Abstract
Soj and Spo0J of the Gram-negative hyperthermophile Thermus thermophilus belong to the conserved ParAB family of bacterial proteins implicated in plasmid and chromosome partitioning. Spo0J binds to DNA near the replication origin and localises at the poles following initiation of replication. Soj oscillates in the nucleoid region in an ATP- and Spo0J-dependent fashion. Here, we show that Soj undergoes ATP-dependent dimerisation in solution and forms nucleoprotein filaments with DNA. Crystal structures of Soj in three nucleotide states demonstrate that the empty and ADP-bound states are monomeric, while a hydrolysis-deficient mutant, D44A, is capable of forming a nucleotide ‘sandwich' dimer. Soj ATPase activity is stimulated by Spo0J or the N-terminal 20 amino-acid peptide of Spo0J. Our analysis shows that dimerisation and activation involving a peptide containing a Lys/Arg is conserved for Soj, ParA and MinD and their modulators Spo0J, ParB and MinE, respectively. By homology to the nitrogenase iron protein and the GTPases Ffh/FtsY, we suggest that Soj dimerisation and regulation represent a conserved biological switch.
Keywords: chromosome segregation, MinCD, ParAB, Soj, Spo0J
Introduction
The equipartitioning of newly replicated chromosomes into the daughter cells is a crucial but poorly understood step in bacterial cell division (Gordon and Wright, 2000). The plasmid-partitioning proteins SopAB of F factor and ParAB of Escherischia coli plasmid P1 are required for faithful DNA segregation (Nordstrom and Austin, 1989; Hiraga, 1992) and their chromosomally encoded homologues, Soj and Spo0J, have been shown to be implicated in the partitioning of chromosomal DNA (Draper and Gober, 2002). Soj and Spo0J may function to orient the oriC regions toward the poles (Sharpe and Errington, 1996; Glaser et al, 1997; Lewis and Errington, 1997; Lin et al, 1997; Mohl and Gober, 1997).
Spo0J (ParB) is a classical helix–turn–helix DNA-binding protein (Khare et al, 2004; Leonard et al, 2004), which binds directly to cis-acting centromere-like elements, parS, located in the origin-proximal region of the chromosome (Lin and Grossman, 1998). Complexes of ParB proteins bound to newly replicated nucleoids have been detected as discrete bipolar foci coincident with oriC in living cells; their duplication and abrupt separation strongly advocate the existence of an active mitotic-like mechanism of DNA segregation in bacteria (Glaser et al, 1997; Gordon et al, 1997; Lin et al, 1997; Mohl and Gober, 1997; Niki and Hiraga, 1997). Hence, the prokaryotic origin-proximal region appears to be the counterpart of the eukaryotic centromere (Wheeler and Shapiro, 1997).
Soj and other ParA proteins are members of a large family of ATPases that include the bacterial cell division regulator MinD, nitrogenase iron protein involved in biological nitrogen fixation and the anion pump ATPase ArsA (Koonin, 1993). Soj has been shown to associate with the promoter regions and inhibit the transcription of several early sporulation genes in vivo, an effect which is antagonised by Spo0J (Ireton et al, 1994; Quisel et al, 1999; Quisel and Grossman, 2000). Soj is believed to repress transcription by binding to single-stranded DNA in the open transcription complex (Cervin et al, 1998; Quisel et al, 1999; Quisel and Grossman, 2000). It is also known to play a role in the formation of condensed Spo0J foci on oriC (Marston and Errington, 1999), but deletion of Soj alone does not seem to have a significant effect on chromosome segregation (Ireton et al, 1994). However, recent work on the roles of the DNA-binding protein RacA and DivIVA of Bacillus subtilis in prespore chromosome segregation has indicated a level of redundancy in the system: specifically, in the absence of Soj and RacA, a ΔdivIVA-like defect in prespore chromosome segregation is observed and deletion of RacA, Soj and Spo0J results in the elimination of the oriC specificity of orientation of the prespore chromosome altogether (Wu and Errington, 2002). Moreover, Soj is required together with Spo0J for stable maintenance of a plasmid bearing a parS site, indicating that it does function in parS-Spo0J-mediated partitioning (Lin and Grossman, 1998). Localisation studies of Soj have shown dynamic oscillation in spo0J+ cells compared with static nucleoid association in a Δspo0J background. It has been deduced that ParB of Caulobacter crescentus acts as a nucleotide exchange factor for ParA, stimulating the rapid exchange of ADP for ATP (Quisel et al, 1999; Easter and Gober, 2002; Figge et al, 2003). The N-terminal regions of ParB of C. crescentus, ParB of plasmid P1 and SopB of F plasmid have all been shown to be the determinants for interaction with ParA/SopA (Radnedge et al, 1998; Figge et al, 2003; Ravin et al, 2003).
In the case of the nitrogenase iron protein, the ATPase acts as a molecular switch during biological nitrogen fixation, coupling nucleotide hydrolysis to electron transfer in a multi-protein complex (Schindelin et al, 1997). Nucleotide hydrolysis is achieved by the formation of a nucleotide ‘sandwich' dimer. It has been postulated that MinD forms a similar dimer dependent on ATP (Hu et al, 2003; Lutkenhaus and Sundaramoorthy, 2003). However, published structures of MinD proteins have shown them to be exclusively monomeric (Cordell and Löwe, 2001; Hayashi et al, 2001; Sakai et al, 2001), even when co-crystallised with the nonhydrolysable ATP analogue, AMPPCP (Hayashi et al, 2001).
The MinD protein of E. coli regulates division site selection in conjunction with the division inhibitor MinC and the topological specificity factor MinE, and exhibits an oscillatory pattern similar to that of Soj (Raskin and de Boer, 1999). MinCD oscillation is dependent on MinE, which stimulates MinD ATPase activity in vitro (Hu and Lutkenhaus, 2001), in a seemingly analogous fashion to Spo0J-dependent oscillation of Soj (Quisel et al, 1999). MinD undergoes ATP- and membrane-dependent self-association by virtue of a conserved C-terminal amphipathic helix which associates with the lipid bilayer (Lutkenhaus and Sundaramoorthy, 2003). Members of the ParA family, including Soj, do not contain this helix and do not associate with the membrane, but have been shown to be dynamically associated with the nucleoid in vivo (Marston and Errington, 1999; Quisel et al, 1999; Ebersbach and Gerdes, 2001).
Here, we present structural and biochemical evidence that Thermus thermophilus Soj forms an ATP-dependent ‘sandwich' dimer, shows dimerisation-dependent DNA-binding activity, forming nucleoprotein filaments, and that ATP hydrolysis is activated by a short N-terminal Spo0J peptide. We present data suggesting that a conserved mechanism of activation most likely exists for Spo0J activation of Soj, ParB activation of ParA and MinE activation of MinD. We hypothesise that the nucleotide-dependent dimerisation of the ATPase acts as a molecular switch, which regulates binding to the nucleoid, the membrane or another surface within the bacterial cell. The emerging parallels between the MinCDE and Soj–Spo0J systems suggest a conserved mechanism of spatiotemporal regulation of protein localisation in bacteria.
Results and discussion
Soj undergoes ATP-dependent dimerisation in solution
We investigated the oligomeric state of Soj in solution by size exclusion chromatography (Figure 1) and sedimentation velocity experiments (Figure 2). Size exclusion chromatography of wild-type Soj indicates that it is a monomer in the absence of nucleotide and in the presence of ADP. In the presence of ATP, wild type Soj eluted as two species with retention times indicative of a dimer–monomer equilibrium (Figure 1A; Table I). A mutant Soj deficient in nucleotide hydrolysis, D44A, was constructed based on the structure of the Soj monomer, which will be discussed later. D44 would be expected to co-ordinate the attacking nucleophile in the ATP-bound state. Soj D44A elutes as a single peak in the absence of nucleotide and in the presence of ADP, with the retention time of the wild-type monomeric protein. In the presence of ATP, Soj D44A elutes as a single peak with a retention time of the dimeric wild-type protein (Figure 1B; Table I).
Figure 1.
