct reads and each genotype had an allele frequency >25%.
Phylogenetic and population structure analyses
Only SNPs with minor allele frequency (MAF) >5% and missing data <10% (a total of 11,434) were used for phylogenetic and population structure analyses. A subset of 2,769 SNPs at fourfold-degenerate sites was used to construct a maximum parsimony phylogenetic tree using PAUP*65 (parameters ‘search=heuristic start=stepwise addseq=random swap=tbr') with 100 bootstrap replications. Two S. tetrandra accessions, Sp42 and Sp43, were used as the outgroup because of their larger genetic distance from the cultivars. Principal component analysis was performed using the 11,434 SNPs with the EIGENSOFT66 smartpca (v13050) program with the parameter ‘-k' set to 10. Population structure analysis was performed using the program STRUCTURE23 (v2.3.4). To determine the most likely group number, STRUCTURE was run 20 times on 1,000 randomly selected SNPs for each K value from 1 to 20. The optimal K was selected using the Evanno method67. After determining the ΔK, the subgroup membership of each accession was determined by 15,000 iterations for each K (K= 2 to 20) using the 11,434 SNPs.
π was used to calculate nucleotide diversities in each Spinacia group. SNPs with genotype information in at least 90% of the accessions were used in the calculation. A sliding window approach was used to calculate π in the assembled spinach genome with a window size of 10 kb and a step size of 1 kb. LD decay was calculated based on the correlation coefficient (r2) of alleles using Haploview68 (v4.2; parameters ‘-dprime -maxdistance 1000 -minMAF 0.05 -hwcutoff 0.001').
Selective sweep and population differentiation
Whole-genome screening of selective sweeps was performed by comparing allele frequency differentiation between S. turkestanica and S. oleracea using XP-CLR27 (v1.0), a method based on modelling the likelihood of multilocus allele frequency differentiation between two populations. Genetic distances between SNPs were interpolated according to their physical distances in our genetic map. XP-CLR was run for each pseudochromosome or scaffold with parameters ‘-w1 0.0005 100 100 -p0 0.7'. Final estimates were tabulated in non-overlapping 10-kb windows across the genome. Adjacent windows with high XP-CLR scores (top 3%) were grouped into a single region, which was then given a score using the maximum of region-wise XP-CLR. Regions with top 1% of XP-CLR scores were considered as potential selected sweeps. We further calculated the π ratios between S. turkestanica and S. oleracea (πw/πc) in 10-kb sliding windows with a step size of 1 kb and identified regions with top 50% highest π ratios. Three selective sweeps derived from the XP-CLR analysis were not shared with those by the π ratio approach and thus excluded from the final results of the putative selected regions.
The population fixation index FST was calculated using a sliding window approach (10-kb sliding windows with a step size of 1 kb) using the R package HIERFSTAT69 (v0.04-22). Genomic regions with top 1% of FST values between S. oleracea and S. turkestanica were identified as potential differentiated regions.
Genome-wide association study of bolting
Bolting trait of 59 spinach accessions (20–30 replications) were evaluated in the greenhouse at Shanghai Normal University in the spring of 2015. Bolting time was monitored daily and determined as the number of days from planting to the first elongation of the floral stem observed in at least half of the used plants for each accession. Correction for population stratification and genome-wide association study for the bolting trait were performed using EIGENSOFT66 (version 6.0.1; parameters ‘-q YES -k 10'), and using a total of 11,434 SNPs (MAF>5% and missing data <10%). SNPs with P<0.0001 were considered significantly associated.
Data availability
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession LZYP00000000. The version described in this paper is version LZYP01000000. Raw sequence reads of genome and transcriptome sequencing have been deposited in NCBI sequence read archive (SRA) under accession number SRP076521. The spinach genome sequence and the annotation are also available at SpinachBase (http://www.spinachbase.org). The authors declare that all other data supporting the findings of this study are available from the corresponding authors on request.
Additional information
How to cite this article: Xu, C. et al. Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat. Commun. 8, 15275 doi: 10.1038/ncomms15275 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Supplementary Figures, Supplementary Tables, Supplementary Note and Supplementary References
Geographic information and statistics of the RNA-Seq libraries for the 120 Spinach accessions. Libraries were sequenced using the single-end mode with the read length of 100 bp.
