Abstract
Selenophosphate, an activated form of selenium that can serve as a selenium donor, is generated by the selD gene product, selenophosphate synthetase (SPS). Selenophosphate is required by several bacteria and by mammals for the specific synthesis of Secys-tRNA, the precursor of selenocysteine in selenoenzymes. Although free selenide can be used in vitro for synthesis of selenophosphate, the physiological system that donates selenium to SPS is incompletely characterized. To detect potential selenium-delivery proteins, two known sulfurtransferases and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; EC 1.2.1.12) were analyzed for ability to bind and transfer selenium. Rhodanese (EC 2.8.1.1) was shown to bind selenium tightly, with only part of the selenium being available as substrate for SPS in the presence of added reductant. 3-Mercaptopyruvate sulfurtransferase (3-MST; EC 2.8.1.2) and GAPDH also bound selenium supplied as selenodiglutathione formed from 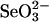 and glutathione. Selenium bound to 3-MST and GAPDH was released more readily than that from rhodanese and also was more available as a substrate for SPS. Although rhodanese retained tightly bound selenium under aerobic conditions, the protein gradually became insoluble, whereas GAPDH containing bound selenium was stable at neutral pH for a long period. These results indicate that 3-MST and GAPDH have more suitable potentials as a physiological selenium-delivery protein than rhodanese. In the presence of a selenium-binding protein, a low level of selenodiglutathione formed from
and glutathione. Selenium bound to 3-MST and GAPDH was released more readily than that from rhodanese and also was more available as a substrate for SPS. Although rhodanese retained tightly bound selenium under aerobic conditions, the protein gradually became insoluble, whereas GAPDH containing bound selenium was stable at neutral pH for a long period. These results indicate that 3-MST and GAPDH have more suitable potentials as a physiological selenium-delivery protein than rhodanese. In the presence of a selenium-binding protein, a low level of selenodiglutathione formed from 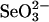 and glutathione could effectively replace the high concentrations of selenide routinely used as substrate in the SPS in vitro assays.
and glutathione could effectively replace the high concentrations of selenide routinely used as substrate in the SPS in vitro assays.
Keywords: 3-mercaptopyruvate sulfurtransferase, glyceraldehyde-3-phosphate dehydrogenase, rhodanese, selenodiglutathione, selenium delivery
Selenocysteine is cotranslationally incorporated into several prokaryotic- and eukaryotic-specific selenoproteins at inframe UGA codons. It has been shown that four genes, selA, selB, selC, and selD, are required for selenocysteine codon recognition and translation (1–3). In these processes, the selenium is derived from an activated selenium donor, selenophosphate, which is generated by the selD gene product, selenophosphate synthetase (SPS) (4, 5). However, the metabolic pathway that provides the selenium to SPS as substrate for selenophosphate biosynthesis remains to be characterized.
The reaction of  with several thiols produces selenotrisulfides. A selenotrisulfide derivative has been postulated as a possible intermediate in the bioconversion of dietary inorganic selenium into bioactive selenocompounds (6). In many bacteria and eukaryotes, glutathione (GSH) is a prime candidate as the thiol compound in vivo, because GSH is the most abundant low-molecular-weight thiol in most cells. Recently, Lindemann and Hintelmann (7) succeeded in detecting selenodiglutathione (GSSeSG) in a yeast extract for the first time. Contrary to previous expectations, this result indicated that GSSeSG may exist as a normal intermediate in the cell under physiological conditions. Although GSSeSG has been considered a key intermediate in the selenium metabolic pathway (6, 8), questions remain regarding stability and the mechanism of specific incorporation of the selenium into selenocysteine. Indeed, other studies indicate that GSSeSG is too labile to serve as a selenium source, especially in the presence of excess GSH under aerobic conditions (9).
with several thiols produces selenotrisulfides. A selenotrisulfide derivative has been postulated as a possible intermediate in the bioconversion of dietary inorganic selenium into bioactive selenocompounds (6). In many bacteria and eukaryotes, glutathione (GSH) is a prime candidate as the thiol compound in vivo, because GSH is the most abundant low-molecular-weight thiol in most cells. Recently, Lindemann and Hintelmann (7) succeeded in detecting selenodiglutathione (GSSeSG) in a yeast extract for the first time. Contrary to previous expectations, this result indicated that GSSeSG may exist as a normal intermediate in the cell under physiological conditions. Although GSSeSG has been considered a key intermediate in the selenium metabolic pathway (6, 8), questions remain regarding stability and the mechanism of specific incorporation of the selenium into selenocysteine. Indeed, other studies indicate that GSSeSG is too labile to serve as a selenium source, especially in the presence of excess GSH under aerobic conditions (9).
