Abstract
Bacterial infections provide diagnostic dilemmas that could be enlightened by modern imaging technologies. We have developed a simple method for imaging bacterial infections in mice that relies on the phosphorylation and trapping of the thymidine kinase (TK) substrate 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5-[125I] iodouracil ([125I]FIAU) within bacteria. FIAU was found to inhibit the growth of WT Escherichia coli but not a TK– strain, indicating that WT E. coli could metabolize this compound. In silico analyses demonstrated that all pathogenic strains of bacteria whose genomes have been sequenced contain a TK gene highly homologous to the E. coli TK. Accordingly, we demonstrated that localized infections caused by representatives of five genera of bacteria could be readily imaged with [125I]FIAU. Such imaging provides a general method for the diagnosis of localized bacterial infections that could be translatable to the clinic.
Keywords: nuclear medicine, nucleoside analogs, single-photon emission computed tomography, thymidine kinase
Recent advances in medical technologies have ironically brought bacterial infections back into the limelight. The large number of individuals immunosuppressed as a result of therapy for cancer, transplantation, or autoimmune disease or as a result of AIDS has resulted in a population susceptible to relatively nonvirulent organisms. Similarly, the increasing use of prosthetic devices has provided new niches for bacterial colonization. These developments, coupled with the emergence of bacterial strains resistant to antibiotics, have elicited an urgent need to develop improved methods for the diagnosis and treatment of bacterial infections (1–3).
Conventional diagnostic methods for infections are contingent on the examination and culture of bacteria recovered from suspicious sites. These methods are time-consuming and insensitive, and the results are often obtained too late to guide clinical decision making (1–3). In particular, it is difficult or impossible to distinguish localized bacterial infections from sites of sterile inflammation. Radiological distinction between infection and neoplasm is also problematic, especially in patients who are neutropenic or immunosuppressed.
For all these reasons, the importance of developing new methods to image bacterial infections is widely recognized. Several intriguing strategies have been put forward in this regard, such as the use of radiolabeled antibiotics, antibodies, antimicrobial or chemotactic peptides, or bacteriophages (1–4). 99mTc-hexamethyl propyleneamine oxime (HMPAO) and 111In-oxine have been used to label leukocytes that home to sites of infection, and radiolabeled antigranulocyte antibodies or 18F-fluorodeoxyglucose have been used for similar purposes (3, 5, 6). Although all these methods benefit from the inherent high sensitivity of nuclear techniques, most have proved either cumbersome, relatively nonspecific, or logistically difficult due to the challenges in manufacturing materials suitable for clinical use (1–3, 5, 6).
The imaging of experimental tumors has improved dramatically in recent years, in part due to the development of high-resolution devices dedicated to imaging small animals and in part due to novel types of chemical sensors (7–12). One of the most powerful strategies involves the engineering of tumor cells with an enzyme that is not present in normal mammalian cells, which then traps a subsequently administered radiolabeled substrate (8–11). One of the best enzymes for this purpose is the herpes simplex virus 1 (HSV1) thymidine kinase (TK), which transfers a γ phosphate group from ATP to the 5′ hydroxyl group of pyrimidine deoxynucleosides (13). Very high sensitivities can be achieved by using HSV1-TK as the exogenously introduced enzyme and a radiolabeled HSV1-TK substrate as the radiotracer. This radiotracer is phosphorylated by transfected cells and accumulates within them, because the charged phosphate prevents its efflux across the plasma membrane (14). Based on the success of this approach for imaging transfected tumor cells, we wondered whether the endogenous TKs of bacteria were sufficiently similar to that of HSV1 to permit bacteria to be imaged in mammalian hosts. The results recorded below demonstrate that prokaryotic TK genes indeed allow conventional HSV1-TK substrates to be used to image a wide range of bacteria, providing a simple method to detect localized infections in animals.
Materials and Methods
Cell Lines. HCT116, HT-29, and CT-26 cells were purchased from American Type Culture Collection and grown in McCoy's 5A Medium (Invitrogen) supplemented with 5% FBS (HyClone) at 37°C with 5% CO2.
