The reaction of the ligand N 2,N 3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide with silver(I) nitrate led to the formation of a three-dimensional coordination polymer.
Keywords: crystal structure, carboxamide, pyrazine, pyridine, silver(I), Ag—Ag bond, three-dimensional coordination polymer, hydrogen bonding
Abstract
The title ligand, C18H16N6O2·2H2O (L1) [N 2,N 3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide], crystallized as a dihydrate. The molecule is U-shaped with the carboxamide groups being cis to one another, making a dihedral angle of 81.6 (5)°. The terminal pyridine rings are inclined to one another by 58.5 (4)°. There is an intramolecular N—H⋯Npyrazine hydrogen bond present, forming an S(5) ring motif. In the crystal, adjacent molecules are linked by N—H⋯Ocarboxamide hydrogen bonds, forming a chain along [001]. A chain of hydrogen-bonded water molecules is linked to the chain of (L1) molecules by O—H⋯N hydrogen bonds, forming columns propagating along the c axis. The columns are linked by C—H⋯O and C—H⋯N hydrogen bonds, forming a three-dimensional supramolecular structure. The reaction of ligand (L1) with silver(I) nitrate led to the formation of a new three-dimensional coordination polymer, {[Ag(C18H16N6O2)]NO3}n, poly[[[μ4-N 2,N 3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] (I). The asymmetric unit is composed of half of one silver ion, located on a twofold rotation axis, half a ligand molecule and half a positionally disordered nitrate anion located about a twofold rotation axis. The full molecule of the ligand is generated by twofold rotational symmetry, with this twofold axis bisecting the Car—Car bonds of the pyrazine ring and the Ag—Ag bond. The carboxamide groups are now trans to one another, making a dihedral angle of 65.8 (4)°. The two terminal pyridine rings are inclined to one another by 6.6 (3)°. Two ligands wrap around an Ag—Ag bond of 3.1638 (11) Å, forming a figure-of-eight-shaped complex molecule. Each silver ion is coordinated by two pyridine N atoms and by two carboxamide O atoms of neighbouring molecules, hence forming a three-dimensional framework. The nitrate anion is linked to the framework by N—H⋯O and C—H⋯O hydrogen bonds.
Chemical context
The title ligand, N 2,N 3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide (L1), is one of a series of ligands synthesized in order to study the superexchange in supramolecular complexes formed using pyrazine carboxamide derivatives and first row transition metal ions (Cati, 2002 ▸; Cati et al., 2004 ▸). To the best of our knowledge, neither the synthesis nor the crystal structure of (L1) have been described previously. It is very similar to the ligand N 2,N 3-bis(pyridin-2-ylmethyl)pyrazine-2,3-dicarboxamide (L2), for which a number of transition metal complexes have been described, including some interesting tetranuclear 2×2 grid-like and square complexes (Hausmann et al., 2003 ▸; Klingele et al., 2007 ▸). Two such complexes, {[Cu4(L2)4](ClO4)4}·5CH3OH·4H2O and {[Ni4(L2)4]Cl4}·5CH3CN·13H2O (Cati et al., 2004 ▸), exhibit anion encapsulation, and magnetic susceptibility measurements indicate that they are weakly anti-ferromagnetic, with J values of −5.87 and −2.64 cm−1, respectively.
A search of the Cambridge Structural Database (CSD; Groom et al., 2016 ▸), indicated that silver nitrate is an excellent metal salt for the formation of multi-dimensional coordination polymers. The silver ion can have multiple coordination geometries and modes, and the nitrate anion has been shown to coordinate to metal ions in a number of different modes, many of which involve bridging metal ions. The properties of the complexes formed are extremely varied. For example, with the tetradendate ligand 1,6-bis(2H-1,2,3-triazol-2-yl)hexane, Huo et al. (2016 ▸) synthesized the three-dimensional coordination polymer, catena-[[μ-2,2′-(butane-1,4-diyl)bis(2H-1,2,3-triazole)]bis(μ-nitrato)disilver]. They showed that it exhibits highly selective and sensitive luminescence sensing of Cr2O7
2− ions in aqueous solution. With the rigid tripodal arene-core-based nitrogen ligand, 1,3,5-tris(pyrazol-1-yl)benzene, Shu et al. (2006 ▸) formed a porous metal–organic framework, viz. catena-[bis(μ3-nitrato-O,O′,O′′)bis(μ3-1,3,5-tris(pyrazol-1-yl)benzene-N,N′,N′′)trisilver(I) nitrate]. The nitrate counter-anions located in the cationic framework can be exchanged reversibly without destruction of the structure. Hence, this compound can act as a zeolite-like porous material for anion exchange.
The title ligand has potentially two bidentate (N,N) and two monodentate (Npyridine) coordination sites. It is therefore an interesting ligand to study its coordination behaviour with silver nitrate, and herein, we describe the solid state structures of ligand (L1), and the new three-dimensional coordination polymer, poly[[[μ4-N 2,N 3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)]nitrate] (I).
Structural commentary
The title ligand (L1) crystallized as a dihydrate, and its molecular structure is illustrated in Fig. 1 ▸. The molecule is U-shaped with the carboxamide groups (C6/N3/C5/O1) being cis to one another, making a dihedral angle of 81.6 (5)°. The terminal pyridine rings (N4/C7–C11) are inclined to one another by 58.5 (4)°. There is an intramolecular N—H⋯N hydrogen bond present, forming an S(5) ring motif (Fig. 1 ▸ and Table 1 ▸).
Figure 1.
A view of the molecular structure of ligand (L1), with atom labelling. Displacement ellipsoids are drawn at the 50% probability level. The intramolecular N—H⋯N contact is shown as a dashed line (see Table 2 ▸).
Table 1. Hydrogen-bond geometry (Å, °) for (L1) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3N⋯N1 | 0.88 (3) | 2.08 (5) | 2.718 (7) | 129 (5) |
| N5—H5N⋯O1i | 0.88 (3) | 2.05 (4) | 2.858 (7) | 151 (6) |
| O1W—H1WA⋯O2W ii | 0.86 | 1.91 | 2.762 (7) | 177 |
| O1W—H1WB⋯N6iii | 0.94 | 1.97 | 2.886 (7) | 164 |
| O2W—H2WA⋯O1W iv | 0.86 | 1.91 | 2.765 (8) | 172 |
| O2W—H2WB⋯N4 | 0.85 | 2.06 | 2.888 (7) | 164 |
| C3—H3⋯O1W v | 0.95 | 2.38 | 3.273 (8) | 156 |
| C4—H4⋯O2W vi | 0.95 | 2.58 | 3.253 (8) | 128 |
| C6—H6A⋯N2vii | 0.99 | 2.57 | 3.515 (8) | 159 |
| C16—H16⋯N4iv | 0.95 | 2.60 | 3.451 (9) | 149 |
Symmetry codes: (i) 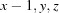 ; (ii)
; (ii)  ; (iii)
; (iii) 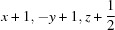 ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  ; (vii)
; (vii)  .
.
