Abstract
Uridine insertion/deletion RNA editing in trypanosomatid mitochondria is a posttranscriptional RNA modification phenomenon required for translation of mitochondrial mRNAs. This process involves guide RNA-mediated cleavage at a specific site, insertion or deletion of Us from the 3′ end of the 5′ mRNA fragment, and ligation of the two mRNA fragments. The Leishmania major RNA ligase-containing complex protein 2 expressed in insect cells has a 3′–5′ exoribonuclease activity and was therefore renamed RNA editing exonuclease 1 (REX1). Recombinant REX1 specifically trims 3′ overhanging Us and stops at a duplex region. Evidence is presented that REX1 is responsible for deletion of the 3′ overhanging Us from the bridged mRNA 5′ cleavage fragment and that RNA editing ligase 1 is responsible for the ligation of the two mRNA cleavage fragments in U-deletion editing. The evidence involves both in vivo down-regulation of REX1 expression in Trypanosoma brucei by RNA interference and the reconstitution of precleaved U-deletion in vitro editing with only two recombinant enzymes: recombinant REX1 and recombinant RNA editing ligase 1.
Keywords: ligase, trypanosomes, REX1, editing
Uridine insertion/deletion RNA editing is a posttranscriptional RNA modification phenomenon that occurs in the mitochondria of kinetoplastid protists (1). The insertion and deletion of Us into transcripts of 12 mitochondrial-encoded cryptogenes is mediated by guide RNAs (gRNAs) that hybridize downstream of the editing sites and recruit several protein complexes that interact via RNA. The RNA ligase-containing complex (L-complex) from both Leishmania sp. and Trypanosoma brucei contains ≈16 proteins, which have been labeled LC-X (for L-complex protein) or MP-X (for mitochondrial protein), respectively (2, 3), by relative gel mobility. These proteins include the LC-2 (MP100) and LC-3 (MP99) “Exo_endo_phos”-Pfam motif proteins (4), the RNA editing ligase 1 (REL1) (5–7) and RNA editing ligase 2 (REL2), and the RNA editing 3′ terminal uridylyltransferase (TUTase) 2 (RET2) proteins, among others. Editing involves an initial cleavage of the mRNA transcript just upstream of the mRNA–gRNA anchor duplex, which is followed by either an addition of Us to the 3′ end of the 5′ fragment or a deletion of non-base-paired Us from the 3′ end of the 5′ fragment. The two fragments are then ligated, thereby extending the mRNA–gRNA duplex. The process repeats at the next upstream editing site, and, after completion of the editing mediated by a single gRNA, in some cases, additional gRNAs hybridize to the edited sequences and mediate overlapping blocks of editing, extending the edited region further upstream (8). The RET2 TUTase in the L-complex was shown to be responsible for the gRNA-mediated addition of Us to the editing sites, and the RET1 TUTase, which is not a component of the L-complex but interacts via RNA, is responsible for the addition of Us to the 3′ end of the gRNAs (9, 10).
Heterologous expression of properly folded active editing enzymes has proven difficult. The only recombinant proteins that have been shown to exhibit enzymatic activity are the two TUTases, RET1 (11) and RET2 (9), and the two RNA ligases, REL1 and REL2 (12, 13) (G.G., A. M. Simpson, X.K., K.R., M. Nebohacova, and L.S., unpublished work).
Materials and Methods
Tandem Affinity Purification (TAP) Isolation of Leishmania major LC-2 Overexpressed in Leishmania tarentolae. L. major RNA editing exonuclease 1 (REX1) was PCR amplified from L. major genomic DNA with the primers 5′-TCCCCCGGGATGCGGGGTGCGCTGGCGCG-3′ and 5′-TCCCCCGGGCAGGACTTGGAACTGCATGC-3′ (restriction sites added are in boldface), and the product was digested with XmaI and cloned. The vector was the pMRP1-TAP vector (14) that was self-ligated after removal of the mitochondrial RNA binding protein 1 insert by BamHI digestion, followed by digestion at its single XmaI site. L. tarentolae cells were transfected and selected for G418 resistance. Mitochondria were isolated as described (15) from late log phase cell cultures (108 cells per ml) in brain heart infusion medium with 10 μg/ml hemin and 100 μg/ml geneticin. The TAP isolation of the tagged REX1 protein was performed as described (16).
Cloning of L. major REX1 in Baculovirus Expression Vector. TAP-tagged L. major REX1 was PCR amplified from the pREX1-TAP vector by using the primers 5′-AGGCCTATGCGGGGTGCGCTGGCGCGTAGCGCATGT-3′ and 5′-TCTAGAGGTTGACTTCCCCGCGGAAT-3′. PCR products were cloned into pCR2.1-TOPO vector (Invitrogen). TAP-tagged REX1 DNA sequence was released by digestion with StuI and XbaI and inserted into the corresponding sites of pFastBac1 (Invitrogen) for baculovirus expression in Sf-9 insect cells as described by the company.