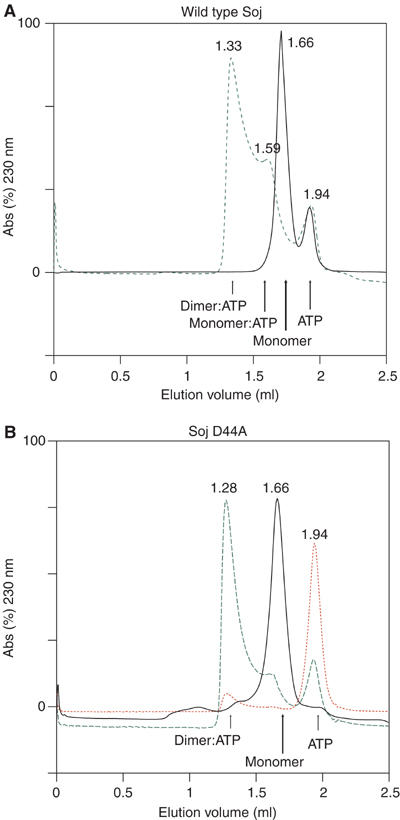
Size exclusion chromatography of T. thermophilus wild-type Soj and Soj D44A in the presence and absence of ATP. The elution volume of each species is indicated in millilitres above the respective trace. (A) In the absence of ATP, wild-type Soj elutes as a monomeric species (black). In the presence of ATP, Soj elutes as a dimer–monomer equilibrium (green). (B) Soj D44A, deficient in nucleotide hydrolysis, also elutes as a monomer in the absence of ATP (black), but elutes almost solely as a dimer in the presence of ATP (green). Red: absorbance at 260 nm indicating the elution volume of ATP and the presence of ATP in the dimeric protein.
Figure 2.

Sedimentation velocity analysis of wild-type Soj (A, E), Soj K20A (B, F), Soj D44A (C, G) and Soj G16V (D, H), without and with ATP, respectively. For each sample, a series of scans (at intervals of ∼15 min) are shown. Also shown are the residuals from fitting a series of 12 scans (at intervals of ∼1.5 min), near the middle of the sedimentation run, to a model for either one component (most runs) or two components specifically for mutant D44A+ATP. This fitting was used to estimate the sedimentation and diffusion coefficients, and hence calculate the molecular mass, for each component and the random distributions of the residuals suggest that the fits are valid.
Table 1.
S20,w and Mr for TTJ wt and mutants
| Without ATP |
+ ATP |
|||
|---|---|---|---|---|
| S20,w (Svedbergs) | Mr (kDa) | S20,w (Svedbergs) | Mr (kDa) | |
| wt | 2.37±0.01 | 29.0±0.5 | 2.77±0.02 | 25.0±1.0 |
| K20A | 2.45±0.01 | 27.0±0.5 | 2.45±0.02 | 24.6±0.9 |
| D44A | 2.31±0.01 | 25.1±0.5 | 2.75±0.05 | 24.9±2.4 |
| 9.17±0.02 | 58.3±1.4 | |||
| G16V | 2.32±0.01 | 27.5±0.7 | 2.94±0.01 | 24.7±0.6 |
Sedimentation of wild-type Soj, Soj K20A and Soj D44A in the absence of nucleotide showed them all to be monomeric, with an estimated molecular weight of 27.0±2.0 kDa (Figure 2A–C). Sedimentation of wild-type Soj and Soj K20A in the presence of ATP gave identical spectra, consistent with the proteins being monomeric (Figure 2E–F). However, sedimentation of Soj D44A in the presence of ATP gave a sedimentation profile indicating the presence of an additional species with a sedimentation velocity corresponding to a doubling of the molecular weight from monomer to dimer (Figure 2G). Two-thirds of Soj D44A was observed to be dimeric and one-third monomeric. The P-loop mutant Soj G16V (Soj G12V in B. subtilis) was observed to sediment as a monomer, despite binding ATP (Figure 2D and H). This mutant is deficient in dimerisation because of a steric clash in the active site.
We conclude that Soj dimerises in solution dependent on ATP and is exclusively monomeric in the absence of nucleotide and in the presence of ADP. ATP hydrolysis actively promotes dissociation of the dimer.
Soj binds nonspecifically to DNA in an ATP-dependent manner
To investigate the ability of Soj to bind to DNA, electrophoretic mobility shift assays were performed. We found that Soj is able to shift a 2.9 kbp plasmid in a concentration-dependent fashion to a maximal shift at which the DNA is saturated with protein (Figure 3). Two bands of DNA are observed in the gel shift assays corresponding to supercoiled plasmid DNA and a smaller fraction of relaxed, open-circle DNA. The ability of Soj to shift DNA is strictly dependent on ATP (Figure 3C and F) and is only weakly observed at saturating protein concentrations in the absence of nucleotide or in the presence of ADP (Figure 3A, B, D and E). Soj K20A, which is unable to bind nucleotide, is unable to shift DNA at any protein concentration (Figure 3G–I). Soj D44A binds to DNA efficiently at all protein concentrations, as judged by the uniform appearance of a single shifted band (Figure 3F). Wild-type Soj shows a high efficiency of binding, but the pattern of shifted bands is characterised by the appearance of smeared bands of lower molecular weight (Figure 3C), an observation most likely attributable to the time-dependent dissociation of Soj as a consequence of ATP hydrolysis. Soj G16V is incapable of effectively shifting DNA in an ATP-dependent manner, confirming that ATP-dependent dimerisation is the critical requirement for assembly of the protein onto DNA (Figure 3J–L). The steric clash between the valine residues in this mutant is only relevant in the ATP-bound conformation; hence, at high protein concentrations in the absence of nucleotide and in the presence of ADP, the protein shows a notable propensity to shift the DNA (Figure 3J–K). This effect, also observed to an extent with the wild-type protein (Figure 3A–B), is most likely a concentration-dependent effect that ‘dimerises' the protein in the absence of nucleotide. We conclude that ATP-dependent dimerisation of Soj is critical for DNA binding.
Figure 3.
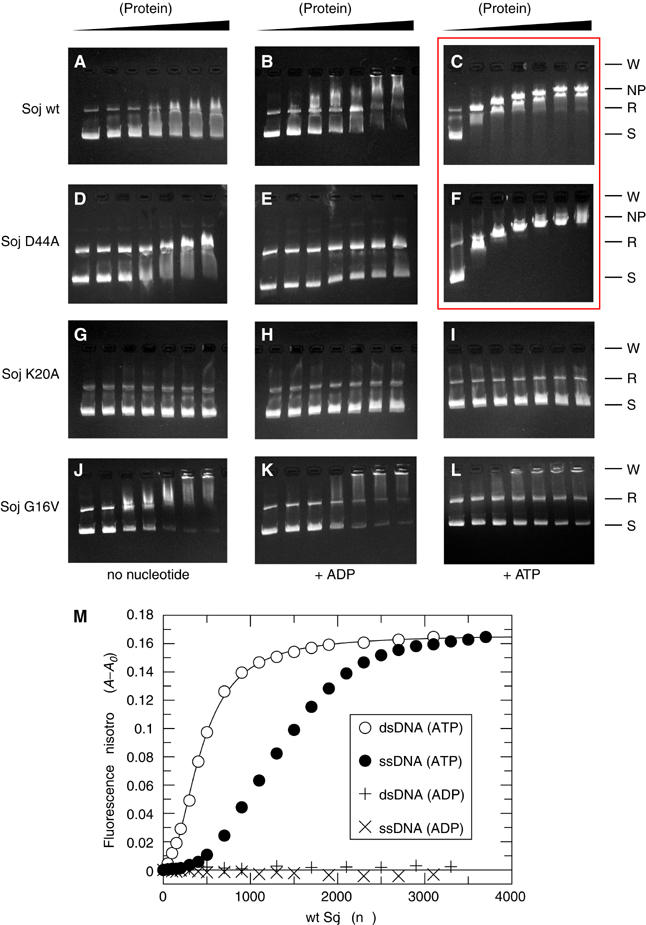
Electrophoretic mobility shift assays of Soj DNA binding. Wild-type Soj, hydrolysis-deficient Soj D44A, nucleotide-binding-deficient Soj K20A and dimerisation-deficient Soj G16V bind to DNA only weakly at saturating protein concentrations in the absence of nucleotide (A, D, G, J, left) and in the presence of ADP (B, E, H, K, middle). Soj K20A and Soj G16V fail to bind DNA in the presence of ATP (I, L), indicating that ATP-dependent dimerisation is necessary for DNA binding. Wild-type Soj and Soj D44A shift DNA in a concentration-dependent fashion until saturation in the presence of ATP (C, F). Hydrolysis of ATP by wild-type Soj results in the presence of multiple shifted bands of various molecular weights (C), while hydrolysis-deficient Soj D44A produces a single shifted band at all protein concentrations (F). Key: W=wells, NP=nucleoprotein filament, R=relaxed, S=supercoiled. (M) Fluorescence anisotropy analysis of ATP-dependent Soj binding to DNA.
