Abstract
To genetically transform plants, Agrobacterium exports its transferred DNA (T-DNA) and several virulence (Vir) proteins into the host cell. Among these proteins, VirE3 is the only one whose biological function is completely unknown. Here, we demonstrate that VirE3 is transferred from Agrobacterium to the plant cell and then imported into its nucleus via the karyopherin α-dependent pathway. In addition to binding plant karyopherin α, VirE3 interacts with VirE2, a major bacterial protein that directly associates with the T-DNA and facilitates its nuclear import. The VirE2 nuclear import in turn is mediated by a plant protein, VIP1. Our data indicate that VirE3 can mimic this VIP1 function, acting as an ‘adapter' molecule between VirE2 and karyopherin α and ‘piggy-backing' VirE2 into the host cell nucleus. As VIP1 is not an abundant protein, representing one of the limiting factors for transformation, Agrobacterium may have evolved to produce and export to the host cells its own virulence protein that at least partially complements the cellular VIP1 function necessary for the T-DNA nuclear import and subsequent expression within the infected cell.
Keywords: Agrobacterium-to-plant protein transport, nuclear import, VirE2, VirE3
Introduction
Agrobacterium genetically transforms plants by transporting a single-stranded copy (T-strand) of the transferred DNA (T-DNA) from its tumor-inducing (Ti) plasmid into the host cell and integrating it into the host cell genome. Most of the molecular reactions of the transformation process are mediated by virulence (Vir) proteins encoded by the Ti plasmid. For example, VirA and VirG sense signals secreted by susceptible host cells and activate the vir genes, VirD1, VirD2 and VirC1 proteins generate the T-strand, which then covalently associates with VirD2, and VirB proteins and VirD4 are required for DNA and protein export from Agrobacterium into the host cell (reviewed in Winans et al, 1994; Christie, 1997; Tzfira and Citovsky, 2000; Tzfira et al, 2000; Zupan et al, 2000).
Agrobacterium infection has traditionally been viewed as a process of T-DNA transport (Zambryski, 1989). Increasing evidence indicates, however, that in addition to T-DNA, a multitude of bacterial proteins, such as VirD2, VirF, and VirE3, are also exported into the host cell, most of them separately from the T-DNA itself (Vergunst et al, 2000, 2003; Schrammeijer et al, 2003; Cascales and Christie, 2004). The roles that these Agrobacterium proteins play—from within the host cell—in genetic transformation are just beginning to emerge. For example, VirF is an F-box protein (Schrammeijer et al, 2001). VirD2 is exported together with its cognate T-strand, and, inside the plant cell, combines with VirE2 into a transfer (T) complex, which is then imported into the host cell nucleus with the help of both VirE2 and VirD2 (reviewed in Tzfira and Citovsky, 2000; Tzfira et al, 2000; Zupan et al, 2000). Interestingly, VirD2 nuclear import is directly mediated by cellular karyopherin α (Ballas and Citovsky, 1997), while nuclear import of VirE2 is facilitated by a host factor VIP1 (Tzfira et al, 2001), which functions as an adaptor between VirE2 and karyopherin α (Tzfira et al, 2002; Citovsky et al, 2004).
VirE3 is encoded by several soil bacteria, such as Agrobacterium tumefaciens, Agrobacterium rhizogenes, and Rhizobium etli (Winans et al, 1987; Kalogeraki et al, 2000), but it shows no homologies to any other known genes. Recent studies suggest that, during transformation, VirE3 is exported into the host yeast (Schrammeijer et al, 2003) and plant cells (Vergunst et al, 2003). By analogy to other proteins exported from different species of pathogenic bacteria to their eukaryotic hosts, VirE3 may have acquired (most probably by convergent evolution) functional features of a host protein required for the infection (Nagai and Roy, 2003). Transported bacterial proteins that belong to this category usually do not exhibit significant sequence similarities with the eukaryotic factor they mimic, making their functional annotation difficult (Nagai and Roy, 2003). Thus, the function of VirE3 remains virtually unknown. Here, we directly demonstrate the Agrobacterium-to-plant cell transport of VirE3 and shed light on the VirE3 function within the host cell by showing that this bacterial virulence protein can substitute for the cellular protein VIP1 in interacting with and facilitating nuclear import of VirE2 via the karyopherin α-mediated pathway and allow subsequent T-DNA expression.
Results
VirE3 is exported from Agrobacterium into plant cells
Traditionally, transport of proteins, including VirE3, from Agrobacterium to plant cells has been shown using a genetic method that infers protein transport from measuring production of transgenic plant calli under selection conditions (Vergunst et al, 2000, 2003). Here, we developed a more direct approach to assay for Agrobacterium-to-plant VirE3 export without applying selection pressure. VirE3 was fused to a chimeric transcriptional activator mGAL4-VP16, containing the yeast Gal4 DNA binding domain fused to the VP16 transcriptional activator from herpes simplex virus, and expressed in Agrobacterium, which was inoculated on Arabidopsis plants that carry a β-glucuronidase (GUS) reporter transgene driven by a mGAL4-VP16-inducible GAL4-UAS promoter (www.plantsci.cam.ac.uk/Haseloff/Home.html). VirE3 transport from Agrobacterium into plant induces the GUS gene expression, which can be detected 48–72 h after inoculation.
Figure 1A shows that inoculation of Arabidopsis leaf segments with Agrobacterium-expressing mGAL4-VP16-VirE3 resulted in GUS expression detected histochemically as indigo–blue areas. In contrast, inoculation with Agrobacterium-expressing mGAL4-VP16 fusion with an unrelated protein, GFP, did not induce the expression of GUS activity (Figure 1B). Similar results were obtained when Arabidopsis root segments were inoculated with Agrobacterium-expressing mGAL4-VP16-VirE3 (Figure 1C) or mGAL4-VP16-GFP (Figure 1D). As expected, no GUS-specific staining was observed using wild-type Arabidopsis, that is, plants lacking the reporter transgene (not shown). These results indicate that VirE3 is exported from Agrobacterium into Arabidopsis leaf and root cells.
Figure 1.
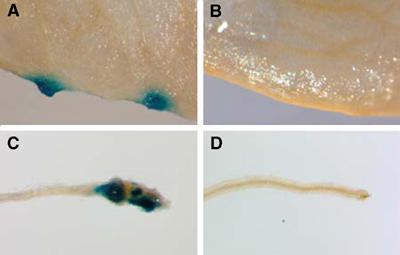
Export of VirE3 from Agrobacterium to Arabidopsis cells. Leaf segments were inoculated with Agrobacterium-expressing mGAL4-VP16-VirE3 (A) or mGAL4-VP16-GFP (B). Root segments were inoculated with Agrobacterium-expressing mGAL4-VP16-VirE3 (C) or mGAL4-VP16-GFP (D).
VirE3 is imported into the cell nucleus and interacts with karyopherin α
To gain the first insight into the VirE3 role within the host cells during Agrobacterium infection, we examined its subcellular localization in plant cells. To this end, VirE3 was tagged with GFP and transiently expressed, following biolistic delivery of its encoding DNA construct, in the epidermal cells of tobacco leaves. In addition, another fluorescent reporter, DsRed2, was expressed from the same DNA construct; free, unfused DsRed2 is known to partition between the cell cytoplasm and the nucleus, conveniently visualizing and identifying both of these cellular compartments (Dietrich and Maiss, 2002; Goodin et al, 2002; Schultheiss et al, 2003). Figure 2A shows that GFP-VirE3 was imported into the plant cell nucleus, displaying a predominantly intranuclear accumulation as determined by confocal microscopy with optical sections through the cell nucleus. As expected, in the same GFP-VirE3-expressing cell, DsRed2 was found both in the cytoplasm and in the nucleus (Figure 2B). The combined image of GFP and DsRed2 fluorescence showed overlapping signal (yellow color) within the cell nucleus, confirming the location of GFP-VirE3 in a distinct subnuclear site (Figure 2C). As the predicted size of the GFP-VirE3 fusion protein (ca. 100 kDa) is substantially larger than the size exclusion limit of the nuclear pore (reviewed by Dingwall and Laskey, 1991; Garcia-Bustos et al, 1991), its accumulation within the nucleus must result from the active process of nuclear import.
