Abstract
Mitotic catastrophe is the response of mammalian cells to mitotic DNA damage. It produces tetraploid cells with a range of different nuclear morphologies from binucleated to multimicronucleated. In response to DNA damage, checkpoints are activated to delay cell cycle progression and to coordinate repair. Cells in different cell cycle phases use different mechanisms to arrest their cell cycle progression. It has remained unclear whether the termination of mitosis in a mitotic catastrophe is regulated by DNA damage checkpoints. Here, we report the presence of a mitotic exit DNA damage checkpoint in mammalian cells. This checkpoint delays mitotic exit and prevents cytokinesis and, thereby, is responsible for mitotic catastrophe. The DNA damage-induced mitotic exit delay correlates with the inhibition of Cdh1 activation and the attenuated degradation of cyclin B1. We demonstrate that the checkpoint is Chk1-dependent.
Cell cycle checkpoints are mechanisms that coordinate the progression of the cell cycle with intracellular and extracellular events that are otherwise unrelated to the cell division processes. In response to DNA damage, an elaborate network of signaling pathways, collectively called the DNA damage checkpoint, is activated to prevent the damaged DNA from being replicated or transmitted to the next generation (for review, see refs. 1 and 2). In the center of this network are two parallel but partially overlapping protein kinase cascades, the ataxia-telangiectasia, mutated-Chk2 and ataxia-telangiectasia, mutated and Rad3-related-Chk1 kinase modules, that transduce the damage signal to downstream effectors involved in cell cycle control, DNA repair, and apoptosis (3). The importance of DNA damage checkpoint control is demonstrated by findings that link defects in the checkpoint to human cancers (3).
The main function of the DNA damage checkpoint is to halt the cell cycle progression. Because the driving force of the cell cycle progression are cyclin-dependent kinases (Cdks), it is not surprising to find that the end result of checkpoint activation usually is the inhibition of Cdks. In G1, DNA damage activates p53, which in turn activates the expression of p21Cip1, a Cdk inhibitor. The induction of p21 is the major mechanism underlying DNA damage-induced G1 arrest (4–6). In addition, cyclin D1 is rapidly destabilized in response to DNA damage during G1, leading to redistribution of p21 from Cdk4 to Cdk2 and, thereby, contributing to the G1 arrest (7). In G2, the DNA damage checkpoint is realized through Chk1-dependent phosphorylation and inactivation of the Cdc25C phosphatase, whose activity is necessary for the activation of Cdc2 (Cdk1) and entry into mitosis (8, 9).
Mitotic catastrophe is a term used to describe the failure of mitosis. In budding yeast, it is caused by chromosome missegregation and followed by cell death (10, 11). DNA damage induces mitotic catastrophe in mammalian cells that results in the formation of cells with either multimicronuclei or two nuclei (12–14). Multimicronucleated cells are nonviable and arise through the formation of nuclear envelopes around clusters of chromosomes or chromosome fragments during a catastrophic mitosis (12–14). The multimicronucleation is likely caused by severe DNA damage that fragmented chromosomes into pieces. Mitotic DNA damage may also induce cytokinesis failure (12, 13), leading to binucleation. The so-formed binucleated cells are arrested in G1 in a p53-dependent manner (12, 13).
It is unclear whether mammalian cells maintain a metaphase DNA damage checkpoint that prevents securin degradation, as in budding yeast, and whether DNA damage-induced mitotic catastrophe is regulated by DNA damage checkpoints. We report here that, in contrast to budding yeast, mammalian securin is not stabilized in response to mitotic DNA damage. Instead, there is a mitotic exit DNA damage checkpoint that delays mitotic exit and prevents cytokinesis, resulting in the formation of binucleated cells.
Materials and Methods
Antibodies and Reagents. We used the following primary antibodies: anti-Chk1 (G-4), anti-cyclinB1 (GNS1), and anti-p55CDC from Santa Cruz Biotechnology; anti-Cdh1 (clone DH01) and anti-securin (clone DCS-280) from NeoMarkers (Fremont, CA); anti-phospho-Chk1 (S345) and anti-phospho-Chk2 from Cell Signaling Technology (Beverly, MA); anti-Brca1 from Calbiochem (OP92); anti-α-tubulin (clone DM1A) from Sigma; anti-Cdc27 (clone 35) from Becton Dickinson Pharmingen; and anti-γ-H2AX from Upstate Biotechnology (Lake Placid, NY).
To produce an RNA interference effect in cultured cells, we used the retrovirus-based vector system, pRETRO-SUPER (15). RNA interference targeting sequences (19-nt) were chosen from the genes of interest, synthesized, and cloned into pRETRO-SUPER. The sequences of the 19 nucleotides are: Cdh1, 5′-tgagaagtctcccagtcag-3′; Brca1, 5′-ctgtgagaactctgaggac-3′; Chk1, 5′-gaagcagtcgcagtgaaga-3′ and 5′-gcgtgccgtagactgtcca-3′ (both gave similar results). For the control, we used a construct designed to silence mouse Id2.
Cell Culture and Treatments. HeLa cells, HeLa cells expressing histone H2B tagged with GFP (16), or BJ cells [from American Type Culture Collection (ATCC)] were cultured in DMEM containing 10% FCS. Prometaphase arrest was achieved by treating cells with 65 ng/ml nocodazole for 18 h. Typically, ≈90% cells were in mitosis at the end of the arrest. In most of the experiments, mitotic cells were shaken off from a nocodazole-arrested population of cells to obtain a pure preparation of mitotic cells. Cells were given an irradiation dose of 10 Gy to introduce DNA damage. Mouse embryonic stem cells were cultured as described in ref. 17. They were arrested for 12 h with nocodazole (65 ng/ml), collected through mechanical detachment, and irradiated with a dose of 10 Gy.
