Abstract
The underlying mechanism of Fusobacterium nucleatum (Fn) in the carcinogenesis of colorectal cancer (CRC) is poorly understood. Here, we examined Fn abundance in CRC tissues, as well as β-catenin, TLR4 and PAK1 protein abundance in Fn positive and Fn negative CRCs. Furthermore, we isolated a strain of Fn (F01) from a CRC tissue and examined whether Fn (F01) infection of colon cancer cells activated β-catenin signaling via the TLR4/P-PAK1/P-β-catenin S675 cascade. Invasive Fn was abundant in 62.2% of CRC tissues. TLR4, PAK1 and nuclear β-catenin proteins were more abundant within Fn-positive over Fn-negative CRCs (P < 0.05). Fn and its lipopolysaccharide induced a significant increase in TLR4/P-PAK1/P-β-catenin S675/C-myc/CyclinD1 protein abundance, as well as in the nuclear translocation of β-catenin. Furthermore, inhibition of TLR4 or PAK1 prior to challenge with Fn significantly decreased protein abundance of P-β-catenin S675, C-myc and Cyclin D1, as well as nuclear β-catenin accumulation. Inhibition of TLR4 significantly decreased P-PAK1 protein abundance, and for the first time, we observed an interaction between TLR4 and P-PAK1 using immunoprecipitation. Our data suggest that invasive Fn activates β-catenin signaling via a TLR4/P-PAK1/P-β-catenin S675 cascade in CRC. Furthermore, TLR4 and PAK1 could be potential pharmaceutical targets for the treatment of Fn-related CRCs.
Keywords: Fusobacterium nucleatum, colorectal cancer, β-catenin signaling, toll-like receptor 4, p21-activated kinase 1
INTRODUCTION
A growing body of evidence suggests a complex link between gut microbiota, immunity, and colorectal carcinogenesis [1]. Recent studies show that a high amount of tissue-associated Fusobacterium nucleatum (Fn) has been connected to an advanced disease stage and poor clinical outcome in colorectal cancer (CRC) [1, 2]. However, the underlying mechanisms associated with this pathogenesis remain undetermined.
Recent studies reveal that Fn, a Gram-negative anaerobe, is associated with host immunity in colon carcinogenesis [3, 4]. It has been shown that Fn can stimulate the host immune response via toll-like receptor 4 (TLR4), which is a Gram-negative lipopolysaccharide (LPS) receptor [5].
It has been reported that high protein abundance of TLR4 in mouse intestinal epithelium induces colitis-associated tumor development [6], whereas TLR4 knock-out mice are protected against this disease [7]. In humans, high protein abundance of TLR4 is associated with more advanced grades of colonic neoplasia and lower overall survival [8, 9]. However, little is known about the exact TLR4 signaling cascade that is activated by gut microbiota in the colon.
It is well known that Wnt signaling pathway activation and β-catenin nuclear accumulation can be observed in approximately 90% of CRCs [10]. Mutations of the adenomatous polyposis coli (APC) or β-catenin genes are the main cause of β-catenin signaling activation in the majority of CRCs [10]. It was recently suggested that, in addition to genetic mutations, other signaling pathways and microenvironmental factors (such as gut flora) may be required to stabilize nuclear β-catenin [11, 12].
In recent years, it has been reported that p21-activated kinase 1 (PAK1) is associated with colon cancer progression and metastasis [13, 14]. PAK1 directly phosphorylates β-catenin at the Ser675 site, leading to a more stable and transcriptionally active β-catenin [15]. Interestingly, a recent study suggests that TLR4 can cause intestinal neoplasia through activation of the β-catenin signaling pathway [16]. Based on these findings, we hypothesized that Fn activates β-catenin signaling through a TLR4/PAK1 cascade in CRCs. A better understanding of signaling pathways activated by Fn may offer new opportunities to target the microbiota for CRC prevention and therapy.
To test this hypothesis, we examined whether Fn infection is associated with β-catenin nuclear accumulation in human CRC tissues and cells, and whether β-catenin accumulation by Fn is induced via the TLR4/PAK1 cascade. To our knowledge, this is the first study to examine the TLR4/PAK1 cascade activated by Fn in human CRC.
RESULTS
Invasive Fn is abundant in CRC tissues using fluorescence in situ hybridization (FISH)
First, we investigated the bacterial community composition of 14 CRC tissues using 16S rRNA sequencing. Firmicutes, Proteobacteria, Bacteroidetes and Fusobacteria were the dominant phyla in most CRCs (Figure 1A). Hierarchically clustered heat map analysis of the highly represented bacterial taxa in the 14 CRCs is shown in Figure 1B. There was a great diversity in the bacterial composition among different CRC tissues. In a subset of CRCs, we detected a higher relative abundance of Fusobacterium, Parvimonas, Porphyromonas, and Gemella, which have been recently suggested as microbial pathogens potentially associated with CRC [17–19].
Figure 1. Biome analysis of human colorectal cancer samples.