Purification of Recombinant L. major REX1–Calmodulin Binding Protein (CBP) Fusion Protein. The TAP isolation of the tagged L. major REX1 protein was performed as described (16). EGTA-eluted L. major REX1–CBP was desalted over an NAP25 column (Amersham Pharmacia) into 50 mM phosphate (pH 7.5) and 10% glycerol and loaded on a HiTrapSP-HP column (Amersham Pharmacia) that was eluted with a two-part linear gradient of 0–0.8 M NaCl over 50 ml, followed by 0.8–2 M NaCl over another 10 ml. The eluted fractions were diluted into 10 mM MgCl2, 500 nM DTT, and 5 mM Tris·HCl (pH 7.9), assayed for exonuclease activity by incubation with 5′ end-labeled RNA for 2 h at 27°C, and terminated by the addition of an equal volume of formamide and 10 mM EDTA. Degradation products were resolved on a 7 M urea and 15% polyacrylamide gel. To determine the relative purification of REX1–CBP, fractions were analyzed on 8–16% polyacrylamide SDS gradient gels (NOVEX, San Diego; Invitrogen). Gels were stained with SYPRO Ruby (Molecular Probes) and imaged by using a BioChemi Bioimaging System (Ultraviolet Products, San Gabriel, CA). TAP-isolated REX1 material used for all other enzymatic reactions was partially purified by size-exclusion chromatography on a Superose 6 column (Amersham Pharmacia) equilibrated with buffer containing 20 mM KCl, 20 mM Hepes (pH 7.5), and 0.2 mM EDTA. Fractions containing recombinant (r) REX1 were pooled and stored at –20°C after the addition of an equal volume of glycerol.
Exonuclease Activity Assay. Recombinant L. major REX1 was incubated with one of four synthetic RNAs terminating in homogeneous 3′ nucleotides: 6-U, 5′-GCUAUGUCUGCUAACUUGUUUUUU-3′; 2-U, 5′-GCACUACACGAUAAAUAUAAAAAGUU-3′; 2-A, 5′-GCACUACACGAUAAAUAUAAAAAGAA-3′; 2-C, 5′-GCACUACACGAUAAAUAUAAAAAGCC-3′; and 3-G, 5′-GCACUACACGAUAAAUAUAAAAAGGG-3′ (Oligos Etc., Guilford, CT; Dharmacon, Lafayette, CO). RNAs were 5′ end-labeled by using T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP and diluted into reaction buffer, and rREX1 was added. Reactions were terminated with formamide solution and analyzed on 7 M urea/15% polyacrylamide gels.
RNA Interference (RNAi). Primers for RNAi of T. brucei REX1 were designed to amplify a 500-bp sequence from the 3′ end of the gene. The PCR primer sets are as follows: 5′-AAGCTTATGGCATTGGCTCAGTCATG-3′ and 5′-TCTAGAAAGCGGACATCTTCTGCTCG-3′; and 5′-CGGGATCCATGGCATTGGCTCAGTCATG-3′ and 5′-GGCCAATTGAAGCGGACATCTTCTGCTCG-3′.
Added restriction sites are shown in boldface. The sequences were amplified from T. brucei genomic DNA and cloned into the pCR2.1-TOPO vector. The inserts were released from the vector with HindIII and XbaI digestion and with MunI and BamHI digestion and were inserted into the pLEW-HX-100GFP (17) vector to form a head-to-head RNAi plasmid under Parp promoter control. The plasmid was transformed into T. brucei 29-13 procyclic cells that contain integrated T7 RNA polymerase and tetracycline repressor (18). The cells were grown at 27°C in SDM-79 medium as described (11). RNAi was induced by addition of 1 μg/ml tetracycline. Mitochondria were isolated as described (15).
RNA Analysis. Total RNA from the T. brucei RNAi cell line was purified by the acid guanidium isothiocyanate method (19). RT-PCR of T. brucei REX1 mRNA was performed with Super-Script II reverse transcriptase according to the commercial protocol (Invitrogen), by using the primers 5′-AAGCTTATGGCATTGGCTCAGTCATG-3′ and 5′-CCCAAGCTTCTAAAGGGCACCAATAATC-3′. RT-PCR of T. brucei REX2 mRNA was performed by using the primers 5′-AAGCTTCAGTCGCTTGCACCTGCTAG-3′ and 5′-AAGCTTAACCACCTGAAACTCAACAAGGAGA-3′.
Glycerol Gradient Sedimentation of Mitochondrial Lysate. Purified mitochondria were lysed with 0.5% Nonidet P-40 in 50 mM Hepes (pH 8.1), 10 mM MgCl2, and 60 mM KCl in the presence of Complete Protease Inhibitor Mixture (Roche Diagnostics). The clarified extract was centrifuged on a 10–30% glycerol gradient in the SW41 rotor (Beckman Coulter) for 20 h at 111,132 × g (30,000 rpm). The fractions were treated with [α-32P]ATP at 27°C for 30 min to autoadenylate the endogenous REL1 and REL2 (20, 21) and mixed with SDS loading buffer for 8–16% denaturing gradient gel electrophoresis, and the gels were blotted for PhosphorImager (Amersham Biosciences) and Western blot analysis.
In Vitro Precleaved Editing Assay. The following chemically synthesized RNA substrates (Oligos Etc.; Qiagen, Chatsworth, CA) were used for the precleaved in vitro editing assay. The 3′ fragment and the gRNAs terminate in dideoxynucleotides to prevent the 3′ addition of Us by TUTase activity: 5′ fragment, 5′-GCACUACACGAUAAAUAUAAAAAG-3′; 5′-UU fragment, 5′-GCACUACACGAUAAAUAUAAAAAGUU-3′ (the 2 Us to be deleted are in italics); 3′ fragment, 5′-AACAUUAUGCUUCUUCGddC-3′;. –2 gRNA, 5′-AAGAAGCAUAAUGUUagCUUUUUAUAUUUAUCGUGUAGUCddG-3′ (guiding nucleotides are in boldface lowercase); –1 gRNA, 5′-A AGA AGCAUA AUGUUaCUUUUUAUAUUUAUCGUGUAGUCddG-3′ (guiding nucleotide is in bold-face lowercase); and 0 gRNA, 5′-AAGAAGCAUAAUGUUCUUUUUAUAUUUAUCGUGU AGUCddG-3′.
