Abstract
This study presents the crystal structure of domain I of the Escherichia coli ribosome recycling factor (RRF) bound to the Deinococcus radiodurans 50S subunit. The orientation of RRF is consistent with the position determined on a 70S-RRF complex by cryoelectron microscopy (cryo-EM). Alignment, however, requires a rotation of 7° and a shift of the cryo-EM RRF by a complete turn of an α-helix, redefining the contacts established with ribosomal components. At 3.3 Å resolution, RRF is seen to interact exclusively with ribosomal elements associated with tRNA binding and/or translocation. Furthermore, these results now provide a high-resolution structural description of the conformational changes that were suspected to occur on the 70S-RRF complex, which has implications for the synergistic action of RRF with elongation factor G (EF-G). Specifically, the tip of the universal bridge element H69 is shifted by 20 Å toward h44 of the 30S subunit, suggesting that RRF primes the intersubunit bridge B2a for the action of EF-G. Collectively, our data enable a model to be proposed for the dual action of EF-G and RRF during ribosome recycling.
Keywords: crystallography, elongation factor G, protein synthesis, ribosome recycling, ribosome recycling factor
Introduction
Ribosome recycling is the final stage of translation and involves the concerted action of the ribosome recycling factor (RRF) and elongation factor G (EF-G) to disassemble the post-termination complex for the next round of translation. RRF is universally conserved in bacteria, but not present in archaea or eukaryotes (with the exception of chloroplast and mitochondrial RRFs). Deletion of frr, the gene encoding RRF, is lethal to Escherichia coli cells (Janosi et al, 1994) and, in the absence of RRF, ribosomes remain bound to the mRNA and initiate spontaneous translation downstream of the stop codon (Ryoji et al, 1981; Janosi et al, 1998). The cellular importance and kingdom distribution of RRF make bacterial ribosome recycling an attractive target for drug design; however, such an undertaking requires an atomic understanding of the ribosomal binding site of RRF.
Although RRF was discovered in the early 1970s (Hirashima and Kaji, 1970; Subramanian and Davis, 1973), the exact mechanism by which RRF mediates ribosome recycling still remains to be fully elucidated: The experiments reported by Fujiwara et al (2004) indicate that RRF action is independent of the ribosomal translocase activity of EF-G, whereas Kaji and co-workers have presented evidence that the translocase function of EF-G is necessary for RRF-mediated release of deacylated tRNA from the ribosome and mRNA release is concomitant with EF-G dissociation (Hirokawa et al, 2002b; Kiel et al, 2003). In their in vitro system, the combined action of EF-G and RRF converts polysomes to monosomes, while the addition of initiation factor 3 (IF3) is required to split the ribosome into the component subunits (Hirokawa et al, 2002b). In contrast, Ehrenberg and co-workers found that the dissociation of 70S ribosomes required EF-G-dependent GTP hydrolysis in the presence of RRF, and that IF3 was instead necessary for the removal of the deacylated tRNA from the programmed small subunit (Karimi et al, 1999).
The structure of RRF has been solved from five different organisms (Selmer et al, 1999; Kim et al, 2000; Toyoda et al, 2000; Yoshida et al, 2001; Nakano et al, 2002, 2003), revealing an L-shaped two-domain molecule, with similar dimensions to tRNA, which led to the suggestion that RRF is a structural and functional tRNA mimic (Selmer et al, 1999). Nuclear magnetic resonance (NMR) and molecular dynamics simulation studies of Aquifex aeolicus RRF (Yoshida et al, 2001, 2003) have demonstrated that the hinge region between domains I and II is flexible enough to allow ∼60° rotational freedom perpendicular to the axis of domain I, while the interdomain angle of 90° remains constant. The relative orientation of domains I and II from all available RRF crystal structures fall within the 60° rotational range (Nakano et al, 2003) and, with the exception of the E. coli RRF structure where a detergent was bound within the hinge region (Kim et al, 2000), also have an interdomain angle of 90°. Genetic-based analysis of the hinge region suggests that the flexibility between domains I and II is important for RRF function in vivo (Toyoda et al, 2000).
Contrary to the concept of molecular mimicry of tRNA by RRF (Selmer et al, 1999), recent studies using hydroxyl radical probing and cryoelectron microscopy (cryo-EM) have revealed that RRF adopts a significantly different orientation on the ribosome to that of a tRNA, such that domain I of RRF interacts predominantly with the large subunit (Lancaster et al, 2002; Agrawal et al, 2004), rather than with the small subunit as predicted by tRNA mimicry. This orientation is in agreement with the observation that domain I alone (RRF-DI) binds to the 50S subunit with an affinity comparable to that of RRF (Nakano et al, 2003). Furthermore, RRF-DI can inhibit the ability of RRF to disassemble polysomes to monosomes in vitro, suggesting that RRF-DI binds in the same position as domain I of the full-length RRF (Nakano et al, 2003).
Here, we present the crystal structure of RRF-DI bound to the large subunit of Deinococcus radiodurans at a resolution of 3.3 Å. Our study confirms the general orientation observed in the low-medium-resolution hydroxyl radical probing and cryo-EM studies (maximum resolution of 12 Å), although in both cases the position of RRF must be rotated by 12–7° and shifted by 7–8 Å, respectively, to be aligned with the position determined here. The atomic details of the interaction of RRF with the large ribosomal subunit reveal that domain I of RRF contacts exclusively elements involved with tRNA binding and/or translocation: (i) nucleotides G2253–G2255 (E. coli numbering is used throughout) of the P loop (H80), which play an important role for the positioning of the tRNA in the P-site, (ii) the base of A2602 present in H93, which has been suggested to guide the CCA ends of the tRNA from the A- to P-site during translocation, (iii) the ribosomal proteins L16 and L27 at the peptidyl-transferase center (PTC), which have been implicated in positioning of tRNAs at the P site and (iv) extensive contacts with H69–H71, components of the intersubunit bridges B2a and B3. Furthermore, this study reveals the structural details of the conformational changes that were suspected by cryo-EM to occur in the large subunit. In particular, a novel conformation for H69 is observed with a consequent shift in the position of the tip of H69 by 20 Å toward h44 of the small subunit. These results provide a structural basis upon which to rationalize the dual action of RRF and EF-G on the ribosome during ribosome recycling.
