Abstract
To successfully colonize the oral cavity, bacteria must directly or indirectly adhere to available oral surfaces. Fusobacterium nucleatum plays an important role in oral biofilm community development due to its broad adherence abilities, serving as a bridge between members of the oral biofilm that cannot directly bind to each other. In our efforts to characterize the molecular mechanisms utilized by F. nucleatum to physically bind to key members of the oral community, we investigated the involvement of F. nucleatum outer membrane proteins in its ability to bind to the pioneer biofilm colonizer, Streptococcus gordonii. Here, we present evidence that in addition to the previously characterized fusobacterial adhesin RadD, the interaction between F. nucleatum ATCC 23726 and S. gordonii V288 involves a second outer membrane protein, which we named coaggregation mediating protein A (CmpA). We also characterized the role of CmpA in dual‐species biofilm formation with S. gordonii V288, evaluated growth‐phase‐dependent as well as biofilm expression profiles of radD and cmpA, and confirmed an important role for CmpA, especially under biofilm growth conditions. Our findings underscore the complex set of specific interactions involved in physical binding and thus community integration of interacting bacterial species. This complex set of interactions could have critical implications for the formation and maturation of the oral biofilms in vivo, and could provide clues to the mechanism behind the distribution of organisms inside the human oral cavity.
Keywords: Adhesin, Fusobacterium nucleatum, interspecies interaction, oral biofilm, Streptococcus gordonii
1. INTRODUCTION
The human oral cavity is home to hundreds of bacterial species (Aas, Paster, Stokes, Olsen, & Dewhirst, 2005; Chen et al., 2010; Dewhirst et al., 2010; Kuramitsu, He, Lux, Anderson, & Shi, 2007; Paster et al., 2001). These organisms have coevolved and form a complex network of physical and metabolic interactions with their neighbors, as well as the human host. These connections promote the development of a dynamic and well‐organized multispecies microbial community, also known as the oral biofilm or dental plaque. Although a core set of organisms can be found in the oral cavity of most individuals, a variable microbiome exists in response to unique individual determinants (Paster, Olsen, Aas, & Dewhirst, 2006; Turnbaugh et al., 2007; Zaura, Keijser, Huse, & Crielaard, 2009). Depending on the combination of species present in the oral cavity, these communities can cause a multitude of oral and systemic diseases including dental caries, periodontal disease, and many others (Zarco, Vess, & Ginsburg, 2012).
To form these multispecies communities, oral bacteria must first, directly or indirectly, attach to a surface in the oral cavity. Oral streptococci are among the first species to attach to the surface of the teeth and they comprise the majority of the early colonizers (Avila, Ojcius, & Yilmaz, 2009; Diaz et al., 2006; Dige, Nilsson, Kilian, & Nyvad, 2007; Dige, Nyengaard, Kilian, & Nyvad, 2009; Nyvad & Kilian, 1987, 1990). Once established, the early colonizers alter their microenvironment and serve as anchors for subsequent colonizers of the dental plaque. The Gram‐negative bacterium Fusobacterium nucleatum is considered an important species in the development and maturation process of dental plaque (Jorth et al., 2014; Kolenbrander & London, 1993) as it contributes to important structural and metabolic changes. Structurally, F. nucleatum binds to numerous species in the oral cavity, serving as a bridge between early‐ and late‐colonizing species (Guo, He, & Shi, 2014; Kolenbrander, Andersen, & Moore, 1989; Kolenbrander, Parrish, Andersen, & Greenberg, 1995). Metabolically, F. nucleatum is a key contributor to butyrate production (Jorth et al., 2014), which has been linked to the development of periodontal disease (Niederman, Buyle‐Bodin, Lu, Robinson, & Naleway, 1997).
Earlier studies have focused on the identification and characterization of molecular components required for the direct cell‐to‐cell interaction among members of the oral community. Of special interest to our laboratory is the characterization of the interactions between F. nucleatum and other members of the dental plaque community. To date, only two F. nucleatum adhesins have been characterized for their role in interspecies interaction: Fap2 and RadD. Fap2 is a galactose‐inhibitable adhesin, which has been implicated in the interaction between F. nucleatum and the periodontal pathogen Porphyromonas gingivalis (Coppenhagen‐Glazer et al., 2015). RadD is an arginine‐inhibitable adhesin required for the interaction between F. nucleatum and multiple Gram‐positive members of the dental plaque, including the early colonizers Actinomyces naeslundii, Streptococcus sanguinis, Streptococcus oralis, and Streptococcus gordonii (Kaplan, Lux, Haake, & Shi, 2009).
In the work described here, we report the identification of a previously uncharacterized adhesin, which we named coaggregation mediating protein A (CmpA), involved in the interaction between F. nucleatum ATCC strain 23726 and S. gordonii V288. Along with RadD, CmpA plays an important role in the ability of F. nucleatum to coaggregate and form dual‐species biofilms with S. gordonii V288.
2. MATERIALS AND METHODS
2.1. Bacteria and culture conditions
All bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise stated, F. nucleatum strains were grown in Columbia broth or on Columbia agar plates (BD Difco, Detroit, MI) supplemented with 5% defibrinated sheep blood (Colorado Serum Company, Denver, CO) under anaerobic conditions (5% H2, 5% CO2, 90% N2) at 37°C. When necessary, Thiamphenicol and Clindamycin (MP Biomedicals, Irvine, CA) at 5 μg/ml and 0.2 μg/ml, respectively, were added to the media. For P. gingivalis growth, Columbia broth was supplemented with hemin and menadione at 5 μg/ml and 1 μg/ml, respectively. Columbia agar plates were supplemented with 5% defibrinated sheep blood. S. sanguinis and S. gordonii were grown in Todd Hewitt (TH) broth or agar plates (BD Difco, Detroit MI) at 37°C under anaerobic conditions. Streptococcus gordonii selection was carried out with 5 μg/ml erythromycin added to the media. Escherichia coli was grown aerobically at 37°C in Luria–Bertani (LB) broth or agar plates (BD Difco, Detroit, MI). Escherichia coli selection was carried out with 100 μg/ml erythromycin or ampicillin added to the media.
Table 1.