The 5′ fragments were 5′ phosphorylated with T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP. Complementary RNAs were annealed by heating and slow cooling. To obtain the bridged nicked substrate, the 3′ fragment, the 5′ fragment, and the 0 gRNA were used. In some reactions, 1 mM pyrophosphate (PPi) was added to inhibit ligase activity. Reactions were performed at 27°C for 2 h in 50 mm Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 1 mM ATP. For the insertion reaction, 2 mM UTP was added.
Mass Spectrometry. Gel bands of interest were excised and digested with trypsin (Promega) (22). The resulting digests were purified with C18 μZipTips (Millipore) and subjected to mass spectrometric analysis on an Applied Biosystems 4700 Proteomics Analyzer, which is a MALDI tandem TOF mass spectrometer (23). The purified sample was mixed 1:1 with α-cyano-4-hydroxycinnamic acid matrix (10 g/liter), and 1.2 μl of the mixture was spotted onto the MALDI target plate. A reflector mode mass spectrum of the digest was first obtained, after which several individual peptide peaks were manually selected for MS/MS analysis. Seven peptide peaks were analyzed in this fashion, and the peptide sequence for each was manually deduced from its MS/MS spectrum. In six of the seven cases, the sequences matched the protein MP100 from L. major, whereas the seventh was a peptide from the TAP tag used.
Results
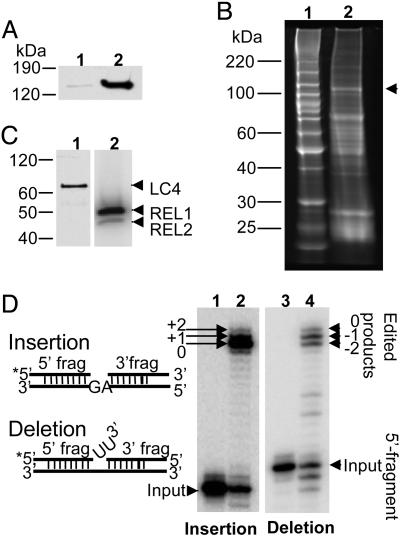
LC-2 (MP100) Is a Bona Fide Component of the L-Complex. Previously, LC-2 (MP100) was tentatively identified as a component of the L-complex by its presence in TAP isolates of several L-complex fusion proteins (16) and its presence in L-complex material purified by column chromatography (24). However, the stoichiometry of the LC-2 protein in TAP-isolated L-complex was not equivalent to that of other proteins (16). To confirm that LC-2 is a component of the L-complex, the L. major LC-2 was cloned as a TAP fusion protein in the pX expression vector, and the plasmid was transfected into L. tarentolae cells (25). Expression and mitochondrial targeting of the LC-2–TAP protein was demonstrated by Western blot analysis by using the peroxidase–antiperoxidase detection method (Fig. 1A). The tagged LC-2 protein was isolated by the TAP affinity procedure and found to copurify with several accepted markers for the L-complex: REL1, REL2, and LC-4 (Fig. 1 B and C). The TAP isolate was also active in in vitro gRNA-mediated U-insertion and U-deletion editing assays, in which two mRNA fragments are bridged by a gRNA at an editing site (precleaved editing assay) (26) (Fig. 1D).
Fig. 1.
Isolation of TAP-tagged LC-2 from mitochondrial lysate of transfected L. tarentolae. (A) Equivalent amounts of cytosol (lane 1) and mitochondrial extract (lane 2) were separated on an 8–16% polyacrylamide SDS gel, which was blotted and probed with the peroxidase–antiperoxidase reagent to detect the TAP-tagged LC-2. (B) Protein composition of TAP-isolated material. Lane 1, molecular mass standards; lane 2, SYPRO Ruby stained gel (arrowhead indicates position of CBP-tagged LC-2). (C) Lane 1, Western blot probed with anti-LC-4 antiserum; the endogenous LC-4 band is indicated by an arrowhead. Lane 2, the TAP-isolated material was incubated with [α-32P]ATP before gel analysis to detect the endogenous REL1 and REL2, which are indicated by arrowheads. Note that REL2 normally labels less than REL1 as a result of being precharged with AMP (29). (D) In vitro precleaved editing activities of the TAP-isolated material. The annealed RNA substrates are shown schematically on the left. Lanes: 1, input RNA for insertion editing; 2, +2-U gRNA-mediated insertion editing; 3, input RNA for deletion editing; 4, –2-U gRNA-mediated deletion editing.
