Abstract
Drosophila peptidoglycan recognition protein LC (PGRP-LC), a transmembrane protein required for the response to bacterial infection, acts at the top of a cytoplasmic signaling cascade that requires the death-domain protein Imd and an IκB kinase to activate Relish, an NF-κB family member. It is not clear how binding of peptidoglycan to the extracellular domain of PGRP-LC activates intracellular signaling because its cytoplasmic domain has no homology to characterized proteins. Here, we demonstrate that PGRP-LC binds Imd and that its cytoplasmic domain is critical for its activity, suggesting that PGRP-LC acts as a signal-transducing receptor. The PGRP-LC cytoplasmic domain is also essential for the formation of dimers, and results suggest that dimerization may be required for receptor activation. The PGRP-LC cytoplasmic domain can mediate formation of heterodimers between different PGRP-LC isoforms, thereby potentially expanding the diversity of ligands that can be recognized by the receptor.
Keywords: Imd, innate immunity, NF-κB, Relish
Pattern-recognition receptors (PRRs) are host innate immune receptors that recognize microbial-specific molecules called pathogen-associated molecular patterns (PAMPs) and activate host defense responses (1). The best-characterized PRRs are the mammalian Toll-like receptor (TLR) family, in which each TLR appears to recognize and mediate the response to a different PAMP. The founding member of the TLR gene family is Drosophila Toll, which is required for the response of Drosophila to fungal and Gram-positive bacterial infections (2–4). In contrast to the mammalian TLRs, Toll does not appear to act as a PRR; pathogen recognition leads to activation of a protease cascade that produces a ligand for Toll (5, 6).
The PRRs that have been identified in Drosophila belong to a different protein family, the peptidoglycan-recognition proteins (PGRPs), which bind bacterial peptidoglycan. The first PGRP to be characterized was a silkworm (Bombyx mori) protein that is required for one branch of the insect immune response: the phenoloxidase cascade that leads to localized melanization at infection sites (7, 8). Subsequently, genes encoding PGRPs of related structure were identified in other animals, from Drosophila to humans (9–11). These proteins all include a peptidoglycan-recognition domain of ≈165 aa that has structural similarity to the peptidoglycan-binding region of lysozyme (12).
The essential role of PGRPs in the Drosophila immune response was revealed by characterization of immunodeficient mutants. Two genes that encode PGRPs, PGRP-SA and PGRP-LC, are required in two different branches of the immune response. PGRP-SA is a soluble protein that is present in the hemolymph (the blood plasma equivalent) and is required for the normal response to Gram-positive bacteria (13). Binding of Gram-positive type peptidoglycan to PGRP-SA (10), together with Gram-negative binding protein 1 (GNBP1) (14), activates the protease cascade that produces the mature ligand for Toll (6, 15). Toll activation promotes nuclear translocation of the Rel/NF-κB proteins Dif and Dorsal, which induce expression of the antimicrobial peptide Drosomycin. PGRP-LC is a transmembrane protein that is required for the response to bacterial infection mediated by the Imd-Relish signaling pathway (16–18). PGRP-LC acts genetically upstream of Imd to promote processing and nuclear localization of Relish, the third Drosophila Rel/NF-κB protein. Activated Relish promotes transcription of genes encoding a distinct set of antibacterial peptides, including Diptericin (Dpt) and CecropinA1 (CecA1) (16, 19). The three major isoforms of the PGRP-LC gene (PGRP-LCa, PGRP-LCx, and PGRP-LCy) are generated by alternative splicing (10, 16, 20). These isoforms share common cytoplasmic and transmembrane domains, but the extracellular PGRP domains are only 39% identical, and they appear to be activated by different forms of peptidoglycan (21).
Despite the genetically defined function of PGRP-LC, the cytoplasmic domain of PGRP-LC has no sequence homology to characterized proteins and its biochemical function is unknown. In this article, we express full-length and truncated forms of PGRP-LC and Imd to define the functions and interactions of protein domains. Our studies demonstrate that activated PGRP-LC is present as multimeric complexes, that the cytoplasmic domain of the protein of PGRP-LC is necessary and sufficient to activate downstream signaling, and that PGRP-LC binds a previously uncharacterized domain of Imd.