Figure 2.
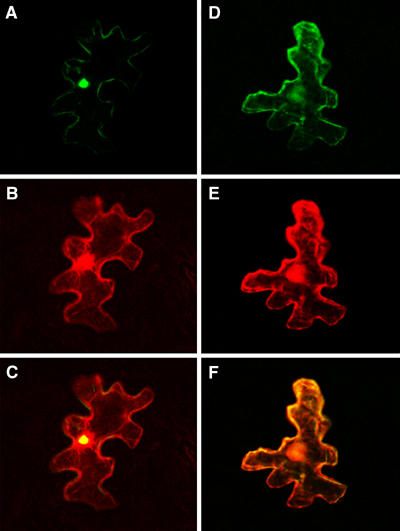
Nuclear import of GFP-VirE3 in tobacco plants requires the presence of the VirE3 NLS sequences: (A) GFP-VirE3; (B) free DsRed2 produced from the GFP-VirE3-expressing construct; (C) merged image; (D) GFP-VirE3-mNLS12; (E) free DsRed2 produced from the GFP-VirE3-mNLS12-expressing construct; and (F) merged image. GFP is in green, DsRed2 is in red, and overlapping GFP and DsRed2 are in yellow. All images are single confocal sections.
The amino-acid sequence of VirE3 (accession number AAC72022.1) contains two potential nuclear localization signals (NLS), 40-KRqtrlespdRKRKY-54 (NLS1) and 80-KRlrvdnpkeltrehgrlRKTKT-102 (NLS2), which correspond to the bipartite NLS motif (with its small and large basic domains indicated in uppercase) conserved in many nuclear proteins (reviewed by Dingwall and Laskey, 1991). Thus, nuclear import of VirE3 may be mediated by karyopherins α, nuclear shuttle receptors known to recognize bipartite NLS-containing proteins and import them into the cell nucleus (reviewed by Chook and Blobel, 2001). Indeed, Figure 3A shows that VirE3 interacted with the Arabidopsis karyopherin α, AtKAPα (Ballas and Citovsky, 1997) in the yeast two-hybrid (Y2H) system (lane 1). In this system, protein interaction is assessed from activation of the HIS3 reporter gene, which allows yeast cells to grow in a histidine-deficient medium. The interaction between VirE3 and AtKAPα was specific because it did not occur with lamin C (Figure 3A, lane 4), a known nonspecific Y2H activator best suited to eliminate false-positive interactions (Bartel et al, 1993). The VirE3–AtKAPα interaction in Y2H was confirmed by induction of another reporter gene, lacZ, encoding β-galactosidase activity (Figure 3B, lane 1).
Figure 3.
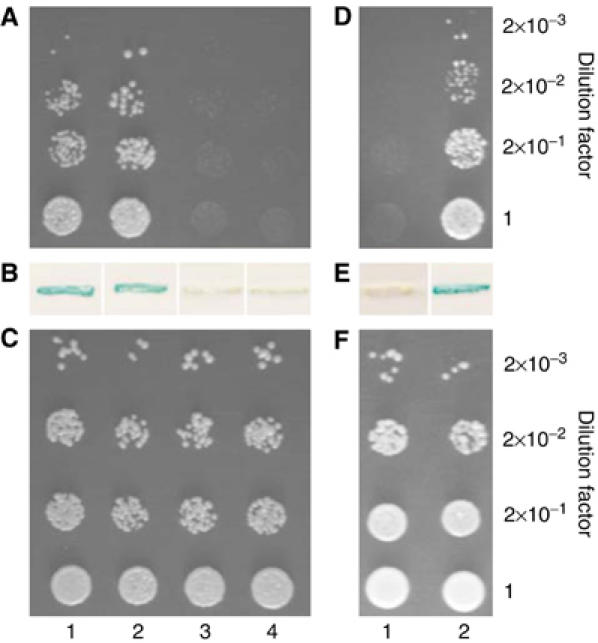
VirE3 interacts with VirE2 and AtKAPα in Y2H assay. (A) Cell growth in the absence of histidine. (B) β-galactosidase assay. (C) Cell growth in the presence of histidine. Lane 1, VirE3+AtKAPα; lane 2, VirE3+VirE2; lane 3, VirE3+VirD2; lane 4, VirE3+lamin C. VirE3-mNLS12 interacts with VirE2, but not with AtKAPα in the Y2H assay. (D) Cell growth in the absence of histidine. (E) β-Galactosidase assay. (F) Cell growth in the presence of histidine. Lane 1, VirE3-mNLS12+AtKAPα; lane 2, VirE3-mNLS12+VirE2.
Next, we directly examined the functional roles of the potential VirE3 NLSs in nuclear import. First, each of the NLS sequences was fused to a reporter enzyme; because NLSs themselves are small peptides, we chose to use a relatively large reporter protein, GUS, to prevent NLS-reporter fusions from diffusing into the cell nucleus. The resulting constructs were transiently expressed in the onion epidermis, a classical system to assay nuclear import of GUS-tagged plant proteins (Shieh et al, 1993). Figure 4 shows that both GUS-NLS1 (panels A and B) and GUS-NLS2 (panels E and F) accumulated in the cell nucleus, colocalizing with the nucleus-specific stain 4′,6-diamidino-2-phenylindole (DAPI). These results indicate that NLS1 and NLS2 of VirE3 are independently active and sufficient to promote nuclear import of the GUS reporter. We substituted several basic residues of both the small and the large basic domains of each of these bipartite NLSs with uncharged alanine or valine residues, resulting in the mutant NLS sequences 40-KaqtrlespdvvaKY-54 (mNLS1) and 80-KalrvdnpkeltrehgrlavTvT-102 (mNLS2). Transient expression experiments demonstrated that GUS-mNLS1 and GUS-mNLS2 remained cytoplasmic (Figure 4, panels C, D and G, H, respectively), indicating that neither mNLS1 nor mNLS2 was able to promote nuclear import of GUS in plant cells.
Figure 4.
VirE3 NLS1 and NLS2 promote nuclear import of GUS reporter in plant cells: (A, B) GUS-NLS1; (C, D) GUS-mNLS1; (E, F) GUS-NLS2; and (G, H) GUS-mNLS2. Panels A, C, E, and G show GUS staining, and panels B, D, F, and H show DAPI staining. Arrows indicate cell nuclei.
We incorporated both NLS mutations in the full-length VirE3 protein. The resulting VirE3-mNLS12 mutant was examined for its ability to bind AtKAPα in the Y2H system and target to the nucleus in plant cells. Coexpression of VirE3-mNLS12 and AtKAPα failed to induce histidine prototrophy (Figure 3D, lane 1) or β-galactosidase activity (Figure 3E, lane 1), indicating that these two proteins do not detectibly interact with each other. This lack of cell growth was not due to protein toxicity because, in the absence of selection, the cells remained viable (Figure 3F, lane 1).
Consistent with the disruption of the VirE3-mNLS12 interaction with AtKAPα, nuclear import of this protein was also compromised. Figure 2D shows that GFP-VirE3-mNLS12 was found largely within the cell cytoplasm, colocalizing with the cytoplasmic pool of the DsRed2 fluorescence, but not with the nuclear DsRed2 (Figure 2E and F). Collectively, these results demonstrate the critical role of the VirE3 NLS1 and NLS2 sequences in nuclear import of this bacterial virulence protein in host cells. Note that small amounts (<10% as quantified based on the intensity of GFP signal on a per cell basis) of VirE3-mNLS12 still detected in the nucleus (Figure 2D) may reflect the presence of other, weaker targeting sequences that augment the major NLS1 and NLS2 signals and that remain marginally active when NLS1 and NLS2 are mutated; similar augmenting NLSs have been reported for numerous other proteins in different organisms (e.g., Braem et al, 2002; Schwemmle et al, 1999).