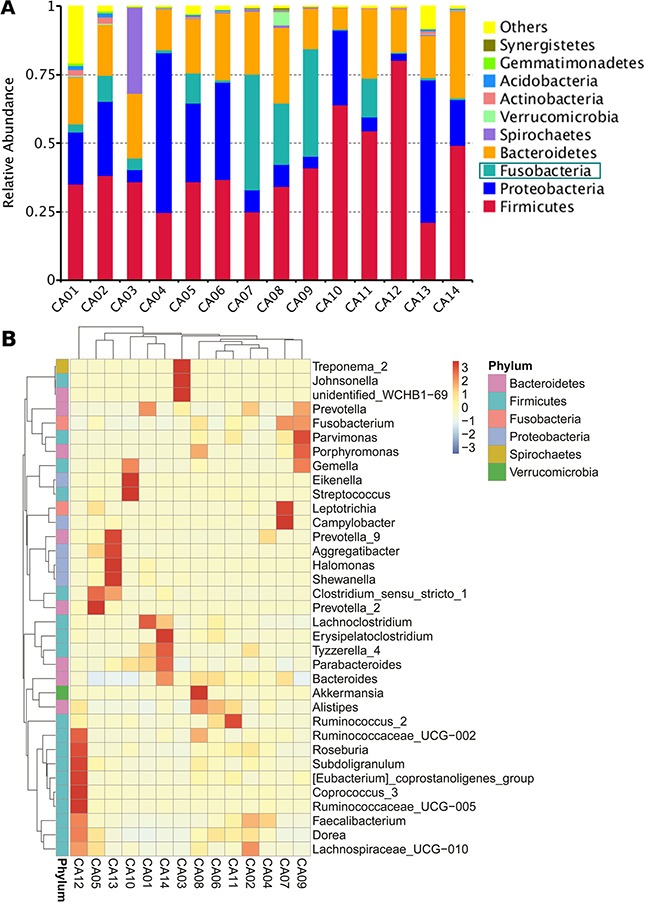
(A) Relative abundance of phyla present in colorectal cancer samples (n = 14). Data shown represent the most abundant phyla, whereas low abundant and unclassified OTUs are grouped as “Others.” (B) Hierarchically clustered heat map analysis of the highly represented bacterial taxa (genus level) in 14 colorectal cancer samples. The relative percentages of the bacterial families are indicated by varying color intensities.
We then examined the abundance of invasive Fn in 98 CRCs using FISH. Positive detection of invasive Fn occurred in 61/98 (62.2%) of the CRC tissues. The clinicopathologic features of Fn-negative vs. Fn-positive lesions are shown in Table 1. There were no significant differences between Fn-negative and Fn-positive CRCs with respect to patient gender, age, histologic grade and lymph node metastasis (P > 0.05). However, Fn was detected at a significantly higher frequency in proximal CRCs than in distal CRCs (P= 0.045).
Table 1. Clinicopathologic characteristics in Fn-negative vs. Fn positive colorectal cancers.
| Fn abundance | Negative(n=37) | Positive(n=61) | P Value |
|---|---|---|---|
| Gender | |||
| Female | 16 | 24 | 0.703 |
| Male | 21 | 37 | |
| Age | |||
| (year) | 59.4 | 58.0 | 0.281 |
| (range) | 36-81 | 40-74 | |
| Location | |||
| Proximal | 16 | 39 | 0.045 |
| Distal | 21 | 22 | |
| Histologic grade | |||
| Well differentiated | 10 | 12 | 0.519 |
| Moderatlely differentiated | 21 | 34 | |
| Poorly differentiated | 6 | 15 | |
| lymph node metastasis | 3 | 7 | 0.738 |
Fn, Fusobacterium nucleatum.
Invasive Fn is associated with activated β-catenin signaling in CRC tissues
We compared β-catenin, TLR4 and PAK1 protein abundance in 30 Fn-positive and 30 Fn-negative CRCs. The nuclear accumulation of β-catenin was more frequent in Fn-positive CRCs (P=0.028), suggesting that activated β-catenin signaling in these CRCs is associated with Fn infection. Similarly, TLR4 and PAK1 proteins were more abundant in Fn-positive CRCs when compared to Fn-negative CRCs (P< 0.05) (Table 2, Figure 2). These findings support the hypothesis that Fn contributes to the carcinogenesis of CRCs by activating β-catenin signaling through the TLR4/PAK1 cascade.
Table 2. Beta-catenin, TLR4 and PAK1 expressions in Fn-positive vs. Fn-negative colorectal cancers.
| Fn-negative (n=30) | Fn-positive (n=30) | P-value | |
|---|---|---|---|
| Beta-catenin | |||
| Plasm accumulation | 25 (83.3) | 28 (93.3) | 0.424 |
| Nuclear accumulation | 3 (10.0) | 10 (33.3) | 0.028 |
| TLR4 | |||
| Positive | 10 (33.3) | 25 (83.3) | 0.000 |
| Strong positive | 4 (13.3) | 14 (46.7) | 0.005 |
| PAK1 | |||
| Positive | 23 (76.7) | 30 (100) | 0.011 |
| Strong positive | 7 (23.3) | 13 (43.3) | 0.100 |
Fn, Fusobacterium nucleatum.
Figure 2. Invasive Fn in CRC tissues is associated with an activated β-catenin signaling pathway and TLR4/PAK1 protein abundance.
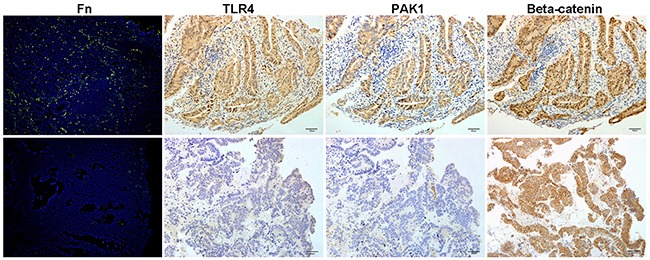
The upper panel shows that abundant invasive Fn in CRC tissue is associated with a high abundance of TLR4 and PAK1 and with β-catenin nuclear accumulation. The lower panel demonstrates that both TLR4 and PAK1 are absent in a Fn-negative CRC, and that cytoplasmic but not nuclear accumulation of β-catenin was seen in the tissue. Invasive Fn was detected by fluorescence in situ hybridization. 200× magnification.