Retrovirus production and infection were performed as described in ref. 15. Stable pools of infected cells were selected and maintained in media containing 2.5 μg/ml puromycin.
Cell Cycle Analysis. Cell cycle distribution was analyzed by using flow cytometry analysis with standard protocols. The percentage of G1, S, and G2 population was measured with the system ii software (Beckman Coulter). For monitoring the progression of mitosis, a HeLa cell line expressing histone H2B tagged with GFP cells on coverslips were arrested with nocodazole for 18 h, irradiated or mock irradiated, released from nocodazole block for various lengths of time, fixed with 4% paraformaldehyde at 4°C, and analyzed under a fluorescence microscope. Chromosomes emitted green fluorescence because of the histone H2B GFP. Cells in anaphase/telophase were scored.
Western Blot Analysis and Immunofluorescence. Western blot analyses were carried out according to standard protocols. The blots were developed by using enhanced chemiluminescence (Amersham Pharmacia Biosciences). For immunofluorescence, the cells were fixed in 4% paraformaldehyde and stained with anti-γ-H2AX (1:500) antibodies.
In the 2D PAGE analysis, the isoelectric focusing was performed with PROTEAN IEF Cell and the 2-D Starter Kit (Bio-Rad) and followed by regular SDS/PAGE.
Results
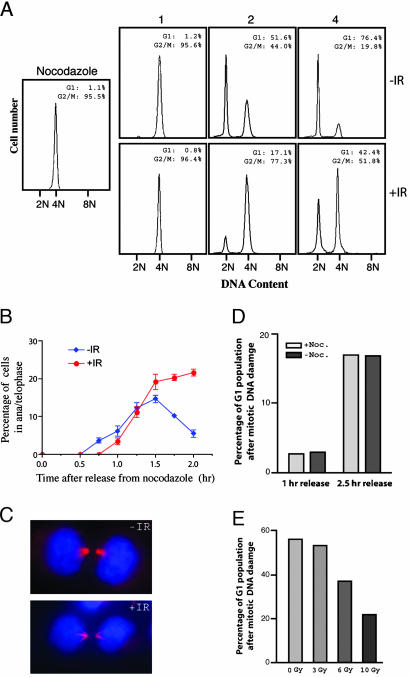
DNA Damage Delays Mitotic Exit in Mammalian Cells. To investigate the mitotic DNA damage response in mammalian cells, we arrested HeLa cells at prometaphase first and then introduced DNA damage in these cells with 10 Gy γ-irradiation. FACS analysis indicated a delay in the appearance of G1 cells in the irradiated sample after removal of the spindle poison (Fig. 1A). At 2 h after the release, there were >50% unirradiated control cells in G1, whereas the number was 17% for the irradiated cells. By 4 h after release, ≈50% of the irradiated cells had not completed mitosis and thus contained 4N DNA content, whereas the number was ≈20% for the unirradiated cells. This delay in the production of G1 cells could be a result of the inability of these cells to initiate anaphase, to exit mitosis, or both. To address that issue, we monitored the progression of mitosis in cells released from a nocodazole arrest. We determined the percentage of cells in anaphase/telophase with and without irradiation. As shown in Fig. 1B, both unirradiated and irradiated cells quickly entered anaphase/telophase after the removal of nocodazole. However, although the unirradiated cells exited mitosis, the proportion of irradiated cells in anaphase/telophase remained high (Fig. 1B), indicating a delay in mitotic exit. We stained the cells with antitubulin antibody to determine precisely where the delay had occurred. As shown in Fig. 1C, it was in late telophase/cytokinesis that the irradiated cells accumulated. Interestingly, the morphology of the midbody in the irradiated cells is very different from that in the unirradiated cells. These results indicate that mitotic DNA damage elicits a delay in cell cycle progression after metaphase-to-anaphase transition but before cytokinesis in mammalian cells. We referred to this checkpoint as mitotic exit DNA damage checkpoint.
Fig. 1.
DNA damage delays mitotic exit. (A) FACS analysis of HeLa cells exiting mitosis with and without having received γ-irradiation. Cells were arrested with nocodazole for 18 h, irradiated, and released from the nocodazole arrest. (B) The progression of mitosis of cells in A was monitored, and the percentages of cells in anaphase/telophase were plotted against time. (C) Micrographs of the late telophase cells that accumulated after mitotic DNA damage. Blue, DAPI; red, tubulin. (D) Mitotic cells collected through mechanical shakeoff without first being arrested by nocodazole displayed the same mitotic exit delay as cells obtained via nocodazole arrest. (E) DNA damage causes mitotic exit delay in normal human fibroblasts (BJ cells). BJ cells were arrested with nocodazole for 18 h, collected through shakeoff, irradiated with different dosages of γ-irradiation, released from the mitotic block, and analyzed with FACS. The size of the G1 population at 24 h after the release is shown.