Methods
DNA Constructs. cDNA clones for imd (GH20785) and PGRP-LCa (LP06704) were purchased from Research Genetics (Huntsville, AL). PGRP-LCx cDNA is described in ref. 16. All imd constructs were subcloned into EcoRI–NotI sites of pMT/V5-HisA vector (Invitrogen) to generate V5 and 6× His C-terminally tagged proteins. The following primer sets were used in PCR amplification of different imd constructs: primers 1 and 5 for the full-length Imd (see the primer list below); primer 2 and 5 for ImdΔ1–135; primers 1 and 3 for ImdΔ136–273; and primers 1 and 4 for ImdΔ173–273. For c-Myc-tagged PGRP-LCa and LCx, constructs were subcloned into EcoRI–PmeI sites of pMT/V5-HisA, which resulted in the deletion of vector-originated V5 and 6× His tags. The following primer sets were used for C-terminal c-Myc tagging: primers 6 and 7 for PGRP-LCa; and primers 6 and 8 for PGRP-LCx. For V5 6× His-tagged full-length and deletion constructs for PGRP-LCa and PGRP-LCx, constructs were subcloned into EcoRI–NotI sites of pMT/V5-HisA. The following sets of primers were used for PCR amplification: primers 6 and 9 for PGRP-LCa; primers 6 and 10 for PGRP-LCx; primers 11 and 10 for PGRP-LCxΔ1–144; primers 12 and 10 for PGRP-LCxΔ1–263; primers 6 and 13 for PGRP-LCxΔ333–500; and primers 6 and 14 for PGRP-LCxΔ394–500. Primer 1 (forward), 5′-CCGA AT TCATGTCA A AGCTCAGGA ACCTGT-3′; primer 2 (forward), 5′-GGTTACGAATTCATGCGAAAGGGTAGCACCAGTACCG-3′; primer 3 (reverse), 5′-GCTAATGCGGCCGCGGCGAGCTGCAGGCGCTCAAGTTT-3′; primer 4 (reverse), 5′-GCTAATGCGGCCGCGGTTGTGACTGCATCATGGCCACA-3′; primer 5 (reverse), 5′-TTGCGGCCGCGGGCTGTTTGTCTTGCGCTTCTCC-3′; primer 6 (forward), 5′-CCGAATTCATGCCTTTTAGCAATGAAACGG-3′; primer 7 (reverse), 5′-GGCCGGGTTTAAACTTATTACAAGTCCTCTTCAGAAATGAGCTTTTGCTCCGACCAATGAGTCCAGTTGGC-3′; primer 8 (reverse), 5′-GGCCGGGTTTAAACTTATTACAAGTCCTCT TCAGA A ATGAGCTTTTGCTCGATTTCGTGTGACCAGTGCGG-3′; primer 9 (reverse), 5′-TTGCGGCCGCCCCGACCA ATGAGTCCAGT TGGCGA AGCT TGC-3′; primer 10 (reverse), 5′-TTGCGGCCGCCCGATTTCGTGTGACCAGTGCGGCCACTT-3′; primer 11 (forward), 5′-CCGAATTCATGAGTTCACACCTGCGCGACCTTAA-3′; primer 12 (forward), 5′-CCGAATTCATGGGAAGTGCGCCGGGCTCCAAACAC-3′; primer 13 (reverse), 5′-TTGCGGCCGCCCGCTATTATCGATGACATCCAAGT-3′; and primer 14 (reverse), 5′-TTGCGGCCGCCCACCCCAGCTGTCCATGTGAAACG-3′.
S2 Cell Culture and Transfection. Maintenance and transient transfection of Drosophila S2 cells were performed by following the manufacturer's suggested protocols with no modification (DES-Inducible kit, Invitrogen). S2 cells were cultured and passaged every 4 or 5 days in Schneider's Drosophila medium supplemented with 10% heat-inactivated FBS, 50 units/ml penicillin G, and 50 μg/ml streptomycin sulfate (Invitrogen). Transient transfection was performed by using the calcium phosphate method with 20 μg of DNA. Induction of genes under the control of the metallothionein promoter was performed by adding CuSO4 to the medium at the final concentration of 500 μM at 48–72 h or 48–96 h after transfection.
Immune Challenge. Escherichia coli cells were cultured overnight until OD600 reached 1.5. E. coli bacteria were washed and resuspended in PBS (pH 7.5) and added to S2 cell culture at an appropriate concentration (see Fig. 1 legend). S2 cells were harvested 6 h after incubation. Injections of adult flies with E. coli were carried out as described in ref. 22.
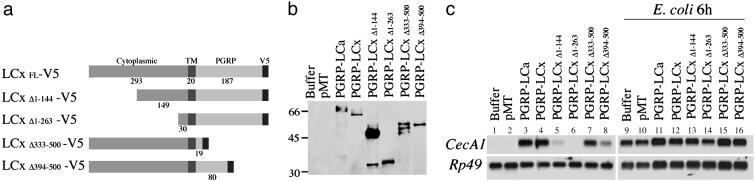
Fig. 1.