While the ATP-dependent polymerisation of Soj on DNA could be a simple charge effect, it is consistent with in vivo localisation studies which show that wild-type Soj dynamically associates with the nucleoid in an Spo0J-dependent fashion and remains statically associated with the nucleoid in a Δspo0J background (Marston and Errington, 1999; Quisel et al, 1999). In addition, Quisel et al showed that the P-loop mutant G12V of B. subtilis does not associate with the nucleoid in a Δspo0J background and its dynamic behaviour is abolished. Furthermore, the MinD protein of E. coli has been shown to undergo surface-dependent polymerisation. MinD assembles into protein filaments in an ATP- and phospolipid-dependent manner (Lackner et al, 2003), supporting in vivo localisation studies which show that MinD localises to the polar membrane (Raskin and de Boer, 1999) and oscillates in membrane-associated coiled structures that extend between the cell poles (Shih et al, 2003). It is worthwhile mentioning at this point that the similarity between the Soj and MinD proteins is so great that, it appears to us, mistakes have been made in the assignment of these proteins, such that there are in fact two published ‘MinD' structures which are more than likely members of the Soj family, lacking the MinD-characteristic C-terminal amphipathic helix which mediates membrane association (Szeto et al, 2002; Hu and Lutkenhaus, 2003).
Soj binds preferentially to double-stranded DNA
To investigate whether Soj binds preferentially to double-stranded or single-stranded DNA, fluorescence anisotropy measurements (Figure 3M) were made using a 5′-fluorescein-labelled oligonucleotide (see Materials and methods). There is no change in the anisotropy of the DNA for Soj in the presence of ADP, indicating that Soj:ADP does not bind to either double-stranded or single-stranded DNA. In the presence of ATP, however, Soj binds to both double-stranded and single-stranded DNA. Binding of Soj to double-stranded DNA can be fitted by the Hill equation for cooperative binding, yielding a Hill coefficient of 2.11. Binding of Soj to single-stranded DNA is approximately 3.5-fold less efficient and cannot be fitted by the Hill equation, although a sigmoidal curve is apparent. We conclude that Soj only binds DNA in the ATP-bound form and that Soj binds preferentially to double-stranded DNA in a cooperative fashion.
Soj does not form polymers in the absence of DNA
To investigate the ability of Soj to form polymers independent of DNA, we pelleted Soj in the presence and absence of nucleotide and in the presence and absence of DNA. Soj was found exclusively in the supernatant in the absence of DNA, irrespective of the presence of ATP (Figure 2). In the presence of DNA, a large fraction of Soj was found in the pellet only in the presence of ATP and this fraction was greater for the hydrolysis-deficient Soj D44A than for the wild-type protein (Figure 4). From these results, we conclude that Soj polymerisation is strictly dependent on ATP and DNA.
Figure 4.

DNA-dependent pelleting of Soj. Samples were performed in duplicates for reliability. Wild-type Soj is found exclusively in the supernatant when centrifuged in the presence of DNA but without ATP. In the presence of ATP, a substantial fraction (one-third) of wild-type Soj is found in the pellet. Hydrolysis-deficient Soj D44A is also found in the pellet in the presence of ATP and DNA, but to a greater extent than wild-type Soj (experiments performed at above saturating protein concentrations: two Soj dimers: one 24 bp binding site), indicating that ATP hydrolysis results in dissociation from the DNA. These results indicate that ATP-dependent dimerisation is a prerequisite for DNA binding.
Soj assembles into nucleoprotein filaments dependent on ATP
Soj was examined for its ability to form filaments that could be visualised by electron miscroscopy in the presence and absence of nucleotide and in the presence and absence of DNA. Figure 5 shows nucleoprotein filaments formed by Soj: (A) no filaments formed in the absence of nucleotide (data shown for wild-type protein, although the same result was obtained with Soj D44A), (B) no filaments formed in the presence of ADP, (C) wild-type Soj forms nucleoprotein filaments in the presence of ATP and linearised pU0J DNA, (D) Soj D44A formed filaments indistinguishable from the wild-type protein, (E) Soj also forms filaments on relaxed, open-circle DNA, (F–H) High-magnification images of the nucleoprotein filament, indicating that they most likely have a regular, perhaps helical structure. Taken together with the results of the pelleting and gel shift assays, we have demonstrated that Soj forms nucleoprotein filaments in an ATP- and DNA-dependent manner. Soj G16V indicates that, critically, it is ATP-dependent dimerisation of Soj, which facilitates DNA binding (electron microscopy not performed with Soj G16V). Furthermore, the fluorescence anisotropy data indicate a cooperative mode of DNA binding, which is consistent with the formation of protein–protein contacts in the nucleoprotein filament.
Figure 5.

Electron micrographs of Soj nucleoprotein filaments. (A) Wild-type Soj+plasmid DNA (pU0J, a 2938 bp pUC19 derivative) in the absence of nucleotide. (B) Wild-type Soj+pU0J+ADP. (C) Wild-type Soj+linearised pU0J+ATP produces nucleoprotein filaments which are indistinguishable from those produced by (D) Soj D44A under the same conditions. (E) Nucleoprotein filaments formed by Soj D44A in the presence of relaxed, open circle pU0J+ATP. (F–H) High-magnification images of single nucleoprotein filaments, indicating a regular, perhaps helical arrangement.
Soj is a P-loop, deviant Walker A ATPase
We have solved the crystal structures of Soj in the empty (1.6 Å) and ADP-bound (2.1 Å) states (Figure 6A and B, Tables II and III). The core of Soj is a twisted arch of stacked β-strands surrounded by α-helices, with one antiparallel and seven parallel β-strands (Figure 6A). The α-helices of Soj are clustered on either side of the twisted arch of β-sheet. On the outside of the arch are helices H3–H9 and within the centre of the arch are helices H1, H2, H10, H11 and H12 (Figure 6A).
Figure 6.
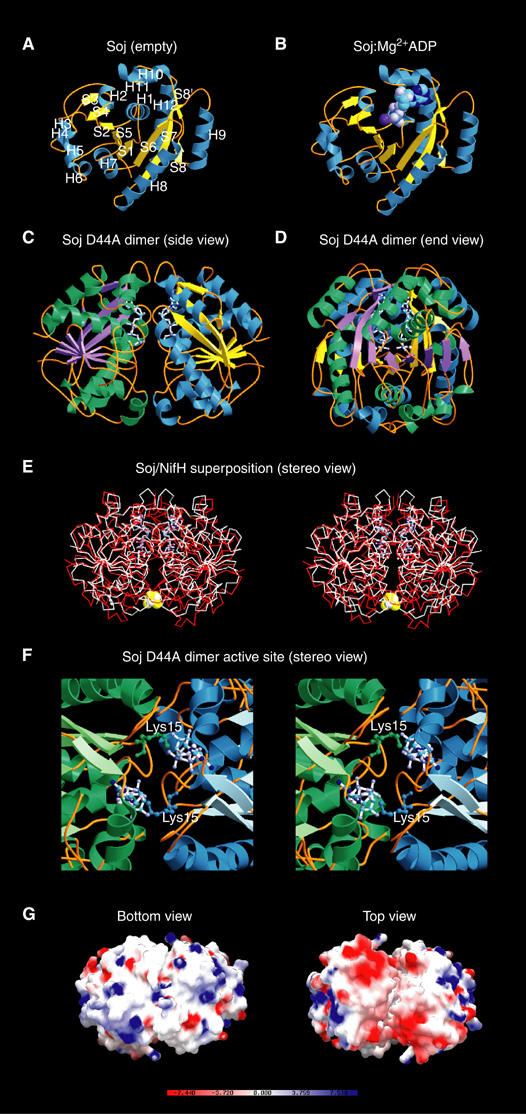
Crystal structures of Soj. (A) Crystal structure of Soj in the empty state at 1.6 Å. The arrangement of the sheet and helices follows that of the MinD family of ATPases (Cordell and Löwe, 2001; Hayashi et al, 2001; Sakai et al, 2001). (B) Structure of Soj:Mg2+ADP at 2.1 Å. Nucleotide binding is coupled to rearrangement of the P-loop. (C) Structure of hydrolysis-deficient Soj D44A in the dimeric state (side view), indicating the symmetrical assembly of the two monomers and the close proximity of the two nucleotides. (D) End view of Soj D44A dimer. (E) Structure of Soj D44A superimposed on the structure of nitrogenase iron protein from A. vinelandii (PDB ID: 1n2c) (Schindelin et al, 1997). The high structural homology between the two proteins indicates that the dimeric structure of Soj is correct. (F) Stereo view of the Soj D44A dimer active site. The nucleotide-binding surface of each monomer contributes to the formation of the active site chamber, which accommodates two molecules of ATP. Each monomer also contributes a universally conserved lysine (Lys15), which stabilises the negative charges on the opposing ATP. (G) Bottom and top views of the electrostatic surface potential maps of the Soj D44A dimer. The top view clearly indicates two patches of negative charge (one on each monomer) and, importantly, the existence of a cleft between the two monomers in which the surfaces are entirely complementary.
