Abstract
EB1 proteins bind to microtubule ends where they act in concert with other components, including the adenomatous polyposis coli (APC) tumor suppressor, to regulate the microtubule filament system. We find that EB1 is a stable dimer with a parallel coiled coil and show that dimerization is essential for the formation of its C-terminal domain (EB1-C). The crystal structure of EB1-C reveals a highly conserved surface patch with a deep hydrophobic cavity at its center. EB1-C binds two copies of an APC-derived C-terminal peptide (C-APCp1) with equal 5 μM affinity. The conserved APC Ile2805–Pro2806 sequence motif serves as an anchor for the interaction of C-APCp1 with the hydrophobic cavity of EB1-C. Phosphorylation of the conserved Cdc2 site Ser2789–Lys2792 in C-APCp1 reduces binding four-fold, indicating that the interaction APC–EB1 is post-translationally regulated in cells. Our findings provide a basis for understanding the dynamic crosstalk of EB1 proteins with their molecular targets in eukaryotic organisms.
Keywords: coiled coil, microtubule plus-end tracking proteins, phosphorylation, protein–protein interaction, +TIP
Introduction
The intrinsic dynamic properties of microtubules (MTs) are important in establishing and maintaining a specific organization of cellular components during the cell cycle. Diverse factors regulate MT dynamics both spatially and temporally. Among these, a number of proteins and protein complexes have been identified that specifically track growing MT plus ends (referred to as +TIP proteins; Carvalho et al, 2003). EB1 (end-binding protein 1) is a member of a highly conserved and ubiquitously expressed family of +TIP proteins that has emerged as a key player in the modulation of MT dynamics in eukaryotic organisms. EB1 proteins are implicated in most if not all MT-based processes including maintenance of cell polarity, regulation of chromosome stability, positioning of the mitotic spindle, and anchoring of MTs to their nucleation sites (Tirnauer and Bierer, 2000; Gundersen, 2002; Carvalho et al, 2003; Galjart and Perez, 2003).
The dynamic crosstalk among +TIP proteins together with the observation that EB1 is always localized at growing MT ends suggests that EB1 is involved in the establishment of macromolecular ‘plus-end complexes' at MT tips (Schroer, 2001; Galjart and Perez, 2003). In budding yeast, the EB1-mediated interaction network involving Kar9p and the myosin molecule Myo2 is required for MT guidance toward the bud along actin cables to ensure proper spindle alignment (Liakopoulos et al, 2003). Thus the role of the EB1 orthologue Bim1p in Saccharomyces cerevisiae is to link different functionalities to the MT plus end allowing them to stably capture specialized cellular targets. Similarly, in fission yeast, the EB1 homologue Mal3p transiently tethers the CLIP-170 homologue Tip1p and the kinesin molecule Tea2p into larger particles whose function is to guide MT growing tips to cell ends (Busch et al, 2004). In Drosophila, the guanine nucleotide exchange factor DRho-GEF2 utilizes EB1-mediated MT dynamics to search for cortical subdomains for directing localized actomyosin contraction (Rogers et al, 2004). It appears very likely that similar EB1-dependent MT capture and guided processes are conserved throughout higher eukaryotes (Gundersen, 2002; Kusch et al, 2003).
EB1 proteins are comprised of conserved amino- and carboxy-terminal domains (Tirnauer and Bierer, 2000; Bu and Su, 2003). The N-terminal domain is necessary and sufficient for MT binding. Its structure, recently solved by X-ray crystallography, revealed a calponin homology (CH) fold (Hayashi and Ikura, 2003). The C-terminal domain (EB1-C) contains a putative coiled coil, which mediates subunit oligomerization (Rehberg and Gräf, 2002). However, the detailed molecular organization of EB1 proteins is still not known.
EB1-C comprises a unique EB1-like sequence motif that acts as a binding site for other +TIP proteins. It interacts with the carboxy terminus of the adenomatous polyposis coli (APC) tumor suppressor (Su et al, 1995; Berrueta et al, 1999; Askham et al, 2000; Mimori-Kiyosue et al, 2000; Nakamura et al, 2001; Bu and Su, 2003; Wen et al, 2004), a well-conserved 2843-residue +TIP phosphoprotein (Trzepacz et al, 1997) with a pivotal function in cell cycle regulation (Dikovskaya et al, 2001; Mimori-Kiyosue and Tsukita, 2001). The transient binding of APC to EB1 may play a central role in spindle positioning and fidelity of chromosome segregation (Fodde et al, 2001). Together with the fact that APC lacks the EB1-binding site in many malignant human colon tumors, it has been speculated that abrogation of the APC–EB1 interaction may contribute to cancer progression (Su et al, 1995; Tirnauer and Bierer, 2000; Fodde et al, 2001). Recent data suggest that the interaction between EB1-C and the carboxy terminus of APC (C-APC) is also implicated in the capturing and stabilization of MTs in vivo (Wen et al, 2004). Interfering with the EB1–APC interaction inhibits fibroblast migration, demonstrating the importance of EB1–APC-mediated stable MT formation in promoting directed cell migration (Wen et al, 2004). Based on the apparent functional conservation, the fact that Kar9p and APC share a sequence site of limited homology (Bienz, 2001), and the finding that the molecules are regulated by post-translational phosphorylation events (Askham et al, 2000; Nakamura et al, 2001; Liakopoulos et al, 2003), it has been proposed that Kar9p may be the functionally related APC molecule in yeast.
Another binding partner of EB1-C is the well-conserved +TIP protein dynactin/p150glued, a component of the large cytoplasmic dynein/dynactin complex (Berrueta et al, 1999; Askham et al, 2002; Bu and Su, 2003; Goodson et al, 2003; Ligon et al, 2003). The EB1–p150glued/dynactin interaction is required for the regulation of MT dynamics and for MT anchoring at its nucleation sites during the formation and maintenance of a radial MT array. It should be noted that a growing body of evidence also suggests that EB1, APC, and the dynein/dynactin complex are functional components of centrosomes, which anchor the MT minus end (Berrueta et al, 1998; Askham et al, 2002; Rehberg and Gräf, 2002; Rogers et al, 2002; Louie et al, 2004). Thus the dynamic crosstalk between these proteins appears also important for MT nucleation and stabilization at centrosomes.