Domains of PGRP-LCx required for antimicrobial peptide gene induction. (a) PGRP-LC deletion constructs. PGRP-LC is a type II transmembrane protein with a previously uncharacterized N-terminal cytoplasmic domain, a transmembrane domain (TM) and a C-terminal PGRP domain. A V5 epitope was added in the C terminus of full-length and deletion constructs. (b) Protein expression in transiently transfected S2 cells was confirmed by Western blotting with anti-V5 antibody. (c) Induction of CecA1 was measured on Northern blots with RNA samples prepared from S2 cells expressing the PGRP-LCx deletion constructs. RNAs were purified from the cells after protein expression and, as shown in lanes 9–16, treatment with E. coli (102-fold dilution of OD600 = 1.5, 6 h of incubation). Rp49 was used as a loading control.
Coimmunoprecipitation and Western Blot Analysis. Lysates were prepared from 10-ml cultures of transiently transfected cells by adding 1 ml RIPA buffer containing 1× PBS, 1% Igepal CA-630 (Sigma–Aldrich), 0.5% sodium deoxycholate, 0.1% SDS with phosphatase inhibitor (100 μM sodium orthovanadate), and protease inhibitors (Roche Applied Science). Lysates were precleared by adding 50 μl of 1:1 slurry of protein A–Sepharose in PBST (PBS, pH 7.5/1% Triton-X-100) containing 1 mg/ml BSA, followed by rocking for 30 min at 4°C and centrifugating the samples for 5 s at 15,000 × g. The resulting supernatants were transferred to fresh tubes, and 50 μl of protein A–Sepharose was added with 2 μg of anti-c-Myc rabbit polyclonal IgG (Research Diagnostics, Flanders, NJ) or 2 μg of anti-V5 mouse monoclonal IgG (Invitrogen). Samples were then incubated overnight at 4°C with rocking. Immunoprecipitates were washed with RIPA buffer four times each for 10 min, separated by 12.5% SDS/PAGE, and transferred to nitrocellulose. Blots were probed with either the anti-V5 or the anti-c-Myc antibodies (1:2,000 dilution) overnight.
Northern Blot Analysis for the Induction of Antimicrobial Peptides. The 5-ml cultures of S2 cells were harvested after protein expression and immune challenge as described above. RNAs were prepared by homogenizing the cells in 1 ml of RNA STAT-60 solution (Tel-Test, Friendswood, TX). Subsequent RNA purification steps were performed by following the manufacturer's instructions. Northern blot analyses were performed as described (22, 23).
Results
The Cytoplasmic Domain of PGRP-LC Is Required for Downstream Signaling. To study the activities and physical properties of the PGRP-LC isoforms, we constructed and expressed V5-tagged and c-Myc-tagged full-length PGRP-LCa and PGRP-LCx in Drosophila S2 cells (a macrophage-like cell line). The predicted sizes of PGRP-LCa and PGRP-LCx proteins were 57 and 55 kDa (16), and each isoform migrated as a cluster of three bands of ≈66 kD (Fig. 1b, lanes 3 and 4), possibly the result of posttranslational modification.
The antibacterial peptide gene CecropinA1 (CecA1), which is induced in response to infection of Drosophila larvae or adults, is also induced by overexpression of PGRP-LCa or PGRP-LCx in transgenic flies (16, 17). Activation of CecA1 by PGRP-LC overexpression requires the same downstream gene products required for the response to bacterial infection. Overexpression of epitope-tagged full-length PGRP-LCa or PGRP-LCx in S2 cells induced expression of CecA1 to the same level as incubation of the cells with E. coli for 6 h (Fig. 1c, lanes 3 and 4 vs. 9 and 10; Myc-tagged PGRP-LCa and PGRP-LCx-induced CecA1 to similar levels).
We used the activation of CecA1 expression in S2 cells as an assay to map the domains of PGRP-LC that are important for activation of downstream signaling. PGRP-LCx encodes a type II transmembrane protein with a 293-aa N-terminal cytoplasmic domain and a 187-aa C-terminal extracellular domain that includes the peptidoglycan-recognition domain (Fig. 1a). Constructs that lacked the N-terminal half of the cytoplasmic region (PGRP-LCΔ1–144) showed greatly reduced induction of CecA1, and constructs that lacked 90% of the cytoplasmic region (PGRP-LCΔ1–263) failed completely to induce CecA1 (Fig. 1c, lanes 5 and 6). In contrast, overexpression of constructs that lacked 90% of the extracellular domain, including the entire peptidoglycan-recognition domain (PGRP-LCΔ333–500), induced CecA1 expression to levels comparable with that seen with full-length PGRP-LC (Fig. 1c, lane 7). Thus, the membranetethered cytoplasmic domain, but not the extracellular PGRP domain, is crucial for activation of the downstream events that lead to CecA1 induction in response to PGRP-LC overexpression.