Alternatively, NifS-like proteins and selenocysteine lyase enzymes, which decompose selenocysteine to elemental selenium and alanine, have been considered as candidates for the control of free selenium levels in vivo (10, 11). Although selenocysteine lyase can mobilize a transfer form of selenium Se* from l-selenocysteine for selenophosphate biosynthesis, for the selenocysteine lyase to be effective as a selenium-delivery protein additional cellular components and proteins seem to be required to assist in the specific delivery of the active form of selenium to SPS. Several proteins have been recognized as sulfur transferases or delivery proteins in various sulfur metabolism pathways (12). These proteins include rhodanese and 3-mercaptopyruvate sulfurtransferase (3-MST). Transport proteins that can effectively bind selenium in the presence of the much higher concentrations of sulfur normally present in cells clearly are required to ensure the effective delivery of selenium in metabolic pathways. However, the selectivity and affinity of sulfurtransferases toward selenium have not been well studied (except for rhodanese), and therefore the possibility remains that a protein of this type could function in selenium-delivery systems. We recently showed that rhodanese, which is a typical sulfurtransferase, binds selenium from  and GSH (13). The selenium presumably bound as a perselenide derivative of the active cysteine could be used as substrate by SPS when DTT was added to the reaction mixture. Furthermore, recent reports have shown that glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and deoxyribose phosphate aldolase actively bind selenium in Escherichia coli (14).
and GSH (13). The selenium presumably bound as a perselenide derivative of the active cysteine could be used as substrate by SPS when DTT was added to the reaction mixture. Furthermore, recent reports have shown that glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and deoxyribose phosphate aldolase actively bind selenium in Escherichia coli (14).
In the present work, we first compared the ability of three proteins of interest to bind selenium under controlled in vitro conditions. Next, the stability of the three proteins with bound selenium was examined in the presence or absence of physiological concentrations of GSH. We also established a more sensitive assay method for SPS activity to estimate the relative availability of various selenium-delivery systems. Finally, we compared the effectiveness of selenium derived from GSSeSG as substrate for SPS in the presence of each selenium-binding protein in this SPS assay.
Materials and Methods
Materials. Bovine liver rhodanese, human erythrocyte GAPDH, Na2 SeO3, ATP, thioctic acid (α-lipoic acid), and AMP were obtained from Sigma. SPS was purified by the procedure of Veres et al. (15). Dihydrolipoic acid (DHLA) was prepared according to the method of Pagani et al. (16)
The 3-mst gene from human liver was cloned into pGEX-2T (pGEX-2T/MS 1.2), and the gene product from the recombinant expression system was purified as the glutathione S-transferase fusion protein. All buffers and reagents were prepared from the highest-grade chemicals available.
Methods. Selenium binding and release of selenium from proteins. Persulfide-free rhodanese (E form) was prepared as described (13). In the present selenium-binding study, rhodanese, 3-MST, and GAPDH were reacted with 0.1 mM 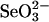 and 0.4 mM GSH in PBS (pH 7.4), containing 1 mM EDTA. After incubation at 37°C for 10 min, reaction mixtures were applied to an FPLC fast desalting column (10 × 100 mm; Amersham Pharmacia) to remove the excess
and 0.4 mM GSH in PBS (pH 7.4), containing 1 mM EDTA. After incubation at 37°C for 10 min, reaction mixtures were applied to an FPLC fast desalting column (10 × 100 mm; Amersham Pharmacia) to remove the excess 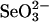 , GSH, and small-molecular-weight products. When the release of selenium bound to proteins was studied, 2 mM DTT was added to the reaction mixture, and after further incubation at 37°C for 10 min, the mixture was applied to the desalting column. Each protein eluted in the flow-through fractions was monitored for protein content by reaction with the Bradford reagent (17) and for selenium by atomic absorption. All fractions of the elution profile also were assayed for selenium to detect low-molecular-weight selenium products.
, GSH, and small-molecular-weight products. When the release of selenium bound to proteins was studied, 2 mM DTT was added to the reaction mixture, and after further incubation at 37°C for 10 min, the mixture was applied to the desalting column. Each protein eluted in the flow-through fractions was monitored for protein content by reaction with the Bradford reagent (17) and for selenium by atomic absorption. All fractions of the elution profile also were assayed for selenium to detect low-molecular-weight selenium products.
Each flow-through fraction from the FPLC column was stored at 4°C for 4 weeks to study the stability of selenium-binding proteins. The protein solutions, 0.5 ml-volumes contained in 1.5-ml screw-capped tubes, were in contact with air as the head space.
Selenium binding in the presence of excess GSH. Rhodanese, 3-MST, and GAPDH were reacted with 0.1 mM  and 4 mM GSH in PBS (pH 7.4) containing 1 mM EDTA. After incubation at 37°C for 10 min, mixtures were applied to an FPLC fast desalting column (10 × 100 mm; Amersham Pharmacia), and the elution patterns of protein and selenium were monitored.
and 4 mM GSH in PBS (pH 7.4) containing 1 mM EDTA. After incubation at 37°C for 10 min, mixtures were applied to an FPLC fast desalting column (10 × 100 mm; Amersham Pharmacia), and the elution patterns of protein and selenium were monitored.