Bacterial strains and plasmids used in this study
| Species | Strains | Description | Source |
|---|---|---|---|
| Escherichia coli | DH10B™ | F‐ mcrA Δ(mrr‐hsdRMS‐mcrBC) Φ80lacZΔM15 Δ lacX74 recA1 araD139 Δ(araleu)7697 galU galK rpsL (StrR) endA1 nupG | Thermo Fisher |
| Fusobacterium nucleatum | ATCC 23726 | ssp. nucleatum wild type | ATCC |
| ΔFn0254 | ATCC 23726 Fn0254::pIP0254 | Kaplan et al. (2009) | |
| ΔFn1526 (radD) | ATCC 23726 Fn1526::pIP1526 | Kaplan et al. (2009) | |
| ΔFn1554 (cmpA) | ATCC 23726 Fn1554::pIP1554 | Kaplan et al. (2009) | |
| ΔFn1893 | ATCC 23726 Fn1893::pIP1983 | Kaplan et al. (2009) | |
| ΔFn2047 | ATCC 23726 Fn2047::pIP2047 | Kaplan et al. (2009) | |
| aim1 | ATCC 23726 aim1::pIPaim1 | Kaplan et al. (2009) | |
| BL83 | ATCC 23726 radD::catP fn1554::Erm | This study | |
| Streptococcus gordonii | ATCC 10558 | S. gordonii wild type | ATCC |
| ATCC 51656 | S. gordonii wild type | ATCC | |
| DL1 | S. gordonii wild type | ATCC | |
| V288 | S. gordonii wild type | ATCC | |
| BL98 | V288 attB::mCherry | This study | |
| Streptococcus sanguinis | ATCC 10556 | WT S. sanguinis | ATCC |
| Porphyromonas gingivalis | 4612 | WT P. gingivalis | Lamont et al. (1995) |
| Plasmid | Description | Purpose | Source |
|---|---|---|---|
| pHS70 | F. nucleatum suicide vector; ermB | Suicide plasmid | S. Haake (unpublished data) |
| pBPL9 | pHS31 with Fn1554_ inserted into SnaBI site | Gene inactivation plasmid | This study |
| pVA8912:mCherry | pVA8912 carrying mCherry under the control of ldh promoter. | Vickerman et al. (2015) |
2.2. Coaggregation assay
Coaggregations were performed in coaggregation buffer (CAB) (150 mmol/L NaCl, 1 mmol/L Tris, 0.1 mmol/L CaCl2, 0.1 mmol/L MgCl2) as previously described with minor modifications (Kaplan et al., 2009). Overnight cultures of S. gordonii and F. nucleatum were diluted 10‐fold into fresh medium in the morning and grown until they reached OD600 1.5 for S. gordonii and OD600 2.0 for F. nucleatum. Bacterial cells were washed with CAB and resuspended, also with CAB. Equal numbers of bacterial cells were combined to a final concentration of 2 × 109 cells ml−1 in a 1.5 ml microcentrifuge tube. To account for autoaggregation, the cells were also incubated in the absence of the binding partner. The tubes were vortexed for 10 s and incubated for 10 min at room temperature. After incubation, the bacterial mixtures were centrifuged at low speed (100g) for 1 min to pellet cellular aggregates, while leaving the nonaggregated bacteria in suspension. The supernatant was then removed without disturbing the pellet, and the optical density of the recovered supernatant was measured at 600 nm. For coaggregation inhibition assays, 50 mmol/L of l‐arginine was added to the reaction tube containing the different F. nucleatum strains and vortexed before addition of the partner strain. Relative coaggregation was determined by subtracting the turbidity of the recovered supernatant after coaggregation from the turbidity of the cell mixture before coaggregation and dividing the results by the turbidity before coaggregation.
2.3. Strain construction
2.3.1. Fusobacterium nucleatum radD cmpA double mutant
An internal gene fragment from cmpA was amplified via PCR using Taq DNA polymerase and the primer pair 1554F’ 5′‐GAATGGCAGGATTTGCTTCA‐3′ and 1554R’ 5′‐TTGGTTAGTTCCCTTTGCGTA‐3′ (Kaplan et al., 2009). The amplicon was subcloned into pJET2.1 (New England Biolabs, Ipswich, MA) according to the manufacturer's protocol, prior to cloning into the F. nucleatum suicide vector pHS70 (see Table 1 for plasmid details). The resulting integration vector, named pBPL9, was transformed into the F. nucleatum radD mutant strain as described previously (Haake, Yoder, Attarian, & Podkaminer, 2000). Mutants were selected on Columbia agar plates containing 5% blood and 0.2 μg/ml clindamycin. Clindamycin resistance was confirmed by restreaking colonies onto Columbia blood plates containing 0.2 μg/ml clindamycin. Thiamphenicol resistance, and therefore radD mutation was confirmed by patching clindamycin‐resistant colonies onto Columbia blood plates containing 5 μg/ml thiamphenicol. Mutants were also confirmed by PCR analysis.
2.3.2. mCherry+ S. gordonii
To construct the mCherry‐expressing S. gordonii strain BL98, we transformed plasmid pVA8912 (Vickerman, Mansfield, Zhu, Walters, & Banas, 2015) into the wild‐type S. gordonii V288 according to previously published protocol (Warren, Lund, Jones, & Hruby, 2007), utilizing competence‐stimulating peptide N‐DVRSNKIRLWWENIFFNKK (Pepmic, Suzhou, China). The mCherry encoding gene was inserted into S. gordonii attB site and is expressed under the control of the ldh promoter (for a full description of the construct, see Vickerman et al., 2015). The mCherry expression had no effect on coaggregation or dual‐species biofilm formation with F. nucleatum 23726.
2.4. Biofilm growth
One milliliter of SHI‐FSMS (50% SHI medium, Tian et al., 2010, 25% filtered saliva, 0.5% mannose, 0.5% sucrose; de Avila et al., 2015) containing 1 × 108 F. nucleatum cells and 5 × 104 S. gordonii cells diluted from overnight cultures were added to the wells of a 24‐well polystyrene culture plates (Thermo Fisher Scientific, Waltham, MA, USA) and incubated overnight under anaerobic conditions (5% H2, 5% CO2, 90% N2) at 37°C. After overnight growth, the planktonic cells were removed and the biofilm was washed three times with 500 μl of pre‐reduced, sterile phosphate‐buffered saline (PBS). The biofilm that remained attached to the wells was either processed for confocal laser scanning microscopy (CLSM) analysis or collected for DNA isolation.
2.5. Confocal laser scanning microscopy
Overnight biofilm samples were fluorescently labeled with the nucleic acid staining dye SYTO9 (Invitrogen, Carlsbad, CA) according to manufacturer's instructions and visualized using a Leica SPE I inverted CLSM (Leica, Wetzlar, Germany). SYTO9 fluorescence was measured using an excitation of 488 nm and emission at 530 nm. mCherry was expressed from the chromosome of S. gordonii and detected using an excitation of 543 nm and emission at 600 nm. Image analysis was carried out with the open‐source image processing software, FIJI (Schindelin et al., 2012).
2.6. Nucleic acid isolation
Genomic DNA was extracted from biofilms using MasterPure™ DNA Purification Kit (Epicenter®, Madison, WI, USA) according to manufacturer's instructions. The concentration of purified bacterial DNA was determined by Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA).
Total RNA was extracted using PureLinK™ RNA Mini kit (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer's instructions. Genomic DNA contamination was removed from total RNA using Turbo DNA‐free™ kit (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer's instructions and confirmed by PCR using 16S rRNA primers Bac1 and Bac2 (Rupf, Merte, & Eschrich, 1999).