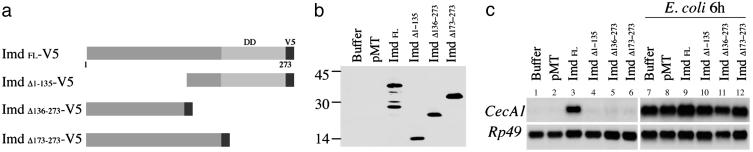
Physical Interactions Between PGRP-LC and Imd, a Putative Adaptor Protein. The death-domain protein Imd functions at the top of the genetically defined cytoplasmic signaling cascade required for the activation of the Relish transcription factor (24–27). Because epistasis analyses demonstrated that Imd acts downstream of the transmembrane protein PGRP-LC (17), we tested whether PGRP-LC and Imd could be components of the same protein complex. V5-tagged Imd expressed in Drosophila S2 cells migrated as five major bands ranging in size from 20 to 40 kDa (Fig. 2b); based on the migration of truncated forms of the protein (see below), it is likely that the 40-kD protein represents full-length Imd (28). Overexpression of epitope-tagged Imd induced CecA1 expression, as had been observed previously in both Imd-overexpressing transgenic flies and S2 cells (28). The Imd protein includes an 80-aa C-terminal death domain and a 173-aa N-terminal region that shows no significant homology to other proteins (28). We expressed three deletion constructs of Imd (Fig. 2a) in S2 cells. The constructs deleted the N-terminal half, the C-terminal half including the death domain, and only the death domain: ImdΔ1–135, ImdΔ136–273 and ImdΔ173–273. Although each deletion construct produced stable protein, none of the deleted versions of Imd induced CecA1 expression (Fig. 2c, lanes 4–6), indicating that both N-terminal and C-terminal domains are essential in this assay.
Fig. 2.
Domains of Imd required for antimicrobial peptide gene induction. (a) Imd deletion constructs. The C-terminal 80-aa death domain (DD) is shown. V5 tag was added in the C terminus of full-length and deletion constructs. (b) Protein expression in transiently transfected S2 cells was confirmed by Western blotting with anti-V5 antibody. (c) Induction of CecA1 was measured on Northern blots. Deletion of the N-terminal half, the C-terminal half including the death domain, or only the death domain all abolished CecA1 induction.
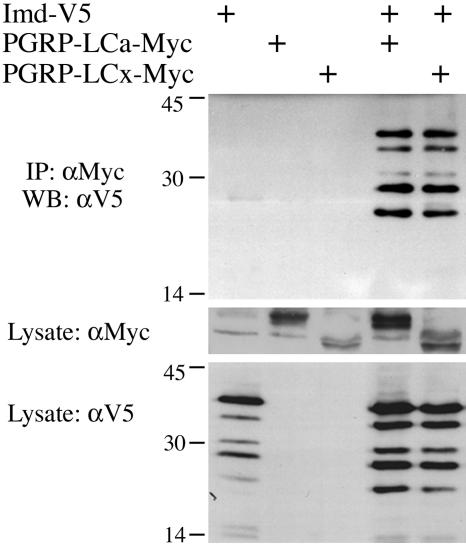
To test for physical associations between PGRP-LC and Imd, we coexpressed epitope-tagged Imd and PGRP-LC isoforms in S2 cells. As shown in Fig. 3, Imd coprecipitated with either PGRP-LCa or PGRP-LCx. Similar results were obtained in reciprocal coimmunoprecipitation assays in which Imd was immunoprecipitated and PGRP-LC was detected in the precipitate (data not shown).
Fig. 3.
Physical interaction of PGRP-LC and Imd. Lysates were prepared from S2 cells transiently transfected with expression vectors for V5-tagged Imd and c-Myc-tagged PGRP-LCa or PGRP-LCx (20 μg each). Anti-c-Myc antibody was used to immunoprecipitate PGRP-LC, and anti-V5 antibody was used to detect Imd on Western blots.