To rule out the possibility that the observed checkpoint is a consequence of arresting cells with nocodazole, we analyzed mitotic cells collected through mechanical shakeoff of a population of cells never exposed to the microtubule-disrupting drug. Comparable delays in mitotic exit (measured by the size of the G1 peak on FACS profiles) were seen in these cells (Fig. 1D). In addition, the delay in mitotic exit could be induced by treating mitotic cells with adriamycin, a different DNA damaging agent (data not shown), indicating that the observed mitotic exit delay is not specific to γ-irradiation. Furthermore, we used normal diploid human fibroblasts (BJ cells from ATCC) in a similar experiment to see whether the checkpoint operates in normal untransformed cells. As shown in Fig. 1E, DNA damage delays the mitotic exit of these cells as well. We also found that DNA damage delays mitotic exit of mouse embryonic stem cells (see below).
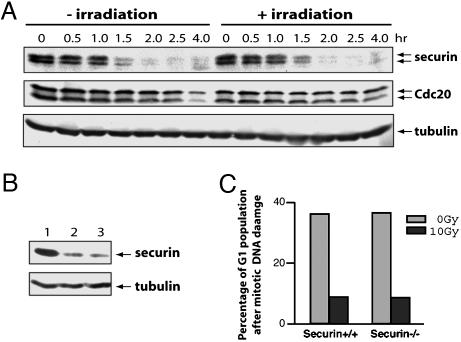
Securin Is Not Stabilized by DNA Damage. The observation that cells entered anaphase after DNA damage suggests that unlike its yeast counterpart, mammalian securin should not be stabilized by DNA damage. Therefore, we analyzed the effect of DNA damage on the levels of the securin protein. As shown in Fig. 2A, after the removal of nocodazole from the cell culture media, securin was rapidly degraded regardless of whether the cells had been irradiated. In agreement with this finding, we could not detect any irradiation-induced changes in the levels of Cdc20 (Fig. 2A), a factor required for the ubiquitination of securin by anaphase-promoting complex (APC) (18–20).
Fig. 2.
Securin is not stabilized by DNA damage. (A) Western blot analysis of securin and Cdc20 in cells treated similarly as in Fig. 1 A. (B) Western blot analysis of securin in cells that were arrested with nocodazole, irradiated, left with the mitotic block for an additional 2 h, and released for 2 h. Lanes: 1, before the release; 2, without irradiation; 3, with irradiation. The gel was run too short to separate the two forms of securin seen in A. (C) Securin is not required for the mitotic exit DNA damage checkpoint. Mouse embryonic stem cells (WT or securin null) were arrested with nocodazole for 12 h, irradiated, released, and analyzed with FACS. The proportion of G1 cells from the unirradiated and irradiated samples were graphed.
Because the length of time that cells spent between irradiation and removal of nocodazole was short (≈20 min), it was formally possible that the cells did not have enough time to respond to the damage or to mount an arrest in metaphase. We tested that possibility by incubating cells with nocodazole up to 2 h after irradiation. Right after the removal of the microtubule poison from both the irradiated and the unirradiated samples, cells entered anaphase irrespective of irradiation (data not shown), and securin was degraded normally (Fig. 2B), indicating that the apparent lack of a metaphase DNA checkpoint is not a result of insufficient time for the checkpoint to be activated.
We examined the mitotic exit damage checkpoint in mouse ES cells that lack securin (17). We found that both WT and securin–/– ES cells responded to mitotic DNA damage similarly, indicating that securin is not an essential part of the mitotic exit DNA damage checkpoint (Fig. 2C).
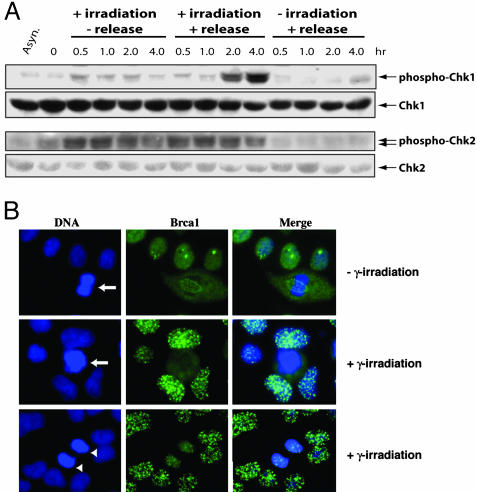
Activation of Chk1 and Chk2 in Mitosis. Mammalian DNA damage checkpoints are controlled by ataxia-telangiectasia, mutated and ataxia-telangiectasia, mutated and Rad3-related kinases and their downstream effector kinases Chk1 and Chk2 (for review, see ref. 2). We determined the phosphorylation status of Chk1 and Chk2 kinases by using phosphorylation specific antibodies against Chk1 (Ser-345) or Chk2 (Ser-33/35) as a measurement of their activation. Chk1 displayed some degree of activation in unirradiated asynchronous and mitotic cells (Fig. 3A). Irradiation did not induce further activation of Chk1 in the prometaphase-arrested cells. However, if the irradiated cells were released from the mitotic block, the level of Chk1 phosphorylation increased and became prominent by 2 h, which is not seen in the unirradiated and released cells (Fig. 3A). This activation of Chk1 could not be attributed to contaminating G2 cells (if any) in our mitotic cell preparation, because if the cells were kept in nocodazole-containing media, no additional activation of Chk1 was observed (Fig. 3A). These data indicate that Chk1 only become activated after cells have passed the metaphase to anaphase transition. In contrast to the little activation of Chk1 in metaphase, Chk2 was readily activated by DNA damage, no matter whether the cells were kept in nocodazole containing media or released from the mitotic block (Fig. 3A). Little activation of Chk2 in unirradiated cells was observed (Fig. 3A). These results indicate that mammalian cells activate Chk1 and Chk2 differentially in response to mitotic DNA damage.