In Vitro Bacterial Susceptibility Assays. 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5-iodouracil (FIAU) (Moravek catalog no. M251) and penciclovir (Moravek catalog no. M972) were purchased from Moravek Biochemicals (Brea, CA). Zidovudine was purchased from Glaxo Wellcome. Bacterial susceptibility tests were performed in 96-well microtiter plates (VWR Scientific), with serial dilutions of drug placed in each well. Each well was inoculated with Escherichia coli (Yale University E. coli Genetic Stock Center, New Haven, CT) and grown in Luria broth (Invitrogen) at 37°C. E. coli TK mutants were generated by selecting for spontaneously resistant colonies on plates containing 1 mg/ml Zidovudine. Ten resistant clones were selected and screened for deletions in the TK gene by using the PCR primers SZ46-eTKKO20F and (5′-TGATGAAAAGTAGAACAGTCG-3′) SZ49-eTK-KO789R (5′-ATCAAGACGCAGCACCATG-3′). One resistant clone was found to contain a deletion in the TK gene and was used for subsequent experiments. As a control for integrity of the DNA of this clone, its 16S rRNA gene was amplified by using the primers SZ-16S-Ecoli993F (5′-ACATCCACGGAAGTTTTCAG-3′) SZ-16S-Ecoli1454R (5′-CCGAAGGTTAAGCTACCTAC-3′).
Tumor Inoculation and Spore Administration. All animal experiments were overseen and approved by the Animal Welfare Committee of The Johns Hopkins University and were in compliance with university standards. Six- to 8-wk-old athymic nu/nu or BALB/c mice, purchased from Harlan Bioproducts for Science (Indianapolis), were used for tumor implantation studies. Five million cells were injected s.c. into the right flank of each mouse. Tumor volume was calculated as length × width2 × 0.5, and mice were treated with Clostridium novyi-NT spores when tumors occupied ≈250 mm3. C. novyi-NT spores were prepared as described (15), and mice were i.v. injected with 300 million spores suspended in 250 μl of PBS.
[125I]FIAU Preparation. This synthesis was based on ref. 16. Briefly, 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranoside)-uracil (300 μg, 1.22 mmol, Moravek) was dissolved in 170 μl of 2 M HNO3. To this solution, 1.5 mCi (1 Ci = 37 GBq) of [I-125] NaI (ICN) was added and the contents heated at 130°C for 45 min. The reaction was quenched with 150 μl of HPLC mobile phase (20:79.9:0.1% MeCN:H2O:triethylamine). The resulting [125I]FIAU was purified by reverse-phase HPLC by using two passages over a Phenomenex Luna C18 semiprep column (10 μm, 4.6 × 250 mm, Phenomenex, Torrance, CA) by using the above-mentioned isocratic mobile phase at a flow rate of 2 ml/min. The product was concentrated under reduced pressure and formulated in 0.9% physiological saline before sterile filtration through a 0.22-μm syringe filter. Formulations were kept at 1 mCi/ml to minimize the injection volume. The final radiochemical yield was ≈50%, the radiochemical purity was >99%, and the specific radioactivity was >2,000 Ci/mmol.
Experimental Infections. E. coli strains or clinical isolates from the Johns Hopkins Hospital Microbiology Laboratory, including Staphylococcus aureus 29213 and 25923, Streptococcus pneumoniae 49619, Enterococcus faecalis 49532, and Staphylococcus epidermidis F362, were used to create experimental infections. Bacteria were grown to log phase in Mueller Hinton Broth with Cations (Remel, Lenexa, KS) or BBL Todd Hewitt Broth (Becton Dickinson). Localized infections were generated by injecting ≈1 × 109 E. coli and ≈1 × 108 of the other bacterial strains into mouse thighs. Morphologic examinations of the infectious lesions in the thighs of mice injected with the TK-deficient (TK–) strain of E. coli showed that they were as intense as those resulting from WT E. coli. To quantify the minimal number of bacteria required to generate signals upon imaging, mice were injected in the thigh with various amounts of S. aureus 25923. One hour later, the mice were killed, the muscles were harvested and homogenized, and the extracts spread on blood agar plates (Becton Dickinson) for colony counting. The plating efficiency of S. aureus 25923 grown in liquid media was found to be >95%.
In Vivo Imaging. Mice were injected with 225 μCi of [125I]FIAU via the tail vein and imaged at various time points thereafter. Before imaging, mice were anesthetized via s.c. administration of acepromazine and ketamine. Each scan took ≈10 min with a dedicated small-animal single-photon emission computed tomography (SPECT)/computed tomography (CT) camera (Gamma Medica X-SPECT, Northridge, CA) in planar acquisition mode using a low-energy high-resolution (LEHR) parallelhole collimator. For each bacterial strain used, at least two mice were injected and imaged; representative images are shown in Figs. 1c, 3, 5, and 6. To obtain SPECT/CT images, animals were first scanned for ≈40 min by using a small-animal SPECT camera in tomographic acquisition mode, using two LEHR parallel-hole collimators. The animals then underwent CT by using appropriate fiducial markers that allowed coregistration.
