The phenotypic and physiological potential of a cell is determined by its transcription program, which, in turn, is determined by the array of transcription factors present in the cell. A sophisticated and detailed understanding of the role of a particular transcription factor requires that all of its target genes be identified. This task is a daunting one, but it has become feasible with the availability of complete genome sequences, coupled with genome-wide analytical methods such as transcription profiling and chromatin immunoprecipitation (ChIP) assays. In a recent study, Harbison et al. (1) used ChIP analysis, together with predicted sequence-recognition motifs, to determine the genomic occupancy of 203 DNA-binding transcription regulators in yeast. Because of the scope of that study, the question of whether all of the targets of a particular transcription regulator function in a particular physiological setting were identified was left unanswered. In a recent issue of PNAS, Galgoczy et al. (2) focus on a specific biological question with the goal of identifying the complete set of target genes for the transcription regulators that determine cell type in the budding yeast Saccharomyces cerevisiae. Cell-type specification in yeast has been studied intensely in a number of laboratories over the last 20 years, and the information gained from those studies served as a touchstone for the current work; that is, certain targets could be anticipated. However, there is ample room for surprise. Some target genes may have eluded discovery, and it is conceivable that the cell-type regulators also make connections to other physiological processes. Indeed, both possibilities are realized in the current study. In their work, Galgoczy et al. (2) used ChIP analysis, as had Harbison et al. (1), but, in addition, they used transcription profiling and phylogenetic comparisons. The application of all three approaches resulted in “overdetermination” of the target gene sets for the yeast cell-type regulators, giving one confidence that the complete sets have been identified.
An understanding of the role of a particular transcription factor requires that all of its target genes be identified.
Yeast Cell Type: The Regulators
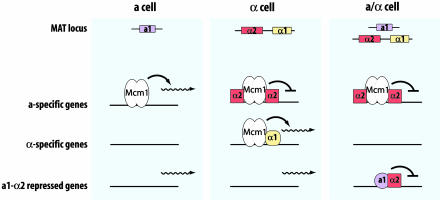
Cell type in yeast is determined by information at the mating-type locus (3) (Fig. 1). MATα encodes two transcription regulators, α1 and α2. α1 is required to activate transcription of genes that impart the “α character,” e.g., genes that encode α-factor pheromone and the receptor for a-factor pheromone. α2 is a negative regulator of genes that give the cell an a character, e.g., genes that encode a-factor pheromone and the receptor for α-factor. MATa, the alternate allele at the mating-type locus, also encodes a transcription regulator, a1, but this regulator plays no role in determination of the a cell type. Instead, the absence of the two MATα-encoded regulators is sufficient to confer the a character; the absence of α1 precludes expression of α-specific genes, and the absence of α2 permits expression of a-specific genes. Altogether, four α-specific and six a-specific genes have been identified. It is unlikely that either of these gene sets contains many undiscovered members crucial for mating, because the introduction of just two a-specific gene products, a-factor and the receptor for α-factor, enables a MATα cell to mate as if it were a MATa cell (4).
Fig. 1.
Cell-type regulation circuitry. The three yeast cell types and the regulatory proteins encoded by the MAT locus are depicted. These proteins, together with Mcm1, which is encoded elsewhere in the genome, govern expression of three sets of genes: a-specific genes, α-specific genes, and a1·α-repressed genes. See text for additional details. Also note that one of the a1·α2-repressed genes is MAT α1. This repression therefore precludes expression of α-specific genes in the a/α cell type. (Figure courtesy of D. Galgoczy and A. D. Johnson.)
An a/α diploid contains both MATα and MATa alleles, and its phenotype is conferred by another regulatory activity, the a1·α2 heterodimer. This heterodimer is a repressor that prevents expression of the negative regulators of a/α traits. Because diploid and haploid cells are physiologically and morphologically distinctive, a1·α2 must control a broad spectrum of traits, including meiosis and sporulation, sensitivity to DNA damaging agents, and the pattern of bud emergence (5–8).
Yeast Cell Type: The Targets
To identify targets of α2 and the a1·α2 heterodimer, Galgoczy et al. (2) carried out three separate analyses: genome-wide ChIP analysis, transcriptional profiling with microarrays, and phylogenetic comparisons by using sequences from four closely related Saccharomyces sensu stricto species. The α2 targets that emerged with each method were the same: six sites that control expression of a-specific genes. (A seventh site was also found, but it controls the activity of a recombinational enhancer involved in mating-type switching.) All six a-specific gene sites had previously been identified by more conventional analyses. Thus, the α2 sites are highly overdetermined, and it is unlikely that they are contaminated by false-positives or false-negatives.
