A second metastable polymorphic form of the antifungal anilinopyrimidine active pyrimethanil was isolated from an attempted co-crystallization experiment with meso-erythriol in dimethyl sulfoxide (DMSO). The origin of the polymorphic behaviour is revealed in that the conformation of each dimer present in the asymmetric unit of the structure is unique and determined by the rotation of the second molecule in the dimer with respect to the first.
Keywords: crystal structure, polymorphism, pyrimethanil, hydrogen bonding
Abstract
A second metastable form of the title compound, C12H13N3 (systematic name: 4,6-dimethyl-N-phenylpyrimidin-2-amine), was isolated from an attempted co-crystallization experiment with meso-erythriol in dimethyl sulfoxide (DMSO). The crystals of form 2 at 120 K are monoclinic, space group P21/n with Z′ = 4 compared to the previously reported triclinic form with Z′ = 2 [Sun et al. (2011 ▸). Acta Chim. Sin. 69, 1909–1914]. The four independent molecules in the asymmetric unit form two discrete dimeric units through a concerted pair of N—H⋯N hydrogen bonds with a graph-set notation of R 2 2(8). The origin of the polymorphic behaviour is revealed in that the conformation of each dimer present in the asymmetric unit of the structure is unique and determined by the rotation of the second molecule in the dimer with respect to the first.
Chemical context
(4,6-Dimethyl-pyrimidin-2-yl)-phenyl-amine, pyrimethanil (1) is a broad spectrum systemic fungicide from the anilinopyrimidine class of agents, which also include cyprodinil and mepanipyrim. It was discovered in 1987 (Buhmann et al., 1988 ▸) and is marketed under the trade name SCALA®. Anilinopyrimidines are used extensively for protection against leaf moulds and other fungi. In a recent paper (Sun et al., 2011 ▸), the synthesis and electronic properties of pyrimethanil were presented, including a discussion on the atomic charges, total energy and frontier orbital energy. As part of this wider study, the crystal structure of pyrimethanil was determined at 295 K and used as an initial starting model in the structural optimization process. The structure was triclinic, space group P
 , with Z′ = 2, with two independent molecules in the asymmetric unit. The two independent molecules form a dimeric structural unit through a concerted pair of N—H⋯N hydrogen bonds with a graph-set notation of
, with Z′ = 2, with two independent molecules in the asymmetric unit. The two independent molecules form a dimeric structural unit through a concerted pair of N—H⋯N hydrogen bonds with a graph-set notation of  (8). We have recently been investigating the co-crystallization behaviour of pyrimethanil in an attempt to modify the physicochemical properties of the bulk solid material to improve its overall performance. During the course of one of the co-crystallization screens, the crystal structure of a second polymorphic crystal form of pyrimethanil was determined on a crystal that was isolated from the reaction product of an attempted co-crystallization experiment with meso-erythriol in dimethylsulfoxide (DMSO). In this communication, we report the single crystal X-ray structure of this second, metastable, monoclinic polymorphic form of pyrimethanil at 120 K.
(8). We have recently been investigating the co-crystallization behaviour of pyrimethanil in an attempt to modify the physicochemical properties of the bulk solid material to improve its overall performance. During the course of one of the co-crystallization screens, the crystal structure of a second polymorphic crystal form of pyrimethanil was determined on a crystal that was isolated from the reaction product of an attempted co-crystallization experiment with meso-erythriol in dimethylsulfoxide (DMSO). In this communication, we report the single crystal X-ray structure of this second, metastable, monoclinic polymorphic form of pyrimethanil at 120 K.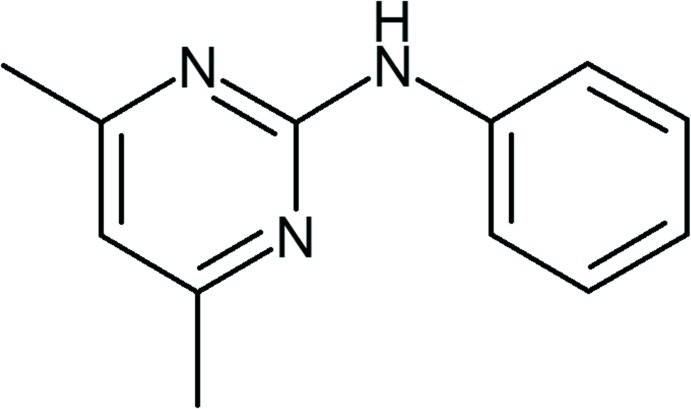
Structural commentary
The crystal structure of form 2 of pyrimethanil is monoclinic, space group P21/n with four independent molecules of pyrimethanil in the asymmetric unit, (Z′ = 4). For clarity, the independent molecules are labelled with suffixes A, B, C and D. The four independent molecules arrange themselves into two dimeric units A–B and C–D, each through a concerted pair of N—H⋯N hydrogen bonds with a graph-set notation of  (8), in a similar arrangement to the dimeric structure found in form 1. Figs. 1 ▸ and 2 ▸ show displacement ellipsoid plots for the two dimers, A–B and C–D and hydrogen-bond distances and angles are given in Table 1 ▸. The phenyl and pyrimidine rings defined by atoms C1–C6 and N2/N3/C7–C10, respectively, for molecules A to D are approximately co-planar. A calculated least-squares plane through the six atoms of the phenyl ring and the six atoms of the pyrimidine ring gave r.m.s. deviations from planarity and a calculated dihedral angle between them as follows: molecule A, 0.0019 Å, 0.0050 Å, 10.8 (1)°; molecule B, 0.0076 Å, 0.0102 Å, 14.8 (1)°; molecule C, 0.0049 Å, 0.0153 Å, 8.2 (1)° and molecule D, 0.0081 Å, 0.0105 Å, 13.5 (1)°. The small variation in the angular range of the dihedral angles appears consistent with that observed for the other pyrimethanil structures discussed below, 7.5-13.1°.
(8), in a similar arrangement to the dimeric structure found in form 1. Figs. 1 ▸ and 2 ▸ show displacement ellipsoid plots for the two dimers, A–B and C–D and hydrogen-bond distances and angles are given in Table 1 ▸. The phenyl and pyrimidine rings defined by atoms C1–C6 and N2/N3/C7–C10, respectively, for molecules A to D are approximately co-planar. A calculated least-squares plane through the six atoms of the phenyl ring and the six atoms of the pyrimidine ring gave r.m.s. deviations from planarity and a calculated dihedral angle between them as follows: molecule A, 0.0019 Å, 0.0050 Å, 10.8 (1)°; molecule B, 0.0076 Å, 0.0102 Å, 14.8 (1)°; molecule C, 0.0049 Å, 0.0153 Å, 8.2 (1)° and molecule D, 0.0081 Å, 0.0105 Å, 13.5 (1)°. The small variation in the angular range of the dihedral angles appears consistent with that observed for the other pyrimethanil structures discussed below, 7.5-13.1°.
Figure 1.
View of the A–B dimer of the asymmetric unit with atom labelling. Ellipsoids are drawn at the 50% probability level. The intermolecular N—H⋯N hydrogen bonds are shown as dashed lines.
Figure 2.