VirE3 interacts with VirE2
What is the role of VirE3 within the host cell during the Agrobacterium-mediated genetic transformation? Potentially, VirE3 may assist the function of the invading Agrobacterium T-complex by interacting with one of its bacterial protein components, VirD2 or VirE2. To test this idea, we utilized the Y2H system to examine possible interactions between VirE3 and VirD2 and VirE2. Figure 3A shows that yeast cells expressing VirE3 and VirE2 exhibited strong growth in the absence of histidine (lane 2); however, VirE3 did not interact with another component of the Agrobacterium T-complex, VirD2 (lane 3). Also, as mentioned above, in negative control experiments, no interaction between VirE3 and lamin C was detected (Figure 3A, lane 4). All protein interactions detected using induction of the HIS3 reporter (cell growth in the absence of histidine, Figure 3A) were confirmed using induction of an independent lacZ reporter (Figure 3B). Under the nonselective conditions, all combinations of the tested proteins resulted in the efficient cell growth, indicating that neither of the tested proteins was toxic to yeast cells (Figure 3C). These observations suggest that VirE3 specifically recognizes and interacts with VirE2 in the Y2H system.
The interaction between VirE3 and VirE2 was then demonstrated in planta. To this end, we utilized a bimolecular fluorescence complementation (BiFC) assay (Hu et al, 2002). In this approach, a molecule of yellow spectral variant of GFP (YFP) is split into two parts, N-terminal (nYFP) and C-terminal (cYFP), neither of which fluoresces on its own. However, when nYFP and cYFP are brought together as fusions with interacting proteins, the fluorescence is restored (Hu et al, 2002). Here, we used BiFC in two different plant species—tobacco and onion—both of which can be transformed by Agrobacterium (Dommisse et al, 1990). For positive control experiments, we utilized VIP1 and VirE2, which are well-known to interact with each other (Tzfira et al, 2001, 2002). Indeed, coexpression of nYFP-tagged VIP1 with cYFP-tagged VirE2 resulted in a strong fluorescent signal of the reconstructed YFP reporter in onion and tobacco tissues (Figure 5A). Similar YFP fluorescence was observed in both plant species when nYFP-tagged VirE3 was coexpressed with cYFP-tagged VirE2 (Figure 5A and B). As expected, absolutely no YFP signal was detected in negative control experiments, in which nYFP-VIP1 or nYFP-VirE3 was coexpressed with unfused cYFP (Figure 5A), or when cYFP-VirE2 was coexpressed with unfused nYFP (data not shown). This lack of background signal, previously reported for BiFC in animal and bacterial systems (Hu et al, 2002; Atmakuri et al, 2003; Tsuchisaka and Theologis, 2004), significantly facilitated identification of cells exhibiting the reconstructed YFP fluorescence in the BiFC assay in planta. Taken together, our data indicate that VirE3 specifically interacts with VirE2 within living yeast and plant cells.
Figure 5.
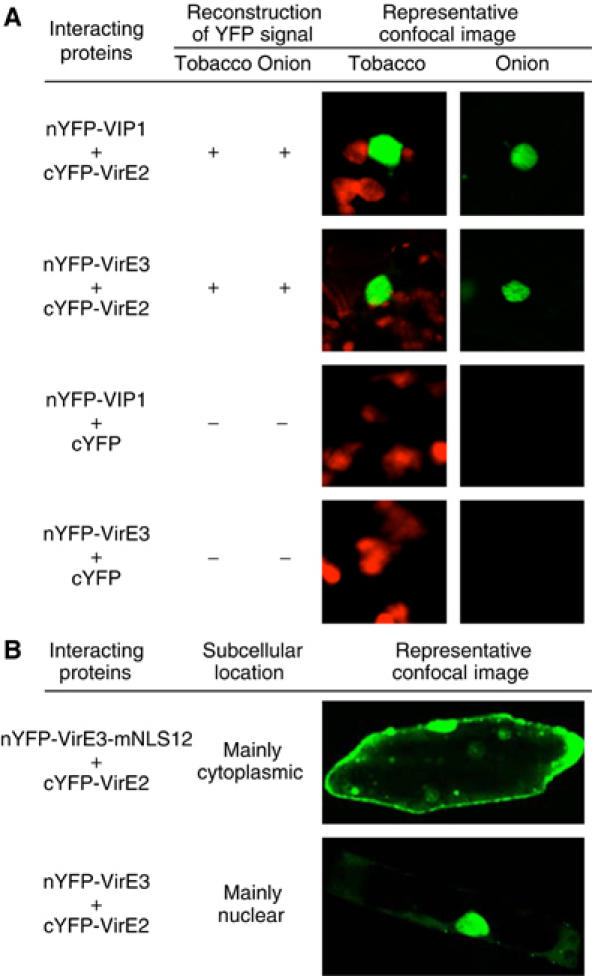
BiFC assay for the VirE3–VirE2 interaction in planta. Positive reconstruction of YFP fluorescence indicates that the signal was observed in numerous (30–100) cells per each bombardment. Negative reconstruction of YFP fluorescence indicates that the signal was never found in any bombarded cells. Representative confocal images of bombarded cells observed in each experimental system are shown. YFP is in green, and plastid autofluorescence is in red. Note that onion cells do not contain chloroplasts. (A) Images focus on tobacco and onion cell areas with reconstructed YFP signal. (B) Images show entire onion cells. All images are single confocal sections.
VirE3 assists nuclear import of VirE2 in mammalian cells
Nuclear import of VirE2 is facilitated by a host cell protein, VIP1, which acts as a molecular adapter between VirE2 and cellular karyopherins α (Tzfira et al, 2001, 2002; Ward and Zambryski, 2001; Citovsky et al, 2004). Since VirE3 exhibited some functional resemblance to VIP1 in terms of its nuclear localization in plant cells and binding to AtKAPα and VirE2, we tested whether or not VirE3 can also assist nuclear import of VirE2. Initially, these experiments were performed in mammalian cells because one of the hallmarks of VIP1 is its ability to promote VirE2 nuclear import in nonplant systems (Tzfira et al, 2001; Citovsky et al, 2004), in which VirE2 is normally cytoplasmic (Guralnick et al, 1996; Rhee et al, 2000; Tzfira and Citovsky, 2001; Tzfira et al, 2001; Citovsky et al, 2004).
Confocal microscopy observations demonstrated that GFP-tagged VirE2 transiently expressed in COS-1 cells was completely excluded from the cell nucleus, remaining confined to the cell cytoplasm (Figure 6A). GFP-VirE3, on the other hand, exhibited nuclear accumulation in these cells (Figure 6B), consistent with its interaction with karyopherin α, which is conserved between plants, yeast, and animals (Hicks et al, 1996; Ballas and Citovsky, 1997). Coexpression of GFP-VirE2 and the intact, unlabeled VirE3 redirected a significant proportion of the fluorescent signal to the cell nucleus (Figure 6C), suggesting that VirE3 may play a role in VirE2 nuclear import. In control experiments and consistent with previous observations (Tzfira et al, 2001), GFP-VIP1 accumulated in the cell nucleus (Figure 6D), whereas the intact VIP1 promoted nuclear import of GFP-VirE2 (Figure 6E); the extent of GFP-VirE2 nuclear import observed in the presence of VirE3 was similar to that observed in the presence of VIP1 (cf. panels C and E in Figure 6). Note that the confocal optical sections with the plane of focus through the cell nucleus detect intranuclear accumulation of the GFP label rather than its perinuclear binding. Potentially, transient expression of GFP-VirE2 and VirE3 from separate plasmids may not have generated protein concentrations necessary for the complete nuclear import of VirE2. Importantly, the VirE3-assisted nuclear uptake of GFP-VirE2 depended on the presence of the functional NLS sequences of VirE3, because the VirE3-mNLS12 mutant, which was compromised for its ability to enter the nucleus of plant (see Figure 2D–F) or animal cells (data not shown) and to interact with karyopherin α (see Figure 3D–F), also failed to lead GFP-VirE2 into the nuclei of COS-1 cells (Figure 6F).