2.7. qPCR
To quantify the relative proportions of each species in the respective dual‐species biofilms, previously designed species‐specific primer pairs were used (Park, Shokeen, Haake, & Lux, 2016). For F. nucleatum ATCC 23726 and its mutant derivatives, a portion of the Fusobacterium‐specific fomA gene was amplified with Fn‐F (forward) 5′AGAGTTTGATCCTGGCTCAG3′ and Fn‐R (reverse) 5′GTCATCGTGCACACAGAATTGCTG3′ primers. For S gordonii, srtA‐F (forward) 5′TATTATGGTGCTGGTACGATGAAAGAGACTC3′ and srtA‐R (Reverse) 5′TATAGATTTTCATACCAGCCTTAGCACGATC3′ primers were chosen to amplify a portion of S. gordonii srtA gene. Primer pairs were tested for possible cross‐reactivity with the other species and for amplification efficiency. The efficiency range observed was between 90% and 100%. Real‐time qPCR was performed using an iCycler Thermal Cycler (Bio‐Rad, Hercules, CA) in a total volume of 20 μl containing 2 μl of 10× iQ SYBR® Green Supermix (Bio‐Rad, Hercules, CA), 0.5 μmol/L each of forward and reverse primers, 7 μl of Millipore water, and 1 μl (10 ng) of template DNA. Amplification and detection were carried out in 96‐well optical plates (Thermo Fisher Scientific, Waltham, MA). Each PCR run was carried out with an initial incubation of 10 min at 95°C followed by 40 cycles of denaturing at 95°C for 15 s and annealing and elongation at 60°C for 1 min. After the 40 cycles of amplification, an additional denaturing step was performed at 95°C for 1 min followed by annealing and elongation at 60°C for 1 min. A melting curve analysis was completed after each run. In addition, gel electrophoresis was utilized during optimization step to determine size and number of amplicons. The DNA concentrations (ng ml−1) were calculated with standard curves obtained by 10‐fold serial dilutions of previously purified and quantified bacterial genomic DNA. Three independent qPCR runs were performed with three technical replicates for each sample to assess reproducibility and inter‐run variability.
2.8. Transcriptional analysis
To determine radD and cmpA expression pattern, 1 μg of total RNA was used for cDNA synthesis using SuperScript® III First‐Strand (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer's protocol. For qRT‐PCR, iQ SYBR® Green Supermix (Bio‐Rad, Hercules, CA, USA) was used for fluorescence detection with the iCycler real‐time PCR system (Bio‐Rad), according to the manufacturer's instructions. radD cDNA was amplified using 5′‐GGATTTATCTTTGCTAATTGGGGAAATTATAG‐3′ forward and 5′‐ACTATTCCATATTCTCCATAATATTTCCCATTAGA‐3′ reverse primers (B. Shokeen, J. Park, and R. Lux unpublished data), and cmpA was amplified using 5′‐TTGGGATCAAGGAAAACATCAATTAGG‐3′ forward and 5′‐ATAATTCCTTTATTATCTCCCATATAAGCAATACC‐3′ reverse primers. Expression levels of rpoB were determined using 5′‐CAAAAACTCATTGAAAGACTTGATTTTGGA‐3′ forward and 5′‐GAATGCTAATTCAAATCCTTTTTCTTCCCT‐3′ reverse primers for normalization of the qRT‐PCR data (B. Shokeen, J. Park, and R. Lux unpublished data).
2.9. Statistical analysis
Student's t‐test was performed to determine statistical significance using Excel 2010 (Microsoft, Seattle, WA, USA).
3. RESULTS
3.1. Multiple F. nucleatum adhesins are involved in its coaggregation with S. gordonii
For detailed characterization of the physical interaction between F. nucleatum and S. gordonii, we collected F. nucleatum cells, ATCC strain 23726, that were in stationary phase, after overnight growth, and cells that were in late exponential phase to perform coaggregation experiments with midexponential phase S. gordonii V288 cells. We observed less coaggregation with stationary phase F. nucleatum cultures (63.03% ± 2.39) when compared to those in late exponential phase (80.85% ± 0.5) (Figure 1a). Surprisingly, the coaggregation defect usually observed for the radD mutant derivative of F. nucleatum was less evident at the late exponential phase, compared to F. nucleatum collected the stationary growth phase (Figure 1a). This phenotype was not evident among other commonly used S. gordonii strains (DL1, ATCC 10558, and ATCC 51656) (Figure 1b). Thus, F. nucleatum coaggregation with S. gordonii V288 might require an additional, and yet uncharacterized, surface adhesin that is likely expressed as cells enter the exponential phase of growth.
Figure 1.
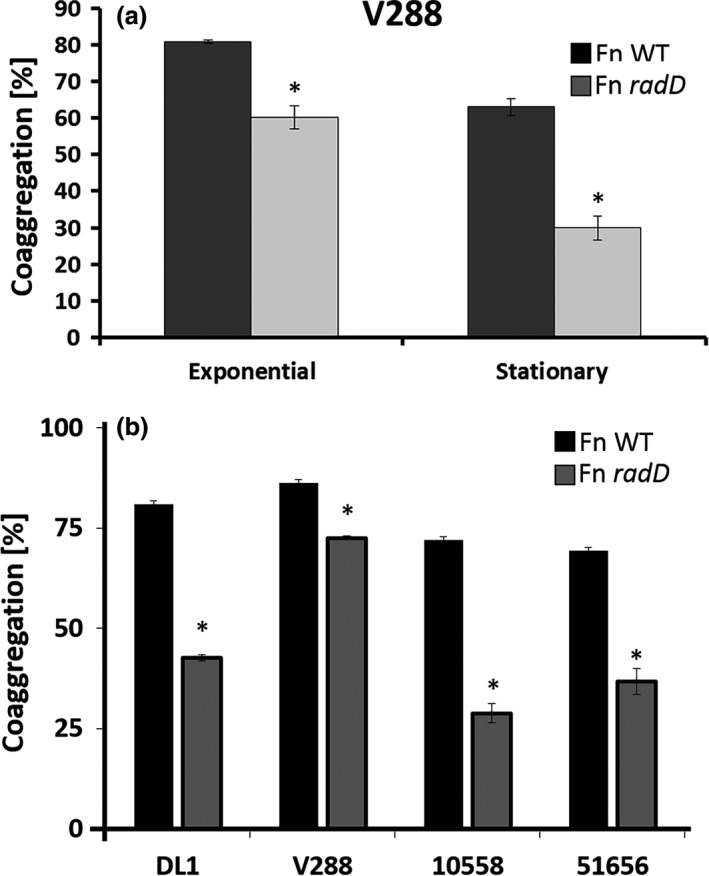
Quantitative coaggregation of (a) wild‐type (WT) Streptococcus gordonii strain V288 and the WT Fusobacterium nucleatum strain ATCC 23726, or the radD mutant derivative, at different phases of growth. (b) Quantitative coaggregation of WT S. gordonii strains (DL1, V288, ATCC 10558, and ATCC 51656) with WT F. nucleatum strain ATCC 23726, or the radD mutant derivative at exponential growth. Data represent means and standard deviation of percent coaggregation of at least three independent experiments. *p < .05 compared to wild‐type control
3.2. Identification of an additional adhesin involved in the interaction between F. nucleatum and S. gordonii
The genome of the sequenced F. nucleatum strain ATCC 25586 encodes at least eight large autotransporter‐like outer membrane proteins (OMPs): Fn0254, Fn0387, Fap2 (Fn1449), RadD (Fn1526), Fn1554, Fn1893, Fn2047, and Aim1 (Fn2058) (Kaplan et al., 2009), of which, six have been shown to bind arginine: Fn0254, RadD, Fn1554, Fn1893, Fn2047, and Aim1 (Kaplan et al., 2009). Since the interaction between F. nucleatum (ATCC 23726) and S. gordonii, including strain V288, is inhibited by the presence of 50 mmol/L arginine (Figure 4a of this manuscript; Kaplan et al., 2009), we hypothesized that the additional F. nucleatum adhesin involved in its interaction with S. gordonii was one of the arginine‐binding adhesins. Therefore, we screened the remaining five previously identified arginine‐binding adhesins (Fn0254, Fn1554, Fn1893, Fn2047, or Aim1) for their ability to coaggregate with S. gordonii V288. The Fn1554 mutant strain of F. nucleatum consistently coaggregated less with S. gordonii V288 than the wild‐type parent strain or any of the other mutants tested (Figure 2). Thus, we concluded that Fn1554 (now named CmpA for coaggregation mediating protein A) and RadD are important F. nucleatum coaggregation mediating proteins involved in its interaction with S. gordonii V288.