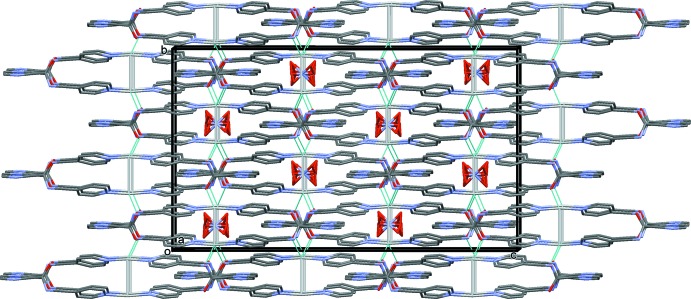
The reaction of the ligand with silver(I) nitrate led to the formation of a three-dimensional coordination polymer (I). The coordination of the ligand to the silver ions is illustrated in Fig. 2 ▸. Selected bond lengths and angles in (I) are given in Table 2 ▸. The asymmetric unit is composed of a silver ion, located on a twofold rotation axis, half a ligand molecule and half a nitrate anion. The full molecule of the ligand is generated by twofold rotational symmetry, with this twofold axis bisecting the C4—C4i bonds of the pyrazine ring and the Ag1—Ag1i bond (Table 2 ▸). The carboxamide groups (C6/N3/C5/O1) are now trans to one another, making a dihedral angle of 65.8 (4)°. The terminal pyridine rings (N4/C7–C11) are inclined to one another by 6.6 (3)°. Two ligands effectively wrap around a Ag—Ag bond of 3.1638 (11) Å, forming a figure-of-eight-shaped molecule, with each silver ion being coordinated by two pyridine N atoms. The silver ions are each further coordinated by the carboxamide O atom, O1, of neighbouring molecules, hence forming a three-dimensional framework, illustrated in Fig. 3 ▸. If one considers that the silver ion, Ag1, is fivefold coordinate (N2O2Agi) then its coordination sphere can be described as distorted trigonal–bipyramidal, with a τ5 value of 0.8 (τ5 = 1 for perfect trigonal–pyramidal geometry and 0 for perfect square-pyramidal geometry; Addison et al., 1984 ▸). However, if one considers the Ag1 ion to be fourfold coordinate, N2O2, with a τ4 value of 0.55, its coordination sphere can be described as intermediate between trigonal–pyramidal and seesaw (τ4 = 1 for a perfect tetrahedral geometry and 0 for a perfect square-planar geometry. For intermediate structures, including trigonal–pyramidal and seesaw, τ4 falls within the range of 0 to 1; Yang et al., 2007 ▸). The nitrate anion that does not coordinate to the silver(I) ion is positionally disordered, and also located about a twofold rotation axis.
Figure 2.
A view of the figure-eight arrangement of the title complex (I), with atom labelling for the asymmetric unit and some symmetry-related atoms (see Table 1 ▸ for details). The unlabelled atoms of the ligand on the left-hand-side of the figure are related to the labelled atoms by twofold rotational symmetry (symmetry operation: −x +  , −y +
, −y +  , z). The nitrate anions have been omitted for clarity.
, z). The nitrate anions have been omitted for clarity.
Table 2. Selected geometric parameters (Å, °) for (I) .
| Ag1—Ag1i | 3.1638 (11) | Ag1—O1ii | 2.814 (5) |
| Ag1—N4 | 2.109 (5) | ||
| N4—Ag1—N4iii | 173.0 (2) | O1ii—Ag1—N4 | 97.80 (15) |
| O1ii—Ag1—O1iv | 109.48 (14) | O1iv—Ag1—N4 | 86.26 (15) |
| Ag1i—Ag1—N4 | 86.51 (14) | Ag1i—Ag1—O1ii | 125.26 (10) |
Symmetry codes: (i) 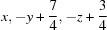 ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Figure 3.
A view along the c axis of the three-dimensional framework of complex (I), showing the Ag⋯O bonds as dashed lines (see Table 2 ▸). The nitrate anions and the C-bound H atoms have been omitted for clarity.
Supramolecular features
In the crystal of ligand (L1), molecules are linked by N—H⋯O(water) hydrogen bonds forming chains propagating along the c-axis direction (Table 1 ▸ and Fig. 4 ▸). Parallel to this chain of molecules is a chain of hydrogen-bonded water molecules (Table 1 ▸ and Fig. 4 ▸), which is linked to the chain of (L1) molecules by O—H⋯N hydrogen bonds, forming columns propagating along the c axis. The columns are linked by C—H⋯O and C—H⋯N hydrogen bonds, forming a three-dimensional supramolecular structure (Table 1 ▸ and Fig. 5 ▸).
Figure 4.
A partial view along direction [111] of the crystal packing of ligand (L1). The hydrogen bonds are shown as dashed lines (see Table 1 ▸)
Figure 5.
A view along the a axis of the crystal packing of ligand (L1). The columns of (L1) molecules, linked by hydrogen bonds involving the water molecules, are indicated by blue circles. The hydrogen bonds are shown as dashed lines (see Table 1 ▸), and for clarity, only the H atoms involved in hydrogen bonding have been included.
In (I), the nitrate anion is situated in the cavities of the three-dimensional framework and is linked to the framework by N—H⋯O and C—H⋯O hydrogen bonds (Table 3 ▸ and Fig. 6 ▸). The nitrate anion in (I) is not essential for forming the three-dimensional structure, although it may act as a template for the formation of the framework (Batten et al., 2009 ▸). This is in contrast to the MOF catena-[bis(μ3-nitrato-O,O′,O′′)bis(μ3-1,3,5-tris(pyrazol-1-yl)benzene-N,N′,N′′)trisilver(I) nitrate] mentioned above (Shu et al., 2006 ▸), in which there are nitrate anions coordinating the silver ions in a μ3 fashion and present also in the framework cavities. There are, of course, other examples reported in the Cambridge Structural Database (Groom et al., 2016 ▸).
Table 3. Hydrogen-bond geometry (Å, °) for (I) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N3—H3N⋯O11v | 0.88 | 1.86 | 2.744 (14) | 178 |
| N3—H3N⋯O13v | 0.88 | 2.26 | 2.875 (13) | 127 |
| C4—H4⋯O11vi | 0.95 | 2.45 | 3.378 (13) | 165 |
| C4—H4⋯O14vii | 0.95 | 2.40 | 3.33 (2) | 168 |
| C9—H9⋯O13viii | 0.95 | 2.50 | 3.224 (14) | 133 |
| C11—H11⋯O13v | 0.95 | 2.51 | 3.154 (13) | 126 |
Symmetry codes: (v)  ; (vi)
; (vi)  ; (vii)
; (vii) 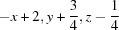 ; (viii)
; (viii) 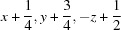 .
.
Figure 6.
A view along the a axis of the crystal packing of complex (I), showing the Ag⋯O bonds as dashed lines (see Table 2 ▸). For clarity, all H atoms have been omitted.
In describing compound (I) as a three-dimensional coordination polymer, we make here the distinction between a coordination polymer and a metal–organic framework. Both have a three-dimensional framework but there are no cavities, even small ones, in the structure of (I). Hence, it should be classed as a three-dimensional coordination polymer according to the IUPAC recommendations on the ‘Terminology of metal–organic frameworks and coordination polymers’ (Batten et al., 2013 ▸).