View of the C–D dimer of the asymmetric unit with atom labelling. Ellipsoids are drawn at the 50% probability level. The intermolecular N—H⋯N hydrogen bonds are shown as dashed lines.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1A—H1AB⋯N3B | 0.91 (3) | 2.11 (3) | 2.997 (3) | 165 (2) |
| N1B—H1BB⋯N3A | 0.97 (3) | 2.08 (3) | 3.022 (3) | 162 (3) |
| N1C—H1CB⋯N3D | 0.94 (3) | 2.05 (3) | 2.975 (3) | 166 (3) |
| N1D—H1DB⋯N3C | 0.92 (3) | 2.08 (3) | 2.987 (3) | 167 (3) |
Supramolecular features
A view of the crystal packing down the a-axis is shown in Fig. 3 ▸. The  (8) hydrogen-bonded rings defined by atoms N3A/C7A/N1A/H1AB/N3B/C7B/N1B/H1BB and N3C/C7C/N1C/H1CB/N3D/C7D/N1D/H1DB for the two dimers are twisted such that each dimer forms a cross pattern, with a dihedral angle of 42.8 (2)° for dimer A–B and 47.5 (2)° for dimer C–D. These dihedral angles are between planes C6A/N1A/C7A and C6B/N1B/C7B for A–B and C6C/N1C/C7C and C6D/N1D/C7D for C–D. The angles are somewhat reduced in magnitude when compared to the equivalent calculation performed for form 1, 55.7 (1)°. Fig. 4 ▸ shows an overlay of the two dimeric units in form 2, dimer A–B is shown in violet and C–D in blue, which reveals the origin of the polymorphic behaviour and in turn the reason why Z′ = 4. In this figure, molecules A and C have been overlaid (r.m.s. deviation = 0.181Å) using the standard routine in Mercury (Macrae et al., 2008 ▸). It can be seen that molecule B in the A–B dimer is rotated 134° with respect to molecule D in the C–D dimer, thus making each dimer unique. It is interesting to note that the dimer found in the structure of form 1 has a similar conformation/orientation to the C–D dimer in the present structure. There are no further significant intermolecular contacts and the crystal packing between dimers appears to be driven largely by van der Waals forces only.
(8) hydrogen-bonded rings defined by atoms N3A/C7A/N1A/H1AB/N3B/C7B/N1B/H1BB and N3C/C7C/N1C/H1CB/N3D/C7D/N1D/H1DB for the two dimers are twisted such that each dimer forms a cross pattern, with a dihedral angle of 42.8 (2)° for dimer A–B and 47.5 (2)° for dimer C–D. These dihedral angles are between planes C6A/N1A/C7A and C6B/N1B/C7B for A–B and C6C/N1C/C7C and C6D/N1D/C7D for C–D. The angles are somewhat reduced in magnitude when compared to the equivalent calculation performed for form 1, 55.7 (1)°. Fig. 4 ▸ shows an overlay of the two dimeric units in form 2, dimer A–B is shown in violet and C–D in blue, which reveals the origin of the polymorphic behaviour and in turn the reason why Z′ = 4. In this figure, molecules A and C have been overlaid (r.m.s. deviation = 0.181Å) using the standard routine in Mercury (Macrae et al., 2008 ▸). It can be seen that molecule B in the A–B dimer is rotated 134° with respect to molecule D in the C–D dimer, thus making each dimer unique. It is interesting to note that the dimer found in the structure of form 1 has a similar conformation/orientation to the C–D dimer in the present structure. There are no further significant intermolecular contacts and the crystal packing between dimers appears to be driven largely by van der Waals forces only.
Figure 3.
View of the crystal packing down the a axis. Only the nitrogen heteroatom H atoms are shown for clarity. The intermolecular N—H⋯N hydrogen bonds (see Table 1 ▸) are shown as dotted lines.
Figure 4.
View of the overlay of dimer A–B (violet) and dimer C–D (blue).
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.38 update February 2017; Groom et al., 2016 ▸) for both the pyrimethanil framework and its protonated counterpart yielded three hits, all of which were genuine examples of the material under investigation. Only one entry was found which related to an example that was not a co-crystal, solvate or salt form and that was for the triclinic, P
 , Z′ = 2, form 1 polymorph (CELNOY; Sun et al., 2011 ▸. The remaining two entries were salt forms where the basic nitrogen atom (N3) had been protonated. These examples are the monochloroacetate (MIRYOC; Li et al., 2008 ▸) and the p-toluenesulfonate (XEZFUE; Li et al., 2007 ▸). One further example, which is not yet available in the current release of the database, is an exciting 1:1 co-crystal of pyrimethanil with a second antifungal active, dithianon (SAJJAR; Pöppler et al., 2017 ▸). This material is currently being marketed under the trade name FABAN®.
, Z′ = 2, form 1 polymorph (CELNOY; Sun et al., 2011 ▸. The remaining two entries were salt forms where the basic nitrogen atom (N3) had been protonated. These examples are the monochloroacetate (MIRYOC; Li et al., 2008 ▸) and the p-toluenesulfonate (XEZFUE; Li et al., 2007 ▸). One further example, which is not yet available in the current release of the database, is an exciting 1:1 co-crystal of pyrimethanil with a second antifungal active, dithianon (SAJJAR; Pöppler et al., 2017 ▸). This material is currently being marketed under the trade name FABAN®.
Synthesis and crystallization
Crystals of form 2 of pyrimethanil were isolated from the reaction product of an attempted co-crystallization screen with meso-erythriol in dimethylsulfoxide (DMSO). The screen consisted of approximately 20 mg of pyrimethanil being dispensed per vial along with 20 volumes of the appropriate solvent, approx. 400 µl, at room temperature. The appropriate coformer (ratio 1:1) was also dispensed into the vials in the same manner along with a further 20 volumes of solvent. For the vials that gave clear solutions, these were filtered through a 4 µm filter to remove any potential seeds that may remain in the solution. The vials were placed in a platform shaker incubator (Heidolph Titramax/Inkubator 1000) and subjected to a series of heating–cooling cycles under shaking from room temperature (RT) to 323 K (8 h cycles; heating to 323 K for 4 h and then cooling to RT for a further 4 h) for a maximum of 48 h. The resulting solutions were then allowed to evaporate slowly over a period of 14 days. The solid materials obtained from the screen were analysed by X-ray powder diffraction and were investigated further if they displayed diffraction patterns that were clearly different from that of form 1 or the coformer itself. Unfortunately, it has not been possible thus far to repeat the above experiment to generate more form 2 material, leading us to conclude that form 2 is a metastable form with respect to form 1.