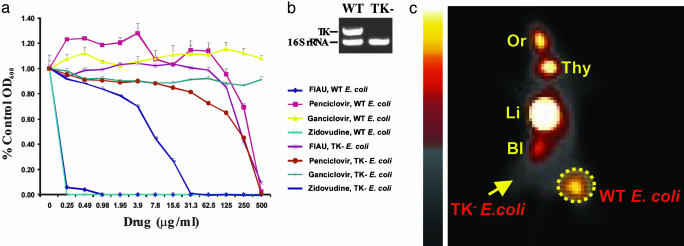
Fig. 1.
In vivo and in vitro experiments with WT and TK– E. coli.(a) Various concentrations of the indicated nucleoside analogs were included in the culture medium, and growth was assessed at 24 h. (b) Results of a multiplex PCR demonstrating the deletion of TK in the selected TK– clone. The WT lane shows the PCR products of the TK and 16S rRNA genes generated from WT E. coli; only the control 16S rRNA gene could be amplified in the selected TK– clone. (c) The right thighs of mice were inoculated with WT E. coli, and the left thigh was inoculated with an equal number of TK– E. coli. Six hours later, mice received an i.v. injection of [125I]FIAU. Sixteen hours after injection of tracer, planar images were taken. Normal tissues that metabolize or excrete FIAU or its iodinated derivatives include the oral mucosa, thyroid, liver/GI tract, and bladder (indicated as Or, Thy, Li, and Bl, respectively). The dotted circle indicates the WT E. coli infectious lesion, and the arrow points to the TK– E. coli infectious lesion.
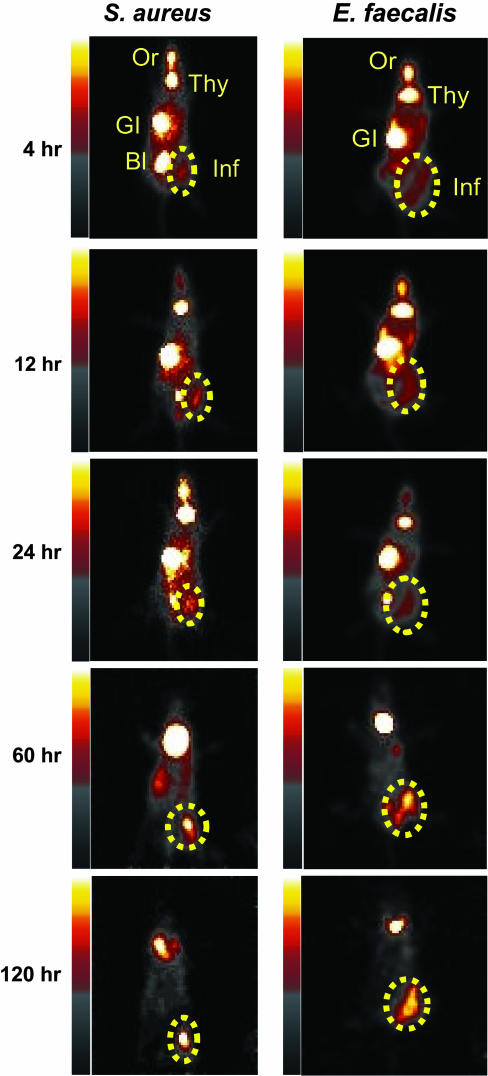
Fig. 3.
In vivo imaging of infections with S. aureus 25923 and E. faecalis 49532. One thigh of each mouse was inoculated with the indicated bacteria. Six hours later, mice received an injection of 225 μCi of [125I]FIAU via the tail vein. Serial imaging was performed at the indicated times thereafter, and planar images are shown. Normal tissues, oral mucosa, thyroid, liver/GI tract, and bladder, that accumulate [125I]FIAU or [125I] are labeled as Or, Thy, GI, and BI, respectively. The dotted circles indicate infectious foci.