Fig. 3.
Activation of DNA damage checkpoint in mitosis. (A) Western blot analyses of the activation of Chk1 and Chk2. Cells were similarly treated as those in Fig. 2 except some cells were not let to leave mitotic block. (B) Immunostaining of Brca1. Asynchronous HeLa cells were irradiated and processed for immunostaining 1 h later. Arrows in B indicate a metaphase cell; arrowheads indicate a telophase cell.
Brca1 Foci Formation Is Absent in Irradiated Metaphase Cells. The difference in the activation of Chk1 and Chk2 prompted us to ask why Chk1 is only activated in cells that have passed the metaphase to anaphase transition. It was shown previously that Chk1 activation requires the function of Brca1, whereas Chk2 does not (21). In response to DNA damage, mammalian cells display characteristic nuclear Brca1 foci (22, 23). It is possible that in metaphase cells Brca1 is unable to form such foci because of the condensed nature of the chromosomes and, thereby, no Chk1 activation. We irradiated unsynchronized HeLa cells and immunostained them 1 h later. Brca1 foci were readily detectable in interphase cells but not in cells with condensed and unseparated chromosomes (Fig. 3B), although metaphase cells can sense the presence of DNA damage as shown by γ-H2AX staining (Fig. 7, which is published as supporting information on the PNAS web site). In the unirradiated mitotic cells, Brca1 forms discrete foci along the structure of the microtubule spindle (Fig. 3B), as has been reported in ref. 24. In agreement with the finding that Chk1 is activated only after the release from metaphase arrest, we found that cells in cytokinesis displayed Brca1 foci (Fig. 3B). p53BP1 displayed essentially the identical staining pattern as Brca1 (data not shown). These results indicate that Brca1 as well as p53BP1 is not recruited to the site of DNA damage in irradiated mitotic cells until cytokinesis, when the chromosomes start decondensing. The lack of the Brca1 focus formation in metaphase may be a result of the highly condensed nature of metaphase chromosomes.
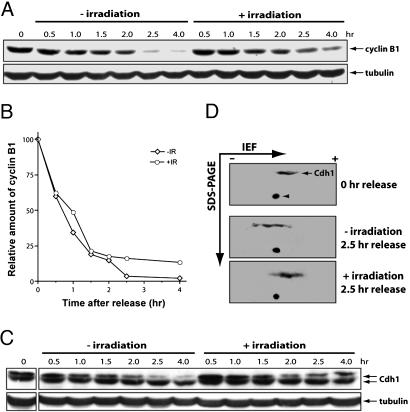
DNA Damage Attenuates Cyclin B1 Degradation. B type cyclins are degraded to allow mitotic exit and cytokinesis (25, 26). The ubiquitination of cyclin B1 can be carried out by both APC/Cdc20 and APC/Cdh1, with APC/Cdh1 acting late in mitosis (27). Because DNA damage delays mitotic exit, we decided to examine its effect on the level of cyclin B1. Up to 2 h after the release from nocodazole block, cyclin B1 was degraded as rapidly in the irradiated as in the unirradiated cells (Fig. 4 A and B). However, after that, although the degradation continued in the unirradiated cells, it slowed down in the irradiated cells. The persistence of cyclin B1 in the irradiated cells correlates with and may underlie the mitotic exit delay induced by DNA damage (Fig. 1 A and B).
Fig. 4.
Mitotic DNA damage induces stabilization of cyclin B1. (A) Western blot analysis of cyclin B1. HeLa cells were arrested with nocodazole, irradiated, and released. Cell lysates were prepared from cells harvested at different time points after the release and analyzed. (B) Quantitation of the data in A. (C) Cell lysates from A were analyzed for Cdh1 with Western blot analysis. Note the persistence of the slow migrating form of Cdh1 in the irradiated samples. (D) The 2.5-h samples from A were also analyzed by 2D gel electrophoresis, followed by Western blotting probed with anti-Cdh1 antibodies. Arrowhead indicates a nonspecific dot that serves nicely as a landmark on the blots.
DNA Damage Inhibits the Activation of Cdh1. The attenuated degradation of cyclin B1 in the irradiated cells suggested that APC/Cdh1 might be inhibited by DNA damage. In budding yeast, the activation of Cdh1 depends on the Cdc14 phosphatase, which is activated by the mitotic exit network (28, 29). This regulation is likely to be conserved in mammals because human Cdh1 is dephosphorylated before mitotic exit and the dephosphorylated form is the active form (30, 31). We analyzed the effect of DNA damage on the phosphorylation status of Cdh1. As shown in Fig. 4C, in the unirradiated cells, the level of the phosphorylated form of Cdh1 gradually decreased as the cells progressed through mitosis. However, this decrease was inhibited by the irradiation. We further analyzed this effect through 2D gel electrophoresis. Because of a large number of Cdk phosphorylation sites in Cdh1 (30), Cdh1 appeared on 2-D gel blots as a line instead of distinct dots (Fig. 4D). When the cells were released from the mitotic block without irradiation, the line shifted toward the anion side (basic side) of the isoelectric focusing gel, indicating the dephosphorylation of Cdh1 (compare Fig. 4D Top and Middle). Irradiation blocked this shift (Fig. 4D Bottom). Taken together from the results of the 1D and 2D gel analyses, we concluded that DNA damage in mitosis inhibits the activation of Cdh1.