Figure 4.
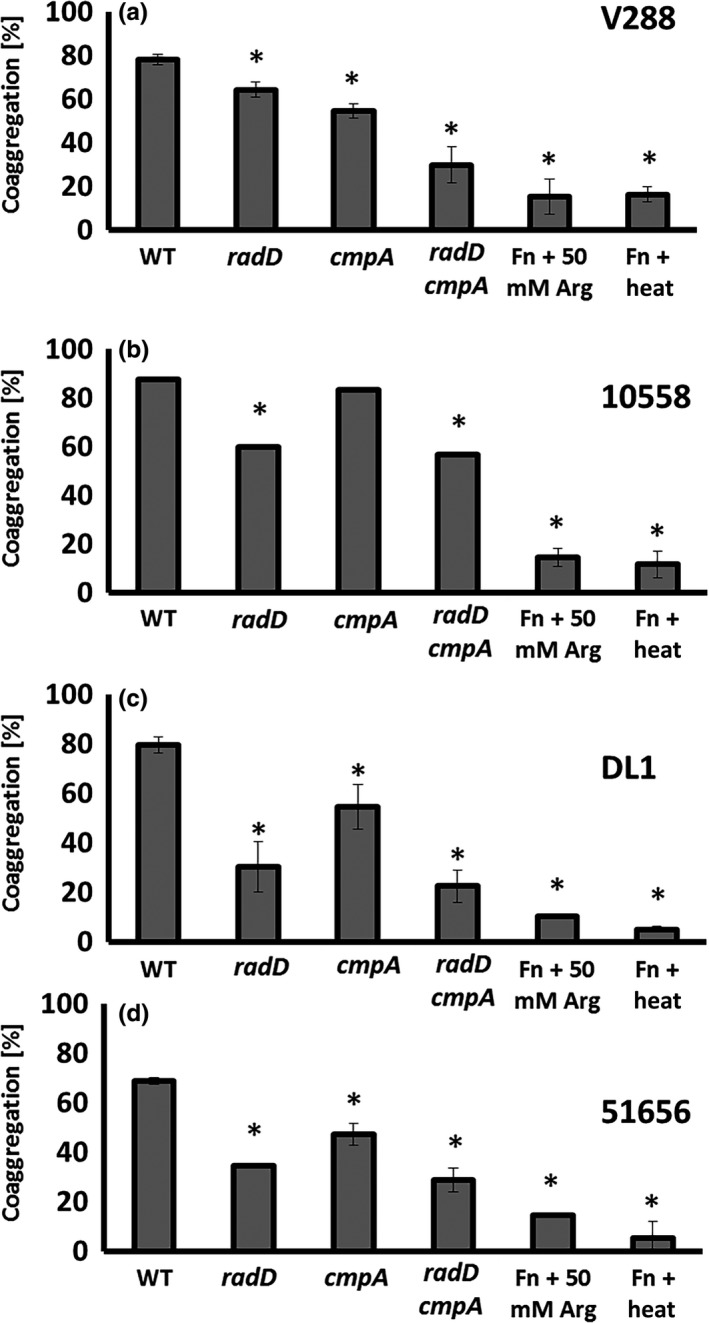
Quantitative coaggregation of wild‐type Streptococcus gordonii strain (a) V288, (b) ATCC 10558, (c) DL1, and (d) ATCC 51656 with Fusobacterium nucleatum strains: Wild‐type (WT) and the mutant derivatives: radD, cmpA, and radD cmpA double mutant. Data represent means and standard deviation of percent coaggregation of at least three independent experiments. *p < .05 compared to wild‐type control
Figure 2.
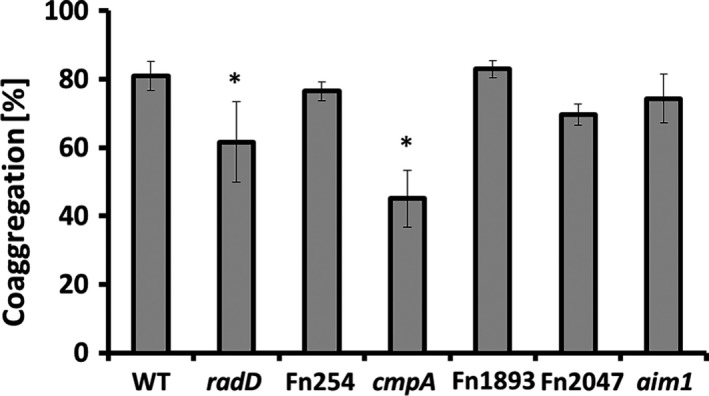
Quantitative coaggregation of wild‐type (WT) Streptococcus gordonii strain V288 with Fusobacterium nucleatum ATCC 23726WT strain and OMP mutant derivatives (radD, Fn254, cmpA, Fn1893, Fn2047, and aim1). Data represent means and standard deviation of percent coaggregation of at least three independent experiments. *p < .05 compared to wild‐type control
The individual absence of the proteins encoded by radD and cmpA only resulted in a partial coaggregation defect with S. gordonii. For further characterization of these OMPs in binding to S. gordonii V288, we constructed a double radD cmpA mutant, by using previously established approaches (Haake et al., 2000; Kaplan et al., 2005, 2009) to inactivate the ORF encoding cmpA in the radD mutant background (Figure 3a and b). Consistent with the idea that cmpA and radD function independently from each and are the two major adhesins involved in the F. nucleatum 23726 interaction with S. gordonii V288, the double mutant had a stronger coaggregation defect than either single mutant, and the defect in coaggregation was similar to that observed when the coaggregation inhibitor arginine was added to the buffer (Figure 4a).