List of plant genome sequences used in the comparative genomic analysis
Spinach disease resistance genes
Genes in the candidate selective sweep regions
Markers and QTLs overlap with or close to (<200 kb) potential selective sweeps and differentiated regions.
Genes in the highly differentiated regions (top 1% FST) between S. turkestanica and S. oleracea.
Acknowledgments
We thank Dr Jim Giovannoni for critical reading of the manuscript and Drs Mingcheng Luo and Tingting Zhu for help in generating BioNano genome maps. This work was supported by grants from the Development and Collaborative Innovation Center of Shanghai (No. ZF1205), Project of Prospering Agriculture with Science and Technology, Shanghai, China (2015, No. 9), Capacity Construction Project of Local Universities, Shanghai, China (No. 14390502700), Set-Sail Plan Project supported by Science and Technology Commission of Shanghai, China (No. 14YF1409400), National Natural Science Foundation of China (No. 31501754) and the United States National Science Foundation (IOS-0923312 and IOS-1539831).
Footnotes
The authors declare no competing financial interests.
Author contributions C.X., C.J., X.C., X.W., C.G., Y.X. and J.D. contributed to plant sample collection, DNA/RNA preparation, library construction and sequencing; C.X., C.J., H.S., Y.Z., X.S. and Z.Z. worked on genome assembly and annotation, and comparative and population genomic analyses; W.L., C.J. and Z.F. contributed to website construction; C.J., C.X., H.S. and Z.F. wrote and revised the manuscript; Quanhua Wang, Z.F., C.J., Quanxi Wang, B.M., S.D. and S.H. conceived strategies, designed experiments and managed projects. All authors read and approved the manuscript.
References
- Morelock T. & Correll J. in Vegetables I (eds Prohens, J. & Nuez, F.) 189–218 (Springer, 2008).
- Khattak J. Z. K., Torp A. M. & Andersen S. B. A genetic linkage map of Spinacia oleracea and localization of a sex determination locus. Euphytica 148, 311–318 (2006). [Google Scholar]
- Andersen S. B. & Torp A. M. in Wild Crop Relatives: Genomic and Breeding Resources (ed. Kole, C.) 273–276 (Springer-Verlag, 2011).
- Ryder E. J. in Leafy Salad Vegetables 195–227 (AVI Publishing Company, 1979).
- Shohag M. J. et al. Natural variation of folate content and composition in spinach (Spinacia oleracea) germplasm. J. Agric. Food Chem. 59, 12520–12526 (2011). [DOI] [PubMed] [Google Scholar]
- Mou B. Evaluation of oxalate concentration in the US spinach germplasm collection. HortScience 43, 1690–1693 (2008). [Google Scholar]
- Solberg S. O., Yndgaard F. & Axelsson J. Nitrate and oxalate in germplasm collections of spinach and other leafy vegetables. Emir. J. Food Agric. 27, 698–705 (2015). [Google Scholar]
- Hu J. G., Mou B. Q. & Vick B. A. Genetic diversity of 38 spinach (Spinacia oleracea L.) germplasm accessions and 10 commercial hybrids assessed by TRAP markers. Genet. Resour. Crop Evol. 54, 1667–1674 (2007). [Google Scholar]
- Khattak J. Z. K., Christiansen J. L., Torp A. M. & Andersen S. B. Genic microsatellite markers for discrimination of spinach cultivars. Plant Breeding 126, 454–456 (2007). [Google Scholar]
- Kuwahara K., Suzuki R., Ito Y., Mikami T. & Onodera Y. An analysis of genetic differentiation and geographical variation of spinach germplasm using SSR markers. Plant Genet. Resour. 12, 185–190 (2013). [Google Scholar]
- Xu C. et al. De novo and comparative transcriptome analysis of cultivated and wild spinach. Sci. Rep. 5, 17706 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm J. C. et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505, 546–549 (2014). [DOI] [PubMed] [Google Scholar]
- Arumuganathan K. & Earle E. D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218 (1991). [Google Scholar]
- Lam E. T. et al. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol. 30, 771–776 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V. & Zdobnov E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- Piegu B. et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 16, 1262–1269 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mas J. et al. The genome of melon (Cucumis melo L.). Proc. Natl Acad. Sci. USA 109, 11872–11877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoche A. E. et al. Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol. 16, 184 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. & Conery J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000). [DOI] [PubMed] [Google Scholar]
- Dangl J. L. & Jones J. D. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833 (2001). [DOI] [PubMed] [Google Scholar]
- Irish B. M., Correll J. C., Feng C., Bentley T. & de los Reyes B. G. Characterization of a resistance locus (Pfs-1) to the spinach downy mildew pathogen (Peronospora farinosa f. sp. spinaciae) and development of a molecular marker linked to Pfs-1. Phytopathology 98, 894–900 (2008). [DOI] [PubMed] [Google Scholar]
- Fujito S. et al. Evidence for a common origin of homomorphic and heteromorphic sex chromosomes in distinct Spinacia species. G3 5, 1663–1673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubatzky V. E. & Yamaguchi M. in World Vegetables: Principles, Production, and Nutritive Values 457–473 (Springer, 1997).