Database survey
A search of the Cambridge Structural Database (Version 5.38, update February 2017; Groom et al., 2016 ▸) for Ag–Ag complexes, excluding silver ion clusters of any kind, gave 321 hits. Limiting the search to Ag–Ag complexes with each silver ion coordinated by two pyridine N atoms, gave 95 hits. The Ag—Ag distances vary between ca 2.6–3.6 Å. One compound, bis[μ2-2,7-di-tert-butyl-9,9-dimethyl-N,N′-bis[(3-pyridyl)methyl]xanthene-4,5-dicarboxamide]disilver bis(trifluoromethanesulfonate) chloroform solvate (HIFKUD; Yue et al., 2007 ▸), is particularly interesting because it too involves a dicarboxamide ligand, viz. N,N′-bis[(3-pyridyl)methy]xanthene-4,5-dicarboxamide), that wraps around an Ag—Ag bond forming a similar figure-of-eight-shaped complex. Here the Ag—Ag bond length is 3.134 (1) Å, slightly shorter than the value of 3.1638 (11) Å observed in (I); Table 2 ▸. A search for the benzene analogue of ligand (L1), N-(4-pyridylmethyl)carbamoyl)benzene, gave only two hits. Both of them are mercury(II) complexes, viz. the binuclear complex bis{μ2-1,2-bis[N-(4-pyridylmethyl)carbamoyl]benzene}tetrakis(trifluoroacetato)dimercury(II) methanol solvate (XAHSIJ; Burchell et al., 2004 ▸) and the two-dimensional network catena-[bis{μ2-1,2-bis[N-(4-pyridylmethyl)carbamoyl]benzene}dichloridomercury(II) 1,2-dichloroethane solvate] (XAHSOP; Burchell et al., 2004 ▸). A search for the benzene analogue of ligand (L2), [N-(2-pyridylmethyl)carbamoyl]benzene, gave zero hits, while that for [N-(3-pyridylmethyl)carbamoyl]benzene gave eight hits. The latter includes the crystal structure of the dihydrate of the ligand itself (PANROM; Ge et al., 2005 ▸) and the structures of seven first-row transition metal one-, two- and three-dimensional coordination polymers.
Synthesis and crystallization
Ligand (L1) was prepared using the same procedure as for ligand (L2) (Cati et al., 2004 ▸). Dimethyl pyrazine-2,3-dicarboxylate (1.96 g, 10 mmol; Alvarez-Ibarra et al., 1994 ▸) and an excess of 4-(aminomethyl)pyridine (3.24 g, 30 mmol) in 35 ml of methanol were heated to reflux and heating was continued for 72 h in a two-necked flask (100 ml). The brown solution that formed was concentrated and 15 ml of water were added, which precipitated quantitatively ligand (L1). The solid was collected by filtration, washed with 10 ml of water and dried in air. Recrystallization in ethanol gave colourless plate-like crystals (yield is quantitative; m.p. 474 K). Spectroscopic data: 1H NMR (400 MHz, DMSO-d6): 9.33 (t, 1H, J hg = 6.1, Hh); 8.86 (s, 1H, Hn = Hm); 8.49 (dd, 2H, J ba = 4.5, J be = 1.5, Hb = Hd); 7.39 (dd, 2H, J ab = 4.5, J eb = 1.5, Ha = He); 4.52 (d, 2H, J gh = 6.1, Hg). 13C NMR (400 MHz, DMSO-d6): 165.8, 150.3, 148.9, 147.6, 145.6, 123.0, 42.2. IR (KBr pellet, cm−1): 3273 (s), 3031 (s), 1675 (vs), 1602 (vs), 1564 (vs), 1520 (vs), 1416 (vs), 1364 (s), 1311 (s), 1292 (s), 1220 (s), 1185 (m), 1164 (m), 1124 (s), 1069 (m), 995 (s), 871 (w), 830 (m), 787 (m), 745 (m), 715 (m), 611 (m), 575 (w), 504 (m), 495 (m), 475 (m). Elemental analysis for [C18H16N6O2]·H2O (M r = 366.39 g mol−1): calculated: C: 59.01 H: 4.95 N: 22.94%; found: C: 59.10 H: 5.05 N: 23.10%.
Complex (I): A solution of (L1) (46 mg; 0.126 mmol) in 6 ml CHCl3 was introduced into a 13 mm diameter glass tube. It was layered with methanol (ca 2 ml) used as a buffer zone. A solution of AgNO3 (21 mg, 0.126 mmol) in MeOH (6 ml) was then added gently to avoid possible mixing. The glass tube was sealed with a perforated parafilm and left at room temperature. Colourless block-like crystals were obtained after a few days (yield 60 mg, 92%). Elemental analysis for AgC18H16N7O5: (M r = 518.25 g mol−1): calculated: C: 41.72 H: 3.11 N: 18.92%; found: C: 41.65 H: 3.09 N: 18.85%.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. For the ligand (L1), the NH and water H atoms were located in difference-Fourier maps and refined with distance restraints: O—H = 0.85 (2) Å, N—H = 0.88 (2) Å with U iso(H) = 1.5U eq(O) and 1.2U eq(N). In the final cycles of refinement, the water H atoms were treated as riding atoms. For complex (I), the NH H atoms were included in calculated positions and treated as riding: N—H = 0.88 Å with U iso(H) = 1.2U eq(N). For both compounds, the C-bound H atoms were included in calculated positions and refined as riding: C—H = 0.95–0.99 Å with U iso(H) = 1.2U eq(C). The nitrate anion is positionally disordered about a twofold rotation axis and was refined with fixed occupancies (N10A and N10B = 0.5, O11 and O13 = 0.5, O12 and O14 = 0.25), and all their ADP’s were made equal to that of atom O11. Using a one-circle image-plate diffraction system it is not possible to measure 100% of the Ewald sphere, particularly for triclinic or monoclinic systems. This is the case for ligand (L1), which crystallized in the monoclinic space group Pc and for which only 94.7% of the Ewald sphere was accessible.
