Abstract
The emergence of functional interactions between nucleic acids and polypeptides was a key transition in the origin of life and remains at the heart of all biology. However, how and why simple non-coded peptides could have become critical for RNA function is unclear. Here we show that putative ancient peptide segments from the cores of both ribosomal subunits, as well as derived homopolymeric peptides comprising lysine or the non-proteinogenic lysine analogues ornithine or diaminobutyric acid, potently enhance RNA polymerase ribozyme (RPR) function irrespective of chirality or chiral purity. Lysine decapeptides enhance RPR function by promoting holoenzyme assembly through primer-template docking, accelerate RPR evolution and enable RPR-catalyzed RNA synthesis at near physiological (≥1 mM) Mg2+ concentrations enabling templated RNA synthesis within membranous protocells. Our results outline how compositionally simple, mixed chirality peptides may have augmented the functional potential of early RNAs and promoted the emergence of the first protocells.
Life is widely believed to have descended from a simpler, primordial biology that lacked DNA and proteins but in which RNA played a central role1. A core component of this “RNA world” would have been an RNA polymerase ribozyme (RPR) to replicate primordial RNA genomes and become encapsulated within membranous compartments to form the first protocells2. Modern day RPR analogues have been developed3,4 some of which can synthesize ribozymes4, aptamers5 or ~200 nucleotide (nt) sequences using favorable RNA templates6, and even amplify short RNA sequences5, but their activity is strictly dependent on very high concentrations of available magnesium ions (optimal [Mg2+] ~ 200 mM)7,8. Such high [Mg2+] not only greatly accelerate RNA degradation9 but cause amphiphile aggregation, precipitation and membrane destabilization10,11, suggesting a fundamental incompatibility of RPR activity with the formation of membranous protocells. However, ribozyme function within the RNA world likely emerged not in isolation but in the context of a complex chemical environment comprising a range of other molecular entities potentially including amino acids, peptides and phospholipids12. Functional cooperation between RNA and peptides in particular appears to be ancient and likely reflects deep evolutionary history. For example, in modern biology the complex and manifold interactions between RNAs and (poly)peptides are central to RNA’s roles in gene expression. This includes ribozymes such as the spliceosome13, ribosome14 and RNase P15, all of which are functionally dependent on association with cognate polypeptides, despite all-RNA catalytic sites. Indeed, previous work had shown that some RNA-binding proteins and derived peptides can promote folding and function even in simple ribozymes16,17 and, in the case of the RNase P ribozyme core critically reduce the requirement for bivalent magnesium ions15. We therefore wondered if RPR function might be similarly enhanced by interaction with peptides, and, in particular, if they might reduce the strict Mg2+ dependency of RPR function.
A critical test of this hypothesis ideally would involve early peptides from the RNA world. As a potential source of ancient peptides we turned to the structure of the ribosome, the most prominent, putative molecular relic from the RNA world. According to accretion models of ribosome evolution18, the structure of the ribosome comprises a quasi-historical record of different stages of ribosomal evolution with Mg2+ ions enriched as counterions in the innermost core of the large ribosomal subunit reflecting the most ancient stages19,20, but progressively replaced during ribosome expansion by the acquisition of unstructured polypeptide “fingers” with a strikingly biased amino acid content21. It is tempting to speculate that such functional cooperation between RNA and peptides conserved within the ribosomal core structure reflects deep evolutionary history of the RNA world and its transition to a RNA-peptide (RNP) world19.
Here we have investigated if the compositional features of these ribosomal core peptide segments, widely considered to be among the most ancient protein sequences on earth22, could confer functions transferable to critical primordial RNA contexts. We identify multiple peptides from both ribosomal subunits that strongly enhance RNA polymerase ribozyme activity. From these peptide sequences we derive a simplified homopolymeric lysine decapeptide (K10), which we find to enhance RPR function at low [Mg2+] regardless of chirality, accelerate RPR evolution and enable RNA-catalyzed templated RNA synthesis within a membranous model protocell.
Results
RPR activation by short ribosomal peptides
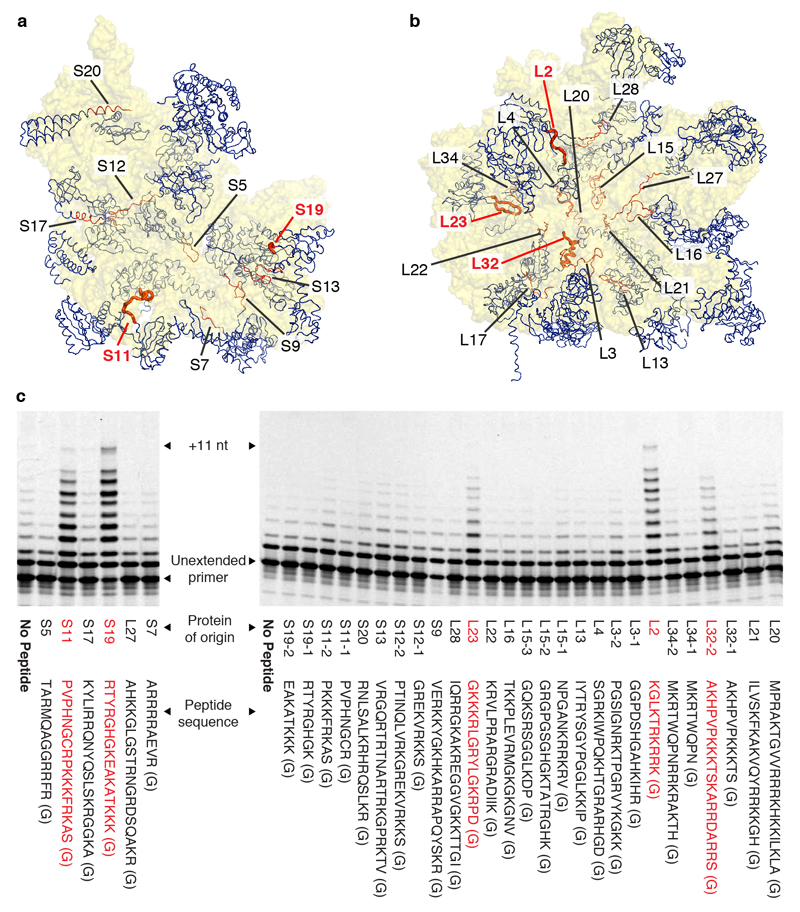
In order to examine if the unusual compositional and structural features of ribosomal core peptides could confer functional benefits transferable to other RNA contexts, we systematically examined peptide segments from ribosomal proteins of the Thermus thermophilus ribosome for their ability to enhance the RNA polymerase activity of the in vitro-evolved Z polymerase ribozyme. We focused on extended peptide segments of ribosomal proteins that reach deep into the cores of both subunits, as judged by the structure of the T. thermophilus 70S ribosome (PDB: 2XQD, 2XQE)23, which exhibit a highly biased amino acid composition (~20% R, K; ~12% G) (Fig. 1a, b). While many peptides showed no (or very weak) effects, we discovered multiple ribosomal peptides that displayed a striking enhancement of RPR activity at suboptimal Mg2+ concentration ([Mg2+] << 200 mM), particularly peptides from ribosomal proteins S11 (PVPHNGCRPKKKFRKAS), S19 (RTYRGHGKEAKATKKK), L2 (KGLKTRKRRK), L23 (GKKKRLGRYLGKRPD), and L32 (AKHPVPKKKTSKARRDARRS) (Fig. 1c, Supplementary Fig. 1).