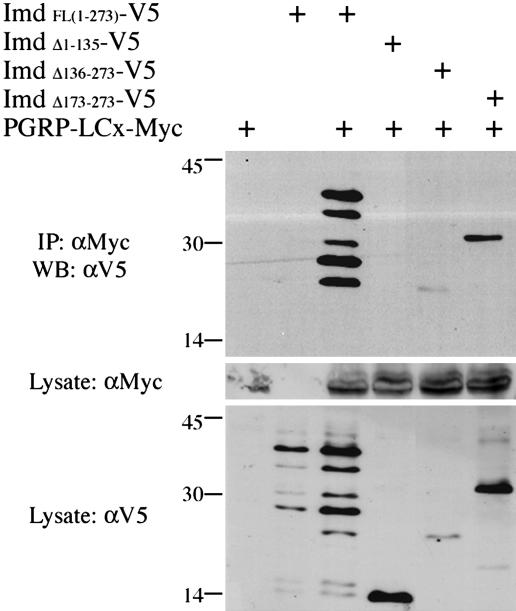
Imd constructs that lacked the C-terminal death domain (Δ173–273) or the C-terminal half of the protein (Δ136–273) still coimmunoprecipitated with PGRP-LCx (Fig. 4, lanes 5 and 6). In contrast, deletion of the N-terminal half of Imd (Δ1–135) abolished its ability to bind PGRP-LCx, defining the Imd N terminus as the essential PGRP-LC interaction domain (Fig. 4, lane 4). Similar results were observed in reciprocal coimmunoprecipitation assays (data not shown).
Fig. 4.
Identification of the domain of Imd that interacts with PGRP-LC. Lysates were prepared from cells transiently transfected with expression vectors for a V5-tagged Imd deletion construct and c-Myc-tagged PGRP-LCx (20 μg each). Anti-c-Myc antibody was used to immunoprecipitate the full-length PGRP-LCx, and anti-V5 antibody was used to detect Imd on Western blots. Deletion of the N-terminal half of Imd abolished the interaction, whereas deletion of the C-terminal half did not prevent interaction, suggesting that the N-terminal part is responsible for the interaction with PGRP-LC.
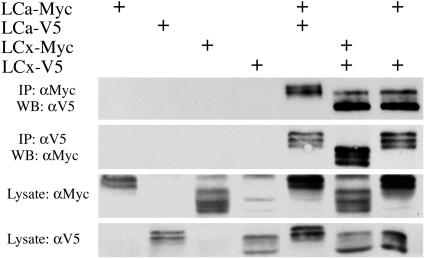
Homotypic and Heterotypic Interactions Between PGRP-LC Isoforms. Many transmembrane receptors, including some TLRs, function as dimers or oligomers (29–31). To test whether PGRP-LC can form multimeric complexes, we examined the ability of epitope-tagged PGRP-LC isoforms to coimmunoprecipitate. As shown in Fig. 5 (lanes 5 and 6), V5-tagged PGRP-LCa coimmunoprecipitated with c-Myc-tagged PGRP-LCa. The same results were obtained with LCx isoform, indicating that both isoforms are capable of homotypic interactions. In addition, coexpressed PGRP-LCa and PGRP-LCx coimmunoprecipitated, indicating that they are capable of heterotypic interaction (Fig. 5, lane 7).
Fig. 5.
Homotypic and heterotypic interactions between PGRP-LCa and PGRP-LCx. Lysates were prepared from cells transiently transfected with expression vectors for the two PGRP-LC isoforms tagged with either V5 or c-Myc (20 μg each) and subjected to coimmunoprecipitation.
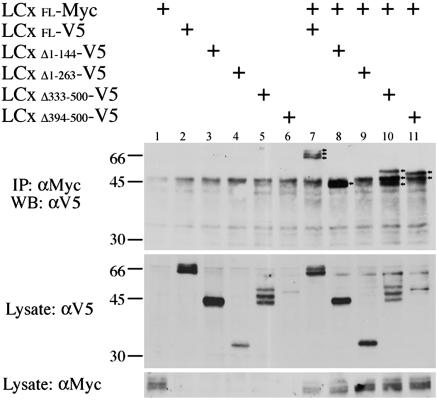
We tested the deletion constructs to identify the domains required for the interaction between PGRP-LCx monomers. Truncated constructs lacking the C-terminal half or 90% of the extracellular domain coimmunoprecipitated with full-length PGRP-LCx as efficiently as the full-length protein (Fig. 6, lanes 10 and 11). Truncated protein that lacked the membrane-distal (N-terminal) half of the cytoplasmic domain could interact with full-length PGRP-LCx as well as the full-length protein (Fig. 6, lane 8). In contrast, deletion of 90% of the cytoplasmic domain abolished the interaction, (Fig. 6, lane 9). Similar results were seen in reciprocal coimmunoprecipitation assays (data not shown). Thus, the membrane-proximal half of the cytoplasmic domain is required for the interaction between monomers.
Fig. 6.