Results
The binding position of domain I of RRF on the 50S subunit
Crystals of the D. radiodurans 50S subunit (D50S)-RRF-DI complex yielded a 3.3 Å structure (see Table I and Materials and methods for details). The unbiased electron density map, based on the native D50S structure (Harms et al, 2001), shows clear density, which can be unambiguously assigned to the three-helix bundle of RRF-DI (Figure 1A). The three helix bundle of domain I of RRF is comprised of noncontiguous sections, namely, the N-terminal α-helix (α1), residues Met1-Gly30 and the two C-terminal α-helices (α3 and α4), residues Thr106-Phe185 (Figure 1B). The three-layer β/α/β sandwich, comprising domain II of RRF, has been replaced by a Gly-Gly-Gly loop bridging the 10 Å gap between Gly30 and Thr106 to create RRF-DI (Nakano et al, 2003). Docking of the two known crystal structures of E. coli RRF (Kim et al, 2000; Nakano et al, 2002) into the density demonstrates the excellent agreement between the free and bound forms of domain I of RRF, and indicates that there are no gross conformational changes in domain I upon ribosome binding (data not shown).
Table 1.
Crystallographic data
| Crystal information | |
| Space group | I222 |
| Unit-cell parameters (Å) | a=168.5, b=405.0, c=693.0 |
| Diffraction data statistics | |
| Resolution (Å) | 30–3.30 (3.36–3.30) |
| Completeness (%) | 95.2 (86.8) |
| Rsym (%) | 10.4 (39.8) |
| I/s(I) | 9.8 (2.3) |
| Refinement statistics | |
| R factor (%) | 27.8 (38.9) |
| Rfree (%) | 33.6 (41.1) |
| Bond distances r.m.s. (Å) | 0.009 |
| Bond angles r.m.s. | 1.3° |
Figure 1.

The binding position of domain I of RRF on the 50S subunit. (A) Stereo view of the 2FoRRF-DI–FcNative electron density map of RRF-DI with the fitted structure of domain I (purple ribbons with cyan side chains) of E. coli RRF (Kim et al, 2000). (B) Sequence alignment of E. coli RRF-DI compared with the full-length E. coli and D. radiodurans RRFs (Swiss-Prot Accession numbers P16174 and Q9RU82, respectively). The regions of α-helical secondary structure are indicated in red. Identical and conservative substitutions are shaded dark and light blue, respectively. The three Gly residues (G) that replace domain II in the RRF-DI protein are in bold typeface, whereas dashes indicate gaps in the protein sequence alignment. A conversion table for the E. coli RRF-DI and full-length RRF can be downloaded from http://www.riboworld.com/pubrel/rrfalign.html. (C) Overall orientation of RRF-DI (purple) on the D. radiodurans 50S subunit. Ribosomal rRNA and proteins are colored gray, except for ribosomal proteins L16 (brown), L11 (cyan) and L27 (pink) and rRNA regions, H69 (orange), H71 (pale blue), H80 (cyan), H93 (yellow) and the H95 (SRL, blue). (D) Superposition of RRF-DI (pink) with positions of A- (green) and P-tRNA (cyan) based on relative positions from 70S·tRNA3 structure (Yusupov et al, 2001). The docking of the two extremes of domain II (RRF-e1, closed in red and RRF-e2, open in blue) from the NMR analysis of A. aeolicus RRF (Yoshida et al, 2001) is also included, as well as the putative position of a P/E-tRNA hybrid site (orange).
RRF-DI is located on the intersubunit side of the 50S subunit, positioned such that the tip is in close proximity to H80 of the PTC and the three-helix bundle extends toward the sarcin–ricin loop (SRL) (Figure 1C). The general orientation of RRF-DI is consistent with the hydroxyl radical probing data of Lancaster et al (2002) and very similar to the cryo-EM reconstruction reported by Agrawal et al (2004). To map the position of RRF determined by hydroxyl radical probing onto the position visualized by cryo-EM, it was necessary to shift the RRF by 5 Å toward the PTC and 13 Å in the direction of ribosomal protein L5. In order to superimpose the RRF position from the cryo-EM onto our position, it is necessary to make an additional 8 Å translation of RRF toward H80 of the PTC as well as a rotation of ∼7° (see Supplementary Figure 1). Accordingly, the discrepancies observed between the latter two RRF positions has consequences for the specific interactions described between the amino acids in RRF and the nucleotides of the 23S rRNA, namely, that the contacts present in the crystal structure are translated by one pass of the α-helix of RRF with respect to the cryo-EM position.
The orientation of RRF excludes the possibility of binding of tRNAs at either A or P sites of the ribosome (Figure 1D). This is consistent with the observation that RRF competes with the binding of NAcPhe-tRNA to the P site of nonprogrammed ribosomes, but not with deacylated tRNA to the E site (Hirokawa et al, 2002a). However, the binding position of RRF-DI does not overlap with the position of the CCA-ends of the A- and P-tRNAs, which would explain why antibiotics that prevent interaction of the CCA-ends of the tRNAs with the ribosome, such as chloramphenicol, lincomycin and clindamycin, do not inhibit RRF binding (Ishino et al, 2000). Biochemical as well as cryo-EM analysis of the RF2-termination complex have revealed that following release of the polypeptide chain, deacylated tRNA remains bound at the P and E sites (Rawat et al, 2003; Marquez et al, 2004). Unless the action of RF3 to dissociate RF2 also promotes slippage of the P-tRNA into the hybrid P/E position, then the overlap between RRF and the P-tRNA suggests that RRF must induce translocation of the P-tRNA to a P/E hybrid site.
RRF interaction at the PTC
Multiple components, both RNA and protein, in proximity to the PTC of the ribosome interact with the tip of domain I of RRF (Figure 2A and B). The long N-terminal extension of ribosomal protein L27 approaches the N-terminal region of α1 and the adjacent loop connecting α3 and α4 of RRF (Figure 3A). The N-terminal extension of L27 is highly flexible and the tip is partially disordered in the native 50S structure (Harms et al, 2001), whereas interaction with RRF-DI promotes a single distinct conformation. Residues 3–5 of the N-terminal end of L27 align along the α3–α4 loop of RRF (Asp145-Glu147), utilizing hydrophobic interactions as well as potential hydrogen bonds between the ɛ-amino group of Lys4 of L27 and the side-chain oxygens of both Asp145 (OD2) and Glu147 (OE1) of RRF (Figure 3A). L27, which is only present in bacterial ribosomes, has a relatively well-conserved N-terminal extension, such that His3 is conserved between E. coli and D. radiodurans, and Lys4 and Lys5 are present in all available L27 sequences. This suggests that L27 probably plays a role in the binding of RRFs of all bacterial species. Hydrogen bonding is also possible between ribosomal protein L16 and α4 of RRF-DI. The side-chain oxygen (OE2) of Glu81, located within the elongated loop extending from the globular domain of L16, comes within hydrogen-bonding distance of NH1 of Arg155, which protrudes from α4 of RRF.
Figure 2.