Figure 1. Ribosomal peptides enhance the activity of an artificial RNA polymerase ribozyme.
a, b Proteins (blue) and their peptide extensions (orange) in the structures of the T. thermophilus 30S and 50S ribosomal subunits. c, PAGE analysis of primer extensions on template TI by the RPR Z at a suboptimal [Mg2+] of 22 mM (left panel, 4°C for 22 days) or 46 mM (right panel, 4°C for 7 days), alone or in the presence of 400 μM of peptides derived from ribosomal proteins (terminating in an extra glycine residue (G) from synthesis). Ribosomal proteins comprising very long unstructured peptide segments give rise to two peptides e.g. L32-1 & L32-2. Peptide segments enhancing RPR function (red) are highlighted in bold in a & b.
Next, we dissected amino acid contributions of one of the peptides, the S11 peptide segment (S11p: PVPHNGCRPKKKFRKAS(G)) in order to better understand critical peptide features responsible for RPR activation. Both N- and C-terminal truncations of S11p retained the ability to enhance RPR activity, provided they contained a central 11 amino acid segment (NGCRPKKKFRK) (Supplementary Fig. 2) enriched in positively charged amino acids, comprising two arginine (R) and four lysine (K) residues. Overall the activating peptide segments (Fig. 1c) displayed a significantly higher K content (22 K of 78 amino acids (28%) vs. 81 K of 434 amino acids (19%) amongst all peptides tested, K≥22 p=0.027), though no enrichment of R content (14 R of 78 amino acids (18%) vs. 80 R of 434 amino acids (18%)), implicating K as a potential key functional component.
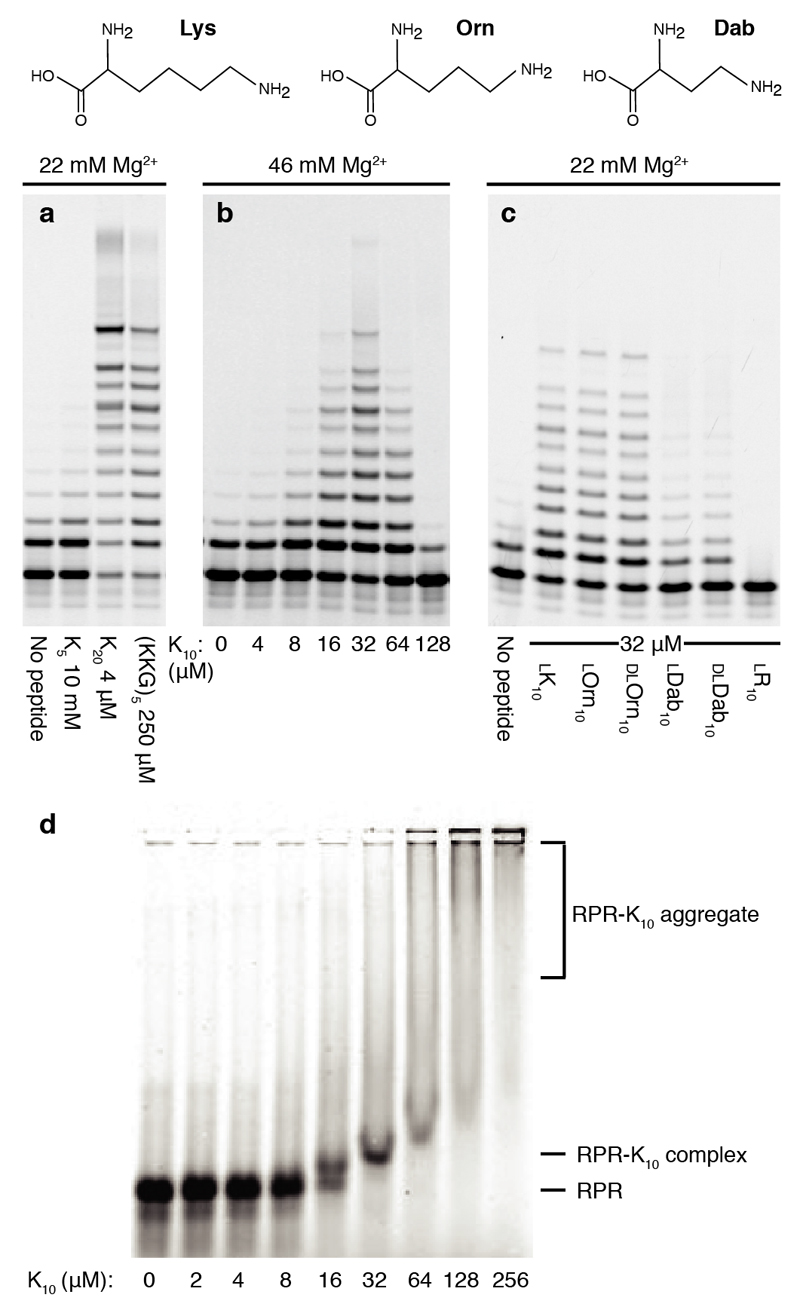
Having identified a range of peptides and established the functional importance of K therein, we sought to establish if the exact ribosomal peptide sequence was of critical functional importance, or if simplified K-rich peptides could recapitulate the activation effect. While a K5 pentapeptide provided no functional benefits, both K20 and (KKG)5 peptides showed strong RPR activation (Fig. 2a), as did a shorter K10 decapeptide (Fig. 2b). Importantly, while being able to substitute in part for Mg2+ as counterion, unlike Mg2+, the K10 peptide appears not to accelerate RNA hydrolysis (Supplementary Fig. 3).