Figure 3.
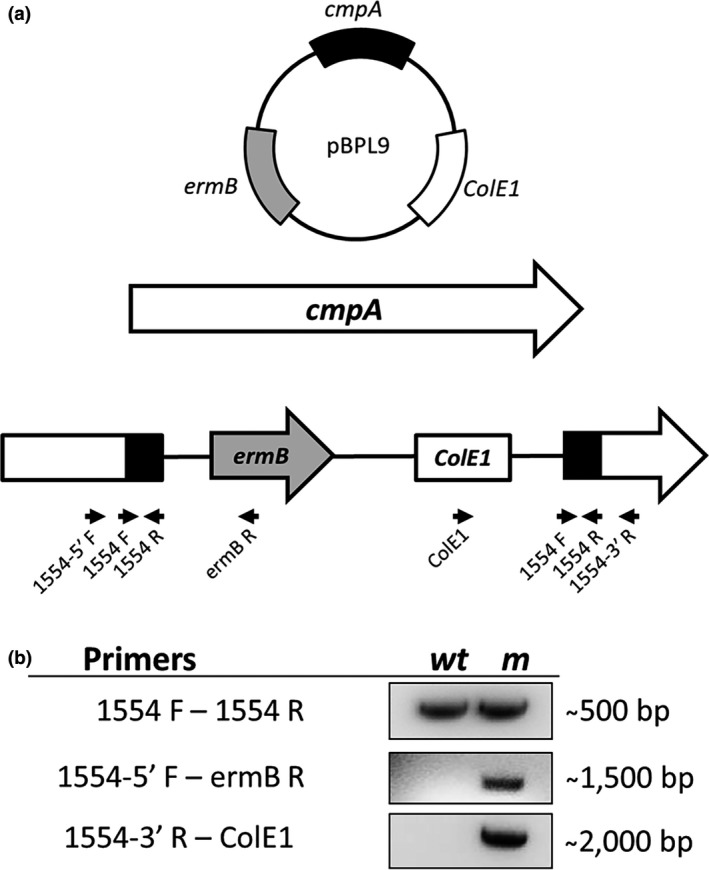
(a) Fusobacterium nucleatum radD cmpA double mutant (strain BL83) was constructed by insertion of the inactivation plasmid pBPL9 into cmpA (Fn1554) in a radD::catP mutant background. Black arrows indicate the location of primers used for mutant construction and analysis. (b) Confirmation of plasmid insertion into cmpA by PCR analysis of the cmpA mutant (m) with wild‐type (WT) as the control
3.3. CmpA coaggregation involvement is S. gordonii strain dependent
In contrast to above findings with S. gordonii V288, we had previously failed to observe a CmpA involvement in the interaction between F. nucleatum 23726 and S. gordonii ATCC10588, which was used as a representative of this oral Streptococcus species in one of our earlier studies (Kaplan et al., 2009). Thus, we decided to test the possible involvement of CmpA in the coaggregation between F. nucleatum and other S. gordonii strains such as ATCC 51656 and DL110558 as well as the previously used ATCC 10588. We found that the interaction of F. nucleatum with S. gordonii ATCC 10558 does not seem to involve CmpA (Figure 4b), whereas strains DL1 and ATCC 51656 presented only a subtle defect in coaggregation (Figure 4c and d).
We also investigated if CmpA was involved in F. nucleatum interaction with the periodontal pathogen P. gingivalis (strain 4612) and with another streptococcal species closely related to S. gordonii and S. sanguinis (ATCC 10556). However, we did not observe any difference in coaggregation compared to the wild‐type strains (data not shown). Thus, to the extent that we have tested, CmpA seems to be largely involved in the specific interaction between F. nucleatum strain 23726 and S. gordonii V288.
3.4. CmpA is required for dual‐species biofilm formation with S. gordonii
The interaction between streptococcal species and F. nucleatum is hypothesized to be an important step in oral biofilm development (Kolenbrander, 1989; Kolenbrander & London, 1993). Previous studies demonstrate that F. nucleatum requires radD to form a dual‐species biofilm with at least one of the oral streptococci species, S. sanguinis (Kaplan et al., 2009; Lancy, Dirienzo, Appelbaum, Rosan, & Holt, 1983). Since our data demonstrate that both RadD and CmpA are involved in the physical binding between F. nucleatum and S. gordonii V288, we investigated whether these adhesins were also involved in dual‐species biofilm formation. To differentiate between S. gordonii and F. nucleatum, we utilized an mCherry‐expressing S. gordonii V288 strain (BL98). While wild‐type F. nucleatum formed a uniform and thick biofilm layer on top of the mCherry‐expressing S. gordonii cells, the adhesin mutants, both single and double mutants, formed an irregular and thinner biofilm, as determined by confocal laser scanning microscopy (Figure 5a). Analysis of three randomly selected biofilm images revealed that the average height and maximum height of the biofilms were 38.21 μm (±11.48) and 241.33 μm (±23.67) for the WT F. nucleatum; 6.88 μm (±1.87) and 163.33 μm (±41.86) for the radD mutant; 8.47 μm (±0.19) and 171.66 μm (±48.41) for the cmpA mutant; and 4.48 μm (±1.04) and 71.66 μm (±34.93) for the double mutant.
Figure 5.
