Abstract
Dihydroxyacetone (Dha) kinases are a sequence-conserved family of enzymes, which utilize either ATP (in animals, plants, bacteria) or the bacterial phosphoenolpyruvate carbohydrate phosphotransferase system (PTS) as a source of high-energy phosphate. The PTS-dependent kinase of Escherichia coli consists of three subunits: DhaK contains the Dha binding site, DhaL contains ADP as cofactor for the double displacement of phosphate from DhaM to Dha, and DhaM provides a phospho-histidine relay between the PTS and DhaL∷ADP. DhaR is a transcription activator belonging to the AAA+ family of enhancer binding proteins. It stimulates transcription of the dhaKLM operon from a sigma70 promoter and autorepresses dhaR transcription. Genetic and biochemical studies indicate that the enzyme subunits DhaL and DhaK act antagonistically as coactivator and corepressor of the transcription activator by mutually exclusive binding to the sensing domain of DhaR. In the presence of Dha, DhaL is dephosphorylated and DhaL∷ADP displaces DhaK and stimulates DhaR activity. In the absence of Dha, DhaL∷ADP is converted by the PTS to DhaL∷ATP, which does not bind to DhaR.
Keywords: AAA+ ATPase, enhancer binding proteins, protein–protein interaction, PTS, transcription
Introduction
Gene expression in prokaryotes is frequently controlled by small molecules, which act as signals of the metabolic state and environmental conditions. In most cases, these molecules exert their influence by binding to transcription factors and receptor proteins. Small molecules are also substrates of enzymes and permeases; yet enzymes and receptors usually have different folds reflecting different structural requirements for catalysis and recognition. Here we report that the two catalytic subunits of the Escherichia coli dihydroxyacetone (Dha) kinase act antagonistically as coactivator and corepressor of the transcription activator DhaR.
The E. coli Dha operon (dhaKLM) encodes the three subunits DhaK, DhaL and DhaM of the Dha kinase. A fourth divergently transcribed gene, dhaR, codes for the transcription regulator DhaR (Figure 1A). DhaR belongs to the family of bacterial enhancer binding proteins (EBP; Buck et al, 2000). It consists of an N-terminal sensing domain (37 kDa), a central AAA+ domain (24 kDa, ATPases associated with diverse cellular activities; Lupas and Martin, 2002) and a C-terminal helix–turn–helix motif (5 kDa) (Figure 1B). The N-terminal sensing domain consists of a GAF (amino acids (aa) 52–189) and a PAS domain (aa 203–265), the ligand and protein interaction domains of two-component system sensor kinases (Ponting and Aravind, 1997; Taylor and Zhulin, 1999). The central AAA+ domain consists of seven highly conserved sequence motifs (C1–C7), which are shared with other functionally unrelated AAA+ proteins (Morett and Segovia, 1993). Orthologs of the E. coli DhaR are encoded adjacent to the dha operons in Citrobacter freundii (71% identity), Klebsiella pneumoniae (70% identity), in a genetic island of meningitic E. coli K1 (32% identity) and in Vibrio parahaemolyticus (34% identity) (Daniel et al, 1995; Huang et al, 2001; Sun et al, 2003). DhaR also features sequence similarity (<30% identity) to AcoR, the regulator of acetoin metabolism in Thermoanaerobacter tengcongensis and Clostridium magnum (Figure 1C).
Figure 1.
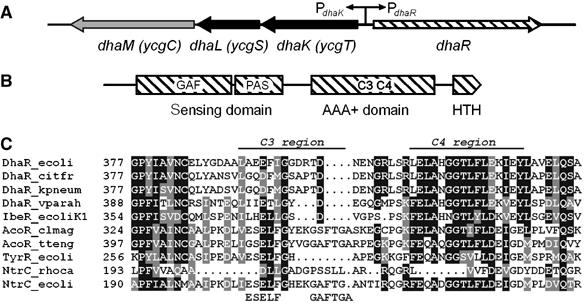
(A) Structure of the Dha operon of E. coli. The three genes encode DhaK (Dha binding subunit, SWISS-PROT entry P76015), DhaL (ADP cofactor binding subunit, P76014) and DhaM (phosphotransferase subunit of the PTS, P37349). DhaR (P76016) encodes the transcription regulator DhaR. (B) Domain structure of DhaR with the N-terminal sensing, the central AAA+ (ATPase) and the C-terminal helix–turn–helix DNA binding domains. PAS and GAF are two conserved folds of the sensing domain. (C) Comparative alignment of the C3 and C4 regions of the central AAA+ domains: DhaR of E. coli (P76016), C. freundii (P45512), K. pneumoniae (MGH 78578, http://genome.wustl.edu/projects/bacterial/kpneumoniae/) and V. parahaemolyticus (Q87SQ5); IbeR of E. coli K1 (Q8VP28); AcoR of C. magnum (Q46141) and T. tengcongensis (Q8RBX1); TyrR (σ70 dependent) of E. coli (P07604), NtrC of R. capsulatus (P09432), NtrC (σ54 dependent) of E. coli (P06713). The GAFTGA motif participates in σ54 binding, and the ESELF sequence is important for the positioning of the GAFTGA loop (Xu et al, 2004).
Dha kinases are a family of sequence-related enzymes, which utilize as phosphate donor either ATP or a phosphoprotein of the phosphoenolpyruvate: sugar phosphotransferase system (PTS). The PTS is an energy-transducing system involved in carbohydrate uptake and control of carbon metabolism, which is ubiquitous in eubacteria but does not occur in archaebacteria and eukaryotes (Postma et al, 1996). ATP-dependent kinases occur in eukaryotes as well as in bacteria. The Dha kinases of C. freundii (DAK) and E. coli (DhaK, DhaL, DhaM) are prototypes of ATP- and PTS-dependent kinases, respectively (Daniel et al, 1995; Gutknecht et al, 2001). ATP-dependent kinases (DAK) consist of two domains. PTS-dependent kinases consist of two subunits (DhaK, DhaL), which are homologous to DAK, and an additional third subunit (DhaM), which is homologous to proteins of the PTS. DhaK (38 kDa) contains the substrate binding site to which Dha and Dha-phosphate are covalently bound in hemiaminal linkage with His-230 (Siebold et al, 2003; Garcia-Alles et al, 2004). DhaL (23 kDa), an eight helix barrel of regular up-down topology, contains a tightly bound ADP as coenzyme (Bächler et al, to be published). This ADP serves as phosphorylation site for the double displacement of phosphate from DhaM to Dha, and thus plays a role analogous to the histidines and cysteines in the proteins of the PTS. In what follows, the nonphosphorylated form of DhaL is designated as DhaL∷ADP and the phosphorylated form as DhaL∷ATP.
Dha is the product of glycerol oxidation in C. freundii and K. pneumoniae (Forage and Lin, 1982; Daniel et al, 1995) and of the transketolase reaction between xylulose-5-phosphate and formaldehyde in methylotrophic yeast (Waites and Quayle, 1981). Free Dha may also arise as the by-product of aldol cleavage by fructose-6-phosphate aldolase (Schurmann and Sprenger, 2001) and in paracatalytic reactions (Lubini and Christen, 1979). It has been shown that Dha and similar short-chain triose sugars have an increased propensity to react with proteins in Maillard type reactions (Tessier et al, 2003), that Dha can induce DNA damage, cell-cycle block and apoptosis (Petersen et al, 2004) and that in yeast Dha kinases are involved in detoxification of Dha (Molin et al, 2003). In animals and plants, Dha kinases may thus have a ‘housecleaning' function by preventing the accumulation of Dha in toxic concentrations.
Lin and co-workers (Jin and Lin, 1984; Paulsen et al, 2000) demonstrated that E. coli growing on Dha had increased Dha kinase activity, and Beutler et al (2001) observed that DhaK and DhaL were upregulated in the proteome of E. coli lacking enzyme I (EI), the master enzyme of the PTS, and that EI is necessary for Dha kinase activity (Gutknecht et al, 2001). Here we provide a molecular explanation for these observations. In brief, DhaL∷ADP is the coactivator of DhaR while DhaL∷ATP is transcription inactive. The former arises when phosphate is transferred from ATP to the inducer Dha, and the latter when ADP is rephosphorylated by DhaM. Binding of DhaK and Dhal∷ADP to DhaR is mutually exclusive. DhaK and DhaL thus are enzyme subunits and at the same time also coregulators of transcription.