Despite substantial progress that has been made in identifying the interaction networks between +TIP proteins, their detailed nature and mechanisms of regulation are poorly defined. In order to provide a structural basis for understanding the important role of EB1 proteins, here we have carried out a biophysical analysis of EB1–APC. We conclude that EB1 proteins assemble into dimeric structures, which is mediated by a parallel coiled coil. Using X-ray crystallography and isothermal titration calorimetry (ITC), we propose that a major interaction site between EB1-C and APC involves the APC dipeptide segment Ile2805–Pro2806. We further found that the interaction is regulated by APC phosphorylation of the conserved cyclin-dependent mitotic kinase Cdc2 target residue Ser2789. Our findings are consistent with the APC sequence segment Val2781–Lys2819 being the major interaction site between APC and EB1 and suggest that specific C-APC phosphorylation represents a mechanism for regulating the EB1–APC complex during the cell cycle.
Results
Molecular organization and overall structure of EB1
The EB1 molecule is comprised of three structural domains (Figure 1A): an N-terminal MT-binding domain (residues 1–133; referred to as domain N), a flexible intermediate domain (residues 134–192; referred to as domain I), and a C-terminal +TIP-binding domain (residues 193–268; referred to as domain C). Its amino-terminal segment (residues 193–225; referred to as subdomain Ca) exhibits a high potential for adopting an α-helical coiled-coil structure (Cohen and Parry, 1990; Juwana et al, 1999) and the very C-terminal acidic subdomain Cb (residues 226–268) is composed of low-complexity sequence.
Figure 1.
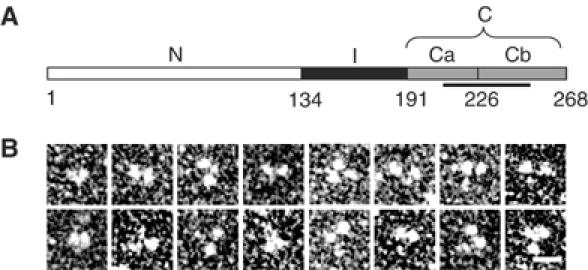
Molecular organization of human EB1. (A) Schematic representation of the domain organization of human EB1. Domains N, I, and C (subdivided in Ca and Cb) are depicted in white, black, and gray, respectively. Corresponding domain boundaries are indicated by residue positions. The unique EB1-like sequence motif is highlighted by a line. (B) High-magnification TEM gallery of glycerol sprayed/rotary metal shadowed EB1 specimens. Scale bar, 10 nm.
The overall structure of EB1 was assessed by analytical ultracentrifugation (AUC), transmission electron microscopy (TEM), and limited proteolysis. Sedimentation velocity and equilibrium AUC experiments yielded an sw,20 value of 3.5 S and an average molecular mass of 62 kDa, respectively (monomer mass of recombinant EB1 is 32 kDa). Inspection of EB1 molecules by TEM after glycerol spraying and rotary metal shadowing suggested that two larger globular domains, ∼4 nm in diameter, are joined via a smaller domain to form a flexible Y-shaped structure (Figure 1B). The dimension of the larger domains is in agreement with the one deduced from the crystal structure of EB1's N-terminal CH domain (Hayashi and Ikura, 2003). Thrombin digestion of EB1 yielded nonspecific cleavage within the sequence Pro145–Pro161 (see Materials and methods), indicating that this segment is flexible. These data establish that EB1 is an elongated dimeric molecule.
Biophysical solution analysis of EB1-C
To test whether the predicted coiled coil is involved in EB1 dimerization, a fragment corresponding to residues Ala193–Glu225 was prepared. The fragment, referred to as CysEB1-Ca, contained an extra Cys residue to permit disulfide bond formation, and two extra Gly residues for flexibility at its N-terminus to allow determination of the orientation of the helical monomers within the coiled coil (Harbury et al, 1993). As shown in Figure 2A, at 5°C and under reducing conditions, CysEB1-Ca displayed a far-ultraviolet (UV) circular dichroism (CD) spectrum characteristic of proteins with a helical content of ∼50%. The stability of CysEB1-Ca under the same solution conditions was assessed by a thermal unfolding experiment recorded by CD at 222 nm. The fragment (0.2 mg/ml) revealed a broad transition, indicating that it unfolds readily with increasing temperature and is largely denatured at 55°C (Figure 2B). Consistent with these findings, sedimentation equilibrium experiments at 5°C and under reducing conditions yielded an average molecular mass of 6.2 kDa (monomer mass of CysEB1-Ca is 4.5 kDa), indicating an equilibrium between monomers and dimers of the fragment. An increase of 30% in helical signal and a sigmoidal shaped melting profile with a Tm centered at 43°C was observed for CysEB1-Ca under oxidizing conditions (Figure 2A and B). Both the CD spectrum and the shape of the unfolding profile are characteristic for stable α-helical coiled-coil structures. Reducing and nonreducing SDS–PAGE revealed protein bands migrating at apparent molecular masses consistent with monomers and disulfide bonded dimers, respectively, of CysEB1-Ca (Figure 2A, inset).
Figure 2.
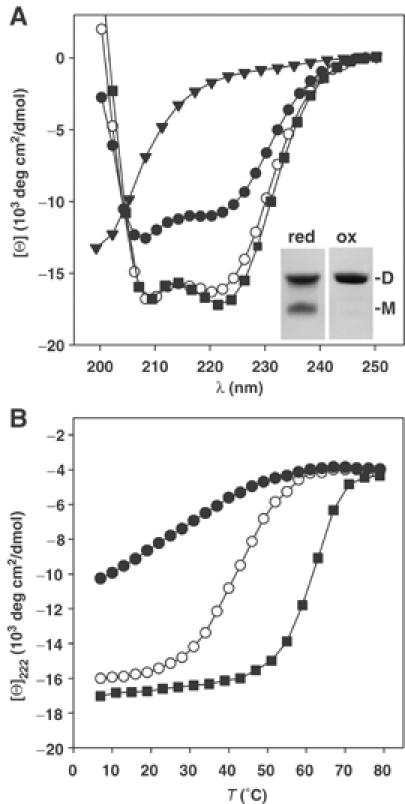
Analysis of EB1-C and fragments. (A) Far-UV CD spectra recorded at 5°C of EB1-C (closed squares), CysEB1-Ca under reducing (closed circles) and oxidizing conditions (open circles), and EB1-Cb (closed triangles). Inset: Coomassie blue-stained tricine–SDS–PAGE of CysEB1-Ca under reducing (red) and oxidizing (ox) conditions. The migration positions for monomers (M) and dimers (D) are indicated. (B) Thermal unfolding profiles recorded by CD at 222 nm. Symbols are the same as for panel A. The CD measurements were carried out in PBS at a protein concentration of 0.2 mg/ml.