- Grubben G. J. H. & Denton O. A. PROTA 2 vegetables PROTA Foundation; Backhuys Publishers [distr.] (2004). [Google Scholar]
- Purugganan M. D. & Fuller D. Q. The nature of selection during plant domestication. Nature 457, 843–848 (2009). [DOI] [PubMed] [Google Scholar]
- Chen H., Patterson N. & Reich D. Population differentiation as a test for selective sweeps. Genome Res. 20, 393–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. et al. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M. & Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25, 2607–2608 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G., Yorke J. A. & Zimin A. QuorUM: an error corrector for Illumina reads. PLoS ONE 10, e0130821 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R. et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 24, 1384–1395 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M., Henkel C. V., Jansen H. J., Butler D. & Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011). [DOI] [PubMed] [Google Scholar]
- Hunt M. et al. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 14, R47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielbasa S. M., Wan R., Sato K., Horton P. & Frith M. C. Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli S. V. & Salzberg S. L. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 27, 334–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton M. et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods 12, 780–786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton J. M. et al. Tools and pipelines for BioNano data: molecule assembly pipeline and FASTA super scaffolding tool. BMC Genomics 16, 734 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Kurtz S. & Willhoeft U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9, 18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y. & Wessler S. R. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38, e199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Keswani U., Ranganathan N., Guibotsy M. L. & Levine D. Exploring repetitive DNA landscapes using REPCLASS, a tool that automates the classification of transposable elements in eukaryotic genomes. Genome Biol. Evol. 1, 205–220 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbiss S., Willhoeft U., Gremme G. & Kurtz S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37, 7002–7013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P., Longden I. & Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- Ossowski S. et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327, 92–94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L. et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Tzvetkova A. & Morgenstern B. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome Biol. 7, S11.1–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata H. & Gotoh O. Benchmarking spliced alignment programs including Spaln2, an extended version of Spaln that incorporates additional species-specific features. Nucleic Acids Res. 40, e161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- Zheng Y. et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 9, 1667–1670 (2016). [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J. & Roos D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M. & Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S. et al. High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb. Protoc. 2011, 940–949 (2011). [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D. L. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4 Sinauer Associates (2002) http://paup.scs.fsu.edu/Cmd_ref_v2.pdf. [Google Scholar]
- Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- Evanno G., Regnaut S. & Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). [DOI] [PubMed] [Google Scholar]
- Goudet J. hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186 (2005). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures, Supplementary Tables, Supplementary Note and Supplementary References
Geographic information and statistics of the RNA-Seq libraries for the 120 Spinach accessions. Libraries were sequenced using the single-end mode with the read length of 100 bp.
List of plant genome sequences used in the comparative genomic analysis
Spinach disease resistance genes
Genes in the candidate selective sweep regions
Markers and QTLs overlap with or close to (<200 kb) potential selective sweeps and differentiated regions.
Genes in the highly differentiated regions (top 1% FST) between S. turkestanica and S. oleracea.
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession LZYP00000000. The version described in this paper is version LZYP01000000. Raw sequence reads of genome and transcriptome sequencing have been deposited in NCBI sequence read archive (SRA) under accession number SRP076521. The spinach genome sequence and the annotation are also available at SpinachBase (http://www.spinachbase.org). The authors declare that all other data supporting the findings of this study are available from the corresponding authors on request.