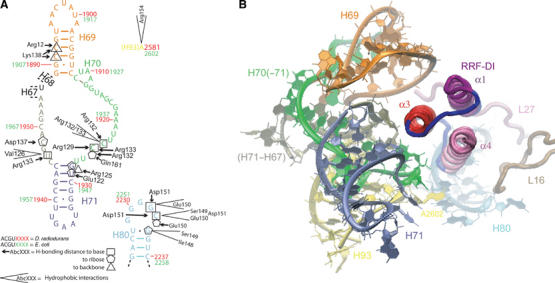
Interactions between domain I of RRF and the 50S subunit. (A) Schematic representation of the interaction of RRF-DI with H69–H71, H80 and H93. The numbering for both E. coli (green) and D. radiodurans (red) is given on the relevant regions of the secondary structure diagram of the 23S rRNA of D. radiodurans (Harms et al, 2001). Arrows indicate hydrogen-bonding distance to the base (square), or backbone interactions with the ribose (pentagon) or phosphate-oxygens (triangle), for each rRNA position. Hydrophobic interactions are indicated with open wedges. The colors of the nucleotides correspond to those presented in (B). (B) Overview of the interactions of RRF-DI with the large ribosomal subunit. Predominant contacts include α3 of RRF-DI with H69 (orange) and position at the base of H71 (green and tan). The relative position of the loop region linking α3 and α4 with H80 (light blue), A2602 in H93 (yellow) and the extensions of ribosomal proteins L16 (brown) and L27 (pink) are illustrated in the background.
Figure 3.
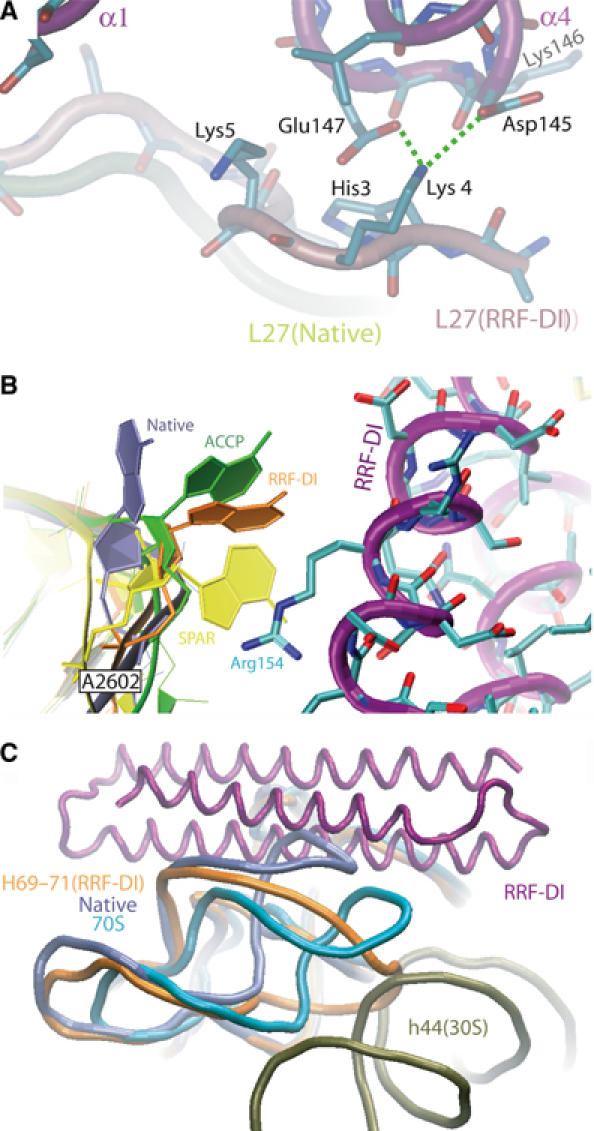
Conformational changes induced upon RRF binding to the ribosome. (A) RRF-DI induces a distinct conformation in L27. The native (yellow) and RRF-DI (pink) conformations of the N-terminal region of L27 are shown. Hydrogen bond interactions between Lys4 of L27 and the side chains of Asp145 and Glu147 within the loop between α3 and α4 of RRF-DI (purple) are indicated with dashed green lines. Hydrophobic interactions are present between Lys5 and the N-terminal of α1 and His3 with the α3–α4 loop, including Lys146 of RRF-DI. (B) Movement of A2602 of H93 upon binding of RRF-DI. Comparison of the orientation of A2602 in the RRF-DI D50S structure (orange) with the native (Harms et al, 2001) (purple), CCA-puromycin- (ACCP, green) and sparsomycin-bound (SPAR, yellow) D50S structures (Bashan et al, 2003). Arg154 of α3 of RRF-DI (purple backbone with cyan side chains) forms hydrophobic interactions with A2602 of H93. (C) Binding of RRF-DI induces a shift in the position of H69. Longitudinal view of RRF-DI (purple), with a superposition of H69–H71 from the RRF-DI bound D50S (orange), native D50S (pale blue; Harms et al, 2001) and 70S·tRNA3 (aqua; Yusupov et al, 2001) structures. The position of h44 (olive) of the 16S rRNA of the small subunit illustrates a potential clash with the position of H69 from the RRF-DI-bound D50S structure.
Two distinct regions of domain V of the 23S rRNA also interact with the tip of RRF-DI. The first involves contact between residues located within the loop connecting α3 and α4 of RRF-DI and the single-stranded loop of H80 (Figure 2A). The loop region of H80 is universally conserved across the three phylogenetic domains (Cannone et al, 2002), and has been termed the P loop (Puglisi et al, 1997), because nucleotides G2251 and G2252 form Watson–Crick base pairs with the terminal C74 and C73, respectively, of a P-tRNA.
Multiple hydrogen bond and hydrophobic interactions are made between Ile148-Asp151 and G2253-G2255. Hydrogen bond interactions are observed between the side-chain oxygens (OD1 and OD2) of Asp151 with the N1 of G2253 and the N3 of C2254, respectively, and also between the O2′ ribose of C2254 and the backbone N of Glu150 (Figure 2A). Additional hydrophobic interactions are observed from Glu150 and Asp151 with G2253/C2254 as well as Ile148 and Ser149 with G2255. Of these contacts, the interaction between Glu150 and G2253 was also predicted by the cryo-EM study (Agrawal et al, 2004).
The second region of rRNA-RRF contact involves hydrophobic interaction between Arg154 in α4 of RRF with A2602, located in H93 of the 23S rRNA. Interestingly, the universally conserved residue A2602 has shifted from its position in the native structure to lie adjacent to the side chain of Arg154 of RRF-DI, with an orientation closely resembling that seen in the ACC-puromycin (ACCP) D50S complex (Bashan et al, 2003) (Figure 3B). A2602 is at the center of an observed rotational symmetry within the PTC and has been proposed to guide the CCA-ends of the tRNAs during translocation (Bashan et al, 2003). Multiple orientations for A2602 have been observed depending on the ligand bound (Figure 3B). For example, the antibiotic sparsomycin, which has been shown to induce EF-G-independent translocation (Fredrick and Noller, 2003), binds at the PTC predominantly through stacking interactions with the base of A2602 (Bashan et al, 2003; Hansen et al, 2003). The binding of RRF seems to have disrupted the interaction between H80 and the CCA-end of the P-tRNA via conformational rearrangement of A2602, which could, in turn, facilitate the aforementioned translocation of the deacylated P-tRNA to the hybrid P/E site.