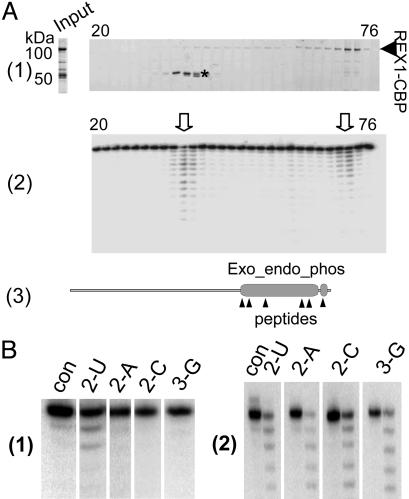
Isolation of Enzymatically Active Recombinant REX1 Protein. The L. major LC-2 gene was expressed as a TAP fusion protein (16, 27) in Sf-9 insect cells by using the baculovirus system. Isolation of the recombinant protein was performed by using the TAP procedure, which yielded a preparation with two major bands at ≈100 and 50 kDa and several minor bands (Fig. 2A, Input). This preparation showed an active 3′–5′ exoribonuclease activity with little specificity for the terminal nucleotide, except a slight preference for U (data not shown). This material was fractionated by ion-exchange chromatography, resulting in the separation of the two major peaks shown in Fig. 2 A1. One peak contained the 100-kDa protein, which is the expected size for the LC-2–CBP protein, and the other contained a doublet at ≈50 kDa. Major peaks of 3′–5′ exoribonuclease activity, obtained by using a 5′ end-labeled single-stranded RNA ending in 6 Us (6-U RNA) as substrate, were associated with each protein peak (Fig. 2 A2), although a minor amount of nuclease activity was spread throughout the gradient. The 50-kDa protein (Fig. 2 A1, *) was shown by mass spectrometry analysis of tryptic peptides to be a fragment of the LC-2 protein containing the nuclease motif. As shown in Fig. 2 A3, six peptides were identified localizing to the Exo_endo_phos and CBP motifs.
Fig. 2.
The 3′–5′ exonuclease activity of recombinant L. major LC-2. (A) The L. major LC-2–TAP fusion protein was expressed in a baculovirus system and affinity purified. The rLC-2–CBP was further purified over a HiTrapSP-HP column developed with a 0–2 M NaCl gradient over 60 ml. (A1) The fractions were analyzed on 8–16% SDS polyacrylamide gels that were stained with SYPRO Ruby. Input, the TAP-isolated material loaded on the column. The bands migrating at ≈50 kDa (*) were identified by mass spectrometry as L. major LC-2 degradation products. The two bands gave almost identical mass spectra, and all identified peptides from these bands corresponded to the C-terminal region of the LC-2–CBP fusion protein. (A2) The fractions were incubated with 5′ end-labeled 6-U RNA substrate, and the products were analyzed on a 7M urea/15% polyacrylamide gel. Two major peaks of exonuclease activity were observed (arrowheads). (A3) Diagram of the LC-2 protein showing the location of the Exo_endo_phos and CBP motifs and the sequenced peptides from the 50-kDa bands. (B1) U specificity of 100-kDa fraction (con, no enzyme control). The single-stranded RNAs used ending in 2 Us, 2 As, 2 Cs, or 2 Gs are indicated above each lane. (B2) Lack of U specificity of the 50-kDa REX1 fraction (con, no enzyme control for each digestion). The single-stranded RNAs used are indicated above each lane.
The 100-kDa protein fraction showed a U specificity of the 3′–5′ exonuclease activity (Fig. 2B1), only digesting the two 3′-terminal Us from a substrate with two 3′-terminal Us, whereas the 50-kDa protein fraction could digest other 3′-terminal nucleotides in addition to U (Fig. 2B2). These data argue strongly that the exonuclease activity was a property of the LC-2 protein rather than from any contaminating insect cell nuclease. Additional evidence for this conclusion from RNAi and in vitro reconstitution analysis will be presented below. These data also explain the apparent lack of U specificity of the unfractionated TAP-purified enzyme preparation and open up other interesting questions concerning the determinants that confer U specificity in the intact enzyme.
We have therefore designated LC-2 as REX1. The T. brucei homologue of the L. major REX1, MP100 (28), was not analyzed experimentally for exonuclease activity, but the high level of sequence similarity and the presence of the conserved Exo_endo_phos motif suggests that it is also a 3′–5′ exonuclease (2, 3). The second T. brucei L-complex protein with an Exo_endo_phos motif, MP99, shares extensive sequence similarity with MP100 and LC-2 but more closely resembles the L. major LC-3 protein, except for the lack of the Exo_endo_phos motif in the latter. We propose to designate MP99 as REX2 on the basis of the presence of this motif. LC-3 will be designated REX2* to indicate the overall similarity with MP99, except for the lack of the motif.
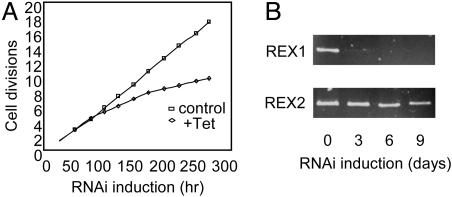
Down-Regulation of Expression of REX1 in T. brucei by Conditional RNAi Affects Cell Growth and Disrupts the L-Complex. A fragment of the REX1 gene was cloned into an RNAi expression vector for transfection of T. brucei 29-13 procyclic cells. Induction of RNAi with tetracycline was effective in an apparent selective degradation of REX1 mRNA, as shown by the RT-PCR results shown in Fig. 3B. The REX1 mRNA was degraded after 3 days of RNAi induction, whereas the closely related REX2 mRNA showed no degradation after 3 days induction and only slight degradation after 9 days induction. The lack of an antibody against REX1 precluded Western blot analysis. The slight effect on the level of REX2 mRNA after 9 days of RNAi induction could be attributable to the high level of sequence similarity of these proteins or to nonspecific effects attributable to breakdown of the L-complex (see below). There was also a slow growth phenotype (Fig. 3A).
Fig. 3.
Down-regulation of REX1 expression in T. brucei procyclics. (A) Growth curve of cells with and without induction of RNAi by addition of tetracycline (Tet). The cultures were diluted daily to maintain log phase growth. (B) RT-PCR of REX1 and REX2 mRNAs in cells down-regulated for REX1 expression.
Only a small effect of REX1 RNAi on in vivo RNA editing was observed, and this was gene specific (data not shown).