Refinement
Crystal data, data collection, and structure refinement details are summarized in Table 2 ▸. The positional coordinates of the N-bound H atoms were all located from a Fourier-difference map and freely refined. All the remaining H atoms were placed geometrically in idealized positions and refined using a riding model (including free rotation about the methyl C—C bond), with C—H = 0.95–0.99 Å and U iso = 1.5U eq(C) for methyl groups and 1.2U eq(C) for other H atoms.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C12H13N3 |
| M r | 199.25 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 120 |
| a, b, c (Å) | 10.5351 (4), 19.1686 (7), 22.1162 (8) |
| β (°) | 102.778 (4) |
| V (Å3) | 4355.6 (3) |
| Z | 16 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.08 |
| Crystal size (mm) | 0.20 × 0.15 × 0.10 |
| Data collection | |
| Diffractometer | Agilent SuperNova, Dual, Cu at zero, Atlas |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku, 2015) |
| T min, T max | 0.960, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 16540, 7552, 4410 |
| R int | 0.053 |
| (sin θ/λ)max (Å−1) | 0.595 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.056, 0.149, 1.00 |
| No. of reflections | 7552 |
| No. of parameters | 565 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.24, −0.29 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989017007563/hb7679sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017007563/hb7679Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017007563/hb7679Isup3.cml
CCDC reference: 1549998
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C12H13N3 | F(000) = 1696 |
| Mr = 199.25 | Dx = 1.215 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.5351 (4) Å | Cell parameters from 2876 reflections |
| b = 19.1686 (7) Å | θ = 2.9–24.5° |
| c = 22.1162 (8) Å | µ = 0.08 mm−1 |
| β = 102.778 (4)° | T = 120 K |
| V = 4355.6 (3) Å3 | Block, colourless |
| Z = 16 | 0.20 × 0.15 × 0.10 mm |
Data collection
| Agilent SuperNova, Dual, Cu at zero, Atlas diffractometer | 7552 independent reflections |
| Radiation source: SuperNova (Mo) X-ray Source | 4410 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.053 |
| Detector resolution: 10.5598 pixels mm-1 | θmax = 25.0°, θmin = 2.9° |
| ω scans | h = −12→12 |
| Absorption correction: multi-scan (CrysAlis PRO; Rigaku, 2015) | k = −22→18 |
| Tmin = 0.960, Tmax = 1.000 | l = −25→26 |
| 16540 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.056 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.149 | w = 1/[σ2(Fo2) + (0.055P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max = 0.002 |
| 7552 reflections | Δρmax = 0.24 e Å−3 |
| 565 parameters | Δρmin = −0.29 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N1A | 0.6124 (2) | 0.75456 (11) | 0.10435 (11) | 0.0263 (6) | |
| H1AB | 0.698 (3) | 0.7460 (13) | 0.1205 (12) | 0.025 (7)* | |
| N2A | 0.4761 (2) | 0.84979 (12) | 0.06807 (10) | 0.0261 (6) | |
| N3A | 0.6823 (2) | 0.86154 (11) | 0.14057 (10) | 0.0256 (5) | |
| C1A | 0.5890 (3) | 0.63127 (14) | 0.10191 (13) | 0.0280 (7) | |
| H1AA | 0.6763 | 0.6288 | 0.1251 | 0.034* | |
| C2A | 0.5192 (3) | 0.57049 (15) | 0.08619 (13) | 0.0328 (7) | |
| H2AA | 0.5586 | 0.5267 | 0.0988 | 0.039* | |
| C3A | 0.3916 (3) | 0.57303 (15) | 0.05197 (12) | 0.0308 (7) | |
| H3AA | 0.3431 | 0.5314 | 0.0410 | 0.037* | |
| C4A | 0.3367 (3) | 0.63763 (15) | 0.03413 (12) | 0.0304 (7) | |
| H4AA | 0.2497 | 0.6399 | 0.0106 | 0.036* | |
| C5A | 0.4056 (2) | 0.69886 (15) | 0.04986 (12) | 0.0276 (7) | |
| H5AA | 0.3659 | 0.7425 | 0.0372 | 0.033* | |
| C6A | 0.5331 (2) | 0.69632 (14) | 0.08422 (12) | 0.0234 (6) | |
| C7A | 0.5858 (3) | 0.82446 (14) | 0.10375 (13) | 0.0245 (7) | |
| C8A | 0.4627 (3) | 0.91995 (15) | 0.06878 (13) | 0.0289 (7) | |
| C9A | 0.5570 (3) | 0.96202 (15) | 0.10415 (13) | 0.0309 (7) | |
| H9AA | 0.5466 | 1.0113 | 0.1040 | 0.037* | |
| C10A | 0.6671 (3) | 0.93098 (14) | 0.13983 (13) | 0.0286 (7) | |
| C11A | 0.3416 (3) | 0.94925 (16) | 0.02772 (14) | 0.0381 (8) | |
| H11A | 0.2651 | 0.9256 | 0.0364 | 0.057* | |
| H11B | 0.3446 | 0.9420 | −0.0158 | 0.057* | |
| H11C | 0.3361 | 0.9993 | 0.0358 | 0.057* | |
| C12A | 0.7746 (3) | 0.97312 (15) | 0.17935 (14) | 0.0375 (8) | |
| H12A | 0.8564 | 0.9470 | 0.1856 | 0.056* | |
| H12B | 0.7536 | 0.9824 | 0.2196 | 0.056* | |
| H12C | 0.7838 | 1.0174 | 0.1586 | 0.056* | |
| N1B | 0.9217 (2) | 0.79655 (12) | 0.22174 (11) | 0.0244 (5) | |