RRF induces conformational change within the intersubunit bridge elements H69–H71
The most extensive contacts between RRF-DI and the 50S subunit are with H69–H71 of domain IV of the 23S rRNA (Figure 2A and B). In the 70S ribosome, H69 and H71 make contact with h44 of the 30S subunit to form intersubunit bridges B2a and B3, respectively (Gabashvili et al, 2000; Yusupov et al, 2001; Gao et al, 2003). Binding of RRF-DI to the 50S induces movement of H69 away from the stalk region, to resemble closer the position observed in the 70S ribosome (Yusupov et al, 2001). However, the loop of H69 in the D50S-RRF-DI structure has a different and more open conformation compared to that in the 70S, such that its tip is shifted by 20 Å toward h44 of the small subunit (Figure 3C and Supplementary Figures 2 and 3). The movement of H69 from its position observed in the native D50S structure is prerequisite for RRF binding to avoid extended clashes between the two components. This may explain why RRF-DI has a higher affinity for 70S ribosomes (KD 0.16 μM) than for 50S subunits (KD 0.52 μM) (Nakano et al, 2003).
The interaction between RRF-DI and H69–H71 occurs predominantly on the α3 side of domain I, such that the opposite α1–α4 interface of RRF-DI remains completely free from contacts (Figure 2A and B). α3 has a multitude of exposed charged residues along one side of the helix that form hydrogen bonds, predominantly with the single-stranded nucleotides located on either side of H71 (Figure 4A) and, to a lesser extent, with the sugar-phosphate backbone of residues located in H69 (Figure 4B). For example, the side chains of Glu122 (OE1) and Arg125 (NH2) appear to form hydrogen bonds with the ribose- and phosphate-oxygens of G1945 and U1946, respectively (H71 in Figure 4A). Similarly, Arg129 can form hydrogen bonds with the ribose O2′ of G1945, and hydrogen bonds are also possible between the side-chain oxygens of Asp137 and the O2′ ribose of C1965.
Figure 4.
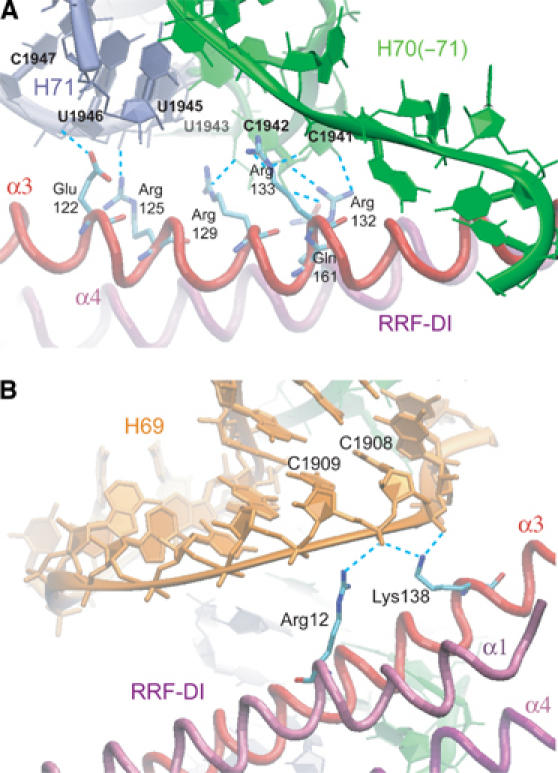
H69 and H71 are the main interaction partners of RRF-DI. (A) Interactions between α3 and α4 of RRF-DI with H67–H71 of the 23S rRNA. Multiple hydrogen bonds (dashed cyan lines) are formed between α3 (red) and α4 (purple) of RRF-DI with H70(–71) (green) and H71 (pale blue). (B) Interactions between α1 and α3 of RRF-DI with H69 of the 23S rRNA. Arg12 (α1) and Lys138 (α3) form hydrogen bonds (dashed cyan lines) with the backbone of nucleotides C1908 and C1909 of H69 (orange).
In addition to interactions with the backbone of the rRNA, base-specific interactions are seen for Arg132 (NH2) and Arg133 (NH1), which are within hydrogen-bonding distance of the O2 of bases C1941 and U1963, respectively. In addition, the NH1 of Arg133 can form hydrogen bonds with the O2 of C1941 (H70–H71 in Figure 4A) and the O2′ of U1963. Interaction with H71 outside of α3 involves Gln161 in α4, the O1E of which is located 3.6 Å from the C1942 (Figure 4A). Contacts to H69 involve the side-chain NH2 of Arg12 in α1 and Lys138 in α3 of RRF-DI, which are within hydrogen-bonding distance of the phosphate-oxygens of C1908 and C1909, as illustrated in Figure 4B.
Many of these interactions are likely to be used for the binding of RRF from other species to their respective ribosomes, since Glu122, Arg129 and Arg132 are universally conserved within all RRF sequences known to date and Arg133 and Lys138 are highly conserved (91–95%). In contrast, Asp137 and Arg12 are less well conserved (50–60%) and are therefore less likely to be crucial for RRF binding. With respect to E. coli RRF, all of the above positions are conserved in D. radiodurans RRF, with the exception that D. radiodurans has Lys, instead of Arg, at positions 12 and 133 (Figure 1B). However, such conservative substitutions would not be expected to disrupt the hydrogen-bonding potential at these positions. With respect to the rRNA, the stretches of nucleotides on either side of H71 (1941–1946 and 1946–1948) are highly conserved (>98%) in all bacteria (Cannone et al, 2002). Although nucleotides within H69, namely C1908 and C1909, are less well conserved, they are identical between E. coli and D. radiodurans, and, more importantly, contacts with these positions utilize only backbone interactions (Figure 4B). This suggests that the binding of D. radiodurans RRF-DI to the D. radiodurans 50S subunit would be virtually identical to that described here for E. coli RRF-DI.
The importance of these interactions for binding of RRF-DI to the ribosome is supported by the fact that mutation of Arg129 to Cys, or Arg132 to Gly, Cys or His, produces nonfunctional E. coli RRFs (Janosi et al, 2000). Consistently, the mutation Arg129Cys would prevent hydrogen bonding to the backbone ribose of G1945 and, similarly, the presence of Gly, Cys or His at position 132 would remove the hydrogen-bonding potential with C1941. Mutation of Arg132Gly in E. coli RRF does not prevent expression or correct folding of the factor, since the crystal structure of this RRF mutant (Nakano et al, 2002) has an identical fold for domain I to that of RRF-DI described here, and also to that of the wild-type E. coli RRF (Kim et al, 2000). Furthermore, both the RRF-Arg132Gly and His mutants are deficient in binding to both 70S ribosomes and 50S subunits, and even in the presence of five-fold excess of the mutant RRFs, binding of wild-type RRF to ribosomes remains unaffected (Nakano et al, 2003), thus emphasizing the importance of the interaction of Arg132 with C1941 for RRF binding and function.