Figure 2. RPR activation by homopolymeric peptides.
Top: the amino acids Lysine (K), Ornithine (Orn) and Diaminobutyric acid (Dab) (top panels) have progressively shorter amino group side chains. Below: PAGE of primer extensions on template TI by the Z RPR over 3 days at 17°C, alone or with indicated peptides of different (a) lengths, (b) concentrations and (c) side chains and chirality. d, EMSA: The fluorescently labeled Z ribozyme (Z-Fluor 647) was incubated with K10 peptide at the indicated concentrations with 22 mM [Mg2+], and resolved on a non-denaturing agarose gel.
We sought to dissect the chemical basis of this RPR activation by varying peptide side-chains and stereochemistry. To our surprise, peptide chirality had little influence upon enhancement, with poly-l-Lysine (ca. K700-K1500) and diastereomeric poly-d-Lysine both boosting Z activity to a comparable extent (Supplementary Fig. 4).
The activating effect was also maintained in both chirally pure (L) or racemic (DL) peptides comprising the non-proteinogenic amino acids ornithine (Orn) and to a lesser extent diaminobutyric acid (Dab) (Fig. 2c, d), in which the side-chain is shortened and the primary amino group is attached to the δ (Orn) or γ (Dab) carbon respectively. In contrast, an arginine R10 homopeptide only inhibited activity, suggesting that RPR activation is not based on a mere counterion effect mediated by positively charged side-chains. Primary amino groups as present in K, Orn and Dab peptides appear to be critical as well as a molecular scaffold of requisite reach (but surprisingly not precise stereochemistry). Indeed, an activating effect can be observed even in the absence of a peptide backbone as in polyethyleneimine, but not with simple polyamines such as spermidine and spermine (Supplementary Fig. 4).
Mechanism of RPR activation
We next sought to understand the mechanistic basis for RPR activation by the lysine decapeptide l-K10. K10 activation follows a bell-shaped concentration curve with optimal activation between 10-50 µM and higher concentrations inhibitory to RPR activity (Fig. 2b). Similar bell-shaped activation curves were observed for other synthetic and ribosomal peptides (Supplementary Fig. 4) with relative activating and inhibitory concentrations dependent on [Mg2+] (Supplementary Fig. 5) suggesting competitive interactions of K10 and Mg2+ ions with the RPR.
We sought to capture the interaction between K10 peptide and the RPR using native electrophoretic mobility shift assays (EMSA), which indeed show that a discrete upshifted RPR-K10 complex of unknown stoichiometry is formed (Fig. 2e) at approximately optimally activating K10 concentrations (K10 : RPR ratio ~20 : 1, although this stoichiometry may not represent complex composition). This complex is dissociated if Mg2+ is present in the gel during electrophoresis, while higher (inhibitory) concentrations of K10 (and R10) merely yielded non-specific aggregates of undefined molecular weights (Supplementary Fig. 6). Although this aggregate formation and inhibition appears to correspond to poly-arginine’s greater ability than poly-lysine to condense DNA in biological contexts24, the activity boost by the K10 and related peptides observed here likely has a different molecular basis.
The EMSA data suggested that RPR activation by K10 involved the formation of a peptide-ribozyme complex; although its role and persistence is unclear, we hypothesized that complex formation might enhance RPR activity by stabilizing some key intermediate in the RPR catalytic cycle. We had previously shown that docking of the primer/template (P/T) duplex with the apo-ribozyme (to form the RPR holoenzyme) is a limiting step in primer extension: RPR constructs loosely-tethered or (like Z) untethered to the P/T require cooperative binding of Mg2+ ions to low-affinity binding sites (i.e. high concentrations of Mg2+ ions) for activity, a requirement which can be partially overcome by tight in cis tethering of the P/T7, which however restricts extension length. Examining the relative degree of activation by K10 peptide in such RPR constructs, we found that while K10 strongly enhanced extension in untethered and to a lesser degree loosely tethered constructs, it provided only negligible activation when the P/T was tightly tethered (Supplementary Fig. 7). Taken together with complex formation detected by EMSA (Supplementary Fig. 6) and the fact that K10 cannot substitute for Mg2+ in RPR catalysis (Supplementary Fig. 7), this supports a mechanistic model whereby K10 exerts its activating effect by effecting P/T docking and holoenzyme formation, presumably through bridging between the P/T duplex to the RPR. Examination of the structural context of stimulatory peptide segments, such as L2 and L32, within the structure of the ribosome illustrates how they connect distinct RNA segments through interactions involving distal K side-chains (Supplementary Fig. 8) providing a potential structural rationalization for the holoenzyme formation and activation effect. Indeed, bridging may be a generic transferable function of such peptides, as K10 could also enhance tRNA maturation by the ribozyme component of RNase P, where R10 peptides again provided no benefit (Supplementary Fig. 9).