| H1BB | 0.837 (3) | 0.8075 (16) | 0.1961 (14) | 0.050 (9)* | |
| N2B | 1.0990 (2) | 0.72567 (11) | 0.21211 (10) | 0.0216 (5) | |
| N3B | 0.9013 (2) | 0.73288 (11) | 0.13366 (10) | 0.0232 (5) | |
| C1B | 0.8653 (3) | 0.85002 (14) | 0.30962 (13) | 0.0295 (7) | |
| H1BA | 0.7786 | 0.8535 | 0.2858 | 0.035* | |
| C2B | 0.8942 (3) | 0.87347 (14) | 0.36999 (13) | 0.0316 (7) | |
| H2BA | 0.8275 | 0.8927 | 0.3876 | 0.038* | |
| C3B | 1.0206 (3) | 0.86906 (14) | 0.40526 (14) | 0.0326 (7) | |
| H3BA | 1.0406 | 0.8848 | 0.4470 | 0.039* | |
| C4B | 1.1167 (3) | 0.84163 (14) | 0.37901 (13) | 0.0299 (7) | |
| H4BA | 1.2033 | 0.8386 | 0.4030 | 0.036* | |
| C5B | 1.0887 (3) | 0.81842 (14) | 0.31799 (13) | 0.0261 (7) | |
| H5BA | 1.1563 | 0.8007 | 0.3002 | 0.031* | |
| C6B | 0.9619 (2) | 0.82115 (13) | 0.28306 (12) | 0.0222 (6) | |
| C7B | 0.9786 (2) | 0.74942 (14) | 0.18899 (12) | 0.0220 (6) | |
| C8B | 1.1475 (2) | 0.68135 (14) | 0.17509 (12) | 0.0229 (6) | |
| C9B | 1.0768 (3) | 0.66375 (14) | 0.11680 (13) | 0.0247 (7) | |
| H9BA | 1.1128 | 0.6339 | 0.0907 | 0.030* | |
| C10B | 0.9522 (3) | 0.69043 (14) | 0.09717 (12) | 0.0233 (6) | |
| C11B | 1.2817 (2) | 0.65336 (15) | 0.20046 (13) | 0.0304 (7) | |
| H11D | 1.3404 | 0.6919 | 0.2169 | 0.046* | |
| H11E | 1.3134 | 0.6298 | 0.1673 | 0.046* | |
| H11F | 1.2791 | 0.6201 | 0.2338 | 0.046* | |
| C12B | 0.8698 (3) | 0.67444 (15) | 0.03397 (12) | 0.0307 (7) | |
| H12D | 0.7828 | 0.6944 | 0.0303 | 0.046* | |
| H12E | 0.8627 | 0.6238 | 0.0283 | 0.046* | |
| H12F | 0.9103 | 0.6947 | 0.0022 | 0.046* | |
| N1C | 1.1500 (2) | 1.08100 (12) | 0.13645 (11) | 0.0274 (6) | |
| H1CB | 1.239 (3) | 1.0913 (16) | 0.1519 (15) | 0.058 (10)* | |
| N2C | 1.0238 (2) | 0.98550 (12) | 0.09055 (10) | 0.0285 (6) | |
| N3C | 1.2273 (2) | 0.97203 (12) | 0.16432 (10) | 0.0254 (5) | |
| C1C | 1.1158 (3) | 1.20305 (14) | 0.13836 (12) | 0.0280 (7) | |
| H1CA | 1.2015 | 1.2066 | 0.1632 | 0.034* | |
| C2C | 1.0416 (3) | 1.26280 (15) | 0.12328 (13) | 0.0333 (7) | |
| H2CA | 1.0768 | 1.3069 | 0.1377 | 0.040* | |
| C3C | 0.9166 (3) | 1.25855 (16) | 0.08734 (13) | 0.0360 (8) | |
| H3CA | 0.8647 | 1.2993 | 0.0776 | 0.043* | |
| C4C | 0.8683 (3) | 1.19384 (17) | 0.06585 (13) | 0.0357 (8) | |
| H4CA | 0.7833 | 1.1907 | 0.0402 | 0.043* | |
| C5C | 0.9410 (3) | 1.13327 (16) | 0.08091 (13) | 0.0314 (7) | |
| H5CA | 0.9056 | 1.0892 | 0.0664 | 0.038* | |
| C6C | 1.0665 (3) | 1.13804 (14) | 0.11756 (12) | 0.0247 (7) | |
| C7C | 1.1299 (3) | 1.01019 (14) | 0.12912 (13) | 0.0252 (7) | |
| C8C | 1.0168 (3) | 0.91572 (15) | 0.08390 (13) | 0.0284 (7) | |
| C9C | 1.1147 (3) | 0.87277 (15) | 0.11533 (13) | 0.0302 (7) | |
| H9CA | 1.1106 | 0.8238 | 0.1089 | 0.036* | |
| C10C | 1.2192 (3) | 0.90258 (14) | 0.15653 (13) | 0.0269 (7) | |
| C11C | 0.8976 (3) | 0.88820 (16) | 0.04020 (13) | 0.0352 (8) | |
| H11G | 0.8728 | 0.9200 | 0.0048 | 0.053* | |
| H11H | 0.8261 | 0.8847 | 0.0618 | 0.053* | |
| H11I | 0.9161 | 0.8419 | 0.0254 | 0.053* | |
| C12C | 1.3266 (3) | 0.85994 (15) | 0.19480 (13) | 0.0332 (7) | |
| H12G | 1.4079 | 0.8867 | 0.2023 | 0.050* | |
| H12H | 1.3372 | 0.8168 | 0.1726 | 0.050* | |
| H12I | 1.3049 | 0.8484 | 0.2345 | 0.050* | |
| N1D | 1.4540 (2) | 1.03911 (12) | 0.24960 (11) | 0.0242 (5) | |
| H1DB | 1.378 (3) | 1.0251 (15) | 0.2234 (13) | 0.039 (9)* | |
| N2D | 1.6334 (2) | 1.10956 (11) | 0.24278 (10) | 0.0240 (5) | |
| N3D | 1.4352 (2) | 1.10768 (11) | 0.16432 (10) | 0.0227 (5) | |
| C1D | 1.3924 (3) | 0.98318 (14) | 0.33508 (13) | 0.0276 (7) | |
| H1DA | 1.3072 | 0.9788 | 0.3099 | 0.033* | |
| C2D | 1.4181 (3) | 0.95921 (14) | 0.39511 (13) | 0.0290 (7) | |
| H2DA | 1.3501 | 0.9391 | 0.4112 | 0.035* | |
| C3D | 1.5421 (3) | 0.96405 (14) | 0.43236 (13) | 0.0304 (7) | |
| H3DA | 1.5596 | 0.9478 | 0.4739 | 0.036* | |
| C4D | 1.6398 (3) | 0.99302 (14) | 0.40792 (13) | 0.0285 (7) | |
| H4DA | 1.7253 | 0.9961 | 0.4331 | 0.034* | |
| C5D | 1.6162 (2) | 1.01761 (14) | 0.34767 (12) | 0.0252 (7) | |
| H5DA | 1.6850 | 1.0368 | 0.3315 | 0.030* | |
| C6D | 1.4905 (3) | 1.01402 (13) | 0.31083 (12) | 0.0227 (6) | |
| C7D | 1.5119 (2) | 1.08765 (13) | 0.21896 (12) | 0.0211 (6) | |
| C8D | 1.6839 (3) | 1.15457 (14) | 0.20693 (13) | 0.0259 (7) | |
| C9D | 1.6144 (3) | 1.17560 (14) | 0.14969 (13) | 0.0264 (7) | |
| H9DA | 1.6521 | 1.2059 | 0.1246 | 0.032* | |
| C10D | 1.4874 (3) | 1.15146 (13) | 0.12938 (12) | 0.0227 (6) | |
| C11D | 1.8201 (3) | 1.17911 (16) | 0.23313 (14) | 0.0356 (8) | |
| H11J | 1.8746 | 1.1393 | 0.2503 | 0.053* | |