Although extensive contacts were predicted between domain I of RRF and H69–H71 by cryo-EM (Agrawal et al, 2004), the specific details of the contacts differ significantly from those described here, for example, the universally conserved Arg132 was proposed to make van der Waals interactions with U1963 (in the loop connecting H67 and H71), whereas we observe hydrogen bonding with C1941 (loop connecting H70–H71). In the cryo-EM structure, Glu122, Val126 and Val130 interact with residues 1915 and 1926 in the loop of H69, while in the X-ray structure, no contacts are made with this region of H69 and Glu122 and Val126 are in closer proximity to H71 (U1946 and U1963). Similarly, Glu122, which was predicted to contact positions in both H69 (1915) and H71 (U1946–C1947), contacts only the latter H71 (to backbone of U1946) in the X-ray structure. Such discrepancies arise from the fact that (i) the X-ray position of RRF-DI is shifted by at least one turn of an α-helix (∼8 Å) toward H80 of the PTC (see Supplementary Figure 1) and (ii) H69 has adopted a new conformation, which could not be resolved in the cryo-EM study (Agrawal et al, 2004; see Supplementary Figure 2). Ultimately, the observed discrepancies probably also reflect the difference in ability to determine and/or envisage molecular interactions at 3.3 or 12 Å resolution.
Insights into the concerted action of RRF and EF-G on the ribosome
By superimposing domain I of the known RRF structures with the position of RRF-DI on the 50S subunit, it is possible to present a model for the full-length RRF on the ribosome. The two most extreme positions of domain II relative to domain I observed by NMR analysis of A. aeolicus RRF (Yoshida et al, 2001) were used for modeling and are referred to hereafter as the closed (RRF-e1) or open (RRF-e2) form depending on whether domain II is located toward, or away from, h44 of the 30S subunit (see Figure 5A). The extended position found in the E. coli RRF (Kim et al, 2000) was not considered here, because it has a detergent bound within the hinge region that is thought to produce a nonphysiological interdomain angle of 110° rather than 90°. This rationale is consistent with the observation that E. coli RRF-Arg132Gly mutant crystallized without detergent had an interdomain angle of 90°, as observed for all other RRFs (Nakano et al, 2002). Independent of which RRF structure was used, domain II overlaps with the position of anticodon stem (but not the loop) of an A-tRNA (Figure 1D), and no contact between domain II of RRF and any component of the 50S subunit are envisaged (Figure 5A). However, positions in domain II are within 10–15 and 20-30 Å from the H43/44 (the L11 binding site) and H95 (SRL), respectively, which correlates with the cleavages in these regions from residues within domain II of RRF (Lancaster et al, 2002).
Figure 5.

Structural model for RRF action on the 70S ribosome. (A) Model for the binding position of RRF on the 70S ribosome. The A. aeolicus RRF structures (Yoshida et al, 2001) containing the most extreme conformations of domain II (RRF-e1, closed conformation in red, and RRF-e2, open conformation in blue, as seen in Figure 1D) were fitted by superimposing domain I with the position of RRF-DI obtained in this study. L11 (cyan), the globular part of S12 (yellow) and h44 of the 16S rRNA (purple) are highlighted, otherwise the 16S and 23S rRNA are colored gray and blue, respectively. The small (30S) and large (50S) subunit ribosomal proteins are colored brown and olive, respectively. (B) The positions of the extremes of domain II in relation to the 30S subunit. Binding of EF-G to the ribosome would force the domain II of RRF-e2 (open, blue) toward the orientation seen in RRF-e1 (closed, red), in closer contact with h44 (purple). Colors of the 30S subunit are as in (A). (C) Model for the interaction between domain II and the hinge region of RRF with domains III and IV of EF-G. The position of EF-G (green) and RRF-e1 (red) are shown. Domain II of RRF is sandwiched between domains III and IV of EF-G (green). H69 (orange) is shown for reference. The gain-of-function mutations, amino-acid substitutions, H504Y and S508F, and deletion of the 4–7 C-terminal residues that restored function to E. coli EF-G in the presence of T. thermophilus RRF (Ito et al, 2002) (blue spheres) are indicated. (D) Overview of the relative orientation of EF-G and the two extreme positions of domain II of RRF. The position of EF-G (green) from the E. coli 70S·EF-G·GDPCP cryo-EM reconstruction (Valle et al, 2003) is docked into the 70S-RRF model, revealing large clashes between domains IV and V of EF-G with domain II of RRF-e2, but less significant overlap is observed between EF-G and the RRF-e1 position of domain II.
Lancaster et al (2002) observed a reduction in the intensity of cleavages within these two regions when deacylated tRNA (bound at the hybrid P/E site, see Figure 1D) was included within the 70S-RRF complex, suggesting either a movement in domain II and/or in the flexible L11 region. Consistent with this flexibility, we have observed in an RRF-70S complex, site-specific crosslinking from Cys39 of E. coli L11 to Tyr45 within domain II of E. coli RRF (T Yoshida, T Ohkubo and Y Kobayashi, unpublished data). Considering the length of the crosslinker, the Cα atoms of Cys39 and Tyr45 must at some stage come within 22 Å of one another. For this to occur, either domain II of RRF must attain an extended conformation, such as that seen in the detergent-bound E. coli RRF structure (Kim et al, 2000), or there must be movement of the N-terminal domain of L11 toward domain II of RRF. Consistent with the latter alternative, the recent cryo-EM reconstruction of E. coli RRF bound to empty 70S ribosomes observed a ∼10 Å movement of the stalk-base, which encompasses L11 and associated binding site on H43/44, toward the bound RRF (Agrawal et al, 2004). The fact that we do not observe any significant difference within the L11 region between RRF-DI bound and native D50S structures suggests that the complete 70S ribosome and/or the complete RRF molecule is required to observe this conformational change.