Results and discussion
Induction of the dha operon with Dha
E. coli MC4100 containing the PdhaK promoter and lacZ in the chromosomal λ attachment site was used to measure dha operon expression (Boyd et al, 2000). Cells were grown in low-phosphate casamino acids minimal medium to which C3 carbohydrates were added as inducers. Dha induced LacZ activity 26-fold (Figure 2A). Glyceraldehyde induced it seven-fold while, glycerol, a structural analog of Dha, had no effect. Induction was confirmed on Western blots (Figure 2B) and the cellular protein concentration quantified using known amounts of purified subunits as standards (Table I). In the noninduced state, the intracellular Dha kinase concentration is of the order of 1 μM. Upon induction, it increases six- to 11-fold. The DhaR concentration was less than 300 molecules of DhaR per cell (the detection limit of the low-titer antiserum was 20 ng DhaR).
Figure 2.
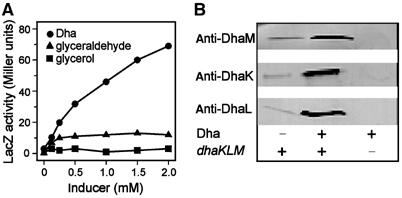
Dha induces dha operon expression. (A) Induction of PdhaK-lacZ activity with Dha (circle), glyceraldehyde (triangle) and glycerol (square). The recombinant PdhaK-lacZ reporter gene was integrated into the chromosome of E. coli MC4100. Cultures were grown for 18 h in a 0.25% casamino acid–MOPS medium in the presence of the indicated concentrations of inducer. (B) Western blot analysis of DhaK, DhaL and DhaM in cell extracts of E. coli MC4100. Cells were grown in LB broth without and with 2 mM Dha. E. coli MC4100ΔdhaKLM was used as negative control. Proteins were identified with polyclonal antisera and a lactoperoxidase-coupled second antibody.
Table 1.
Cellular content of DhaK, DhaL and DhaM subunits
| Number of protein monomers/dimers (concentration in μMa) | |||
|---|---|---|---|
| Proteins | −Dhaa | +Dhab | Fold induction |
| DhaK2 | 676±120 (1.9) | 3775±335 (10.4) | 5.6 |
| DhaL | 733±280 (2.0) | 4800±980 (13.2) | 6.5 |
| DhaM2 | 230±53 (0.6) | 2520±485 (7.0) | 11.0 |
| aA dry weight of 2.8 × 10−13 g/cell and a volume 0.18 μm3/cell were used for calculations (Neidhardt et al, 1996). | |||
| bCells were assayed after 18 h of growth in LB medium, without and with 2 mM Dha. | |||
DhaR is an activator of the dhaKLM operon and a repressor of its own synthesis
The dhaKLM operon and the dhaR gene are divergently transcribed from a 190-bp-long intergenic region (Figure 1A). Disruption of the dhaR gene resulted in the complete disappearance of PdhaK-lacZ reporter gene activity, and the dha operon could no longer be induced with Dha (Figure 3A). Conversely, PdhaR-lacZ reporter gene activity, which is low when DhaR is produced, became constitutively high after disruption of dhaR (Figure 3B). Expression of dhaR under the control of the noninduced (leaky) araC promoter on a low-copy-number plasmid restored inducibility of the dhaKLM operon and repression of the dhaR gene. While dhaKLM expression was inducible with Dha, expression of dhaR was not. DhaRΔ1–311, a mutant without the sensing domain, could no longer activate dhaKLM expression, and autorepression of dhaR was leaky. This indicates that DhaRΔ1–311 retains a reduced affinity for the operator but can no longer be activated. E. coli DhaR could be replaced by DhaR from C. freundii (Figure 3A). The dha operator sequences from the two organisms could also be exchanged indicating that DhaR of E. coli and C. freundii are orthologs (results not shown).
Figure 3.
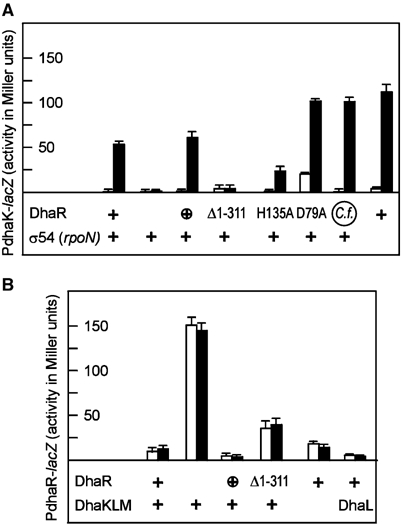
DhaR is the activator of dhaKLM operon (A) and the repressor of dhaR gene transcription (B). Protein expression from the chromosomal gene is indicated with +, and expression from a low-copy-number plasmid with ⊕. The point mutant and truncated proteins were expressed from a low-copy-number plasmid. E. coli strains were grown without (open bars) and with 2 mM Dha (induced, solid bars). (A) Activation of the PdhaK-lacZ reporter gene. DhaR with a signalling domain is necessary for induction by Dha. σ54 is dispensable for activation of dhaKLM. (B) Repression of the PdhaR-lacZ reporter gene. Repression of the dhaR gene by DhaR is not affected by Dha and the Dha kinase subunits. The average standard deviation for all values larger than 10 Miller units is 7%. The complete genotypes are given in Table II.
The N-terminal sensing domain of DhaR features conserved aspartyl (Asp-79) and histidyl (His-135) residues (Figure 4), which, in principle, could be phosphorylated by a sensor histidine kinase or a component of the PTS, for instance by DhaM. However, no 32P-labelled DhaR could be detected in sodium dodecylsulfate (SDS) gels by autoradiography or protein pull-down experiments (results not shown). LacZ activity remained inducible in cells producing the D79A and H135A mutants of DhaR. D79A merely displayed an overall two- to three-fold increased LacZ activity, while H135A had a slightly reduced activity (Figure 3A). This indicates that the sensing domain is an activator of the AAA+ domain and suggests that DhaR activity is not controlled by protein phosphorylation.
Figure 4.

Conserved sequence motives from sensing domains of DhaR homologs. The conserved Asp-79 and His-135 are indicated with asterisks. Abbreviations are as in Figure 1C. STC, signal-transduction and transcriptional-control protein of Clostridium beijerinckii (P26047).
EBPs typically bind to DNA enhancer elements upstream of σ54-dependent promoters (Studholme and Dixon, 2003). However, disruption of rpoN, the gene encoding σ54, did not impair induction of the dha operon by DhaR (Figure 3A). The AAA+ domain of DhaR indeed does not contain two sequence motives (ESELF and GAFTGA), which are conserved in the EBPs that interact with σ54 (Figure 1C). Finally, the dhaR dhaK intergenic region does not comprise a sequence similar to the consensus of a σ54 promoter (Reitzer and Schneider, 2001). Taken together, this indicates that DhaR like TyrR of E. coli and NtrC of Rhodobacter capsulatus (Studholme and Dixon, 2003) and LevR of Lactobacillus casei (Maze et al, 2004) activates an Eσ70 rather than a Eσ54 complex.
Phosphotransferase activity of the PTS is essential for repression of the dha operon
Disruption of ptsI, the gene for EI of the PTS, increased LacZ production 30-fold (Figure 5), confirming the observed upregulation of DhaK and DhaL in the proteome of E. coli ΔptsI (Beutler et al, 2001). Disruption of dhaM had the same effect (Figure 5). Repression was restored by plasmid-encoded DhaM and EI. DhaM contains three histidines, which sequentially transfer phosphate from phospho-HPr of the PTS to the Dha kinase (Gutknecht et al, 2001). All three are essential for repression of dha by DhaM, indicating that the same phospho-relay is utilized for control as well as catalytic activity (Figure 5). The ΔptsIΔdhaR double mutant had no reporter gene activity (Figure 5), indicating that the ΔdhaR deletion is dominant over the ΔptsI deletion and that DhaR and EI of the PTS contribute to the same regulation pathway. EI and DhaM thus are negative regulators, and DhaR is a positive regulator (activator) of dha operon transcription. A functional phosphotransferase relay is essential for complete repression of the dha operon. The absence of the inducer Dha is necessary but not sufficient for repression.
Figure 5.