RPR evolution with peptide
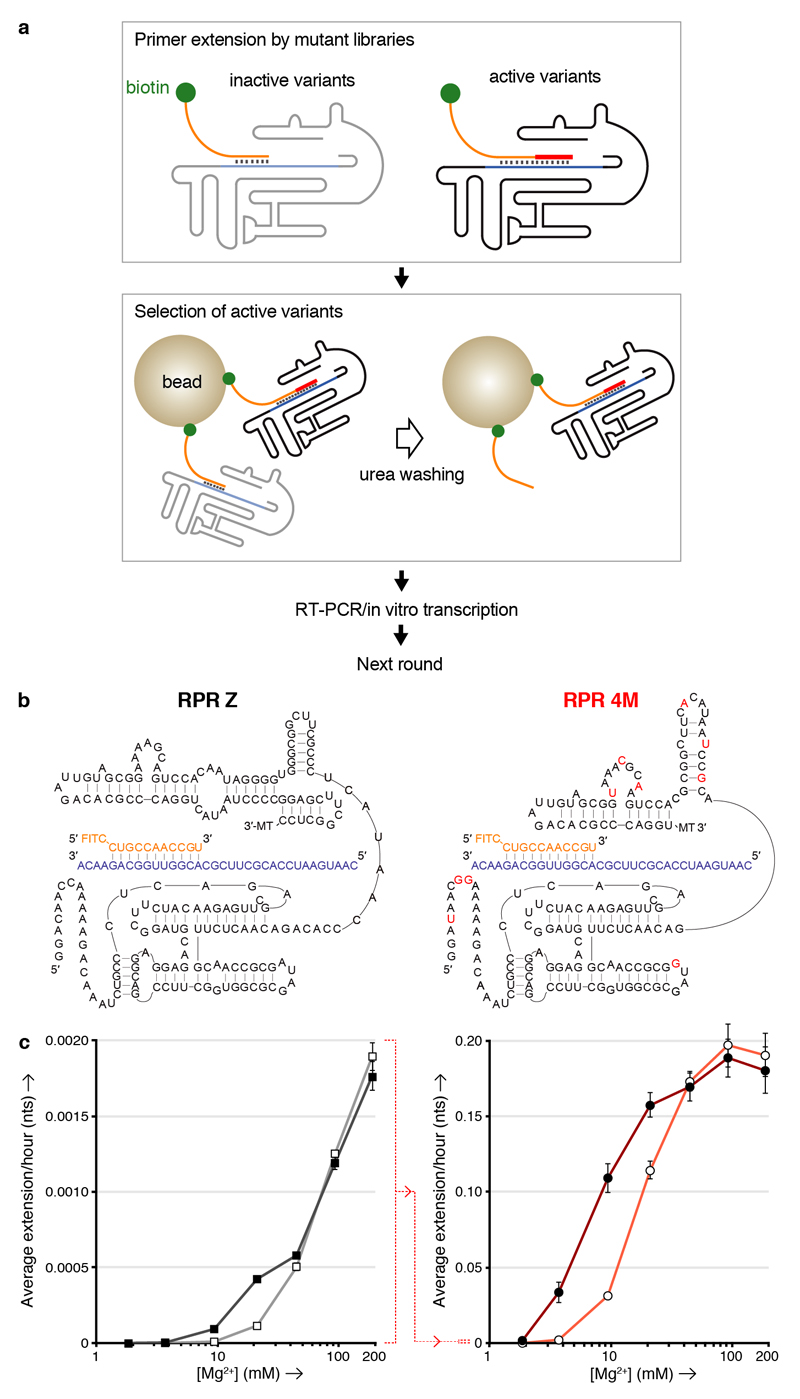
To better understand the impact of K10 on RPR functional potential, we also investigated in vitro RPR evolution in the presence and absence of peptide cofactor, starting from a shorter version of the Z RPR with a truncated accessory domain (Zd)25, which exhibits a similar activity and K10 dependence compared to Z (Supplementary Fig. 10). To this end we developed a rapid in vitro selection system for RPR function, called tethered self-tagging (TST), conceptually similar to the CST method developed for the evolution of proteinaceous polymerases26 (Fig. 3a, Supplementary Fig. 11). TST test selections at low [Mg2+] (25 mM) revealed a striking impact of K10 peptide on RPR evolution with selections in the presence of peptide progressing almost an order of magnitude faster, requiring less total incubation time for all 7 rounds of selection than the first round in the absence of peptide (Supplementary Fig. 12). Such peptide-mediated enhancements could also sensitise initially-weak activities to recovery using in vitro evolution.
Figure 3. K10 and [Mg2+] dependence of RPR activity by Z and the evolved 4M.
a, Outline of the TST selection scheme, showing how in cis-primer extension by a ribozyme prevents ribozyme loss during a denaturing wash on beads, allowing it to be recovered and amplified. b, Putative secondary structures of the Z and 4M ribozymes surrounding the primer (orange)/template (purple) duplex, showing 4M selected mutations in red (3’ MT = tail sequence from selection construct). c, Average primer extension per hour on template TI by the Z (left) or 4M (right) RPRs in 5 d (left) or 4 h (right) reactions at varying [Mg2+], with 6 μM K10 (filled symbols) or without it (open symbols) (error bars represent S. D., N = 3; note the change in scale indicated between the two panels (dotted red line)).
Encouraged by these efficiency gains, we carried out TST selections in the presence of K10 peptide under progressively lower [Mg2+] conditions, yielding RPR variants increasingly capable of RNA synthesis under such challenging conditions (Supplementary Fig. 13). Selection at much lower [Mg2+] (2 mM) yielded the RPR variant 4M (Fig. 3b), which exhibited a greatly reduced Mg2+ dependence displaying clear activity with boosts from K10 peptide at [Mg2+] between 2 - 30 mM. Furthermore, 4M outperformed Z by approximately two orders of magnitude over all Mg2+ concentrations both with and without K10 peptide (Fig. 3c, Supplementary Fig. 14). 4M mutations (including a number of insertions) cluster near the 5’ end, and in the accessory domain within a purine-rich loop (thought to be positioned near the active site25), and within the linker connecting the core to the accessory domain potentially reshaping this linker region into a new hairpin domain (Fig. 3b).
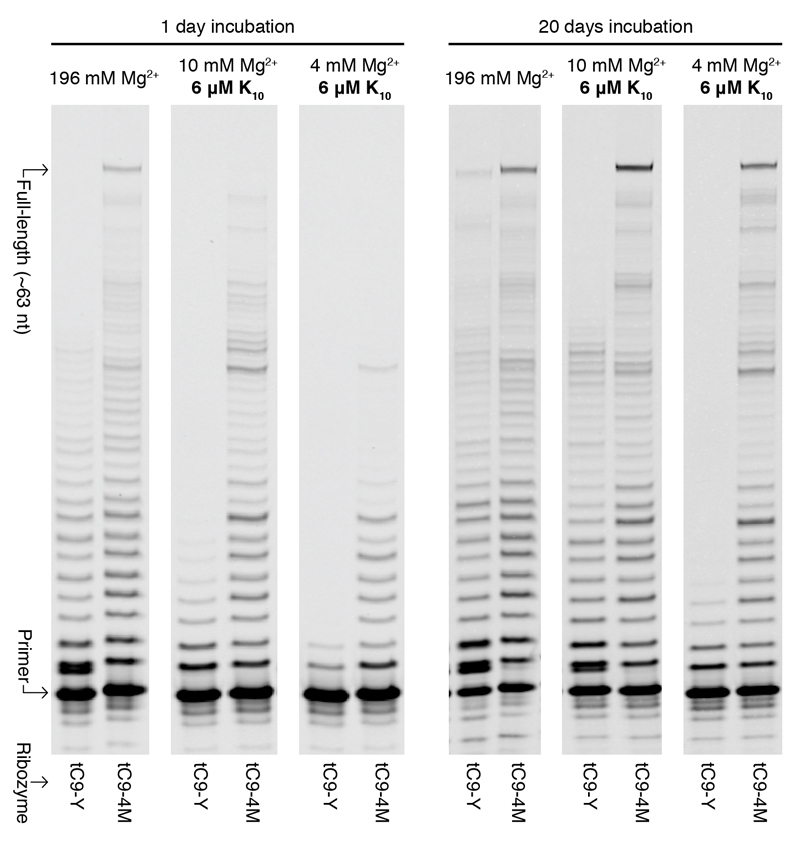
Next we appended a previously described 5’ sequence (5’-GUCAUUGA8 (tC9)6) that boosts long RNA synthesis, yielding tC9-4M which could perform long RNA synthesis down to 2 mM Mg2+ in the presence of K10 or at a near-physiological [Mg2+] of 1 mM in the presence of molecular crowding agents like PEG (Supplementary Fig. 15). tC9-4M greatly outperformed a previous top RPR, the fast and accurate tC9-Y6, under all conditions, synthesizing at high [Mg2+] (196 mM) more full-length (~63 nt) RNA products (0.8%) in a single day than tC9-Y did in 20 days (0.4%) (Fig. 4a, Supplementary Fig. 16).