Figure 6.
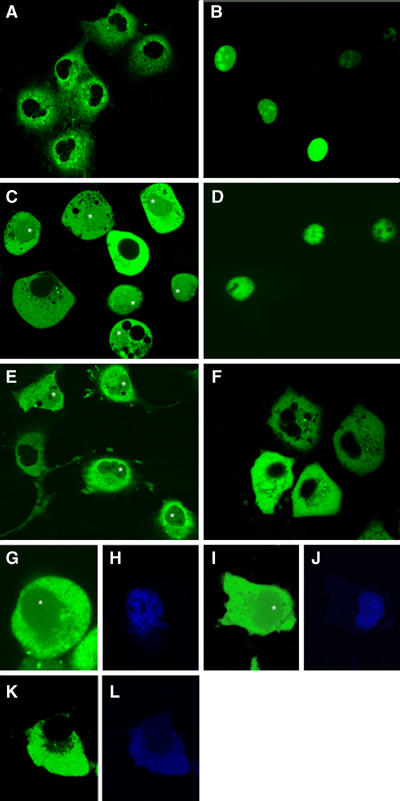
VirE3 facilitates nuclear import of VirE2 in COS-1 cells: (A) GFP-VirE2; (B) GFP-VirE3; (C) GFP-VirE2+VirE3; (D) GFP-VIP1; (E) GFP-VirE2+VIP1; (F) GFP-VirE2+VirE3-mNLS12; (G, H) YFP-VirE2+CFP-VIP1; (I, J) YFP-VirE2+CFP-VirE3; and (K, L) YFP-VirE2+CFP-VirE3-mNLS12. YFP is in green (G, I, and K), and CFP is in blue (H, J, and L). Asterisks indicate cell nuclei that contain VirE2. All images are single confocal sections.
To examine expression levels of VIP1, VirE3, and VirE3-mNLS12 in nuclear import experiments, these proteins were tagged with CFP and coexpressed with YFP-VirE2. Figure 6 shows that CFP-VIP1 and CFP-VirE3, but not CFP-VirE3-mNLS12, were able to facilitate nuclear import of YFP-VirE2 in COS-1 cells (panels G, I, and K, respectively). Quantification of the YFP signal on a per cell basis demonstrated that intact as well as CFP-tagged VIP1 and VirE3 redirected comparable amounts (30–40%) of GFP-VirE2 or YFP-VirE2 to the cell nucleus (Table I). These results are consistent with the observations that tagged VIP1 and VirE3 interacted with VirE2 and localized to the cell nucleus in the BiFC assay in planta (see Figure 5). Quantification of the YFP fluorescence also showed that YFP-VirE2 was expressed to a similar degree in all transfection systems (Table I). Importantly, quantification of the CFP signal indicated that VIP1, VirE3, and VirE3-mNLS12 accumulated to comparable levels in the transfected cells, with CFP-VIP1 and CFP-VirE3 found almost exclusively in the cell nucleus and CFP-VirE3-mNLS12 showing predominantly cytoplasmic localization together with minor nuclear accumulation (Figure 6H, J, L and Table I; see also Figure 2D).
Table 1.
Quantification of coexpression of fluorescently tagged proteins in COS-1 cells
| YFP signal (% of total per cell) |
YFP signal (pixels per cell) | CFP signal (% of total per cell) |
CFP signal (pixels per cell) | |||
|---|---|---|---|---|---|---|
| Cytoplasm | Nucleus | Cytoplasm | Nucleus | |||
| YFP-VirE2+CFP-VIP1 | 70 (4) | 30 (4) | 165 (10) | 10 (3) | 90 (3) | 244 (12) |
| YFP-VirE2+CFP-VirE3 | 59 (2) | 41 (2) | 159 (8) | 2 (1) | 98 (1) | 205 (13) |
| YFP-VirE2+CFP-VirE3-mNLS12 |
95 (1) |
5 (1) |
174 (11) |
91 (1) |
9 (2) |
211 (11) |
| The data are derived from three experiments with 50–100 YFP-expressing cells per experiment; average values are given, and standard error values are indicated in parentheses. | ||||||
The importance of VirE3 NLS for VirE2 nuclear import was further demonstrated in planta using BiFC, which not only detects protein interactions but also identifies the subcellular location of the interacting proteins. Following coexpression, nYFP-VirE3-mNLS12 still bound cYFP-VirE2, but was severely compromised in its ability to promote VirE2 nuclear import in tobacco (not shown) and onion cells (Figure 5B), resulting in predominantly cytoplasmic localization of the VirE3-mNLS12–VirE2 complexes as compared to the largely nuclear location of the VirE3–VirE2 complexes.
VirE3 complements the VIP1 function in nuclear import of VirE2 in planta
The VIP1-like function of VirE3 was examined directly in tobacco plants in which the VIP1 gene expression was knocked down by antisense technology (Tzfira et al, 2001). Note that while these VIP1 antisense plants are highly recalcitrant to Agrobacterium infection, they are not completely resistant (Tzfira et al, 2001) and can be modified genetically. Thus, the VIP1 antisense plants were retransformed with Agrobacterium to produce double-transgenic plants that express the VIP1 cDNA in the antisense orientation and the virE3 gene in the sense orientation. A total of seven independently transformed lines were produced and tested for their ability to support nuclear import of VirE2. A detailed analysis of one representative line is described here. These double-transgenic plants were first examined for the presence of sense and antisense VIP1 RNA and sense VirE3 RNA using quantitative RT–PCR and strand-specific oligonucleotide primers (Ni et al, 1998; Tzfira et al, 2001). Figure 7 shows that, as expected, control, wild-type tobacco plants produced the sense (panel A, lane 1), but not antisense (panel B, lane 1) VIP1 RNA or VirE3 transcript (panel C, lane 1). VIP1 antisense plants possessed much lower amounts of the sense VIP1 transcript (Figure 7A, lane 2), generated antisense VIP1 RNA (Figure 7B, lane 2), but did not produce VirE3 RNA (Figure 7C, lane 2). Figure 7 also shows that the double-transgenic VIP1 antisense/VirE3 line contained significantly reduced amounts of the sense VIP1 RNA (panel A, lane 3), generated the antisense VIP1 RNA (panel B, lane 3), and accumulated the VirE3 sense RNA (panel C, lane 3). In control experiments, analysis of actin-specific transcripts generated comparable amounts of RT–PCR products in all samples, indicating similar efficiencies of the RT–PCR reactions (Figure 7D).
Figure 7.