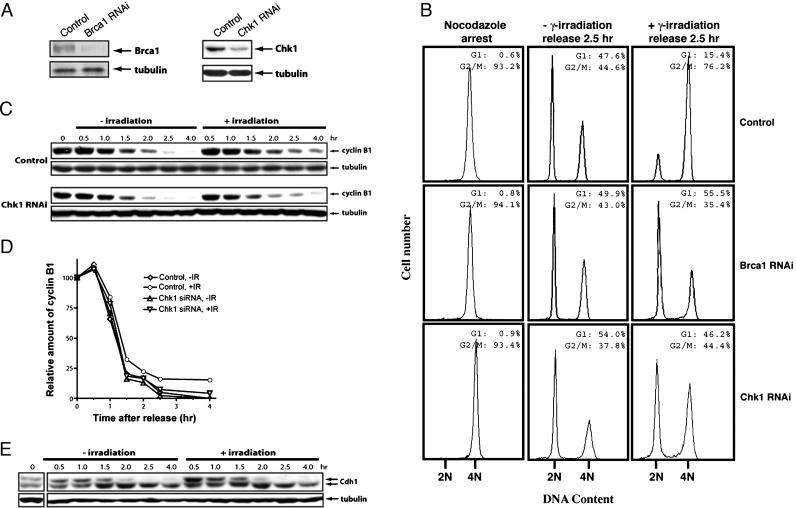
The Mitotic Exit DNA Damage Checkpoint Is Brca1- and Chk1-Dependent. To determine whether Brca1 or Chk1 mediates the mitotic exit delay in response to DNA damage, we generated cells that express Brca1 or Chk1 at a reduced level via small interfering RNA-mediated gene silencing (Fig. 5A). The knockdown cells are viable and do not display any discernable growth defects, but they did display defects in the G2/M DNA damage checkpoint as reported previously. We analyzed the mitotic DNA damage response in these cells. As shown in Fig. 5B, more Brca1 and Chk1 RNA interference cells exited mitosis than the control cells after irradiation, whereas mitotic exit was not affected in the unirradiated control and knockdown cells. When the levels of cyclin B1 were examined, we saw the same attenuation of cyclin B1 degradation in the irradiated control cells as before, but the attenuation was lost in Chk1 knockdown cells (Fig. 5 C and D). We also found that in the Chk1 knockdown cells, DNA damage no longer inhibited Cdh1 dephosphorylation or activation (Fig. 5E). Although Chk2 may provide some function when Chk1 is compromised, we found that Chk2 knockdown does not affect the response to mitotic DNA damage (data not shown).
Fig. 5.
Brca1 and Chk1 mediate the mitotic exit DNA damage checkpoint. (A) Western blot analysis of the levels of Brca1 (Left) and Chk1 (Right) in the asynchronously growing control and knockdown cells. (B) FACS profiles of the control (Top), Brca1 knockdown (Middle), and Chk1 (Bottom) knockdown cells. The cells were arrested with nocodazole and mitotic cells were collected, irradiated, and released for 2.5 h. (C) Western blot analysis of cyclin B1 in the control (Upper) and Chk1 (Lower) knockdown cells. The cells were treated the same way as in B. (D) Quantitation of the data in C. (E) Western blot analysis of Cdh1 in cells from C. Notice the disappearance of the slower migrating form of Cdh1 in the irradiated samples.
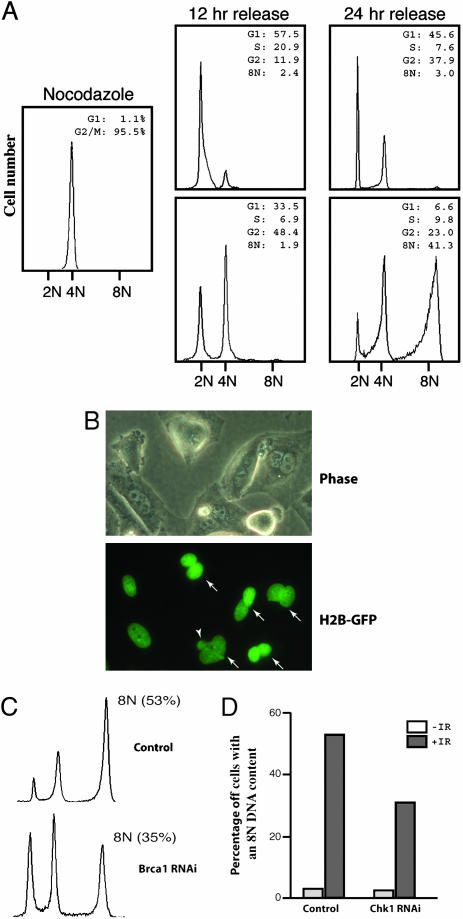
Mitotic DNA Damage-Induced Binucleation Depends on the Function of Mitotic Exit DNA Damage Checkpoint. Long-term observation of the irradiated mitotic cells indicated that the 4N DNA-containing cells (Fig. 1A) delayed not just mitotic exit but also failed to complete mitoses and become 2N G1 cells because, by 12 h after the damage and release, there were still ≈50% of the cells with a 4N DNA content (Fig. 6A), similar to the proportion at 4 h after the release (Fig. 1A). These 4N DNA-containing cells started new DNA synthesis without cytokinesis and reached the G2 phase with an 8N DNA content by 24 h (Fig. 6A). Microscopic examination indicated that DNA damage induced the formation of binucleated cells (Fig. 6B). Some binucleated cells also contained micronuclei, and sometimes the two nuclei were fused together forming an odd-shaped multilobulated nucleus (Fig. 6 B and Fig. 8 Left, which is published as supporting information on the PNAS web site). The binucleated cells started to appear by 4 h after the release. By 12 h, ≈50% of the irradiated population were binucleated (Fig. 8 Center), accounting for those showing 4N DNA content on the FACS profile. In contrast, there were <10% of binucleated cells in the unirradiated control population (Fig. 8 Center). Binucleation, as well as altered nuclear morphologies, could also be observed in similarly treated BJ cells (Fig. 8 Left and Center).