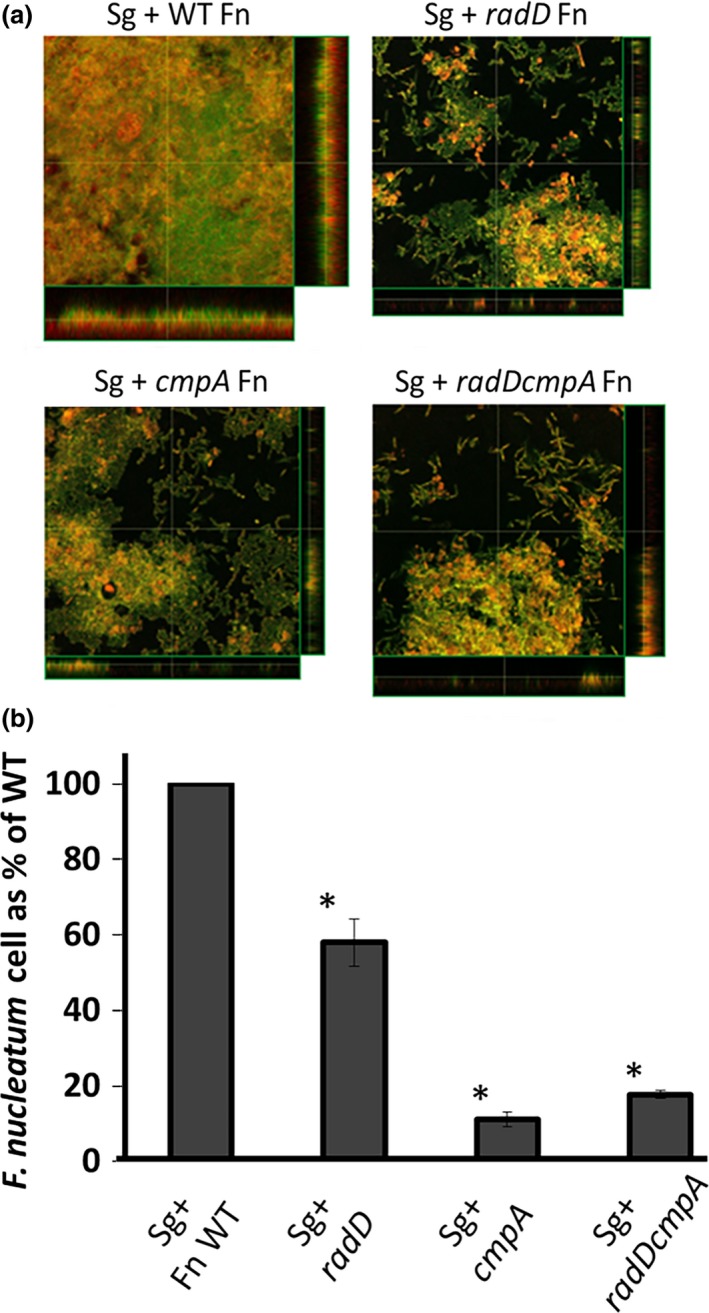
Streptococcus gordonii and Fusobacterium nucleatum dual‐species biofilm: (a) Confocal laser scanning microscopy of syto9‐stained dual‐species biofilm after overnight incubation. S. gordonii (Sg) cells constitutively express mCherry from their chromosome and appear orange/yellow on the images. Wild‐type (WT) F. nucleatum (Fn) and its mutant derivatives (radD, cmpA, and radD cmpA double mutant) are strained by syto9‐only and appear green on the images. (b) The presence of the Fn mutant strains in the Sg‐Fn dual‐species biofilm is displayed as the percentage of Fn cells normalized to the number of attached Sg cells/well, compared to that measured with WT Fn. Cellular ratios were determined by measuring DNA concentration by qPCR, targeting the F. nucleatum gene fomA and the S. gordonii gene srtA. At least three independent experiments were performed per strain combination. Each value represents means and standard deviation of at least three independent experiments. *p < .05 compared to wild‐type control
qPCR analysis of the total DNA extracted from the dual‐species biofilm revealed a decrease in the ratio of F. nucleatum to S. gordonii cells, as measured by their respective relative DNA concentration, when radD or cmpA mutants were used, compared to wild‐type cells (Figure 5b). Similar results were observed for the radD cmpA double mutant (Figure 5b).
3.5. radD and cmpA expression pattern
To further characterize radD and cmpA, we measured their mRNA level in wild‐type F. nucleatum throughout planktonic growth in Columbia broth from the early exponential to the stationary phase as well as from a single time point from the overnight biofilm. Both radD and cmpA mRNA level increased as the cultures entered stationary phase, but quickly decreased in the following time points (Figure 6a and b) implicating a growth‐dependent regulation of adhesins expression. Under overnight biofilm conditions, radD expression was about threefold lower while cmpA expression was about threefold higher compared to the planktonic cells collected from the same wells (Figure 6c).
Figure 6.
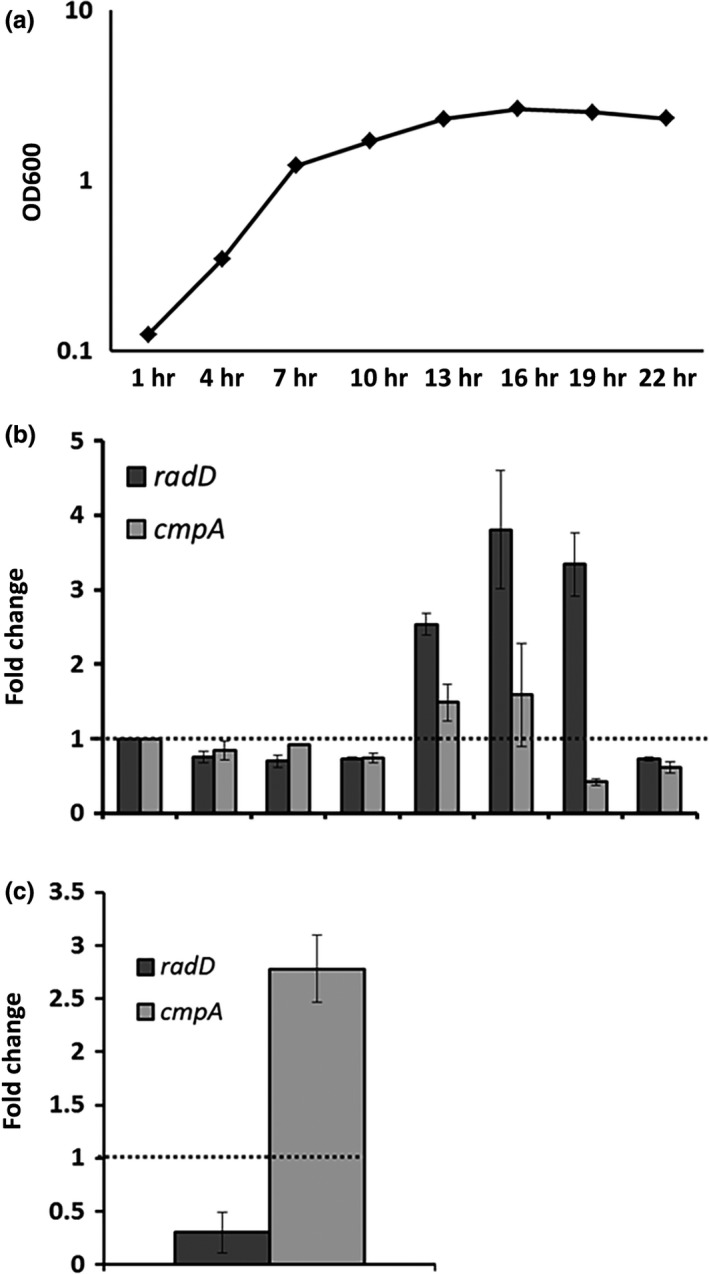
radD and cmpA expression: WT Fusobacterium nucleatum was grown in Columbia broth for 25 hr. Cell samples were collected every 3 hr and (a) OD600 was measured, as well as (b) radD and cmpA expression by qRT‐PCR. Gene expression was normalized to rpoB and compare to the first time point. The dashed line was added to aid in the comparison. (c) radD and cmpA expression were also measured in cells from Fn biofilm grown overnight in SHI‐FSMS. Gene expression was normalized to rpoB and compare to planktonic cells grown under the same conditions. Each value represents means and standard deviation of at least three independent experiments. To aid with visualization, the dashed line represents no change
4. DISCUSSION
The organization of the oral microbial community is thought to involve a complex network of interactions, often mediated by surface adhesins. As part of our long‐term effort to characterize the physical interaction between F. nucleatum and other members of the oral microbial community, we investigated, at the molecular level, the interaction between F. nucleatum 23726 and one of its early‐colonizer partners, S. gordonii. Here, we provide evidence that the physical interaction between F. nucleatum 23726 and S. gordonii V288 is mediated by at least two arginine‐inhibitable adhesins, RadD and CmpA.