SPS activity. Recombinant Haemophilus influenzae SPS that had been produced in E. coli was purified (18) and assayed for activity with the radiochemical method by using TLC as described (5, 13). In the present study, activity of SPS in vitro in the presence of a selenium-binding protein was assayed by using HPLC equipped with a UV detector. The AMP product and ATP in the supernatant solution were separated and determined as described below.
SPS assay with selenium-binding protein. The SPS assay was performed anaerobically at 37°C in a reaction mixture containing 100 mM N-tris(hydroxymethyl)methylglycine (Tricine)·KOH (pH 8.0), 2 mM DHLA, 8 mM MgCl2, 20 mM KCl, 2 mM ATP, 5–10 μM SPS (≈0.182–0.364 mg/ml), 0.1 mM 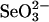 , and 4.0 mM GSH. After 30 min, the reaction was terminated by the addition of 1.2 M HClO4 followed by neutralization with KOH. A 10-μl aliquot of the supernatant of the incubation mixture was injected into a reverse-phase C18 column (4.5 × 150 mm; LiChrosphere RP-18e, Merck) that had been preequilibrated with the mobile-phase, 0.1 M triethylamine-containing phosphate buffer (pH 8.0):methanol = 97:3. A standard curve for ≈0.1–2.0 nmol of AMP was constructed from stock solutions of sodium AMP (10–200 μM). A flow rate of 1.0 ml/min was used with a running time of 20 min. The retention times and peak areas were monitored at 260 nm. The AMP produced from ATP in enzyme assay mixtures was estimated by extrapolation from values of area on the calibration curve.
, and 4.0 mM GSH. After 30 min, the reaction was terminated by the addition of 1.2 M HClO4 followed by neutralization with KOH. A 10-μl aliquot of the supernatant of the incubation mixture was injected into a reverse-phase C18 column (4.5 × 150 mm; LiChrosphere RP-18e, Merck) that had been preequilibrated with the mobile-phase, 0.1 M triethylamine-containing phosphate buffer (pH 8.0):methanol = 97:3. A standard curve for ≈0.1–2.0 nmol of AMP was constructed from stock solutions of sodium AMP (10–200 μM). A flow rate of 1.0 ml/min was used with a running time of 20 min. The retention times and peak areas were monitored at 260 nm. The AMP produced from ATP in enzyme assay mixtures was estimated by extrapolation from values of area on the calibration curve.
Analysis of selenium and protein. Protein concentration was determined by using the Bradford procedure (17). Selenium was analyzed by atomic absorption spectroscopy with a graphite furnace atomizer (Z-8000; Hitachi, Tokyo).
Results
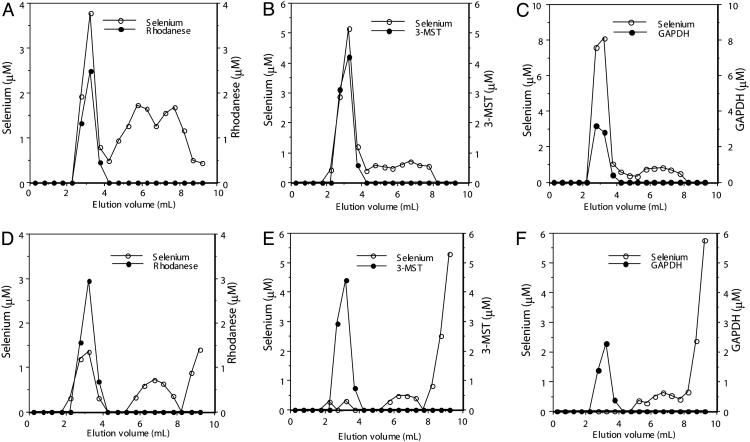
Selenium Binding and Release of Selenium from Protein. As shown in Fig. 1, after a 10-min reaction with 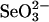 and GSH, selenium coeluted with rhodanese (Fig. 1 A), 3-MST (Fig. 1B), and GAPDH (Fig. 1C) in the flow-through fractions at the indicated volumes. In contrast, elution of selenium was not observed in the flow-through fractions containing 3-MST (Fig. 1E) and GAPDH (Fig. 1F) when DTT was added subsequent to the initial reaction with selenite and GSH. However, in the case of rhodanese, the release of bound selenium was incomplete (Fig. 1D), indicating that rhodanese binds selenium more tightly than 3-MST and GAPDH.
and GSH, selenium coeluted with rhodanese (Fig. 1 A), 3-MST (Fig. 1B), and GAPDH (Fig. 1C) in the flow-through fractions at the indicated volumes. In contrast, elution of selenium was not observed in the flow-through fractions containing 3-MST (Fig. 1E) and GAPDH (Fig. 1F) when DTT was added subsequent to the initial reaction with selenite and GSH. However, in the case of rhodanese, the release of bound selenium was incomplete (Fig. 1D), indicating that rhodanese binds selenium more tightly than 3-MST and GAPDH.