Figure 4. Robust RNA synthesis during long incubations.
PAGE of primer extensions by RPRs tC9-Y and tC9-4M tethered to the long repeat template I-6.
Over long incubations at low [Mg2+] (4 and 10 mM), tC9-4M with K10 peptide yielded 2.3% and 4.6% full-length products (the latter a higher yield than the 2.9% it made at optimal high [Mg2+] (196 mM), presumably due to reduced RPR and template degradation), whilst tC9-Y was barely active under these conditions. tC9-4M could also synthesize RNAs longer than itself (177 nt) at low [Mg2+] (10 mM) with K10, though provided much better yields (2.4% of ~195 nt) at optimal high [Mg2+] (196 mM) (vs. 0.37% observed previously for tC9-Y under optimal conditions5) (Supplementary Fig. 17).
Finally, deep sequencing of total extension products showed that tC9-4M exhibits an essentially identical overall fidelity compared to tC9-Y (> 97.5% per position)5 and that neither low [Mg2+] nor K10 peptide significantly alter RPR fidelity (Fig. 4b, Supplementary Fig. 18).
Peptide-mediated RNA synthesis in protocells
Encapsulation of RNA synthesis and replication within compartments is widely considered an important transition in early evolution as well as a key stepping stone in generating a simple synthetic cell2,27, but high [Mg2+] can disrupt membrane integrity and cause precipitation of some membrane-forming amphiphiles. Simple ribozymes with low [Mg2+] requirements such as the hammerhead endonuclease ribozyme have been shown to operate at a reduced rate in membranous protocells formed from fatty acids (indicating ~ 1 mM free Mg2+)11,28, but the exceptionally high [Mg2+] requirement for RPR activity had previously rendered enzymatic RNA synthesis fundamentally incompatible with such membranous compartments.
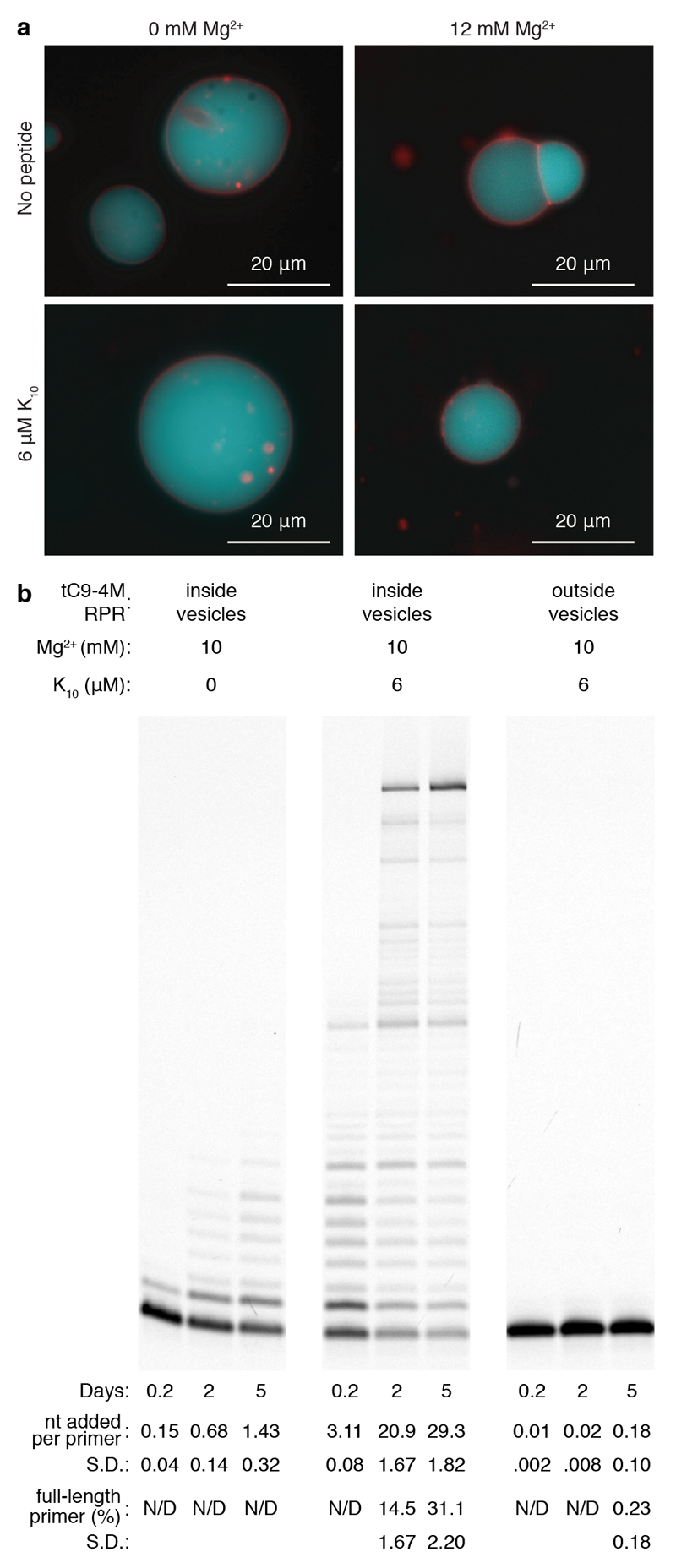
To begin to explore if the reduced Mg2+ requirements of tC9-4M in combination with K10 peptide could allow enzymatic RNA synthesis within model membranous compartments, we generated stable vesicles formed from the modern membrane lipid 1-palmitoyl-2-oleoylphosphatidylcholine (POPC), which could encapsulate RNA even in the presence of 10 mM Mg2+ with and without K10 peptide (Fig. 5a). Only in the presence of K10 peptide, we observed robust templated full-length RNA synthesis by tC9-4M within POPC compartments (Fig. 5b, and Supplementary Fig. 19 (variant protocol)). When only the primer / template and K10 were encapsulated and tC9-4M was added later to these preformed vesicles, no RNA synthesis was observed, confirming that the POPC vesicles were intact and able to compartmentalize macromolecules without leakage whilst still supporting RNA synthesis exclusively within the vesicular lumen.