Fn activates β-catenin signaling in colon cancer cells
We directly cultured Fn anaerobically from frozen tumor sections of a right-sided colon cancer and obtained a single isolate Fn (F01) (Supplementary Figure 1). The Fn (F01) sequence has been submitted to GenBank (accession number: SUB1766768 Seq01 KX692281). SW480 and Caco-2 cells were challenged with Fn (F01) to investigate the direct influence of invasive Fn on colon cancer cells. Fn (ATCC10953) from the oral cavity and nonpathogenic E. coli were used as controls. High protein abundance of β-catenin (total β-catenin and P-β-catenin S675) and its target genes, C-myc and Cyclin D1, were detected in SW480 cells (Figure 3A–3E, Supplementary Figure 2A-2C) and Caco-2 cells (Supplementary Figure 3) infected with Fn (F01). Fn (F01) challenge in SW480 cells stimulated the nuclear translocation of both total β-catenin and β-catenin S675 (Figure 4). Fn (ATCC10953) also stimulated the protein abundance of β-catenin S675 and its target genes C-myc and Cyclin D1 (Figure 3F–3J, Supplementary Figure 2D-2F). However, E coli seldom caused an increase in β-catenin S675, C-myc and Cyclin D1 protein abundance; although it significantly increased TLR4 abundance (Figure 3K–3O, Supplementary Figure 2G-2I). These results suggest that Fn from both colon cancer tissue and oral cavity origin can activate the β-catenin signaling pathway in colon cancer cells.
Figure 3. Fn activates the β-catenin signaling pathway in SW480 cells through the TLR4/P-PAK1/P-β-catenin S675 cascade.
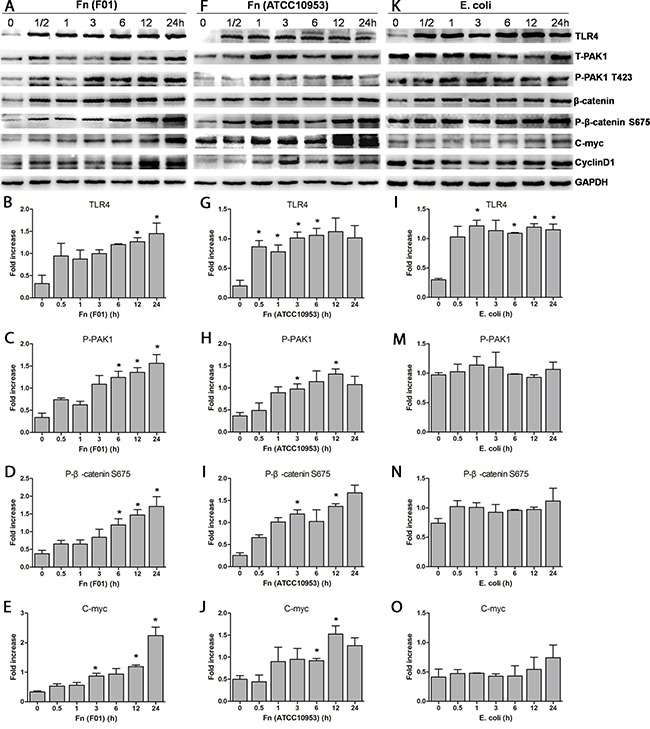
(A-E) Western blots showing that the levels of TLR4, P-PAK1, P-β-catenin S675 and C-myc gradually increase when SW480 cells are challenged with Fn (F01) over increasing time periods. (F-J) The levels of TLR4, P-PAK1, P-β-catenin S675 and C-myc also gradually increase when SW480 cells are challenged with Fn (ATCC10953) over increasing time periods. (K-O) The levels of P-PAK1, P-β-catenin S675 and C-myc do not significantly increase when SW480 cells are challenged with E. coli for increasing time periods, although TLR4 protein significantly increases. T-PAK1, total PAK1; P-PAK1, phosphorylated PAK1. Bar diagrams represent the results obtained after densitometric scanning from three different experiments. Bars represent the mean ± SD. *, P< 0.05, compared with control group (0 h).
Figure 4. Fn activates nuclear accumulation of β-catenin and P-β-catenin S675 in SW480 cells.
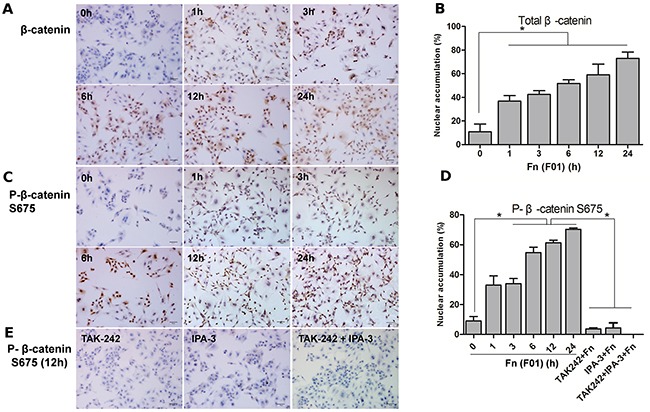
(A, B) Fn (F01) significantly induces nuclear accumulation of total β-catenin over increasing time periods in SW480 cells. (C, D) Fn (F01) significantly induces nuclear accumulation of P-β-catenin S675 over increasing time periods in SW480 cells. (D, E) TAK-242 and IPA-3 inhibited the nuclear translocation of P-β-catenin S675 induced by Fn (F01) in SW480 cells. Three random 200× magnification fields per sample were evaluated, and the average percentage of β-catenin nuclear accumulation per field was calculated. Bars represent the mean ± SD. *, P< 0.05.
Fn activates β-catenin signaling via LPS and involves TLR4/PAK1 cascade
To investigate if Fn activates β-catenin signaling through its LPS, we challenged SW480 cells with LPS extracted from cultured Fn (F01). LPS induced high protein abundance of P-β-catenin S675 and its target genes C-myc and Cyclin D1, with similarly increasing trends (Figure 5). These findings indicate that the LPS of Fn is the main activator of β-catenin signaling in colon cancer cells.
Figure 5. LPS of Fn could be the main reason for activation of the β-catenin signaling pathway through the TLR4/P-PAK1/P-β-catenin S675 cascade.