Fig. 1.
Elution profiles of products formed from reaction of selenium-binding proteins with GSH and 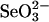 . Reaction mixtures (100 μl), containing PBS (pH 7.4), 0.1 mM
. Reaction mixtures (100 μl), containing PBS (pH 7.4), 0.1 mM  , 0.4 mM GSH, and 10 μM rhodanese E form (A and C), 3-MST (B and E), or GAPDH (C and F) were incubated at 37°C for 30 min under anaerobic conditions. After additional incubation with 2 mM DTT (D–F) or without DTT (A–C) at 37°C, reaction mixtures were applied to a gel filtration column (1.0 × 10 cm) and eluted with PBS (pH 7.4). Aliquots of each fraction were assayed to determine selenium and protein contents as described in Materials and Methods.
, 0.4 mM GSH, and 10 μM rhodanese E form (A and C), 3-MST (B and E), or GAPDH (C and F) were incubated at 37°C for 30 min under anaerobic conditions. After additional incubation with 2 mM DTT (D–F) or without DTT (A–C) at 37°C, reaction mixtures were applied to a gel filtration column (1.0 × 10 cm) and eluted with PBS (pH 7.4). Aliquots of each fraction were assayed to determine selenium and protein contents as described in Materials and Methods.
Stability of Selenium-Binding Protein. To examine the relative stability of the three proteins in their selenium-bound forms, the flow-through fraction of each protein from the FPLC column was stored at 4°C for 4 weeks under aerobic conditions. The results of this experiment are summarized in Table 1. Although most of the selenium initially bound to 3-MST had dissociated during storage, the 3-MST protein was still soluble and active. In contrast, selenium-bound rhodanese gradually aggregated along with the selenium. Part of the selenium-bound GAPDH was insoluble, but >50% of the selenium initially bound was recovered together with a soluble form of the protein.
Table 1. Stability of selenium-binding proteins.
| Remaining amount, %
|
Binding ratio (Se:protein)
|
||
|---|---|---|---|
| Protein | Selenium | Protein | |
| Rodanese | 30.9 | 67.3 | 0.5 |
| 3-MST | 31.9 | 94.8 | 0.3 |
| GAPDH | 56.6 | 76.1 | 1.4 |
Each flow-through fraction (0.5 ml) from the FPLC column was stored in a 1.5-ml sample tube under aerobic conditions at 4°C for 4 weeks. Concentrations of selenium and protein in soluble supernatant fractions obtained after centrifugation are represented as the percentage of the corresponding values obtained before sample storage.
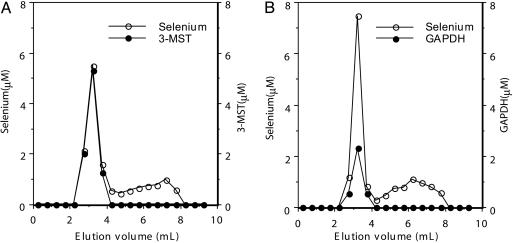
Effect of Physiological GSH Levels on Selenium-Binding Protein. GSSeSG is produced most effectively and stably when the ratio of  is 4:1. In the presence of increasing levels of GSH in the millimolar concentration range, the amount of selenium bound to rhodanese showed a tendency to decrease, as reported previously (13). However, in the presence of the physiological GSH concentration range and 0.1 mM
is 4:1. In the presence of increasing levels of GSH in the millimolar concentration range, the amount of selenium bound to rhodanese showed a tendency to decrease, as reported previously (13). However, in the presence of the physiological GSH concentration range and 0.1 mM  , the amount of selenium bound to 3-MST did not change (Fig. 2A) as compared to that observed with 0.4 mM GSH and 0.1 mM
, the amount of selenium bound to 3-MST did not change (Fig. 2A) as compared to that observed with 0.4 mM GSH and 0.1 mM 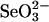 (Fig. 1B). GAPDH also bound an equivalent amount of selenium at the same ratio in the presence of 4 mM GSH and 0.1 mM
(Fig. 1B). GAPDH also bound an equivalent amount of selenium at the same ratio in the presence of 4 mM GSH and 0.1 mM 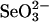 (Fig. 2B) as compared to that observed with 1/10th the concentration of GSH (Fig. 1C). Furthermore, the selenium-bound forms of 3-MST and GAPDH were stable in the presence of large excesses of GSH for at least 2 h at 37°C.
(Fig. 2B) as compared to that observed with 1/10th the concentration of GSH (Fig. 1C). Furthermore, the selenium-bound forms of 3-MST and GAPDH were stable in the presence of large excesses of GSH for at least 2 h at 37°C.
Fig. 2.