Mapping of the region of PGRP-LCx required for homotypic interactions. Lysates derived from cells transiently transfected with expression vectors for a V5-tagged deletion construct and the c-Myc-tagged full-length PGRP-LCx (20 μg each) were prepared and subjected to coimmunoprecipitation. Anti-c-Myc antibody was used to immunoprecipitate the full-length PGRP-LCx and anti-V5 antibody was used to detect the deletion constructs of PGRP-LCx by Western blotting. Note that deletion of the extracellular domain did not significantly affect the interaction, whereas deletion of the cytoplasmic domain abolished it (arrows mark IP products in lanes 7, 8, 10, and 11). Deletion of the distal half of the cytoplasmic domain did not prevent the interaction (lane 8), suggesting that the proximal half of the cytoplasmic domain is crucial for the interaction.
Discussion
The data presented here demonstrate that PGRP-LC is an essential component of a signal-transducing receptor complex that presumably binds peptidoglycan PAMPs and initiates the cellular response to this PAMP. The other PGRP required in a Drosophila immune response, PGRP-SA, is a small, soluble protein that circulates in the hemolymph and is required for activation of a hemolymph protease cascade (13). In contrast, PGRP-LC is a transmembrane protein that acts as a PRR that directly couples pathogen recognition to intracellular signaling.
All isoforms of PGRP-LC share a common cytoplasmic domain that has no similarity to other proteins. Nevertheless, the deletion analysis presented here defines three functions for the previously uncharacterized cytoplasmic domain of PGRP-LC. First, the membrane-proximal half of the PGRP-LC cytoplasmic domain is required for formation of multimeric receptor complexes. Interactions between PGRP-LC isoforms have been detected (21), but it was not known which domains mediated interaction. As noted previously, the ability to form heteromeric complexes may broaden the spectrum of PAMPs that can be recognized. Second, PGRP-LC can form a complex with the cytoplasmic protein Imd. Our experiments demonstrate that the N-terminal domain of Imd is essential for its interaction with PGRP-LC, although that interaction need not be direct. Third, the PGRP-LC cytoplasmic domain is necessary and sufficient for activation of downstream signaling, as assayed by expression of CecA1. The results suggest that activation of signaling could involve the following sequence of events: binding of bacterial proteoglycan to PGRP-LC induces receptor multimerization (which can be mimicked by overexpression); and multimerization of the receptor activates the protein complex that includes both PGRP-LC and Imd, ultimately leading to activation of the IκB kinase (Ird5) and Relish activation.
Although PGRP-LC has no homology to TLRs, our data highlight parallels between the PGRP-LC/Imd and TLR pathways. TLR signaling requires a membrane complex that includes the TLR and death-domain protein MyD88; similarly, signaling from PGRP-LC occurs in a complex that includes the death-domain protein Imd. TLR activity causes activation of an IκB kinase complex that phosphorylates the inhibitor protein IκB (32); similarly, activation of PGRP-LC leads to activation of an IκB kinase complex that phosphorylates the Rel-Ank protein Relish. The adaptor molecule MyD88 has an N-terminal death domain and the C-terminal TIR domain (33–37). The MyD88 TIR domain binds the TIR domain of TLR and the MyD88 death domain binds the death domain of the kinase IRAK, thereby linking the membrane receptor to downstream events. Although the cytoplasmic domain of PGRP-LC is not similar to any other protein, our data suggest that it is required for interaction with the previously uncharacterized N-terminal domain of Imd. The C-terminal half (Δ132–273) of Imd contains a death domain and can interact with another death-domain protein, dFADD (27), which is required for downstream signaling and could be also a part of the receptor complex.
In animals, the immune-responsive tissues are the fat body, blood cells (38, 39), and epidermis (40–43). Although the interactions described here occur in Schneider cells, a hemocyte line, it is likely that PGRP-LC and Imd interact in the fat body and possibly the epidermis as well. Imd and PGRP-LC mutants have very similar effects on the antimicrobial peptide response in flies, and most of the antimicrobial peptides are produced in the fat body. Overexpression of PGRP-LC in the fat body is sufficient to drive antimicrobial peptide induction, indicating that the fat-body cells are ready to initiate the signaling upon the presence of active PGRP-LC (16, 17). The response to septic wounding that was used to define the phenotypes of both Imd and PGRP-LC mutants (16–18) can occur in the absence of blood cells (44). Imd also acts in the epidermis to allow a local response to septic wounding (42), and it is likely that PGRP-LC also acts at that site. Although we have provided information about the biochemical mechanism of action of PGRP-LC, additional work will be required to define the diversity of functions of PGRP-LC in systemic immune responses.