Table 4. Experimental details.
| (L1) | (I) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C18H16N6O2·2H2O | [Ag(C18H16N6O2)]NO3 |
| M r | 384.40 | 518.25 |
| Crystal system, space group | Monoclinic, P c | Orthorhombic, F d d d |
| Temperature (K) | 153 | 153 |
| a, b, c (Å) | 4.3677 (6), 14.0232 (12), 15.1816 (18) | 14.9776 (16), 17.3228 (12), 29.570 (4) |
| α, β, γ (°) | 90, 96.153 (16), 90 | 90, 90, 90 |
| V (Å3) | 924.50 (19) | 7672.1 (14) |
| Z | 2 | 16 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.10 | 1.10 |
| Crystal size (mm) | 0.50 × 0.15 × 0.05 | 0.40 × 0.30 × 0.20 |
| Data collection | ||
| Diffractometer | Stoe IPDS 1 | Stoe IPDS 1 |
| Absorption correction | Multi-scan (MULABS; Spek, 2009 ▸) | Multi-scan (MULABS; Spek, 2009 ▸) |
| T min, T max | 0.763, 1.000 | 0.985, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 7134, 3388, 1693 | 13548, 1721, 1038 |
| R int | 0.107 | 0.096 |
| (sin θ/λ)max (Å−1) | 0.615 | 0.600 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.051, 0.122, 0.77 | 0.045, 0.107, 0.90 |
| No. of reflections | 3388 | 1721 |
| No. of parameters | 260 | 141 |
| No. of restraints | 8 | 0 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.21, −0.25 | 0.68, −0.65 |
Supplementary Material
Crystal structure: contains datablock(s) L1, I, Global. DOI: 10.1107/S2056989017006387/pj2044sup1.cif
Structure factors: contains datablock(s) L1. DOI: 10.1107/S2056989017006387/pj2044L1sup2.hkl
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017006387/pj2044Isup3.hkl
Supporting information file. DOI: 10.1107/S2056989017006387/pj2044L1sup4.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
(L1) N2,N3-Bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide . Crystal data
| C18H16N6O2·2H2O | F(000) = 404 |
| Mr = 384.40 | Dx = 1.381 Mg m−3 |
| Monoclinic, Pc | Mo Kα radiation, λ = 0.71073 Å |
| a = 4.3677 (6) Å | Cell parameters from 3264 reflections |
| b = 14.0232 (12) Å | θ = 2.0–25.9° |
| c = 15.1816 (18) Å | µ = 0.10 mm−1 |
| β = 96.153 (16)° | T = 153 K |
| V = 924.50 (19) Å3 | Plate, colourless |
| Z = 2 | 0.50 × 0.15 × 0.05 mm |
(L1) N2,N3-Bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide . Data collection
| Stoe IPDS 1 diffractometer | 3388 independent reflections |
| Radiation source: fine-focus sealed tube | 1693 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.107 |
| φ rotation scans | θmax = 25.9°, θmin = 2.0° |
| Absorption correction: multi-scan (MULABS; Spek, 2009) | h = −5→5 |
| Tmin = 0.763, Tmax = 1.000 | k = −17→16 |
| 7134 measured reflections | l = −18→18 |
(L1) N2,N3-Bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide . Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.051 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.122 | w = 1/[σ2(Fo2) + (0.0453P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 0.77 | (Δ/σ)max < 0.001 |
| 3388 reflections | Δρmax = 0.21 e Å−3 |
| 260 parameters | Δρmin = −0.25 e Å−3 |
| 8 restraints | Extinction correction: (SHELXL2016; Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.036 (6) |
(L1) N2,N3-Bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(L1) N2,N3-Bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.7750 (11) | 0.9108 (3) | 0.4491 (3) | 0.0358 (13) | |
| O2 | 0.5478 (11) | 0.9994 (3) | 0.2578 (3) | 0.0351 (12) | |
| N1 | 0.4263 (14) | 1.1135 (3) | 0.5363 (4) | 0.0321 (15) | |
| N2 | 0.1716 (14) | 1.1331 (4) | 0.3588 (4) | 0.0330 (15) | |
| N3 | 0.7147 (13) | 0.9491 (3) | 0.5921 (3) | 0.0258 (14) | |
| H3N | 0.612 (13) | 0.998 (3) | 0.609 (4) | 0.031* | |
| N4 | 0.4923 (14) | 0.5966 (4) | 0.6225 (4) | 0.0403 (16) | |
| N5 | 0.1699 (14) | 0.9119 (4) | 0.3107 (4) | 0.0313 (14) | |
| H5N | 0.002 (11) | 0.906 (5) | 0.337 (4) | 0.038* | |
| N6 | −0.0999 (16) | 0.5615 (4) | 0.3180 (4) | 0.0436 (17) | |
| C1 | 0.4668 (15) | 1.0457 (4) | 0.4743 (4) | 0.0267 (17) | |
| C2 | 0.3401 (16) | 1.0564 (4) | 0.3878 (4) | 0.0258 (16) | |
| C3 | 0.1396 (19) | 1.1999 (5) | 0.4209 (5) | 0.043 (2) | |
| H3 | 0.030625 | 1.256737 | 0.403978 | 0.051* | |
| C4 | 0.2586 (18) | 1.1888 (4) | 0.5079 (5) | 0.0360 (19) | |
| H4 | 0.219526 | 1.236935 | 0.549295 | 0.043* | |
| C5 | 0.6613 (15) | 0.9624 (4) | 0.5039 (4) | 0.0280 (16) | |
| C6 | 0.9192 (16) | 0.8748 (4) | 0.6312 (5) | 0.0290 (17) | |
| H6A | 0.985533 | 0.891453 | 0.693707 | 0.035* | |
| H6B | 1.105193 | 0.872221 | 0.599314 | 0.035* | |
| C7 | 0.7688 (16) | 0.7778 (4) | 0.6278 (5) | 0.0307 (17) | |
| C8 | 0.8016 (18) | 0.7151 (4) | 0.5602 (5) | 0.0376 (19) | |
| H8 | 0.917205 | 0.733033 | 0.513344 | 0.045* | |
| C9 | 0.669 (2) | 0.6270 (5) | 0.5600 (5) | 0.046 (2) | |
| H9 | 0.702062 | 0.584555 | 0.513239 | 0.056* | |
| C10 | 0.4698 (19) | 0.6573 (5) | 0.6889 (5) | 0.044 (2) | |
| H10 | 0.357252 | 0.636975 | 0.735668 | 0.053* | |
| C11 | 0.5967 (17) | 0.7473 (5) | 0.6950 (5) | 0.0374 (19) | |
| H11 | 0.568042 | 0.787610 | 0.743743 | 0.045* | |