By aligning the 5.5 Å crystal structure of the Thermus thermophilus 70S·tRNA3 complex (Yusupov et al, 2001) with the D50S-RRF complex, it is possible to model the position of domain II of RRF relative to the 30S subunit (Figure 5A). Taking into account the interdomain flexibility, domain II of RRF in the closed conformation comes into close proximity of ribosomal protein S12 and the decoding center at the top of h44 (Figure 5B). Since RRF works in conjunction with EF-G to disassemble the post-termination ribosome complex, we performed a docking of EF-G into our RRF-70S model, using the position of EF-G·GDPCP bound on the E. coli 70S ribosome determined by cryo-EM (Gao et al, 2003; Valle et al, 2003). This docking revealed that domains IV and V of EF-G partially overlap with the position of domain II of RRF (Figure 5D); however, the overlap is significantly reduced when the most closed orientation of domain II is used (RRF-e1). In this case, domain IV of EF-G, together with H69, form a tight binding pocket for RRF (Figure 5C), such that binding of EF-G to empty ribosomes would prevent subsequent binding of RRF, as observed in RRF/EF-G·GDPNP competition experiments (Kiel et al, 2003). The resulting structural model suggests that during ribosome recycling, when both factors are present on the ribosome, domain II of RRF makes contact with domain IV of EF-G, whereas the hinge region of RRF nestles against domain III of EF-G (Figure 5C). This model is consistent with the proposed sites of interaction between EF-G and RRF based on genetic complementation studies of these factors from T. thermophilus and E. coli (Ito et al, 2002). The gain-of-function mutations were identified as two substitutions (H504Y and S508F) located within positions of domain IV, as well as deletion of the C-terminal 4–7 amino acids of EF-G, all of which are in close proximity to domain II of the closed form of RRF (Figure 5C). In this position RRF still has a slight overlap with EF-G, suggesting that upon EF-G binding, domain II of RRF will be pushed toward h44, as illustrated in Figure 5B.
Discussion
The tRNA-like shape of RRF led to the proposal that RRF not only structurally mimics a tRNA and therefore binds to the ribosome in an analogous way but also functionally mimics a tRNA and is translocated through the ribosome by EF-G (Selmer et al, 1999). Subsequently, hydroxyl radical probing and cryo-EM reconstructions of RRF-70S ribosome complexes revealed that RRF-DI binds to the 50S in a position that obliquely transverses the A- and P-tRNAs, thus ruling out any possible structural mimicry of tRNA by RRF (Lancaster et al, 2002; Agrawal et al, 2004). In addition, both studies put forward a new model for ribosome recycling, whereby EF-G acts through RRF on bridges B2a and B3 to facilitate dissociation of the post-termination complex. These studies, at a maximum resolution of 12 Å, have created the ideal basis to further investigate at higher resolution the mechanism of binding of RRF on the ribosome and evaluate the suggested structural model.
The position of RRF on the large subunit determined here by X-ray crystallography is very similar to the positions derived from the hydroxyl radical probing (Lancaster et al, 2002) and cryo-EM (Agrawal et al, 2004) studies. However, small differences in orientation are observed (see Supplementary Figure 1), such that to be aligned with the X-ray crystallography orientation of RRF, the cryo-EM and hydroxyl radical probing RRF positions need to be translated by 8–7 Å and rotated by 7° and 12°, respectively. These changes in orientation have some important consequences with respect to the interaction of specific residues of RRF with the amino acids of the ribosomal proteins and nucleotides of the 23S rRNA.
A detailed characterization of the interactions between RRF and the large subunit provides support for the translocation of RRF through the ribosomal particle, despite its different positioning: all the contacts observed between RRF and the large subunit encompass elements involved with either the correct positioning of the tRNA on the ribosome or translocation of tRNA through the ribosome. These include nucleotides of the P loop (H80), which plays an important role for the positioning of the P-tRNA and the nucleotide A2602 present in H93, which has been suggested to guide the translocation of tRNAs from the A- to P-site (Bashan et al, 2003). In addition, RRF contacts L16 and L27, both of which have been associated with proper placement of tRNA ligands at the PTC (Wower et al, 1998; Harms et al, 2001). L27 has been crosslinked from the A- and P-site-bound tRNAs (Wower et al, 1998 and references therein) and N-terminal deletions of L27 suggest that it is the first 3–6 amino acids that can come within close proximity of the aminoacyl moiety of the P-tRNA (R Zimmermann and A Mankin, personal communication, 2004). It may be of relevance to note that there is no L27 homologue present in archaeal or eukaryotic ribosomes (L21e occupies the equivalent space but has no sequence or structural similarity to L27 and does not have the extension that reaches into the PTC), consistent with the absence of RRF in the cytoplasm of these organisms.
The most extensive interaction that RRF has with the large subunit is, however, through the helices H69 and H71, which are the large subunit components of intersubunit bridges B2a and B3, respectively, involved in the translocation of tRNA (Valle et al, 2003; Spahn et al, 2004). Bridge B2a, which is positioned between the A- and P-tRNAs in the 70S·tRNA3 structure (Yusupov et al, 2001), has acquired a new position in the cryo-EM reconstruction of the RRF-70S complex, being shifted by 7 Å toward the E site. According to the authors, rigid-body fitting of X-ray structure of H69 from native 50S subunits into the shifted density was not possible (Agrawal et al, 2004), leading to the suggestion that H69 had adopted a new conformation. Interestingly, in our 50S-RRF complex, H69 has also undergone a substantial conformational change, resulting in the repositioning of the tip of H69 and movement by 20 Å toward the small ribosomal subunit. In this position on a 70S ribosome, the tip of H69 would clash with its counter-bridge element h44 of the small subunit (Figure 3C and Supplementary Figure 2).
We believe that the observed movement and novel fold of H69 induced by RRF binding to the 50S subunit represent the same conformational changes that RRF would try to induce in a 70S ribosome. However, the extent of conformational change that H69 can undergo in the 70S ribosome depends on h44, which could either restrain or follow the movement of H69. In agreement with the latter possibility, a better fit of H69 into the cryo-EM density maps of the RRF-70S (Agrawal et al, 2004) can be obtained when using the new conformation of H69 observed in our structure than when the conformations from the native 50S (Harms et al, 2001), 70S cryo-EM (Gao et al, 2003) or crystal (Yusupov et al, 2001) structures are used (see Supplementary Figure 2). In our opinion, the rearrangement of bridge B2a observed by cryo-EM (Agrawal et al, 2004) can therefore be explained by the RRF-induced conformational change of H69 that causes a subsequent shift in h44. From the fact that the nucleotides of H69 that contact h44 are shifted and rotated (by ∼20 Å and more than 30°) relative to their position on the 70S ribosome, we expect, in addition, that either the bridging nucleotides of h44 have changed conformation or that the site of interaction between the two helices is now different (Supplementary Figure 3). In this respect, we note that the RRF-induced movement of H69 would bring the tip into contact with the noncanonical base pair G1487–A1413 of h44 (Supplementary Figure 3), the same base pair that has also been associated with conformational changes in h44 induced by binding of IF1 to the 30S subunit (Carter et al, 2001).