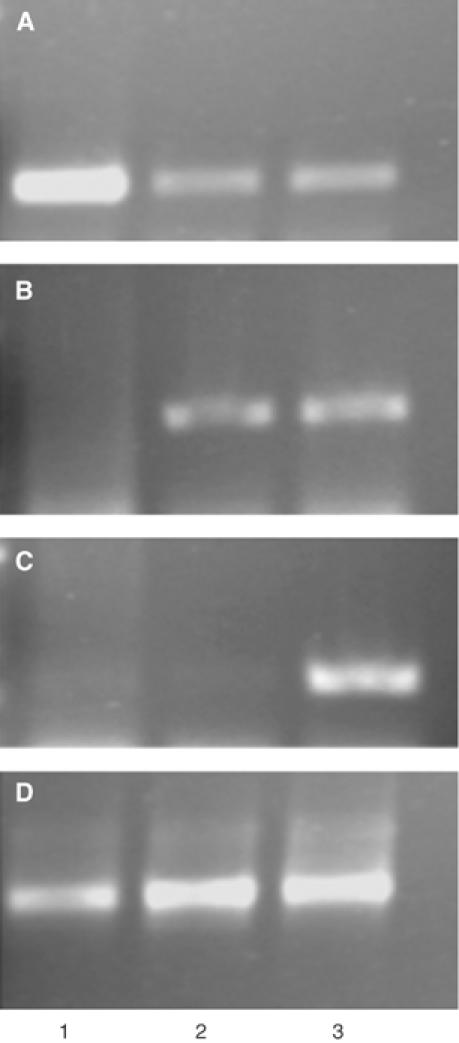
Quantitative RT–PCR analysis of VIP1 antisense/VirE3 double-transgenic plants. (A) Detection of sense VIP1 RNA-specific product (290 bp). Lanes 1–3, wild-type plants, VIP1 antisense plants, and VIP1 antisense/VirE3 double-transgenic plants, respectively. (B) Detection of antisense VIP1 RNA-specific product (290 bp) in the samples shown in panel A. (C) Detection of sense VirE3 RNA-specific product (210 bp) in the same samples shown in panel A. (D) Detection of sense actin RNA-specific product (500 bp) in the samples shown in panel A.
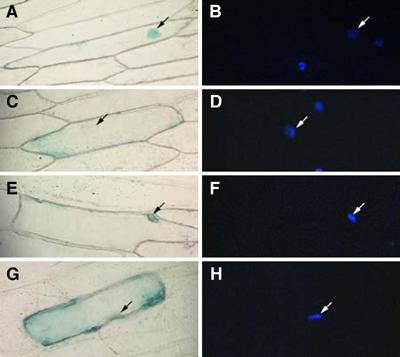
Next, the transgenic plant lines were examined for nuclear import of VirE2 tagged with GUS reporter and transiently expressed in leaf epidermal cells following biolistic delivery of its coding DNA construct. Figure 8 shows that, as expected, GUS-VirE2 accumulated in the cell nuclei of the wild-type tobacco plants (panel A); the location of the nucleus was confirmed by DAPI staining (panel B). Consistent with previous observations (Tzfira et al, 2001), the parental VIP1 antisense plants did not promote efficient nuclear import of GUS-VirE2, displaying predominantly cytoplasmic accumulation of this protein (Figure 8C and D) and confirming that VIP1 is required for nuclear import of VirE2. However, GUS-VirE2 efficiently accumulated in the cell nuclei of the double-transgenic plants (Figure 8E and F), indicating that VirE3 expression compensated for the lack of VIP1 and restored VirE2 nuclear import in VIP1 antisense plants. In control experiments, free GUS expressed in all plant lines remained cytoplasmic (not shown, but see Table II).
Figure 8.
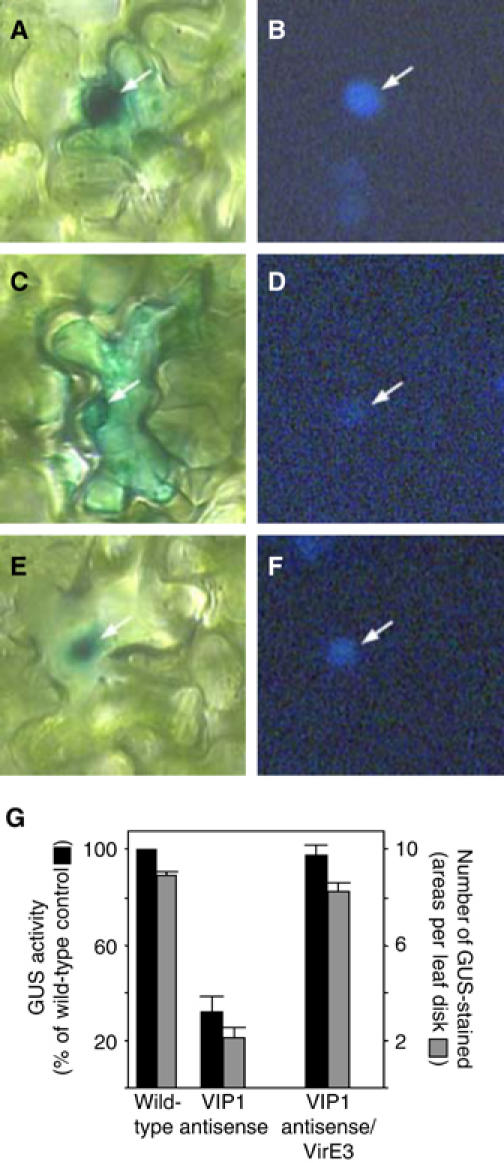
Restoration of GUS-VirE2 nuclear import and T-DNA gene expression in VIP1 antisense/VirE3 double-transgenic plants. (A, B) GUS-VirE2 expressed in wild-type plants. (C, D) GUS-VirE2 expressed in VIP1 antisense plants. (E, F) GUS-VirE2 expressed in VIP1 antisense/VirE3 double-transgenic plants. Panels A, C, and E represent GUS staining, and panels B, D, and F represent DAPI staining. Arrows indicate cell nuclei. (G) Expression of GUS activity contained within Agrobacterium T-DNA. Black bars indicate total GUS activity, with that in control, wild-type plants defined as 100%, and gray bars indicate the number of GUS-stained areas per inoculated leaf disk. All data represent average values of three independent experiments (20 disks each) with indicated standard deviation values.
Table 2.
Effect of transgenic VirE3 expression on nuclear localization of GUS-VirE2 in VIP1 antisense plants
| GUS activity (% of total per cell) |
||
|---|---|---|
| Nucleus | Cytoplasm | |
| Wild-type plants | ||
| GUS-VirE2 | 97 (2) | 3 (1) |
| GUS alone | 0 | 100 (6) |
| VIP1 antisense plants | ||
| GUS-VirE2 | 20 (6) | 80 (2) |
| GUS alone | 0 | 100 (6) |
| VIP1 antisense/VirE3 plants | ||
| GUS-VirE2 | 89 (4) | 11 (5) |
| GUS alone |
0 |
100 (4) |
| The data are derived from 20 independent measurements of GUS-positive cells; average values are given, and standard error values are indicated in parentheses. | ||
We then quantified on a per cell basis the nuclear and cytoplasmic content of GUS-VirE2 in the wild-type, VIP1 antisense, and VIP1 antisense/VirE3 double-transgenic plants. Table II demonstrates that, while free GUS reporter remained cytoplasmic in all plant lines, virtually all (97%) GUS-VirE2 accumulated in the nuclei of the expressing cells in the wild-type plants. In contrast, only 20% of GUS-VirE2 was imported into the cell nuclei of the VIP1 antisense plants. That VirE2 nuclear import was not blocked completely suggests that either a small fraction of VirE2 enters the plant cell nucleus by another, VIP1-independent pathway, or that residual amounts of VIP1 in the antisense plants may still promote this low level of import. Nuclear import of GUS-VirE2 in the VIP1 antisense plants was restored to almost wild-type levels (89%) by transgenic expression of VirE3 (Table II). These observations further support the cellular VIP1-like role of the bacterial VirE3 protein in nuclear transport of VirE2.
VirE3 complements the VIP1 function in T-DNA expression in Agrobacterium-infected tissues
If VirE3 compensates for low levels of VIP1 during nuclear import of VirE2 and, by implication, the T-complex, its expression in the VIP1 antisense plants should also promote Agrobacterium-mediated genetic transformation of these plants. To test this notion, we focused on early T-DNA expression, which represents the transformation stage that immediately follows nuclear import. The efficiency of T-DNA expression was determined by inoculating leaf disks derived from the wild-type, VIP1 antisense, and VIP1 antisense/VirE3 double-transgenic plants with Agrobacterium carrying on its T-DNA a uidA gene encoding GUS reporter (Tzfira et al, 2001).