| C12 | 0.3639 (17) | 0.9848 (4) | 0.3138 (4) | 0.0297 (17) | |
| C13 | 0.1367 (17) | 0.8444 (4) | 0.2372 (5) | 0.0337 (18) | |
| H13A | 0.332934 | 0.841179 | 0.210178 | 0.040* | |
| H13B | −0.024426 | 0.867753 | 0.191430 | 0.040* | |
| C14 | 0.0529 (17) | 0.7469 (4) | 0.2655 (4) | 0.0303 (17) | |
| C15 | −0.1501 (17) | 0.6908 (5) | 0.2141 (5) | 0.0362 (18) | |
| H15 | −0.242090 | 0.714271 | 0.158778 | 0.043* | |
| C16 | −0.224 (2) | 0.6001 (5) | 0.2413 (5) | 0.049 (2) | |
| H16 | −0.368187 | 0.563552 | 0.204091 | 0.058* | |
| C17 | 0.1048 (19) | 0.6151 (5) | 0.3680 (6) | 0.042 (2) | |
| H17 | 0.200289 | 0.589113 | 0.421904 | 0.050* | |
| C18 | 0.1843 (19) | 0.7075 (5) | 0.3446 (5) | 0.0389 (18) | |
| H18 | 0.327922 | 0.743333 | 0.382699 | 0.047* | |
| O1W | 0.7645 (12) | 0.5986 (3) | 0.9268 (3) | 0.0503 (15) | |
| H1WA | 0.624209 | 0.595327 | 0.962023 | 0.075* | |
| H1WB | 0.768245 | 0.545186 | 0.889768 | 0.075* | |
| O2W | 0.3065 (12) | 0.4183 (3) | 0.5379 (3) | 0.0478 (14) | |
| H2WA | 0.133161 | 0.418283 | 0.504471 | 0.072* | |
| H2WB | 0.327355 | 0.474826 | 0.557396 | 0.072* |
(L1) N2,N3-Bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.051 (3) | 0.039 (3) | 0.021 (3) | 0.009 (2) | 0.019 (3) | 0.000 (2) |
| O2 | 0.052 (3) | 0.040 (2) | 0.015 (3) | −0.005 (2) | 0.015 (2) | −0.0003 (19) |
| N1 | 0.055 (4) | 0.024 (3) | 0.018 (3) | 0.001 (3) | 0.007 (3) | −0.002 (2) |
| N2 | 0.050 (4) | 0.030 (3) | 0.019 (3) | 0.006 (3) | 0.004 (3) | 0.003 (2) |
| N3 | 0.041 (4) | 0.025 (3) | 0.012 (3) | 0.004 (3) | 0.009 (3) | −0.001 (2) |
| N4 | 0.047 (4) | 0.032 (3) | 0.041 (4) | 0.001 (3) | 0.001 (4) | 0.007 (3) |
| N5 | 0.038 (4) | 0.034 (3) | 0.023 (3) | −0.010 (3) | 0.006 (3) | −0.006 (2) |
| N6 | 0.054 (4) | 0.033 (3) | 0.044 (4) | −0.005 (3) | 0.007 (4) | −0.003 (3) |
| C1 | 0.035 (5) | 0.029 (3) | 0.018 (4) | −0.002 (3) | 0.009 (3) | 0.001 (3) |
| C2 | 0.034 (4) | 0.025 (3) | 0.020 (4) | 0.000 (3) | 0.008 (3) | 0.000 (3) |
| C3 | 0.061 (6) | 0.027 (4) | 0.040 (5) | 0.011 (4) | 0.003 (4) | 0.003 (3) |
| C4 | 0.060 (5) | 0.025 (4) | 0.023 (4) | 0.007 (4) | 0.009 (4) | −0.003 (3) |
| C5 | 0.037 (4) | 0.031 (4) | 0.017 (4) | −0.001 (3) | 0.006 (3) | 0.008 (3) |
| C6 | 0.036 (4) | 0.031 (4) | 0.020 (4) | 0.006 (3) | 0.001 (3) | 0.000 (3) |
| C7 | 0.032 (5) | 0.030 (4) | 0.027 (4) | 0.013 (3) | −0.007 (4) | −0.002 (3) |
| C8 | 0.056 (5) | 0.034 (4) | 0.022 (5) | 0.001 (4) | 0.001 (4) | −0.001 (3) |
| C9 | 0.066 (6) | 0.031 (4) | 0.040 (5) | 0.005 (4) | 0.001 (5) | −0.005 (3) |
| C10 | 0.057 (6) | 0.037 (4) | 0.039 (5) | 0.007 (4) | 0.009 (4) | 0.007 (4) |
| C11 | 0.053 (5) | 0.035 (4) | 0.024 (4) | 0.002 (4) | 0.006 (4) | −0.001 (3) |
| C12 | 0.047 (5) | 0.027 (4) | 0.015 (4) | 0.007 (3) | −0.002 (4) | 0.001 (3) |
| C13 | 0.047 (5) | 0.029 (3) | 0.025 (4) | −0.003 (3) | 0.003 (4) | −0.007 (3) |
| C14 | 0.038 (5) | 0.029 (3) | 0.025 (5) | 0.005 (3) | 0.006 (4) | −0.005 (3) |
| C15 | 0.047 (5) | 0.035 (4) | 0.028 (4) | 0.005 (4) | 0.007 (4) | −0.003 (3) |
| C16 | 0.063 (6) | 0.040 (4) | 0.042 (5) | −0.008 (4) | 0.000 (5) | −0.004 (4) |
| C17 | 0.052 (5) | 0.038 (4) | 0.038 (5) | 0.001 (4) | 0.012 (4) | 0.003 (4) |
| C18 | 0.041 (5) | 0.040 (4) | 0.036 (5) | −0.003 (4) | 0.002 (4) | −0.006 (3) |
| O1W | 0.066 (4) | 0.036 (3) | 0.051 (4) | −0.006 (3) | 0.013 (3) | −0.001 (2) |
| O2W | 0.061 (4) | 0.036 (3) | 0.047 (3) | 0.000 (2) | 0.007 (3) | 0.001 (2) |
(L1) N2,N3-Bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide . Geometric parameters (Å, º)
| O1—C5 | 1.245 (7) | C6—H6B | 0.9900 |
| O2—C12 | 1.248 (7) | C7—C8 | 1.370 (9) |
| N1—C4 | 1.331 (8) | C7—C11 | 1.398 (9) |
| N1—C1 | 1.364 (8) | C8—C9 | 1.364 (10) |
| N2—C3 | 1.346 (9) | C8—H8 | 0.9500 |
| N2—C2 | 1.351 (8) | C9—H9 | 0.9500 |
| N3—C5 | 1.347 (8) | C10—C11 | 1.378 (10) |
| N3—C6 | 1.456 (8) | C10—H10 | 0.9500 |
| N3—H3N | 0.88 (3) | C11—H11 | 0.9500 |
| N4—C10 | 1.330 (9) | C13—C14 | 1.491 (8) |
| N4—C9 | 1.354 (10) | C13—H13A | 0.9900 |
| N5—C12 | 1.325 (8) | C13—H13B | 0.9900 |
| N5—C13 | 1.458 (8) | C14—C15 | 1.366 (10) |
| N5—H5N | 0.88 (3) | C14—C18 | 1.389 (10) |
| N6—C17 | 1.340 (10) | C15—C16 | 1.386 (9) |
| N6—C16 | 1.344 (10) | C15—H15 | 0.9500 |
| C1—C2 | 1.377 (9) | C16—H16 | 0.9500 |
| C1—C5 | 1.485 (8) | C17—C18 | 1.397 (10) |
| C2—C12 | 1.518 (9) | C17—H17 | 0.9500 |
| C3—C4 | 1.375 (10) | C18—H18 | 0.9500 |
| C3—H3 | 0.9500 | O1W—H1WA | 0.8569 |
| C4—H4 | 0.9500 | O1W—H1WB | 0.9371 |
| C6—C7 | 1.510 (9) | O2W—H2WA | 0.8649 |
| C6—H6A | 0.9900 | O2W—H2WB | 0.8483 |
| C4—N1—C1 | 115.9 (6) | C7—C8—H8 | 119.9 |
| C3—N2—C2 | 114.8 (6) | N4—C9—C8 | 123.9 (7) |
| C5—N3—C6 | 122.5 (5) | N4—C9—H9 | 118.1 |
| C5—N3—H3N | 99 (4) | C8—C9—H9 | 118.1 |
| C6—N3—H3N | 139 (5) | N4—C10—C11 | 125.2 (7) |
| C10—N4—C9 | 115.0 (6) | N4—C10—H10 | 117.4 |
| C12—N5—C13 | 122.6 (6) | C11—C10—H10 | 117.4 |
| C12—N5—H5N | 128 (4) | C10—C11—C7 | 118.3 (6) |