Table 2.
Crystallographic data
| Crystal | λ (Å) | Resol. (Å) | I/σIa | Rmb (%) | Multipl.c | Compl.d (%) |
|---|---|---|---|---|---|---|
| 1. Soj (apoprotein), space group P43212, a=b=61.35 Å, c=124.53 Å | ||||||
| KAu(CN)2 | 0.9202 | 1.8 | 14.3(3.8) | 0.094 | 7.0 | 99.8 |
| Na2WO4 | 0.9793 | 1.8 | 10.8(3.1) | 0.072 | 3.7 | 99.9 |
| NATI | 0.9393 | 1.6 | 14.0(4.3) | 0.096 | 7.0 | 99.8 |
| 2. Soj:Mg2+ADP, space group P43212, a=b=61.38 Å, c=126.66 Å | ||||||
| NATI | 1.4880 | 2.1 | 14.3(4.0) | 0.055 | 3.6 | 99.2 |
| 3. Soj D44A:Mg2+ATP, space group P212121, a=87.43 Å, b=95.91 Å, c=123.94 Å | ||||||
| NATI |
0.934 |
1.8 |
14.8(6.2) |
0.067 |
4.0 |
99.9 |
| aSignal-to-noise ratio for the highest resolution of intensities. | ||||||
| bRm: ∑h∑I∣I(h,I)−I(h)∣/∑h∑II(h,I) where I(h,I) are symmetry-related intensities and I(h) is the mean intensity of the reflection with unique index h. | ||||||
| cMultiplicity for unique reflections. | ||||||
| dCompleteness for unique reflections. | ||||||
Table 3.
Refinement statistics
| Soj (apoprotein) | Soj:Mg2+ADP | Soj D44A:Mg2+ATP | |
|---|---|---|---|
| Model | 5-247, 254 H2O | 5-247, ADP | Chains A-D, 5-247, |
| 4 ATP, 1279 H2O | |||
| Diffraction data | NATI, 1.6 Å | NATI, 2.1 Å | NATI, 1.8 Å |
| R-factor, R-freea | 0.229, 0.252 | 0.218, 0.266 | 0.1815, 0.2156 |
| B average/bondedb | 29.66 Å2/2.70 Å2 | 50.01/0.75 Å2 | 20.29 Å2/2.46 Å2 |
| Geometry bonds/anglesc | 0.006 Å/1.37° | 0.009 Å/1.15° | 0.005 Å/1.29° |
| Ramachandrand | 92.1%/0.0% | 92.7%/0.0% | 92.0%/0.0% |
| PDB IDe | 1wcv | 2bej | 2bek |
| aIn all, 5% of reflections were randomly selected for determination of the free R-factor, prior to any refinement. | |||
| bTemperature factors averaged for all atoms and r.m.s.d. of temperature factors between bonded atoms. | |||
| cR.m.s.d. from ideal geometry for bond lengths and restraint angles (Engh, 1991). | |||
| dPercentage of residues in the ‘most favoured' region of the Ramachandran plot and percentage of outliers (PROCHECK; Laskowski et al, 1993). | |||
| eProtein Data Bank identifiers for co-ordinates. | |||
In the Soj apo-protein, the P-loop adopts an extended conformation between Q14 and V18, which partially occludes the active site. Upon ADP binding, the P-loop undergoes a conformational change, which involves tightening of the GGVG turn (not shown). ADP sits in a cavity at the molecular surface created by residues G17–T22, I206–A213, M178 and Y235 (Figure 6B). The side chain of M178 lies across the plane of the adenine base, with a hydrophobic stacking distance of 3.6 Å. Mutation of the corresponding residue in ParA of plasmid P1, M314I, gave a weak segregation defect of alternating segregation and mis-segregation in successive generations (Li et al, 2004), a defect most likely attributable to a reduced ATPase activity of the protein.
A DALI structural similarity search (Holm and Sander, 1995) finds many nucleotide-binding proteins which resemble Soj. The most similar are the MinD proteins from Archaeoglobus fulgidus (PDB entry 1HYQ) and Pyrococcus horikoshii (PDB entry 1ION), with Z-scores of 27.2 and 26.8, respectively. A. fulgidus MinD has a root-mean-square deviation (r.m.s.d.) of superimposed Cα of 2.4 Å over 232 equivalent residues, while P. horikoshii MinD has an r.m.s.d. of 2.2 Å over 243 equivalent residues. The nitrogenase iron protein from Clostridium pasteurianum (PDB entry 1CP2), dethiobiotin synthase (PDB entry 1BYI) and the arsenite-translocating ATPase fragment (PDB entry 1F48) gives Z-scores of 20.2, 12.3 and 11.5, respectively. All of these proteins are dimeric ATPases, except MinD, which has been observed as a monomeric ATPase in all three published crystal structures (Cordell and Löwe, 2001; Hayashi et al, 2001; Sakai et al, 2001). An explanation for these observations most likely lies in the fact that the two subunits of MinD which would constitute the dimeric ATPase are not covalently linked, relying on ATP binding for dimer formation, but dissociating upon ATP hydrolysis. In contrast, the subunits of both nitrogenase iron protein (NifH) and ArsA are covalently linked. NifH is a homodimer in which the two subunits are covalently linked by a 4Fe:4S cluster; formation of the ADP·AlF4− transition state results in rotation of the subunits towards their nucleotide-bound interfaces, generating a more compact dimer (Schindelin et al, 1997). ArsA is twice the sizes of NifH, MinD and Soj, but consists of two similar domains connected by a short linker such that each ArsA monomer is functionally a pseudodimer (Zhou et al, 2000).
Soj also bears significant resemblance to the bacterial GTPase, Ffh, a homologue of the eukaryotic signal recognition particle (SRP54) (Freymann et al, 1997). Indeed, the similarities extend as far as heterodimerisation of Ffh with the GTPase FtsY, the bacterial homologue of the α subunit of the SRP receptor (SR), constituting a nucleotide ‘sandwich' dimer (Egea et al, 2004; Focia et al, 2004). Interestingly, the similarity of Soj to Ffh provides a unique insight into the putative nature of the N-terminal domain of ParA family members, which is not found in members of the chromosomal (Soj) family. Based on a sequence alignment of plasmid ParA proteins and Ffh homologues, we propose that the N-terminal domain of ParA forms a four-helix bundle structurally homologous to that of the N domain of Ffh. A secondary structure prediction of plasmid P1 ParA supports this possibility, with secondary structure elements of Ffh N domain (Freymann et al, 1997) almost exactly overlapping predicted secondary structure elements of ParA. Given that the precise function of the N domain of the SRP is not known at this stage, it would be premature to speculate about the role of such a domain in ParA, although it has been demonstrated that the N-termini of P1 and P7 ParA proteins are required for autoregulation of transcription of the par operon (Hayes et al, 1994).