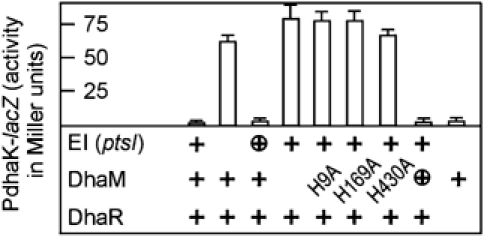
DhaM and EI of the PTS negatively control dhaKLM transcription. Expression of EI and DhaM from the chromosomal gene is indicated with +, and expression from a low-copy-number plasmid with ⊕. The DhaM point mutants were encoded by low-copy-number plasmids. The dha operon is constitutively active in the absence of a functional DhaM or EI, and inactive in the absence of DhaR. The average standard deviation for all values larger than 10 Miller units is 8.4%.
The catalytic DhaL and DhaK subunits are coactivators and corepressors of DhaR–controlled gene expression
The dhaK, dhaL and dhaM genes were interrupted, and the mutants complemented with plasmids coding for DhaK, DhaL and DhaM. To avoid gene-dosage effects, the proteins were expressed from low–copy-number plasmids (3–4 copies; Lutz and Bujard, 1997) under the control of the PLtetO-1 promoter induced with 100 μg/ml anhydrotetracyline. Reporter gene expression was assayed in the absence and presence of the inducer Dha (Figure 6, open and solid bars) and in ptsI cells, which did not produce EI of the PTS (gray bars). Induction with Dha or deletion of ptsI resulted in activation of reporter gene expression (Figure 6, column a). Disruption of dhaM resulted in constitutive activation of LacZ activity (c). Disruption of dhaKL resulted in constitutive repression, which could not be relieved either by induction with Dha or by interruption of ptsI (d). Inducibilty was restored when DhaK and DhaL were expressed from a plasmid (e and l). The difference between (a) and (e) most probably reflects the dosage difference between chromosome- and plasmid-borne genes. The H230K mutant of DhaK, which cannot bind Dha (Siebold et al, 2003), repressed LacZ activity but could no longer be induced with Dha (f).
Figure 6.
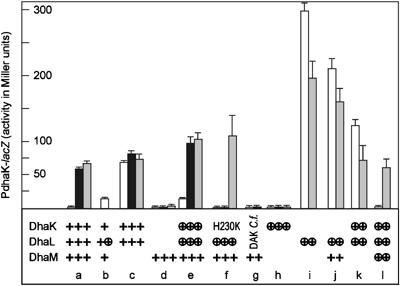
DhaL is an activator and DhaK is a repressor of dha transcription. Expression of DhaK, DhaL and DhaM from the chromosomal gene is indicated with +, and expression from a low-copy-number plasmid with ⊕. The DhaK H230K mutant and the ATP-dependent Dha kinase (DAK) from C. freundii were expressed from low-copy-number plasmids. E. coli ptsI+ were grown without (open bars) and with 2 mM Dha (solid bars). E. coli ΔptsI lacking EI of the PTS were grown without Dha (gray bars). The average standard deviation for all values larger than 10 Miller units is 12%.
Surprisingly, expression of DhaL alone sufficed to strongly activate the dha operon, while DhaK alone had no effect at all (Figure 6, columns h and i). Superactivation by DhaL was four times stronger than physiological induction with Dha and the induction elicited by interruption of ptsI and dhaM (compare i with a and c). This overshooting activation was attenuated by coexpression of DhaM (j) or DhaK (k) and fully neutralized when DhaM and DhaK were expressed together (l, compare with a), suggesting that the activity of DhaL is antagonized by DhaK and DhaM. A mild activation by DhaL was also seen in wild-type cells containing an extra copy of dhaL on a plasmid (b). It will be shown below that DhaK and DhaL bind to DhaR in a mutually exclusive manner and that DhaL∷ADP is inactivated by DhaM-mediated phosphorylation.
Interruption of ptsI attenuated the DhaL-mediated overshoot of reporter gene activity (i–k, compare gray with open bars), which is the opposite of the effect in wild-type cells (a and e). Addition of glucose to the medium had a similar effect (results not shown). Repression of gene activity by glucose or a ptsI interruption is indicative of catabolite repression control. Inactivation of EI (ptsI) thus must have two antagonistic effects on the regulation of the dha operon. On the one hand, it increases dha expression by leaving DhaL∷ADP in the transcriptionally active form, and on the other hand, it decreases activation by catabolite repression. Dha activity stimulated to its maximum by DhaL∷ADP (i–k) can only be decreased (directly or indirectly) by catabolite repression, while fully repressed dha activity (a and e) can only increase with rising concentrations of DhaL∷ADP.
Addition of the inducer Dha (Figure 6, solid bars) had no effect on the constitutively high dha activity of dhaM mutants (c). Similarly, Dha did not further stimulate the strong dha activity of ptsI mutants and of the superactive DhaL mutants (results not shown). The ATP-dependent Dha kinase (DAK) of C. freundii did not activate transcription (Figure 6, column g). This indicates that unlike the DhaRs of C. freundii and E. coli (Figure 3A), the ATP- and PTS-dependent kinases are not exchangeable. A fusion protein between E. coli DhaK and DhaL, which displayed 10% of wild-type kinase activity, also had no transcription control activity (results not shown), suggesting that DhaL and DhaK must be independent subunits to act as coactivator and repressor. The autorepressor function of DhaR is not at all affected by the subunits of the Dha kinase (Figure 3B). DhaR remains autorepressing after deletion of the dhaKLM genes as well as in the presence of the activating DhaL subunit.
In summary, the genetic analyses of dhaKLM and dhaR transcription show the following: (i) DhaR is an autonomous autorepressor of its own synthesis, (ii) DhaR and DhaL are positive regulators of the dhaKLM operon, (iii) phosphorylated DhaM and DhaK are negative regulators, (iv) Dha is the inducer and (v) there is no evidence for DhaR being controlled by protein phosphorylation. Hence, DhaR activity is assumed to be controlled by protein–protein interaction.
DhaK and DhaL form protein complexes with the DhaR sensing domain
Full-length DhaR with a carboxy-terminal histidine tag and the sensing domain DhaR(N) (residues 1–318) with an amino-terminal histidine tag were purified. Full-length DhaR, which formed inclusion bodies, was sparingly soluble after purification and contaminated with proteolytic carboxy-terminal fragments (Figure 7A, lane 1). Therefore, the better soluble N-terminal sensing domain DhaR(N) was used for the experiments shown in Figures 7B and 9. Protein complexes between histidine-tagged DhaR and kinase subunits were affinity captured with Ni2+-NTA beads (pull-down) and characterized by gel electrophoresis (Figure 7).
Figure 7.
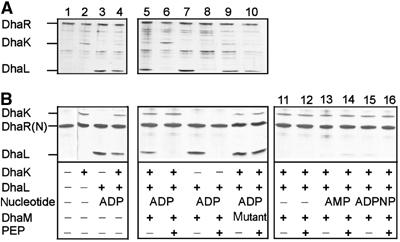
Association of DhaK and DhaL with full-length DhaR (A) and DhaR(N) signalling domain (B) is modulated by the phosphorylation state of DhaL∷ADP. Protein mixtures were preincubated, and complexes containing hexahistidine-tagged DhaR or DhaR(N) were affinity captured with Ni2+-NTA-beads, analyzed by denaturing polyacrylamide gel electrophoresis and visualized by silver staining. Lane 1, pull-down of purified DhaR with background; lanes 2 and 3, binary complexes of DhaK and DhaL∷ADP with DhaR; lane 4, competition between DhaK and DhaL∷ADP for DhaR; lanes 5–10, the PTS-dependent phosphorylation of DhaL∷ADP reduces DhaL affinity for DhaR. Phosphorylation of DhaL∷ADP requires PEP and active DhaM (lanes 6 and 8). The mutant DhaM(H9A, H169A, H430A) is not active (lanes 9 and 10). Lanes 11–16, DhaL apoenzyme (without ADP) and apoenzyme complemented with AMP or ADPPNP do not bind to DhaR (shown only with DhaR(N). The concentrations were as follows: DhaR (full length) 100 nM, DhaR(N) 100 nM, DhaK 300 nM, DhaL∷ADP 400 nM, apo-DhaL 400 nM, ADP 1 μM, AMP 1 μM, ADPNP 10 μM, DhaM 5 nM, PEP 1 mM, and EI and HPr 5 nM.
Figure 9.