| H11K | 1.8195 | 1.2133 | 0.2660 | 0.053* | |
| H11L | 1.8553 | 1.2008 | 0.2002 | 0.053* | |
| C12D | 1.4050 (3) | 1.17113 (14) | 0.06759 (12) | 0.0280 (7) | |
| H12J | 1.3215 | 1.1895 | 0.0731 | 0.042* | |
| H12K | 1.3897 | 1.1299 | 0.0408 | 0.042* | |
| H12L | 1.4499 | 1.2069 | 0.0484 | 0.042* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1A | 0.0219 (14) | 0.0189 (13) | 0.0347 (15) | 0.0032 (11) | −0.0007 (12) | −0.0004 (11) |
| N2A | 0.0253 (13) | 0.0271 (14) | 0.0271 (14) | 0.0056 (10) | 0.0085 (12) | 0.0052 (11) |
| N3A | 0.0276 (13) | 0.0204 (13) | 0.0298 (14) | 0.0016 (10) | 0.0081 (12) | −0.0037 (11) |
| C1A | 0.0240 (15) | 0.0266 (16) | 0.0311 (17) | 0.0024 (13) | 0.0011 (14) | 0.0006 (14) |
| C2A | 0.0337 (18) | 0.0251 (16) | 0.0398 (19) | 0.0014 (14) | 0.0085 (16) | −0.0032 (14) |
| C3A | 0.0364 (18) | 0.0294 (17) | 0.0262 (17) | −0.0101 (14) | 0.0059 (15) | −0.0022 (14) |
| C4A | 0.0252 (16) | 0.0427 (19) | 0.0214 (16) | −0.0045 (14) | 0.0009 (14) | 0.0023 (14) |
| C5A | 0.0265 (16) | 0.0295 (17) | 0.0251 (17) | 0.0034 (13) | 0.0021 (14) | 0.0050 (13) |
| C6A | 0.0224 (15) | 0.0254 (16) | 0.0221 (16) | 0.0013 (12) | 0.0041 (13) | −0.0002 (13) |
| C7A | 0.0283 (16) | 0.0245 (16) | 0.0232 (17) | 0.0037 (13) | 0.0110 (14) | 0.0022 (13) |
| C8A | 0.0303 (16) | 0.0313 (18) | 0.0291 (18) | 0.0097 (14) | 0.0152 (15) | 0.0083 (14) |
| C9A | 0.0382 (18) | 0.0221 (16) | 0.0357 (19) | 0.0080 (14) | 0.0150 (16) | 0.0023 (14) |
| C10A | 0.0340 (16) | 0.0251 (16) | 0.0310 (18) | 0.0019 (14) | 0.0164 (15) | −0.0032 (14) |
| C11A | 0.0343 (17) | 0.0355 (18) | 0.044 (2) | 0.0104 (14) | 0.0087 (16) | 0.0124 (16) |
| C12A | 0.0437 (19) | 0.0260 (17) | 0.043 (2) | −0.0022 (14) | 0.0105 (17) | −0.0082 (15) |
| N1B | 0.0214 (13) | 0.0265 (13) | 0.0235 (14) | 0.0034 (11) | 0.0011 (12) | −0.0061 (11) |
| N2B | 0.0212 (12) | 0.0215 (12) | 0.0229 (13) | 0.0011 (10) | 0.0062 (11) | 0.0006 (10) |
| N3B | 0.0262 (12) | 0.0225 (13) | 0.0203 (13) | −0.0026 (10) | 0.0038 (11) | −0.0029 (11) |
| C1B | 0.0302 (16) | 0.0261 (16) | 0.0338 (18) | 0.0027 (13) | 0.0105 (15) | −0.0036 (14) |
| C2B | 0.0387 (18) | 0.0273 (17) | 0.0317 (19) | 0.0012 (14) | 0.0138 (16) | −0.0061 (14) |
| C3B | 0.046 (2) | 0.0250 (16) | 0.0273 (17) | −0.0012 (14) | 0.0084 (16) | −0.0056 (14) |
| C4B | 0.0303 (16) | 0.0299 (17) | 0.0280 (18) | −0.0013 (13) | 0.0034 (14) | −0.0032 (14) |
| C5B | 0.0274 (16) | 0.0236 (15) | 0.0281 (17) | 0.0008 (13) | 0.0081 (14) | −0.0024 (13) |
| C6B | 0.0251 (16) | 0.0171 (14) | 0.0234 (16) | −0.0022 (12) | 0.0030 (14) | −0.0005 (12) |
| C7B | 0.0221 (15) | 0.0205 (14) | 0.0240 (16) | −0.0028 (12) | 0.0062 (14) | 0.0005 (13) |
| C8B | 0.0239 (15) | 0.0225 (15) | 0.0233 (16) | 0.0022 (12) | 0.0076 (14) | 0.0053 (13) |
| C9B | 0.0284 (16) | 0.0231 (15) | 0.0250 (17) | 0.0006 (12) | 0.0109 (14) | −0.0038 (13) |
| C10B | 0.0288 (15) | 0.0207 (15) | 0.0212 (16) | −0.0027 (12) | 0.0073 (14) | 0.0024 (13) |
| C11B | 0.0278 (16) | 0.0327 (17) | 0.0294 (17) | 0.0072 (13) | 0.0032 (14) | −0.0010 (14) |
| C12B | 0.0320 (16) | 0.0324 (17) | 0.0267 (17) | 0.0003 (14) | 0.0043 (14) | −0.0022 (14) |
| N1C | 0.0240 (14) | 0.0253 (14) | 0.0301 (15) | −0.0001 (11) | −0.0002 (12) | −0.0008 (11) |
| N2C | 0.0263 (13) | 0.0323 (14) | 0.0247 (14) | −0.0054 (11) | 0.0005 (12) | 0.0002 (11) |
| N3C | 0.0243 (13) | 0.0256 (14) | 0.0248 (14) | −0.0040 (10) | 0.0022 (11) | 0.0030 (11) |
| C1C | 0.0257 (15) | 0.0320 (17) | 0.0253 (17) | −0.0002 (13) | 0.0035 (14) | 0.0034 (14) |
| C2C | 0.0367 (18) | 0.0299 (17) | 0.0337 (18) | 0.0038 (14) | 0.0086 (16) | 0.0043 (15) |
| C3C | 0.0395 (19) | 0.0359 (19) | 0.0348 (19) | 0.0111 (15) | 0.0130 (17) | 0.0109 (15) |
| C4C | 0.0289 (17) | 0.051 (2) | 0.0263 (18) | 0.0053 (15) | 0.0037 (15) | 0.0039 (16) |
| C5C | 0.0263 (16) | 0.0366 (18) | 0.0310 (18) | 0.0012 (14) | 0.0057 (15) | −0.0008 (14) |
| C6C | 0.0254 (16) | 0.0293 (17) | 0.0195 (16) | 0.0028 (13) | 0.0055 (14) | 0.0032 (13) |
| C7C | 0.0232 (16) | 0.0297 (17) | 0.0218 (16) | −0.0043 (13) | 0.0030 (14) | 0.0020 (13) |
| C8C | 0.0285 (16) | 0.0322 (18) | 0.0250 (17) | −0.0099 (14) | 0.0071 (14) | 0.0019 (14) |
| C9C | 0.0362 (17) | 0.0253 (16) | 0.0283 (17) | −0.0101 (14) | 0.0056 (15) | 0.0017 (14) |
| C10C | 0.0293 (16) | 0.0268 (17) | 0.0254 (17) | −0.0035 (13) | 0.0079 (14) | 0.0033 (13) |
| C11C | 0.0334 (17) | 0.0402 (19) | 0.0289 (17) | −0.0138 (14) | 0.0003 (15) | −0.0041 (15) |