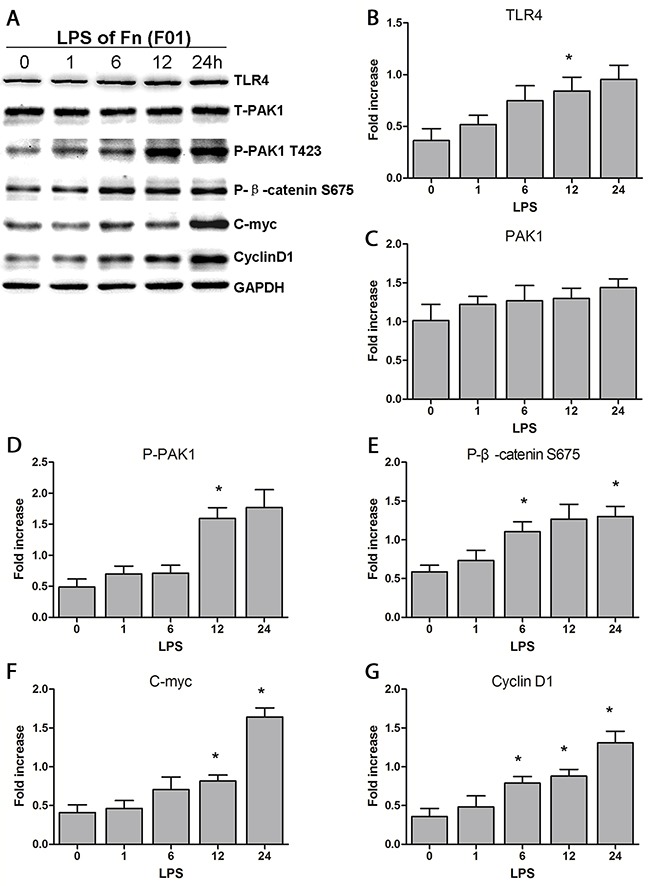
(A-G) Western blots showing that TLR4, P-PAK1, P-β-catenin S675, C-myc and Cyclin D1 levels gradually increase when SW480 cells are challenged with LPS extracted from Fn (F01) over increasing time periods. Bar diagrams represent the results obtained after densitometric scanning from three different experiments. Bars represent the mean ± SD. *, P< 0.05, as compared with control group (0 h).
Moreover, in cells challenged with Fn (F01), Fn (ATCC10953) and the LPS of Fn (F01), elevations of both TLR4 and phosphor-PAK1 (P-PAK1) proteins levels began at 0.5h or 1h, and showed similar trends of growth over time (Figure 3, Figure 5). These findings indicate that activated β-catenin signaling by Fn infection in colon cancer involve a TLR4/P-PAK1 cascade.
Fn activating β-catenin signaling might involve TLR4/P-PAK1 interaction
An Fn (F01) induced TLR4 increase in SW480 cells did not change as a result of pretreatment with the TLR4 inhibitor TAK-242 or the PAK1 inhibitor IPA-3. However, pretreatment of SW480 cells with TAK-242 significantly decreased P-PAK1 protein abundance, suggesting that in this instance, P-PAK1 activation could depend on TLR4. Furthermore, pretreatment with either TAK-242 or IPA-3, or both, prior to challenge with Fn (F01) led to a significant decrease of P-β-catenin S675, C-myc and Cyclin D1 with similar trends (Figure 6A–6O, Supplementary Figure 4). Additionally, a significant decrease of nuclear β-catenin accumulation was observed (Figure 4D and 4E). These results suggest that the TLR4/PAK1 cascade has a role in Fn-challenged β-catenin signaling activation in colon cancer cells. Using immunoprecipitation (IP), we observed for the first time an interaction between TLR4 and P-PAK1 (Figure 6P), suggesting that TLR4 is possibly an upstream activator of P-PAK1.
Figure 6. Activation of the β-catenin signaling pathway by Fn (F01) can be inhibited by both the TLR4 inhibitor (TAK-242) and PAK1 inhibitor (IPA-3).
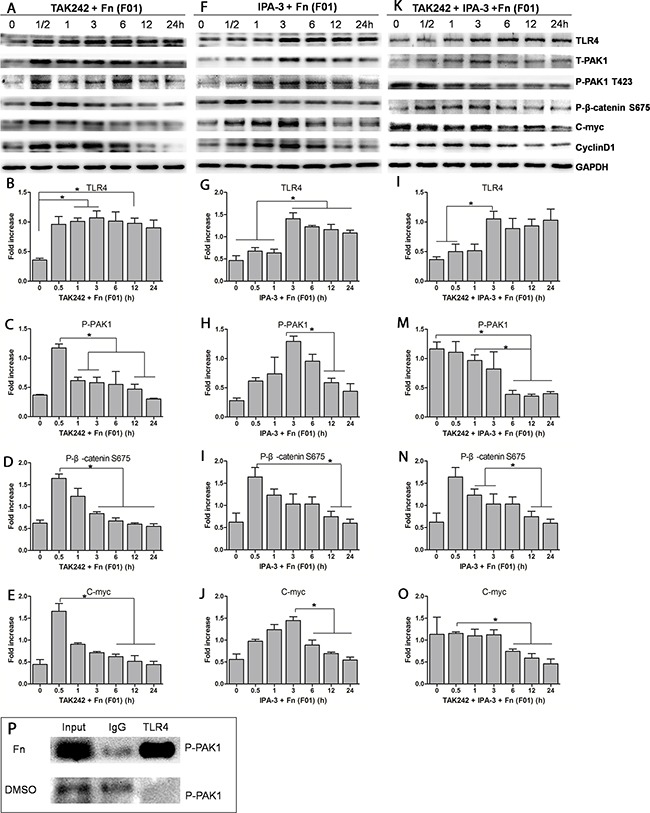
(A-E) Western blots showing that P-PAK1, P-β-catenin S675 and C-myc levels significantly decrease when SW480 cells are treated with TAK-242 prior to Fn (F01) challenge. (F-J) Western blots showing that P-PAK1, P-β-catenin S675 and C-myc levels significantly decrease when SW480 cells are treated with IPA-3 prior to Fn (F01) challenge. (K-O) Western blots showing that P-PAK1, P-β-catenin S675 and C-myc levels significantly decrease when SW480 cells are treated with both TAK-242 and IPA-3 prior to Fn (F01) challenge. (P) Cell lysates from Fn (F01) challenged SW480 cells were subjected to immunoprecipitation with anti-TLR4 agarose beads. DMSO was used as controls. Total lysates (Input), IgG and anti-TLR4 immunoprecipitations were subjected to western blots with P-PAK1 antibodies. Bar diagrams represent the results obtained after densitometric scanning from three different experiments. Bars represent the mean ± SD. *, P< 0.05.