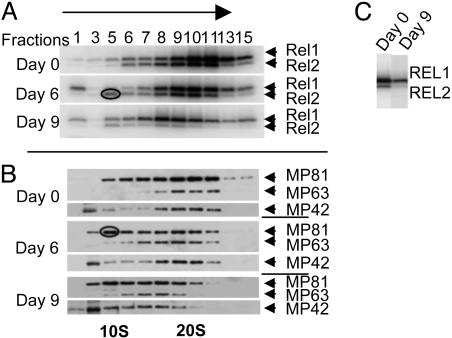
The effect of down-regulation of REX1 expression on the L-complex was monitored by glycerol gradient analysis of mitochondrial extracts. By using autoadenylation of REL1 and REL2 (Fig. 4A) and immunodetection of MP81, MP63, and MP42 (Fig. 4B) as markers for the L-complex, a significant steady decrease in the S-value of this complex was observed with increasing time of RNAi. There was also an ≈70% decrease in the abundance of L-complex REL1 and an ≈80% decrease in the abundance of L-complex REL2 (Fig. 4C) and an appearance of both REL2 (Fig. 4A, circled) and MP81 (Fig. 4B, circled) in gradient fraction 5, suggesting release of the REL2 subcomplex that contains REL2, MP81, and RET2 (12). A REL1 band was also seen in fraction 1 in the RNAi-induced cells, and we attribute it to release of free REL1 due to breakdown of the L-complex. It is interesting that free REL1 is released, whereas REL2 is released only in the REL2 subcomplex, which perhaps suggests a differential sensitivity of the ligases in the L-complex to down-regulation of REX1.
Fig. 4.
Effect of down-regulation of REX1 expression on stability of the L-complex. (A) Equivalent amounts of clarified mitochondrial lysates from uninduced and tetracycline-induced REX1–RNAi cells were fractionated on 10–30% glycerol gradients, and the fractions were subjected to autoadenylation with [α-32P]ATP and electrophoresed in an 8–16% polyacrylamide SDS gel. The gel was blotted onto a membrane and exposed by using a PhosphorImager to visualize the endogenous REL1 and REL2 bands. (B) The blot was reacted with antibodies against MP81, MP63, and MP42. The REL2 and MP81 bands apparently released from the L-complex in the form of the REL2 subcomplex are indicated by circles in A and B, respectively. (C) In a separate experiment, the L-complex peak fraction from uninduced, 9-day RNAi-induced cells were autoadenylated, and equal amounts were separated in a denaturing gel.
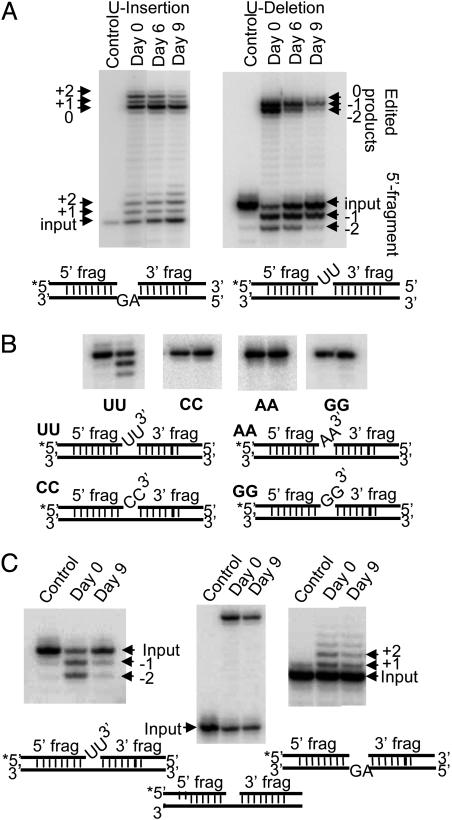
Down-Regulation of Expression of REX1 Preferentially Affects in Vitro Precleaved U-Deletion Editing. The peak L-complex fractions of mitochondrial extracts from cells down-regulated for REX1 expression were used for in vitro precleaved editing assays. There was a 78% inhibition of +2-U guided U-insertion editing after 9 days of RNAi (Fig. 5A Left), but there was a 98% inhibition of –2-U guided U-deletion editing after 9 days of RNAi (Fig. 5A Right). This decrease in the ligated edited product was accompanied by a decrease in the relative abundance of the 5′ fragment with two Us deleted.
Fig. 5.
Effect of down-regulation of REX1 expression on in vitro precleaved editing of the 20S L-complex fraction. (A Left) Peak L-complex fractions from glycerol gradients of mitochondrial lysates from cells uninduced or induced with tetracycline for REX1 RNAi for 6 or 9 days were used for in vitro precleaved U-insertion assays. (Right) The same fractions were used for in vitro precleaved U-deletion assays. The RNA substrates are diagrammed below. The positions of the 3′-extended 5′ fragments and the ligated products are indicated. (B) U specificity of exonuclease trimming of 3′ overhang. Model RNA substrates 3′ terminating in 2 Us, 2 As, 2 Cs, or 3 Gs (indicated below the gels) were digested with the peak L-complex fraction from uninduced cells in the presence of 1 mM PPi to inhibit ligation. (C Left) Peak L-complex fractions from glycerol gradients of mitochondrial lysates from cells uninduced (Day 0) or induced with tetracycline for REX1 RNAi for 9 days (Day 9) were incubated with the –2-U-deletion substrate shown below the panel in the presence of 1 mm PPi to inhibit ligation. (Center) Ligation of a bridged nicked substrate shown below the panel by L-complex fractions from uninduced (Day 0) and 9-day RNAi-induced cells (Day 9). (Right) 3′ addition of Us to 5′ fragment of +2-U-insertion substrate shown below the panel by L-complex fractions from cells uninduced (Day 0) and 9-day RNAi induced (Day 9) in the presence of 1 mM PPi to inhibit ligation.