Hydrolysis-deficient Soj forms a nucleotide ‘sandwich' dimer
We have crystallised the hydrolysis-deficient mutant of Soj, Soj D44A, in the ATP-bound state (Tables II and III), showing conclusively for the first time the ability of this family of proteins to form a nucleotide ‘sandwich' dimer (Figure 6C–D) which is structurally homologous to the nitrogenase iron protein dimer (Schindelin et al, 1997) (Figure 6E). The dimer interface is made between the nucleotide-binding surfaces of each monomer and is stabilised by networks of H-bonds between residues on adjacent chains and water-mediated hydrogen bonds. The ‘signature' lysine, K15, of each chain stabilises the α and γ phosphates of the ATP moiety bound by the adjacent chain; the amino group is in 2.96 Å co-ordination distance to the α-phosphate and 2.76 Å from the γ-phosphate. The P-loops and switch II motifs of each chain are found very close to each other in the dimer structure (Figure 6F). In the region of the P-loops, the amide nitrogens of G16 and G17 from adjacent chains stabilise the γ-phosphate of one ATP moiety.
The dimeric structure of Soj complexed with ATP contains a cleft on both sides of the dimer, which extends part of the way down the dimer interface and in which the two surfaces are complementary in terms of both shape and charge distribution. The cleft itself is almost exclusively lined by hydrophobic residues contributed by each monomer (Figure 6G). A comparison of the active sites of Soj and NifH indicates that the nucleotides adopt different conformations. The two ATP molecules in the Soj active site chamber adopt a ‘kinked' conformation in which the γ-phosphate is accommodated without structural rearrangement of the switch II region and K15 of the opposing subunit stabilises the α- and β-phosphates. In the NifH active site, ADP adopts an extended conformation in which the conserved lysine (K10) of the opposing subunit stabilises the β-phosphate and the switch II region undergoes a conformational change, placing the P-loop G16 of each subunit ∼4 Å apart compared to ∼10 Å in the nucleotide-free protein. This results in a rotation of ∼13° of each monomer towards the subunit interface, closing the dimer into a more compact structure in the complex. AlF4− in the NifH dimer structure mimics the transition state of the hydrolysis reaction by adopting a planar conformation in which octahedral co-ordination is completed by the terminal oxygen of the β-phosphate of ADP and the attacking nucleophilic water, which is co-ordinated by the side-chain carboxyl oxygen of D39 (D44 in Soj). Based on the structure of NifH with molybdenum iron protein, we suggest that formation of the transition state closes the dimer interface such that Soj is competent for interaction with Spo0J.
The effects of several mutations of the Walker A box have been reported for B. subtilis Soj. Wild-type Soj oscillates within the nucleoid region of the cell over a time course of several minutes, but in a Δspo0J background, Soj remains statically associated with the nucleoid (Quisel et al, 1999). The mutation K16Q in B. subtilis Soj has been observed to abolish the in vivo oscillatory behaviour of Soj, its association with the nucleoid and its response to Spo0J, consistent with a lack of nucleotide binding (Quisel et al, 1999). This observation is supported by the equivalent mutation, K20A, in T. thermophilus Soj, which abolishes nucleotide binding. A second mutant, D125A, in B. subtilis Soj is also predicted not to bind nucleotide. A third mutation, G12V, however exhibits a different localisation pattern. Like wild-type Soj, Soj G12V localises to the cell poles in Spo0J+ cells, but, unlike wild-type Soj, it also localises to the poles in a spo0J null mutant (Quisel et al, 1999). Mutation of the equivalent residue, G16V, in the structure of dimeric T. thermophilus Soj indicates a steric clash between V16 of each monomer and P121 of the switch II region, V16 of the adjacent monomer and the γ-phosphate of ATP, which would prevent dimerisation of Soj. Soj G16V sediments as a monomer in the presence of ATP (Figure 2H), confirming that it is deficient in dimerisation, and from this we conclude that nucleoprotein filament formation is dependent on ATP-mediated dimerisation.
Soj ATPase is activated by the N-terminal 20 amino acids of Spo0J
The ATPase activity of wild-type Soj was assayed using the malachite green method of Chan et al (1986). Wild-type Soj displays low basal ATPase activity (Figure 7A). Soj K20A displays no detectable ATPase activity, a consequence of its failure to bind nucleotide. We found that wild-type Soj can be moderately stimulated by Spo0J, yielding an increase in phosphate production by a factor of three (Figure 7A). The activation of Soj by Spo0J is enhanced in the presence of both 19 bp duplex parS DNA and pU0J plasmid DNA, both of which result in an increase in ATPase activity of almost an order of magnitude over the basal activity (Figure 7A). The presence of double-stranded DNA in the absence of Spo0J fails to elicit an increase in the hydrolysis rate, indicating that the DNA-bound form of Spo0J is a more effective activator than unbound Spo0J and that DNA binding has no effect on Soj activity. Solution studies of Spo0J place the primary dimerisation determinant in the C-terminal 60 amino acids of the protein, but biochemical and structural studies have also shown that the N-terminal and central DNA-binding domains dimerise, a requirement for HTH-mediated DNA binding (Leonard et al, 2004). We hypothesise that Spo0J undergoes DNA-dependent dimerisation of its N-termini and that this is coupled to activation of ATP hydrolysis by Soj.
Figure 7.
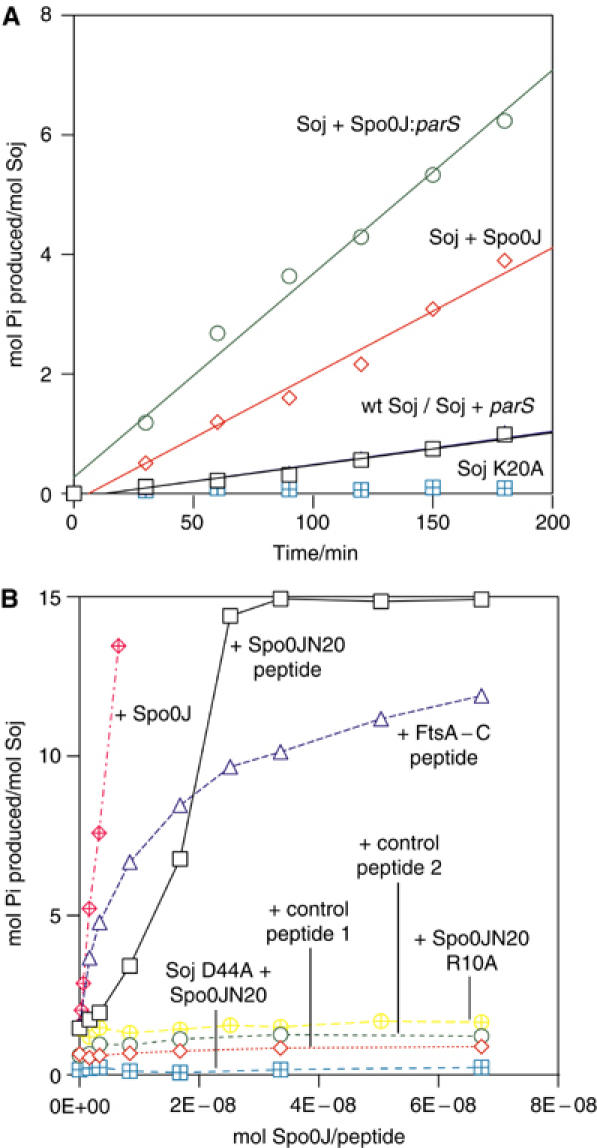
(A) Time course activation of Soj ATPase activity. Wild-type Soj displays low basal ATPase activity (black squares). Soj is stimulated approximately three-fold by Spo0J (red diamonds) and by almost an order of magnitude by Spo0J in the presence of parS DNA (green circles). Soj K20A is devoid of ATPase activity. (B) Spo0J activation of Soj ATPase activity. Spo0J strongly stimulates Soj ATPase activity at nanomolar concentrations (red hatched diamonds). Soj can also be stimulated by Spo0JN20, a 20 amino-acid peptide from the extreme N-terminus of Spo0J (black squares), although the peptide exhibits only a modest 8% of the activation stimulated by an equimolar amount of full-length Spo0J. Soj is not stimulated by either of the control peptides (red diamonds and green circles) and hydrolysis-deficient Soj D44A is not stimulated by Spo0JN20 (blue hatched squares). The Spo0JN20 R10A peptide, which has the putative catalytic arginine mutated to alanine, fails to stimulate Soj (yellow hatched diamonds), indicating that this residue is critical for activation of ATP hydrolysis. Interestingly, a 19 amino-acid peptide representing the conserved extreme C-terminus of FtsA also strongly stimulates Soj, but to a lesser extent than Spo0JN20. The kinetics of activation by Spo0JN20 and full-length Spo0J indicate possible cooperativity of binding and activation, consistent with dimeric Spo0J binding dimeric Soj.