DISCUSSION
As a newly identified CRC-associated pathogen, the specific underlying pathogenic mechanism of Fn, is poorly understood. Here, we report that Fn activates the β-catenin signaling pathway via a TLR4/P-PAK1 cascade in the carcinogenesis of CRC.
It was recently argued that PAK1 activity is required for full β-catenin signaling in the carcinogenesis of CRC [15, 20]. PAKs are involved in the innate immune response, and accumulating evidence implicates PAK1 in colon inflammation and cancer [21]. Previous studies have shown that PAK1 can enhance β-catenin levels and activity through S675 phosphorylation in CRC cells [15]. Phosphorylation at Thr423 is critical for the maintenance of PAK1 activation [22]. Our study demonstrates that Fn significantly increases P-PAK1 protein levels, leading to β-catenin S675 phosphorylation and nuclear accumulation, whereas inhibition of PAK1 greatly decreases the level of P-β-catenin S675. This indicates that P-PAK1 is an upstream component necessary for maintaining Fn-challenged transcriptional activation of β-catenin.
We further investigated the possible mechanism by which Fn modulates PAK1/β-catenin signaling. A recent study reported that adhesin FadA, identified from Fn, binds to E-cadherin, thereby mediating Fn attachment of and invasion into colon cancer cells, subsequently resulting in activation of β-catenin signaling [4]. Another study shows that Fn invasion into host cells and cytokine production are independent of surface TLRs [23]. In the present study, however, Fn challenge significantly increased the abundance of TLR4 and P-PAK1 proteins. Pretreatment with TAK-242, a TLR4 inhibitor, prior to challenge with Fn led to the significant decrease of P-PAK1 abundance. Furthermore, a decrease in nuclear β-catenin accumulation, and reduced C-myc and CyclinD1 protein levels were also observed. These data suggest that P-PAK1/β-catenin signaling activation is likely dependent on TLR4. Furthermore, we observed for the first time an interaction between TLR4 and P-PAK1 using IP, suggesting that TLR4 is possibly the upstream element responsible for activating PAK1; however, the mechanisms of interaction between TLR4 and P-PAK1 require further investigation.
TLR4 recognizes the LPS of Gram-negative bacteria [24]. In the present study, LPS extracted from Fn (F01) significantly increased the protein abundance of TLR4, P-PAK1, β-catenin S675, C-myc and Cyclin D1. Moreover, β-catenin signaling can be inhibited by both TLR4 and PAK1 inhibitors. Collectively, our results indicate that Fn may primarily activate β-catenin signaling through its LPS, subsequently involving a TLR4/PAK1 cascade in colon cancer cells.
As a heterogeneous group of organisms, Fn is a common resident of both the oral cavity and the human gut mucosa [25]. In the present study, both a gut-derived Fn (F01) strain from a CRC patient and a Fn (ATCC10953) strain originating from the oral cavity was able to induce PAK1-mediated transcriptional activation of β-catenin. A recent study reported the abundance of some oral bacteria in the stool of patients with CRC, suggesting an oral-gut translocation route [26]. Thus, some oral bacteria such as Fn, can become opportunistic pathogens upon invasion of the gut microbiota, thereby indicating that improved oral hygiene may be useful in the prevention and treatment of CRCs. However, whether the present F01 strain, originating from CRC tissue, displays a more invasive phenotype than those originating from the oral cavity or from healthy controls, requires further investigation. Moreover, our data show that TAK-242 and IPA-3 are very effective in blocking β-catenin signaling in colon cancer cells challenged by Fn. Therefore, TLR4 and PAK1 could be putative pharmaceutical targets for the treatment of Fn-related CRCs.
Recently it has been argued that instead of a single pathogenic bacterial species, it may be multiple species that contribute to CRC [27]. In our study, in addition to Fn, we found that Porphyromonas, Parvimonas, and Gemella spp. were also abundant in some CRC tissues. Other large-scale studies have identified an over-abundance of Fn within CRC tissues. Furthermore, they have observed a significant co-occurrence of Fusobacterium, Leptotrichia and Campylobacter species [28,29]. The presence of these pathogens in CRCs could be attributed to a multi-species environment. Moreover, these other microbial species may be contributing to the potentiation of tumorigenesis and the β-catenin signaling.
Taken together, our data suggest that invasive Fn activates β-catenin signaling via a TLR4/P-PAK1/P-β-catenin S675 cascade in the carcinogenesis of CRC; TLR4 and PAK1 could be potential pharmaceutical targets for the treatment of Fn-related CRCs.