Figure 5. Ribozyme-catalyzed RNA synthesis in phospholipid protocells.
a, Fluorescence micrograph of POPC vesicles containing RNA and NTPs, forming across a range of conditions with and without Mg2+ and K10 peptide; the membrane is stained with Rhodamine G (red) and encapsulated RNA is labeled with fluorescein (blue). b, PAGE of extensions at 17°C upon the long repeat template I-6 encapsulated within POPC vesicles, by tC9-4M added externally or within the vesicles. Quantification (N=4) of extension per primer and full-length extension are displayed beneath.
Discussion
Prebiotic chemistry provides increasingly clear indications that early RNA replication arose not in isolation but in a chemically diverse environment, which very likely included building blocks for simple peptides and lipids12. Here we have explored to what extent short peptides can interact with and augment the function of RNA polymerase ribozymes (RPRs) and can facilitate primordial RNA replication and the formation of model protocells.
Our results show that unstructured peptide segments from several ribosomal proteins, thought to be among the most ancient evolved coded protein sequences in biology22, are able to potently enhance the activity of an evolutionarily unrelated RNA polymerase ribozyme (RPR). It has been suggested that the replacement of Mg2+ by peptides in the outer and less ancient layers of the ribosome may reflect a transition from inorganic metal ions to peptides as counterions during ribosomal evolution19. Our data indeed indicates how these peptides might have promoted such a transition in proto-ribosomal function and how similar peptides might have helped early RPRs (and other ribozymes) achieve a similar shift from a high to a low [Mg2+] regime. Such a transition is crucial for the emergence of self-replication and cellular evolution, as high Mg2+ concentrations, while helpful in promoting RNA folding and catalysis, are destructive both to RNA stability and to the integrity of prebiotically plausible membranes - placing conflicting demands upon plausible solute compositions for early RNA replication. While membranes can be protected to some extent by chelating agents such as citrate, which has been shown to sequester Mg2+ in a form still able to promote non-enzymatic RNA synthesis from phosphorimidazolides29, citrate-chelated Mg2+ is inaccessible for RPR catalysis (Supplementary Fig. 20).
From the salient sequence features of activating ribosomal peptides we derived a maximally simple homopolymeric lysine decapeptide K10 that could relieve the dependency of RPRs on high [Mg2+] and potentiate RPR function, stability and evolution, enabling templated RNA synthesis down to near-physiological (≥1 mM) Mg2+ concentrations. In analogy to peptides’ function within the ribosomal core, and the ability of other peptides to promote simple ribozyme assembly17, the K10 peptide does not substitute for Mg2+ in catalysis but rather appears to act as a molecular bridge and counterion promoting RPR holoenzyme formation, i.e. the docking of the polyanionic RNA primer-template duplex to the polyanionic RPR apoenzyme. The ability of peptides to fulfill this highly Mg2+-dependent role may be a critical element in realizing ribozyme-catalyzed RNA synthesis in primordial protocells based on fatty acid or related amphiphiles, in addition to the more Mg2+-tolerant model POPC phospholipid membranes examined here.
A long-standing question has been whether and how early peptides could have conferred a beneficial heritable phenotype in the absence of encoded synthesis. The simple peptides described herein confer a critical phenotypic and adaptive advantage irrespective of chirality (or chiral purity) which is dependent largely on length and composition rather than sequence. Furthermore, other roles are emerging for other compositionally biased peptides e.g. in promoting template accessibility or membrane localization30,31. Unlike peptides with a defined sequence, such compositionally simple peptides would not depend on encoded synthesis for heredity but could plausibly emerge from prebiotic sources via a biased chemical (or non-specific enzymatic) peptide synthesis pathway. As proposed, they could become generally heritable in the form of peptidyl-transferase ribozymes with single amino acid specificity32 akin to the modern-day D-Ala-D-Ala ligase enzymes involved in bacterial cell wall synthesis. Such dedicated (homo)-peptide synthetase ribozymes would provide a conceptually simple linkage of peptide-encoded phenotypes to RNA-encoded genotypes in the absence of translation, and offer a plausible origin for a peptidyl-transferase ribozyme before its recruitment into a nascent proto-ribosomal system of encoded protein synthesis33,34.
While lysine is not usually considered to be among the most common amino acids emerging from prebiotic chemistry12, we find that decapeptides formed from the non-proteinogenic Lys analogues Orn and Dab (proposed as components of an early genetic code35) are functional substitutes and provide similar RPR activation. It has been postulated that the earliest amino acids incorporated into a primeval genetic code must have included the amino acids with the highest ‘catalytic potential’36 and amino acids bearing side chain primary amino groups (such as the above) certainly hold unique functional potential and might have benefited early protocellular entities in ways other than enhancing RPR function and RNA replication. For example, K-rich peptides have been shown to be capable of forming primitive catalysts with decarboxylase and retro-aldolase activities37,38 potentially providing simple metabolic functions, or can promote liquid-liquid demixing and coacervate formation at higher concentrations39, potentially a key mechanism for prebiotic substrate concentration and protocellular (and subcellular) molecular organization40.
In summary, our results suggest that the advent of RNA self-replication could have been promoted by simple mixed chirality peptides that reduced RNA dependence on inorganic counterions, accelerated RNA evolution and enabled partition of RNA replication within membranous compartments, a prerequisite for Darwinian evolution. The ancient molecular symbiosis between nucleic acids and peptides, a molecular memory of which remains captured within the structure of the ribosome, may thus have been a key factor in the emergence of the first protocells.