At 2 days after inoculation, Gus expression was quantified by two approaches: a fluorimetric assay to determine the overall enzymatic activity in the transformed tissue and a histochemical assay to determine the number of GUS-expressing areas per transformed leaf disk. Since the uidA gene contained on the T-DNA lacked regulatory sequences for expression in bacteria (Janssen and Gardner, 1990), our measurements represented the GUS activity directed by the T-DNA after its transfer to the plant rather than its potentially leaky expression in Agrobacterium. Figure 8G shows that, as expected (Tzfira et al, 2001), VIP1 antisense plants inoculated with Agrobacterium displayed approximately 25–35% GUS expression as compared to the wild-type plants. Transgenic expression of VirE3 restored this decreased susceptibility of VIP1 antisense plants to Agrobacterium-mediated transformation to 85–95% of the wild-type level (Figure 8G). These results lend additional support for the biological role of VirE3 in the transformation process.
Discussion
We developed a simple experimental system to detect directly protein export from Agrobacterium to plant cells. Using this approach, we demonstrated export of VirE3 into the leaf and root cells of Arabidopsis. Our results also suggest that, once in the host cell cytoplasm, VirE3 likely interacts with the plant karyopherin α proteins, which mediate its import into the cell nucleus. VirE3 nuclear import in plant cells required the presence of two major and independently active bipartite NLS sequences located within the N-terminal portion of the protein.
Besides its ability to target to the cell nucleus, VirE3 specifically recognized and interacted with VirE2. This interaction was demonstrated in vitro, in Y2H, and in planta. For protein interactions in plant cells, we adapted a BiFC assay for use in plant tissues. This assay allowed us to demonstrate the formation of VirE3–VirE2 heterodimers directly in living plant cells with virtually no background signal.
These observations provided a clue regarding a possible role of VirE3 during Agrobacterium-mediated genetic transformation. One characteristic feature of VirE2, a known major protein component of the T-complex (reviewed by Tzfira et al, 2000; Zupan et al, 2000), is that its nuclear import is plant-specific, occurring in plant but not in animal or yeast cells (Citovsky et al, 1992a, 1994, 2004; Guralnick et al, 1996; Rhee et al, 2000; Tzfira and Citovsky, 2001). This is because the VirE2 nuclear import is most likely facilitated by a VirE2-binding protein VIP1, which is found in plant but not in animal or yeast cells (Tzfira et al, 2001). That VirE3 resembles VIP1 in its capacity for nuclear import as well as the ability to bind VirE2 suggests that it also may fulfill the VIP1 function during Agrobacterium infection, that is, act as an ‘adapter' molecule between VirE2 and karyopherin α, ‘piggy-backing' VirE2 into the host cell nucleus. Indeed, VirE3 coexpressed with VirE2 in mammalian cells redirected a significant proportion of VirE2 into the cell nucleus. Furthermore, while VirE2 remained predominantly cytoplasmic in VIP1 antisense plants with knocked-down levels of VIP1, its nuclear import was restored following transgenic expression of VirE3. Thus, VirE3 is capable of facilitating nuclear accumulation of VirE2 in planta. This VirE3 function is reminiscent of that of the bacterial wilt avirulence protein PopP2 that interacts with the Arabidopsis RRS1 disease resistance proteins and ‘piggy-backs' it into the plant cell nucleus (Deslandes et al, 2003).
The effect of VirE3 expression on the subcellular localization of VirE2 in mammalian and in plant cells is consistent with formation of VirE2–VirE3 complexes in yeast and in plant cells, representing additional biological evidence for functional interaction between these two proteins within the cellular environment. This VirE3–VirE2 interaction is likely relevant for Agrobacterium infection because transgenic expression of VirE3 in the VIP1 antisense plants significantly elevated their susceptibility to genetic transformation by Agrobacterium. Thus, VirE3 also complements the function of VIP1 during the Agrobacterium infection of plant cells.
That the biological role of VirE3 during the Agrobacterium-mediated genetic transformation is similar to the role of a host factor explains previous observations that VirE3 was not absolutely essential for Agrobacterium infection in some plant species (Winans et al, 1987; Kalogeraki et al, 2000). This functional redundancy between the bacterial and the host factors also makes biological sense. VIP1 is important for nuclear import of VirE2 in the host cells and, thus, for Agrobacterium infection; however, VIP1 is not an abundant cellular protein, potentially representing one of the limiting factors for transformation, especially for low inocula of Agrobacterium (Tzfira et al, 2001, 2002). We propose that Agrobacterium has evolved to produce and export to the host cells its own virulence protein, VirE3, that, at least partially, complements the VIP1 function. This idea is consistent with the early observations that VirE3 represents one of the host range factors of Agrobacterium (Hirooka and Kado, 1986), potentially compensating for the absence or very low levels of VIP1 in some plants. This strategy of Agrobacterium to improve its infection efficiency may represent a more general ability of infectious microorganisms to encode and export protein functions, most probably acquired by convergent evolution (Nagai and Roy, 2003), similar to those normally provided by the host cell.
In addition, that VirE3 mimics a host function essential for the infection suggests that other Agrobacterium virulence proteins, which have not been studied because early experiments showed that they are dispensable for the bacterial tumorigenicity (e.g., Hirooka and Kado, 1986; Stachel and Nester, 1986; Winans et al, 1987), may represent redundant but, in fact, critical cellular activities for the Agrobacterium-mediated process of genetic transformation.
Materials and methods
Agrobacterium-to-plant cell protein transport assay
VP16 transcriptional activator fused to the GAL4 DNA binding domain modified for optimal expression in Arabidopsis (mGAL4) was obtained from Dr J Haseloff (University of Cambridge, UK) and cloned as SalI–KpnI polymerase chain reaction (PCR)-amplified fragment under the virB promoter in the pE2431 plasmid (obtained from Dr S Gelvin, Purdue University, IN). Then, VirE3 (Winans et al, 1987) derived from the pURK228 cosmid (Knauf and Nester, 1982) and GFP were PCR-amplified as KpnI–EcoRI fragments and fused to the C-terminus of mGAL4-VP16 in pE2431. High-fidelity Pfu DNA polymerase (Invitrogen) was used in all PCR reactions, and DNA constructs were verified by sequencing.
Agrobacterium strain GV3101 containing the pMP90 helper plasmid and constructs expressing mGAL4-VP16-VirE3 or mGAL4-VP16-GFP was treated for 8 h at 28°C with 100 μM acetosyringone (AS) to induce vir gene expression and then cocultivated at 22°C with 1 × 1 cm leaf segments of Arabidopsis plants carrying GUS reporter transgene under mGAL4-VP16-inducible GAL4-UAS promoter (obtained from Dr J Haseloff). After 48 h, the leaf segments were stained with X-Gluc (Nam et al, 1999) for histochemical detection of GUS. For protein export into roots, the Arabidopsis plants were grown for 3 weeks on the B5 agar, and roots were removed and maintained for 3 days on callus induction medium (CIM) before inoculation with Agrobacterium. GUS activity was assayed after 3 days of cocultivation on CIM in the presence of 100 μM AS.
Y2H protein interaction assay
LexA fusions of AtKAPα, VirE2, and VirD2 in pSTT91 (TRP1+; Sutton et al, 2001) have been described (Tzfira et al, 2002). virE3 (Winans et al, 1987) or virE3-mNLS12 (see below) genes were cloned as PCR-amplified EcoRI–BamHI fragments into pGAD424 (LEU2+, Clontech), producing fusions with the GAL4 activation domain. Protein interaction, indicated by histidine prototrophy, was assayed in the Saccharomyces cerevisiae strain TAT7 (L40 (Hollenberg et al, 1995)-ura3) (SenGupta et al, 1996) by growing cells for 2 days at 30°C on a leucine-, tryptophan-, and histidine-deficient medium. Alternatively, cells were grown on a leucine- and tryptophan-deficient medium, and the β-galactosidase activity was assayed on nitrocellulose filters (Hollenberg et al, 1995).