Fig. 6.
DNA damage-induced mitotic catastrophe depends on mitotic exit DNA damage checkpoint. (A) FACS analysis of the long-term released HeLa cells before and after γ-irradiation. HeLa cells were arrested with nocodazole for 18 h, and the mitotic cells were collected through mechanical detachment, irradiated, released, and analyzed at 2.5 h. (B) Micrographic pictures showing the binucleated histone H2B-GFP cells taken at 12 h after the irradiation and release. Arrows indicate binucleated cells, and the arrowhead indicates a micronucleus. (C) FACS profiles at 48 h of the control (Upper) and Brca1 knockdown (Lower) cells. The cells were treated and analyzed the same way as in A. (D) A diagram showing the accumulation of DNA damage-induced 8N DNA-containing cells in the control and Chk1-depleted cells 48 h after irradiation and release. The cells were treated and analyzed the same way as in A.
It has been reported that mitotic catastrophe results in a p53-dependent tetraploid G1 arrest (12, 13). However, the binucleated/tetraploid HeLa cells could not stay arrested, probably because they lack p53 function, entered the cell cycle again, and became 8N DNA-containing by 24 h after the release (Fig. 6A and Fig. 8 Right). In contrast, BJ cells do not become octoploid (the tetraploid G2 state) after mitotic DNA damage (Fig. 8 Right) nor do the p53-proficient HCT116 cells (12, 13).
These data indicate that DNA damage not just delays mitotic exit but blocks it, leading to the formation of binucleated cells, in other words, mitotic catastrophe. Because more Chk1 or Brca1 knockdown than the control cells completed mitosis after DNA damage when compared with the control cells (Fig. 5B), fewer of the knockdown cells became octoploid by 24 h after the release (Fig. 6 C and D). Thus, when the function of Chk1 or Brca1 was compromised, more cells escaped the DNA damage-induced mitotic catastrophe, demonstrating that the mitotic exit DNA damage checkpoint regulates mitotic catastrophe.
Discussion
DNA damage, especially double-strand breaks (DSBs), may cause very different consequence in mitosis than in other phases of the cell cycle. For instance, because the chromosomes are being pulled apart, the broken chromosome ends generated by DSBs won't be able to stay in close proximity for repairing and rejoining during mitosis. In fact, repairing DNA damage while the DNA is condensed and is being pulled apart might even be mechanically impossible. Thus, loss or gain of the broken arms is a real risk for cells that have suffered mitotic DNA damage or entered mitosis with damaged DNA. Indeed, lagging chromosomes could often been seen in anaphase/telophase cells after DNA damage. To solve the problem, cells undergo catastrophic mitosis after a delay in mitotic exit and become binucleated to avoid the risk of missegregation and the generation of aneuploid daughter cells. The tetraploid/binucleated cells are then prevented from cycling again in a p53- and p21-dependent manner (12, 13), thereby preserving genomic integrity. If the mitotic DNA damage is too severe, cells may choose to die through apoptosis or necrosis (14).
There have been several reports describing the effects of mitotic DNA damage in mammalian cells, including the delay of mitotic exit and metaphase arrest (32–34). However, it had never been demonstrated that the observed effects are caused by DNA damage checkpoint activation nor had the connection between mitotic DNA damage checkpoint and mitotic catastrophe been established. By showing the dependence of the DNA damage-induced mitotic exit delay on Brca1 and Chk1 function, we demonstrate that this delay is a genuine DNA damage checkpoint response, thus establishing the existence of a mitotic exit DNA damage checkpoint in mammalian cells. We have shown that the activation of mitotic exit DNA damage checkpoint correlates with the inhibition of the activation of Cdh1. When the checkpoint function was compromised, fewer cells responded to mitotic DNA damage with mitotic exit delays and, thus, fewer mitotic catastrophes were observed in Chk1 or Brca1 depleted cells, indicating that the mitotic exit DNA damage checkpoint regulates DNA damage-induced mitotic catastrophe.
Mitotic catastrophe is a well documented phenomenon that occurs in tumor cells treated with DNA damaging agents (14, 35–38) and is thought to be the primary means of tumor cell death resulted from DNA damaging therapeutic agents (14). These anticancer agents can also suppress tumor growth by inducing senescence-like growth arrest (39–41). The senescence-like phenotype is largely p53-dependent (40). It is therefore possible that this senescence-like growth arrest is actually what follows mitotic catastrophe or binucleation. Indeed, Chang et al. (41) found that the so-arrested colon cancer cells (HCT116) were tetraploid.