These data suggest that subdomain Ca mediates the parallel in-register assembly of two EB1 monomers. However, they further reveal that the sequence Ala193–Glu225 of EB1 does not form a stable coiled coil in isolation. To assess the effect of the C-terminal flanking subdomain Cb on the stability of the coiled coil, a fragment comprising residues Asp191–Tyr268 of EB1, referred to as EB1-C, was analyzed. The far-UV CD spectrum recorded from EB1-C was characteristic for predominantly (∼70%) helical proteins (Figure 2A). The reversible sigmoidal shaped melting profile with a Tm centered at 63°C obtained at a protein concentration of 0.2 mg/ml demonstrates that the EB1-C molecule is very stable (Figure 2B). Sedimentation equilibrium experiments yielded an average molecular mass of 20 kDa consistent with the formation of a dimeric structure (monomer mass of EB1-C is 9.2 kDa). To probe the possibility that the short 43-residue-long subdomain Cb forms a stable structure, the C-terminal EB1 peptide segment Leu226–Tyr268, referred to as EB1-Cb, was analyzed by CD. As shown in Figure 2A, the spectrum is characteristic for peptides with little if any secondary structure.
Together, these findings demonstrate that coiled coil and C-terminus together constitute a stable folding unit.
Crystal structure of EB1-C
The crystal structure of EB1-C was determined by single isomorphous replacement combined with anomalous scattering (SIRAS) using a thiomersal derivative. The asymmetric unit of the crystal contains two largely helical monomers, denoted A and B, which are related by a noncrystallographic two-fold symmetry axis (Figure 3). Each 80-residue monomer starts with a long smoothly curved helix (α1, residues 191–230), which is followed by a hairpin connection leading to a short second helix (α2, residues 237–248) running antiparallel to α1. For the residues C-terminal of α2 (residues 250–268), interpretable density is only observed for segment 250–258 and only in monomer B. In agreement with the CD analysis, this suggests that the C-terminal region is largely disordered in solution. The observed ordering of the nine-residue segment in monomer B is a consequence of the crystal packing (also, see below). The two parallel α1 helices of the EB1-C dimer wrap around each other in a slightly left-handed supercoil up to residue Leu221. With the exception of Arg214 and Tyr217, the residues occupying the core a and d positions of the four heptad repeats between Ala193 and Leu221 (Figure 4A) pack in the typical ‘knobs-into-hole' fashion, consistent with a coiled-coil structure (Harbury et al, 1993). Beyond Leu221, the two α1 helices diverge into a fork-like structure. The two α2 helices run antiparallel to helices α1 and form a similar fork in the opposite orientation and rotated by 90°. As a result, two helical segments from each monomer (residues 219–229 and 237–247) form a four-helix bundle (Figure 3). The side chains forming the hydrophobic core of this bundle (equivalent residue pairs are 221/239, 224/242, and 227/245) are highly conserved (Figure 4A). The large hydrophobic surface buried in this bundle is expected to significantly contribute to the observed stability (Figure 2) of the dimeric structure of EB1-C (Supplementary Figure 1).
Figure 3.
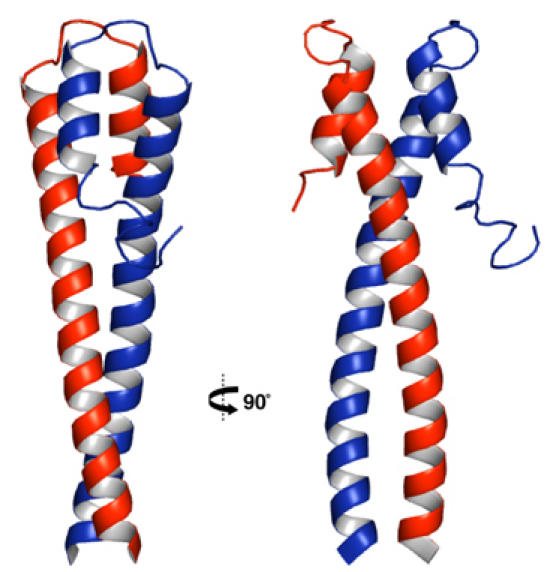
Overall crystal structure of EB1-C in cartoon representation and in two orientations 90° apart. Monomers A and B are colored in red and blue, respectively.
Figure 4.
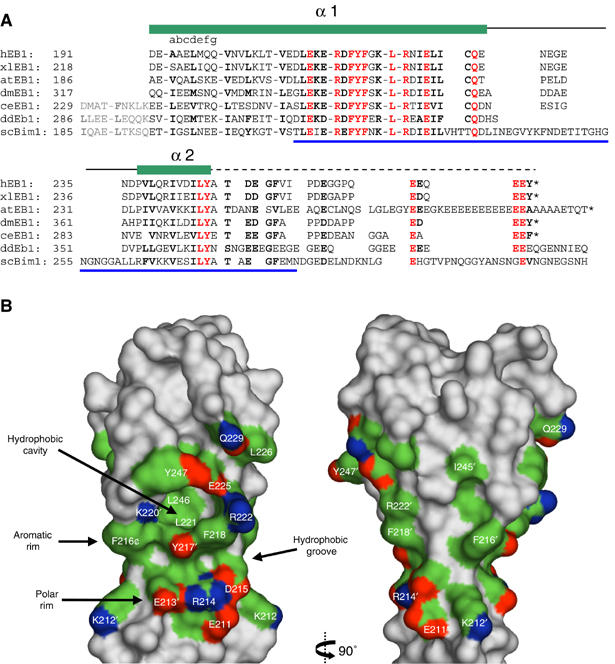
The EB1-like structural motif. (A) Structure-based sequence alignment of C-terminal EB1 domains from different species. Sequence identities and accession numbers are as follows: h, Homo sapiens, SwissProt: Q15691; xl, Xenopus laevis, GenBank: AAH68630; at, Arabidopsis thaliana, GenBank: BAB11500; dm, Drosophila melanogaster, TrEMBL: Q9V9A6; ce, Caenorhabditis elegans, GenBank: NP_507526; dd, Dictyostelium discoideum, TrEMBL: Q8WQ86; sc, Saccharomyces cerevisiae, SwissProt: P40013. Helices α1 and α2 (green bars), the loop α1–α2 (black line), the carboxy-terminal flexible tail (dashed line), and the EB1-like sequence motif (blue line) are indicated. The N-terminal heptad repeats (abcdefg) are shown as blocks of seven amino-acid residues. Predicted N-terminal heptad repeat sequence extensions in unicellular organisms are shown in gray. Conserved residues are depicted in red (invariant) and bold (highly conserved). (B) Surface views of the EB1-like structural motif as seen perpendicular to the coiled-coil axis from two orientations 90° apart (same views as in Figure 3). The flexible peptide segments starting from Thr249 have been removed in both monomers for clarity. Highly conserved and surface accessible amino-acid residue side chains are indicated and colored according to the atom type: blue, nitrogen; red, oxygen; green, carbon. The positions of the polar and aromatic rims and of the hydrophobic cavity are indicated by arrows.