MATERIALS AND METHODS
Sample collection
All tissue samples were obtained from patients undergoing colonoscopy or surgery at the Affiliated Hospital of Southwest Medical University (Sichuan, China) between February 2015 and December 2015. Fresh CRC tissue specimens (n= 14) were obtained for bacterial 16s rRNA amplification and analysis, with 7 of the samples collected from the proximal location and 7 from the distal location. The average age of the 14 CRC patients was 57.2 years. Formalin-fixed, paraffin-embedded CRC tissues (n= 98) were obtained from the pathology department archives of the same hospital for FISH analysis and immunohistochemical staining. Clinicopathologic data for each patient was obtained from the hospital records. Informed consent was obtained from all participants and the project was approved by the institutional review board.
Cell culture and reagents
The human colon cancer cell lines SW480 and Caco-2 were grown in DMEM high glucose, supplemented with 10% fetal bovine serum (FBS, Bovogen), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Beyotime, china) in a 37°C, 5% CO2 controlled incubator. A stock solution of the TLR4 antagonist TAK-242 (MedChemexpress, USA) was prepared in DMSO, and then further diluted with DMEM to yield a final concentration of 1 μM. TLR4 was inhibited with 1 μM TAK-242 for 1 hour prior to Fn or LPS stimulation. PAK1 was inhibited by IPA-3 (MedChemexpress, USA) (10 μM final concentration in DMEM) for 1 hour prior to stimulation.
DNA extraction, 16S rRNA amplification, sequencing and data analysis
Total genomic DNA was extracted from 4 mm3 fresh CRC samples using TIANamp Bacteria DNA Kit (Tiangen, China) according to the manufacturer's protocol. The DNA was diluted to a 1 ng/μL working stock using sterile water. 16S rRNA amplification, sequencing and data analysis were performed as previously described [30]. PCRs were conducted using the 515F/806R primer set [31], which amplifies the V4 region of the 16S rRNA gene.
PCR products were mixed in equidensity ratios. The pooled PCR products were purified with GeneJET Gel Extraction Kit (Thermo Scientific, USA). Sequencing libraries were generated using NEB Next® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following manufacturer's recommendations, and index codes were added. The library quantity and quality was assessed using the Qubit@ 2.0 Fluorometer (Thermo Scientific, USA) and the Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on an Illumina HiSeq platform, generating 250 bp paired-end reads. Sequences were analyzed using the QIIME [32] software package.
Microbial FISH analysis
5-μm-thick sections were prepared using samples obtained from the pathology department archives and hybridized as described in our previous study [33]. The sequence of the FITC-labeled Fn-targeted probe, FUS664, was: 5′- CTT GTA GTT CCG C(C /T) TAC CTC -3′ [34]. Five random 200 × magnification fields per sample were evaluated by an observer blind to sample status; the number of bacteria per field was calculated. Cases were considered Fn-positive when FUS664 probe-associated bacteria were visualized, on average, more than 10 times per field.
Immunohistochemical staining
Indirect immunohistochemistry was performed with formalin-fixed, paraffin-embedded tissue sections. Primary antibodies used in the study were as follows: TLR4 (1:100 dilution; Santa Cruz, CA), PAK1 (1:100; Cell Signaling), and β-Catenin (1:100; Cell Signaling). After application of the appropriate secondary antibodies, the labeled antigens were visualized by the development of brown pigment via a standard 3,3′- diaminobenzidine (DAB) protocol. Slides were then counterstained lightly with hematoxylin. Staining without primary antibody was performed as a negative control. Immunoreactivities were estimated as described in our previous report [35].
Western blot and IP assay
Total cellular protein was isolated from cultured cells with a protein extraction solution (Beyotime, china). Proteins were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes at 0.35 A (1 h, 4°C) using a wet-blotting apparatus (Bio-Rad). Membranes were blocked with 5% skim milk in PBST for 2 hours at room temperature, and then incubated overnight at 4°C with the primary antibodies diluted in PBST as follows: TLR4 (1:500), PAK1 (1:1000), phosphorylated-PAK1 Thr 423 (1:500; Santa Cruz), β-Catenin (1:1000), P-β-Catenin Ser 675 (1:1000; Cell Signaling), C-myc (1:1500; Proteintech), Cyclin D1 (1:200; Beyotime) and GAPDH (1:10000; Bioworld). Membranes were then incubated with appropriate secondary antibodies for 1 h at room temperature. Blots were quantified by densitometry using Quantity One 4.5.0 software (Bio-Rad Laboratories, Inc.).
The IP assay was conducted as previously described using an IP Kit (Beyotime, China) [36]. SW480 cells were stimulated with Fn (F01) and a total cell lysate was obtained. SW480 cells treated with DMSO were used as a control. Total protein was immunoprecipitated using an anti-TLR4 antibody (Santa Cruz) complexed to protein A/G. The immunoprecipitated proteins were then analyzed by western blot with an antibody to detect P-PAK1 T423.
Fn culture and LPS extraction
Fn culture was performed as previously described [37]. Frozen 6 mm3 sections of proximal colon cancer tissue were thawed and immediately placed into 500 μL of sterilized PBS, washed to remove loosely associated bacteria, and then homogenized using a sterile mortar and pestle. This suspension was directly poured into fastidious anaerobe broth (FAB), supplemented with vancomycin (4 ug/mL) and neomycin sulfate (30 ug/mL), and then incubated for 1 d in a humidified anaerobic chamber (85% N2, 10% H2, and 5% CO2 at 37°C). One hundred milliliter aliquots of this broth were spread onto pre-reduced brain heart infusion agar (BHI) plates supplemented with 5% defibrinated sheep blood (DSB), josamycin (3 ug/mL), vancomycin (4 ug/mL), neomycin sulfate (30 ug/mL), norfloxaxin (1 ug/mL), haemin (5 ug/mL) and menadione (1 ug/mL). Plates were inspected every 2 days for growth, and all colonies were picked and streak-purified further onto pre-reduced BHI plates supplemented with haemin (5 ug/mL), menadione (1 ug/mL), and 5% DSB. Single colonies were examined by phase microscopy, looking for slender rods or needle-shaped cells that are characteristic of Fn. DNA was extracted from positively identified isolates using TIANamp Bacteria DNA Kit. Aliquots were used as template DNA for a PCR with primers and conditions as described by Park et al [38]. An amplicon size of 171 bp confirmed that the isolate belonged to Fn. Next, verified isolates underwent a PCR to partially amplify the 16S rRNA gene for sequencing and confirmation of speciation using the same DNA template; primers and conditions are as defined by Zhao et al [39]. This product (1499 bp) was sent to Sangon Biotech (Shanghai, China) for sequence analysis, and obtained traces that confirmed Fn as the species. The isolated strain was obtained from the tumor specimen from patient number 01, and thus named F01. The strain was stored at −80°C in FAB + glycerol (50% v/v).