At the moment, we do not know exactly how the mitotic exit DNA damage checkpoint delays mitotic exit. In budding yeast, there are two mitotic DNA damage checkpoints, one that institutes a Pds1-dependent metaphase arrest (42–48) and the other blocks mitotic exit through the activation of Bub2 (49, 50), a negative regulator of the mitotic exit network. It is possible that there is an unidentified mammalian Bub2 homologue that regulates mitotic exit, and its activity is modulated by Chk1. Furthermore, it is unclear whether the checkpoint also facilitates the termination of mitosis. Binucleation is a natural process during animal development. For example, cardiac myocytes undergo a last round of cell cycle without cytokinesis, producing binucleated cells (51). These cells actually attempt cytokinesis as they do form a contractile ring but eventually quit dividing (52, 53). Therefore, there must be mechanism(s) that cells use to terminate mitosis and become binucleated, and this same mechanism(s) may be used by the Chk1-mediated mitotic exit DNA damage checkpoint to induce mitotic catastrophe.
Supplementary Material
Acknowledgments
We thank Dr. S. Sazer for critical reading of the manuscript; Dr. G. Nolan (Stanford University, Stanford, CA) for providing retrovirus packaging cell lines; Dr. J. Jin (Harvard Medical School, Boston) and Dr. Y. Wang (Florida State University, Tallahassee) for providing reagents and helpful discussions; Dr. Jun Qin (Baylor College of Medicine) for providing many useful reagents; and J. P. York for excellent technical support. P.Z. is supported by grants from the National Institutes of Health and the March of Dimes Foundation.
Abbreviations: APC, anaphase-promoting complex; Cdk, cyclin-dependent kinase.
References
- 1.Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 2.Nyberg, K. A., Michelson, R. J., Putnam, C. W. & Weinert, T. A. (2002) Annu. Rev. Genet. 36, 617–656. [DOI] [PubMed] [Google Scholar]
- 3.Shiloh, Y. (2003) Nat. Rev. Cancer 3, 155–168. [DOI] [PubMed] [Google Scholar]
- 4.Deng, C., Zhang, P., Harper, J. W., Elledge, S. J. & Leder, P. (1995) Cell 82, 675–684. [DOI] [PubMed] [Google Scholar]
- 5.Almasan, A., Linke, S. P., Paulson, T. G., Huang, L. C. & Wahl, G. M. (1995) Cancer Metastasis Rev. 14, 59–73. [DOI] [PubMed] [Google Scholar]
- 6.Elledge, S. J. (1996) Science 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- 7.Agami, R. & Bernards, R. (2000) Cell 102, 55–66. [DOI] [PubMed] [Google Scholar]
- 8.Peng, C. Y., Graves, P. R., Thoma, R. S., Wu, Z., Shaw, A. S. & Piwnica-Worms, H. (1997) Science 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez, Y., Wong, C., Thoma, R. S., Richman, R., Wu, Z., Piwnica-Worms, H. & Elledge, S. J. (1997) Science 277, 1497–1501. [DOI] [PubMed] [Google Scholar]
- 10.Molz, L., Booher, R., Young, P. & Beach, D. (1989) Genetics 122, 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinert, T. A. & Hartwell, L. H. (1988) Science 241, 317–322. [DOI] [PubMed] [Google Scholar]
- 12.Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J. P., Sedivy, J. M., Kinzler, K. W. & Vogelstein, B. (1998) Science 282, 1497–1501. [DOI] [PubMed] [Google Scholar]
- 13.Andreassen, P. R., Lacroix, F. B., Lohez, O. D. & Margolis, R. L. (2001) Cancer Res. 61, 7660–7668. [PubMed] [Google Scholar]
- 14.Roninson, I. B., Broude, E. V. & Chang, B. D. (2001) Drug Resist. Updat. 4, 303–313. [DOI] [PubMed] [Google Scholar]
- 15.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Cancer Cell 2, 243–247. [DOI] [PubMed] [Google Scholar]
- 16.Kanda, T., Sullivan, K. F. & Wahl, G. M. (1998) Curr. Biol. 8, 377–385. [DOI] [PubMed] [Google Scholar]
- 17.Mei, J., Huang, X. & Zhang, P. (2001) Curr. Biol. 11, 1197–1201. [DOI] [PubMed] [Google Scholar]
- 18.Visintin, R., Prinz, S. & Amon, A. (1997) Science 278, 460–463. [DOI] [PubMed] [Google Scholar]
- 19.Fang, G., Yu, H. & Kirschner, M. W. (1998) Mol. Cell 2, 163–171. [DOI] [PubMed] [Google Scholar]
- 20.Fang, G., Yu, H. & Kirschner, M. W. (1998) Genes Dev. 12, 1871–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yarden, R. I., Pardo-Reoyo, S., Sgagias, M., Cowan, K. H. & Brody, L. C. (2002) Nat. Genet. 30, 285–289. [DOI] [PubMed] [Google Scholar]