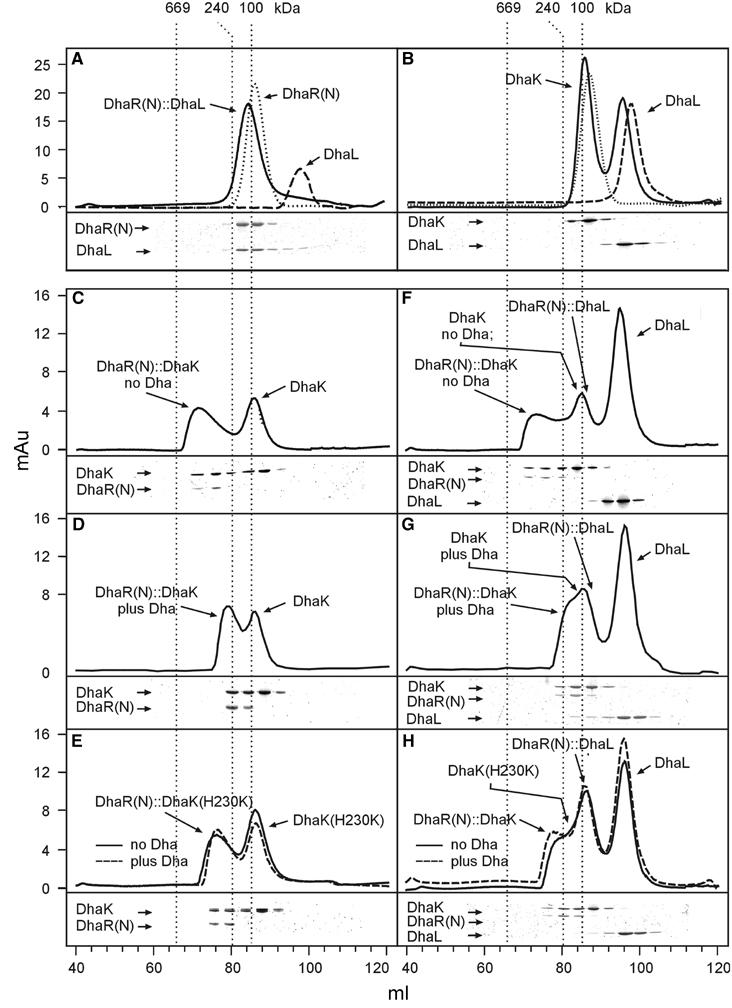
Characterization of DhaL∷DhaR(N), DhaK∷DhaR(N) and DhaK∷DhaL binary complexes by gel filtration chromatography. Fractions were analyzed by gel electrophoresis. The gels shown refer to the chromatograms in solid lines. (A) DhaL∷DhaR(N) complex (solid, 20 μM, 1:1 subunit ratio), dimeric DhaR(N) alone (dotted, 20 μM) and DhaL alone (dashed, 20 μM). (B) DhaK and DhaL (solid, 40 μM, 1:1 subunit ratio), dimeric DhaK alone (dotted, 40 μM) and DhaL alone (dashed, 40 μM). DhaK and DhaL together migrate slightly faster than each subunit alone because of a weak association (Kd ∼0.15 μM; Garcia-Alles et al, 2004). (C–E) Binary complexes between DhaR(N) (3 μM) and DhaK (15 μM) without (C) and with 0.1 mM Dha (D), and between DhaR(N) and the DhaK(H230K) mutant (E: solid, without Dha; dashed, with Dha). (F–H) Complex formation in the presence of all three subunits, DhaR(N) (3 μM), DhaL (30 μM) and DhaK (15 μM). (F) A large DhaR∷DhaK complex forms in the absence of Dha. Excess DhaK and DhaL follow behind. A DhaR∷DhaL complex forms in the presence of Dha (G) and in the presence of DhaK(230K) (H). Notice how DhaL is shifted from the elution volume of the monomer (98 ml) to the elution volume of the DhaL∷DhaR(N) complex (82 ml), which partially overlaps with the volume of the DhaK dimer (86 ml). The gel filtration column was calibrated with thyroglobulin (669 kDa), catalase (240 kDa) and hexokinase (100 kDa). For details, see text.
Full-length DhaR and DhaR(N) captured both DhaK and DhaL∷ADP with comparable affinity (Figure 7, lanes 2–4). Phosphorylation of DhaL∷ADP by DhaM and phosphoenolpyruvate (PEP, in the presence of EI and HPr) resulted in dissociation of the DhaR∷DhaL complex (compare lanes 7 and 8) and the concomitant association of more DhaK (compare Figure 7A, lanes 5 and 6). In the presence of inactive DhaM (H9A, H169A, H430A), DhaL∷ADP was not phosphorylated and the DhaL∷DhaR(N) complex remained stable (lanes 9 and 10). The addition of Dha had no effect on the interaction between DhaK and DhaR as long as Dha could not be phosphorylated (results not shown).
To further elucidate the function of ADP, the nucleotide was removed by gel filtration of DhaL in the presence of EDTA, and apo-DhaL was then reconstituted with AMP and ADPNP. The phosphorylation reaction was started by the addition of PEP. Neither apo-DhaL nor DhaL supplemented with AMP and ADPNP (adenylylimidodiphosphonate) bound to DhaR(N) (Figure 7B, lanes 11–16). Both nucleotides have been shown to form a complex with DhaL, but DhaL∷AMP cannot be phosphorylated by DhaM and ADPPNP cannot be hydrolyzed (Bächler et al, to be published). Identical results were obtained in pull-down experiments performed without DhaK (results not shown).
The observation that DhaK does not interfere with complex formation between DhaR(N) and DhaL∷ADP in vitro is at variance with the genetic analysis where interruption of dhaK resulted in DhaL-dependent superactivation of the dha operon in vivo (Figure 6, columns i and j). In vivo, DhaL is expected to be in the DhaL∷ATP form (if EI and DhaM are active and Dha is absent) and thus without affinity for DhaR. The inconsistency between in vitro and in vivo can be explained by the different DhaR/DhaL ratios. In the pull-down experiment, DhaL and DhaR(N) are present in equimolar amounts and DhaL is kept in the phosphorylated form by DhaM-mediated rephosphorylation. The small proportion of DhaL∷ADP, which might be present in the steady state (due to the intrinsic ATPase activity of DhaL; Bächler et al, to be published) and which might bind to DhaR(N), is below the limit of gel electrophoretic detection (Figure 7). In vivo, however, DhaL is present in a large excess over DhaR, and even a small proportion of DhaL∷ADP might suffice to saturate DhaR in the absence of competing DhaK.
The pull-down experiments and the genetic analysis (above) suggest that binding of DhaL and DhaK to DhaR(N) are mutually exclusive. To confirm this, the soluble complexes between DhaR(N), DhaK and DhaL were further analyzed by native protein gel electrophoresis (Schagger and Pfeiffer, 2000). Two complexes of low electrophoretic mobility formed when DhaR(N) and DhaK were present in a 1:1 ratio. These complexes disappeared when one subunit was in excess over the other (Figure 8A). The DhaR(N)∷DhaK complex could be dissociated with less than a four-fold molar excess of DhaL∷ADP (Figure 8B), confirming that binding of DhaL∷ADP and DhaK to DhaR(N) are mutually exclusive. The DhaL∷DhaR(N) complex could not be detected, because DhaL∷ADP dissociated during electrophoresis. This complex could, however, be characterized by gel filtration, as shown below (Figure 9A).
Figure 8.
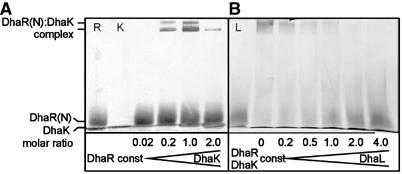
Native protein gel electrophoresis of DhaK∷DhaR complexes. (A) Titration of DhaR(N) (5 μM constant concentration) with DhaK (0, 0.1, 1.0, 5.0 and 10 μM). (B) Dissociation of the 1:1 DhaK∷DhaR(N) complex (5 μM constant concentration) with increasing concentrations of DhaL∷ADP (0, 1.0, 2.5, 5.0, 10 and 20 μM). Lanes R, K, L, 5 μM DhaR(N), 10 μM DhaK, 20 μM DhaL.