| C13—N5—H5N | 106 (4) | C10—C11—H11 | 120.8 |
| C17—N6—C16 | 116.6 (6) | C7—C11—H11 | 120.8 |
| N1—C1—C2 | 120.9 (6) | O2—C12—N5 | 123.9 (6) |
| N1—C1—C5 | 116.8 (6) | O2—C12—C2 | 119.7 (6) |
| C2—C1—C5 | 122.2 (5) | N5—C12—C2 | 116.2 (6) |
| N2—C2—C1 | 123.1 (5) | N5—C13—C14 | 112.5 (6) |
| N2—C2—C12 | 111.4 (6) | N5—C13—H13A | 109.1 |
| C1—C2—C12 | 125.5 (6) | C14—C13—H13A | 109.1 |
| N2—C3—C4 | 122.5 (7) | N5—C13—H13B | 109.1 |
| N2—C3—H3 | 118.7 | C14—C13—H13B | 109.1 |
| C4—C3—H3 | 118.7 | H13A—C13—H13B | 107.8 |
| N1—C4—C3 | 122.6 (6) | C15—C14—C18 | 116.6 (6) |
| N1—C4—H4 | 118.7 | C15—C14—C13 | 121.9 (6) |
| C3—C4—H4 | 118.7 | C18—C14—C13 | 121.5 (6) |
| O1—C5—N3 | 123.0 (6) | C14—C15—C16 | 121.0 (7) |
| O1—C5—C1 | 120.7 (6) | C14—C15—H15 | 119.5 |
| N3—C5—C1 | 116.3 (5) | C16—C15—H15 | 119.5 |
| N3—C6—C7 | 112.7 (6) | N6—C16—C15 | 122.9 (7) |
| N3—C6—H6A | 109.1 | N6—C16—H16 | 118.6 |
| C7—C6—H6A | 109.1 | C15—C16—H16 | 118.6 |
| N3—C6—H6B | 109.1 | N6—C17—C18 | 123.0 (8) |
| C7—C6—H6B | 109.1 | N6—C17—H17 | 118.5 |
| H6A—C6—H6B | 107.8 | C18—C17—H17 | 118.5 |
| C8—C7—C11 | 117.3 (6) | C14—C18—C17 | 119.9 (7) |
| C8—C7—C6 | 121.6 (6) | C14—C18—H18 | 120.1 |
| C11—C7—C6 | 121.1 (6) | C17—C18—H18 | 120.1 |
| C9—C8—C7 | 120.3 (7) | H1WA—O1W—H1WB | 113.1 |
| C9—C8—H8 | 119.9 | H2WA—O2W—H2WB | 105.0 |
| C4—N1—C1—C2 | 0.2 (9) | C7—C8—C9—N4 | 2.1 (12) |
| C4—N1—C1—C5 | 178.1 (6) | C9—N4—C10—C11 | 3.4 (12) |
| C3—N2—C2—C1 | 0.9 (10) | N4—C10—C11—C7 | −1.4 (12) |
| C3—N2—C2—C12 | 179.9 (6) | C8—C7—C11—C10 | −0.5 (11) |
| N1—C1—C2—N2 | 0.3 (10) | C6—C7—C11—C10 | −178.9 (6) |
| C5—C1—C2—N2 | −177.6 (6) | C13—N5—C12—O2 | −4.5 (10) |
| N1—C1—C2—C12 | −178.5 (6) | C13—N5—C12—C2 | 171.7 (6) |
| C5—C1—C2—C12 | 3.6 (10) | N2—C2—C12—O2 | 78.2 (8) |
| C2—N2—C3—C4 | −2.5 (11) | C1—C2—C12—O2 | −102.8 (8) |
| C1—N1—C4—C3 | −1.8 (10) | N2—C2—C12—N5 | −98.1 (7) |
| N2—C3—C4—N1 | 3.2 (12) | C1—C2—C12—N5 | 80.8 (9) |
| C6—N3—C5—O1 | 2.1 (10) | C12—N5—C13—C14 | 148.9 (6) |
| C6—N3—C5—C1 | −175.4 (5) | N5—C13—C14—C15 | 141.5 (7) |
| N1—C1—C5—O1 | −161.3 (6) | N5—C13—C14—C18 | −40.3 (9) |
| C2—C1—C5—O1 | 16.6 (9) | C18—C14—C15—C16 | 1.4 (10) |
| N1—C1—C5—N3 | 16.2 (8) | C13—C14—C15—C16 | 179.6 (6) |
| C2—C1—C5—N3 | −165.8 (6) | C17—N6—C16—C15 | −0.6 (11) |
| C5—N3—C6—C7 | −79.3 (7) | C14—C15—C16—N6 | −0.9 (12) |
| N3—C6—C7—C8 | 94.4 (7) | C16—N6—C17—C18 | 1.6 (11) |
| N3—C6—C7—C11 | −87.3 (8) | C15—C14—C18—C17 | −0.4 (10) |
| C11—C7—C8—C9 | 0.1 (11) | C13—C14—C18—C17 | −178.7 (6) |
| C6—C7—C8—C9 | 178.5 (7) | N6—C17—C18—C14 | −1.2 (11) |
| C10—N4—C9—C8 | −3.8 (11) |
(L1) N2,N3-Bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H3N···N1 | 0.88 (3) | 2.08 (5) | 2.718 (7) | 129 (5) |
| N5—H5N···O1i | 0.88 (3) | 2.05 (4) | 2.858 (7) | 151 (6) |
| O1W—H1WA···O2Wii | 0.86 | 1.91 | 2.762 (7) | 177 |
| O1W—H1WB···N6iii | 0.94 | 1.97 | 2.886 (7) | 164 |
| O2W—H2WA···O1Wiv | 0.86 | 1.91 | 2.765 (8) | 172 |
| O2W—H2WB···N4 | 0.85 | 2.06 | 2.888 (7) | 164 |
| C3—H3···O1Wv | 0.95 | 2.38 | 3.273 (8) | 156 |
| C4—H4···O2Wvi | 0.95 | 2.58 | 3.253 (8) | 128 |
| C6—H6A···N2vii | 0.99 | 2.57 | 3.515 (8) | 159 |
| C16—H16···N4iv | 0.95 | 2.60 | 3.451 (9) | 149 |
Symmetry codes: (i) x−1, y, z; (ii) x, −y+1, z+1/2; (iii) x+1, −y+1, z+1/2; (iv) x−1, −y+1, z−1/2; (v) x−1, −y+2, z−1/2; (vi) x, y+1, z; (vii) x+1, −y+2, z+1/2.
(I) Poly[[[µ4-N2,N3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] . Crystal data
| [Ag(C18H16N6O2)]NO3 | Dx = 1.795 Mg m−3 |
| Mr = 518.25 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Fddd | Cell parameters from 6115 reflections |
| a = 14.9776 (16) Å | θ = 1.9–25.9° |
| b = 17.3228 (12) Å | µ = 1.10 mm−1 |
| c = 29.570 (4) Å | T = 153 K |
| V = 7672.1 (14) Å3 | Block, colourless |
| Z = 16 | 0.40 × 0.30 × 0.20 mm |
| F(000) = 4160 |
(I) Poly[[[µ4-N2,N3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] . Data collection
| Stoe IPDS 1 diffractometer | 1721 independent reflections |
| Radiation source: fine-focus sealed tube | 1038 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.096 |
| φ rotation scans | θmax = 25.3°, θmin = 2.7° |
| Absorption correction: multi-scan (MULABS; Spek, 2009) | h = −17→17 |
| Tmin = 0.985, Tmax = 1.000 | k = −20→20 |
| 13548 measured reflections | l = −35→35 |
(I) Poly[[[µ4-N2,N3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.045 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.107 | H-atom parameters constrained |
| S = 0.90 | w = 1/[σ2(Fo2) + (0.0575P)2] where P = (Fo2 + 2Fc2)/3 |
| 1721 reflections | (Δ/σ)max = 0.003 |
| 141 parameters | Δρmax = 0.68 e Å−3 |
| 0 restraints | Δρmin = −0.65 e Å−3 |
(I) Poly[[[µ4-N2,N3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] . Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell esds are taken into account in the estimation of distances, angles and torsion angles |