To examine the specificity of the 3′–5′ exonuclease activity in the gradient-purified L-complex, several RNA substrates with 3′ overhangs of 2 Us, 2 As, 2 Cs, or 2 Gs were used, and inorganic PPi was added to inhibit ligase activity. As shown in Fig. 5B, the 3′–5′ exonucleolytic digestion of the L-complex fraction from cells not induced for REX1 RNAi was specific for a 3′ oligo-U single-strand overhang. No trimming was observed of 3′ terminal A, C, or G residues.
In Fig. 5C Left, the –2-U precleaved RNA substrate was incubated in the presence of PPi with the peak L-complex gradient fraction from cells uninduced or induced for REX1 RNAi for 9 days. The results again show an inhibition of the trimming of the 3′ overhanging Us from the 5′ fragment after down-regulation of REX1 expression. These data suggest that REX1 is the major exonuclease involved in this reaction and raise the question of the role of the second exonuclease REX2.
Control experiments on the effect of REX1 down-regulation on ligation of a nicked bridged substrate and on the RET2-mediated 3′ addition of Us (9) in the absence of ligation are shown in Fig. 5C (Center and Right, respectively). There was a slight effect on ligation and a slight effect on 3′ U addition to the 5′ fragment in the presence of pyrophosphate after 9 days of RNAi induction. These effects are probably nonspecific because of the disruption of the L-complex caused by loss of REX1.
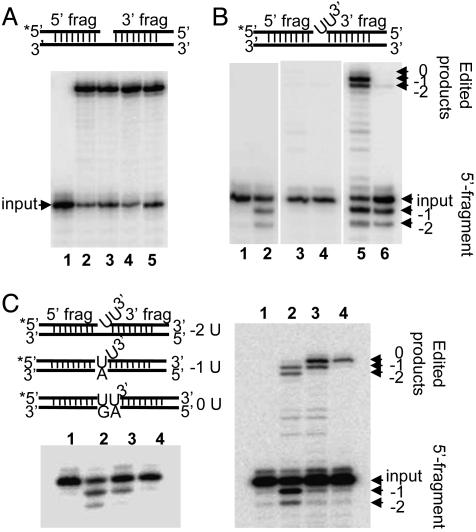
Reconstitution of Precleaved gRNA-Mediated in Vitro U-Deletion Editing with Recombinant REX1 and REL1. Both rREL1 and rREL2 obtained by expression in a baculovirus system (see Materials and Methods) were equally capable of efficiently ligating a nicked bridged substrate, as shown in Fig. 6A (lanes 2 and 3). Addition of rREX1 had no effect on either ligation activity (Fig. 6A, lanes 4 and 5). However, rREL1 and rREL2 were both incapable of ligating a bridged substrate with two 3′ overhanging Us (Fig. 6B, lanes 3 and 4).
Fig. 6.
Reconstitution of in vitro precleaved U-deletion editing with two recombinant proteins. Recombinant L. major REX1, recombinant L. tarentolae REL1, and recombinant L. tarentolae REL2 were used for this assay. (A) Ligation of a 5′ end-labeled (*) bridged nicked RNA substrate, shown above the panel. The RNA was mixed with 0.3 pmol of rREL1 or 0.1 pmol of rREL2 in the presence or absence of rREX1. Lanes: 1, mock reaction without enzymes; 2, rREL1; 3, rREL2; 4, rREL1 plus rREX1; 5, rREL2 plus rREX1. (B) The –2-U-deletion substrate shown above the panel was mixed with rREX1 alone, rREL1 alone, or rREL2 alone and also with mixtures of rREL1 or rREL2 and rREX1. Lanes: 1, mock reaction without enzymes; 2, rREX1; 3, rREL1, 4, rREL2; 5, rREL1 plus rREX1; 6, rREL2 plus rREX1. (C Left) Lanes: 1, input RNA; 2, the –2-U substrate RNA plus rREX1; 3, the –1-U substrate RNA plus rREX1; 4, the 0-U substrate RNA plus rREX1. (Right) gRNA-mediated U-deletion editing. Lanes: 1, mock reaction without enzymes but with the –2-U gRNA; 2, –2-U gRNA-mediated production of –1-U and –2-U ligated products (arrows); 3, –1-U gRNA-mediated production of –1-U ligated product (arrow); 4, 0-U gRNA-mediated production of no U ligated product (arrow). The 5′ fragments are indicated, as are the ligated products in each lane. The substrate RNAs used for both panels are diagrammed on the left.
As expected, rREX1 by itself removed the two 3′ overhanging Us from the 5′ fragment (Fig. 6B, lane 2). However, a mixture of rREX1 and rREL1 efficiently removed the 3′ Us and ligated the edited products (Fig. 6B, lane 5), whereas a mixture of rREX1 and rREL2 removed the two 3′ Us but was much less efficient at ligating the two fragments (≈4% of the activity of the rREL1 plus rREX1 assay) (Fig. 6B, lane 6). Addition of recombinant LC-4 (MP63) or rREL2 with rREX1 did not affect the ligation activity of rREL1 with the –2-U substrate (data not shown).