In conclusion, DhaL∷ADP and DhaK bind to the sensing domain of DhaR in a mutually exclusive manner. PTS-dependent phosphorylation of DhaL∷ADP inhibits binding of DhaL. This is congruent with the genetic analysis, which showed that the dha operon was activated under conditions that favor the formation of DhaL:ADP, namely addition of the substrate Dha and interruption of the PTS-dependent phosphorylation cascade.
Stoichiometry of the complexes between DhaR sensing domain, DhaK and DhaL
Gel filtration on a calibrated Superdex-200 column was used to characterize the stoichiometry of the complexes between DhaL, DhaK and DhaR(N). Full-length DhaR in 500 mM NaCl at pH 9.3 eluted as a homodimer but precipitated under the conditions where complexes with DhaL and DhaK might form (not shown). The better soluble sensing domain, DhaR(N), formed stable complexes with DhaL and DhaK (Figure 9). In the presence of DhaR(N), DhaL eluted together with DhaR(N) in the relative elution volume corresponding to 125 kDa, a molecular weight compatible with DhaR(N)2DhaL1 stoichiometry (Figure 9A). In contrast, DhaL did not form a complex of comparable stability with DhaK (Figure 9B). DhaK and DhaL eluted as separated peaks slightly ahead of the isolated subunits, as expected of a weak complex that dissociated during gel filtration. DhaK and DhaR(N) in a 5:1 molar ratio formed a complex that eluted in a skewed peak (Figure 9C) with a maximum corresponding to an estimated molecular weight of 466 kDa. This and the DhaK to DhaR(N) ratio point to a dimer [DhaR(N)2∷DhaK2]2 of DhaR(N)2∷DhaK2 protomers. Addition of Dha reduced the estimated mass of the complex to 200 kDa, while DhaR(N) and DhaK remained associated (Figure 9C). Binding of Dha dissociated the dimer into protomers but did not dissociate the protomer into DhaR(N) and DhaK subunits. This resistance of the complex between DhaR(N) and DhaK was also noticed in pull-down assays performed in the presence of Dha (results not shown). DhaK(H230K) and DhaR(N) formed a complex that eluted in an intermediate volume (280 kDa) between those of the wild-type with and without Dha (Figure 9, compare E with C and D), suggesting that protomers containing DhaK(H230K) are in a monomer–dimer equilibrium. Addition of Dha had no effect (Figure 9E), as expected of a DhaK mutant, which does not bind Dha (Siebold et al, 2003).
Finally, the complexes formed in the presence of all three subunits were analyzed (Figure 9F–H). Similar to the conditions in vivo (Table I), DhaL and DhaK were present in molar excess over DhaR(N), all other conditions being identical with those of Figure 9C–E. The most salient feature of Figure 9F–H is that DhaK and DhaL never occur together in fractions containing high-molecular-weight complexes between DhaR(N) and DhaK. This indicates that heterotrimeric complexes between the three subunits do not form and is consistent with mutually exclusive binding of DhaL and DhaK to DhaR(N). In the absence of Dha, DhaL eluted late as monomer and far behind the 466 kDa DhaR(N)∷DhaK complex (Figure 9F). In the presence of Dha (Figure 9G), DhaL eluted earlier than the monomer but still behind the forward edge of the DhaR(N)2∷DhaK2 protomer. This indicates that DhaL formed a DhaR(N)2DhaL1 complex (as in Figure 9A) by displacing DhaK from DhaR(N) but did not bind to the DhaR(N)2∷DhaK2 protomer. In the presence of DhaK(H230K), the fraction of monomeric protomers is small and does not increase upon the addition of Dha. Hence, only a small amount of DhaR(N)2DhaL1 could form (Figure 9, compare H with F and G).
The oligomeric composition of the binary complexes was further characterized by equilibrium sedimentation. The DhaK dimer had a molecular mass of 75 kDa (theoretical 79 kDa) and the DhaR(N) dimer a mass of 69 kDa (theoretical 74 kDa). DhaR(N) and DhaL formed a complex of 96 kDa corresponding to DhaR(N)2∷DhaL1 stoichiometry. The DhaK∷DhaR(N) complex had a mass of 580 kDa corresponding to a [DhaR(N)2DhaK2]4 tetramer. A complex of a comparable size could be identified by gel filtration when the DhaR(N) to DhaK ratio was 1:1 instead of 5:1 (results not shown). The molecular weights estimated from the relative elution volumes of gel filtration are consistently larger than those obtained by analytical ultracentrifugation, but they afford the same, most probable subunit stoichiometry.
All gel filtration experiments were performed with the isolated sensor domain. Therefore, it cannot be excluded that the full-length DhaR in complex with the operator sequence might display additional oligomerization states that are not observable with the sensor domain alone. However, the fact that a DhaR mutant without a sensor domain has reduced autorepressor activity (Figure 3B) suggests that sensor domain-dependent dimerization is decisive for strong binding to the operator.
Conclusion
The novelty of transcription regulation by the PTS-dependent Dha kinase is that (i) two catalytic subunits of a metabolic enzyme form a coactivator/corepressor complex with a transcription activator and (ii) transcription activation is coupled to the enzymatic turnover of the substrate. DhaK and DhaL∷ATP are the catalytic subunits that bind Dha and phosphorylate it, respectively. DhaL∷ADP is then rephosphorylated by DhaM, a multiphosphoryl protein of the PTS (Figure 10A). DhaR is a transcription activator from the family of enhancer binding proteins. The strong autorepression suggests that the occupancy of the operator by DhaR is quite high (Figure 3B). DhaK binds to the sensing domain of DhaR (Figure 10B) and thereby keeps DhaR in a transcription inactive state. Dephosphorylated DhaL∷ADP displaces DhaK and activates DhaR. DhaK and DhaL function as sensor and discriminator subunits for Dha, glyceraldehyde and other chemically reactive short-chain carbonyl compounds (Garcia-Alles et al, 2004). The double-check mechanism of binding and turnover increases the selectivity such that compounds that only bind but are not phosphorylated would not turn on the dha operon. So far, only a few enzymes with double roles in catalysis and transcription control have been described. Mammalian glyceraldehyde 3-phosphate dehydrogenase is part of a coactivator complex that confers redox dependence on the transcription of histone genes (Zheng et al, 2003). Glutamine synthetase (GS) of Bacillus subtilis, when feedback-inhibited by glutamine or AMP, inhibits binding of the TnrA repressor to DNA (Wray et al, 2001). The ATPase subunit of the maltose transporter (MalK), the β cystathionine-lyase MalY and the acetyl esterase Aes negatively control the transcriptional activator MalT of the E. coli maltose regulon (Boos and Shuman, 1998; Schreiber et al, 2000; Joly et al, 2002; Schlegel et al, 2002).
Figure 10.
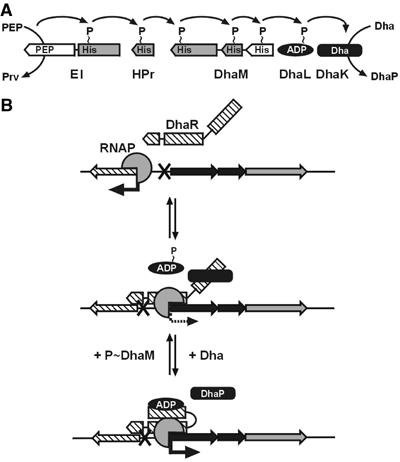
Model of transcription control by the E. coli Dha kinase. (A) Phosphoryl flow from PEP and the general PTS proteins EI and HPr to DhaM and DhaL∷ADP of the Dha kinase. PTS proteins are shown as rectangular boxes. The Dha kinase subunits, which are homologous to the domains of the ATP-dependent kinases, are in black. Sequence-related domains of DhaM and PTS domains/subunits are in gray. (B) Model of transcription control by DhaR. DhaR is an (auto)repressor of the dhaR gene and an activator of the dhaKLM operon. DhaL∷ADP is a coactivator of DhaR, and DhaK is an antagonist of DhaL∷ADP. Dha is the inducer that by binding to DhaK reduces the affinity of DhaK for DhaR, and by dephosphorylation of DhaL∷ATP increases the affinity of DhaL∷ADP for DhaR. RNAP, RNA polymerase.