Effect of physiological concentration of GSH on the selenium-binding state of 3-MST and GAPDH. Reaction mixtures (100 μl) containing PBS (pH 7.4), 4 mM GSH, 0.1 mM 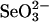 , and 10 μM 3-MST (A) or GAPDH (B) were incubated for 10 min at 37°C. Reaction mixtures were applied to a gel filtration column, and aliquots of each fraction were assayed for selenium and protein as described in Materials and Methods.
, and 10 μM 3-MST (A) or GAPDH (B) were incubated for 10 min at 37°C. Reaction mixtures were applied to a gel filtration column, and aliquots of each fraction were assayed for selenium and protein as described in Materials and Methods.
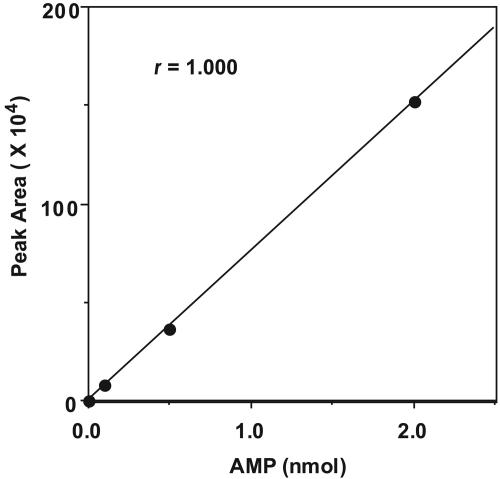
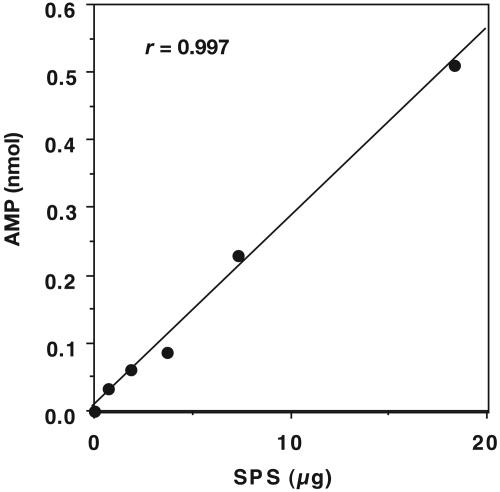
Development of an HPLC Assay for SPS. Nucleotides separated by HPLC can be detected at low concentrations by on-line UV-absorbancy measurements at 260 nm (19). The formation of products from the SPS reaction was monitored by using a reverse-phase HPLC column with an ion-pair mobile phase. An isocratic solvent system was found to be sufficient for separation of excess ATP substrate from the small amount of AMP product on a C18 column. Retention times were 6.7 min (AMP) and 15.0 min (ATP). The calibration curve for AMP over a 20-fold concentration range was linear, and as indicated in Fig. 3, nanomolar amounts of product were detected and quantitated. Fig. 4 illustrates that the formation of AMP was proportional to the amount of enzyme present over the range of 0.73–18.3 μg of SPS.
Fig. 3.
Calibration curve for AMP determination by HPLC. Reaction mixtures (100 μl) contained 50 mM Tricine·KOH (pH 8.0), 2 mM DHLA, 8 mM MgCl2, 50 mM KCl, 0.1 mM Mg triplex, 2 mM ATP, and varying concentrations of AMP. After the addition of HClO4 and KOH, a 10-μl aliquot of the mixture was injected into the C18 column. The area of each AMP peak from a given run is plotted versus the total nanomoles of AMP injected. HPLC conditions are described in Materials and Methods.
Fig. 4.
Linearity of AMP formation dependent on SPS concentration. Reaction mixtures containing from 0.73 to 18.3 μg of enzyme (0.2–5.0 μM), 50 mM Tricine·KOH (pH 8.0), 2 mM DHLA, 8 mM MgCl2, 50 mM KCl, 0.1 mM Mg triplex, 2 mM ATP, 0.1 mM 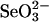 , and 4 mM GSH were incubated at 37°C for 30 min under anaerobic conditions. Reactions were terminated by the addition of 1.2 M HClO4 followed by neutralization with KOH. The amount of AMP produced was determined by reverse-phase HPLC using a standard curve as described in Materials and Methods.
, and 4 mM GSH were incubated at 37°C for 30 min under anaerobic conditions. Reactions were terminated by the addition of 1.2 M HClO4 followed by neutralization with KOH. The amount of AMP produced was determined by reverse-phase HPLC using a standard curve as described in Materials and Methods.