A primary interest of this structure determination was to gain insight into the spatial arrangement of the conserved residues of the unique EB1-like sequence motif (Figure 4A). Remarkably, most of the invariant and highly conserved residues form an extended and contiguous surface patch at the interface between the two monomer segments forming the four-helix bundle (Figure 4B). The charged side chains of Glu213′ (a prime is used to discriminate one monomer from the other), Arg214, and Asp215 form a polar rim. Similarly, the side chains of Phe216′, Tyr217′, and Phe218 form an aromatic rim. A wedge-like, predominantly hydrophobic groove is produced between the two rims. In the center of the surface patch, a prominent deep hydrophobic cavity is observed with the Leu221 and Leu246 residues at its floor. One wall is formed by the aromatic rim and the remaining walls are formed by the side chains of the residues Lys220′, Arg222, Glu225, Tyr247, and Ala248. Since the residues involved in the formation of these structural features are highly conserved throughout species and distributed along the entire length of the EB1-like sequence motif, they establish the sequence-to-structure relationship of the Conserved Domain Database sequence motif pfam03271.
For further description, we will refer to cavity A or B depending on whether its floor residues belong to monomer A or B. In the crystal structure, the 19 C-terminal residues are disordered in monomer A but partly ordered in monomer B. The length and sequence of the C-terminal EB1 peptide segment following α2 is less conserved but contains several invariant acidic residues (Figure 4A). Inspection of the structure and interactions of the nine additional residues ordered in monomer B revealed that this peptide segment interacts with both hydrophobic cavities albeit in a distinct manner. It is folded such that it forms an intramolecular interaction by occupying cavity B with its Thr249 side chain and an intermolecular contact by occupying cavity A of a neighboring dimer with its Ile255 side chain. The cavity B/threonine interaction is less attractive as a model for a putative tight interaction. It cannot be energetically very favorable, as it is not observed for monomer A. The cavity A/isoleucine interaction in turn may mimic a similar, biologically relevant interaction (see below). The tripeptide Val254B–Ile255B–Pro256B (from a neighboring dimer in the crystal) packs smoothly to the strictly conserved surface around cavity A (Figure 5A), and the main-chain carbonyl oxygen of Ile255B makes a hydrogen bond to the hydroxyl oxygen of the invariant side chain of Tyr217 (Figure 5B).
Figure 5.
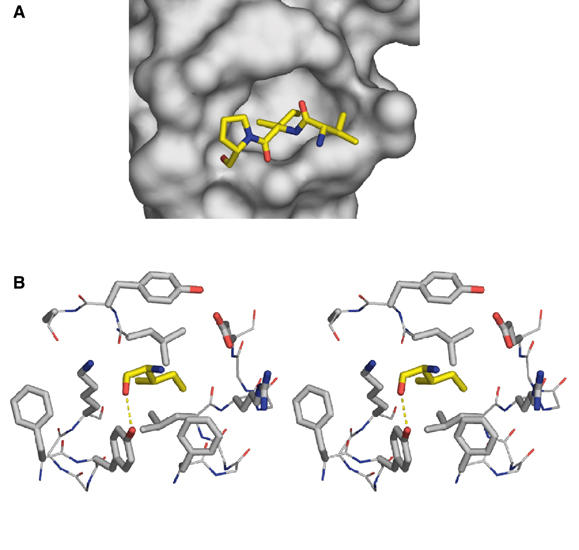
Peptide binding to the hydrophobic cavity of EB1-C. (A) Interaction of the tripeptide segment Val254B–Ile255B–Pro256B (in stick representation) originating from a neighboring EB1-C dimer in the crystal with cavity A (in surface representation). (B) Stereo view of cavity A with the cavity occupying Ile255 (yellow). All side chains are represented as thick stick models. The cavity-forming residues are as follows: left wall, Lys220′ and Phe216′; right wall, Arg222; lower wall, Phe216, Tyr217′, and Phe218; upper wall, Ala248, Tyr247, and Glu225; floor, Leu221 and Leu246. The intermolecular hydrogen bond formed between the main-chain carbonyl oxygen of Ile255 and the side-chain hydroxyl oxygen of Tyr217 is indicated. Residue side chains are colored according to the atom type: blue, nitrogen; red, oxygen; gray (for cavity-forming residues) or yellow (for Ile255), carbon.
Interaction of EB1-C with APC-derived C-terminal peptides
Based on pull-down experiments, the EB1–APC interaction has been mapped to a 40-residue C-terminal segment of APC (Bu and Su, 2003). The characteristic dimeric structure of EB1-C suggests that it may bind two copies of APC simultaneously. To test this hypothesis, a high sensitivity ITC binding study was performed with EB1 fragments and APC-derived C-terminal peptides. Figure 6B (inset of left panel) shows the exothermic ITC profile obtained at 25°C by titrating a 90 μM (monomer concentration) solution of EB1-C with the 39-mer human APC peptide Val2781–Lys2819 (1000 μM; referred to as C-APCp1). Analysis of the data showed that the best fit is obtained with a model that assumes n=1 independent and equal binding sites on the EB1-C monomer for C-APCp1. The fit (Figure 6B, left panel) yielded the equilibrium dissociation constant KD=5.1±0.2 μM.
Figure 6.