LPS extraction from the Fn culture was performed using an LPS extraction kit (Intron Biotechnology) according to the manufacturer's instructions.
Immunocytochemistry
The SW480 cells cultured on glass slides were treated with Fn (F01, MOI 1:100) for 12 hours. Cells were then fixed in 4 % paraformaldehyde for 20 min at 4°C. After being permeabilized by incubation with 0.5 % Triton X-100 for 15 min at room temperature, cells were incubated with 0.3% hydrogen peroxide in methanol for 10 min. The slides were incubated with primary antibodies β-Catenin (1:100) and P-β-Catenin S675 (1:100) for overnight at 4°C. Glass slides were incubated with secondary antibody for 1 h at 37°C. The labelled antigens were visualized by the development of brown pigment via DAB. Slides were then counterstained lightly with hematoxylin. Staining without primary antibody was used as a negative control.
Three random 200× magnification fields per sample were evaluated and the average percentage of β-Catenin and P-β-Catenin S675 nuclear accumulation per field were calculated respectively.
Scanning electron microscopy (SEM)
Cultured Fn (F01) was fixed with 4% glutaraldehyde overnight at 4°C, dehydrated through a graduated series of ethanol, then spread on silver paper, critical point dried and coated with gold palladium. Specimens were prepared for viewing under a JEOL-6380LV emission scanning electron microscopy (JEOL, Japan).
Fn (ATCC10953) and nonpathogenic E. coli culture
Fn (ATCC10953) was graciously supplied by Dr. Junqiang Jiang of the Affiliated Dental Hospital of Southwest Medical University, and nonpathogenic E. coli was isolated from healthy human stool for use as a control. Fn (ATCC10953) was incubated for 4 days in FAB under anaerobic conditions at 37°C. E. coli was cultured for 24 hours in Luria-Bertani broth at 37°C.
Bacterial infection and LPS stimulation of colon cancer cells
Fn (F01) and Fn (ATCC10953) were cultured with FAB, supplemented with vancomycin (4 ug/mL) and neomycin sulfate (30 ug/mL) in the anaerobic system at 37°C for 4-5 days, until they reached an OD540nm of 0.8, corresponding to 109 CFU/mL. The bacteria were diluted to 107CFU/mL, corresponding to a multiplicity of infection (MOI) of 100 [40].
The SW480 and Caco-2 cells were separately seeded in 6-well plates and incubated for 24 to 48 hours (until they reached 60% of the well) at 37°C. Cells were washed with PBS, and media was replaced with DMEM high glucose, supplemented with 10% FBS. The cells were then infected with bacteria (MOI 1:100) for 30 minutes, 1, 3, 6, 12 and 24 hours. For LPS stimulation, the SW480 cells were incubated with 0.1 ug/mL Fn LPS for 1, 6, 12 and 24 hours. No bacteria or LPS were added to the control plates. The plates were shaken gently and incubated at 37°C in an incubator with controlled 5% CO2. At each time point, media was removed and cells were washed twice with PBS. Total protein was extracted from cells for western blot.
Statistical analysis
Data are presented as the mean and standard deviation for continuous variables and as proportions for categorical variables. Data were analyzed using one-way ANOVA, followed by Bonferroni test for multiple comparisons. Differences in categorical variables were determined by the Chi-square or Fisher's exact tests, as appropriate. Differences were considered significant if P < 0.05. All significance tests were two-tailed. All statistical tests were performed using SPSS software Version 13.0 (SPSS Inc., Chicago, IL, USA).
SUPPLEMENTARY MATERIALS FIGURES
Acknowledgments
The authors thank M.D. Junqiang Jiang for providing Fusobacterium nucleatum (ATCC10953), and Haisu Wan for Fusobacterium nucleatum culture assistance.
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
GRANT SUPPORT
M.D. Yan Peng and M.D. Xiangsheng Fu are supported by Natural Science Foundation of Sichuan Science and Technology Agency under Award No. 201565.