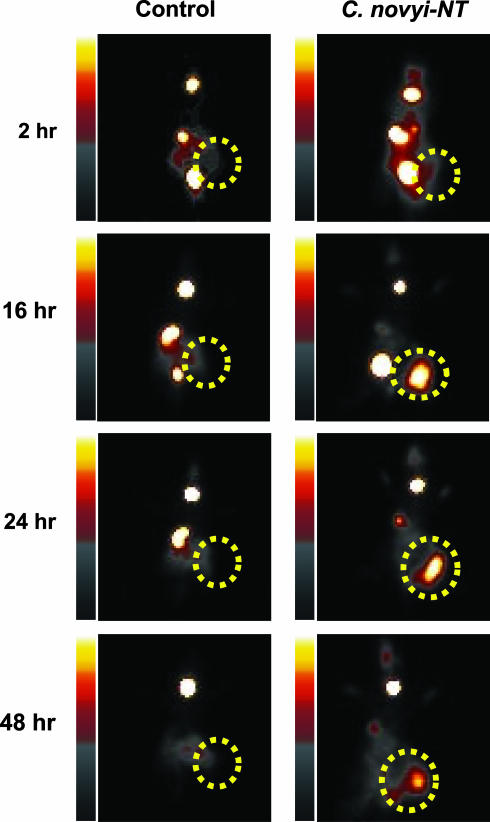
Fig. 5.
Scintigraphic image of a tumor after bacteriolytic therapy. BALB/c mice harboring CT-26 mouse colon tumors were left untreated (Control) or were treated with a single i.v. injection of 300 million C. novyi-NT spores (C. novyi-NT). Twenty-four hours later, 225 μCi of [125I]FIAU was administered via the tail vein, and planar images were taken at the indicated time points. The dotted circles indicate the tumors.
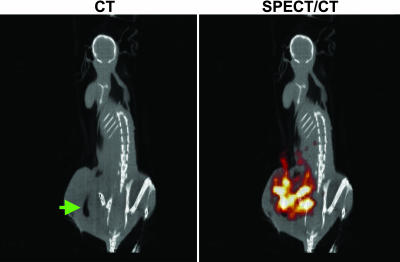
Fig. 6.
SPECT/CT of a tumor after bacteriolytic therapy. A BALB/c mouse harboring a CT-26 mouse tumor was treated with C. novyi-NT and injected with [125I]FIAU, as described in Fig. 5. The animal was scanned by using SPECT with CT coregistration 24 h after injection of [125I]FIAU. The green arrow indicates gas within the tumor caused by the infection, and the red/orange signal represents [125I]FIAU uptake by C. novyi-NT within the tumor.
Biodistribution. Imaging experiments showed that high signal-to-noise ratios in infectious foci could be consistently obtained 24 h after [125I]FIAU administration, so this time point was chosen for detailed analyses. Biodistribution studies were performed in mice injected with ≈1 × 108 S. aureus 25923 into one thigh. Six hours later, the mice were injected with 2 μCi of [125I]FIAU, and, after another 24 h, the mice were killed, their organs were harvested, and radioactivity was determined.
Results
Susceptibility of E. coli to Nucleoside Analogs. To determine whether endogenous bacterial TK could provide a reporter enzyme suitable for imaging, we first examined the susceptibility of E. coli to a variety of common nucleoside analogs in vitro. We assumed growth inhibition indicated that the nucleoside analog was a substrate for the E. coli TK and could thereby serve as an imaging reporter when radiolabeled. Representative results are shown in Fig. 1a. E. coli proved resistant to ganciclovir and penciclovir but quite sensitive to FIAU and Zidovudine.
To determine whether the TK gene was responsible for this sensitivity, we created a derivative of E. coli in which the TK gene was deleted (see Materials and Methods). PCR was used to demonstrate the absence of the TK gene in this derivative (Fig. 1b). The TK– strain was moderately resistant to Zidovudine and highly resistant to FIAU (Fig. 1a). Because FIAU can be radiolabeled by using commercially available reagents and has been successfully used to image tumor cells transfected with HSV1-TK (reviewed in ref. 14), we elected to test its potential for imaging bacterial infections.
In Vivo Imaging of E. coli Infections. [125I]FIAU was synthesized by standard methods and injected i.v. into animals 6 h after intramuscular inoculations of bacteria into the thighs of mice. Fig. 1c shows a whole-body planar scintigram that demonstrates the uptake of [125I]FIAU within the thighs of mice harboring WT E. coli bacteria. Signals from the infectious lesions could be seen as early as 2 h after injection of [125I]FIAU and were optimal ≈16 h after injection. Infections of the same mice inoculated with TK– E. coli in the opposite thighs showed no discernable uptake of [125I]FIAU (Fig. 1c).