The search for a1·α2 targets was somewhat less straightforward. The combination of all three approaches was required to eliminate false-positive genes and sites that had been identified by one of the other approaches. For example, transcriptional profiling revealed genes that are directly repressed by a1·α2 but also yielded genes that are indirectly repressed. Similarly, the a1·α2 ChIP experiment did not give the same degree of IP fragment enrichment as did the α2 experiment, perhaps because chromatin complexes containing a1·α2 heterodimers were less effectively recognized by anti-α2 antibodies than the complexes containing α2 homodimers bound at α2 sites. This reduced enrichment led to the preliminary isolation of DNA fragments that ultimately proved to lack recognizable a1·α2-binding sequences and were false-positives. Nevertheless, although each method has shortcomings, when used together, the three analyses identified all 19 of the previously reported genes that are directly regulated by the a1·α2 heterodimer. A recent, related study by Nagaraj et al. (9) also identified a number of a1·α2 sites and reached a similar conclusion, namely, that combining two or more forms of data are a powerful tool for identification of a full set of coregulated genes.
To identify α1 sites in the genome, Galgoczy et al. (2) relied on transcriptional profiling and phylogenetic comparisons alone, because the lack of suitable antibodies to α1, or tagged versions of α1, precluded ChIP analysis.
Even with this limitation, the analysis was able to identify all known α-specific genes, and another one as well (see below).
Puzzles remain in the transcriptional circuitry that generates the three yeast cell types.
Did any surprises emerge from this work? Are there lessons for the application of these strategies to similar problems in more complex, multicellular organisms? In fact, there were several surprises. First, a previously uncharacterized α-specific gene was identified. Deletion of this gene does not lead to an obvious mating defect, but, given its conservation among the S. sensu stricto species, it seems likely that it plays a role in mating that is yet to be discerned. Second, some genes not thought to be under a1·α2 control were shown to be so. For example, HOG1, which encodes a mitogen-activated protein kinase that is part of an osmosensing signal transduction pathway (10), was shown to be transcriptionally repressed by a1·α2. The biological significance of this repression was supported by the demonstration that a/α diploids were more sensitive to osmostress than were either a or α haploids. Third, and perhaps most provocative, Galgoczy et al. (2) identified seven phylogenetically conserved a1·α2 sites that are significantly occupied in a/α cells but do not seem to control transcription of an adjacent gene. Perhaps there are as-yet-unannotated transcripts controlled by a1·α2. The position of one of the seven sites suggests that it could regulate a known haploid-specific IME4 antisense transcript (11). A provocative possibility is that one (or more) of the other six sites also regulates production of an antisense transcript.
As these examples illustrate, intriguing puzzles remain in the transcriptional circuitry that generates the three yeast cell types. Equally exciting is the prospect of applying this three-pronged genomic analysis to the problem of cell-type determination in multicellular organisms. Here, the problem becomes more difficult because of increased genomic complexity, but the combinatorial power of the genome-wide ChIP, transcriptional profiling, and phylogenetic comparisons may be up to the challenge.
See companion article on page 18069 in issue 52 of volume 101.
References
- 1.Harbison, C. T., Gordon, D. B., Lee, T. I., Rinaldi, N. J., Macisaac, K. D., Danford, T. W., Hannett, N. M., Tagne, J. B., Reynolds, D. B., Yoo, J., et al. (2004) Nature 431, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galgoczy, D. J., Cassidy-Stone, A., Llinás, M., O'Rourke, S. M., Herskowitz, I., DeRisi, J. L. & Johnson, A. D. (2004) Proc. Natl. Acad. Sci. USA 101, 18069–18074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herskowitz, I., Rine, J. & Strathern, J. (1992) in The Molecular and Cellular Biology of the Yeast Saccharomyces (Cold Spring Harbor Lab. Press, Woodbury, NY), Vol. 2, pp. 583–657. [Google Scholar]
- 4.Bender, A. & Sprague, G. F., Jr. (1989) Genetics 121, 463–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heude, M. & Fabre, F. (1993) Genetics 133, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covitz, P. A., Herskowitz, I. & Mitchell, A. P. (1991) Genes Dev. 5, 1982–1989. [DOI] [PubMed] [Google Scholar]
- 7.Fujita, A., Oka, C., Arikawa, Y., Katagai, T., Tonouchi, A., Kuhara, S. & Misumi, Y. (1994) Nature 372, 567–570. [DOI] [PubMed] [Google Scholar]
- 8.Valencia, M., Bentele, M., Vaze, M. B., Herrmann, G., Kraus, E., Lee, S. E., Schar, P. & Haber, J. E. (2001) Nature 414, 666–669. [DOI] [PubMed] [Google Scholar]
- 9.Nagaraj, V. H., O'Flanagan, R. A., Bruning, A. R., Mathias, J. R., Vershon, A. K. & Sengupta, A. M. (2004) BMC Genomics 5, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gustin, M. C., Albertyn, J., Alexander, M. & Davenport, K. (1998) Microbiol. Mol. Biol. Rev. 62, 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah, J. C. & Clancy, M. J. (1992) Mol. Cell. Biol. 12, 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]