The involvement of CpmA in the interaction between F. nucleatum 23726 and S. gordonii was most relevant for strain V288 compared to other S. gordonii strains tested (DL1, ATCC 10558, and ATCC 51656) (Figure 4a–d). The difference in coaggregation phenotype between S. gordonii V288 and DL1 was surprising, since these two strains are derived from the same original isolate, S. gordonii Challis. This has made us wonder how easily S. gordonii could alter its adhesion properties. These data also explain why we failed to observe a CmpA involvement in our previously published screen, which used strain ATCC 10558 as the S. gordonii representative strain for identification of F. nucleatum adhesins (Kaplan et al., 2009). Most significantly, these data add an additional layer of complexity to the interaction between F. nucleatum and S. gordonii. Similar complexity seems to be present in the interaction between F. nucleatum and P. gingivalis; while Fap2 appears to be a major adhesin for the interaction of F. nucleatum with P. gingivalis (Coppenhagen‐Glazer et al., 2015; Park et al., 2016), RadD plays an additional role in binding to strain 4612 (Park et al., 2016). It is worth mentioning that neither RadD nor Fap2 is involved in the interaction between F. nucleatum and P. gingivalis strain ATCC 33277, implicating the existence of at least one more F. nucleatum adhesin involved in the F. nucleatum–P. gingivalis interaction (Park et al., 2016).
While it is widely accepted that the initial interaction between oral bacteria can be assessed in vitro by measuring the ability of their planktonic cells to coaggregate, in the oral cavity, the ability of individual cells or group of cells to integrate into a biofilm is crucial for their survival and maintenance in the oral cavity (Kolenbrander, Palmer, Periasamy, & Jakubovics, 2010). Cells that integrate into a biofilm undergo significant physiological changes compared to their planktonic counterparts. These changes include, but are not limited to (1) gene and protein expression patterns, (2) metabolic preferences, and (3) replication rates (Cook, Costerton, & Lamont, 1998; Resch, Rosenstein, Nerz, & Gotz, 2005). Our results demonstrate that during formation of dual‐species biofilm with S. gordonii V288 in vitro, both RadD and CmpA are key players (Figure 5a and b). Interestingly, while cmpA expression was increased under biofilm conditions, radD expression was decreased (Figure 6c), suggesting that these proteins are likely to be involved in different physiological processes under biofilm conditions. Perhaps, RadD could play a more important role in the initial binding, while CmpA could be more involved in the subsequent stages of biofilm development.
Fusobacterium nucleatum encodes at least eight autotransporter‐like OMPs with molecular weights greater than 200 kDa. RadD, Fap2, and Aim1 have been previously characterized as interspecies adhesins (Coppenhagen‐Glazer et al., 2015; Kaplan et al., 2009) and apoptosis‐inducing proteins (Kaplan et al., 2005). The involvement of CmpA in interspecies interaction leaves five OMPs to be characterized. These autotransporter‐like OMPs possess very similar characteristics and are predicted to contain a core β‐barrel structure (Kaplan et al., 2009). In other bacteria, this structure possesses multiple activities that function in adherence and biofilm formation (Charbonneau & Mourez, 2007; Korotkova et al., 2006; Laarmann, Cutter, Juehne, Barenkamp, & ST Geme, 2002). The wide array of adherence properties found in F. nucleatum strains could be mediated by a combination of OMPs with varying degrees of affinity for different species and/or strains of bacteria present in the oral cavity.
In summary, the data presented here support the existence of a potentially complex interaction network between F. nucleatum and S. gordonii, which seems to be mediated by varying degrees of preferences for different F. nucleatum adhesins. This variation in adhesin preference could have a profound impact on community composition and species distribution in the oral microbiome, especially if such phenotype is also observed in the interaction among other members of the oral microbial community.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGMENTS
We thank members of the Shi and Lux laboratories for discussion and/or critical reading of the manuscript. We also thank Dr. Batbileg Bor for helping with CSLM. Additionally, we thank Drs. Susan Kinder Haake and M. Margaret Vickerman for kindly providing us with the pHS70 and pVA812::mCherry plasmids, respectively.
Lima BP, Shi W, Lux R. Identification and characterization of a novel Fusobacterium nucleatum adhesin involved in physical interaction and biofilm formation with Streptococcus gordonii . MicrobiologyOpen. 2017;6:e444 https://doi.org/10.1002/mbo3.444
REFERENCES
- Aas, J. A. , Paster, B. J. , Stokes, L. N. , Olsen, I. , & Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity. Journal of Clinical Microbiology, 43, 5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila, M. , Ojcius, D. M. , & Yilmaz, O. (2009). The oral microbiota: Living with a permanent guest. DNA and Cell Biology, 28, 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbonneau, M. E. , & Mourez, M. (2007). Functional organization of the autotransporter adhesin involved in diffuse adherence. Journal of Bacteriology, 189, 9020–9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, T. , Yu, W. H. , Izard, J. , Baranova, O. V. , Lakshmanan, A. , & Dewhirst, F. E . (2010). The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford), 2010, baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, G. S. , Costerton, J. W. , & Lamont, R. J. (1998). Biofilm formation by Porphyromonas gingivalis and Streptococcus gordonii . Journal of Periodontal Research, 33, 323–327. [DOI] [PubMed] [Google Scholar]
- Coppenhagen‐Glazer, S. , Sol, A. , Abed, J. , Naor, R. , Zhang, X. , Han, Y. W. , & Bachrach, G. (2015). Fap2 of Fusobacterium nucleatum is a galactose‐inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infection and Immunity, 83, 1104–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Avila, E. D. , Lima, B. P. , Sekiya, T. , Torii, Y. , Ogawa, T. , Shi, W. , & Lux, R. (2015). Effect of UV‐photofunctionalization on oral bacterial attachment and biofilm formation to titanium implant material. Biomaterials, 67, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst, F. E. , Chen, T. , Izard, J. , Paster, B. J. , Tanner, A. C. , Yu, W. H. , … Wade, W. G. (2010). The human oral microbiome. Journal of Bacteriology, 192, 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, P. I. , Chalmers, N. I. , Rickard, A. H. , Kong, C. , Milburn, C. L. , Palmer, R. J. Jr , & Kolenbrander, P. E . (2006). Molecular characterization of subject‐specific oral microflora during initial colonization of enamel. Applied and Environment Microbiology, 72, 2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dige, I. , Nilsson, H. , Kilian, M. , & Nyvad, B. (2007). In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. European Journal of Oral Sciences, 115, 459–467. [DOI] [PubMed] [Google Scholar]