As RRF requires the action of EF-G to complete its function, we fitted the crystal structure of EF-G on the RRF-50S structure according to the position observed in the 70S·EF-G·GDPCP complex (Gao et al, 2003; Valle et al, 2003). Using domain I of RRF as the reference to dock domain II of RRF revealed that the closed form (RRF-e1) has the least overlap with EF-G (Figure 5C and D). However, even in this position some clashes between domain II of RRF and domains III and IV of EF-G are still present, suggesting that upon EF-G binding, domain II of RRF will be pushed even further toward the 30S subunit. In fact, the plane of flexibility is such that domain II of RRF will close against the 30S subunit, contacting the last turn of h44. Therefore, RRF appears to have a double action on h44, indirectly through the interaction of domain I with H69 that bridges with h44, as well as a more direct one via domain II.
In the model proposed by Kaji and co-workers, RRF is translocated through the ribosome by the action of EF-G (Selmer et al, 1999; Hirokawa et al, 2002b). Consistently, in our RRF-70S structural model all the necessary elements to translocate RRF seem to be in place: (i) EF-G can be accommodated in the same position as observed in the 70S-EF-G complex (Gao et al, 2003; Valle et al, 2003) and (ii) RRF contacts exclusively elements associated with tRNA binding and translocation. However, the position and conformation of the intersubunit bridge B2a in the 70S·tRNA3 complex (Yusupov et al, 2001) and our structural model could have different consequences for the translocation of RRF compared with tRNA. In the RRF-70S structural model, the already altered interaction between H69 and h44 could be further strained by EF-G action, leading to a complete dissociation into subunits, even before the completion of the translocation event as observed in the experiments of Ehrenberg and co-workers (Karimi et al, 1999). In this respect, the buffer conditions used would play an important role, weakening or reinforcing the interaction between the subunits.
Although there remains some controversy as to the involvement of the translocase activity of EF-G during ribosome recycling (Fujiwara et al, 2004), we have evaluated the situation in which translocation is brought to a completion without subunit dissociation (Kiel et al, 2003), by modeling the translocation of RRF through the 70S ribosome. A comparison of the contacts of the A- and P-tRNAs with the ribosome reveals that RRF contacts only nucleotides of the rRNA associated with P-tRNA binding. This resemblance of the P- rather than A-tRNA by RRF is also seen when trying to model the translocation of RRF through the ribosome: when a matrix is applied to RRF that translocates the A-tRNA to the P-site, the final position of RRF has little overlap with the P/E-tRNA and has some clashes with the large subunit (Supplementary Figure 4A). In contrast, when a matrix that translocates the P-tRNA to the E-site is applied to RRF, RRF occupies a position with minimal clashes in the intersubunit cavity and the tip of domain I of RRF superimposes the acceptor-stem loop of the P/E-tRNA (Supplementary Figure 4B). This result suggests that if the latter translocation of RRF occurs on the ribosome, it would result in the full destabilization of a P/E-tRNA releasing it from the ribosome.
In conclusion, in this study we could observe at 3.3 Å resolution, the details of the interaction between RRF and the ribosome, obtaining a structural base upon which to understand the action of RRF during ribosome recycling and opening the possibility for further study toward the rational design of drugs that target this essential process in bacteria. The conformational changes induced on the ribosome by RRF binding have revealed furthermore how the large subunit can trigger the movement of a critical intersubunit bridge (B2a). It will be interesting to see if the same types of conformational changes in H69 are also at the heart of other fundamental processes associated with this ribosomal component, such as subunit association and translocation of the tRNAs.
Materials and methods
Crystallography
Domain I of E. coli RRF was prepared as described previously (Nakano et al, 2003). D. radiodurans 50S subunit crystals were prepared as described (Schlünzen et al, 2001), and soaked in a solution containing 5–10 μM RRF-DI for 24 h, prior to freezing. Preliminary tests were performed at BW6 (HASYLAB/DESY). Data were collected at 100 K from shock frozen crystals at ID29 of the European Synchrotron Radiation Facility/European Molecular Biology Laboratory (ESRF/EMBL) and X06SA at the Swiss Light Source (SLS). Data were recorded on MAR345 or Quantum 4 detectors and processed with HKL2000 (Otwinowski and Minor, 1997) and the CCP4 package (Bailey, 1994).
Modeling and docking
The native structure of the 50S subunit was refined against the structure factor amplitudes of the 50S-RRF-DI complex, using rigid-body refinement as implemented in CNS (Brunger et al, 1998). For the calculation of the free R-factor, 5% of the data were omitted during refinement. The position of RRF-DI was determined from sigmaA-weighted difference maps. Further refinement was carried out using CNS (Brunger et al, 1998) (see Table I for refinement statistics). Ribosome-RRF-DI interactions were originally determined with LigPlot (Wallace et al, 1995). Placement of the 30S subunit (PDB code 1p6g; Gao et al, 2003) and EF-G (PDB code 1pn6; Valle et al, 2003) to the 50S structure were performed by least-squares alignments of the 50S subunit (PDB code 1p85; Gao et al, 2003) of the EM reconstruction of E. coli ribosomes (Gao et al, 2003) onto the D. radiodurans 50S structure (Harms et al, 2001).
Coordinates and figures
3D figures were produced with VMD (Humphrey et al, 1996) and RIBBONS (Carson, 1997) and rendered using POVRAY. Final coordinates have been deposited in the Protein Data Bank under Accession number 1Y69.
Supplementary Material
Supplementary Material
Acknowledgments
We would like to thank Barbara Schmidt and Uwe Vogel for technical assistance and Dr Sean Connell, Professor Christian Spahn, Dr S Uchiyama, Dr H Nakano, Mr M Nishimura and Professor Knud Nierhaus for helpful discussions. Furthermore, we are grateful to Dr Rajendra Agrawal for providing a prerelease of the density map for the cryo-EM reconstruction of the RRF-70S complex and Professor Harry Noller for making available the RRF-70S model derived from hydroxyl radical probing. These studies could not have been performed without the expert assistance of the staff, especially Dr Takashi Tomizaki (SLS) and Dr William Shepard (ESRF), at the synchrotron facilities: X06SA/SLS, ID29/ESRF/EMBL and BW6/HASYLAB/DESY.