REFERENCES
- 1.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–98. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, Kostic AD, Giannakis M, Bullman S, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, Kostic AD, Giannakis M, Watanabe H, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653–61. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu H, Redline RW, Han YW. Fusobacterium nucleatum induces fetal death in mice via stimulation of TLR4-mediated placental inflammatory response. J Immunol. 2007;179:2501–08. doi: 10.4049/jimmunol.179.4.2501. [DOI] [PubMed] [Google Scholar]
- 6.Fukata M, Shang L, Santaolalla R, Sotolongo J, Pastorini C, España C, Ungaro R, Harpaz N, Cooper HS, Elson G, Kosco-Vilbois M, Zaias J, Perez MT, et al. Constitutive activation of epithelial TLR4 augments inflammatory responses to mucosal injury and drives colitis-associated tumorigenesis. Inflamm Bowel Dis. 2011;17:1464–73. doi: 10.1002/ibd.21527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukata M, Chen A, Vamadevan AS, Cohen J, Breglio K, Krishnareddy S, Hsu D, Xu R, Harpaz N, Dannenberg AJ, Subbaramaiah K, Cooper HS, Itzkowitz SH, Abreu MT. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. 2007;133:1869–81. doi: 10.1053/j.gastro.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cammarota R, Bertolini V, Pennesi G, Bucci EO, Gottardi O, Garlanda C, Laghi L, Barberis MC, Sessa F, Noonan DM, Albini A. The tumor microenvironment of colorectal cancer: stromal TLR-4 expression as a potential prognostic marker. J Transl Med. 2010;8:112. doi: 10.1186/1479-5876-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang EL, Qian ZR, Nakasono M, Tanahashi T, Yoshimoto K, Bando Y, Kudo E, Shimada M, Sano T. High expression of Toll-like receptor 4/myeloid differentiation factor 88 signals correlates with poor prognosis in colorectal cancer. Br J Cancer. 2010;102:908–15. doi: 10.1038/sj.bjc.6605558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 11.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–36. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun J. Enteric bacteria and cancer stem cells. Cancers (Basel) 2010;3:285–97. doi: 10.3390/cancers3010285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molli PR, Li DQ, Murray BW, Rayala SK, Kumar R. PAK signaling in oncogenesis. Oncogene. 2009;28:2545–55. doi: 10.1038/onc.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter JH, Douglass LE, Deddens JA, Colligan BM, Bhatt TR, Pemberton JO, Konicek S, Hom J, Marshall M, Graff JR. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin Cancer Res. 2004;10:3448–56. doi: 10.1158/1078-0432.CCR-03-0210. [DOI] [PubMed] [Google Scholar]
- 15.Zhu G, Wang Y, Huang B, Liang J, Ding Y, Xu A, Wu W. A Rac1/PAK1 cascade controls β-catenin activation in colon cancer cells. Oncogene. 2012;31:1001–12. doi: 10.1038/onc.2011.294. [DOI] [PubMed] [Google Scholar]
- 16.Santaolalla R, Sussman DA, Ruiz JR, Davies JM, Pastorini C, España CL, Sotolongo J, Burlingame O, Bejarano PA, Philip S, Ahmed MM, Ko J, Dirisina R, et al. TLR4 activates the β-catenin pathway to cause intestinal neoplasia. PLoS One. 2013;8:e63298. doi: 10.1371/journal.pone.0063298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Xu X, Chen N, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 18.Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, Hercog R, Koch M, Luciani A, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, Su L, Li X, Li X, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 20.Park MH, Kim DJ, You ST, Lee CS, Kim HK, Park SM, Shin EY, Kim EG. Phosphorylation of β-catenin at serine 663 regulates its transcriptional activity. Biochem Biophys Res Commun. 2012;419:543–49. doi: 10.1016/j.bbrc.2012.02.056. [DOI] [PubMed] [Google Scholar]
- 21.Dammann K, Khare V, Gasche C. Tracing PAKs from GI inflammation to cancer. Gut. 2014;63:1173–84. doi: 10.1136/gutjnl-2014-306768. [DOI] [PubMed] [Google Scholar]
- 22.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 23.Quah SY, Bergenholtz G, Tan KS. Fusobacterium nucleatum induces cytokine production through Toll-like-receptor-independent mechanism. Int Endod J. 2014;47:550–59. doi: 10.1111/iej.12185. [DOI] [PubMed] [Google Scholar]
- 24.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–88. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 25.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–78. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 26.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–60. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abreu MT, Peek RM., Jr Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146:1534–1546. e1533. doi: 10.1053/j.gastro.2014.01.001. https://doi.org/10.1053/j.gastro.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, Jenab M, Prehn JH, Hughes DJ. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–90. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 29.Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, Allen-Vercoe E, Holt RA. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo J, Fu X, Liao H, Hu Z, Long L, Yan W, Ding Y, Zha L, Guo Y, Yan J, Chang Y, Cai J. Potential use of bacterial community succession for estimating post-mortem interval as revealed by high-throughput sequencing. Sci Rep. 2016;6:24197. doi: 10.1038/srep24197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–24. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–36. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Chen Y, Fu X, Zhou X, Peng Y, Shi L, Chen T, Wu Y. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int J Cancer. 2016;139:1318–26. doi: 10.1002/ijc.30168. [DOI] [PubMed] [Google Scholar]
- 34.Loy A, Arnold R, Tischler P, Rattei T, Wagner M, Horn M. probeCheck—a central resource for evaluating oligonucleotide probe coverage and specificity. Environ Microbiol. 2008;10:2894–98. doi: 10.1111/j.1462-2920.2008.01706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Fu X, Zhang W, Xiao L, Qiu Y, Peng Y, Shi L, Chen X, Zhou X, Deng M. Wnt signaling pathway is activated in right colon serrated polyps correlating to specific molecular form of β-catenin. Hum Pathol. 2013;44:1079–88. doi: 10.1016/j.humpath.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Liu N, Wang S, Wang L, Zhao J, Su L, Zhang Y, Zhang S, Xu Z, Zhao B, et al. Identification of a small molecule targeting annexin A7. Biochim Biophys Acta. 2013;1833:2092–2099. doi: 10.1016/j.bbamcr.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 37.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SN, Lim YK, Kook JK. Development of quantitative real-time PCR primers for detecting 42 oral bacterial species. Arch Microbiol. 2013;195:473–82. doi: 10.1007/s00203-013-0896-4. [DOI] [PubMed] [Google Scholar]
- 39.Zhao SY, Jiang DY, Xu PC, Zhang YK, Shi HF, Cao HL, Wu Q. An investigation of drug-resistant Acinetobacter baumannii infections in a comprehensive hospital of East China. Ann Clin Microbiol Antimicrob. 2015;14:7. doi: 10.1186/s12941-015-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendes RT, Nguyen D, Stephens D, Pamuk F, Fernandes D, Van Dyke TE, Kantarci A. Endothelial Cell Response to Fusobacterium nucleatum. Infect Immun. 2016;84:2141–48. doi: 10.1128/IAI.01305-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