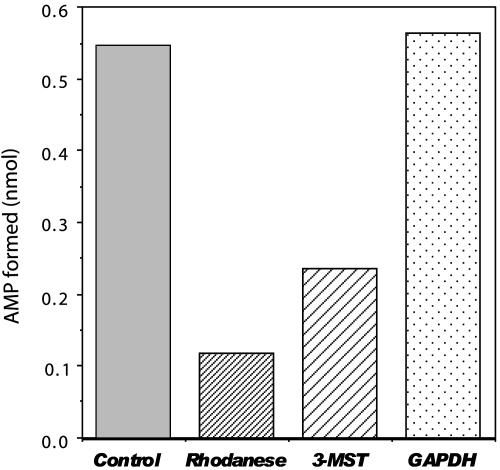
SPS Assays with Selenium-Binding Proteins Using HPLC. To test the availability of selenium provided by a selenium-binding protein as substrate for selenophosphate biosynthesis, assays were performed in the presence of each selenium-binding protein, 0.1 mM  , 4 mM GSH, and 2 mM DHLA instead of the high levels of
, 4 mM GSH, and 2 mM DHLA instead of the high levels of 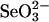 (1.5 mM) and DTT (25 mM) used routinely. In the in vitro SPS assay, the selenium-dependent formation of AMP from ATP is equimolar to orthophosphate and monoselenophosphate, the two other products (15, 18). In reaction mixtures containing GSH,
(1.5 mM) and DTT (25 mM) used routinely. In the in vitro SPS assay, the selenium-dependent formation of AMP from ATP is equimolar to orthophosphate and monoselenophosphate, the two other products (15, 18). In reaction mixtures containing GSH,  , and a binding protein instead of a high selenide level, AMP production could be determined by the HPLC assay method presented here (Fig. 5). In this assay, selenium bound to GAPDH and 3-MST was more available as substrate than that derived from rhodanese, and addition of DHLA accelerated the selenium-dependent hydrolysis of ATP. In the coupled assays with 3-MST and rhodanese, the observed AMP production was lower than in the control assay, in which a high concentration of selenide was used. However, with only 1/15th the amount of selenium, supplied as substrate from GAPDH, the amount of AMP formed in the presence of 2 mM DHLA was the same as in the control assay with 1.5 mM
, and a binding protein instead of a high selenide level, AMP production could be determined by the HPLC assay method presented here (Fig. 5). In this assay, selenium bound to GAPDH and 3-MST was more available as substrate than that derived from rhodanese, and addition of DHLA accelerated the selenium-dependent hydrolysis of ATP. In the coupled assays with 3-MST and rhodanese, the observed AMP production was lower than in the control assay, in which a high concentration of selenide was used. However, with only 1/15th the amount of selenium, supplied as substrate from GAPDH, the amount of AMP formed in the presence of 2 mM DHLA was the same as in the control assay with 1.5 mM 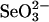 and 25 mM DTT.
and 25 mM DTT.
Fig. 5.
SPS assay with selenium-binding protein. Assays were performed anaerobically at 37°C in 50 mM Tricine·KOH (pH 8.0), 2 mM DHLA, 8 mM MgCl2, 50 mM KCl, 0.1 mM Mg triplex, 2 mM ATP, 0.1 mM  , 4 mM GSH, 5 μM SPS, and selenium-binding protein as indicated. The standard assay mixture contained 1.5 mM selenide and 25 mM DTT in place of GSH, DHLA, and a selenium-binding protein. After a 30-min incubation, reactions were terminated, and the reaction products were separated by reverse-phase HPLC and detected as described in Materials and Methods. The AMP formed in this assay was calculated from a standard curve for AMP prepared as described in Materials and Methods. Measurements were performed in duplicate, and the data are presented as means.
, 4 mM GSH, 5 μM SPS, and selenium-binding protein as indicated. The standard assay mixture contained 1.5 mM selenide and 25 mM DTT in place of GSH, DHLA, and a selenium-binding protein. After a 30-min incubation, reactions were terminated, and the reaction products were separated by reverse-phase HPLC and detected as described in Materials and Methods. The AMP formed in this assay was calculated from a standard curve for AMP prepared as described in Materials and Methods. Measurements were performed in duplicate, and the data are presented as means.
Discussion
SPS, the selD gene product, catalyzes the synthesis of monoselenophosphate, AMP, and orthophosphate in a 1:1:1 ratio from ATP and selenide in vitro. SPS from E. coli (15) and a recombinant form of the closely related enzyme from H. influenzae (18) have been characterized. In the presence of high levels of free selenide and DTT in the in vitro assay system, the apparent Km value for selenide is 20 μM. This value is far above the optimal concentration of selenium needed for growth of various bacterial species and cultured mammalian cells.
Therefore, we considered the possibility that transfer of selenium from a selenotrisulfide or a perselenide intermediate to a potential donor protein could form a relatively stable selenium protein adduct, which would function as a selenotransferase to SPS. In a previous study, we tested rhodanese, a member of the sulfur transferase family, as a possible model for selenium binding and delivery to SPS (13). The results of this study indicated that the stability and availability of selenium bound to proteins are important properties that must be assessed for successful selenium-delivery system participation.