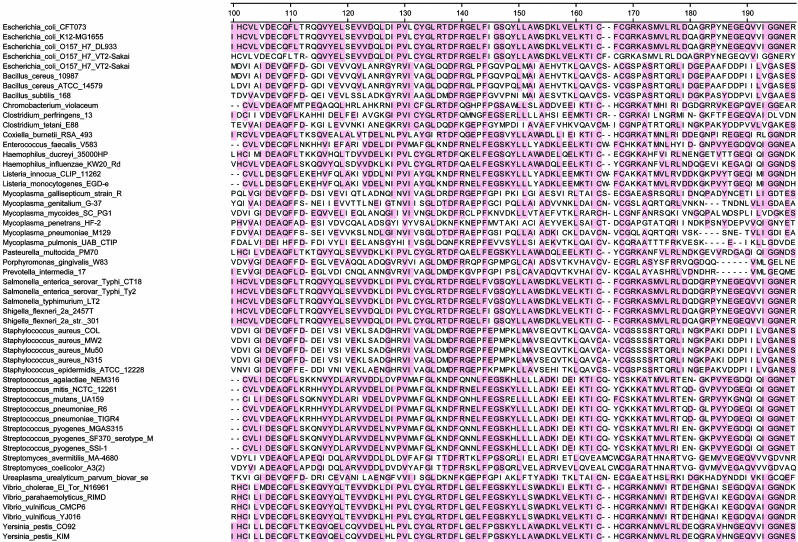
In Silico Analysis of Bacterial TK. We next performed an in silico assessment of TK genes in all 53 pathogenic bacteria whose genomes have been sequenced and made publicly available. This assessment revealed that every one of these bacterial species possessed TK genes. Moreover, the homology between these TK genes was striking, with a clear consensus within the kinase catalytic domain (Fig. 2). Each of the 53 bacteria contained at least 25 residues that were identical to those of the consensus. In contrast, this consensus sequence was not found in mammalian TKs, presumably accounting for the differential capacities of the mammalian enzymes to phosphorylate substrates such as FIAU.
Fig. 2.
TK genes in pathogenic bacteria. For each bacterial species whose genome has been sequenced, the central 100 residues of TK, containing most of the catalytic domain, are listed. Shaded residues represent those present in >50% of the species, as defined by a clustalw algorithm (28). Human and mouse TK sequences could not be well aligned with these conserved residues in bacteria.
Imaging Infections Caused by Pathogenic Bacteria. In light of this high sequence conservation, we expected that [125I]FIAU could be used as a tracer for pathogenic bacteria in general. We randomly selected four patient-derived strains identified in the Johns Hopkins Hospital Microbiological Laboratory to test this expectation. The identities and clinical properties of the selected strains are listed in Table 1. Infectious foci due to E. faecalis, S. aureus, S. epidermidis, and S. pneumoniae could all be readily imaged with [125I]FIAU (Fig. 3 and Fig. 7, which is published as supporting information on the PNAS web site). Robust signals could be observed as early as 4 h after administration of [125I]FIAU. Time-course studies showed that [125I] remained in the infected tissues for long time periods, presumably because [125I]FIAU was incorporated into the DNA of the bacteria. In contrast to the maintenance of this bacterial signal, the background signal in noninfected tissues gradually decreased, presumably because of continuing metabolism and excretion of [125I]FIAU (17, 18). This resulted in very high signal-to-noise ratios by 48 h after administration of the tracer.
Table 1. Bacterial strains imaged after i.m. injection.
| Organism | Clinical significance |
|---|---|
| E. coli | Adult and infantile diarrhea, urinary tract infection, pneumonia, meningitis, and abscess |
| E. faecalis 49532 | Nosocomial infection including vancomycin-resistant enterococci, urinary tract infection, endocarditis, abscess, and meningitis |
| S. pneumoniae 49619 | Pneumonia, meningitis, sinusitis, osteomyelitis, and sepsis |
| S. aureus 29213 and 25293 | Cellulitis, indwelling medical device infection, diabetic ulcer, postsurgical wounds, osteomyelitis, endocarditis, meningitis, mastitis, phlebitis, pneumonia, boils, furuncles, and impetigo |
| S. epidermidis F362 | Endocarditis, cellulitis, urinary tract infection, and indwelling medical device infection |
Representative infections in humans caused by bacteria used in this study.