DhaL∷ADP and DhaK are functionally analogous to the IIB and IIC domains of PTS transporters (for review, see Postma et al, 1996) where IIB transfers phosphate to sugars being translocated by the IIC domains. Some IIB domains interact with DNA binding proteins. Dephosphorylated IICBGlc, for instance, forms a complex with the repressor Mlc and by sequestering it promotes expression of glucose-related genes (Lee et al, 2000; Tanaka et al, 2000; Nam et al, 2001; Plumbridge, 2002). The phosphorylated IIB domain of BglF can transfer phosphate to and thereby inactivate the antiterminator BglG. In the absence of a phosphorylatable substrate, transcription of the bgl operon is thus aborted (Gorke, 2003).
What then is the biological function of the PTS-dependent Dha kinase? The answer may be found in the observations that (i) E. coli DhaR can control the gene for the ATP-dependent C. freundii kinase, (ii) the ATP-dependent C. freundii kinase cannot control E. coli DhaR activity and (iii) the genome of K. pneumoniae contains genes for a PTS-dependent Dha kinase, for DhaR and for a C. freundii-like ATP-dependent kinase. Based on this, we suggest that in K. pneumoniae, the PTS-dependent kinase functions as ‘sensor-kinase' to control expression of the ATP-dependent ‘metabolic kinase'. In E. coli, DhaK and DhaL function as enzyme subunits that feedback-control their own expression. By switching from ATP to the PTS as a source of high-energy phosphate, the Dha kinase becomes integrated in and put under the control of the PTS, an energy-transducing system involved in carbohydrate uptake and control of carbon metabolism (for reviews, see Saier et al, 1996; Deutscher et al, 2002). It is conceivable that hemiaminal formation, the mechanism to discriminate between Dha and glycerol (Garcia-Alles et al, 2004), has been invented only once in evolution and was then used for both catalysis and signalling. Its use for different biological tasks thus is an interesting example of parsimony in evolution.
Materials and methods
Plasmids and bacterial strains
The relevant properties of plasmids and E. coli strains are given in Table II. The details of their construction are provided in Supplementary data. Standard procedures were used for plasmid purification, restriction analysis, ligation and transformation (Sambrook et al, 1989). Genomic DNA of E. coli W3110 and C. freundii (gift of Dr R Daniel, University of Göttingen) was used as template for PCR amplifications. The genes were inactivated with PCR products according to Datsenko and Wanner (2000) as detailed in Supplementary data.
Table 2.
Strains and plasmids
| Strain/plasmid | Relevant genotype or structures | Source/reference |
|---|---|---|
| Bacterial strains | ||
| MC4100 | F-, araD139, Δ(argF-lac)U169, prsL150, relA1, deoC1, rbsR, fthD5301, fruA25, λ- | Lab stock |
| TH074 | as MC4100 but ΔptsI | Hesterkamp and Erni (1999) |
| DHB6521 | F-, λ-, λs, Δlac(MS265), mel, NalAr, supF58 (=su III+) | Boyd et al (2000) |
| CBZ | MC4100 attBλ∷PdhaK E. coli-lacZ-bla | This work |
| CBZΔI | TH074 attBλ∷PdhaK E. coli-lacZ-bla | This work |
| CBZΔR | As CBZ but ΔdhaR | This work |
| CBZΔKL | As CBZ but ΔdhaKL | This work |
| CBZΔIΔKL | As CBZΔI but ΔdhaKL | This work |
| CBZΔKLM | As CBZ but ΔdhaKLM | This work |
| CBZΔIΔKLM | As CBZΔI but ΔdhaKLM | This work |
| CBZΔM | As CBZ but ΔdhaM | This work |
| CBZΔrpoN | As CBZ but ΔrpoN | This work |
| CBZΔIΔR | As CBZΔI but ΔdhaR | This work |
| CBZΔIΔM | As CBZΔI but ΔdhaM | This work |
| CBΔKLM | As MC4100 but ΔdhaKLM | This work |
| DH5αZ | F-, λ-, end A1, hsdR17, hsdM+, supE44, thi1, recA96, relA1 Δ(argF lacZYA)U169, Φ80d, Δ(lacZ)M15, tetR+ | Lutz et al (1997) |
| Plasmids | ||
| pBR322 | bla, tetRA, oriR ColE1 | Lab stock |
| pET28 | PT7, lacIq, kan, oriR ColE1, His tag | Novagen |
| pZS*24-MCS-1 | Plac/ara-1, oriR SC101*, kan | Lutz et al (1997) |
| pZE21-MCS-1 | PLtetO-1, oriR colE1, kan | Lutz et al (1997) |
| pZA31-Luc | luc under control of PLtetO-1 (oriR, p15A, cat) | Lutz et al (1997) |
| pJF K(H230K)L | dhaK(H230K)L under control of Ptac (oriR ColE1, bla) | This work |
| pJF K H6 | dhaK with C-terminal H6 tag under control of Ptac (oriR ColE1, bla) | This work |
| pJF L H6 | dhaL with C-terminal H6 tag under control of Ptac (oriR ColE1, bla) | This work |
| pJF M H6 | dhaM with C-terminal H6 tag under control of Ptac (oriR ColE1, bla) | This work |
| pJFM(H9A,H169A, H430A) | dhaM (H9A, H169A, H430A) under control of Ptac (oriR ColE1, bla) | This work |
| pJFRH6 | dhaR with C-terminal H6 tag under control of Ptac (oriR ColE1, bla) | This work |
| pAC R Ec cterm | dhaR (aa 312–642) under control of ParaB, (oriR pACYC177, kan) | This work |
| pAC K | dhaK under control of ParaB, (oriR pACYC177, kan) | This work |
| pAC M | dhaM under control of ParaB, (oriR pACYC177, kan) | This work |
| pAC M(H9A) | dhaM H9A under control of ParaB, (oriR pACYC177, kan) | This work |
| pAC M(H169A) | dhaM H169A under control of ParaB, (oriR pACYC177, kan) | This work |
| pAC M(H430A) | dhaM H430A under control of ParaB, (oriR pACYC177, kan) | This work |
| pAC ptsI | ptsI under control of ParaB, (oriR pACYC177, kan) | This work |
| pAC R Ec | dhaR E. coli under control of ParaB, (oriR pACYC177, kan) | This work |
| pAC R Cf | dhaR C. freundii under control of ParaB, (oriR pACYC177, kan) | This work |
| pZStetR MCS | PLtetO-1, (MCS, tetR, oriR SC101*, cat) | This work |
| pZS KLM | dhaKLM under control of PLtetO-1, (tetR, oriR SC101*, cat) | This work |
| pZS KL | dhaKL under control of PLtetO-1, (tetR, oriR SC101*, cat) | This work |
| pZS L | dhaL under control of PLtetO-1, (tetR, oriR SC101*, cat) | This work |
| pZS K(H230K)L | dhaK (H230 K)L under control of PLtetO-1, (tetR, oriR SC101*, cat) | This work |
| pZS KL cf | dhaKL of C. freundii under control of PLtetO-1, (tetR, oriR SC101*, cat) | This work |
| pET28 R(N) | dhaR (aa 1–318) under control of PT7 (lacIq, oriR pBR322, kan) | This work |
| pBR PdhaK-lacZ | PdhaK E. coli-lacZ, (oriR ColE1, kan) | This work |
| pBR PdhaR-lacZ | PdhaR E. coli-lacZ, (oriR ColE1, kan) | This work |
Cell growth and media
Cells were grown at 37°C Luria–Bertani (LB) broth containing appropriate antibiotics (100 μg/ml ampicillin, 25 μg/ml chloramphenicol, 50 μg/ml kanamycin). To study gene regulation with PdhaK -lacZ and PdhaR -lacZ reporter strains, a carbon-limited MOPS medium was used: 75 mM morpholine-propansulfonic acid (MOPS) pH 7.5, 0.25% casamino acids (carbon-limited culture), 1 mM K2HPO4, 34 mM NaCl, 40 mM KCl, 20 mM (NH4)2SO4, 1 μM FeSO4, 3 mM MgSO4, 1 μM ZnCl2, 10 μM CaCl2. MOPS agar plates contained 1% (w/v) casamino acids. The PLtetO-1 promoter of pZS plasmids was induced with 100 ng/ml anhydro-tetracycline.