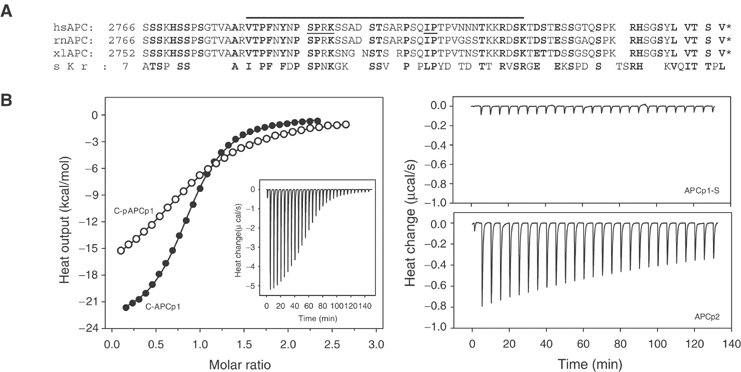
Binding of APC-derived C-terminal peptides to EB1-C. (A) Sequence alignment of carboxy-terminal APC segments from different species. Sequence identities and accession numbers are as follows: hs, Homo sapiens, SwissProt: P25054; rn, Rattus norvegicus, SwissProt: P70478; xl, Xenopus laevis, TrEMBL: P70039; sc, Saccharomyces cerevisiae, SwissProt: P32526. Conserved residues are depicted in bold. The thick line indicates the C-APCp1 peptide sequence (Val2781–Lys2819). The hsAPC Ile2805–Pro2806 dipeptide segment that was targeted for substitution and the Cdc2 consensus site Ser2789–Lys2792 are underlined. (B) ITC binding study. Left panel: Integrated heats of reaction (symbols) with the best fit to the data (lines). The fits were obtained with n=1 (referred to the EB1-C monomer) and yielded equilibrium dissociation constants of KD=5.1 (ΔH0app=−21.1 kcal/mol binding sites) and 17.9 μM (ΔH0app=−19.5 kcal/mol binding sites) for C-APCp1 (closed symbols) and C-pAPCp1 (phosphorylated at Ser2789; open symbols), respectively. Inset: Raw data obtained for 30 10 μl injections of C-APCp1 (1000 μM) into the sample cell containing 90 μM EB1-C. Right upper panel: Raw data obtained for C-APCp1-S (800 μM) into EB1-C (90 μM). Right lower panel: Raw data obtained for C-APCp2 (970 μM) into EB1-C (90 μM). The measurements were carried out at 25°C in PBS. The concentrations of EB1-C refer to the monomer.
To test whether an intact EB1-like structural motif is required for C-APCp1 binding, a similar ITC experiment was carried out with the stable dimeric CysEB1-Ca fragment under oxidizing conditions. The X-ray structure of EB1-C suggests that in CysEB1-Ca the upper and lower walls of the hydrophobic cavity are compromised, while the aromatic and polar rims are expected to be preserved (Figure 4B). No significant binding was observed by titrating a CysEB1-Ca with C-APCp1 under the same conditions used to assess the interaction between EB1-C and C-APCp1 (not shown). The interaction between the monomeric EB1-Cb fragment with C-APCp1 was probed by CD spectroscopy. The concentration-independent far-UV CD spectrum recorded at 5°C from C-APCp1 was characteristic for peptides with little if any secondary structure. No significant change in secondary structure was observed by incubating equimolar amounts of C-APCp1 with EB1-Cb, indicating that the peptides do not interact (not shown). Together, these findings suggest that an intact and dimeric EB1-like structural motif is important for the EB1–APC binding reaction.
Remarkably, within the C-APCp1 sequence, there is a single isoleucine followed by a proline (Ile2805 and Pro2806; Figure 6A) reminiscent of the tripeptide segment Val254–Ile255–Pro256 bound to the EB1-C cavity A (Figure 5). To test whether the Ile2805–Pro2806 dipeptide is critical for binding, an APC-p1 peptide variant in which these two residues were mutated to serine (referred to as C-APCp1-S) was analyzed by ITC. As shown in Figure 6B (right upper panel), binding of the mutant C-APCp1-S peptide to EB1-C is abolished. In contrast, a truncated 17-residue C-APCp1 variant (Ser2797–Thr2813; referred to as C-APCp2), which contains the isoleucine–proline dipeptide in its center, still specifically interacts with EB1-C although with reduced millimolar affinity (estimated KD of 300–400 μM; Figure 6B, right lower panel).
These findings demonstrate that the dipeptide segment Ile2805–Pro2806 plays an important role in anchoring C-APCp1 to EB1-C. However, the data obtained on the shorter C-APCp2 peptide suggest that additional C-APCp1 residues besides Ile2805–Pro2806 are necessary to increase the binding affinity. The carboxy terminus of APC is the target of Cdc2 in vivo at multiple sites (Trzepacz et al, 1997; Askham et al, 2000). Notably, the APC sequence segment Ser2789–Lys2792 represents such a Cdc2 consensus site (Figure 6A). To assess whether phosphorylation of Ser2789 affects binding of C-APCp1 to EB1-C, a phosphorylated peptide variant, referred to as C-APCp1-pSer2789, was analyzed by ITC (Figure 6B, left panel). The fit to the data was obtained with n=1 independent and equal binding sites on the EB1-C monomer and yielded the equilibrium dissociation constant KD=17.9±0.2 μM.
Discussion
Despite the central role of EB1 proteins in many if not all MT-based processes, their detailed molecular organization is still not known. Our biophysical and structural data establish that human EB1 is a very stable dimeric protein with a parallel coiled coil. The crystal structure of EB1-C reveals that the coiled-coil-mediated dimerization is essential for the formation of a highly conserved surface patch comprising a deep hydrophobic cavity and two rims, one polar and the other aromatic. The C-terminal partially conserved 19-residue EB1 tails are intrinsically disordered. The high sequence conservation of the EB1-like motif strongly suggests that all EB1 orthologues assemble into dimeric structures as depicted in Figure 7.
Figure 7.
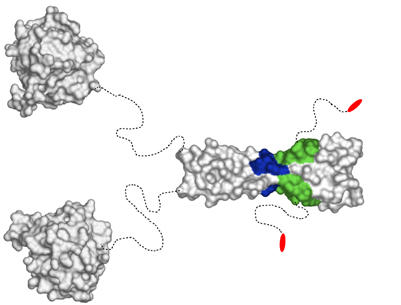
Structural organization of EB1 proteins and mapped +TIP-binding sites. The structures of the globular N-terminal MT-binding (PDB entry 1PA7) and C-terminal EB1 domains are depicted as surface views. Dashed and curved lines schematize flexible peptide segments of the dimeric molecule. The binding sites for APC (hydrophobic cavity and polar rim) and dynactin/p150glued (hydrophobic cavity, polar rim, and C-terminal dipeptide segment) are highlighted by colors: green, hydrophobic groove; blue, polar rim; red, C-terminal dipeptide (see also Figure 4B).