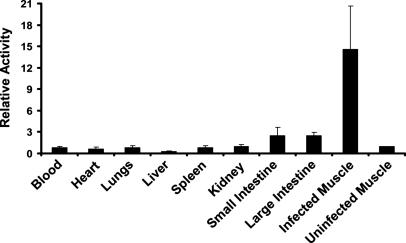
For quantitative distribution measurements, mouse thighs were infected with S. aureus 25923 and [125I]FIAU was administered 6 h later. Tissues were harvested after another 24 h and radioactivity measured. As shown in Fig. 4, the infected muscle contained much higher levels of [125I]FIAU than the other tissues, with the ratio of radioactivity in infected thighs to uninfected (contralateral) thighs exceeding 14:1.
Fig. 4.
Biodistribution. The right thigh of each of four mice was inoculated with S. aureus 25923. Six hours later, mice were injected i.v. with 2 μCi of [125I]FIAU. Mice were killed 24 h later, and radioactivity was measured in the indicated organs. Values were normalized to those found in the left (uninfected) thigh. Bars and brackets indicate means and standard deviations, respectively. The fraction of the total injected dose (ID) that was present in the uninfected thighs (ID/g) was 0.028 ± 0.013% per gram of tissue (mean ± standard deviation). The relative activity shown on the y axis was determined by normalizing the radioactivity in each tissue to that in the corresponding uninfected muscle.
To determine the minimal number of bacteria that could be imaged with this approach, we injected various numbers of S. aureus 25923 into mouse thighs. One hour later, the thigh tissue was excised, homogenized, and spread on blood agar plates. The 1-h time point was chosen because this was the earliest time point at which injections of [125I]FIAU consistently produced discernable scintigraphic images of infectious foci. We found that as few as 2 × 106 colony-forming units per gram of muscle tissue produced discernable signals.
Imaging Intratumoral Infections. We next attempted to image infectious foci that were created by a process other than i.m. injection. It has been shown that the spores of anaerobic bacteria, when systemically administered to mice, germinate only within tumor tissues (19–22). C. novyi-NT is a derivative of C. novyi that is devoid of its major systemic toxin gene and can therefore be safely delivered to animals (15, 23). When injected i.v. into mice bearing tumors, <1% of the spores localize within tumors, the remainder being sequestered in the spleen and liver (L.D., unpublished data). The few spores localized within the tumor germinate rapidly, achieving a density of ≈108 per gram of tissue by 24 h.
It is not known whether C. novyi contains a TK gene homologous to the other bacteria tested in this study, because its genomic sequence has not been determined. Nevertheless, based on the high conservation of prokaryotic TK genes shown in Fig. 2, we assumed that C. novyi-NT would contain a TK gene suitable for imaging with an FIAU probe. This assumption was confirmed by imaging. BALB/c mice bearing CT-26 mouse colon tumors were treated with a single i.v. injection of C. novyi-NT and [125I]FIAU was administered 24 h later. Serial images showed that the tumors could be visualized as early as 16 h after injection of tracer, with maximum uptake observed 24–48 h after injection of [125I]FIAU. No uptake was observed in tumors that had not been treated with C. novyi-NT (Fig. 5). Similar results were obtained in nude mice harboring HCT116 and HT-29 colon cancer xenografts (data not shown).
Because planar γ camera imaging is limited in its ability to reveal anatomical detail, SPECT/CT imaging was also performed. As observed in tumor-bearing rabbits treated with C. novyi-NT spores (24), areas of gas produced by the bacteria within CT-26 tumors could also be visualized upon CT, providing definitive evidence for infection (Fig. 6). Coregistration of CT images with corresponding SPECT images demonstrated that bacterial germination and tracer uptake were limited to the tumor region (Fig. 6). Untreated mice showed no signs of gas or tracer uptake within their tumors (data not shown).
Discussion
As noted in the Introduction, a variety of experimental techniques have been proposed for imaging infectious lesions (1–6). The approach described above fulfills most of the criteria established for an ideal tracer for imaging infections (3), i.e., it displays efficient accumulation and good retention in infectious foci, minimal accumulation in nontarget organs, no toxicity, the potential for early diagnosis, ready availability, low cost, simple low-hazard preparation, and a low radiation burden. Based on its mechanism of action and the results presented in Fig. 5, it can differentiate infection from nonmicrobial inflammation such as occurs in response to tumors.