Biochemical studies of P1 ParB, C. crescentus ParB and F plasmid SopB have mapped the ParA/SopA interaction determinant to the extreme N-terminus of the protein (Surtees and Funnell, 1999; Figge et al, 2003; Ravin et al, 2003). Alignments of putative Spo0J proteins indicated to us that the region responsible for Soj activation lay in the first 20 amino acids, given the high conservation observed. This region of seemingly flexible nature is not visible in the structure of Spo0J (Leonard et al, 2004). We found that the hydrolysis rate of Soj could be strongly and specifically stimulated by this N-terminal 20 amino-acid peptide, Spo0JN20 (Figure 7B). The peptide did not activate Soj to the same extent as Spo0J, exhibiting 8% activation compared with full-length Spo0J at equimolar concentrations (Figure 7B). This is in agreement with our observation that the context of the N-termini is important for activation. Additional protein–protein contacts may be made in the case of full-length Spo0J, thereby increasing affinity for the binding site, and there is likely an entropic effect of both ligands being supplied by a dimeric Spo0J molecule rather than a monomeric ligand binding to two sites. Soj was not activated by two control peptides (Figure 7B). Interestingly, the peptide CASVGSWIKRLNSWLRKEF exhibited moderate stimulation of Soj (75% when compared with TTJN20) (Figure 7B). This 19 amino-acid peptide represents the extreme C-terminus of E. coli FtsA, conservation of which was first recognised by Löwe and van den Ent (Löwe and van den Ent, 2001), and which is also disordered in the crystal structure (van den Ent and Lowe, 2000). Alignments of the extreme C-terminus of FtsA and the extreme N-termini of both Spo0J and MinE indicate weak but recognisable homology, and the possible conservation of a putative catalytic basic residue (Figure 8). Indeed, mutation of this residue (R10) to alanine in the Spo0JN20 peptide results in a complete abrogation of the activating property of the peptide (Figure 7B). Deletion mutants of FtsA lacking five or more residues from the C-terminus are biologically inactive in E. coli, generating filamentous cells which fail to form septal rings (Yim et al, 2000).
Figure 8.
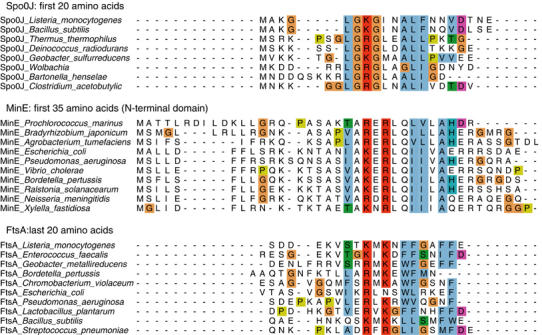
Sequence alignment of putative activating peptides of Spo0J, MinE and FtsA. Alignment of the N-terminal regions of Spo0J and MinE proteins with the C-terminus of FtsA proteins reveals a remarkable sequence homology, which includes a universally conserved basic (lys/arg) residue (arginine 10 in T. thermophilus Spo0J). The functional homology of these putative activating peptides is further strengthened by the observations that E. coli FtsA C-terminus can stimulate Soj and mutation of the arginine 10 to alanine in the Spo0JN20 peptide abrogates stimulation of ATP hydrolysis (Figure 7).
Overexpression of MinE allows division site assembly at the poles, giving rise to a mini-cell phenotype. Specifically, the N-terminal 1–22 amino acids of MinE are responsible for counteracting the effect of the division inhibitor MinCD and suppressing the filamentous phenotype observed when MinCD is induced in the absence of wild-type MinE (Zhao et al, 1995). Furthermore, the N-terminus of MinE has also been shown to contain the interaction determinant for MinD ATPase (Ma et al, 2003).
We speculate that Spo0J, FtsA and MinE may share a conserved mechanism of nucleotide hydrolysis activation of their respective interaction partners. There are two possibilities for the mechanism of activation: the first involves activation of ATP hydrolysis, the result of which is destabilisation of the dimer and a shift of the equilibrium towards the monomeric form. The second mechanism involves nucleotide exchange of the monomer following ATP hydrolysis, the result of which is a shift in the equilibrium to the ATP-bound, dimeric state. We find the mechanism of activation rather than nucleotide exchange more attractive because it involves interaction of a dimeric activator (Spo0J, MinE) containing two ligands with dimeric ATPase (Soj, MinD respectively) containing two binding sites, and couples dimerisation of the ATPase to protein:protein complex assembly, as is observed for the interaction of NifH with molybdenum iron protein (Schindelin et al, 1997).
The structure of the nitrogenase iron protein in complex with molybdenum iron protein shows that the ATP-bound state, and hence ‘closed' conformation of the dimer, is necessary for productive interaction with its binding partner (Schindelin et al, 1997). We hypothesise that the ATP-dependent dimerisation of Soj acts as an identical molecular switch which regulates its putative interaction with Spo0J, and, by extension, that dimerisation of MinD regulates its interaction with MinC/MinE. We predict that the identification of a hydrolysis-deficient mutant which is constitutively dimeric in the presence of ATP will be a useful tool in the characterisation of physiologically relevant protein:protein complexes and their intracellular functions. ATPases of this deviant Walker A family play fundamental roles in a diverse range of cellular processes, from nitrogen fixation and anion extrusion to bacterial division site selection and plasmid and chromosome segregation.
Finally, the similarities between MinDE and Soj/Spo0J are wide-ranging. MinD and Soj oscillate (jump) between places in the cell: MinD oscillates by binding to the membrane, while Soj oscillates by binding to the nucleoid. MinD and Soj have the same three-dimensional structure apart from the amphiphatic helix on MinD that is involved in membrane binding, and most likely form the same ATP-dependent dimer. MinD and Soj ATPase activity is activated by a short, disordered peptide located on a dimeric binding partner (MinE N-terminus, Spo0J N-terminus, respectively) and both MinD and Soj bind to extended surfaces (MinD: membrane, Soj: DNA) and binding is regulated by dimerisation. Both the MinDE and the Soj/Spo0J system are involved in accurate positioning of molecules (MinC and region of the nucleoid, respectively). The same oscillatory mechanisms involving surface-assisted polymerisation may be used to position the septum and the origins of replication.
Materials and methods
Protein expression and purification
Soj from T. thermophilus HB27 (ATCC BAA-163D) was cloned into pHis17 to generate a C-terminally hexa-histidine-tagged protein. Vectors expressing the mutant proteins Soj G16V, K20A and D44A were constructed using the QuikChange protocol (Stratagene). Single colonies of C41 (DE3) were used to inoculate 2 × 65 ml cultures of 2 × TY, 0.4% glucose and 100 μg/ml ampicillin, and grown overnight at 37°C. The cultures were used to inoculate 12 l of 2 × TY+0.4% glucose, which was induced with 1 mM IPTG when OD600=0.6 and the temperature reduced to 25°C. The cells were grown overnight. Soj proteins were purifed by NiNTA followed by heparin affinity chromatography and gel filtration on a Sephacryl S200 column (Amersham Biosciences), equilibrated in TEN+100 mM NaCl, pH 8.5. The wild-type and mutant proteins eluted as a single peak at a position, indicating them to be monomeric.
Crystallisation and data collection
1152 crystallisation conditions were screened using in-house nanolitre crystallisation robotics (Stock et al, 2004). Crystals of the native protein were grown using the sitting drop vapour diffusion technique using 6% PEG 3350, 0.15 M NaCl and 0.4 M KI as the crystallisation solution. Drops composed of 1 μl protein at 2 mg/ml and 1 μl crystallisation solution were incubated overnight at 19°C. Crystals grew in space group P43212 with cell dimensions a=b=61.35 Å and c=124.53 Å, and were frozen in mother liquor plus 25% glycerol. Heavy metal derivatives were made by adding KAu(CN)2 or Na2WO4 solutions to the drop to a final concentration of 4 mM. The drops were incubated overnight at 19°C and the crystals flash frozen as for the natives. The native and derivative data sets were collected at ID29 ESRF, Grenoble, France.