Acknowledgments
We thank Drs. K. Hong (Albert Einstein College of Medicine) and M. Povelones (Stanford University, Stanford, CA) for S2 cells; E. Espinoza, S. Guzman, and W.-S. Chen for technical support; C. A. Brennan and N. Matova for comments on the manuscript; K.V.A. laboratory members for helpful discussions; and T. R. Clandinin for intellectual support. This work was supported by National Institutes of Health Grant AI45149 (to K.V.A.) and the Lita Annenberg Hazen Foundation.
Author contributions: K.-M.C. and K.V.A. designed research; K.-M.C. and H.L. performed research; K.-M.C. contributed new reagents/analytic tools; K.-M.C., H.L., and K.V.A. analyzed data; and K.-M.C. and K.V.A. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PGRP, peptidoglycan-recognition protein; PAMP, pathogen-associated molecular pattern; TLR, Toll-like receptor; PRR, pattern-recognition receptor.
References
- 1.Janeway, C. A., Jr., (1989) Cold Spring Harbor Symp. Quant. Biol. 54, 1–13. [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M. & Hoffmann, J. A. (1996) Cell 86, 973–983. [DOI] [PubMed] [Google Scholar]
- 3.Leulier, F., Rodriguez, A., Khush, R. S., Abrams, J. M. & Lemaitre, B. (2000) EMBO Rep. 1, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutschmann, S., Kilinc, A. & Ferrandon, D. (2002) J. Immunol. 168, 1542–1546. [DOI] [PubMed] [Google Scholar]
- 5.Levashina, E. A., Langley, E., Green, C., Gubb, D., Ashburner, M., Hoffmann, J. A. & Reichhart, J. M. (1999) Science 285, 1917–1919. [DOI] [PubMed] [Google Scholar]
- 6.Ligoxygakis, P., Pelte, N., Hoffmann, J. A. & Reichhart, J. M. (2002) Science 297, 114–116. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida, H., Kinoshita, K. & Ashida, M. (1996) J. Biol. Chem. 271, 13854–13860. [DOI] [PubMed] [Google Scholar]
- 8.Ochiai, M. & Ashida, M. (1999) J. Biol. Chem. 274, 11854–11858. [DOI] [PubMed] [Google Scholar]
- 9.Kang, D., Liu, G., Lundstrom, A., Gelius, E. & Steiner, H. (1998) Proc. Natl. Acad. Sci. USA 95, 10078–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Werner, T., Liu, G., Kang, D., Ekengren, S., Steiner, H. & Hultmark, D. (2000) Proc. Natl. Acad. Sci. USA 97, 13772–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, C., Xu, Z., Gupta, D. & Dziarski, R. (2001) J. Biol. Chem. 276, 34686–34694. [DOI] [PubMed] [Google Scholar]
- 12.Kim, M. S., Byun, M. & Oh, B. H. (2003) Nat. Immunol. 4, 787–793. [DOI] [PubMed] [Google Scholar]
- 13.Michel, T., Reichhart, J. M., Hoffmann, J. A. & Royet, J. (2001) Nature 414, 756–759. [DOI] [PubMed] [Google Scholar]
- 14.Pili-Floury, S., Leulier, F., Takahashi, K., Saigo, K., Samain, E., Ueda, R. & Lemaitre, B. (2004) J. Biol. Chem. 279, 12848–12853. [DOI] [PubMed] [Google Scholar]
- 15.Weber, A. N., Tauszig-Delamasure, S., Hoffmann, J. A., Lelievre, E., Gascan, H., Ray, K. P., Morse, M. A., Imler, J. L. & Gay, N. J. (2003) Nat. Immunol. 4, 794–800. [DOI] [PubMed] [Google Scholar]
- 16.Choe, K.-M., Werner, T., Stoven, S., Hultmark, D. & Anderson, K. V. (2002) Science 296, 359–362. [DOI] [PubMed] [Google Scholar]
- 17.Gottar, M., Gobert, V., Michel, T., Belvin, M., Duyk, G., Hoffmann, J. A., Ferrandon, D. & Royet, J. (2002) Nature 416, 640–644. [DOI] [PubMed] [Google Scholar]
- 18.Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R. A. (2002) Nature 416, 644–648. [DOI] [PubMed] [Google Scholar]
- 19.Stoven, S., Ando, I., Kadalayil, L., Engstrom, Y. & Hultmark, D. (2000) EMBO Rep. 1, 347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werner, T., Borge-Renberg, K., Mellroth, P., Steiner, H. & Hultmark, D. (2003) J. Biol. Chem. 278, 26319–26322. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko, T., Goldman, W. E., Mellroth, P., Steiner, H., Fukase, K., Kusumoto, S., Harley, W., Fox, A., Golenbock, D. & Silverman, N. (2004) Immunity 20, 637–649. [DOI] [PubMed] [Google Scholar]
- 22.Wu, L. P., Choe, K.-M., Lu, Y. & Anderson, K. V. (2001) Genetics 159, 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, New York), 2nd Ed.