Although we were able to detect readily various bacterial infections using [125I]FIAU, nucleosides with positron emitters would enable even higher sensitivity. In fact, [124I]FIAU, the positron-emitting analog of [125I]FIAU, has been successfully used in humans to image brain tumors in patients undergoing ganciclovir therapy after gene therapy, with favorable imaging and safety profiles (25). This suggests that the approach described herein for imaging routine bacterial infections could be readily translated to the clinic.
The results raise several issues for future research. First, successful imaging has so far been demonstrated in only seven bacterial species representing five different genera. Although the TK homologies listed in Fig. 2, coupled with the imaging data presented above, suggest that radiolabeled FIAU will be applicable to most if not all pathogenic bacteria, this must be formally demonstrated before the onset of clinical trials. Second, it is not clear that FIAU is the optimum nucleoside for imaging based on prokaryotic TKs. Although radiolabeled FIAU has many advantages, including ease of synthesis, it is possible that other nucleoside derivatives will be more efficiently incorporated into bacterial DNA or have more favorable properties. Because a large number of pyrimidine analogs have been synthesized as potential anticancer and anti-HIV therapeutics, this possibility can easily be explored in future work. Finally, the sensitivity of E. coli to FIAU, coupled with the uptake of FIAU by several other bacteria, suggests that TK substrates could be used not only for imaging bacteria but also for therapy. Indeed, it has been shown that nucleoside analogues are potent inhibitors of the growth of mycoplasma and mycobacterial strains (26, 27). Recognition of the widespread presence and unique substrate specificities of bacterial TKs may therefore prove useful for the design of novel diagnostic as well as therapeutic modalities.
Supplementary Material
Acknowledgments
We thank E. Latice Watson for assistance with animal experiments. This work was supported by the Miracle Foundation, the Clayton Fund, and National Institutes of Health Grants CA 062924, CA 43460, CA 92871, and CA 103175.
Author contributions: C.B., L.D., S.Z., K.W.K., B.V., and M.G.P. designed research; C.B., C.A.F., Y.W., N.A., J.F., and L.H.D. performed research; C.B., C.A.F., I.C., Y.W., J.D., K.W.K., B.V., and M.G.P. contributed new reagents/analytic tools; C.B., L.D., N.A., L.H.D., S.Z., K.W.K., B.V., and M.G.P. analyzed data; and C.B., S.Z., K.W.K., B.V., and M.G.P. wrote the paper.
Abbreviations: HSV1, herpes simplex virus 1; TK, thymidine kinase; [125I]FIAU, 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5-[125I]iodouracil; SPECT, single-photon emission computed tomography; CT, computed tomography.
References
- 1.Lupetti, A., Welling, M. M., Pauwels, E. K. & Nibbering, P. H. (2003) Lancet Infect. Dis. 3, 223–229. [DOI] [PubMed] [Google Scholar]
- 2.Signore, A., Annovazzi, A., Corsetti, F., Capriotti, G., Chianelli, M., De Winter, F. & Scopinaro, F. (2002) BioDrugs 16, 241–259. [DOI] [PubMed] [Google Scholar]
- 3.Rennen, H. J., Boerman, O. C., Oyen, W. J. & Corstens, F. H. (2001) Eur. J. Nucl. Med. 28, 241–252. [DOI] [PubMed] [Google Scholar]
- 4.Rusckowski, M., Gupta, S., Liu, G., Dou, S. & Hnatowich, D. J. (2004) J. Nucl. Med. 45, 1201–1208. [PubMed] [Google Scholar]
- 5.Peters, A. M. (1998) Br. J. Radiol. 71, 252–261. [DOI] [PubMed] [Google Scholar]