Materials and Methods
Primer extension assays
Standard primer extension reactions by the RNA polymerase ribozymes were performed with 250 nM each of ribozyme, template and 5’-FITC-labeled primer in the extension buffer containing 50 mM Tris•HCl buffer (pH 8.3), 0–200 mM MgCl2, 0.5 mM of each NTP, and at 17°C (unless indicated otherwise). Each NTP chelates 1-2 Mg2+ ions (with KDs at 37°C of 30 μM and 25 mM)41, and the predicted unchelated accessible Mg2+ concentrations (free [Mg2+]) are displayed. Initially, RNAs (ZLT, template TI (5’-CAAUGAAUCCACGCUUCGCACGGUUGGCAGAACA-3’), and primer FITC11 (5’-FITC-CUGCCAACCGU-3’) unless indicated otherwise) were annealed in H2O (50ºC for 5 min, 17ºC for 10 min). Then they were mixed with the extension buffer, including peptides (oligomer concentrations indicated). In some conditions where extension by the ribozyme was particularly limited (Supplementary Figs 2, 4, 10, 15), 8% PEG-6000 was added to the extension buffer to accelerate the primer extension by molecular crowding. After the incubation time indicated in the figure legends, the reactions were stopped by adding three volumes of stop buffer (75 mM EDTA, 8M Urea, and 10× (RNA) or 100× (DNA) excess of competing oligonucleotide able to hybridize to the template sequence), heated to 94ºC for 4 min, and resolved by urea-PAGE gels (20% PA, 8M urea). The gels were analyzed using a Typhoon Trio Scanner (GE Healthcare). Quantification of extension products was performed as previously described6, with the exception of those in Fig. 5b where bands beyond +11 were grouped into 11 nt ‘bins’ weighted by their average length. All nucleic acid sequences used are described in the three Supplementary Tables.
Formation and observation of POPC giant vesicles
Giant vesicles were prepared by the droplet transfer method42. First, an Eppendorf tube (2 mL) was filled with 0.5 mL of the “lower solution” containing 50 mM Tris-HCl (pH 8.3), 500 mM glucose and 2 μM Rhodamine 6G (Sigma-Aldrich). Then the interfacial phase (0.5 mL mineral oil (Sigma-Aldrich) containing 0.5 mM 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, Anatrace)) was added on the lower solution. The Eppendorf tube was left at room temperature for a few minutes to let POPC make a single layer between the outer solution and interfacial phase. Finally, a water/oil emulsion was prepared by pipetting the “inner phase” (50 μL, 4 μM RNA primer FITC11, 50 mM Tris HCl (pH 8.3), 0−12 mM MgCl2, 500 mM sucrose, ± 6 μM K10 peptide) in 1.0 mL of 0.5 mM POPC in mineral oil and added gently on the interfacial phase. The tube was immediately centrifuged at 300 g for 10 min at RT. After centrifugation, mineral oil and debris between mineral oil and aqueous phase were removed, and the aqueous phase was centrifuged again (2000 g, 5 min, RT). Then supernatant was removed and POPC vesicles were washed with 200 μL of 50 mM Tris HCl (pH 8.3), 500 mM glucose and vesicles were collected by centrifugation (2000 g, 2 min, RT). Finally vesicles were suspended in 50 μL of the “outer phase” (50 mM Tris HCl (pH 8.3), 0−12 mM MgCl2, 500 mM sucrose, ± 6 μM K10 peptide) and observed under the fluorescent microscope.
RNA primer extension in POPC giant vesicles
12.5 pmol of tC9-4MMT and the RNA primer/template (FITC11/I-6) were annealed in water separately. Then the primer/template was mixed with reaction buffer (50 mM Tris HCl (pH 8.3), 12 mM MgCl2, 0.5 mM each NTP, 500 mM sucrose, ± 6 μM K10 peptide) with/without tC9-4MMT to make 50 μl of the inner phase. Then vesicles were formed as described above. The vesicles were washed three times with 200 μL of 50 mM Tris HCl (pH 8.3), 500 mM glucose and suspended in 50 μl of the outer phase (50 mM Tris HCl (pH 8.3), 12 mM MgCl2, 0.5 mM each NTP, 500 mM sucrose, ± 6 μM K10 peptide) with/without tC9-4MMT. Then, the vesicles were incubated at 17ºC for the periods indicated. After incubation, the vesicles were washed three times with 200 μL of 50 mM Tris HCl (pH 8.3), 500 mM glucose, and collected by centrifugation. The vesicles were mixed with 3 volumes of the stop buffer (95 % formamide, 7.5 mM EDTA, and 10× excess of competing RNA able to hybridize to the template sequence), heated to 94ºC for 4 min, and resolved by urea-PAGE gels (10% PA, 8M urea). The gels were analyzed using a Typhoon Trio Scanner (GE Healthcare). A variant vesicle preparation method (Supplementary Materials & Methods) using paraffin oil and higher POPC concentrations also yielded in-vesicle extension (Supplementary Fig. 19)43.
Supplementary Material
Acknowledgments
The authors thank S. James for assistance with fidelity data processing. This work was supported by postdoctoral fellowships from JSPS (Japanese Society for the Promotion of Science) and HFSP (Human Frontiers Science Program) (S.T.) and by the Medical Research Council (J.A., P.H., program no. MC_U105178804). Correspondence and requests for materials should be addressed to P.H.