Co-crystals of Soj:ADP were obtained by adding 1 mM ATPγS and 2 mM MgSO4 to the protein solution prior to screening of crystallisation conditions. Crystals containing ADP (ATPγS apparently partly hydrolysed) grew in 8% PEG 550-MME+8% PEG 20 K, 0.1 M sodium acetate, pH 5.5 and 0.2 M KSCN. The crystals were frozen under the same cryoprotectant as the natives and also belong to space group P43212 with unit cell dimensions a=b=61.38 Å and c=126.66 Å. A native data set to 2.1 Å resolution was collected on beam line 14-2 at the SRS facility, Daresbury Laboratory, UK.
Crystals of the Soj D44A dimer were grown by adding 250 mM CHES, pH 10.0, 1 mM ATP and 5 mM MgSO4 to the protein solution and concentrating it from 2 to 8 mg/ml prior to screening of crystallisation conditions. Crystals were obtained in 200 mM imidazole, pH 7.6, and 10–20% isopropanol. Crystals grew in space group P212121 with unit cell dimensions a=87.43 Å, b=95.91 Å and c=123.94 Å. A native data set to 1.8 Å resolution was collected on beam line ID14-1 at the ESRF, Grenoble, France.
All crystals were indexed and integrated using the MOSFLM package and further processed using the CCP4 package. The structure of Soj apoprotein was solved by mulitple isomorphous replacement (MIR), while Soj:ADP and the Soj D44A dimer were solved by molecular replacement. REFMAC (Murshudov et al, 1999) was used for TLS refinement of Soj:ADP.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed in a Beckman Optima XL-A analytical ultracentrifuge with an An60-Ti rotor, with the samples in various buffers as described in the text.
Sedimentation velocity was at 60 000 or 50 000 rev min−1, 5.0°C, with scans of the single cell taken at 0-min intervals (to obtain scans as closely spaced as possible; in practice about 1.5 min apart). Adjacent sets of data were analysed by the method of Stafford (1994, 1997) using the program DCDT+ (Philo, 2000).
Size exclusion chromatography
Size exclusion chromatography was performed on a calibrated Superdex 200 3.2. Precision Column (Amersham Biosciences). Samples were applied in a volume of 10 μl at 2 mg/ml. The column was equilibrated in 50 mM CHES, 100 mM NaCl, 5 mM MgSO4±0.5 mM ATP, pH 10.0 (the high pH was required to prevent precipitation of the protein upon the addition of ATP; we checked that the protein eluted as a single peak consistent with a monomer in the absence of ATP at the same elution volume as in 50 mM Tris–HCl, pH 8.5).
DNA-binding assays
The ability of wild-type Soj, Soj D44A, Soj K20A and Soj G16V to bind to double-stranded, plasmid DNA was assayed in the absence of nucleotide and in the presence of Mg2+ADP or Mg2+ATP. Binding reactions were performed in a volume of 10 μl in 50 mM Tris–HCl, pH 8.5, 5 mM MgSO4. Each reaction contained 200 fmol of pUC19 and 0–100 pmol Soj±0.75 mM ADP or ATP. Reactions were incubated for 10 min at 25°C, mixed with gel loading buffer, run on a 1% agarose gel in 0.5 × TB+1 mM MgSO4 buffer and stained with ethidium bromide.
Fluorescence anisotropy
Fluorescence anisotropy measurements were collected using a Perkin-Elmer LS50B luminescence spectrometer. A 5′-fluoresceinated oligonucleotide (5′-AAAACAAACCCAAAACAAACCC-3′) was used as a fluorescein-labelled single-stranded DNA and annealed to its complementary, unlabelled oligonucleotide to create fluorescein-labelled double-stranded DNA. The binding buffer was 20 mM Tris (pH 8.5), 100 mM NaCl, 5 mM MgSO4, 1 mM ADP or ATP. Wild-type Soj was serially titrated into the cuvette, which contained 10 nM 5′-fluoresceinated DNA. The measurements were performed at 298 K. The data were plotted and the curves fitted using the program GraFit.
Pelleting assays
Wild-type Soj and Soj D44A were pelleted in the presence and absence of ATP and in the presence and absence of pU0J DNA. Reactions were performed in a volume of 30 μl. Each reaction contained 750 pmol wild-type Soj/Soj D44A, 50 mM Tris–HCl, pH 8.5, 5 mM MgSO4±1 mM ATP. For pelleting in the presence of pU0J DNA, reactions contained 1.5 pmol of pU0J. Samples were centrifuged at 100 000 r.p.m. for 1 h at ambient temperature. The supernatants were carefully removed and mixed with an equal volume of SDS gel loading buffer. The pellets were washed with 30 μl buffer and then solubilised in 30 μl SDS gel loading buffer. An equal volume of buffer was then added to normalise the concentrations of components in the supernatant and pellet. A volume of 30 μl of each sample was then run on a 12.5% denaturing polyacrylamide gel.
Electron microscopy
Supercoiled or XmnI digested pU0J DNA (40–400 ng) was incubated with Soj, Soj K20A or Soj D44A (0–5 μg) in 50 mM Tris–HCl, pH 8.5, 5 mM MgSO4±0.75 mM ATP. The complexes were incubated at 25°C for 10 min, after which they were applied to glow discharged carbon-coated grids for 30 s. The grids were washed with one drop of distilled water, and stained with three drops of 2% uranyl acetate before being blotted to dryness. Images were taken on a Philip EM208 electron microscope at × 50 000 magnification.
ATPase activity assays
The ATPase activity of Soj was assayed by the spectrophotometric detection of inorganic phosphate following termination of the reaction with an acidic solution containing malachite green reagent (1:1:2:2 ratio of 5.72%. ammonium molybdate, (w/v) in 6 N HCl, 2.32% (w/v) polyvinyl alcohol (Sigma), 0.08712% (w/v) malachite green (Sigma) and distilled water, respectively). Reactions were performed in a volume of 50 μl containing 50 mM HEPES, pH 8.0 at 37°C for 3.5 h. Each reaction contained 1.82 nmol Soj and 50 nmol ATP. Spo0J activation was assayed by the addition of 0–2 nmol T. thermophilus Spo0J to the reaction. Activation by Spo0J was assayed in the presence of an equimolar amount of parS duplex and in the presence of the 2.9 kbp plasmid pUOJ (Leonard et al, 2004), such that there was an equimolar amount of 24 bp binding sites. The ability of the N-terminal 20 amino acids of Spo0J to activate Soj was assayed by incubation of the ATPase with increasing concentrations of the 20 amino-acid peptide MSRKPSGLGRGLEALLPKTG (Spo0JN20) and the mutant peptide MSRKPSGLGAGLEALLPKTG (Spo0JN20R10A). Control reactions contained the peptides PEGDIPAIYR and ILFPEGDIPAIYRYGL. The sequence of the E. coli FtsA C-terminal peptide (FtsA-C) was CASVGSWIKRLNSWLRKEF. Reactions were terminated by the addition of 200 μl malachite green reagent. The colour was allowed to stabilise for 5 min before the absorbance was measured at 630 nm. A calibration curve was constructed using 0–13 nmol inorganic phosphate standards and samples were normalised for acid hydrolysis of ATP by the malachite green reagent.
Acknowledgments
We thank the staff at ID14-1 and ID29 of ESRF (Grenoble, France) and the staff of ID14-1 of Daresbury Synchrotron Radiation Source (UK) for assistance with data collection. We thank Henrique Ferreira and Jeff Errington (Sir William Dunn School of Pathology, Oxford, UK) for providing us with plasmid pU0J.