The highly conserved surface patch of EB1-C strongly supports a functional selection, which we propose to be driven by binding to APC. Our binding data suggest that (1) an intact EB1-C dimer interface is essential for the interaction, (2) the central deep hydrophobic cavity formed by the highly conserved side chains of helices α1 and α2′ plays a central role in the binding reaction, and (3) EB1-C can simultaneously bind two APC-derived carboxy-terminal 39-mer peptides with equal 5 μM affinity. Remarkably, mutating the Ile2805 and Pro2806 APC residues to serine completely abolishes binding of C-APCp1 to EB1-C, demonstrating the essential role of this dipeptide segment in the interaction. However, the very high dissociation constant obtained for the 17-mer C-APCp2 peptide demonstrates that additional interactions with the surrounding conserved EB1-C surface are needed to increase binding affinity. Consistent with this conclusion, a mutational study targeting Glu211, Glu213, and Asp215 indicates that EB1-C's polar rim (Figure 4B) may also be critical for the formation of the EB1–APC complex (Wen et al, 2004).
The extensive presence of low-complexity sequence stretches in C-APC together with the fact that the EB1-binding site is preserved in a short sequence segment (Val2781–Lys2819; Figure 6A) makes an intrinsically disordered structure for the carboxy termini of APC appear very likely. This suggests that the EB1–APC interaction is established between a folded domain and a flexible polypeptide chain segment reminiscent of the one formed between EB1-C and C-APCp1. The deep hydrophobic cavity seen in the EB1-C crystal structure is thus expected to serve as a binding site into which the APC Ile2805 side chain is anchored like the tripeptide segment bound to cavity A (Figure 5). Consistent with this conclusion, mutating Lys220 and Arg222 to alanine, which is expected to compromise the left and right walls, respectively, of the EB1-C cavity (Figures 4B and 5), abolishes the EB1–APC interaction, inhibits stable MT formation in vivo, and reduces cell migration (Wen et al, 2004). Notably, the APC Ile2805–Pro2806 sequence motif is strictly conserved in the rat and frog APC orthologues (Figure 6A). In the budding yeast APC-related Kar9p molecule, whose carboxy-terminal domain is also predicted to be intrinsically disordered, the isoleucine is conservatively replaced by a leucine residue (Leu510). The isoleucine side chain fits optimally into the cavity but the one of a leucine appears equally suitable. The role of the invariant proline residue may be to restrict the number of possible conformations the polypeptide backbone can assume upon binding.
The moderate but specific affinity of C-APCp1 for EB1-C is consistent with the observed dynamic and transient nature of EB1–APC in vivo (Mimori-Kiyosue and Tsukita, 2001). The ∼four-fold decreased binding of the phosphorylated C-pAPCp1 peptide suggests that the site encompassing Ser2789 is implicated in the regulation of the interaction. It appears likely that further downregulation of the APC affinity for EB1 is mediated by phosphorylation of additional serine residues present in the sequence segment Val2781–Lys2819 (Trzepacz et al, 1997; Askham et al, 2000; Bu and Su, 2003). However, Ser2789 is a conserved Cdc2 phosphorylation site (Trzepacz et al, 1997) that may play an important role in modulating the association of APC and EB1 during mitosis (Askham et al, 2000). Moreover, phosphorylation of Ser2789 inhibits the EB1–APC-promoted MT polymerization activity in vitro (Nakamura et al, 2001). In S. cerevisiae, Cdc28 (the yeast Cdc2 orthologue) phosphorylation of the corresponding conserved Kar9p Ser496 residue is critical for asymmetric loading of the yeast molecule onto MTs at the spindle pole and for proper MT capture and guidance (Liakopoulos et al, 2003). In analogy to EB1–APC, immunoprecipitation experiments suggest that the effect of Cdc28 phosphorylation is to inhibit the ability of Kar9p to interact with the EB1 orthologue Bim1p (Liakopoulos et al, 2003). Together, these findings support our conclusion that the APC sequence segment Val2781–Lys2819 represents a major interaction site between APC and EB1 in vivo. They further suggest that specific C-APC phosphorylation is a mechanism for regulating the EB1–APC complex during the cell cycle.
The structural organization of EB1-C (Figure 7) also rationalizes functional data obtained on the EB1–dynactin/p150glued interaction. A fragmentation analysis suggests that the last two conserved carboxy-terminal EB1 residues, Glu267 and Tyr268, are essential for the association with the p150glued CAP-Gly domain (Bu and Su, 2003). However, a mutational study indicates that both polar rim and hydrophobic cavity also contribute to complex formation (Wen et al, 2004). The fact that the APC–EB1 and EB1–dynactin/p150glued interactions appear to be mediated by partially overlapping parts of EB1-C is consistent with the observation that EB1 may form mutually exclusive complexes with these proteins under certain conditions (Askham et al, 2002).
In conclusion, our analysis reveals that a central aspect of both interactions, APC–EB1 and EB1–dynactin/p150glued, is the involvement of a flexible polypeptide chain segment and a folded binding site. Such an interaction mode is well suited to recruit different molecular functionalities to a common location without imposing undesired structural constraints on the resulting assemblies. Moreover, as expected from in vivo observations, such regulated (e.g., via phosphorylation events) protein–protein interactions provide a basis for understanding the dynamic crosstalk among +TIP proteins at growing MT ends, the anchoring of MT tips to cellular structures such as centrosomes, and the delivery of proteins to the cell periphery (Carvalho et al, 2003; Galjart and Perez, 2003).
Materials and methods
Construction of expression plasmids
The EST clone IMAGE:3922022 was used as a template for PCR amplification of the complete coding sequence of the human EB1 gene. The amplified product was ligated into a modified pET-15b (Novagen) bacterial expression vector at the BamHI site. For the EB1-derived fragments, the cloned full-length EB1 cDNA was used as a template for PCR amplification. The PCR products were ligated into the bacterial expression vectors pPEP-T (Brandenberger et al, 1996) for CysEB1-Ca, into pHisTrx (Frank et al, 2002) for EB1-Cb, and into a modified pET-15b for EB1-C at the BamHI–EcoRI sites, respectively. The inserted sequences of all constructs were verified by Sanger dideoxy DNA sequencing.
Protein and peptide preparations
The Escherichia coli host strains BL21(DE3) (Stratagene) and JM109(DE3) (Promega) were used for expression. Bacteria were grown at 37°C in LB medium containing 100 mg/l ampicillin. Bacterial cultures were induced at OD600=0.8 by adding IPTG to 1 mM and incubating at 37°C for 4 h. Affinity purification of the 6xHis-tagged fusion proteins by immobilized metal affinity chromatography on Ni2+-Sepharose (Amersham) was performed under native conditions at room temperature as described in the manufacturer's instructions.