Footnotes
Data availability. All relevant data such as ribozyme and peptide sequences, primer and template sequences and detailed experimental conditions are included within the manuscript, specifically in Supplementary Tables and Supplementary Materials & Methods,
Contributions S.T. and P.H. conceived and designed the experiments. S.T. performed all experiments together with J. A. for selection design, fidelity measurement and peptide assays. All authors discussed the results, and jointly wrote and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.The RNA World. third edn. Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 2.Szostak JW, Bartel DP, Luisi PL. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 3.Johnston WK, Unrau PJ, Lawrence MS, Glasner ME, Bartel DP. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- 4.Wochner A, Attwater J, Coulson A, Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–212. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- 5.Horning DP, Joyce GF. Amplification of RNA by an RNA polymerase ribozyme. Proc Natl Acad Sci U S A. 2016;113:9786–9791. doi: 10.1073/pnas.1610103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attwater J, Wochner A, Holliger P. In-ice evolution of RNA polymerase ribozyme activity. Nature chemistry. 2013;5:1011–1018. doi: 10.1038/nchem.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attwater J, et al. Chemical fidelity of an RNA polymerase ribozyme. Chemical Science. 2013;4:2804–2814. doi: 10.1039/c3sc50574j. [DOI] [Google Scholar]
- 8.Muller UF, Bartel DP. Improved polymerase ribozyme efficiency on hydrophobic assemblies. RNA. 2008;14:552–562. doi: 10.1261/rna.494508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Attwater J, Wochner A, Pinheiro VB, Coulson A, Holliger P. Ice as a protocellular medium for RNA replication. Nat Commun. 2010;1 doi: 10.1038/ncomms1076. [DOI] [PubMed] [Google Scholar]
- 10.Monnard PA, Apel CL, Kanavarioti A, Deamer DW. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: implications for a prebiotic aqueous medium. Astrobiology. 2002;2:139–152. doi: 10.1089/15311070260192237. [DOI] [PubMed] [Google Scholar]
- 11.Chen IA, Salehi-Ashtiani K, Szostak JW. RNA catalysis in model protocell vesicles. J Am Chem Soc. 2005;127:13213–13219. doi: 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nature chemistry. 2015;7:301–307. doi: 10.1038/nchem.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valadkhan S, Mohammadi A, Jaladat Y, Geisler S. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci U S A. 2009;106:11901–11906. doi: 10.1073/pnas.0902020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 15.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 16.Coetzee T, Herschlag D, Belfort M. Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev. 1994;8:1575–1588. doi: 10.1101/gad.8.13.1575. [DOI] [PubMed] [Google Scholar]
- 17.Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bokov K, Steinberg SV. A hierarchical model for evolution of 23S ribosomal RNA. Nature. 2009;457:977–980. doi: 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- 19.Hsiao C, Mohan S, Kalahar BK, Williams LD. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol. 2009;26:2415–2425. doi: 10.1093/molbev/msp163. [DOI] [PubMed] [Google Scholar]
- 20.Klein DJ, Moore PB, Steitz TA. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith TF, Lee JC, Gutell RR, Hartman H. The origin and evolution of the ribosome. Biology direct. 2008;3:16. doi: 10.1186/1745-6150-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alva V, Soding J, Lupas AN. A vocabulary of ancient peptides at the origin of folded proteins. eLife. 2015;4:e09410. doi: 10.7554/eLife.09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeRouchey J, Hoover B, Rau DC. A Comparison of DNA Compaction by Arginine and Lysine Peptides: A Physical Basis for Arginine Rich Protarnines. Biochemistry. 2013;52:3000–3009. doi: 10.1021/bi4001408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang QS, Cheng LK, Unrau PJ. Characterization of the B6.61 polymerase ribozyme accessory domain. RNA. 2011;17:469–477. doi: 10.1261/rna.2495011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinheiro VB, et al. Synthetic genetic polymers capable of heredity and evolution. Science. 2012;336:341–344. doi: 10.1126/science.1217622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attwater J, Holliger P. A synthetic approach to abiogenesis. Nat Methods. 2014;11:495–498. doi: 10.1038/nmeth.2893. [DOI] [PubMed] [Google Scholar]
- 28.Engelhart AE, Adamala KP, Szostak JW. A simple physical mechanism enables homeostasis in primitive cells. Nature chemistry. 2016;8:448–453. doi: 10.1038/nchem.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adamala K, Szostak JW. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 2013;342:1098–1100. doi: 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamat NP, Tobe S, Hill IT, Szostak JW. Electrostatic Localization of RNA to Protocell Membranes by Cationic Hydrophobic Peptides. Angew Chem Int Ed Engl. 2015;54:11735–11739. doi: 10.1002/anie.201505742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia TZ, Fahrenbach AC, Kamat NP, Adamala KP, Szostak JW. Oligoarginine peptides slow strand annealing and assist non-enzymatic RNA replication. Nature chemistry. 2016 doi: 10.1038/nchem.2551. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Cech TR. Evolution of biological catalysis: ribozyme to RNP enzyme. Cold Spring Harbor symposia on quantitative biology. 2009;74:11–16. doi: 10.1101/sqb.2009.74.024. [DOI] [PubMed] [Google Scholar]
- 33.Krupkin M, et al. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos Trans R Soc Lond B Biol Sci. 2011;366:2972–2978. doi: 10.1098/rstb.2011.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrov AS, et al. History of the ribosome and the origin of translation. Proc Natl Acad Sci U S A. 2015;112:15396–15401. doi: 10.1073/pnas.1509761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartman H, Smith TF. The evolution of the ribosome and the genetic code. Life. 2014;4:227–249. doi: 10.3390/life4020227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szathmary E. The origin of the genetic code: amino acids as cofactors in an RNA world. Trends Genet. 1999;15:223–229. doi: 10.1016/s0168-9525(99)01730-8. [DOI] [PubMed] [Google Scholar]
- 37.Johnsson K, Allemann RK, Widmer H, Benner SA. Synthesis, structure and activity of artificial, rationally designed catalytic polypeptides. Nature. 1993;365:530–532. doi: 10.1038/365530a0. [DOI] [PubMed] [Google Scholar]
- 38.Muller MM, Windsor MA, Pomerantz WC, Gellman SH, Hilvert D. A rationally designed aldolase foldamer. Angew Chem Int Ed Engl. 2009;48:922–925. doi: 10.1002/anie.200804996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koga S, Williams DS, Perriman AW, Mann S. Peptide-nucleotide microdroplets as a step towards a membrane-free protocell model. Nature chemistry. 2011;3:720–724. doi: 10.1038/nchem.1110. [DOI] [PubMed] [Google Scholar]
- 40.Mann S. Systems of creation: the emergence of life from nonliving matter. Acc Chem Res. 2012;45:2131–2141. doi: 10.1021/ar200281t. [DOI] [PubMed] [Google Scholar]
- 41.Thomen P, et al. T7 RNA polymerase studied by force measurements varying cofactor concentration. Biophys J. 2008;95:2423–2433. doi: 10.1529/biophysj.107.125096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miele Y, Bánsági T, Jr, Taylor AF, Stano P, Rossi F. In: Advances in Artificial Life, Evolutionary Computation and Systems Chemistry Vol. 587 Engineering Enzyme-Driven Dynamic Behaviour in Lipid Vesicles. Rossi F, et al., editors. Ch. 18. Springer International; 2016. pp. 197–208. [Google Scholar]
- 43.Fujii S, et al. Liposome display for in vitro selection and evolution of membrane proteins. Nature Protocols. 2014;9:1578–1591. doi: 10.1038/nprot.2014.107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.