BiFC protein interaction assay in planta
PCR amplification was used to dissect YFP into two parts: the 5′ part, termed nYFP, that terminated at codon 174, and the 3′ part, termed cYFP, that began with the ATG codon preceding the YFP codon 175. nYFP and cYFP were cloned into the NcoI–BamHI sites of pRTL2-GUS (Restrepo et al, 1990), replacing GUS and producing pRTL2-nYFP and pRTL2-cYFP. The tested genes were fused to the C-terminus of nYFP or cYFP by insertion into the BamHI site of pRTL2-nYFP or into the NcoI site of pRTL2-cYFP, respectively. The resulting constructs were mixed (1:1 w/w), and cobombarded into tobacco leaves or onion scales (see below), followed by incubation for 28 h at 25°C to allow expression of the transfected DNA and reconstruction of the functional YFP. All fluorescence microscopy was performed using a Zeiss LSM 5 Pascal confocal laser-scanning microscope. Experiments were repeated at least three times, with the entire bombarded leaf area examined in each experiment.
Mutagenesis of VirE3 NLS sequences
Fusions of VirE3 NLS1 or NLS2 to the C-terminus of GUS were produced by PCR using reverse primers 5′CGCGGATCCTCAATACTTTCGCTTCCTATCGGG GCTCTCCAGTCTGGTTTGCCGCTTTAATTGTTTGC CTCCCTGC3′ or 5′CGCGGATCCTCACGTCTTGGTTTTGCGAAGTCT ACCGTGCTCACGCGTTAATTCTTTTGGGTTGTCTA CTCGAAGCCGTTTTAATTGTTTGCCTCCCTGC3′, encoding the NLS1 and NLS2, respectively. GUS fusions with mNLS1 and mNLS2 were made using reverse primers 5′CGCGGATCCTCAATACTTTGCCACCACATCGGG GCTCTCCAGTCTGGTTTGCGCCTTTAATTGTTTGC CTCCCTGC3′ and 5′CGCGGATCCTCACGTCACGGTTACGGCAAGTCT ACCGTGCTCACGCGTTAATTCTTTTGGGTTGTCTA CTCGAAGCGCTTTTAATTGTTTGCCTCCCTGC3′, respectively. The resulting fragments were cloned into the NcoI–BamHI sites of pRTL2-GUS instead of GUS, producing pRTL2-GUS-NLS1, pRTL2-GUS-NLS2, pRTL2-GUS-mNLS1, and pRTL2-GUS-mNLS2. Both mutations were introduced into the full-length VirE3 using the BD Transformer™ Site-Directed Mutagenesis Kit, resulting in VirE3-mNLS12.
VirE3-mediated nuclear import of VirE2 in mammalian cells
For expression of intact proteins, PCR-amplified VIP1 or virE3 and virE3-mNLS12 sequences were cloned into the XhoI-BamHI or KpnI-BamHI sites of pCB6 (Tzfira et al, 2001), respectively. For GFP, YFP, and CFP tagging, PCR-amplified virE2, virE3, virE3-mNLS12, or VIP1 sequences were cloned into the EcoRI–BamHI or SalI-BamHI, respectively. Sites of pEGFP-C1, pEYFP-C1, or pECFP-C1 (Clontech). The indicated combinations of these expression constructs were introduced into 24-h-old COS-1 cells using FuGENE 6 (Roche), and subcellular localization of fluorescently tagged proteins was examined 24 h after transfection under a confocal microscope (Tzfira et al, 2001). For quantification of nuclear import as well as expression levels of YFP- and CFP-tagged proteins on a per cell basis, the signal intensity of total intracellular, intranuclear, or cytoplasmic fluorescence was determined using the recorded confocal images. Experiments were repeated at least three times, with 50–100 expressing cells examined in each independent experiment.
Bombardment and nuclear import of VirE3 and VirE2 in plant tissues
For expression from a single plasmid, GFP-VirE3 or GFP-VirE3-mNLS12 from pEGFP-C1-VirE3 or pEGFP-C1-VirE3-mNLS12 were first cloned into the NcoI–BamHI sites of pRTL2-GUS instead of GUS, and then the 35S promoter-GFP-VirE3-terminator or 35S promoter-GFP-VirE3-mNLS12-terminator cassettes were transferred into the SphI site of pGDR, which already contained a DsRed2 gene driven by the 35S promoter (Goodin et al, 2002), resulting in pGDR-GFP-VirE3 and pGDR-GFP-VirE3-mNLS12. The GUS-VirE2-expressing construct has been described (Citovsky et al, 1992b).
DNA (50 μg) was adsorbed onto 10 mg of 1-μm gold particles (Bio-Rad, CA) and bombarded at 150 psi into the leaf epidermis of greenhouse-grown Nicotiana tabacum plants or into the epidermis of onion scales using a Helios gene gun (PDS-1000/He, Bio-Rad), followed by incubation for 16 h at 25°C. For detection of fluorescently tagged proteins, the bombarded tissues were directly viewed under a confocal microscope. DsRed2 represents a useful marker for confocal microscopy analysis of nuclear import, because it does not require excitation by UV light and, thus, can be visualized using a conventional He/Ne laser, and, in addition to staining the cell nucleus, it allows to see the outlines of the entire plant cell, otherwise invisible on a confocal image.
For detection of GUS-tagged proteins, the samples were stained histochemically (Tzfira et al, 2001). The intensity of indigo dye formed during GUS assay in the cytoplasm and nucleus was quantified on a per cell basis by photodensitometry of the recorded images as described (Citovsky et al, 1992b; Howard et al, 1992; Tzfira et al, 2001). All experiments were repeated at least four times, with 10–20 expressing cells examined in each experiment.
Generation of VIP1 antisense/VirE3 double-transgenic plants
virE3 was inserted as a PCR-amplified EcoRI–BamHI fragment into pRTL2 (Restrepo et al, 1990), and the entire expression cassette was cloned as a HindIII fragment into the binary vector pBAR with a BASTA selection marker. The resulting pBAR-VirE3 construct in the Agrobacterium strain EHA105 was used to transform VIP1 antisense tobacco plants (produced and characterized previously, see Tzfira et al, 2001) as described. Double-transgenic plants expressing VIP1 in antisense orientation and VirE3 in sense orientation were selected on a kanamycin- and BASTA-containing medium and maintained and propagated in sterile conditions on an MS medium (Murashige and Skoog, 1962) with no exogenous growth regulators; 30-day-old plants were transferred to soil and grown at 25°C for 16/8 h photoperiod at 200 μmol photons m−2 s−1.
Quantitative RT–PCR
Total RNA was extracted from 2 g of leaf tissue, treated with RQ1 RNase-free DNase and M-MLV reverse transcriptase with strand-specific primers (i.e.,forward or reverse to detect antisense or sense transcripts, respectively) derived from VIP1, VirE3, and tobacco actin (GenBank accession number X63603), PCR-amplified (Ni et al, 1998), and the products detected by ethidium bromide staining. VIP1 forward and reverse primers, 5′CGAACGGTGTTGTTCCTCCTAATTCTCTT3′ and 5′GCTCAGCAAGTCTATCACC3′, respectively, generated a 290-bp product; VirE3 forward and reverse primers, 5′GGTGGACGGGACTTATCGAG3′ and 5′TTAGAAACCTCTGGAGGTGGAACG3′, respectively, generated a 210-bp product; and actin forward and reverse primers, 5′TCACTGAAGCACCTCTTAACC3′ and 5′CAGCTTCCATTCCAATCATTG3′, respectively, generated a 500-bp product.