| C12C | 0.0367 (17) | 0.0281 (17) | 0.0327 (18) | −0.0041 (14) | 0.0033 (15) | 0.0062 (14) |
| N1D | 0.0219 (13) | 0.0243 (13) | 0.0228 (14) | −0.0046 (11) | −0.0032 (12) | 0.0054 (11) |
| N2D | 0.0226 (13) | 0.0231 (13) | 0.0264 (13) | −0.0004 (10) | 0.0059 (11) | −0.0020 (11) |
| N3D | 0.0238 (12) | 0.0205 (12) | 0.0235 (13) | 0.0034 (10) | 0.0048 (11) | 0.0039 (11) |
| C1D | 0.0222 (15) | 0.0250 (16) | 0.0329 (18) | −0.0002 (12) | 0.0006 (14) | 0.0049 (14) |
| C2D | 0.0318 (17) | 0.0255 (16) | 0.0310 (18) | −0.0005 (13) | 0.0095 (15) | 0.0070 (14) |
| C3D | 0.0424 (19) | 0.0229 (16) | 0.0258 (17) | 0.0080 (14) | 0.0076 (16) | 0.0027 (13) |
| C4D | 0.0258 (16) | 0.0296 (17) | 0.0278 (17) | 0.0061 (13) | 0.0007 (14) | −0.0003 (14) |
| C5D | 0.0201 (15) | 0.0281 (16) | 0.0256 (17) | −0.0009 (12) | 0.0012 (14) | −0.0007 (13) |
| C6D | 0.0303 (16) | 0.0163 (14) | 0.0211 (16) | 0.0021 (12) | 0.0052 (14) | −0.0001 (12) |
| C7D | 0.0229 (15) | 0.0171 (14) | 0.0238 (16) | 0.0021 (12) | 0.0059 (14) | −0.0031 (13) |
| C8D | 0.0270 (15) | 0.0265 (16) | 0.0254 (17) | −0.0021 (13) | 0.0083 (14) | −0.0026 (14) |
| C9D | 0.0288 (16) | 0.0274 (16) | 0.0259 (17) | −0.0020 (13) | 0.0119 (15) | 0.0000 (13) |
| C10D | 0.0289 (16) | 0.0177 (14) | 0.0222 (16) | 0.0043 (12) | 0.0072 (14) | −0.0033 (12) |
| C11D | 0.0291 (16) | 0.0438 (19) | 0.0348 (19) | −0.0069 (14) | 0.0089 (15) | 0.0015 (15) |
| C12D | 0.0337 (16) | 0.0231 (16) | 0.0269 (17) | 0.0004 (13) | 0.0059 (14) | 0.0012 (13) |
Geometric parameters (Å, º)
| N1A—C7A | 1.368 (3) | N1C—C7C | 1.378 (3) |
| N1A—C6A | 1.407 (3) | N1C—C6C | 1.408 (3) |
| N1A—H1AB | 0.91 (3) | N1C—H1CB | 0.94 (3) |
| N2A—C7A | 1.338 (3) | N2C—C7C | 1.334 (3) |
| N2A—C8A | 1.353 (3) | N2C—C8C | 1.346 (3) |
| N3A—C10A | 1.340 (3) | N3C—C10C | 1.343 (3) |
| N3A—C7A | 1.354 (3) | N3C—C7C | 1.357 (3) |
| C1A—C2A | 1.380 (4) | C1C—C2C | 1.385 (4) |
| C1A—C6A | 1.398 (4) | C1C—C6C | 1.389 (4) |
| C1A—H1AA | 0.9500 | C1C—H1CA | 0.9500 |
| C2A—C3A | 1.390 (4) | C2C—C3C | 1.382 (4) |
| C2A—H2AA | 0.9500 | C2C—H2CA | 0.9500 |
| C3A—C4A | 1.387 (4) | C3C—C4C | 1.384 (4) |
| C3A—H3AA | 0.9500 | C3C—H3CA | 0.9500 |
| C4A—C5A | 1.383 (4) | C4C—C5C | 1.391 (4) |
| C4A—H4AA | 0.9500 | C4C—H4CA | 0.9500 |
| C5A—C6A | 1.390 (4) | C5C—C6C | 1.392 (4) |
| C5A—H5AA | 0.9500 | C5C—H5CA | 0.9500 |
| C8A—C9A | 1.380 (4) | C8C—C9C | 1.382 (4) |
| C8A—C11A | 1.501 (4) | C8C—C11C | 1.501 (4) |
| C9A—C10A | 1.385 (4) | C9C—C10C | 1.387 (4) |
| C9A—H9AA | 0.9500 | C9C—H9CA | 0.9500 |
| C10A—C12A | 1.504 (4) | C10C—C12C | 1.497 (4) |
| C11A—H11A | 0.9800 | C11C—H11G | 0.9800 |
| C11A—H11B | 0.9800 | C11C—H11H | 0.9800 |
| C11A—H11C | 0.9800 | C11C—H11I | 0.9800 |
| C12A—H12A | 0.9800 | C12C—H12G | 0.9800 |
| C12A—H12B | 0.9800 | C12C—H12H | 0.9800 |
| C12A—H12C | 0.9800 | C12C—H12I | 0.9800 |
| N1B—C7B | 1.375 (3) | N1D—C7D | 1.371 (3) |
| N1B—C6B | 1.410 (3) | N1D—C6D | 1.408 (3) |
| N1B—H1BB | 0.97 (3) | N1D—H1DB | 0.92 (3) |
| N2B—C7B | 1.338 (3) | N2D—C7D | 1.339 (3) |
| N2B—C8B | 1.356 (3) | N2D—C8D | 1.357 (3) |
| N3B—C10B | 1.339 (3) | N3D—C10D | 1.339 (3) |
| N3B—C7B | 1.349 (3) | N3D—C7D | 1.352 (3) |
| C1B—C2B | 1.378 (4) | C1D—C2D | 1.374 (4) |
| C1B—C6B | 1.397 (3) | C1D—C6D | 1.397 (3) |
| C1B—H1BA | 0.9500 | C1D—H1DA | 0.9500 |
| C2B—C3B | 1.389 (4) | C2D—C3D | 1.384 (4) |
| C2B—H2BA | 0.9500 | C2D—H2DA | 0.9500 |
| C3B—C4B | 1.378 (4) | C3D—C4D | 1.380 (4) |
| C3B—H3BA | 0.9500 | C3D—H3DA | 0.9500 |
| C4B—C5B | 1.389 (4) | C4D—C5D | 1.383 (4) |
| C4B—H4BA | 0.9500 | C4D—H4DA | 0.9500 |
| C5B—C6B | 1.388 (4) | C5D—C6D | 1.394 (4) |
| C5B—H5BA | 0.9500 | C5D—H5DA | 0.9500 |
| C8B—C9B | 1.381 (4) | C8D—C9D | 1.375 (4) |
| C8B—C11B | 1.500 (4) | C8D—C11D | 1.500 (4) |
| C9B—C10B | 1.386 (4) | C9D—C10D | 1.392 (4) |
| C9B—H9BA | 0.9500 | C9D—H9DA | 0.9500 |
| C10B—C12B | 1.505 (4) | C10D—C12D | 1.496 (4) |
| C11B—H11D | 0.9800 | C11D—H11J | 0.9800 |
| C11B—H11E | 0.9800 | C11D—H11K | 0.9800 |
| C11B—H11F | 0.9800 | C11D—H11L | 0.9800 |
| C12B—H12D | 0.9800 | C12D—H12J | 0.9800 |
| C12B—H12E | 0.9800 | C12D—H12K | 0.9800 |
| C12B—H12F | 0.9800 | C12D—H12L | 0.9800 |
| C7A—N1A—C6A | 131.9 (2) | C7C—N1C—C6C | 131.4 (3) |
| C7A—N1A—H1AB | 111.3 (17) | C7C—N1C—H1CB | 111 (2) |
| C6A—N1A—H1AB | 116.8 (16) | C6C—N1C—H1CB | 117 (2) |
| C7A—N2A—C8A | 115.7 (3) | C7C—N2C—C8C | 116.1 (3) |
| C10A—N3A—C7A | 116.2 (3) | C10C—N3C—C7C | 116.3 (2) |
| C2A—C1A—C6A | 121.0 (3) | C2C—C1C—C6C | 120.8 (3) |
| C2A—C1A—H1AA | 119.5 | C2C—C1C—H1CA | 119.6 |