Determination of β-galactosidase (LacZ) activity
LacZ activity was determined by the method of Miller (1992) in a microtiter plate. Cultures were grown overnight at 37°C in carbon-limited MOPS medium (containing 2 mM Dha where indicated), chilled on ice and diluted 1:2 to 1:10 in buffer Z (Miller, 1992) in a final volume of 1 ml. Cells were permeabilized with two drops of chloroform, one drop of 0.1% SDS and 15 s vortexing. In all, permeabilized cells (110 μl/well) were incubated for 5 min at 28°C, and 30 μl o-nitrophenyl-β-D-galactopyranoside (ONPG), 4 mg/ml in buffer Z) was added to start the reaction, and absorbance at 420 nm was monitored continuously in a Spectramax 250 Plate reader. The measured absorbance values were converted into Miller units (MU=1.81(−0.015+1.86 × ODmicrotiter 420, correction for omitted alkalization with Na2CO3 and optical path length). The listed LacZ activities are the averaged measurements of at least three independent cultures and assays.
Protein purification
E. coli MC4100 transformed with plasmid pJFRH6 was used to overproduce full-length DhaR with a C-terminal hexahistidine tag. E. coli BL21(DE3) transformed with plasmid pET28 R(N) was used to overproduce the N-terminal sensing domain DhaR(N) with an N-terminal hexahistidine tag. Cells were grown in 1 l of LB medium in an Erlenmeyer flask on a rotary shaker at 37°C. When the cells had reached A550=0.8, protein expression was induced with 0.2 mM isopropyl-β-D-thiogalactopyranoside, and incubation was continued for 20 h at 18°C. Cells were harvested by centrifugation (7500 g for 20 min at 4°C), resuspended in 2.5 ml/g wet weight of buffer A (20 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM β-mercaptoethanol, 10% glycerol, 0.2 mM PMSF) and lysed by two passages through a French pressure cell (1000 p.s.i.). Cell debris was removed by low-speed centrifugation (12 000 × g, 10 min, 4°C), membranes by high-speed centrifugation (360 000 g, 1 h, 4°C), and the supernatant containing DhaR or DhaR(N) was mixed with Ni2+ NTA affinity resin (equilibrated with buffer A) and shaken for 0.5 h at 20°C. The resin was washed with two changes of buffer A containing 25 and 40 mM imidazole, respectively. The protein was eluted with 200 mM imidazole in buffer A, and DhaR and DhaR(N) containing fractions were pooled and dialyzed against buffer B (20 mM Tris–HCl pH 8.0, 300 mM NaCl, 1 mM DTT, 10% glycerol, 2 mM EDTA). The yield of DhaR(N) was ∼30 mg of pure DhaR(N) per liter of cell culture. The yield of full-length DhaR was ∼1 mg of 80% pure protein, which was contaminated with proteolytic carboxy-terminal fragments and could not be concentrated without precipitation.
DhaK, DhaK(H230K), DhaL, DhaM and DhaM(H9A, H169A, H430A) were purified as described by Gutknecht et al (2001), and EI and HPr were purified as described by Mao et al (1995) with one important modification: all buffers used for DhaL purification were supplemented with 1 mM MgCl2 and 0.01 mM ADP, and EDTA was omitted. This prevents the loss of the ADP cofactor and it greatly stabilized DhaL against denaturation/aggregation.
Gel electrophoresis and Western blot analysis
Blue native protein gel electrophoresis. Complexes between DhaR(N), DhaK and DhaL were formed as described in Affinity purification. Proteins were separated on a blue native protein gel (Schagger and Pfeiffer, 2000) (5–13% polyacrylamide gradient (acrylamide:bisacrylamide, 48:1.5, 85 V, 4 h, 4°C)) and silver stained.
Denaturing gel electrophoresis. Protein samples were mixed with one-fifth volume of 6 × sample loading buffer, boiled for 5 min and separated on an SDS–17.5% polyacrylamide gel. Gels were stained with RuBP (ruthenium II tris (bathophenanthroline disulfonate)) and scanned with a fluorescence scanner (Fuji FLA-3000) (Lamanda et al, 2004). The fluorescence intensity per mol of DhaL and DhaK was 100 and 120% of the intensity per mol of DhaR(N), respectively, as determined with calibration gels of purified proteins.
Western blot analysis. E. coli MC4100 were grown overnight in LB medium without or with 2 mM Dha. Strain CBΔKLM was used as negative control. Washed cells were lysed by sonication, cell debris removed by Eppendorf centrifugation and the supernatant separated on a 17.5% polyacrylamide gel. Proteins were semidry electrotransferred (Bio-Rad) to nitrocellulose, and DhaK, DhaL and DhaM detected with subunit-specific rat antisera, horseradish peroxidase-conjugated anti-rat IgG (DAKO) and 4-chloro-1-naphthol staining. The protein amount was determined by densitometry and comparison of the staining intensity with three standard curves obtained with known amounts of purified DhaK, DhaL and DhaM, which were separated in parallel. The number of molecules per cell was calculated from the amount of protein on the blot and the cell volume-derived dry weight (Neidhardt and Umbarger, 1996). The dry weight was determined as follows: cells from 2 ml of culture were collected on glass fiber filters (GF/F Whatman) under suction, washed with 2 × 5 ml water, dried and weighed.
Affinity purification (pull-down) assays
Complexes were formed by incubation of DhaK and DhaL with DhaR(N) or DhaR in 0.4 ml buffer C (10 mM HEPES, pH 7.5, 40 mM NaCl, 5 mM MgCl2, 2.0 mM DTT, 0.05% Triton X-100) for 20 min at 20°C. A 50 μl portion of a 50% (v/v) suspension of Ni2+-NTA resin in buffer C was added to each sample and the incubation continued for 30 min on a rocking platform. The Ni2+-NTA resin was collected by Eppendorf centrifugation (1 min, 3000 r.p.m.), washed four times with 0.5 ml of buffer C, and the bound proteins were eluted with two times 25 μl 50 mM EDTA. The eluted subunits were separated by gel electrophoresis in SDS and silver stained.
Gel filtration
Complexes between DhaR(N), DhaK and DhaL, as specified in the figure legends, were formed by incubation in 1 ml of buffer D (10 mM HEPES pH 7.5, 150 mM NaCl, 6 mM MgCl2, 2 mM DTT, 100 μM ADP) for 10 min at 20°C. Protein complexes were separated on a Superdex 200 16/60, column (Pharmacia) equilibrated with buffer D containing 10 μM ADP at a flow rate of 1 ml/min and at 20°C. Fractions (2 ml) were collected. Aliquots (25 μl) were used for gel electrophoretic analysis.
Analytical ultracentrifugation
Molecular mass (Mr) and sedimentation coefficients (s20,w) of DhaL, DhaK, DhaR(N), DhaL∷DhaR(N) and DhaK∷DhaR(N) were determined by sedimentation velocity (SV) and sedimentation equilibrium (SE) centrifugation using a Beckman XL-A analytical ultracentrifuge equipped with an optical absorbance system. The rotor speeds for SV were 52 000 r.p.m. (DhaL), 54 000 r.p.m. (DhaR(N), DhaK) and 48 000 r.p.m. (DhaR(N)∷DhaK complex), and the temperature was 20°C. Protein concentration was 0.5 mg/ml in buffer E (10 mM HEPES pH 7.5, 40 mM NaCl, 6 mM MgCl2, 0.5 mM DTT). SE was measured at protein concentrations between 0.4 and 0.75 mg/ml in buffer D. The rotor speeds were 24 000 r.p.m. (DhaL), 16 000 r.p.m. (DhaR(N), DhaK) and 9000 r.p.m. (DhaR(N)∷DhaL and DhaR(N)∷DhaK complexes), and the temperature was 20°C. A partial specific volume (v) of 0.73 cm3/g was used. In experiments with DhaL, 50 μM ADP was added to buffer D.
Supplementary Material
Supplementary Material
Acknowledgments
The C. freundii genomic DNA was a generous gift from Dr Rolf Daniel (University of Göttingen). This work was supported by the Swiss National Science Foundation grant 3100A0-105247.