For separation of the recombinant proteins from the N-terminal carrier polypeptides, proteins were dialyzed against thrombin cleavage buffer (20 mM Tris–HCl, pH 8.4, 150 mM NaCl, 2.5 mM CaCl2). Proteolytic cleavage was carried out for 8–24 h at room temperature using human thrombin (Sigma) at a concentration of 5 U/mg recombinant protein. Unexpectedly, nonspecific cleavage was observed for the full-length EB1 protein. N-terminal amino-acid sequencing and mass spectrometric analyses of processed protein fragment bands revealed that cleavage occurred within the EB1 sequence Pro145–Pro161. The processed poypeptides were separated from the oligohistidine containing polypeptide tags by reapplication to immobilized metal affinity columns.
The homogeneity of the recombinant proteins was confirmed by either 15% SDS–PAGE or tricine–SDS–PAGE (Schagger and von Jagow, 1987) and their identities were assessed by mass spectral analyses. The identity of the EB1-Cb fragment was further confirmed by N-terminal sequencing. Protein samples were dialyzed against PBS (5 mM sodium phosphate buffer, pH 7.4, 150 mM NaCl).
The N-acetylated and C-amidated APC-derived peptides were assembled on an automated continuous-flow synthesizer employing standard methods. The purity of the peptides (∼95%) was verified by reversed-phase analytical HPLC and their identities were assessed by mass spectral analysis.
Exact concentrations of protein and peptide solutions were determined by tyrosine absorbance at 276 nm in 6 M GuHCl (Edelhoch, 1967).
Biophysical characterization
For CD spectroscopy, protein samples were in PBS. Reducing buffer conditions were obtained by supplementing samples with 1 mM DTT. Far-UV CD spectra and thermal unfolding profiles were recorded on a J-810 spectropolarimeter (Jasco Inc.) equipped with a temperature-controlled quartz cell of 0.1 cm path length. The spectra shown are the averages of five accumulations. The data were evaluated with the Jasco and Sigma Plot (Jandel Scientific) software. A ramping rate of 1°C/min was used to record the thermal unfolding profiles. The apparent midpoints of the transitions, Tm's, were taken as the maximum of the derivative d[Θ]222/dT.
AUC was performed on an Optima XL-A analytical ultracentrifuge (Beckman Instruments) equipped with an An-60ti rotor. The recombinant EB1 molecules were analyzed in PBS and protein concentrations were adjusted to 0.1–0.5 mg/ml in PBS supplemented with 1 mM DTT. Sedimentation velocity experiments were performed at 54 000 r.p.m. in a 12 mm epon double-sector cell. Sedimentation coefficients were corrected to water by the standard procedure (Eason, 1986). Sedimentation equilibrium runs were performed at 15 000 and 21 000 r.p.m. for EB1 and at 36 000 r.p.m. for CysEB1-Ca. For all samples, a partial specific volume of 0.73 ml/g was assumed.
Electron micrographs were taken in a Philips Morgagni TEM operated at 80 kV equipped with a Megaview III CCD camera. Protein samples (0.5 mg/ml) in PBS were supplemented with glycerol to a final concentration of 30%. The samples were subsequently sprayed onto freshly cleaved mica and rotary shadowed in a BA 511 M freeze-etch apparatus (Balzers) with platinum/carbon at an elevation angle of 3–5° (Fowler and Aebi, 1983). It should be noted that the TEM and AUC experiments on full-length EB1 were carried out with the uncleaved 6xHis-tagged protein.
High-sensitivity ITC experiments to assess the interaction between EB1-C and APC-derived peptides were performed at 25°C in PBS using a VP-ITC calorimeter (Microcal Inc., Northampton, MA). For each experiment, the sample cell (volume 1.4 ml) was filled with an ∼100 μM EB1-C solution. A 300 μl syringe was filled with an ∼1 mM APC peptide solution (present in the same buffer as EB1-C). The reference cell contained water. Typically, 10 μl of APC peptide aliquots from the stirred syringe (305 r.p.m.) were injected 30 times into the sample cell. At each injection, APC peptide was bound to EB1-C, leading to a characteristic heat signal. Integration of the individual calorimeter traces yielded the heat of binding, hi, of each injection step. The binding isotherms were fitted via a nonlinear least squares minimization method to determine the binding stoichiometry, n, the equilibrium binding constant, KD, and the apparent change in enthalpy, ΔH0.
Crystal structure determination
EB1-C crystals grew within 1 week from a 15–20 mg/ml stock solution at 20°C using the sitting drop method. The crystals exhibited space group P21 (no. 4) and were grown using a reservoir solution containing 50 mM sodium citrate at pH 4.0–4.5, 10% PEG 3350, and 4% γ-butyrolactone (Sigma). Data sets were collected using CuKα radiation produced by an Enraf-Nonius FR591 rotating anode generator. A total of 360 rotation images of 0.5° were recorded on a MAR345 imaging plate. The structure of EB1-C was solved by SIRAS using a mercury derivative obtained by overnight soaks of crystals in the presence of 1 mM thiomersal. Iterative rounds of model building and maximum likelihood refinement resulted in a complete 1.54 Å resolution model for the EB1-C dimer. The final model converged at an R/Rfree of 0.18/0.22 with very good stereochemistry. Data sets and refinement statistics are given in Supplementary Table 1. Figures were prepared with the program PyMOL (De-Lano Scientific LLC, San Carlos, CA, www.pymol.org). The structure of EB1-C has been deposited in the Protein Data Bank (PDB entry 1WU9).
Supplementary Material
Supplementary Figure
Supplementary Table
Acknowledgments
We are indebted to Drs Y Barral, D Fitzgerald, RA Kammerer, and D Liakopoulos for insightful discussions, to Dr I Jelesarov for assistance with the ITC experiments, and to A Lustig for performing the AUC analysis. We are grateful to the Interdisciplinary Center for Microscopy of the University of Basel and to the Institute for Molecular Biology and Biophysics of ETH Zürich for access to the TEM and CD spectropolarimeter, respectively. This work was supported by the Swiss National Science Foundation grant #31-64978.01 (to MOS) and by the Eidgenössische Technische Hochschule Zürich grant #TH-42/02-2 (to FKW and MOS).