Varying the number of guiding nucleotides in the gRNA changed the number of Us deleted in the edited products produced with the mixture of rREL1 and rREX1, as shown in Fig. 6C Right. When a gRNA that could base pair with one of the two 3′ Us was used, there was a complete inhibition of the –2-U ligated product with no effect on the –1-U ligated product (Fig. 6C Right, lane 3). Use of a gRNA that could base pair with both Us led to the disappearance of both the –2-U and –1-U ligated products (Fig. 6C Right, lane 4). This evidence strongly suggests that REX1, in addition to being U specific, stops at a duplex region. This conclusion was confirmed by incubation of the same RNA substrates with rREX1 alone (Fig. 6 Left). Two 3′ over-hanging Us are trimmed (Fig. 6C Left, lane 2), and base pairing of one or two of these Us yielded the trimming of a single overhanging U (Fig. 6C, lane 3) or no Us, respectively (Fig. 6C Left, lane 4).
Discussion
Our results indicate that REX1 is the major 3′–5′ exonuclease responsible for U-deletion activity. After overexpression in an insect cell line, affinity-purified CBP-tagged rREX1 was shown to be a U-specific 3′–5′ exoribonuclease that digests a 3′ single-stranded oligo-U overhang and stops at a duplex region, as is predicted to occur in the in vivo editing reaction. Furthermore, depletion of REX1 from the L-complex by RNAi fairly selectively affected U-deletion precleaved editing in vitro. Finally, gRNA-directed precleaved U-deletion editing could be reconstituted in vitro by a mixture of the two purified recombinant proteins from L. tarentolae, rREX1 and rREL1 (RNA ligase 1). Our results are consistent with previous proposals based on differential adenylate sensitivity of in vitro U-insertion and U-deletion editing using gradient-purified fractions (29, 30).
The observed low effect of REX1 RNAi on the relative abundance of edited transcripts was consistent with the slow growth nonlethal phenotype. This low effect could be attributable to the lower relative extent of U-deletion editing compared with U-insertion editing, to the turnover rate of the REX1 enzyme, or to the presence of a second exonuclease, REX2 (MP99). It remains to be seen what is the role of REX2, which is present in the REL1 subcomplex in T. brucei and the Leishmania REX2*, a homologue of the T. brucei REX2 that lacks the exonuclease domain. Previous results indicated that a 5S gradient fraction from a REL1–TAP-isolated fraction of T. brucei, which contained at least MP63 (LC-4) and REX2 (MP99) in addition to REL1, supported precleaved U-deletion in vitro editing. In contrast, a 5S fraction from a REL2–TAP isolated fraction did not support U-deletion editing (12). However, both of these fractions were not well characterized in terms of their components. It is also possible that REX2 is active as a U-specific editing exonuclease in vitro but not in vivo.
We propose that, in vivo, REX1 has assumed the role of REX2, at least in Leishmania in which REX2* is apparently defective and perhaps in both species, and transiently interacts with the REL1 subcomplex in the U-deletion editing reaction. Of course, the situation in vivo is likely to be more complex in that both REL1 and REL2 interact with at least two other L-complex proteins, and these interactions may affect their specificity and activities.
We also conclude that REL1 is the RNA ligase that is responsible for the ligation of mRNA fragments that have had the overhanging 3′ Us removed from the 5′ fragment by the REX1 exonuclease. The strongest evidence for REL1 being the ligase involved in U-deletion editing is that rREL1 is solely capable of ligating a bridged RNA substrate that was 3′-trimmed by rREX1 activity. The inability of rREL2 to ligate this substrate in the presence of rREX1 suggests an interaction between rREL1 and the rREX1-trimmed RNA that does not occur in the case of rREL2, although preliminary results indicate no detectable interaction between rREL1 and rREX1 in a coupled transcription–translation system (data not shown). There appears to be a fundamental difference between rREL1 and rREL2 in regard to the ability to ligate the product of the rREX1 exonuclease trimming reaction, the nature of which should prove interesting.
The biological role of REL2 in editing is still enigmatic. The inability to ligate RNA-bridged fragments trimmed by rREX1 in vitro and the association in vivo of REL2 with the RET2 TUTase that is responsible for the insertion of Us at editing sites (12) make the idea that REL2 is involved only in U-insertion editing attractive (13, 30). However, loss of REL2 from the L-complex has no effect on viability or on editing (31), whereas loss of REL1 affects both U-deletion and U-insertion editing (31), possibly suggesting that REL1 is involved in ligation at both U-deletion and U-insertion editing sites.
Acknowledgments
We thank all members of the L.S. laboratory for advice and discussion. We thank Ken Stuart (Seattle Biomedical Research Institute, Seattle) for the gift of monoclonal antibodies against MP81, MP63, and MP42. This work was supported in part by National Institutes of Health Grant AI09102 (to L.S.)
Author contributions: X.K., K.R., and L.S. designed research; X.K., K.R., G.G., A.M.F., and S.Z. performed research; X.K., G.G., A.M.F., and S.Z. contributed new reagents/analytic tools; X.K., K.R., G.G., A.M.F., and L.S. analyzed data; and L.S. wrote the paper.
Abbreviations: gRNA, guide RNA; L-complex, RNA ligase-containing complex; REL1/2, RNA editing ligase 1/2; TUTase, terminal uridylyltransferase; RET1/2, RNA editing 3′ TUTase 1/2; REX1/2, RNA editing exonuclease 1/2; r, recombinant; TAP, tandem affinity purification; CBP, calmodulin binding protein; RNAi, RNA interference; PPi, pyrophosphate.