References
- Agrawal R, Sharma M, Kiel M, Hirokawa G, Booth T, Spahn C, Grassucci R, Kaji A, Frank J (2004) Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc Natl Acad Sci USA 101: 8900–8905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S (1994) The CCP4 suite: programs for protein crystallography. Collaborative Computational Project, Number 4. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HA, Kossoy E, Kessler M, Yonath A (2003) Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell 11: 91–102 [DOI] [PubMed] [Google Scholar]
- Brunger A, Adams P, Clore G, DeLano W, Gros P, Grosse-Kunstleve R, Jiang J, Kuszewski J, Nilges M, Pannu N, Read R, Rice L, Simonson T, Warren G (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M (1997) Ribbons. Methods Enzymol 277: 493–505 [PubMed] [Google Scholar]
- Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501 [DOI] [PubMed] [Google Scholar]
- Fredrick K, Noller HF (2003) Catalysis of ribosomal translocation by sparsomycin. Science 300: 1159–1162 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Ito K, Yamami T, Nakamura Y (2004) Ribosome recycling factor disassembles the post-termination ribosomal complex independent of the ribosomal translocase activity of elongation factor G. Mol Microbiol 53: 517–528 [DOI] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CMT, Grassucci RA, Svergun DI, Frank J, Penczek P (2000) Solution structure of the E. coli 70S ribosome at 11.5 Angstrom resolution. Cell 100: 537–549 [DOI] [PubMed] [Google Scholar]
- Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, van Roey P, Agrawal RK, Harvey SC, Sali A, Chapman MS, Frank J (2003) Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113: 789–801 [DOI] [PubMed] [Google Scholar]
- Hansen JL, Moore PB, Steitz TA (2003) Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol 330: 1061–1075 [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688 [DOI] [PubMed] [Google Scholar]
- Hirashima A, Kaji A (1970) Factor dependent breakdown of polysomes. Biochem Biophys Res Commun 41: 877–883 [DOI] [PubMed] [Google Scholar]
- Hirokawa G, Kiel M, Muto A, Kawai G, Igarashi K, Kaji H, Kaji A (2002a) Binding of ribosome recycling factor to ribosomes, comparison with tRNA. J Biol Chem 277: 35847–35852 [DOI] [PubMed] [Google Scholar]
- Hirokawa G, Kiel MC, Muto A, Selmer M, Raj VS, Liljas A, Igarashi K, Kaji H, Kaji A (2002b) Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J 21: 2272–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graphics 14: 33–38 [DOI] [PubMed] [Google Scholar]
- Ishino T, Atarashi K, Uchiyama S, Yamami T, Saihara Y, Yoshida T, Hara H, Yokose K, Kobayashi Y, Nakamura Y (2000) Interaction of ribosome recycling factor and elongation factor EF-G with E. coli ribosomes studied by the surface plasmon resonance technique. Genes Cells 5: 953–963 [DOI] [PubMed] [Google Scholar]
- Ito K, Fujiwara T, Toyoda T, Nakamura Y (2002) Elongation factor G participates in ribosome disassembly by interacting with ribosome recycling factor at their tRNA-mimicry domains. Mol Cell 9: 1263–1272 [DOI] [PubMed] [Google Scholar]
- Janosi L, Mori H, Sekine Y, Abragan J, Janosi R, Hirokawa G, Kaji A (2000) Mutations influencing the frr gene coding for ribosome recycling factor (RRF). J Mol Biol 295: 815–829 [DOI] [PubMed] [Google Scholar]
- Janosi L, Mottagui-Tabar S, Isaksson L, Sekine Y, Ohtsubo E, Zhang S, Goon S, Nelken S, Shuda M, Kaji A (1998) Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J 17: 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janosi L, Shimizu I, Kaji A (1994) Ribosome recycling factor (ribosome releasing factor) is essential for bacterial growth. Proc Natl Acad Sci USA 91: 4249–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Pavlov M, Buckingham R, Ehrenberg M (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell 3: 601–609 [DOI] [PubMed] [Google Scholar]
- Kiel M, Raj V, Kaji H, Kaji A (2003) Release of ribosome-bound ribosome recycling factor by elongation factor G. J Biol Chem 278: 48041–48050 [DOI] [PubMed] [Google Scholar]
- Kim KK, Min K, Suh SW (2000) Crystal structure of the ribosome recycling factor from Escherichia coli. EMBO J 19: 2362–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster L, Kiel MC, Kaji A, Noller HF (2002) Orientation of ribosome recycling factor from directed hydroxyl radical probing. Cell 111: 129–140 [DOI] [PubMed] [Google Scholar]
- Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH (2004) Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell 118: 45–55 [DOI] [PubMed] [Google Scholar]
- Nakano H, Uchiyama S, Yoshida T, Ohkubo T, Kato H, Yamagata Y, Kobayashi Y (2002) Crystallization and preliminary X-ray crystallographic studies of a mutant of ribosome recycling factor from Escherichia coli, Arg132Gly. Acta Crystallogr D 58: 124–126 [DOI] [PubMed] [Google Scholar]
- Nakano H, Yoshida T, Uchiyama S, Kawachi M, Matsuo H, Kato T, Ohshima A, Yamaichi Y, Honda T, Kato H, Yamagata Y, Ohkubo T, Kobayashi Y (2003) Structure and binding mode of a ribosome recycling factor (RRF) from mesophilic bacterium. J Biol Chem 278: 3427–3436 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Puglisi EV, Green R, Noller HF, Puglisi JD (1997) Structure of a conserved RNA component of the peptidyl transferase centre. Nat Struct Biol 4: 775–778 [DOI] [PubMed] [Google Scholar]
- Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J (2003) A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421: 87–90 [DOI] [PubMed] [Google Scholar]
- Ryoji M, Berland R, Kaji A (1981) Reinitiation of translation from the triplet next to the amber termination codon in the absence of ribosome-releasing factor. Proc Natl Acad Sci USA 78: 5973–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlünzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F (2001) Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413: 814–821 [DOI] [PubMed] [Google Scholar]
- Selmer M, Al-Karadaghi S, Hirakawa G, Kaji A, Liljas A (1999) Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science 286: 2349–2352 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J (2004) Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Davis B (1973) Release of 70 S ribosomes from polysomes in Escherichia coli. J Mol Biol 74: 45–56 [DOI] [PubMed] [Google Scholar]
- Toyoda T, Tin OF, Ito K, Fujiwara T, Kumasaka T, Yamamoto M, Garber MB, Nakamura Y (2000) Crystal structure combined with genetic analysis of the Thermus thermophilus ribosome recycling factor shows that a flexible hinge may act as a functional switch. RNA 6: 1432–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J (2003) Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM (1995) Ligplot—a program to generate schematic diagrams of protein ligand interactions. Protein Eng 8: 127–134 [DOI] [PubMed] [Google Scholar]
- Wower I, Wower J, Zimmermann R (1998) Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J Biol Chem 273: 19847–19852 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Uchiyama S, Nakano H, Kashimori H, Kijima H, Ohshima T, Saihara Y, Ishino T, Shimahara H, Yokose K, Ohkubo T, Kaji A, Kobayashi Y (2001) Solution structure of the ribosome recycling factor from Aquifex aeolicus. Biochemistry 40: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Oka S, Uchiyama S, Nakano H, Kawasaki T, Ohkubo T, Kobayashi Y (2003) Characteristic domain motion in the ribosome recycling factor revealed by 15N NMR relaxation experiments and molecular dynamics simulations. Biochemistry 42: 4101–4107 [DOI] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