- 6.Palestro, C. J., Love, C., Tronco, G. G. & Tomas, M. B. (2000) Radiographics 20, 1649–1660. [DOI] [PubMed] [Google Scholar]
- 7.Weissleder, R. & Ntziachristos, V. (2003) Nat. Med. 9, 123–128. [DOI] [PubMed] [Google Scholar]
- 8.Blasberg, R. G. & Gelovani, J. (2002) Mol. Imaging 1, 280–300. [DOI] [PubMed] [Google Scholar]
- 9.Luker, G. D. & Piwnica-Worms, D. (2001) Acad. Radiol. 8, 4–14. [DOI] [PubMed] [Google Scholar]
- 10.Gambhir, S. S. (2002) Nat. Rev. Cancer 2, 683–693. [DOI] [PubMed] [Google Scholar]
- 11.Herschman, H. R. (2003) Science 302, 605–608. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman, J. M. (2001) Curr. Opin. Oncol. 13, 148–153. [DOI] [PubMed] [Google Scholar]
- 13.Eriksson, S., Munch-Petersen, B., Johansson, K. & Eklund, H. (2002) Cell Mol. Life Sci. 59, 1327–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blasberg, R. G. & Tjuvajev, J. G. (2003) J. Clin. Invest. 111, 1620–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang, L. H., Bettegowda, C., Huso, D. L., Kinzler, K. W. & Vogelstein, B. (2001) Proc. Natl. Acad. Sci. USA 98, 15155–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs, A., Braunlich, I., Graf, R., Lercher, M., Sakaki, T., Voges, J., Hesselmann, V., Brandau, W., Wienhard, K. & Heiss, W. D. (2001) J. Nucl. Med. 42, 467–475. [PubMed] [Google Scholar]
- 17.Deng, W. P., Yang, W. K., Lai, W. F., Liu, R. S., Hwang, J. J., Yang, D. M., Fu, Y. K. & Wang, H. E. (2004) Eur. J. Nucl. Med. Mol. Imaging 31, 99–109. [DOI] [PubMed] [Google Scholar]
- 18.Toyohara, J., Hayashi, A., Sato, M., Gogami, A., Tanaka, H., Haraguchi, K., Yoshimura, Y., Kumamoto, H., Yonekura, Y. & Fujibayashi, Y. (2003) Nucl. Med. Biol. 30, 687–696. [DOI] [PubMed] [Google Scholar]
- 19.Matsumura, H., Takeuchi, A. & Kano, Y. (1997) Biosci. Biotechnol. Biochem. 61, 1211–1212. [DOI] [PubMed] [Google Scholar]
- 20.Low, K. B., Ittensohn, M., Le, T., Platt, J., Sodi, S., Amoss, M., Ash, O., Carmichael, E., Chakraborty, A., Fischer, J., et al. (1999) Nat. Biotechnol. 17, 37–41. [DOI] [PubMed] [Google Scholar]
- 21.Liu, S. C., Minton, N. P., Giaccia, A. J. & Brown, J. M. (2002) Gene Ther. 9, 291–296. [DOI] [PubMed] [Google Scholar]
- 22.Theys, J., Barbe, S., Landuyt, W., Nuyts, S., Van Mellaert, L., Wouters, B., Anne, J. & Lambin, P. (2003) Curr. Gene Ther. 3, 207–221. [DOI] [PubMed] [Google Scholar]
- 23.Dang, L. H., Bettegowda, C., Agrawal, N., Cheong, I., Huso, D., Frost, P., Loganzo, F., Greenberger, L., Barkoczy, J., Pettit, G. R., et al.. (2004) Cancer Biol. Ther. 3, 326–337. [DOI] [PubMed] [Google Scholar]
- 24.Agrawal, N., Bettegowda, C., Cheong, I., Geschwind, J. F., Drake, C. G., Hipkiss, E. L., Tatsumi, M., Dang, L. H., Diaz, L. A., Jr., Pomper, M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs, A., Voges, J., Reszka, R., Lercher, M., Gossmann, A., Kracht, L., Kaestle, C., Wagner, R., Wienhard, K. & Heiss, W. D. (2001) Lancet 358, 727–729. [DOI] [PubMed] [Google Scholar]
- 26.Carnrot, C., Wehelie, R., Eriksson, S., Bolske, G. & Wang, L. (2003) Mol. Microbiol. 50, 771–780. [DOI] [PubMed] [Google Scholar]
- 27.Manallack, D. T., Pitt, W. R., Herdewijn, P., Balzarini, J., De Clercq, E., Sanderson, M. R., Sohi, M., Wien, F., Munier-Lehmann, H., Haouz, A., et al. (2002) J. Enzyme Inhib. Med. Chem. 17, 167–174. [DOI] [PubMed] [Google Scholar]
- 28.Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. G. & Thompson, J. D. (2003) Nucleic Acids Res. 31, 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.