- 22.Scully, R., Chen, J., Plug, A., Xiao, Y., Weaver, D., Feunteun, J., Ashley, T. & Livingston, D. M. (1997) Cell 88, 265–275. [DOI] [PubMed] [Google Scholar]
- 23.Wang, B., Matsuoka, S., Carpenter, P. B. & Elledge, S. J. (2002) Science 298, 1435–1438. [DOI] [PubMed] [Google Scholar]
- 24.Lotti, L. V., Ottini, L., D'Amico, C., Gradini, R., Cama, A., Belleudi, F., Frati, L., Torrisi, M. R. & Mariani-Costantini, R. (2002) Genes Chromosomes Cancer 35, 193–203. [DOI] [PubMed] [Google Scholar]
- 25.King, R. W., Deshaies, R. J., Peters, J. M. & Kirschner, M. W. (1996) Science 274, 1652–1659. [DOI] [PubMed] [Google Scholar]
- 26.Morgan, D. O. (1999) Nat. Cell Biol. 1, E47–E53. [DOI] [PubMed] [Google Scholar]
- 27.Hagting, A., Den Elzen, N., Vodermaier, H. C., Waizenegger, I. C., Peters, J. M. & Pines, J. (2002) J. Cell Biol. 157, 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCollum, D. & Gould, K. L. (2001) Trends Cell Biol. 11, 89–95. [DOI] [PubMed] [Google Scholar]
- 29.Yeong, F. M., Lim, H. H. & Surana, U. (2002) Bioessays 24, 659–666. [DOI] [PubMed] [Google Scholar]
- 30.Kramer, E. R., Scheuringer, N., Podtelejnikov, A. V., Mann, M. & Peters, J. M. (2000) Mol. Biol. Cell 11, 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bembenek, J. & Yu, H. (2001) J. Biol. Chem. 276, 48237–48242. [DOI] [PubMed] [Google Scholar]
- 32.Iwai, M., Hara, A., Andoh, T. & Ishida, R. (1997) FEBS Lett. 406, 267–270. [DOI] [PubMed] [Google Scholar]
- 33.Smits, V. A., Klompmaker, R., Arnaud, L., Rijksen, G., Nigg, E. A. & Medema, R. H. (2000) Nat. Cell Biol. 2, 672–676. [DOI] [PubMed] [Google Scholar]
- 34.Mikhailov, A., Cole, R. W. & Rieder, C. L. (2002) Curr. Biol. 12, 1797–1806. [DOI] [PubMed] [Google Scholar]
- 35.Bernhard, E. J., McKenna, W. G. & Muschel, R. J. (1999) Cancer J. Sci. Am. 5, 194–204. [PubMed] [Google Scholar]
- 36.Jonathan, E. C., Bernhard, E. J. & McKenna, W. G. (1999) Curr. Opin. Chem. Biol. 3, 77–83. [DOI] [PubMed] [Google Scholar]
- 37.Tounekti, O., Pron, G., Belehradek, J., Jr., & Mir, L. M. (1993) Cancer Res. 53, 5462–5469. [PubMed] [Google Scholar]
- 38.Lock, R. B. & Stribinskiene, L. (1996) Cancer Res. 56, 4006–4012. [PubMed] [Google Scholar]
- 39.Chang, B. D., Broude, E. V., Dokmanovic, M., Zhu, H., Ruth, A., Xuan, Y., Kandel, E. S., Lausch, E., Christov, K. & Roninson, I. B. (1999) Cancer Res. 59, 3761–3767. [PubMed] [Google Scholar]
- 40.Chang, B. D., Xuan, Y., Broude, E. V., Zhu, H., Schott, B., Fang, J. & Roninson, I. B. (1999) Oncogene 18, 4808–4818. [DOI] [PubMed] [Google Scholar]
- 41.Chang, B. D., Swift, M. E., Shen, M., Fang, J., Broude, E. V. & Roninson, I. B. (2002) Proc. Natl. Acad. Sci. USA 99, 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agarwal, R., Tang, Z., Yu, H. & Cohen-Fix, O. (2003) J. Biol. Chem. 278, 45027–45033. [DOI] [PubMed] [Google Scholar]
- 43.Gardner, R., Putnam, C. W. & Weinert, T. (1999) EMBO J. 18, 3173–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen-Fix, O. & Koshland, D. (1999) Genes Dev. 13, 1950–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cohen-Fix, O. & Koshland, D. (1997) Proc. Natl. Acad. Sci. USA 94, 14361–14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez, Y., Bachant, J., Wang, H., Hu, F., Liu, D., Tetzlaff, M. & Elledge, S. J. (1999) Science 286, 1166–1171. [DOI] [PubMed] [Google Scholar]
- 47.Wang, H., Liu, D., Wang, Y., Qin, J. & Elledge, S. J. (2001) Genes Dev. 15, 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tinker-Kulberg, R. L. & Morgan, D. O. (1999) Genes Dev. 13, 1936–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Y., Hu, F. & Elledge, S. J. (2000) Curr. Biol. 10, 1379–1382. [DOI] [PubMed] [Google Scholar]
- 50.Hu, F., Wang, Y., Liu, D., Li, Y., Qin, J. & Elledge, S. J. (2001) Cell 107, 655–665. [DOI] [PubMed] [Google Scholar]
- 51.Soonpaa, M. H., Kim, K. K., Pajak, L., Franklin, M. & Field, L. J. (1996) Am. J. Physiol. 271, H2183–H2189. [DOI] [PubMed] [Google Scholar]
- 52.Li, F., Wang, X. & Gerdes, A. M. (1997) J. Mol. Cell. Cardiol. 29, 1553–1565. [DOI] [PubMed] [Google Scholar]
- 53.Li, F., Wang, X., Bunger, P. C. & Gerdes, A. M. (1997) J. Mol. Cell. Cardiol. 29, 1541–1551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.