- Dige, I. , Nyengaard, J. R. , Kilian, M. , & Nyvad, B. (2009). Application of stereological principles for quantification of bacteria in intact dental biofilms. Oral Microbiology and Immunology, 24, 69–75. [DOI] [PubMed] [Google Scholar]
- Guo, L. , He, X. , & Shi, W. (2014). Intercellular communications in multispecies oral microbial communities. Frontiers in Microbiology, 5, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake, S. K. , Yoder, S. C. , Attarian, G. , & Podkaminer, K. (2000). Native plasmids of Fusobacterium nucleatum: Characterization and use in development of genetic systems. Journal of Bacteriology, 182, 1176–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorth, P. , Turner, K. H. , Gumus, P. , Nizam, N. , Buduneli, N. , & Whiteley, M. (2014). Metatranscriptomics of the human oral microbiome during health and disease. MBio, 5, e01012–e01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, C. W. , Lux, R. , Haake, S. K. , & Shi, W. (2009). The Fusobacterium nucleatum outer membrane protein RadD is an arginine‐inhibitable adhesin required for inter‐species adherence and the structured architecture of multispecies biofilm. Molecular Microbiology, 71, 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, C. W. , Lux, R. , Huynh, T. , Jewett, A. , Shi, W. , & Haake, S. K. (2005). Fusobacterium nucleatum apoptosis‐inducing outer membrane protein. Journal of Dental Research, 84, 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander, P. E. (1989). Surface recognition among oral bacteria: Multigeneric coaggregations and their mediators. Critical Reviews in Microbiology, 17, 137–159. [DOI] [PubMed] [Google Scholar]
- Kolenbrander, P. E. , Andersen, R. N. , & Moore, L. V. (1989). Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infection and Immunity, 57, 3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander, P. E. , & London, J. (1993). Adhere today, here tomorrow: Oral bacterial adherence. Journal of Bacteriology, 175, 3247–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander, P. E. , Palmer, R. J. Jr. , Periasamy, S. , & Jakubovics, N. S . (2010). Oral multispecies biofilm development and the key role of cell‐cell distance. Nature Reviews Microbiology, 8, 471–480. [DOI] [PubMed] [Google Scholar]
- Kolenbrander, P. E. , Parrish, K. D. , Andersen, R. N. , & Greenberg, E. P. (1995). Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infection and Immunity, 63, 4584–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkova, N. , Cota, E. , Lebedin, Y. , Monpouet, S. , Guignot, J. , Servin, A. L. , … Moseley, S. L. (2006). A subfamily of Dr adhesins of Escherichia coli bind independently to decay‐accelerating factor and the N‐domain of carcinoembryonic antigen. Journal of Biological Chemistry, 281, 29120–29130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramitsu, H. K. , He, X. , Lux, R. , Anderson, M. H. , & Shi, W. (2007). Interspecies interactions within oral microbial communities. Microbiology and Molecular Biology Reviews, 71, 653–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laarmann, S. , Cutter, D. , Juehne, T. , Barenkamp, S. J. , & ST Geme, J. W . (2002). The Haemophilus influenzae Hia autotransporter harbours two adhesive pockets that reside in the passenger domain and recognize the same host cell receptor. Molecular Microbiology, 46, 731–743. [DOI] [PubMed] [Google Scholar]
- Lamont, R. J. , Chan, A. , Belton, C. M. , Izutsu, K. T. , Vasel, D. , & Weinberg, A. (1995). Porphyromonas gingivalis invasion of gingival epithelial cells. Infection and Immunity, 63, 3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancy, P. Jr. , Dirienzo, J. M. , Appelbaum, B. , Rosan, B. , & Holt, S. C . (1983). Corncob formation between Fusobacterium nucleatum and Streptococcus sanguinis . Infection and Immunity, 40, 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederman, R. , Buyle‐Bodin, Y. , Lu, B. Y. , Robinson, P. , & Naleway, C. (1997). Short‐chain carboxylic acid concentration in human gingival crevicular fluid. Journal of Dental Research, 76, 575–579. [DOI] [PubMed] [Google Scholar]
- Nyvad, B. , & Kilian, M. (1987). Microbiology of the early colonization of human enamel and root surfaces in vivo. Scandinavian Journal of Dental Research, 95, 369–380. [DOI] [PubMed] [Google Scholar]
- Nyvad, B. , & Kilian, M. (1990). Comparison of the initial streptococcal microflora on dental enamel in caries‐active and in caries‐inactive individuals. Caries Research, 24, 267–272. [DOI] [PubMed] [Google Scholar]
- Park, J. , Shokeen, B. , Haake, S. K. , & Lux, R. (2016). Characterization of Fusobacterium nucleatum ATCC 23726 adhesins involved in strain‐specific attachment to Porphyromonas gingivalis . International Journal of Oral Science, 8, 138–144. [Google Scholar]
- Paster, B. J. , Boches, S. K. , Galvin, J. L. , Ericson, R. E. , Lau, C. N. , Levanos, V. A. , … Dewhirst, F. E. (2001). Bacterial diversity in human subgingival plaque. Journal of Bacteriology, 183, 3770–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster, B. J. , Olsen, I. , Aas, J. A. , & Dewhirst, F. E. (2006). The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000, 42, 80–87. [DOI] [PubMed] [Google Scholar]
- Resch, A. , Rosenstein, R. , Nerz, C. , & Gotz, F. (2005). Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Applied and Environment Microbiology, 71, 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupf, S. , Merte, K. , & Eschrich, K. (1999). Quantification of bacteria in oral samples by competitive polymerase chain reaction. Journal of Dental Research, 78, 850–856. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , … Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y. , He, X. , Torralba, M. , Yooseph, S. , Nelson, K. E. , Lux, R. , … Shi, W. (2010). Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Molecular Oral Microbiology, 25, 357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh, P. J. , Ley, R. E. , Hamady, M. , Fraser‐Liggett, C. M. , Knight, R. , & Gordon, J. I. (2007). The human microbiome project. Nature, 449, 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman, M. M. , Mansfield, J. M. , Zhu, M. , Walters, K. S. , & Banas, J. A. (2015). Codon‐optimized fluorescent mTFP and mCherry for microscopic visualization and genetic counterselection of streptococci and enterococci. Journal of Microbiol Methods, 116, 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, T. K. , Lund, S. A. , Jones, K. F. , & Hruby, D. E. (2007). Comparison of transformation protocols in Streptococcus gordonii and evaluation of native promoter strength using a multiple‐copy plasmid. Canadian Journal of Microbiology, 53, 417–426. [DOI] [PubMed] [Google Scholar]
- Zarco, M. F. , Vess, T. J. , & Ginsburg, G. S. (2012). The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Diseases, 18, 109–120. [DOI] [PubMed] [Google Scholar]
- Zaura, E. , Keijser, B. J. , Huse, S. M. , & Crielaard, W. (2009). Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiology, 9, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]