(I) Poly[[[µ4-N2,N3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Ag1 | 0.87500 | 0.96632 (4) | 0.37500 | 0.0464 (2) | |
| O1 | 0.9796 (3) | 0.8101 (3) | 0.14979 (13) | 0.0483 (14) | |
| N1 | 0.9673 (3) | 0.8836 (3) | 0.04722 (14) | 0.0447 (18) | |
| N3 | 1.0135 (3) | 0.9361 (3) | 0.14261 (16) | 0.050 (2) | |
| N4 | 0.9359 (3) | 0.9589 (3) | 0.31086 (16) | 0.0427 (17) | |
| C1 | 0.9207 (3) | 0.8777 (4) | 0.08584 (15) | 0.0337 (16) | |
| C4 | 0.9204 (4) | 0.8805 (4) | 0.00937 (17) | 0.0453 (19) | |
| C5 | 0.9745 (3) | 0.8715 (4) | 0.12935 (16) | 0.0340 (18) | |
| C6 | 1.0689 (4) | 0.9393 (4) | 0.18277 (18) | 0.051 (2) | |
| C7 | 1.0197 (4) | 0.9446 (3) | 0.22692 (19) | 0.0437 (19) | |
| C8 | 1.0640 (4) | 0.9276 (4) | 0.26675 (19) | 0.045 (2) | |
| C9 | 1.0221 (4) | 0.9354 (4) | 0.3075 (2) | 0.046 (2) | |
| C10 | 0.8921 (3) | 0.9745 (3) | 0.27201 (19) | 0.043 (2) | |
| C11 | 0.9302 (4) | 0.9686 (4) | 0.23033 (19) | 0.0447 (19) | |
| O11 | 0.9918 (8) | 0.0666 (8) | 0.0912 (4) | 0.089 (3) | 0.500 |
| O12 | 1.0655 (16) | 0.1323 (15) | 0.1063 (6) | 0.089 (3) | 0.250 |
| O13 | 0.9326 (9) | 0.0862 (7) | 0.1489 (4) | 0.089 (3) | 0.500 |
| O14 | 0.9869 (18) | 0.1233 (19) | 0.1570 (7) | 0.089 (3) | 0.250 |
| N10A | 0.9614 (15) | 0.12500 | 0.12500 | 0.089 (3) | 0.500 |
| N10B | 0.9960 (16) | 0.12500 | 0.12500 | 0.089 (3) | 0.500 |
| H4 | 0.95046 | 0.88600 | −0.01873 | 0.0540* | |
| H3N | 1.00563 | 0.97838 | 0.12661 | 0.0600* | |
| H6A | 1.10904 | 0.98449 | 0.18023 | 0.0610* | |
| H6B | 1.10697 | 0.89253 | 0.18345 | 0.0610* | |
| H8 | 1.12416 | 0.91024 | 0.26574 | 0.0540* | |
| H9 | 1.05428 | 0.92399 | 0.33436 | 0.0560* | |
| H10 | 0.83154 | 0.99047 | 0.27385 | 0.0520* | |
| H11 | 0.89687 | 0.98062 | 0.20392 | 0.0530* |
(I) Poly[[[µ4-N2,N3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Ag1 | 0.0474 (4) | 0.0397 (4) | 0.0521 (4) | 0.0000 | 0.0035 (4) | 0.0000 |
| O1 | 0.048 (2) | 0.054 (3) | 0.043 (2) | 0.000 (2) | −0.005 (2) | 0.010 (2) |
| N1 | 0.047 (3) | 0.057 (4) | 0.030 (2) | 0.003 (3) | 0.004 (2) | −0.004 (2) |
| N3 | 0.078 (4) | 0.035 (4) | 0.037 (3) | 0.002 (3) | −0.015 (3) | −0.006 (2) |
| N4 | 0.042 (3) | 0.039 (3) | 0.047 (3) | 0.001 (2) | −0.009 (2) | −0.006 (2) |
| C1 | 0.037 (2) | 0.035 (3) | 0.029 (3) | 0.005 (3) | 0.002 (2) | −0.010 (3) |
| C4 | 0.059 (3) | 0.047 (4) | 0.030 (3) | 0.009 (4) | 0.006 (2) | 0.000 (3) |
| C5 | 0.030 (2) | 0.046 (4) | 0.026 (3) | 0.005 (2) | 0.010 (2) | −0.007 (4) |
| C6 | 0.062 (4) | 0.057 (5) | 0.033 (3) | −0.003 (3) | −0.008 (3) | −0.006 (3) |
| C7 | 0.051 (3) | 0.036 (4) | 0.044 (3) | −0.004 (3) | −0.012 (3) | −0.008 (3) |
| C8 | 0.046 (3) | 0.044 (4) | 0.045 (4) | 0.007 (3) | −0.006 (3) | −0.004 (3) |
| C9 | 0.051 (4) | 0.051 (4) | 0.037 (3) | 0.009 (3) | −0.008 (3) | 0.000 (3) |
| C10 | 0.037 (4) | 0.041 (4) | 0.052 (3) | −0.004 (3) | −0.010 (3) | −0.006 (3) |
| C11 | 0.050 (3) | 0.045 (4) | 0.039 (3) | −0.001 (3) | −0.015 (3) | −0.008 (3) |
| O11 | 0.112 (6) | 0.088 (6) | 0.067 (4) | 0.0000 | 0.0000 | 0.028 (4) |
| O12 | 0.112 (6) | 0.088 (6) | 0.067 (4) | 0.0000 | 0.0000 | 0.028 (4) |
| O13 | 0.112 (6) | 0.088 (6) | 0.067 (4) | 0.0000 | 0.0000 | 0.028 (4) |
| O14 | 0.112 (6) | 0.088 (6) | 0.067 (4) | 0.0000 | 0.0000 | 0.028 (4) |
| N10A | 0.112 (6) | 0.088 (6) | 0.067 (4) | 0.0000 | 0.0000 | 0.028 (4) |
| N10B | 0.112 (6) | 0.088 (6) | 0.067 (4) | 0.0000 | 0.0000 | 0.028 (4) |
(I) Poly[[[µ4-N2,N3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] . Geometric parameters (Å, º)
| Ag1—Ag1i | 3.1638 (11) | C8—C9 | 1.365 (8) |
| Ag1—N4 | 2.109 (5) | C10—C11 | 1.362 (8) |
| Ag1—O1ii | 2.814 (5) | O11—N10A | 1.493 (14) |
| Ag1—O1iii | 2.814 (5) | O11—N10B | 1.424 (13) |
| Ag1—N4iv | 2.109 (5) | O11—O12 | 1.65 (3) |
| O1—C5 | 1.226 (8) | O12—N10B | 1.19 (3) |
| N1—C1 | 1.342 (6) | O12—N10A | 1.66 (3) |
| N1—C4 | 1.323 (7) | O13—N10A | 1.066 (15) |
| N3—C5 | 1.322 (8) | O13—N10B | 1.36 (2) |
| N3—C6 | 1.450 (7) | O13—O14 | 1.06 (3) |
| N4—C9 | 1.357 (8) | O14—N10B | 0.96 (2) |
| N4—C10 | 1.350 (7) | O14—N10A | 1.02 (2) |
| C1—C5 | 1.522 (6) | C4—H4 | 0.9500 |
| C1—C1v | 1.372 (6) | C6—H6B | 0.9900 |
| C4—C4v | 1.373 (9) | C6—H6A | 0.9900 |
| N3—H3N | 0.8800 | C8—H8 | 0.9500 |
| C6—C7 | 1.502 (8) | C9—H9 | 0.9500 |
| C7—C8 | 1.384 (8) | C10—H10 | 0.9500 |
| C7—C11 | 1.407 (8) | C11—H11 | 0.9500 |
| N4—Ag1—N4iv | 173.0 (2) | C6—C7—C8 | 119.5 (5) |
| O1ii—Ag1—O1iii | 109.48 (14) | C6—C7—C11 | 123.2 (5) |
| Ag1i—Ag1—N4 | 86.51 (14) | C7—C8—C9 | 120.7 (6) |
| O1ii—Ag1—N4 | 97.80 (15) | N4—C9—C8 | 122.1 (5) |
| O1iii—Ag1—N4 | 86.26 (15) | N4—C10—C11 | 123.5 (5) |
| Ag1i—Ag1—O1ii | 125.26 (10) | C7—C11—C10 | 119.1 (5) |
| Ag1i—Ag1—O1iii | 125.26 (10) | C4v—C4—H4 | 119.00 |
| Ag1i—Ag1—N4iv | 86.51 (14) | N1—C4—H4 | 119.00 |
| O1ii—Ag1—N4iv | 86.26 (15) | N3—C6—H6B | 108.00 |
| O1iii—Ag1—N4iv | 97.80 (15) | C7—C6—H6A | 108.00 |
| Ag1vi—O1—C5 | 114.9 (3) | H6A—C6—H6B | 107.00 |
| C1—N1—C4 | 116.2 (5) | C7—C6—H6B | 108.00 |