- 24.Vidal, S., Khush, R. S., Leulier, F., Tzou, P., Nakamura, M. & Lemaitre, B. (2001) Genes Dev. 15, 1900–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khush, R. S., Cornwell, W. D., Uram, J. N. & Lemaitre, B. (2002) Curr. Biol. 12, 1728–1737. [DOI] [PubMed] [Google Scholar]
- 26.Leulier, F., Vidal, S., Saigo, K., Ueda, R. & Lemaitre, B. (2002) Curr. Biol. 12, 996–1000. [DOI] [PubMed] [Google Scholar]
- 27.Naitza, S., Rosse, C., Kappler, C., Georgel, P., Belvin, M., Gubb, D., Camonis, J., Hoffmann, J. A. & Reichhart, J. M. (2002) Immunity 7, 575–581. [DOI] [PubMed] [Google Scholar]
- 28.Georgel, P., Naitza, S., Kappler, C., Ferrandon, D., Zachary, D., Swimmer, C., Kopczynski, C., Duyk, G., Reighhart, J. M., Hoffman, J. A., et al. (2001) Dev. Cell 1, 503–514. [DOI] [PubMed] [Google Scholar]
- 29.Wyllie, D. H., Kiss-Toth, E., Visintin, A., Smith, S. C., Boussouf, S., Segal, D. M., Duff, G. W. & Dower, S. K. (2000) J. Immunol. 165, 7125–7132. [DOI] [PubMed] [Google Scholar]
- 30.Hajjar, A. M., O'Mahony, D. S., Ozinsky, A., Underhill, D. M., Aderem, A., Klebanoff, S. J. & Wilson, C. B. (2001) J. Immunol. 166, 15–19. [DOI] [PubMed] [Google Scholar]
- 31.Bulut, Y., Faure, E., Thomas, L., Equils, O. & Arditi, M. (2001) J. Immunol. 167, 987–994. [DOI] [PubMed] [Google Scholar]
- 32.Anderson, K. V. (2000) Curr. Opin. Immunol. 12, 13–19. [DOI] [PubMed] [Google Scholar]
- 33.Muzio, M., Ni, J., Feng, P. & Dixit, V. M. (1997) Science 278, 1612–1615. [DOI] [PubMed] [Google Scholar]
- 34.Wesche, H., Henzel, W. J., Shillinglaw, W., Li, S. & Cao, Z. (1997) Immunity 7, 837–847. [DOI] [PubMed] [Google Scholar]
- 35.Medzhitov, R., Preston-Hurlburt, P., Kopp, E., Stadlen, A., Chen, C., Ghosh, S. & Janeway, C. A., Jr., Mol. Cell 2, 253–258. [DOI] [PubMed]
- 36.Horng, T. & Medzhitov, R. (2001) Proc. Natl. Acad. Sci. USA 98, 12654–12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun, H., Bristow, B. N., Qu, G. & Wasserman, S. A. (2002) Proc. Natl. Acad. Sci. USA 99, 12871–12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agaisse, H., Petersen, U. M., Boutros, M., Mathey-Prevot, B. & Perrimon, N. (2003) Dev. Cell 5, 441–450. [DOI] [PubMed] [Google Scholar]
- 39.Brennan, C. A. & Anderson, K. V. (2004) Annu. Rev. Immunol. 22, 457–483. [DOI] [PubMed] [Google Scholar]
- 40.Ferrandon, D., Jung, A. C., Criqui, M., Lemaitre, B., Uttenweiler-Joseph, S., Michaut, L., Reichhart, J. & Hoffmann, J. A. (1998) EMBO J. 17, 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzou, P., Ohresser, S., Ferrandon, D., Capovilla, M., Reichhart, J. M., Lemaitre, B., Hoffmann, J. A. & Imler, J. L. (2000) Immunity 13, 737–748. [DOI] [PubMed] [Google Scholar]
- 42.Onfelt Tingvall, T., Roos, E. & Engstrom, Y. (2001) EMBO Rep. 2, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramet, M., Lanot, R., Zachary, D. & Manfruelli, P. (2002) Dev. Biol. 241, 145–156. [DOI] [PubMed] [Google Scholar]
- 44.Basset, A., Khush, R. S., Braun, A., Gardan, L., Boccard, F., Hoffmann, J. A. & Lemaitre, B. (2000) Proc. Natl. Acad. Sci. USA 97, 3376–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]