| C6A—C1A—H1AA | 119.5 | C6C—C1C—H1CA | 119.6 |
| C1A—C2A—C3A | 120.3 (3) | C3C—C2C—C1C | 120.3 (3) |
| C1A—C2A—H2AA | 119.8 | C3C—C2C—H2CA | 119.8 |
| C3A—C2A—H2AA | 119.8 | C1C—C2C—H2CA | 119.8 |
| C4A—C3A—C2A | 118.6 (3) | C2C—C3C—C4C | 118.8 (3) |
| C4A—C3A—H3AA | 120.7 | C2C—C3C—H3CA | 120.6 |
| C2A—C3A—H3AA | 120.7 | C4C—C3C—H3CA | 120.6 |
| C5A—C4A—C3A | 121.6 (3) | C3C—C4C—C5C | 121.6 (3) |
| C5A—C4A—H4AA | 119.2 | C3C—C4C—H4CA | 119.2 |
| C3A—C4A—H4AA | 119.2 | C5C—C4C—H4CA | 119.2 |
| C4A—C5A—C6A | 119.8 (3) | C4C—C5C—C6C | 119.1 (3) |
| C4A—C5A—H5AA | 120.1 | C4C—C5C—H5CA | 120.4 |
| C6A—C5A—H5AA | 120.1 | C6C—C5C—H5CA | 120.4 |
| C5A—C6A—C1A | 118.7 (3) | C1C—C6C—C5C | 119.3 (3) |
| C5A—C6A—N1A | 125.4 (3) | C1C—C6C—N1C | 115.8 (2) |
| C1A—C6A—N1A | 115.8 (2) | C5C—C6C—N1C | 124.9 (3) |
| N2A—C7A—N3A | 126.8 (2) | N2C—C7C—N3C | 126.6 (3) |
| N2A—C7A—N1A | 120.6 (3) | N2C—C7C—N1C | 120.7 (3) |
| N3A—C7A—N1A | 112.5 (2) | N3C—C7C—N1C | 112.7 (2) |
| N2A—C8A—C9A | 121.5 (3) | N2C—C8C—C9C | 121.3 (3) |
| N2A—C8A—C11A | 116.2 (3) | N2C—C8C—C11C | 116.0 (3) |
| C9A—C8A—C11A | 122.2 (3) | C9C—C8C—C11C | 122.7 (3) |
| C8A—C9A—C10A | 118.6 (3) | C8C—C9C—C10C | 118.7 (3) |
| C8A—C9A—H9AA | 120.7 | C8C—C9C—H9CA | 120.6 |
| C10A—C9A—H9AA | 120.7 | C10C—C9C—H9CA | 120.6 |
| N3A—C10A—C9A | 121.1 (3) | N3C—C10C—C9C | 120.8 (3) |
| N3A—C10A—C12A | 117.0 (3) | N3C—C10C—C12C | 116.8 (3) |
| C9A—C10A—C12A | 121.9 (3) | C9C—C10C—C12C | 122.5 (3) |
| C8A—C11A—H11A | 109.5 | C8C—C11C—H11G | 109.5 |
| C8A—C11A—H11B | 109.5 | C8C—C11C—H11H | 109.5 |
| H11A—C11A—H11B | 109.5 | H11G—C11C—H11H | 109.5 |
| C8A—C11A—H11C | 109.5 | C8C—C11C—H11I | 109.5 |
| H11A—C11A—H11C | 109.5 | H11G—C11C—H11I | 109.5 |
| H11B—C11A—H11C | 109.5 | H11H—C11C—H11I | 109.5 |
| C10A—C12A—H12A | 109.5 | C10C—C12C—H12G | 109.5 |
| C10A—C12A—H12B | 109.5 | C10C—C12C—H12H | 109.5 |
| H12A—C12A—H12B | 109.5 | H12G—C12C—H12H | 109.5 |
| C10A—C12A—H12C | 109.5 | C10C—C12C—H12I | 109.5 |
| H12A—C12A—H12C | 109.5 | H12G—C12C—H12I | 109.5 |
| H12B—C12A—H12C | 109.5 | H12H—C12C—H12I | 109.5 |
| C7B—N1B—C6B | 130.8 (2) | C7D—N1D—C6D | 130.6 (3) |
| C7B—N1B—H1BB | 106.7 (18) | C7D—N1D—H1DB | 108.0 (17) |
| C6B—N1B—H1BB | 122.1 (17) | C6D—N1D—H1DB | 121.4 (17) |
| C7B—N2B—C8B | 115.8 (2) | C7D—N2D—C8D | 115.7 (2) |
| C10B—N3B—C7B | 116.6 (2) | C10D—N3D—C7D | 116.9 (2) |
| C2B—C1B—C6B | 120.7 (3) | C2D—C1D—C6D | 120.5 (3) |
| C2B—C1B—H1BA | 119.6 | C2D—C1D—H1DA | 119.8 |
| C6B—C1B—H1BA | 119.6 | C6D—C1D—H1DA | 119.8 |
| C1B—C2B—C3B | 120.3 (3) | C1D—C2D—C3D | 120.8 (3) |
| C1B—C2B—H2BA | 119.9 | C1D—C2D—H2DA | 119.6 |
| C3B—C2B—H2BA | 119.9 | C3D—C2D—H2DA | 119.6 |
| C4B—C3B—C2B | 119.3 (3) | C4D—C3D—C2D | 118.7 (3) |
| C4B—C3B—H3BA | 120.4 | C4D—C3D—H3DA | 120.6 |
| C2B—C3B—H3BA | 120.4 | C2D—C3D—H3DA | 120.6 |
| C3B—C4B—C5B | 120.9 (3) | C3D—C4D—C5D | 121.6 (3) |
| C3B—C4B—H4BA | 119.5 | C3D—C4D—H4DA | 119.2 |
| C5B—C4B—H4BA | 119.5 | C5D—C4D—H4DA | 119.2 |
| C6B—C5B—C4B | 120.0 (2) | C4D—C5D—C6D | 119.4 (2) |
| C6B—C5B—H5BA | 120.0 | C4D—C5D—H5DA | 120.3 |
| C4B—C5B—H5BA | 120.0 | C6D—C5D—H5DA | 120.3 |
| C5B—C6B—C1B | 118.8 (3) | C5D—C6D—C1D | 119.0 (2) |
| C5B—C6B—N1B | 124.8 (2) | C5D—C6D—N1D | 124.6 (2) |
| C1B—C6B—N1B | 116.4 (2) | C1D—C6D—N1D | 116.5 (2) |
| N2B—C7B—N3B | 126.7 (2) | N2D—C7D—N3D | 126.3 (2) |
| N2B—C7B—N1B | 120.6 (3) | N2D—C7D—N1D | 120.6 (3) |
| N3B—C7B—N1B | 112.7 (2) | N3D—C7D—N1D | 113.0 (2) |
| N2B—C8B—C9B | 121.3 (2) | N2D—C8D—C9D | 121.8 (2) |
| N2B—C8B—C11B | 116.6 (2) | N2D—C8D—C11D | 116.0 (3) |
| C9B—C8B—C11B | 122.1 (2) | C9D—C8D—C11D | 122.2 (2) |
| C8B—C9B—C10B | 118.7 (2) | C8D—C9D—C10D | 118.5 (2) |
| C8B—C9B—H9BA | 120.6 | C8D—C9D—H9DA | 120.7 |
| C10B—C9B—H9BA | 120.6 | C10D—C9D—H9DA | 120.7 |
| N3B—C10B—C9B | 120.9 (3) | N3D—C10D—C9D | 120.7 (3) |
| N3B—C10B—C12B | 117.3 (2) | N3D—C10D—C12D | 117.1 (2) |
| C9B—C10B—C12B | 121.8 (2) | C9D—C10D—C12D | 122.2 (2) |
| C8B—C11B—H11D | 109.5 | C8D—C11D—H11J | 109.5 |
| C8B—C11B—H11E | 109.5 | C8D—C11D—H11K | 109.5 |
| H11D—C11B—H11E | 109.5 | H11J—C11D—H11K | 109.5 |
| C8B—C11B—H11F | 109.5 | C8D—C11D—H11L | 109.5 |
| H11D—C11B—H11F | 109.5 | H11J—C11D—H11L | 109.5 |
| H11E—C11B—H11F | 109.5 | H11K—C11D—H11L | 109.5 |
| C10B—C12B—H12D | 109.5 | C10D—C12D—H12J | 109.5 |
| C10B—C12B—H12E | 109.5 | C10D—C12D—H12K | 109.5 |
| H12D—C12B—H12E | 109.5 | H12J—C12D—H12K | 109.5 |
| C10B—C12B—H12F | 109.5 | C10D—C12D—H12L | 109.5 |