GAPDH, a homotetramer, is a well known glycolytic enzyme. A recent study that showed the ability of GAPDH to bind selenium in vivo suggested a possible additional biological role for this abundant enzyme (14). The selenium-binding capacity of GAPDH was characterized under in vitro conditions in the present study. Selenium supplied in the form of GSSeSG, the product of the reaction of GSH with 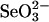 , was bound in a 4:1 ratio to GAPDH and in a 1:1 ratio to each protein subunit. Binding of selenium to GAPDH seems to occur by covalent attachment to a protein thiol residue as judged by ease of release after reduction with DTT or DHLA. The biological importance of this selenium-binding property of GAPDH is unknown, but in view of the wide distribution and abundance of this enzyme in organisms, it could serve as a significant source of selenium for synthesis of selenophosphate, the selenium donor required for biosynthesis of selenocysteine-containing enzymes.
, was bound in a 4:1 ratio to GAPDH and in a 1:1 ratio to each protein subunit. Binding of selenium to GAPDH seems to occur by covalent attachment to a protein thiol residue as judged by ease of release after reduction with DTT or DHLA. The biological importance of this selenium-binding property of GAPDH is unknown, but in view of the wide distribution and abundance of this enzyme in organisms, it could serve as a significant source of selenium for synthesis of selenophosphate, the selenium donor required for biosynthesis of selenocysteine-containing enzymes.
3-MST, a member of the sulfurtransferase family with high homology to rhodanese, is localized in cytosolic and mitochondrial fractions (20). As shown previously for rhodanese, 3-MST bound selenium in a 1:1 ratio, and this binding ability was lost after alkylation of its cysteine residue (data not shown). Although the binding site of selenium seems to be a reactive cysteine in both cases, selenium bound to 3-MST was more readily released after reduction with DTT than selenium bound to rhodanese (Fig. 1).
GSSeSG, which is relatively unstable under aerobic conditions, gradually decomposed to form elemental selenium at neutral pH even in the presence of excess GSH. However, liberation of elemental selenium was not observed from any of the proteins containing bound selenium used in the present study within the first 24 h. Moreover, we found that soluble selenium derivatives formed from 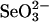 and excess GSH were comparably stable under anaerobic conditions, and the selenium remained in an available form in the SPS system for at least 24 h when the reaction mixture was stored at 4°C (data not shown). Under aerobic conditions, selenium-bound rhodanese was gradually denatured and precipitated, but GAPDH was comparably stable when bound to selenium. As reported (13), selenium binding to rhodanese tended to decrease when GSSeSG was produced in the presence of excess GSH (>8-fold rather than 4-fold) to
and excess GSH were comparably stable under anaerobic conditions, and the selenium remained in an available form in the SPS system for at least 24 h when the reaction mixture was stored at 4°C (data not shown). Under aerobic conditions, selenium-bound rhodanese was gradually denatured and precipitated, but GAPDH was comparably stable when bound to selenium. As reported (13), selenium binding to rhodanese tended to decrease when GSSeSG was produced in the presence of excess GSH (>8-fold rather than 4-fold) to 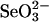 . It is interesting to note that from the experiments in the present study, it is clear that the ratio of bound selenium to 3-MST and to GAPDH remained constant even if a 40-fold (4 mM) excess of GSH was added to the reaction mixture containing 0.1 mM
. It is interesting to note that from the experiments in the present study, it is clear that the ratio of bound selenium to 3-MST and to GAPDH remained constant even if a 40-fold (4 mM) excess of GSH was added to the reaction mixture containing 0.1 mM  . These results indicate that selenium produced from a small amount of
. These results indicate that selenium produced from a small amount of 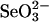 and a physiological concentration of GSH could serve as an adequate source for continued 3-MST and GAPDH binding. Based on the fact that selenium is released more readily from 3-MST than from rhodanese and also that selenium bound to 3-MST is more available as a substrate for SPS (Fig. 5) than from rhodanese, 3-MST has a greater potential as a selenium-delivery protein. In the present study, we used DHLA, a physiological dithiol, instead of DTT as reductant. DHLA has an affinity to rhodanese (21) and reacts with
and a physiological concentration of GSH could serve as an adequate source for continued 3-MST and GAPDH binding. Based on the fact that selenium is released more readily from 3-MST than from rhodanese and also that selenium bound to 3-MST is more available as a substrate for SPS (Fig. 5) than from rhodanese, 3-MST has a greater potential as a selenium-delivery protein. In the present study, we used DHLA, a physiological dithiol, instead of DTT as reductant. DHLA has an affinity to rhodanese (21) and reacts with  to form a stable intramolecular selenotrisulfide (22). Our results indicated that selenium was released from selenium-binding proteins more effectively with DHLA than with DTT under the conditions indicated for Fig. 1.
to form a stable intramolecular selenotrisulfide (22). Our results indicated that selenium was released from selenium-binding proteins more effectively with DHLA than with DTT under the conditions indicated for Fig. 1.