References
- 1.Estévez, A. M. & Simpson, L. (1999) Gene 240, 247–260. [DOI] [PubMed] [Google Scholar]
- 2.Simpson, L., Aphasizhev, R., Gao, G. & Kang, X. (2004) RNA 10, 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worthey, E. A., Schnaufer, A., Mian, I. S., Stuart, K. & Salavati, R. (2003) Nucleic Acids Res. 31, 6392–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S. R., Griffiths-Jones, S., Howe, K. L., Marshall, M. & Sonnhammer, E. L. (2002) Nucleic Acids Res. 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnaufer, A., Panigrahi, A. K., Panicucci, B., Igo, R. P., Salavati, R. & Stuart, K. (2001) Science 291, 2159–2161. [DOI] [PubMed] [Google Scholar]
- 6.Rusche, L. N., Huang, C. E., Piller, K. J., Hemann, M., Wirtz, E. & Sollner-Webb, B. (2001) Mol. Cell. Biol. 21, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McManus, M. T., Shimamura, M., Grams, J. & Hajduk, S. L. (2001) RNA 7, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maslov, D. A. & Simpson, L. (1992) Cell 70, 459–467. [DOI] [PubMed] [Google Scholar]
- 9.Aphasizhev, R., Aphasizheva, I. & Simpson, L. (2003) Proc. Natl. Acad. Sci. USA 100, 10617–10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst, N. L., Panicucci, B., Igo, R. P., Jr., Panigrahi, A. K., Salavati, R. & Stuart, K. (2003) Mol. Cell 11, 1525–1536. [DOI] [PubMed] [Google Scholar]
- 11.Aphasizhev, R., Sbicego, S., Peris, M., Jang, S. H., Aphasizheva, I., Simpson, A. M., Rivlin, A. & Simpson, L. (2002) Cell 108, 637–648. [DOI] [PubMed] [Google Scholar]
- 12.Schnaufer, A., Ernst, N. L., Palazzo, S. S., O'Rear, J., Salavati, R. & Stuart, K. (2003) Mol. Cell 12, 307–319. [DOI] [PubMed] [Google Scholar]
- 13.Huang, C. E., Cruz-Reyes, J., Zhelonkina, A. G., O'Hearn, S., Wirtz, E. & Sollner-Webb, B. (2001) EMBO J. 20, 4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aphasizhev, R., Aphasizheva, I., Nelson, R. E. & Simpson, L. (2003) RNA 9, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braly, P., Simpson, L. & Kretzer, F. (1974) J. Protozool. 21, 782–790. [DOI] [PubMed] [Google Scholar]
- 16.Aphasizhev, R., Aphasizheva, I., Nelson, R. E., Gao, G., Simpson, A. M., Kang, X., Falick, A. M., Sbicego, S. & Simpson, L. (2003) EMBO J. 22, 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LaCount, D. J., Bruse, S., Hill, K. L. & Donelson, J. E. (2000) Mol. Biochem. Parasitol. 111, 67–76. [DOI] [PubMed] [Google Scholar]
- 18.Wirtz, E., Leal, S., Ochatt, C. & Cross, G. A. (1999) Mol. Biochem. Parasitol. 99, 89–101. [DOI] [PubMed] [Google Scholar]
- 19.Chomczynski, P. & Sacchi, N. (1987) Anal. Biochem. 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 20.Peris, M., Simpson, A. M., Grunstein, J., Liliental, J. E., Frech, G. C. & Simpson, L. (1997) Mol. Biochem. Parasitol. 85, 9–24. [DOI] [PubMed] [Google Scholar]
- 21.Sabatini, R. & Hajduk, S. L. (1995) J. Biol. Chem. 270, 7233–7240. [DOI] [PubMed] [Google Scholar]
- 22.Bienvenut, W. V., Deon, C., Pasquarello, C., Campbell, J. M., Sanchez, J. C., Vestal, M. L. & Hochstrasser, D. F. (2002) Proteomics 2, 868–876. [DOI] [PubMed] [Google Scholar]
- 23.Jimenez, A. A., Huang, L., Qiu, Y. & Burlingame, A. L. (1998) in Current Protocols in Protein Science, eds. Coligan, J. E., Dunn, B. M., Ploegh, H. L., Speicher, D. W. & Wingfield, P. T. (Wiley, New York), pp. 16.4.1–16.4.5.
- 24.Panigrahi, A. K., Allen, T. E., Stuart, K., Haynes, P. A. & Gygi, S. P. (2003) J. Am. Soc. Mass Spectrom. 14, 728–735. [DOI] [PubMed] [Google Scholar]
- 25.LeBowitz, J. H., Coburn, C. M., McMahon-Pratt, D. & Beverley, S. M. (1990) Proc. Natl. Acad. Sci. USA 87, 9736–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igo, R. P., Palazzo, S. S., Burgess, M. L., Panigrahi, A. K. & Stuart, K. (2000) Mol. Cell. Biol. 20, 8447–8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M. & Seraphin, B. (2001) Methods 24, 218–229. [DOI] [PubMed] [Google Scholar]
- 28.Panigrahi, A. K., Schnaufer, A., Ernst, N. L., Wang, B., Carmean, N., Salavati, R. & Stuart, K. (2003) RNA 9, 484–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cruz-Reyes, J., Rusche, L., Piller, K. J. & Sollner-Webb, B. (1998) Mol. Cell 1, 401–409. [DOI] [PubMed] [Google Scholar]
- 30.Cruz-Reyes, J., Zhelonkina, A. G., Huang, C. E. & Sollner-Webb, B. (2002) Mol. Cell. Biol. 22, 4652–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao, G. & Simpson, L. (2003) J. Biol. Chem. 278, 27570–27574. [DOI] [PubMed] [Google Scholar]