References
- Cervin MA, Spiegelman GB, Raether B, Ohlsen K, Perego M, Hoch JA (1998) A negative regulator linking chromosome segregation to developmental transcription in Bacillus subtilis. Mol Microbiol 29: 85–95 [DOI] [PubMed] [Google Scholar]
- Chan KM, Delfert D, Junger KD (1986) A direct colorimetric assay for Ca2+-stimulated ATPase activity. Anal Biochem 157: 375–380 [DOI] [PubMed] [Google Scholar]
- Cordell SC, Löwe J (2001) Crystal structure of the bacterial cell division regulator MinD. FEBS Lett 492: 160–165 [DOI] [PubMed] [Google Scholar]
- Draper GC, Gober JW (2002) Bacterial chromosome segregation. Annu Rev Microbiol 56: 567–597 [DOI] [PubMed] [Google Scholar]
- Easter J Jr, Gober JW (2002) ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol Cell 10: 427–434 [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Gerdes K (2001) The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc Natl Acad Sci USA 98: 15078–15083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM (2004) Substrate twinning activates the signal recognition particle and its receptor. Nature 427: 215–221 [DOI] [PubMed] [Google Scholar]
- Engh RA (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr Sect A 47: 392–400 [Google Scholar]
- Figge RM, Easter J, Gober JW (2003) Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol Microbiol 47: 1225–1237 [DOI] [PubMed] [Google Scholar]
- Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM (2004) Heterodimeric GTPase core of the SRP targeting complex. Science 303: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann DM, Keenan RJ, Stroud RM, Walter P (1997) Structure of the conserved GTPase domain of the signal recognition particle. Nature 385: 361–364 [DOI] [PubMed] [Google Scholar]
- Glaser P, Sharpe ME, Raether B, Perego M, Ohlsen K, Errington J (1997) Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev 11: 1160–1168 [DOI] [PubMed] [Google Scholar]
- Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A (1997) Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90: 1113–1121 [DOI] [PubMed] [Google Scholar]
- Gordon GS, Wright A (2000) DNA segregation in bacteria. Annu Rev Microbiol 54: 681–708 [DOI] [PubMed] [Google Scholar]
- Hayashi I, Oyama T, Morikawa K (2001) Structural and functional studies of MinD ATPase: implications for the molecular recognition of the bacterial cell division apparatus. EMBO J 20: 1819–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F, Radnedge L, Davis MA, Austin SJ (1994) The homologous operons for P1 and P7 plasmid partition are autoregulated from dissimilar operator sites. Mol Microbiol 11: 249–260 [DOI] [PubMed] [Google Scholar]
- Hiraga S (1992) Chromosome and plasmid partition in Escherichia coli. Annu Rev Biochem 61: 283–306 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1995) Dali: a network tool for protein structure comparison. Trends Biochem Sci 20: 478–480 [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J (2001) Topological regulation of cell division in E. coli. Spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell 7: 1337–1343 [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J (2003) A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Mol Microbiol 47: 345–355 [DOI] [PubMed] [Google Scholar]
- Hu Z, Saez C, Lutkenhaus J (2003) Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J Bacteriol 185: 196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Gunther NWt, Grossman AD (1994) spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol 176: 5320–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare D, Ziegelin G, Lanka E, Heinemann U (2004) Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat Struct Mol Biol 11: 656–663 [DOI] [PubMed] [Google Scholar]
- Koonin EV (1993) A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol 229: 1165–1174 [DOI] [PubMed] [Google Scholar]
- Lackner LL, Raskin DM, de Boer PA (2003) ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J Bacteriol 185: 735–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Moss DS, Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231: 1049–1067 [DOI] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Lowe J (2004) Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol Microbiol 53: 419–432 [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Errington J (1997) Direct evidence for active segregation of oriC regions of the Bacillus subtilis chromosome and co-localization with the SpoOJ partitioning protein. Mol Microbiol 25: 945–954 [DOI] [PubMed] [Google Scholar]
- Lin DC, Grossman AD (1998) Identification and characterization of a bacterial chromosome partitioning site. Cell 92: 675–685 [DOI] [PubMed] [Google Scholar]
- Lin DC, Levin PA, Grossman AD (1997) Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci USA 94: 4721–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dabrazhynetskaya A, Youngren B, Austin S (2004) The role of Par proteins in the active segregation of the P1 plasmid. Mol Microbiol 53: 93–102 [DOI] [PubMed] [Google Scholar]
- Löwe J, van den Ent F (2001) Conserved sequence motif at the C-terminus of the bacterial cell-division protein FtsA. Biochimie 83: 117–120 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J, Sundaramoorthy M (2003) MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol 48: 295–303 [DOI] [PubMed] [Google Scholar]
- Ma LY, King G, Rothfield L (2003) Mapping the MinE site involved in interaction with the MinD division site selection protein of Escherichia coli. J Bacteriol 185: 4948–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Errington J (1999) Dynamic movement of the ParA-like Soj protein of B. subtilis and its dual role in nucleoid organization and developmental regulation. Mol Cell 4: 673–682 [DOI] [PubMed] [Google Scholar]
- Mohl DA, Gober JW (1997) Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88: 675–684 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr 55 (Part 1): 247–255 [DOI] [PubMed] [Google Scholar]
- Niki H, Hiraga S (1997) Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90: 951–957 [DOI] [PubMed] [Google Scholar]
- Nordstrom K, Austin SJ (1989) Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet 23: 37–69 [DOI] [PubMed] [Google Scholar]
- Philo JS (2000) A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem 279: 151–163 [DOI] [PubMed] [Google Scholar]
- Quisel JD, Grossman AD (2000) Control of sporulation gene expression in Bacillus subtilis by the chromosome partitioning proteins Soj (ParA) and Spo0J (ParB). J Bacteriol 182: 3446–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel JD, Lin DC, Grossman AD (1999) Control of development by altered localization of a transcription factor in B. subtilis. Mol Cell 4: 665–672 [DOI] [PubMed] [Google Scholar]
- Radnedge L, Youngren B, Davis M, Austin S (1998) Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J 17: 6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin NV, Rech J, Lane D (2003) Mapping of functional domains in F plasmid partition proteins reveals a bipartite SopB-recognition domain in SopA. J Mol Biol 329: 875–889 [DOI] [PubMed] [Google Scholar]
- Sakai N, Yao M, Itou H, Watanabe N, Yumoto F, Tanokura M, Tanaka I (2001) The three-dimensional structure of septum site-determining protein MinD from Pyrococcus horikoshii OT3 in complex with Mg-ADP. Structure (Camb) 9: 817–826 [DOI] [PubMed] [Google Scholar]
- Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC (1997) Structure of ADP × AIF4(−)-stabilized nitrogenase complex and its implications for signal transduction. Nature 387: 370–376 [DOI] [PubMed] [Google Scholar]
- Sharpe ME, Errington J (1996) The Bacillus subtilis soj-spo0J locus is required for a centromere-like function involved in prespore chromosome partitioning. Mol Microbiol 21: 501–509 [DOI] [PubMed] [Google Scholar]
- Shih YL, Le T, Rothfield L (2003) Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA 100: 7865–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford WF (1997) Sedimentation velocity spins a new weave for an old fabric. Curr Opin Biotechnol 8: 14–24 [DOI] [PubMed] [Google Scholar]
- Stafford WF III (1994) Boundary analysis in sedimentation velocity experiments. Methods Enzymol 240: 478–501 [DOI] [PubMed] [Google Scholar]
- Stock D, Perisic O, Lowe L (2004) Robotic nanolitre protein crystallisation at the MRC Laboratory of Molecular Biology. Prog Biophys Mol Biol (In press). doi:10.1016/j.pbiomolbio.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Surtees JA, Funnell BE (1999) P1 ParB domain structure includes two independent multimerization domains. J Bacteriol 181: 5898–5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Rothfield LI, King GF (2002) Membrane localization of MinD is mediated by a C-terminal motif that is conserved across eubacteria, archaea, and chloroplasts. Proc Natl Acad Sci USA 99: 15693–15698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent F, Lowe J (2000) Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J 19: 5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L (1997) Bacterial chromosome segregation: is there a mitotic apparatus? Cell 88: 577–579 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (2002) A large dispersed chromosomal region required for chromosome segregation in sporulating cells of Bacillus subtilis. EMBO J 21: 4001–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim L, Vandenbussche G, Mingorance J, Rueda S, Casanova M, Ruysschaert JM, Vicente M (2000) Role of the carboxy terminus of Escherichia coli FtsA in self-interaction and cell division. J Bacteriol 182: 6366–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CR, de Boer PA, Rothfield LI (1995) Proper placement of the Escherichia coli division site requires two functions that are associated with different domains of the MinE protein. Proc Natl Acad Sci USA 92: 4313–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Radaev S, Rosen BP, Gatti DL (2000) Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J 19: 4838–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]