References
- Askham JM, Moncur P, Markham AF, Morrison EE (2000) Regulation and function of the interaction between the APC tumour suppressor protein and EB1. Oncogene 19: 1950–1958 [DOI] [PubMed] [Google Scholar]
- Askham JM, Vaughan KT, Goodson HV, Morrison EE (2002) Evidence that an interaction between EB1 and p150(Glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol Biol Cell 13: 3627–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrueta L, Kraeft SK, Tirnauer JS, Schuyler SC, Chen LB, Hill DE, Pellman D, Bierer BE (1998) The adenomatous polyposis coli-binding protein EB1 is associated with cytoplasmic and spindle microtubules. Proc Natl Acad Sci USA 95: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrueta L, Tirnauer JS, Schuyler SC, Pellman D, Bierer BE (1999) The APC-associated protein EB1 associates with components of the dynactin complex and cytoplasmic dynein intermediate chain. Curr Biol 9: 425–428 [DOI] [PubMed] [Google Scholar]
- Bienz M (2001) Spindles cotton on to junctions, APC and EB1. Nat Cell Biol 3: E67–E68 [DOI] [PubMed] [Google Scholar]
- Brandenberger R, Kammerer RA, Engel J, Chiquet M (1996) Native chick laminin-4 containing the beta 2 chain (s-laminin) promotes motor axon growth. J Cell Biol 135: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W, Su LK (2003) Characterization of functional domains of human EB1 family proteins. J Biol Chem 278: 49721–49731 [DOI] [PubMed] [Google Scholar]
- Busch KE, Hayles J, Nurse P, Brunner D (2004) Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev Cell 6: 831–843 [DOI] [PubMed] [Google Scholar]
- Carvalho P, Tirnauer JS, Pellman D (2003) Surfing on microtubule ends. Trends Cell Biol 13: 229–237 [DOI] [PubMed] [Google Scholar]
- Cohen C, Parry DA (1990) Alpha-helical coiled coils and bundles: how to design an alpha-helical protein. Proteins 7: 1–15 [DOI] [PubMed] [Google Scholar]
- Dikovskaya D, Zumbrunn J, Penman GA, Nathke IS (2001) The adenomatous polyposis coli protein: in the limelight out at the edge. Trends Cell Biol 11: 378–384 [DOI] [PubMed] [Google Scholar]
- Eason R (1986) Centrifugation, a practical approach. In Analytical Ultracentrifugation, Rickwood D (ed) pp 251–286. IRL Press: Oxford (UK) and Washington, DC [Google Scholar]
- Edelhoch H (1967) Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH, Clevers H (2001) Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol 3: 433–438 [DOI] [PubMed] [Google Scholar]
- Fowler WE, Aebi U (1983) Preparations of single molecules and supramolecular complexes for high-resolution metal shadowing. J Ultrastruct Res 83: 319–334 [DOI] [PubMed] [Google Scholar]
- Frank S, Schulthess T, Landwehr R, Lustig A, Mini T, Jeno P, Engel J, Kammerer RA (2002) Characterization of the matrilin coiled-coil domains reveals seven novel isoforms. J Biol Chem 277: 19071–19079 [DOI] [PubMed] [Google Scholar]
- Galjart N, Perez F (2003) A plus-end raft to control microtubule dynamics and function. Curr Opin Cell Biol 15: 48–53 [DOI] [PubMed] [Google Scholar]
- Goodson HV, Skube SB, Stalder R, Valetti C, Kreis TE, Morrison EE, Schroer TA (2003) CLIP-170 interacts with dynactin complex and the APC-binding protein EB1 by different mechanisms. Cell Motil Cytoskeleton 55: 156–173 [DOI] [PubMed] [Google Scholar]
- Gundersen GG (2002) Evolutionary conservation of microtubule-capture mechanisms. Nat Rev Mol Cell Biol 3: 296–304 [DOI] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262: 1401–1407 [DOI] [PubMed] [Google Scholar]
- Hayashi I, Ikura M (2003) Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J Biol Chem 278: 36430–36434 [DOI] [PubMed] [Google Scholar]
- Juwana JP, Henderikx P, Mischo A, Wadle A, Fadle N, Gerlach K, Arends JW, Hoogenboom H, Pfreundschuh M, Renner C (1999) EB/RP gene family encodes tubulin binding proteins. Int J Cancer 81: 275–284 [DOI] [PubMed] [Google Scholar]
- Kusch J, Liakopoulos D, Barral Y (2003) Spindle asymmetry: a compass for the cell. Trends Cell Biol 13: 562–569 [DOI] [PubMed] [Google Scholar]
- Liakopoulos D, Kusch J, Grava S, Vogel J, Barral Y (2003) Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell 112: 561–574 [DOI] [PubMed] [Google Scholar]
- Ligon LA, Shelly SS, Tokito M, Holzbaur EL (2003) The microtubule plus-end proteins EB1 and dynactin have differential effects on microtubule polymerization. Mol Biol Cell 14: 1405–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie RK, Bahmanyar S, Siemers KA, Votin V, Chang P, Stearns T, Nelson WJ, Barth AI (2004) Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J Cell Sci 117: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S (2000) The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol 10: 865–868 [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Tsukita S (2001) Where is APC going? J Cell Biol 154: 1105–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Zhou XZ, Lu KP (2001) Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. Curr Biol 11: 1062–1067 [DOI] [PubMed] [Google Scholar]
- Rehberg M, Gräf R (2002) Dictyostelium EB1 is a genuine centrosomal component required for proper spindle formation. Mol Biol Cell 13: 2301–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD (2002) Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol 158: 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Hacker U, Turck C, Vale RD (2004) Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr Biol 14: 1827–1833 [DOI] [PubMed] [Google Scholar]
- Schagger H, von Jagow G (1987) Tricine–sodium dodecyl sulfate–polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem 166: 368–379 [DOI] [PubMed] [Google Scholar]
- Schroer TA (2001) Microtubules don and doff their caps: dynamic attachments at plus and minus ends. Curr Opin Cell Biol 13: 92–96 [DOI] [PubMed] [Google Scholar]
- Su LK, Burrell M, Hill DE, Gyuris J, Brent R, Wiltshire R, Trent J, Vogelstein B, Kinzler KW (1995) APC binds to the novel protein EB1. Cancer Res 55: 2972–2977 [PubMed] [Google Scholar]
- Tirnauer JS, Bierer BE (2000) EB1 proteins regulate microtubule dynamics, cell polarity, and chromosome stability. J Cell Biol 149: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzepacz C, Lowy AM, Kordich JJ, Groden J (1997) Phosphorylation of the tumor suppressor adenomatous polyposis coli (APC) by the cyclin-dependent kinase p34. J Biol Chem 272: 21681–21684 [DOI] [PubMed] [Google Scholar]
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG (2004) EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol 6: 820–830 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure
Supplementary Table