Agrobacterium T-DNA gene expression
To assay early T-DNA gene expression, 9-mm discs from mature leaves of 1-month-old plants were cocultivated for 48 h at 25°C on a hormone-free MS medium with Agrobacterium strain EHA105 (A600=0.5) harboring a GUS-expressing binary vector pKIWI105 (Janssen and Gardner, 1990). For quantitative analysis, 20 disks per experimental condition were either combined, ground, and assayed for GUS activity using the fluorescent substrate 4-methylumbelliferyl β-D-galactoside or stained histochemically, and the number of GUS-positive areas per disks determined (Nam et al, 1999; Tzfira et al, 2001).
Acknowledgments
We thank Drs Gelvin, Goodin, and Haseloff for the kind gifts of pE2431, pGDR, and mGAL4-VP16 system, respectively. The work in our lab is supported by grants from the NIH, NSF, USDA, BARD, and BSF to VC, and by grants from the BARD and HFSP to TT.
References
- Atmakuri K, Ding Z, Christie PJ (2003) VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol 49: 1699–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Citovsky V (1997) Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc Natl Acad Sci USA 94: 10723–10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel P, Chien C, Sternglanz R, Fields S (1993) Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14: 920–924 [PubMed] [Google Scholar]
- Braem CV, Kas K, Meyen E, Debiec-Rychter M, Van De Ven WJ, Voz ML (2002) Identification of a karyopherin alpha 2 recognition site in PLAG1, which functions as a nuclear localization signal. J Biol Chem 277: 19673–19678 [DOI] [PubMed] [Google Scholar]
- Cascales E, Christie PJ (2004) Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304: 1170–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Blobel G (2001) Karyopherins and nuclear import. Curr Opin Struct Biol 11: 703–715 [DOI] [PubMed] [Google Scholar]
- Christie PJ (1997) Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol 179: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Kapelnikov A, Oliel S, Zakai N, Rojas MR, Gilbertson RL, Tzfira T, Loyter A (2004) Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J Biol Chem 279: 29528–29533 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Warnick D, Zambryski PC (1994) Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc Natl Acad Sci USA 91: 3210–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Wong ML, Shaw A, Prasad BVV, Zambryski PC (1992a) Visualization and characterization of tobacco mosaic virus movement protein binding to single-stranded nucleic acids. Plant Cell 4: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Zupan J, Warnick D, Zambryski PC (1992b) Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256: 1802–1805 [DOI] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C, Maiss E (2002) Red fluorescent protein DsRed from Discosoma sp. as a reporter protein in higher plants. BioTechniques 32: 286–293 [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA (1991) Nuclear targeting sequences—a consensus? Trends Biochem Sci 16: 478–481 [DOI] [PubMed] [Google Scholar]
- Dommisse EM, Leung DWM, Shaw ML, Conner AJ (1990) Onion is a monocotyledonous host for Agrobacterium. Plant Sci 69: 249–257 [Google Scholar]
- Garcia-Bustos J, Heitman J, Hall MN (1991) Nuclear protein localization. Biochim Biophys Acta 1071: 83–101 [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO (2002) pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J 31: 375–383 [DOI] [PubMed] [Google Scholar]
- Guralnick B, Thomsen G, Citovsky V (1996) Transport of DNA into the nuclei of Xenopus oocytes by a modified VirE2 protein of Agrobacterium. Plant Cell 8: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks GR, Smith HMS, Lobreaux S, Raikhel NV (1996) Nuclear import in permeabilized protoplasts from higher plants has unique features. Plant Cell 8: 1337–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka T, Kado CI (1986) Location of the right boundary of the virulence region on Agrobacterium tumefaciens plasmid pTiC58 and a host specifying gene next to the boundary. J Bacteriol 168: 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H (1995) Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol 15: 3813–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E, Zupan J, Citovsky V, Zambryski PC (1992) The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell 68: 109–118 [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Gardner RC (1990) Localized transient expression of GUS in leaf discs following cocultivation with Agrobacterium. Plant Mol Biol 14: 61–72 [DOI] [PubMed] [Google Scholar]
- Kalogeraki VS, Zhu J, Stryker JL, Winans SC (2000) The right end of the vir region of an octopine-type Ti plasmid contains four new members of the vir regulon that are not essential for pathogenesis. J Bacteriol 182: 1774–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf VC, Nester EW (1982) Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8: 45–54 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nagai H, Roy CR (2003) Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol 5: 373–383 [DOI] [PubMed] [Google Scholar]
- Nam J, Mysore KS, Zheng C, Knue MK, Matthysse AG, Gelvin SB (1999) Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol Gen Genet 261: 429–438 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee Y, Gurel F, Gafni Y, Dingwall C, Citovsky V (2000) A genetic system for detection of protein nuclear import and export. Nat Biotechnol 18: 433–437 [DOI] [PubMed] [Google Scholar]
- Schrammeijer B, Dulk-Ras Ad A, Vergunst AC, Jurado Jacome E, Hooykaas PJ (2003) Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res 31: 860–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrammeijer B, Risseeuw E, Pansegrau W, Regensburg-Tuïnk TJG, Crosby WL, Hooykaas PJJ (2001) Interaction of the virulence protein VirF of Agrobacterium tumefaciens with plant homologs of the yeast Skp1 protein. Curr Biol 11: 258–262 [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R (2003) Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J 36: 589–601 [DOI] [PubMed] [Google Scholar]
- Schwemmle M, Jehle C, Shoemaker T, Lipkin WI (1999) Characterization of the major nuclear localization signal of the Borna disease virus phosphoprotein. J Gen Virol 80: 97–100 [DOI] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M (1996) A three-hybrid system to detect RNA–protein interactions in vivo. Proc Natl Acad Sci USA 93: 8496–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh MW, Wessler SR, Raikhel NV (1993) Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol 101: 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachel SE, Nester EW (1986) The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium. EMBO J 5: 1445–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton A, Heller RC, Landry J, Choy JS, Sirko A, Sternglanz R (2001) A novel form of transcriptional silencing by Sum1-1 requires Hst1 and the origin recognition complex. Mol Cell Biol 21: 3514–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Theologis A (2004) Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc Natl Acad Sci USA 101: 2275–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Citovsky V (2000) From host recognition to T-DNA integration: the function of bacterial and plant genes in the Agrobacterium–plant cell interaction. Mol Plant Pathol 1: 201–212 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Citovsky V (2001) Comparison between nuclear import of nopaline- and octopine-specific VirE2 protein of Agrobacterium in plant and animal cells. Mol Plant Pathol 2: 171–176 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Rhee Y, Chen M-H, Citovsky V (2000) Nucleic acid transport in plant–microbe interactions: the molecules that walk through the walls. Annu Rev Microbiol 54: 187–219 [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V (2001) VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J 20: 3596–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V (2002) Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis VIP1 gene. Proc Natl Acad Sci USA 99: 10435–10440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CMT, Regensburg-Tuink TJ, Hooykaas PJJ (2000) VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290: 979–982 [DOI] [PubMed] [Google Scholar]
- Vergunst AC, van Lier MCM, den Dulk-Ras A, Hooykaas PJJ (2003) Recognition of the Agrobacterium VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol 133: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DV, Zambryski PC (2001) The six functions of Agrobacterium VirE2. Proc Natl Acad Sci USA 98: 385–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans SC, Allenza P, Stachel SE, McBride KE, Nester EW (1987) Characterization of the virE operon of the Agrobacterium Ti plasmid pTiA6. Nucleic Acids Res 15: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans SC, Mantis NJ, Chen CY, Chang CH, Han DC (1994) Host recognition by the VirA, VirG two-component regulatory proteins of Agrobacterium tumefaciens. Res Microbiol 145: 461–473 [DOI] [PubMed] [Google Scholar]
- Zambryski PC (1989) Agrobacterium–plant cell DNA transfer. In Mobile DNA, Berg DE, Howe MM (eds), pp 309–333. Washington, DC: American Society for Microbiology [Google Scholar]
- Zupan J, Muth TR, Draper O, Zambryski PC (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J 23: 11–28 [DOI] [PubMed] [Google Scholar]