| C5—N3—C6 | 121.9 (5) | N3—C6—H6A | 108.00 |
| Ag1—N4—C9 | 119.7 (4) | C7—C8—H8 | 120.00 |
| Ag1—N4—C10 | 122.9 (3) | C9—C8—H8 | 120.00 |
| C9—N4—C10 | 117.4 (5) | C8—C9—H9 | 119.00 |
| N1—C1—C5 | 116.7 (4) | N4—C9—H9 | 119.00 |
| N1—C1—C1v | 121.6 (4) | N4—C10—H10 | 118.00 |
| C1v—C1—C5 | 121.6 (4) | C11—C10—H10 | 118.00 |
| N1—C4—C4v | 122.1 (5) | O11—N10A—O13 | 98.0 (8) |
| C5—N3—H3N | 119.00 | O13—N10A—N10B | 113.9 (13) |
| C6—N3—H3N | 119.00 | O11—N10A—N10B | 72.3 (9) |
| O1—C5—N3 | 124.1 (5) | O11—N10B—N10A | 87.5 (11) |
| O1—C5—C1 | 120.7 (6) | O11—N10B—O13 | 89.0 (10) |
| N3—C5—C1 | 115.2 (5) | O13—N10B—N10A | 45.8 (9) |
| N3—C6—C7 | 115.7 (5) | C7—C11—H11 | 120.00 |
| C8—C7—C11 | 117.3 (5) | C10—C11—H11 | 120.00 |
| Ag1i—Ag1—N4—C9 | −71.9 (5) | N1—C1—C5—O1 | −106.9 (6) |
| Ag1i—Ag1—N4—C10 | 106.2 (4) | N1—C1—C5—N3 | 73.3 (7) |
| O1ii—Ag1—N4—C9 | 163.0 (5) | C1v—C1—C5—O1 | 68.8 (8) |
| O1ii—Ag1—N4—C10 | −19.0 (5) | C1v—C1—C5—N3 | −111.0 (7) |
| O1iii—Ag1—N4—C9 | 53.8 (5) | N1—C1—C1v—N1v | 5.5 (11) |
| O1iii—Ag1—N4—C10 | −128.1 (5) | N1—C1—C1v—C5v | −170.1 (6) |
| Ag1vi—O1—C5—N3 | −89.1 (5) | C5—C1—C1v—N1v | −170.1 (6) |
| Ag1vi—O1—C5—C1 | 91.1 (5) | C5—C1—C1v—C5v | 14.4 (10) |
| C4—N1—C1—C5 | 172.9 (6) | N1—C4—C4v—N1v | 4.5 (11) |
| C4—N1—C1—C1v | −2.9 (10) | N3—C6—C7—C8 | 163.0 (6) |
| C1—N1—C4—C4v | −1.9 (10) | N3—C6—C7—C11 | −19.0 (9) |
| C6—N3—C5—O1 | 1.7 (8) | C6—C7—C8—C9 | 177.0 (6) |
| C6—N3—C5—C1 | −178.5 (4) | C11—C7—C8—C9 | −1.1 (9) |
| C5—N3—C6—C7 | −79.7 (7) | C6—C7—C11—C10 | −177.5 (6) |
| Ag1—N4—C9—C8 | 178.2 (5) | C8—C7—C11—C10 | 0.5 (9) |
| C10—N4—C9—C8 | 0.1 (9) | C7—C8—C9—N4 | 0.9 (10) |
| Ag1—N4—C10—C11 | −178.8 (5) | N4—C10—C11—C7 | 0.4 (9) |
| C9—N4—C10—C11 | −0.7 (9) |
Symmetry codes: (i) x, −y+7/4, −z+3/4; (ii) x−1/4, y+1/4, −z+1/2; (iii) −x+2, y+1/4, z+1/4; (iv) −x+7/4, y, −z+3/4; (v) −x+7/4, −y+7/4, z; (vi) −x+2, y−1/4, z−1/4.
(I) Poly[[[µ4-N2,N3-bis(pyridin-4-ylmethyl)pyrazine-2,3-dicarboxamide]silver(I)] nitrate] . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N3—H3N···O11vii | 0.88 | 1.86 | 2.744 (14) | 178 |
| N3—H3N···O13vii | 0.88 | 2.26 | 2.875 (13) | 127 |
| C4—H4···O11viii | 0.95 | 2.45 | 3.378 (13) | 165 |
| C4—H4···O14ix | 0.95 | 2.40 | 3.33 (2) | 168 |
| C9—H9···O13x | 0.95 | 2.50 | 3.224 (14) | 133 |
| C11—H11···O13vii | 0.95 | 2.51 | 3.154 (13) | 126 |
Symmetry codes: (vii) x, y+1, z; (viii) −x+2, −y+1, −z; (ix) −x+2, y+3/4, z−1/4; (x) x+1/4, y+3/4, −z+1/2.
Funding Statement
This work was funded by Swiss National Science Foundation grant . University of Neuchâtel. grant .
References
- Addison, A. W., Rao, T. N., Reedijk, J., van Rijn, J. & Verschoor, G. C. (1984). J. Chem. Soc. Dalton Trans. pp. 1349–1356.
- Alvarez-Ibarra, C., Cuervo-Rodríguez, R., Fernández-Monreal, M. C. & Ruiz, M. P. (1994). J. Org. Chem. 59, 7284–7291.
- Batten, S. R., Champness, N. R., Chen, X. M., Garcia-Martinez, J., Kitagawa, S., Öhrström, L., O’Keeffe, M., Suh, M. P. & Reedijk, J. (2013). Pure Appl. Chem. 5, 1715–1724.
- Batten, S. R., Neville, S. M. & Turner, D. R. (2009). Coordination Polymers, Design, Analysis and Applications. Cambridge: Royal Society of Chemistry.
- Burchell, T. J., Eisler, D. J. & Puddephatt, R. J. (2004). Inorg. Chem. 43, 5550–5557. [DOI] [PubMed]
- Cati, D. (2002). PhD thesis, University of Neuchâtel, Switzerland.
- Cati, D. S., Ribas, J., Ribas-Ariño, J. & Stoeckli-Evans, H. (2004). Inorg. Chem. 43, 1021–1030. [DOI] [PubMed]
- Ge, C.-H., Kou, H.-Z., Wang, R.-J., Jiang, Y.-B. & Cui, A.-L. (2005). Acta Cryst. E61, o2024–o2026.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hausmann, J., Jameson, G. B. & Brooker, S. (2003). Chem. Commun. pp. 2992–2993. [DOI] [PubMed]
- Huo, J. Z., Su, X. M., Wu, X. X., Liu, Y. Y. & Ding, B. (2016). CrystEngComm, 18, 6640–6652.
- Klingele (née Hausmann), J., Boas, J. F., Pilbrow, J. R., Moubaraki, B., Murray, K. S., Berry, K. J., Hunter, K. A., Jameson, G. B., Boyd, P. D. W. & Brooker, S. (2007). Dalton Trans. pp. 633–645. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Shu, M., Tu, C., Xu, W., Jin, H. & Sun, J. (2006). Cryst. Growth Des. 6, 1890–1896.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2004). IPDSI Bedienungshandbuch. Stoe & Cie GmbH, Darmstadt, Germany.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Yang, L., Powell, D. R. & Houser, R. P. (2007). Dalton Trans. pp. 955–964. [DOI] [PubMed]
- Yue, N. L. S., Jennings, M. C. & Puddephatt, R. J. (2007). Eur. J. Inorg. Chem. pp. 1690–1697.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) L1, I, Global. DOI: 10.1107/S2056989017006387/pj2044sup1.cif
Structure factors: contains datablock(s) L1. DOI: 10.1107/S2056989017006387/pj2044L1sup2.hkl
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017006387/pj2044Isup3.hkl
Supporting information file. DOI: 10.1107/S2056989017006387/pj2044L1sup4.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report