References
- Beutler R, Kämpfer U, Schaller J, Erni B (2001) Heterodimeric dihydroxyacetone kinase from a ptsI mutant of Escherichia coli. Microbiology 147: 249–250 [DOI] [PubMed] [Google Scholar]
- Boos W, Shuman HA (1998) Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev 62: 204–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D, Weiss DS, Chen JC, Beckwith J (2000) Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J Bacteriol 182: 842–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD (2000) The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J Bacteriol 182: 4129–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, Stuertz K, Gottschalk G (1995) Biochemical and molecular characterization of the oxidative branch of glycerol utilization by Citrobacter freundii. J Bacteriol 177: 4392–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Galinier A, Martin-Verstraete I (2002) Carbohydrate uptake and metabolism. In Bacillus Subtilis and Its Closest Relatives, Sonenshein AL, Hoch JA, Losick R (eds) pp 129–150. Washington, DC: ASM Press [Google Scholar]
- Forage RG, Lin EC (1982) DHA system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J Bacteriol 151: 591–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Alles LF, Siebold C, Nyffeler TL, Flukiger-Bruhwiler K, Schneider P, Burgi HB, Baumann U, Erni B (2004) Phosphoenolpyruvate- and ATP-Dependent dihydroxyacetone kinases: covalent substrate-binding and kinetic mechanism. Biochemistry 43: 13037–13045 [DOI] [PubMed] [Google Scholar]
- Gorke B (2003) Regulation of the Escherichia coli antiterminator protein BglG by phosphorylation at multiple sites and evidence for transfer of phosphoryl groups between monomers. J Biol Chem 278: 46219–46229 [DOI] [PubMed] [Google Scholar]
- Gutknecht R, Beutler R, Garcia Alles LF, Baumann U, Erni B (2001) The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J 20: 2480–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesterkamp T, Erni B (1999) A reporter gene assay for inhibitors of the bacterial phosphoenolpyruvate: sugar phosphotransferase system. J Mol Microbiol Biotechnol 1: 309–317 [PubMed] [Google Scholar]
- Huang SH, Chen YH, Kong G, Chen SH, Besemer J, Borodovsky M, Jong A (2001) A novel genetic island of meningitic Escherichia coli K1 containing the ibeA invasion gene (GimA): functional annotation and carbon-source-regulated invasion of human brain microvascular endothelial cells. Funct Integr Genomics 1: 312–322 [DOI] [PubMed] [Google Scholar]
- Jin RZ, Lin ECC (1984) An inducible phosphoenolpyruvate: dihydroxyacetone phosphotransferase system in Escherichia coli. J Gen Microbiol 130: 83–88 [DOI] [PubMed] [Google Scholar]
- Joly N, Danot O, Schlegel A, Boos W, Richet E (2002) The Aes protein directly controls the activity of MalT, the central transcriptional activator of the Escherichia coli maltose regulon. J Biol Chem 277: 16606–16613 [DOI] [PubMed] [Google Scholar]
- Lamanda A, Zahn A, Röder D, Langen H (2004) Improved ruthenium II tris (bathophenantroline disulfonate) staining and destaining protocol for a better signal-to-background ratio and improved baseline resolution. Proteomics 4: 599–608 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Boos W, Bouché JP, Plumbridge J (2000) Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J 19: 5353–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubini DG, Christen P (1979) Paracatalytic modification of aldolase: a side reaction of the catalytic cycle resulting in irreversible blocking of two active-site lysyl residues. Proc Natl Acad Sci USA 76: 2527–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Martin J (2002) AAA proteins. Curr Opin Struct Biol 12: 746–753 [DOI] [PubMed] [Google Scholar]
- Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q, Schunk T, Flükiger K, Erni B (1995) Functional reconstitution of the purified mannose phosphotransferase system of Escherichia coli into phospholipid vesicles. J Biol Chem 270: 5258–5265 [DOI] [PubMed] [Google Scholar]
- Maze A, Boel G, Poncet S, Mijakovic I, Le Breton Y, Benachour A, Monedero V, Deutscher J, Hartke A (2004) The Lactobacillus casei ptsHI47T mutation causes overexpression of a LevR-regulated but RpoN-independent operon encoding a mannose class phosphotransferase system. J Bacteriol 186: 4543–4555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JF (1992) A Short Course in Bacterial Genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Molin M, Norbeck J, Blomberg A (2003) Dihydroxyacetone kinases in Saccharomyces cerevisiae are involved in detoxification of dihydroxyacetone. J Biol Chem 278: 1415–1423 [DOI] [PubMed] [Google Scholar]
- Morett E, Segovia L (1993) The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol 175: 6067–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam TW, Cho SH, Shin D, Kim JH, Jeong JY, Lee JH, Roe JH, Peterkofsky A, Kang SO, Ryu S, Seok YJ (2001) The Escherichia coli glucose transporter enzyme IICBGlc recruits the global repressor Mlc. EMBO J 20: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Umbarger HE (1996) Chemical composition of Escherichia coli. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds) pp 13–16. Washington, DC: ASM Press [Google Scholar]
- Paulsen IT, Reizer J, Jin RZ, Lin EC, Saier MH (2000) Functional genomic studies of dihydroxyacetone utilization in Escherichia coli. Microbiology 146: 2343–2344 [DOI] [PubMed] [Google Scholar]
- Petersen AB, Wulf HC, Gniadecki R, Gajkowska B (2004) Dihydroxyacetone, the active browning ingredient in sunless tanning lotions, induces DNA damage, cell-cycle block and apoptosis in cultured HaCaT keratinocytes. Mutat Res 560: 173–186 [DOI] [PubMed] [Google Scholar]
- Plumbridge J (2002) Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr Opin Microbiol 5: 187–193 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Aravind L (1997) PAS: a multifunctional domain family comes to light. Curr Biol 7: R674–R677 [DOI] [PubMed] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR (1996) Phosphoenolpyruvate: carbohydrate phosphotransferase systems. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds) pp 1149–1174. Washington, DC: ASM Press [Google Scholar]
- Reitzer L, Schneider BL (2001) Metabolic context and possible physiological themes of sigma(54)-dependent genes in Escherichia coli. Microbiol Mol Biol Rev 65: 422–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Ramseier TM, Reizer J (1996) Regulation of carbon utilization. In Escherichia coli and Salmonella: Cellular and Molecular Biology, Neidhardt FC, Curtiss R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds) pp 1325–1343. Washington, DC: ASM Press [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Schagger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel A, Danot O, Richet E, Ferenci T, Boos W (2002) The N terminus of the Escherichia coli transcription activator MalT is the domain of interaction with MalY. J Bacteriol 184: 3069–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Steegborn C, Clausen T, Boos W, Richet E (2000) A new mechanism for the control of a prokaryotic transcriptional regulator: antagonistic binding of positive and negative effectors. Mol Microbiol 35: 765–776 [DOI] [PubMed] [Google Scholar]
- Schurmann M, Sprenger GA (2001) Fructose-6-phosphate aldolase is a novel class I aldolase from Escherichia coli and is related to a novel group of bacterial transaldolases. J Biol Chem 276: 11055–11061 [DOI] [PubMed] [Google Scholar]
- Siebold C, Garcia-Alles LF, Erni B, Baumann U (2003) A novel mechanism of covalent substrate binding in the X-ray structure of the DhaK subunit of the Escherichia coli dihydroxyacetone kinase. Proc Natl Acad Sci USA 100: 8188–8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme DJ, Dixon R (2003) Domain architectures of sigma54-dependent transcriptional activators. J Bacteriol 185: 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Van Den HJ, Soucaille P, Qu Y, Zeng AP (2003) Comparative genomic analysis of Dha regulon and related genes for anaerobic glycerol metabolism in bacteria. Biotechnol Prog 19: 263–272 [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kimata K, Aiba H (2000) A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J 19: 5344–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BL, Zhulin IB (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63: 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier FJ, Monnier VM, Sayre LA, Kornfield JA (2003) Triosidines: novel maillard reaction products and crosslinks from the reaction of triose sugars with lysine and arginine residues. Biochem J 369: 705–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites MJ, Quayle JR (1981) The interrelation transketolase and dihydroxyacetone synthase activities in the methylotrophic yeast Candida boidinii. J Gen Microbiol 124: 309–316 [DOI] [PubMed] [Google Scholar]
- Wray LV Jr, Zalieckas JM, Fisher SH (2001) Bacillus subtilis glutamine synthetase controls gene expression through a protein–protein interaction with transcription factor TnrA. Cell 107: 427–435 [DOI] [PubMed] [Google Scholar]
- Xu H, Kelly MT, Nixon BT, Hoover TR (2004) Novel substitutions in the sigma54-dependent activator DctD that increase dependence on upstream activation sequences or uncouple ATP hydrolysis from transcriptional activation. Mol Microbiol 54: 32–44 [DOI] [PubMed] [Google Scholar]
- Zheng L, Roeder RG, Luo Y (2003) S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114: 255–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