| H12D—C12B—H12F | 109.5 | H12J—C12D—H12L | 109.5 |
| H12E—C12B—H12F | 109.5 | H12K—C12D—H12L | 109.5 |
| C6A—C1A—C2A—C3A | 0.4 (4) | C6C—C1C—C2C—C3C | 0.3 (4) |
| C1A—C2A—C3A—C4A | 0.1 (4) | C1C—C2C—C3C—C4C | −1.2 (4) |
| C2A—C3A—C4A—C5A | −0.3 (4) | C2C—C3C—C4C—C5C | 1.7 (4) |
| C3A—C4A—C5A—C6A | 0.2 (4) | C3C—C4C—C5C—C6C | −1.2 (4) |
| C4A—C5A—C6A—C1A | 0.3 (4) | C2C—C1C—C6C—C5C | 0.2 (4) |
| C4A—C5A—C6A—N1A | −178.5 (2) | C2C—C1C—C6C—N1C | −179.2 (2) |
| C2A—C1A—C6A—C5A | −0.5 (4) | C4C—C5C—C6C—C1C | 0.2 (4) |
| C2A—C1A—C6A—N1A | 178.3 (2) | C4C—C5C—C6C—N1C | 179.6 (2) |
| C7A—N1A—C6A—C5A | 9.4 (4) | C7C—N1C—C6C—C1C | 173.8 (3) |
| C7A—N1A—C6A—C1A | −169.3 (3) | C7C—N1C—C6C—C5C | −5.6 (4) |
| C8A—N2A—C7A—N3A | 1.0 (4) | C8C—N2C—C7C—N3C | −3.5 (4) |
| C8A—N2A—C7A—N1A | −176.9 (2) | C8C—N2C—C7C—N1C | 176.3 (2) |
| C10A—N3A—C7A—N2A | −1.8 (4) | C10C—N3C—C7C—N2C | 4.3 (4) |
| C10A—N3A—C7A—N1A | 176.3 (2) | C10C—N3C—C7C—N1C | −175.6 (2) |
| C6A—N1A—C7A—N2A | −16.4 (4) | C6C—N1C—C7C—N2C | 12.0 (4) |
| C6A—N1A—C7A—N3A | 165.3 (2) | C6C—N1C—C7C—N3C | −168.1 (2) |
| C7A—N2A—C8A—C9A | 0.2 (3) | C7C—N2C—C8C—C9C | −0.2 (4) |
| C7A—N2A—C8A—C11A | 178.5 (2) | C7C—N2C—C8C—C11C | −179.7 (2) |
| N2A—C8A—C9A—C10A | −0.4 (4) | N2C—C8C—C9C—C10C | 2.9 (4) |
| C11A—C8A—C9A—C10A | −178.7 (2) | C11C—C8C—C9C—C10C | −177.7 (2) |
| C7A—N3A—C10A—C9A | 1.4 (3) | C7C—N3C—C10C—C9C | −1.2 (3) |
| C7A—N3A—C10A—C12A | −178.7 (2) | C7C—N3C—C10C—C12C | 179.6 (2) |
| C8A—C9A—C10A—N3A | −0.4 (4) | C8C—C9C—C10C—N3C | −2.1 (4) |
| C8A—C9A—C10A—C12A | 179.7 (2) | C8C—C9C—C10C—C12C | 177.1 (2) |
| C6B—C1B—C2B—C3B | 0.4 (4) | C6D—C1D—C2D—C3D | −1.1 (4) |
| C1B—C2B—C3B—C4B | 0.6 (4) | C1D—C2D—C3D—C4D | −0.4 (4) |
| C2B—C3B—C4B—C5B | −0.1 (4) | C2D—C3D—C4D—C5D | 0.6 (4) |
| C3B—C4B—C5B—C6B | −1.5 (4) | C3D—C4D—C5D—C6D | 0.9 (4) |
| C4B—C5B—C6B—C1B | 2.5 (4) | C4D—C5D—C6D—C1D | −2.4 (4) |
| C4B—C5B—C6B—N1B | −177.4 (2) | C4D—C5D—C6D—N1D | 178.1 (2) |
| C2B—C1B—C6B—C5B | −1.9 (4) | C2D—C1D—C6D—C5D | 2.5 (4) |
| C2B—C1B—C6B—N1B | 177.9 (2) | C2D—C1D—C6D—N1D | −177.9 (2) |
| C7B—N1B—C6B—C5B | 20.7 (4) | C7D—N1D—C6D—C5D | −21.4 (4) |
| C7B—N1B—C6B—C1B | −159.1 (2) | C7D—N1D—C6D—C1D | 159.0 (2) |
| C8B—N2B—C7B—N3B | 1.1 (4) | C8D—N2D—C7D—N3D | −2.2 (4) |
| C8B—N2B—C7B—N1B | −177.7 (2) | C8D—N2D—C7D—N1D | 176.3 (2) |
| C10B—N3B—C7B—N2B | −2.7 (4) | C10D—N3D—C7D—N2D | 2.9 (4) |
| C10B—N3B—C7B—N1B | 176.2 (2) | C10D—N3D—C7D—N1D | −175.6 (2) |
| C6B—N1B—C7B—N2B | −9.0 (4) | C6D—N1D—C7D—N2D | 13.2 (4) |
| C6B—N1B—C7B—N3B | 172.0 (2) | C6D—N1D—C7D—N3D | −168.1 (2) |
| C7B—N2B—C8B—C9B | 1.4 (3) | C7D—N2D—C8D—C9D | −0.5 (4) |
| C7B—N2B—C8B—C11B | −179.2 (2) | C7D—N2D—C8D—C11D | −179.7 (2) |
| N2B—C8B—C9B—C10B | −2.2 (4) | N2D—C8D—C9D—C10D | 2.1 (4) |
| C11B—C8B—C9B—C10B | 178.5 (2) | C11D—C8D—C9D—C10D | −178.7 (2) |
| C7B—N3B—C10B—C9B | 1.8 (3) | C7D—N3D—C10D—C9D | −1.1 (3) |
| C7B—N3B—C10B—C12B | −176.9 (2) | C7D—N3D—C10D—C12D | 177.2 (2) |
| C8B—C9B—C10B—N3B | 0.5 (4) | C8D—C9D—C10D—N3D | −1.3 (4) |
| C8B—C9B—C10B—C12B | 179.1 (2) | C8D—C9D—C10D—C12D | −179.5 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1A—H1AB···N3B | 0.91 (3) | 2.11 (3) | 2.997 (3) | 165 (2) |
| N1B—H1BB···N3A | 0.97 (3) | 2.08 (3) | 3.022 (3) | 162 (3) |
| N1C—H1CB···N3D | 0.94 (3) | 2.05 (3) | 2.975 (3) | 166 (3) |
| N1D—H1DB···N3C | 0.92 (3) | 2.08 (3) | 2.987 (3) | 167 (3) |
References
- Buhmann, U., Westermann, J., Baumert, D., Pieroh, E., Cliff, G. R. & Richards, I. C. (1988). US Patent US4783459.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Li, J.-C., Feng, Y.-H., Lin, Q. & Zhang, D.-L. (2007). Acta Cryst. E63, o1162–o1163.
- Li, J.-C., Qiu, X.-Q., Feng, Y.-H. & Lin, Q. (2008). Acta Cryst. E64, o318. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Pöppler, A.-C., Corlett, E. K., Pearce, H., Seymour, M. P., Reid, M., Montgomery, M. G. & Brown, S. P. (2017). Acta Cryst. C73, 149–156. [DOI] [PMC free article] [PubMed]
- Rigaku (2015). CrysAlis PRO. Rigaku Oxford Diffraction, Oxfordshire, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Sun, X.-H., Li, J., Liu, Y. & Ma, H.-X. (2011). Acta Chim. Sin. 69, 1909–1914.
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989017007563/hb7679sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017007563/hb7679Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017007563/hb7679Isup3.cml
CCDC reference: 1549998
Additional supporting information: crystallographic information; 3D view; checkCIF report