For the in vitro SPS assay, we developed an HPLC method for AMP determination. SPS in the picomole range can be quantitated by using this assay as indicated by a linear response of enzyme activity to protein concentration. This sensitive and accurate procedure offers an alternative to the commonly used assay, which is time-consuming and requires radioactive compound. The availability of selenium to the SPS system from the three selenium-bound proteins studied was GAPDH > 3-MST > rhodanese. However, it is unclear whether these results reflect their potential physiological function.
In conclusion, 3-MST and GAPDH exhibit affinity to active selenium derived from GSSeSG (–1 selenium valency) under physiological concentrations of GSH, and the selenium-bound proteins can exist as stable soluble forms. Because selenium bound to 3-MST and GADPH is rapidly releasable in the presence of a physiological dithiol, DHLA, these proteins may have roles as selenium transferases or in the brief storage of excess selenium in the cell.
Author contributions: Y.O., G.M.L., and T.C.S. designed research; and Y.O. and K.I. performed research.
Abbreviations: SPS, selenophosphate synthetase; GSH, glutathione; GSSeSG, selenodiglutathione; 3-MST, 3-mercaptopyruvate sulfurtransferase; GAPDH, glyceraldehyde-3-phosphatedehydrogenase; DHLA, dihydrolipoicacid; Tricine, N-tris(hydroxymethyl)methylglycine.
References
- 1.Zinoni, F., Birkmann, A., Stadtman, T. C. & Bock, A. (1986) Proc. Natl. Acad. Sci. USA 83, 4650–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leinfelder, W., Forchhammer, K., Zinoni, F., Sawers, G., Mandrand-Berthelot, M.-A. & Bock, A. (1988) J. Bacteriol. 170, 540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bock, A. & Stadtman, T. C. (1998) Biofactors 1, 245–250. [PubMed] [Google Scholar]
- 4.Leinfelder, W., Forchhammer, K., Veprek, B., Zchelem, E. & Bock, A. (1990) Proc. Natl. Acad. Sci. USA 87, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veres, Z., Tsai, L., Scholz, T. D., Politino, M., Balaban, R. S. & Stadtman, T. C. (1992) Proc. Natl. Acad. Sci. USA 89, 2975–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganther, H. E. (1999) Carcinogenesis 20, 1657–1666. [DOI] [PubMed] [Google Scholar]
- 7.Lindemann, T. & Hintelmann, H. (2002) Anal. Chem. 74, 4602–4610. [DOI] [PubMed] [Google Scholar]
- 8.Turner, R. J., Weiner, J. H. & Taylor, D. E. (1998) Biometals 11, 223–227. [DOI] [PubMed] [Google Scholar]
- 9.Ganther, H. E. (1971) Biochemistry 10, 41089–41098. [DOI] [PubMed] [Google Scholar]
- 10.Esaki, N., Nakamura, T., Tanaka, H. & Soda, K. (1982) J. Biol. Chem. 257, 4386–4391. [PubMed] [Google Scholar]
- 11.Lacourciere, G. M., Mihara, H., Kurihara, T., Esaki, N. & Stadtman, T. C. (1998) J. Biol. Chem. 275, 23769–23777. [DOI] [PubMed] [Google Scholar]
- 12.Westley, J. (1980) in Enzymatic Basis of Detoxication, ed. Jacoby, W. B. (Academic, New York), Vol. 2, pp. 245–262. [Google Scholar]
- 13.Ogasawara, Y., Lacourciere, G. M. & Stadtman, T. C. (2001) Proc. Natl. Acad. Sci. USA 98, 9494–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacourciere, G. M., Levine, R. L. & Stadtman, T. C. (2002) Proc. Natl. Acad. Sci. USA 99, 9150–9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veres, Z., Kim, I. Y., Scholz, T. D. & Stadtman, T. C. (1994) J. Biol. Chem. 269, 10597–10603. [PubMed] [Google Scholar]
- 16.Pagani, S., Bonomi, F. & Cerletti, P. (1984) Eur. J. Biochem. 142, 361–366. [DOI] [PubMed] [Google Scholar]
- 17.Bradford, M. M. (1976) Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 18.Lacourciere, G. M. & Stadtman, T. C. (1999) Proc. Natl. Acad. Sci. USA 96, 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stocci, V., Cucchiarini, L., Canestrani, F., Piacentinim, M. P. & Fornaini, G. (1987) Anal. Biochem. 167, 181–190. [DOI] [PubMed] [Google Scholar]
- 20.Nagahara, N., Okazaki, T. & Nishino, T. (1995) J. Biol. Chem. 270, 16230–16235. [DOI] [PubMed] [Google Scholar]
- 21.Cianci, M., Gliubich, F., Zanotti, G. & Berni, R. (2000) Biochim. Biophys. Acta 31, 103–108. [DOI] [PubMed] [Google Scholar]
- 22.Self, W. T., Tsai, L. & Stadtman, T. C. (2000) Proc. Natl. Acad. Sci. USA 97, 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]